Abstract
Purpose
CD4+T cells are essential in the pathogenesis of allergic asthma. We have previously demonstrated that microRNA-1165-3p (miR-1165-3p) was significantly reduced in T-helper type (Th) 2 cells and that miR-1165-3p was a surrogate marker for atopic asthma. Little is known about the mechanisms of miR-1165-3p in the regulation of Th2-dominated allergic inflammation. We aimed to investigate the associations between Th2 differentiation and miR-1165b-3p in asthma as well as the possible mechanisms.
Methods
CD4+ naïve T cells were differentiated into Th1 or Th2 cells in vitro. MiR-1165-3p was up-regulated or down-regulated using lentiviral systems during Th1/Th2 differentiation. In vivo, the lentiviral particles with the miR-1165-3p enhancer were administered by tail vein injection on the first day of a house dust mite -induced allergic airway inflammation model. Allergic inflammation and Th1/Th2 differentiation were routinely monitored. To investigate the potential targets of miR-1165-3p, biotin-microRNA pull-down products were sequenced, and the candidates were further verified with a dual-luciferase reporter assay. The roles of a target protein phosphatase, Mg2+/Mn2+-dependent 1A (PPM1A), in Th2 cell differentiation and allergic asthma were further explored. Plasma PPM1A was determined by ELISA in 18 subjects with asthma and 20 controls.
Results
The lentivirus encoding miR-1165-3p suppressed Th2-cell differentiation in vitro. In contrast, miR-1165-3p silencing promoted Th2-cell development. In the HDM-induced model of allergic airway inflammation, miR-1165-3p up-regulation was accompanied by reduced airway hyper-responsiveness, serum immunoglobulin E, airway inflammation and Th2-cell polarization. IL-13 and PPM1A were the direct targets of miR-1165-3p. The expression of IL-13 or PPM1A was inversely correlated with that of miR-1165-3p. PPM1A regulated the signal transducer and activator of transcription and AKT signaling pathways during Th2 differentiation. Moreover, plasma PPM1A was significantly increased in asthmatic patients.
Conclusions
MiR-1165-3p negatively may regulate Th2-cell differentiation by targeting IL-13 and PPM1A in allergic airway inflammation.
Keywords: MicrdoRNA, Th2, allergy, IL-13, asthma; inflammation
INTRODUCTION
Atopic asthma is characterized by pulmonary inflammation, airway hyper-responsiveness and airway remodeling.1 After recognizing a presented allergen, CD4+ T cells differentiate into diverse subsets, initiating and regulating the allergic cascade reactions. T-helper type (Th) 2 cells, releasing interleukin (IL)-4, IL-5 and IL-13, play a foundational role in the pathogenesis of allergic asthma.2 IL-4 promotes differentiation and immunoglobulin (Ig) E class-switching of plasma cells, whereas IL-13 mediates airway hyper-responsiveness and mucus hyper-production. IL-5 is highly specific for eosinophil activation and recruitment, leading to eosinophilic inflammation.3 Adoptive transfer of Th2 cells from allergic mice is sufficient to induce eosinophilic pulmonary inflammation, suggesting that Th2 cells are largely indispensable for the onset of atopic asthma.
CD4+-cell differentiation is regulated by various cytokines and transcription factors. As the master transcription factors, T-bet and GATA-3 control the fate of Th1 and Th2 cells, respectively.4 In addition, activation of the signal transducer and activator of transcription (STAT) 1 and STAT5 regulates the development of Th1 and Th2 cells, respectively.5 STAT1 promoted the expression of T-bet, therefore favoring Th1 differentiation.5 IL-2-driven STAT5 activation is essential in Th2 differentiation, which may be independent of GATA-3.6
MicroRNAs (miRNAs) are a class of small noncoding RNAs that function in the posttranscriptional regulation of complimentary messenger RNA (mRNA) stability and translation.7 The roles of miRNAs in asthma pathogenesis have been well documented.8 However, only a few studies have focused on Th2 cells in asthma. In an adoptive transfer asthma model, miR-19 promoted Th2 differentiation and exacerbated airway inflammation.9 Mice deficient in miR-24 and miR-27 in T cells enhanced Th2-cell responses and airway inflammation in an OVA asthma model; both miR-24 and miR-27 targeted IL-4.10
Previously, we have identified a novel miRNA, miR-1165-3p (previously termed novel miRNA-11), which was significantly decreased in the sorted CD4+ T cells from a murine model of asthma and in Th2 cells.11 Moreover, serum miR-1165-3p levels were significantly elevated in asthmatic patients when compared to those of healthy controls.12 In this study, we further demonstrated that miR-1165-3p inhibited Th2 cell differentiation and alleviated type 2 inflammation in an asthma model.
MATERIALS AND METHODS
Animals
Specific pathogen-free female C57BL/6J mice aged 6 to 8 weeks were obtained from Nanjing Medical University (Nanjing, China). All experiments that involved animal and tissue samples were performed in accordance with the guidelines and procedures approved by the Institutional Animal Care and Use Committee of Nanjing Medical University (IACUC No. 1709011).
House dust mite (HDM) sensitization/challenge protocol
C57BL/6J mice were randomly divided into 4 groups: control, HDM, HDM treated with miR-1165-3p blank and HDM treated with miR-1165-3p enhancer. Mice in the latter 3 groups were exposed intratracheally to whole-body HDM extract from Dermatophagoides pteronyssinus (Greer Laboratories, Lenoir, NC, USA) according to the established 14-day model.13 Mice received 100 μg HDM in 40 μL normal saline (NS) on day 0 and 10 μg HDM in 40 μL NS on days 7–11 intratracheally under isoflurane anesthesia to induce allergic lung inflammation. Mice exposed to 40 μL NS according to the HDM protocol served as healthy controls.
