Abstract
Aim:
This study aimed to investigate the association between endodontic clinical signs and symptoms and the presence of Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia employing polymerase chain reaction (PCR).
Materials and Methods:
Microbial samples were obtained from 60 cases with necrotic pulp with primary teeth infections. DNA extracted from samples were analyzed for endodontic pathogens by using species-specific primers.
Results:
P. gingivalis/T. denticola were detected in 15 symptomatic teeth associated with periapical lesions. T. forsythia/T. denticola were found in 16 symptomatic teeth associated with pain and swelling. P. gingivalis was detected in 9 teeth which were associated with pain, 2 with tenderness on percussion, and 15 with periapical lesions. Statistically significant associations were found between T. forsythia as well as T. denticola in relation to clinical findings of pain and swelling. (P < 0.05). Red complex bacteria showed no statistical significant association with the presence of signs and symptoms.
Conclusion:
Prevalence of P. gingivalis, T. denticola, and T. forsythia suggested association of these bacteria with symptomatic infected pulp and periradicular diseases.
Keywords: P. gingivalis, red complex, T. denticola, T. forsythia
Introduction
Bacterial microflora in endodontic infections are affected by infectious origin, ecological niche, and host defense. The bacterial genera which are found in infected root canals include Gram-negative anaerobes such as Prevotella, Veillonella, Porphyromonas; Gram-positive anaerobes such as Peptostreptococcus, Lactobacillus, Actinomyces; Gram-negative facultative such as Eikenella, Haemophilus and Gram-positive facultative such as Lactobacillus, Actinobacillus, Propionibacterium, etc., These organisms produce clinical symptoms such as pain, swelling, tender on percussion, and sinus formation.[1]
Bacterial invasion of root canal often leads to an inflammatory cascade at the root apex that causes apical periodontitis. Root canal treatment aims to resolve the inflammation at the root apex by inactivation of these residing bacteria and the elimination of the endotoxins produced by the microorganisms. However, the complex root canal system and microorganisms residing in resilient biofilm communities impede sufficient cleaning, which can lead to persistent apical periodontitis.[2,3] An infected root canal comprises a unique microenvironment housing selective microflora. These organisms grow in planktonic forms or as aggregates as well as in biofilms. The microbial composition in the root canal system is an interesting area of research nowadays. Novel technologies such as immunological assays and molecular methods (PCR) methods were developed in recent years.[4]
Sequencing methods based on 16S rRNA are important tools for the identification of both non-cultivable and cultivable pathogenic bacteria. The 16S rDNA gene amplification from bacterial DNA extracted which is followed by cloning and gene sequencing helps in bacterial characterization. Though it is possible to detect genetic material from dead bacterial cells from extirpated dental pulp, the DNase produced by living microbes can degrade their DNA. Thus, bacteria detected in such cases represent endodontic pathogens in an acute infection. Bacterial population associated with primary endodontic infection is predominantly, Gram-negative bacteria though Gram-positive species such as Peptostreptococcus stomatitis, Parvimonas micra, Eubacterium spp. has also been detected in an earlier study.[5]
The present study was done to analyze association between clinical signs and symptoms and the presence of red-complex bacteria in an infected root canal.
Materials and Methods
A total of 60 symptomatic teeth were selected. Inclusion criteria for case selection included no previous treatment, necrotic pulp, or periapical periodontitis or abscess or wet canals. Ethical approval was obtained from the institutional ethical committee on 02/01/2019.
Collection of samples
Samples were collected using sterile paper points after exposure of root canals under rubber dam isolation. For the negative control group, samples were obtained by moistening 10 paper points in sterile normal saline. The paper points were thereafter transferred to Eppendorf tubes containing Tris-EDTA (ethylenediaminetetraacetic acid) (TE) buffer and were stored in −70°C for PCR.