To explore whether miR-1165-3p was involved in Th2 differentiation and asthma pathogenesis, lenti-miR-green fluorescent protein (GFP)-miR-1165-3p virus (enhancer) or lenti-miR-GFP control virus (Abm, Zhengjiang, China) at 5 × 107 in 100 μL NS per mouse were administered by tail vein injection before HDM challenge on day 7. The mice were sacrificed 3 days after the final challenge, and bronchoalveolar lavage fluid (BALF) and lung tissues were collected for analyses.
Measurement and analysis of airway responsiveness
Mice were anesthetized with 70 mg/kg pentobarbital and 1.8 g/kg urethane, followed by 0.5 mg/kg pancuronium bromide and the mice were tracheotomized 72 hours after the final challenge.14 Airway hyperreactivity (AHR) was measured in response to increasing doses of acetylcholine via a FinePointe RC system (Buxco Research Systems, Wilmington, NC, USA) under general anesthesia as described before.15
Bronchoalveolar lavage fluid and serum analysis
After AHR measurement, whole blood was collected without anticoagulants and incubated for 1 hour at 37°C; serum was isolated by centrifugation at 2,000 ×g for 30 minutes. The tracheae were exposed, and BALF was collected by lavage with ice-cold phosphate-buffered saline (PBS, 400 μL × 3; 85%-90% of the lavage volume was recovered) via a tracheal catheter. BALF from each mouse was centrifuged at 1,000 rpm for 10 minutes at 4°C, and the total number of inflammatory cells in BALF was examined by flow cytometry analysis. The supernatant of BALF was collected, divided into 4 equal portions and frozen at −80°C for enzyme-linked immunosorbent assay (ELISA).
Lung histology
The lung specimens were fixed in 5% formalin, paraffin-embedded and cut into 5-µm sections that were then stained with either hematoxylin-eosin or periodic acid-Schiff (PAS) before microscopic analysis. The severity of peribronchial inflammation was graded semiquantitatively for the following features: 0, normal; 1, few cells; 2, a ring of inflammatory cells 1 cell layer deep; 3, a ring of inflammatory cells 2–4 cells deep; and 4, a ring of inflammatory cells 4 cells deep. The numerical scores for the abundance of PAS-positive mucus-containing cells in each airway were determined as follows: 0, <0.5% PAS-positive cells; 1, 5%-25%; 2, 25%-50%; 3, 50%-75%; and 4, >75%.16
In vitro mouse primary T-cell polarization
Naive CD4+ T cells purified from mice were cultured for Th-cell differentiation as described before.17,18 Briefly, CD4+ T cells were isolated from the spleen and lymph nodes of C57/B6 mice using a naive CD4+ T cell isolation kit (130-104-453, Miltenyi Biotec, Bergisch Gladbach, Germany) according to the instructions provided by the manufacturer. Naïve CD4+ T cells (2 × 105) were plated onto 96-well tissue culture plates (Thermo Fisher Scientific, Basingstoke, UK) precoated with 5 μg/mL anti-CD3e antibody (16-0031-85, eBioscience, San Diego, CA, USA) and 2 μg/mL anti-CD28 antibody (16-0281-85, eBioscience) on day 0. Th1-cell cultures contained 20 ng/mL murine IL-2 (96-212-12-5, PeproTech, Offenbach, Germany), 20 ng/mL murine IL-12 (96-210-12-10, PeproTech) and 10 μg/mL anti-IL-4 antibody (16-7041-85, eBioscience). Th2-cell cultures contained 20 ng/mL murine IL-2 (96-212-12-5, PeproTech), 100 ng/mL murine IL-4 (96-214-14-20, PeproTech), 10 μg/mL anti-IL-12 (16-7123-85, eBioscience) and 10 μg/mL anti-interferon (IFN)-γ (16-7311-85, eBioscience). After 2 days, the cells were re-cultured for 3 additional days in the same culture medium with cytokines but without anti-CD3e or anti-CD28 antibody stimulation. Lenti-miRa-GFP-miR-1165-3p virus (enhancer), Lenti-III-mir-GFP control virus (blank), Lentil-III-miR-off-control virus (scramble), Lenti-miRa-off-miR-1165-3p virus (silencer) were added on day 1 (multiplicity of infection, MOI = 15), and cells were harvested on day 5 for quantitative polymerase chain reaction (qPCR), western blotting or flow cytometry, while cell supernatants were collected for ELISA.
ELISA
The levels of IL-4, IL-5, IFN-γ (431105, 430805, 431205, Biolegend, San Diego, CA, USA), eotaxin-1 (FMS-ELM070, FCMACS, Nanjing, China) and IL-13 (96-900-K207, PeproTech) in BALF, lung homogenates or cell supernatants, total IgE (555248, BD Biosciences, San Jose, CA, USA) in sera, or protein phosphatase, Mg2+/Mn2+-dependent 1A (PPM1A; HM11307, Bio-Swamp, Wuhan, China) and total IgE (E-EL-H2161C, Elabscience, Wuhan, China) in human plasma were measured using commercial ELISA kits according to the instructions provided by the manufacturer.