Genomic DNA isolation
Isolation of genomic DNA was performed according to the protocol described by Brezezinska-Blaszezyk et al. (2018).[6] The paper points were then thawed on ice followed by vortexing for 3 min. Then, the paper points were removed and the samples were centrifuged at 200 rotation per minute (RPM) for 5 min. Pellets obtained were suspended in 100 μl of HCl buffer (pH 8.5). Microbial DNA was isolated using genomic mini-kit (A and A Biotech., Gdynia, Polana). The cells were lysed with proteinase K and DNA obtained was suspended in 100 μl of TE buffer (pH 7.4). The obtained DNA content was measured using a spectrophotometer.
Polymerase chain reaction methodology
The universal primers were designed as per 16S rRNA genes provided by GenBank, Primer Premier 5. These were specifically designed to target sequences unique toward the tested organisms. PCR primers used were:
Porphyromonas gingivalis
Forward primer: AGG CAG CTT GCC ATA CTG CG
Reverse primer: ACT GTT AGC AAC TAC CGA TGT (Bioserve India Pvt. Ltd.)
Treponema denticola
Forward primer: TAA TAC CGA ATG TGC TCA TTT ACA T
Reverse primer: TCA AAG AAG CAT TCC CTC TTC TTC TTA (Bioserve India Pvt. Ltd.)
Tannerella forsythysis
Forward primer: GCG TAT GTA ACC TGC CCG CA
Reverse primer: TGC TTC AGT GTC AGT TAT ACC T (Bioserve India Pvt. Ltd.)
The specificity of the probes was assessed using the BLAST program. DNA template (5 μl) was added to the PCR mixture with a final volume of 25 μl. Reaction mixture comprised 2.5 μl of 10X PCR buffer, 1 μl Mg2+, 2 μl dNTP (deoxyribonucleotide triphosphate), 0.5 μl of forward and reverse primers each and 2.5 U of Taq polymerase.
PCR cycle
The PCR program cycle was run under 94°C for 4 min; 30 cycles at 94°C for 30 s, 55°C for 30 s, 72°C for 30 s and finally, 72°C for 10 min. Visualization of bands was done under UV illumination using 1% agarose gel electrophoresis employing ethidium bromide.
Data analysis
Data collected from each sample were recorded in Microsoft Excel Worksheet 2016 and analyzed using version 21.0 of the Statistical Package for Social Sciences (IBM Corporation, Armonk, New York, USA).
Results
Clinical data
Of the studied samples, 35 had pain, 29 had swelling, 06 had pus discharge, 08 were tender on percussion and 25 had periapical lesions. P. gingivalis was found in 09 teeth associated with pain, 02 associated with tenderness on percussion and 15 associated with periapical lesions. T. denticola was found in 16 teeth associated with pain and swelling, 05 with tenderness on percussion and 15 with periapical lesions. Tannerella forsythia was detected in 16 teeth associated with pain and swelling and, 02 teeth associated with tenderness on percussion. Red complex bacteria were detected in 05 teeth associated with pain and swelling, 06 with pus discharge, 03 with tenderness on percussion, and 10 with periapical lesions [Table 1 and Graphs 1-5].
Table 1.
Distribution of study samples according to clinical signs and symptoms and presence of Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia individually or as a “Red complex”
| Clinical signs and symptoms | P. gingivalis | T. denticola | T. forsythia | Red complex |
|---|---|---|---|---|
| Pain | 09 | 16 | 16 | 05 |
| Swelling | - | 16 | 16 | 05 |
| Pus discharge | - | - | - | 06 |
| Tenderness on percussion | 02 | 05 | 02 | 03 |
| Periapical lesion | 15 | 15 | - | 10 |
Graph 1.
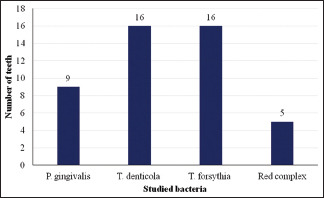
Studied bacteria and number of teeth with pain
Graph 5.
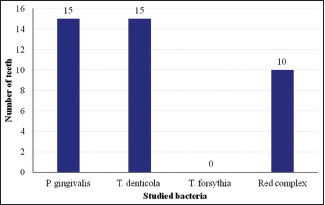
Studied bacteria and number of teeth with periapical lesions
Graph 2.
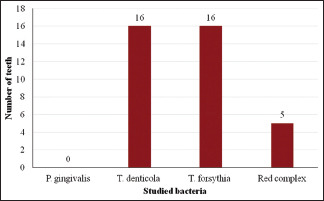
Studied bacteria and number of teeth with swelling
Graph 3.
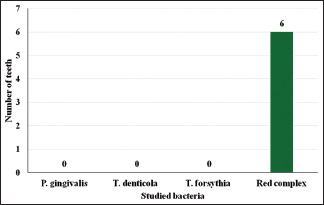
Studied bacteria and number of teeth with pus discharge
Graph 4.
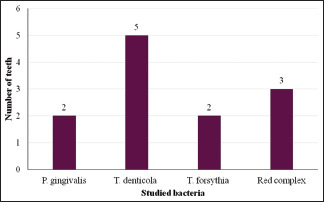
Studied bacteria and number of teeth with tenderness on percussion
Statistically significant associations were observed between T. forsythia as well as T. denticola in relation to clinical findings of pain and swelling (P < 0.05). Other bacteria did not show any significant association with clinical findings. No bacterial DNA was detected in negative controls.
Discussion
Endodontic-origin microorganisms such as P. gingivalis, T. denticola, T. forsythia, and Solobacterium moorei have been linked with osteomyelitis, bacterial endocarditis, brain abscess, obesity, and preterm low-birth-weight.[5] Endodontic infections can progress to systemic infections that are characterized by fever and lymphadenopathy. This reflects the adaptations of microbes to different ecological niches varying from oxygenated to anaerobic environment.[7]
Endodontic flare-ups can occur due to the synergistic presence of Fusobacterium nucleatum, Prevotella spp., and Porphyromonas sp. by increasing the periapical inflammation.[8]
Apical periodontitis has a heterogeneous etiology due to interindividual variations in endodontic microflora. Molecular detection techniques such as species-specific PCR and checkerboard DNA-DNA hybridization assay can identify culture-difficult species from endodontic samples. 16S rRNA gene clone library analysis has revealed that nearly 45–50% of microbial taxa were not cultivable and, hence, was underdiagnosed. Cataloging of these bacterial species provides the 16S rRNA gene sequence.[9] The bacterial 16S rRNA comprises nine hypervariable regions that demonstrate uniform diversity in sequence among various bacterial species which can be used for bacterial identification. These conserved sequences can be used to design universal primers for bacterial DNA amplification.[10]
PCR technique is more sensitive than traditional culture techniques for microbiological identification, especially in refractory cases.[11]
Gram-negative bacteria are found to predominate the microbial population in root canal infections. The lipopolysaccharides are the most important virulent factors which cause clinical symptoms and pathological changes in endodontic infections, due to inflammatory mediators such as IL-1α, IL-1β, TNF-α, PGE2, and matrix metalloproteinases. The microbial population in closed canals is anaerobic while in open canals, it is facultative. The bacterial community shows considerable interindividual variation, thus, influencing the treatment protocols as well.[12]
Gomes et al.[13] using nested PCR on necrotic pulp samples analyzed 46%, 38%, and 22% of positive cases of P. gingivalis, T. denticola, and T. forsythia, respectively.
P. gingivalis is a coccobacillus belonging to family Porphyromonadaceae, formerly it was known as “Bacteroides gingivalis.“[14]
Seol et al.[15] have reported P. gingivalis in 22.5% of abscesses using multiplex PCR. In the current study, 15 teeth with periapical lesions were positive for this organism. Rocas et al.[16] have reported this organism in 70% of cases with endodontic abscesses.