Flow cytometry for cell counting
Inflammatory cells were counted and analyzed by flow cytometry. Cells were counted by counting beads (01-1234-42, eBioscience). After stained with CD16/CD32 Fc receptor (FcR)-blocking antibody (14-0161-85, eBioscience) and fixable viability dye eFluor™ 506 (65-0866-14, eBioscience) according to the manufacturer's instructions, neutrophils (CD45+Ly6G+F4/80−), eosinophils (CD45+SiglecF+F4/80−), macrophages (CD45+SiglecF+F4/80+) and lymphocytes (CD45+CD3+) in BAL fluid were measured by flow cytometry.9,19,20
Staining nuclear protein for flow cytometry
The spleen specimens from animal experiments were prepared as single-cell suspensions. After erythrocyte lysis, differentiated Th1/Th2 cells were stained with CD16/CD32 FcR-blocking antibody, fixable viability dye eFluor™ 506 and anti-CD4-APC (17-0041-83, eBioscience). For intracellular staining, cells were fixed and permeabilized (00-5523-00, eBioscience) according to the manufacturer's instructions, and then intracellular products were stained. Flow cytometry was performed with anti-GATA-PE-Cy7 (560405, BD Biosciences) and anti-T-bet-PE-Cy7 (25-5825-80, eBioscience). A FACS Stratedigm (BD Biosciences) was used for flow cytometry, and data were analyzed by FlowJo software (Treestar, Woodburn, OR, USA).
Reverse transcription and qPCR analysis
Total RNA was prepared from differentiated T cells using an miRNeasy Mini Kit (217004, Qiagen, Hilden, Germany). The mRNAs were reverse transcribed with 5X All-In-One RT MasterMix (G490, Abm, Zhenjinag, China), while miRNAs were reverse transcribed by a Mir-X miRNA First-Strand Synthesis Kit (638313, TaKaRa Bio, Kusatsu, Japan). The qPCR analysis was performed using a CFX96 system (Bio-Rad Laboratories, Hercules, CA, USA) in conjunction with SYBR Advantage qPCR Premix (639676, TaKaRa Bio). The cycling conditions were 95°C for 30 seconds, followed by 95°C for 5 seconds and 60°C for 30 seconds for up to 40 cycles and dissociation at 95°C for 5 seconds, 60°C for 30 seconds and a final extension with 95°C for 15 seconds. The relative abundance of gene targets was determined by the comparative cycle threshold (CT) number normalized to β-actin. The relative abundance of miRNA was determined by the comparative CT number normalized to U6. The primers used are shown in Supplementary Table S1.
Western blot analysis
Total cellular protein was collected following lysis in RIPA Lysis Buffer (P0013B, Beyotime Biotech, Beijing, China) with PMSF (ST506, Beyotime Biotech) on ice and centrifugation for 10 minutes at 12,000 rpm at 4°C. The supernatant was then transferred to a new tube and was denatured in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (P0015, Beyotime Biotech) with heating at 100°C for 10 minutes. The supernatant was then stored at −80°C. The proteins were separated by 12% SDS-PAGE. After electrophoresis, the separated proteins were transferred to 0.45-µm polyvinylidene difluoride membranes (INIP86300, Merck Millipore, Burlington, MA, USA) using a wet transfer method. Nonspecific sites were blocked with 5% nonfat milk in tris-Buffered Saline-Tween 20 (25 mM Tris [pH 7.5], 150 mM NaCl and 0.1% Tween 20) for 2 hours, and the blots were incubated with primary antibodies, including anti-β-actin (4970L, Cell Signaling Technology, Danvers, MA, USA), anti-GATA3 (ab106625, Abcam, Cambridge, England), anti-T-bet (ab91109, Abcam), anti-PPM1a (ab14824, Abcam), and Phospho-Stat Antibody Sampler Kit (9914, Cell Signaling Technology), Stat antibody sampler kit (9939, Cell Signaling Technology), Phospho-AKT (Ser473) (4060, Cell Signaling Technology), and AKT antibody (4691,Cell Signaling Technology)overnight at 4°C. HRP-linked Anti-rabbit IgG and HRP-linked anti-mouse IgG, (7074, 7076, Cell Signaling Technology) were used to detect antibody binding. After treating the membranes with Immobilon Western Chemiluminescent HRP Substrate (WBKLS0500, Merck Millipore), the binding of specific antibodies was visualized using a Syngene G:BOX Imaging System and was analyzed with ImageJ.
Biotin-miRNA pull-down assays
NIH-3T3 cells were seeded in a 10-cm dish at a density that allowed them to grow for 24 hours without reaching complete confluency. A final concentration of Biotin-miR-1165-3p mimics or Biotin-miRNC (GenePharma, Shanghai, China) of 50 µM was used to transfect NIH-3T3 cells using the standard protocol for Hieff Trans™ Liposomal Transfection Reagent (40802ES03, YEASEN, Shanghai, China). Forty-eight hours after washing twice with ice-cold PBS and cross-linking in a UV cross-linker at 1,200 mJ/cm2 for 2 minutes,21 the cells were lysed in cell lysis buffer (9803S, Cell Signaling Technology) supplemented with a protease inhibitor cocktail (539134, Merck Millipore) for 20 minutes, after which the lysates were sonicated for 2 minutes at 20% amplitude. Lysates were cleared of cell debris by centrifugation at 13,000 rpm for 10 minutes. For a 10-cm plate, 50 µL of Dynabeads M-280 Streptavidin beads (11205D, Thermo Fisher Scientific) were activated according to the manufacturer's protocol. The beads were blocked with 10 µg/mL RNase-free BSA and yeast transfer RNA (AM7119, ThermoFisher Scientific) for 3 hours at 4°C. After washing the beads with cell lysis buffer, lysates that had previously been prepared were incubated with the blocked beads and were incubated at 4°C overnight, followed by RNA extraction. Total RNA bound to the streptavidin beads was extracted by using an miRNeasy Mini Kit. The amount of pull-down RNA was quantified by RNA-seq.