Treponema are strict anaerobes, motile, Gram-negative and helically shaped bacterial rods.[17,18] Among all intra-oral Treponema spp, only T. denticola, T. socransky, T. vincentii, and T. pectinovorum can be readily cultivated.
In the present study T. forsythia as well as T. denticola showed significant association, only with clinical findings of pain and swelling however, these pathogenic bacteria were also found in teeth samples with tenderness on percussion and periapical lesions. The association of the red complex with clinical signs and symptoms was found to be non-significant.
Guven et al.[19] performed a study on pulpal samples collected from 20 primary molars with primary endodontic infection. Clinical signs included spontaneous pain, tenderness on percussion, swelling, and tooth mobility. This study found a positive and significant correlation between T. denticola, P. gingivalis, and T. forsythia. However, no statistically significant correlation was found between any of the clinical signs and any particular bacteria. This is a contrast to current study results.
Sanghavi et al.[20] conducted a study to evaluate the correlation between endodontic clinical signs and symptoms and the presence of red complex microorganisms such as P. gingivalis, T. denticola, and T. forsythia using PCR assay. It was concluded that P. gingivalis, T. denticola, and T. forsythia were prevalent in the examined samples that suggested their relationship to the etiology of periradicular diseases.
Treponema spp are highly fastidious, Gram-negative motile spirochetes found in periodontal pockets and root canals with primary infection. Noberga et al.[21] studied the prevalence of oral Treponemas in teeth with endodontic treatment failures and periapical pathologies using samples from 40 root canals using nested PCR technique. The study revealed a high incidence of Treponema spp in acute endodontic conditions indicating high pathogenicity.
In a study conducted by Foschi,[22] 56% of teeth with apical periodontitis showed a positive association with T. denticola.
Foschi et al.[23] performed a study to analyze the role of T. denticola as a mono-infection as well as a part of “red complex“ infection in the causation of endodontic infections in 6–8 weeks old severe combined immunodeficiency mice. Study results demonstrated periapical bone resorption in T. denticola mono-infection when compared to the red complex.
Cardoso et al.[24] conducted a study to correlate the bacterial diversity and levels of endotoxins produced by bacteria found in primary endodontic infection with the volume of root canal determined by using cone-beam computed tomography. Culturable bacteria and endotoxins were detected in 100% of the root canal samples and larger canals hold higher levels of the bacterial population. The different clinical features were positively correlated with the presence of the bacterial population.
T. denticola and T. forsythias have been shown to predispose immunocompromised subjects such as in Down's syndrome to early-onset periodontitis.[25]
T. denticola has been associated with an increase in periodontal destruction, therefore, increasing tooth loss.[10] It is very difficult to isolate and identify T. denticola from clinical samples using culture technique, therefore, the 16S rRNA-based PCR method is used for determining their presence.[26]
P. gingivalis is an asaccharolytic, nonmotile Gram-negative organism requiring an anaerobic environment for growth. It is a late colonizer and possesses numerous virulence factors such as lipopolysaccharides, capsular polysaccharide, fimbriae, and gingipains.[27]
Cao et al.[27] assessed 80 teeth with pulpal necrosis and primary endodontic infection using universal bacterial primers based upon 16S rRNA sequences. This technique demonstrated that both P. gingivalis and P. endodontalis showed significant association (P < 0.005 and P < 0.05, respectively) with the presence of sinus tracts along with abscesses.
Pattanshetty et al.[28] investigated the presence of selective anaerobic microorganisms in 100 primary root canals of symptomatic and asymptomatic non-vital teeth with periapical pathosis using multiplex PCR. It was concluded that significant differences exist in the bacterial composition between asymptomatic and symptomatic primary endodontic infections. T. denticola was present in 21 (42%) and 29 (58%) samples in the symptomatic and asymptomatic groups, respectively. T. forsythia, P. gingivalis, and F. nucleatum were significantly high (P < 0.05) in the symptomatic group, whereas Prevotella intermedia was significantly high (P < 0.05) in the asymptomatic group.
T. forsythia was first isolated at “Forsythe Institute“ in subjects with progressive periodontitis. It was originally assigned to the genus “Bacteroides.“ It exhibits slow growth under fastidious requirements. Subgingival T. forsythia is most likely a source in endodontic samples.[21]
In a study by Lacevic et al.[29] it was observed that levels of T. forsythia in primary endodontic infection and in periodontal lesion were significantly decreased with the increase of patients age.
T. denticola was detected in 15/60 teeth samples with periapical lesions of which five were tender on percussion. The red complex was detected in 10% (6/60) samples taken from acute periradicular abscess samples. These observations are suggestive of the role of “red complex“ in the pathogenesis of acute periradicular abscesses. These findings have been corroborated by Sanghavi et al.[30] using the multiplex PCR technique.
Siqueira and Rocas analyzed five stages of experimental tools for analyzing microorganisms found within acute apical abscesses: 1) culture techniques; 2) molecular tools such as PCR and checkerboard hybridization assay; 3) PCR in addition to cloning and sequencing of targeted amplicons; 4) PCR or DNA hybridization including reverse checkerboard hybridization, and 5) next-generation sequencing.[31,32]
These novel culture-independent methods using DNA amplification of 16S rDNA followed by cloning and sequencing have been used in determining the bacterial diversities.
Implications for clinical practice
Bacteria associated with endodontic infections trigger inflammatory responses in the immune cells, which in later stages of the disease, cause loss of both soft and hard tissue structures supporting the teeth. The red complex bacteria have been characterized as infectious agents of endodontic disease. Early diagnosis and identification of these microorganisms can help in treating such infections. Increased awareness of oral health and the identification of various disease-causing pathogens help to rule out the risk factors involved in oral diseases. Certain therapies have shown promising results that further needs evaluation in prospective clinical trials. Elimination of these pathogens from the site of infection remains a perplexing task, which demands the use of antibiotics.[33]
Conclusion
The high prevalence of P. gingivalis, T. denticola, and T. forsythia in the present study suggests that these bacteria can be correlated with etiopathogenesis of periapical abscesses. The red complex was detected in a few samples with endodontic signs. Further studies are required for identifying a relationship between root canal microbiota and periradicular diseases.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Haapasalo M. Black-pigmented Gram-negative anaerobes in endodontic infections. FEMS Immunol Med Microbiol. 1993;6:213–7. doi: 10.1111/j.1574-695X.1993.tb00329.x. [DOI] [PubMed] [Google Scholar]
- 2.Wolf TG, Paqué F, Zeller M, Willershausen B, Briseño-Marroquín B. Root canal morphology and configuration of 118 mandibular first molars by means of micro–computed tomography: An ex vivo study. J Endod. 2016;42:610–4. doi: 10.1016/j.joen.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Persoon IF, Buijs MJ, Özok AR, Crielaard W, Krom BP, Zaura E, et al. The mycobiome of root canal infections is correlated to the bacteriome. Clin Oral Invest. 2017;21:1871–81. doi: 10.1007/s00784-016-1980-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pecuikine V, Maneliene R, Balcikouyte E, Drukteinis S, Rutkunas V. Microorganisms in root canal infections: A review. Stomatologiya. 2008;10:4–9. [PubMed] [Google Scholar]