RNA-seq and data analysis
RNA from biotin-tagged miR-1165-3p mimics or miRNC pull-down assays were subjected to RNA deep sequencing, which was performed by the RiboBio Institute (Guangzhou, China). RNA-seq libraries were prepared using an Illumina RNA-Seq Preparation Kit and sequenced by a Hiseq3000 PE150 (Illumina, San Diego, CA, USA). TopHat output data were analyzed by Cufflinks to estimate RPKM values for known transcripts and to analyze differentially expressed transcripts.22 Differentially expressed genes were selected by the logFC > 1 and P value < 0.05.
Bioinformatics analysis
The target genes of miR-1165-3p were predicted using miR and a, PITA and RNAhybrid.23,24,25 To create a visual representation, we further screened and sorted the predicted results and used the predicted results of the 3 software programs as the candidate target bases of the miRNA. The intersection of the candidate mRNAs and the different mRNAs in RNA-seq were determined, and a miRNA-mRNA regulatory network was constructed using Cytoscape 3.6.26
Transfection
During the differentiation period, Th2 cells were transfected with either a PPM1A overexpression vector (MR205969, OriGene Tech, Rockville, MD, USA) or an empty vector (PS100001, OriGene Tech) using a Neon Transfection system (MPK1025, Thermo Fisher Scientific)27 on days 1 and 3. Transfections were performed using 2–3 × 107 cells/mL in 10 µL “R buffer” with 1 µg plasmid. The optimal setting used was 1,550 V with three 10-minute pulses. After transfection, the mRNA and protein expression levels of PPM1A were determined by quantitative reverse transcription polymerase chain reaction (qRT-PCR) and western blotting on day 5, respectively.
Dual-luciferase reporter assay
3′ UTR dual-luciferase plasmids, including mutant plasmids, were constructed using plenti-UTR-Dual-Luc (Abm, Zhengjia, China). MiR-1165-3p mimics and the relevant control were purchased from GenePharma. One day before transfection, HEK293T cells were plated on 24-well plates. Blank plasmids and reporter constructs (50 ng) containing the mutant or wild-type 3′ UTR of PPM1A or IL-13 were cotransfected with miR-1165-3p into cells according to the manufacturer's protocol of Hieff Trans™ Liposomal Transfection Reagent. Luciferase activities were assessed using a dual-luciferase reporter assay system (E1910, Promega, Madison, WI, USA) according to the manufacturer's instructions at 48 hours after transfection.
Patient selection and clinical data
Based on GINA 2017 and the Guidelines for the Prevention and Control of Bronchial Asthma of China 2016, a total of 20 asthmatic patients were recruited from the Department of Respiratory and Critical Care Medicine at the First Affiliated Hospital of Nanjing Medical University between September and November of 2017. A total of 18 healthy subjects without current or previous asthma or allergic disease diagnoses were also recruited. Written informed consent was obtained from all of the subjects, and this study was approved by the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University (2017-SR-298). Venous blood samples (approximately 5 mL) were collected in anticoagulant tubes. Samples were then centrifuged at 2,000 rpm for 10 minutes; the plasma was collected and stored at −80°C for future use.
Statistical analysis
The data are expressed as the mean ± standard error of the mean. All of the tests were performed using Prism 7.00 (GraphPad Software, San Diego, CA, USA). The results were analyzed by one-way analysis of variance for repeated measures, followed by Dunnett's post hoc test to determine differences among multiple comparisons. The significance level was set at P < 0.05.
RESULTS
MiR-1165-3p inhibits Th2 differentiation in vitro
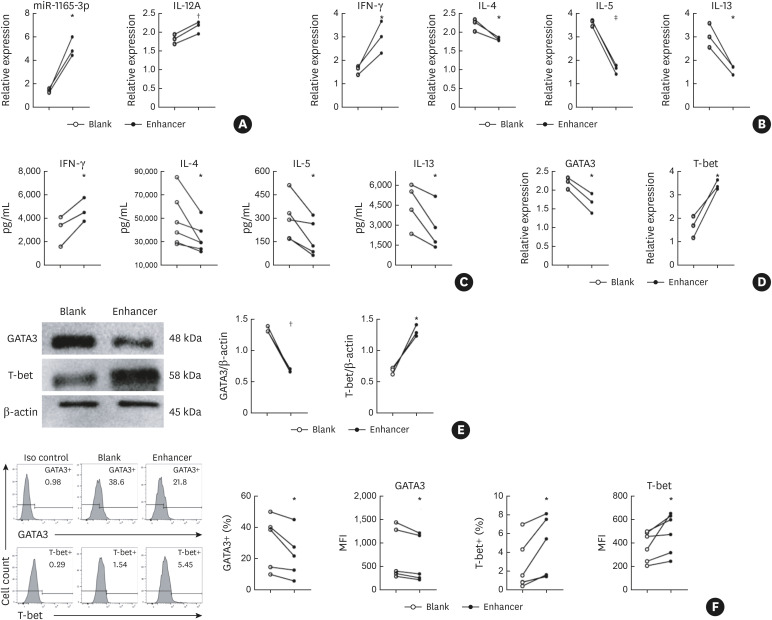
Naive CD4+ T cells purified from the spleens of mice were cultured for Th-cell differentiation as described before.11 Upon infection with lentivirus-encoding miR-1165-3p (Supplementary Fig. S1), miR-1165-3p was significantly increased in Th2 cells (Fig. 1A). To analyze the effects of miR-1165-3p on Th2 differentiation, we first measured the cytokines produced and released by CD4+ T cells. As shown in Fig. 1B and C, type 1 cytokines (IFN-γ and IL-12) were increased and type 2 cytokines (IL-4, IL-5 and IL-13) were decreased, suggesting that miR-1165-3p elevation was accompanied by reduced Th2 differentiation. To further determine whether the master transcription factors T-bet and GATA-3 were involved, we quantified T-bet and GATA-3 using qRT-PCR, western blotting and intracellular flow cytometry (Fig. 1D and F). As expected, miR-1165-3p significantly increased T-bet expression and decreased GATA-3 expression. Overall, miR-1165-3p suppressed type 2 cytokines (IL-4, IL-5 and IL-13) and the Th2 master transcription factor GATA-3, implying that miR-1165-3p was a negative modulator of Th2 differentiation.
Fig. 1. MiR-1165-3p inhibited Th2 differentiation. CD4+ T cells were cultured in Th2-cell conditions for 5 days and were transfected with miR-1165-3p enhancer or blank lentivirus on day 1. (A) MiR-1165-3p was significantly increased in Th2 cells infected with lentivirus-encoding miR-1165-3p (enhancer), (B, C) analysis of type 1 cytokines (IFN-γ and IL-12) and type 2 cytokines (IL-4, IL-5 and IL-13). (D-F) The Th1 and Th2 master transcription factors T-bet and GATA-3 were measured by qRT-PCR, western blotting and intracellular flow cytometry. Data are representative of independent experiments with similar results (n = 3–6).
MiR-1165-3p, microRNA-1165-3p; Th, T helper type; IFN, interferon; IL, interleukin; qRT-PCR, quantitative reverse transcription polymerase chain reaction; MFI, mean fluorescence intensity.
*P < 0.05; †P < 0.01; ‡P < 0.001.
Down-regulation of miR-1165-3p promotes Th2 differentiation
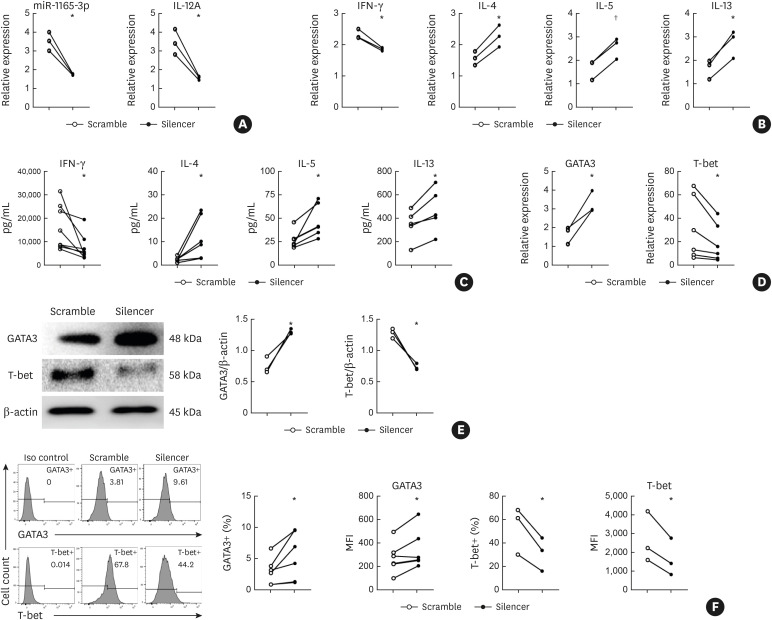
As miR-1165-3p was increased in Th1 cells,11 we wondered whether down-regulation of miR-1165-3p would antagonize Th1 differentiation and therefore favor Th2 differentiation. As expected, lentivirus-encoding short hairpin RNA for miR-1165-3p significantly decreased miR-1165-3p in Th1 cells (Fig. 2A). In addition to decreased miR-1165-3p, type 1 cytokines (IFN-γ and IL-12) were reduced, and type 2 cytokines (IL-4, IL-5 and IL-13) were up-regulated as measured by qRT-PCR (Fig. 2B) and verified by ELISA (Fig. 2C). Accordingly, the Th1 master transcription factor T-bet was significantly decreased and the Th2 master transcription factor GATA-3 was significantly increased (Fig. 2D and F), implying that miR-1165-3p may promote Th1 cell differentiation and suppress Th2-cell differentiation.
Fig. 2. MiR-1165-3p silencing promoted Th2 differentiation. CD4+ T cells were cultured in Th1-cell conditions for 5 days and were infected with miR-1165-3p silencer or scramble lentivirus on day 1. (A) MiR-1165-3p significantly decreased in Th1 cells infected with lentivirus-encoding short hairpin RNA for miR-1165-3p (silencer) (B-C) analysis of type 1 cytokines (IFN-γ and IL-12) and type 2 cytokines (IL-4, IL-5 and IL-13). (D-F) The Th1 and Th2 master transcription factors T-bet and GATA-3 were measured by qRT-PCR, western blotting and intracellular flow cytometry. Data are representative of independent experiments with similar results. (n = 3–6).
MiR-1165-3p, microRNA-1165-3p; Th, T helper type; IFN, interferon; IL, interleukin; qRT-PCR, quantitative reverse transcription polymerase chain reaction; MFI, mean fluorescence intensity.
*P < 0.05; †P < 0.01.