- 5.Nobrega LMM, Montagner F, Ribeiro AC, Mayer MAP, de Almeida Gomes BPF. Bacterial diversity of symptomatic primary endodontic infection by clonal analysis. Braz Oral Res. 2016;30:e103. doi: 10.1590/1807-3107BOR-2016.vol30.0103. [DOI] [PubMed] [Google Scholar]
- 6.Brzezinska-Blaszczyk E, Pawlowska E, Ploszay T, Witas H, Godzik U, Agier J. Presence of archae and selected bacteria in infected root canal system. Can J Microbiol. 2018;64:317–26. doi: 10.1139/cjm-2017-0531. [DOI] [PubMed] [Google Scholar]
- 7.Hsiao WWL, Li KL, Liu Z, Jones C, Fraser-Liggelt CM, Forrad AF. Microbial transformation from normal oral microbiota to acute endodontic infections. BMC Genomics. 2012;13:345. doi: 10.1186/1471-2164-13-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh H. Microbiology of endodontic infection. J Dent Oral Health. 2016;215:44–8. [Google Scholar]
- 9.Rocas IN, Siqueira JF. Root canal microbiota of teeth with chronic apical periodontitis. J Clin Microbiol. 2008;46:3599–606. doi: 10.1128/JCM.00431-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su CY, Shigashi H, Nishimura R, Ohta K, Suguyama M. Detection of oral bacteria on the tongue dorsum using PCR amplification of 16S ribosomal RNA and its association with systemic diseases. Biomed Rep. 2019;10:70–6. doi: 10.3892/br.2018.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rolph HJ, Lennon A, Riggio MP, Saunders WP, MacKenzie D, Colderao L, et al. Molecular identification of microorganisms from endodontic infections. J Clin Microbiol. 2001;39:3282–9. doi: 10.1128/JCM.39.9.3282-3289.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Almeida Gomes BPF, Herrera DR. Etiologic role of root canal infection in apical periodontitis and its relationship with clinical symptomatology. Braz Oral Res. 2018;32(Suppl 1):e69. doi: 10.1590/1807-3107bor-2018.vol32.0069. [DOI] [PubMed] [Google Scholar]
- 13.Gomes PFA, Montagner F, Jacinto RC, Zaia AA, Ferraz CCR, Souza-Filho FJ. Polymerase chain reaction of Porphyromonas gingivalis, Treponema denticola and Tannerella forsythia in primary endodontic infections. J Endod. 2007;33:1049–52. doi: 10.1016/j.joen.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 14.Radhakrishnan P, Anbalagan R, Barani R, Mani M, Seshadri KG, Srikanth P. Sequencing of Porphyromonas gingivalis from saliva in patients with periodontitis and type 2 diabetes mellitus. Int J Med Microbiol. 2019;37:54–9. doi: 10.4103/ijmm.IJMM_18_409. [DOI] [PubMed] [Google Scholar]
- 15.Seol JH, Cho BH, Chung CP, Bae KS. Multiplex polymerase chain reaction detection of black-pigmented bacteria in infections of endodontic origin. J Endod. 2006;32:110–4. doi: 10.1016/j.joen.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 16.Roças IN, Baumgartner JC, Xia T, Siqueira JF., Jr Prevalence of selected bacterial named species and uncultivated phylotypes in endodontic abscesses from two geographic locations. J Endod. 2006;32:1135–8. doi: 10.1016/j.joen.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Ohyama Y, Ogawa T. Detection and quantification of oral treponemes in subgingival plaque by real time-PCR. J Clin Microbiol. 2002;40:3334–40. doi: 10.1128/JCM.40.9.3334-3340.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robertson D, Smith AJ. The microbiology of the acute dental abscess. J Med Microbiol. 2009;58:155–62. doi: 10.1099/jmm.0.003517-0. [DOI] [PubMed] [Google Scholar]
- 19.Guven Y, Ustrin N, Aksakal SD, Topcuoglu N, Aktoren O, Kulekci G. Assessment of the endodontic microbiota of abscessed primary teeth using microarray technology. Int J Dent Res. 2018;29:781–6. doi: 10.4103/ijdr.IJDR_19_18. [DOI] [PubMed] [Google Scholar]