MiR-1165-3p alleviates allergic inflammation in asthma
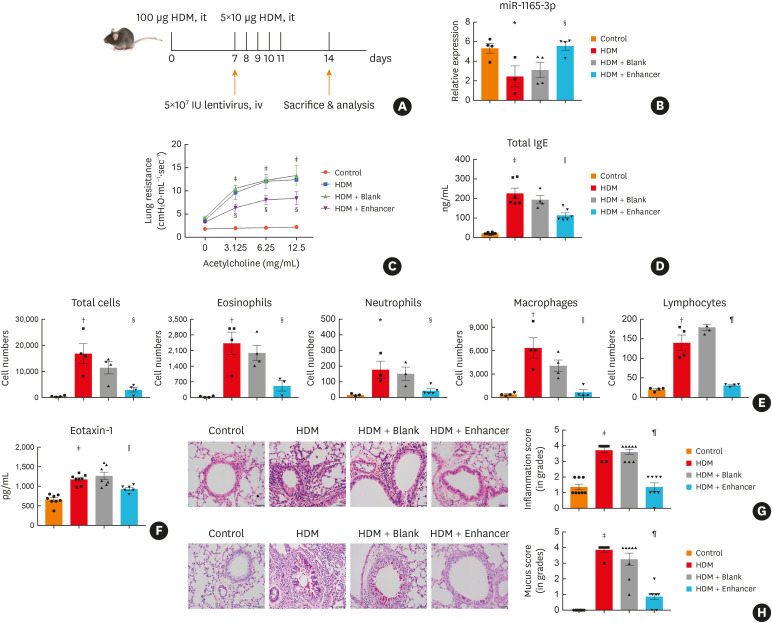
Given the critical role of type 2 immune responses in the pathogenesis of asthma, we next investigated whether miR-1165-3p could alter type 2 inflammation in airways in vivo. We designed a LentimiRa-GFP-miR-1165-3p virus (enhancer) or Lenti-III-mir-GFP control virus (blank), and approximately 5 × 107 lentiviral particles were injected through the tail vein into a mouse on day 7 in an HDM acute asthma model (Fig. 3A).13 Compared to control virus, enhancer virus significantly increased miR-1165-3p in the lung (Fig. 3B). In parallel, airway hyper-responsiveness and total serum IgE were significantly decreased in the asthma model mice treated with miR-1165-3p enhancer virus (Fig. 3C and D). Furthermore, eosinophils infiltration in BALF (Fig. 3E) and eotaxin-1 was also decreased in the asthmatic mouse treated with miR-1165-3p enhancer lentivirus (Fig. 3F). Inflammatory response in the lung parenchyma (Fig. 3G) was comparably significantly alleviated in the mouse treated with miR-1165-3p enhancer lentivirus. Moreover, goblet cell hyperplasia and mucus secretion in the lumen of the bronchioles were significantly relieved (Fig. 3H). In summary, with the up-regulation of miR-1165-3p in the lung, type 2 pulmonary inflammation in a mouse model of acute asthma was significantly alleviated, suggesting that miR-1665-3p might negatively regulate the differentiation of Th2 cells.
Fig. 3. MiR-1165-3p inhibited allergic airway inflammation. (A) Experimental design of the study. (B) The level of miR-1165-3p in different groups. (C) Mice inhaled increasing doses of acetylcholine (0–12.5 mg/mL), and AHR was measured. (D) The concentration of IgE was measured by ELISA. (E) Total and differential cell counts in bronchoalveolar lavage fluid were determined by flow cytometry. (F) The concentrations of eotaxin-1 in lung homogenates were measured with ELISA (G) Lung sections were stained with hematoxylin and eosin stain to analyze the infiltration of inflammatory cells, and the severity of peribronchial inflammation was graded semiquantitatively. (H) Lung sections were stained with PAS to assess goblet cell hyperplasia, PAS-positive and PAS-negative epithelial cells were counted, and the percentage of PAS-positive cells per bronchiole was calculated. Scale bar, 50 μm (n = 3–8).
HDM, house dust mite; miR-1165-3p, microRNA-1165-3p; ELISA, enzyme-linked immunosorbent assay; AHR, airway hyperreactivity; IgE, immunoglobulin E; PAS, periodic acid-Schiff.
*P < 0.05, †P < 0.01, ‡P < 0.001, HDM vs. control group; §P < 0.05, ‖P < 0.01, ¶P < 0.001, HDM + enhancer vs. HDM + blank group.
MiR-1165-3p represses Th2 cells in vivo
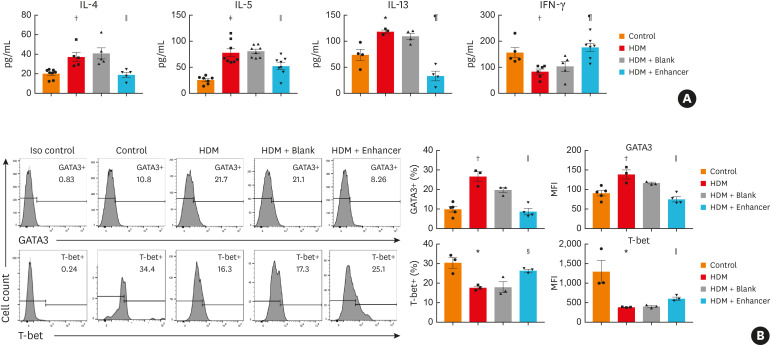
To explore whether Th2 cells were inhibited in the asthma model mice treated with miR-1165-3p enhancer lentivirus, we quantified cytokines in BALF and infiltrated CD4+ T cells in the lung parenchyma. As shown in Fig. 4A, miR-1165-3p significantly decreased the type 2 cytokines IL-4, IL-5 and IL-13. Intracellular transcription factor staining revealed that CD4+GATA-3+ (Th2) cells were significantly reduced from the allergic airway inflammation mouse model infected with miR-1165-3p enhancer lentivirus, while CD4+T-bet+ (Th1) cells were increased (Fig. 4B). In summary, miR-1165-3p suppressed Th2 cell differentiation in vitro and in an allergic airway inflammation mouse model.
Fig. 4. MiR-1165-3p repressed Th2 cell function in vivo. (A) The concentrations of different cytokines in bronchoalveolar lavage fluid were measured with enzyme-linked immunosorbent assay. (B) The percentages and MFI of GATA3+ or T-bet+ cells in CD4+ cells from the spleen were determined by flow cytometry (n = 3–8).
IL, interleukin; IFN, interferon; miR-1165-3p, microRNA-1165-3p; HDM, house dust mite; MFI, mean fluorescence intensity.
*P < 0.05, †P < 0.01, ‡P < 0.001, HDM vs. control group; §P < 0.05, ‖P < 0.01, ¶P < 0.001, HDM+ enhancer vs. HDM + blank group.
PPM1A and IL-13 are the direct targets of miR-1165-3p
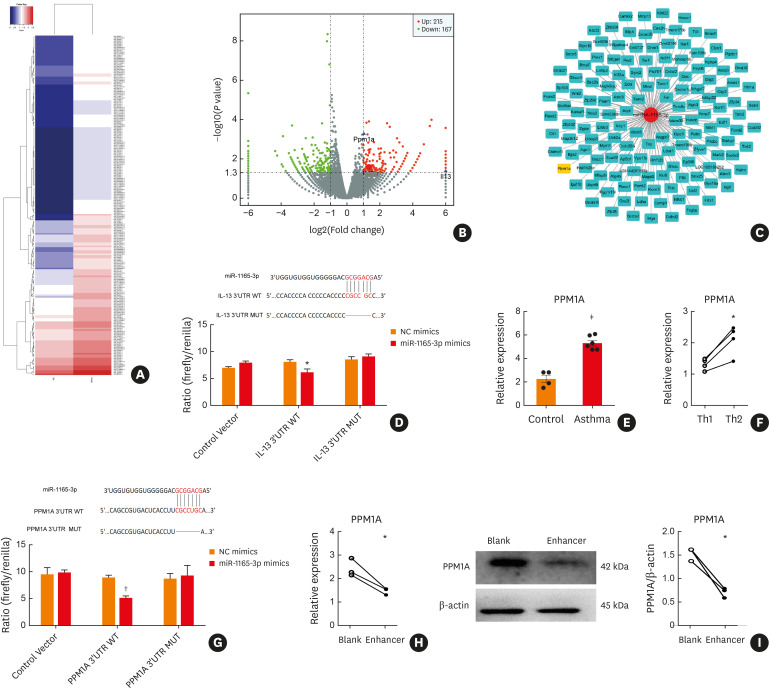
To identify mRNAs directly targeted by miR-1165-3p, we used a biotinylated miRNA-mRNA pull-down approach.21 Considering cell number and transfection efficiency, we took NIH-3T3 as a replace.28 NIH-3T3 cells were transfected with biotinylated miR-1165-3p or negative control. RNAs pulled down by biotinylated miR-1165-3p or the negative control miRNA were subjected to RNA-seq analysis. Compared with the negative control, 159 mRNAs (including IL-13, PPM1A and other potential targets) were enriched in the bio-miR-1165-3p pull-down (Fig. 5A and B). The intersection of different bioinformatics analyses suggested that PPM1A was significantly associated with miR-1665-3p (Fig. 5C).
Fig. 5. MiR-1165-3p targeted IL-13 and PPM1A. (A) Heat map analysis of differentially expressed mRNA between the biotin-miR-1165-3p and NC groups. Blue indicates low expression, and red indicates high expression. (B) Volcano plot assessment of mRNA expression in NIH-3T3 cells between the biotin-miR-1165-3p and NC groups. The left vertical line represents a fold change of ≤−2 (down-regulated), the right vertical line represents a fold change of ≥2 (up-regulated) and the horizontal line represents a P value of ≤0.05. (C) The miRNA-mRNA network was constructed according to the intersection of the bioinformatics data analyses and biotin-miRNA pull-down. (D) Sequence alignment of putative miR-1165-3p binding sites in the 3′ UTR of IL-13. A deletion mutation corresponding to miR-1165-3p target sites is shown. Ratios of repressed luciferase activity of cells in the presence of WT, empty vector or different mutated 3′ UTRs transfected with miR-1165-3p compared to cells transfected with NC mimics (n = 3). (E) The mRNA level of PPM1A in CD4+ T cells from asthmatic or control mice (n = 3–6). (F) The mRNA level of PPM1A in Th1 or Th2 cells (n = 3–6). (G) Sequence alignment of putative miR-1165-3p binding sites in the 3′ UTR of PPM1A. A deletion mutation corresponding to miR-1165-3p target sites is shown. Ratios of repressed luciferase activity of cells in the presence of WT, empty vector or different mutated 3′ UTRs transfected with miR-1165-3p compared to cells transfected with NC mimics. (n = 3). (H) The mRNA level of PPM1A in Th2 cells transfected with miR-1165-3p enhancer or blank lentivirus (n = 3–6). (I) PPM1A in Th2 cells transfected with miR-1165-3p enhancer or blank lentivirus was measured by Western blotting (n = 3–6).