- 20.Sanghavi TH, Shah N, Shah RR, Sanghavi A. Investigate the correlation between clinical sign and symptoms and the presence of P gingivalis, T denticola, and T forsythia individually or as a “Red complex“ by a multiplex PCR method. J Conserv Dent. 2014;17:555–60. doi: 10.4103/0972-0707.144604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noberga LMM, Delboul MG, Martinho FC, Zaia AA, Ferraz CCR, Gomes PFA. Treponema diversity in root canals with endodontic failure. Eur J dent. 2013;7:61–8. [PMC free article] [PubMed] [Google Scholar]
- 22.Foschi F, Cavrini F, Montebugnoli L, Stashenko P, Sambri V, Prati C. Detection of bacteria in endodontic samples by polymerase chain reaction assays and association with defined clinical signs in Italian patients. Oral Microbiol Immunol. 2005;20:289–95. doi: 10.1111/j.1399-302X.2005.00227.x. [DOI] [PubMed] [Google Scholar]
- 23.Bostanci N, Belibasakis GN. Porphorymonas gingivalis: An invasive and evasive opportunistic oral pathogen. FEMS Microbiol Lett. 2012;333:1–9. doi: 10.1111/j.1574-6968.2012.02579.x. [DOI] [PubMed] [Google Scholar]
- 24.Cardoso FGDR, Martinho FC, Ferreira NS, Prado RFD, Manhães-Júnior LRC, Rocco MA, et al. Correlation between volume of root canal, cultivable bacteria, bacterial complexes and endotoxins in primary infection. Braz Dent J. 2019;30:117–22. doi: 10.1590/0103-6440201902239. [DOI] [PubMed] [Google Scholar]
- 25.Ahmed AN, Ramakrishnan R, Victor DJ. Identification of Tannerella forsythia and Treponema denticola in down syndrome subjects and healthy subjects with periodontal disease- A PCR study. Bromed Pharmacol J. 2018;11:525–30. [Google Scholar]
- 26.Chaudhary V, Bhat K, Rao S, Kugayi M, Ingalagi P. Detection of Treponema denticola by PCR in patients with different periodontal status. Int J Curr Micrbiol Appl Sci. 2013;2:172–8. [Google Scholar]
- 27.Cao H, Qi Z, Jiang H, Zhao J, Liu Z, Tang Z. Detection of Porphyromonas endodontalis, Porphyromonas gingivalis and Prevotella intermedia in primary endodontic infections in a Chinese population. Int End J. 2012;46:773–81. doi: 10.1111/j.1365-2591.2012.02035.x. [DOI] [PubMed] [Google Scholar]
- 28.Pattanshetty S, Kotrashetti VS, Bhat K, Nayak RS, Somannavar P, Pujar M, et al. Multiplex polymerase chain reaction detection of selected bacterial species from symptomatic and asymptomatic non-vital teeth with primary endodontic infections. J Invest Clin Dent. 2018;9:e12312. doi: 10.1111/jicd.12312. [DOI] [PubMed] [Google Scholar]
- 29.Lacevic A, Foschi F, Pojskic L, Pojskic N, Bajrovic K, Ijrad J. Correlation of periodontal pathogens in concurrent periodontal diseases. Oral Biol Dent. 2015;3:5–11. [Google Scholar]
- 30.Foschi F, Izard J, Sasaki H, Sambri V, Prati C, Muller R, et al. Treponema denticola in disseminating endodontic infections. J Dent Res. 2006;85:761–5. doi: 10.1177/154405910608500814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siqueira JF, Rocas IN. Microbiology and treatment of endodontic infections. In: Hargreaves K, Berman L, editors. Cohen's Pathway to the Pulp. 10th ed. St. Louis MO: Mosby Inc; 2011. pp. 559–604. [Google Scholar]
- 32.George N, Flamiatos E, Kawasaki K, Kim N, Carriere C, Phan B, et al. Oral microbiota species in acute apical endodontic abscesses. J Oral Microbiol. 2016;8:30989. doi: 10.3402/jom.v8.30989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohanty R, Asopa SJ, Joseph MD, Singh B, Rajguru JP, Saidath K, et al. Red complex: Polymicrobial conglomerate in oral flora: A review. J Family Med Prim Care. 2019;8:3480–6. doi: 10.4103/jfmpc.jfmpc_759_19. [DOI] [PMC free article] [PubMed] [Google Scholar]