IL, interleukin; mRNA, messenger RNA; miR-1165-3p, microRNA-1165-3p; PPM1A, protein phosphatase, Mg2+/Mn2+-dependent 1A; miRNA, microRNA; NC, negative control; Th, T helper type; WT, wild-type.
*P < 0.05; †P < 0.01; ‡P < 0.001.
IL-13 had a putative binding site for miR-1165-3p within its 3′-UTR, which was further confirmed using a luciferase reporter assay (Fig. 5D). As IL-13 was elevated in asthma and Th2 cells, PPM1A was also increased in the mouse model of allergic airway inflammation (Fig. 5E) and in vitro Th2 cells (Fig. 5F). In the luciferase reporter assay, cotransfection with the luciferase construct (containing the 3′-UTR of PPM1A) and the miR-1165-3p mimic resulted in a decrease in luciferase signal compared to mutant plasmids or empty plasmids (Fig. 5G), implying that PPM1A was one of the direct targets of miR-1165-3p. Furthermore, PPM1A was significantly reduced in Th2 cells infected with miR-1165-3p enhancer lentivirus (Fig. 5H and I). Collectively, IL-13 and PPM1A were direct targets of miR-1165-3p in Th2 differentiation.
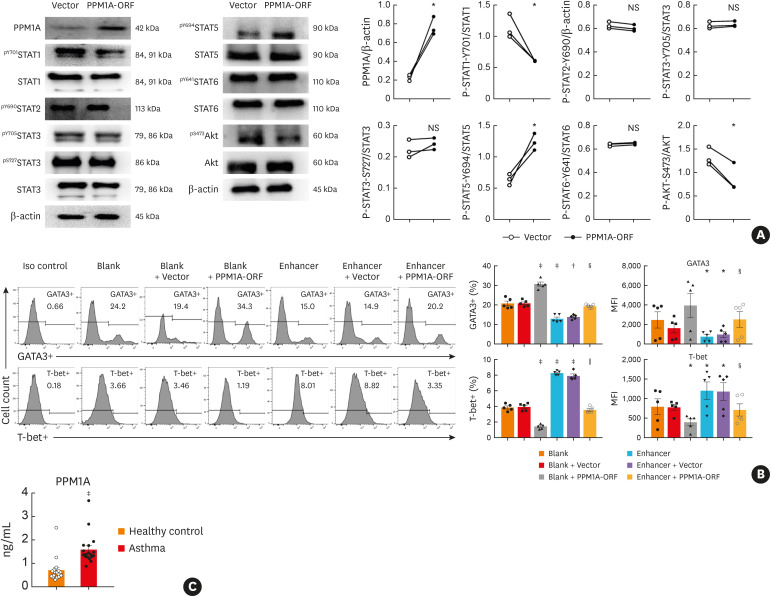
PPM1A regulates Th2 differentiation via STAT and AKT signaling.
PPM1A is a type of Ser/Thr protein phosphatase, which negatively regulate STAT1 and AKT during the differentiation of bone marrow monocytes to macrophages.29 Therefore, Th2 cells were transfected with PPM1A-ORF, which increased PPM1A expression. Furthermore, the phosphorylation of STAT1 and AKT was repressed. In contrast, STAT5 phosphorylation was increased. STAT2, STAT3 and STAT6 showed no difference in the Th2 cells electro-transfected with PPM1A coding plasmid or blank control plasmid (Fig. 6A).
Fig. 6. PPM1A regulated Th2 differentiation through STAT and AKT signaling pathways. (A) PPM1A expression was determined by western blot analysis in cells transfected with either pcMV6-entry (vector) or PPM1A-ORF. STAT and AKT were analyzed by western blot. β-actin was included as an internal control (n = 3–6). PPM1A-ORF vs. vector; (B) Flow cytometry analysis of GATA-3 expression or T-bet expression in Th2-polarized CD4+ T cells infected with miR-1165-3p enhancer lentivirus and/or transfected with PPM1A-ORF. Data are representative of independent experiments with similar results (n = 3–6) (C) PPM1A in the plasma of the asthmatic and control groups was quantified (n = 18–20).
PPM1A, protein phosphatase, Mg2+/Mn2+-dependent 1A; STAT, signal transducer and activator of transcription; NS, not significant; miR-1165-3p, microRNA-1165-3p.
*P < 0.05, †P < 0.01, ‡P < 0.001 vs. blank group; §P < 0.05, ‖P < 0.01, enhancer + PPM1A-ORF vs. enhancer + vector.
To better explore the roles of miR-1165-3p and PPM1A in Th2 differentiation, we co-transfected Th2 cells with miR-1165-3p enhancer lentivirus and PPM1A-ORF plasmid (Fig. 6B). PPM1A overproduction in Th2 cells infected with miR-1165-3p lentivirus at least partially restored the expression of the master transcription factor GATA-3, further suggesting that PPM1A may be required for the differentiation of Th2 cells. To add the clinical significance of our study, we sampled plasma from allergic asthmatic patients, in which total IgE was significantly increased (Supplementary Fig. S2). In these allergic asthmatic patients, PPM1A was also elevated (Fig. 6C), which may further support the importance of PPM1A for Th2 cells and the pathogenesis of allergic asthma.
DISCUSSION
The functions of miRNAs in Th2 differentiation in allergic asthma remain largely elusive. In this study, we reported that the over-expression of miR-1165-3p decreased the production of IL-4, IL-5 and IL-13. Furthermore, GATA-3 was reduced in CD4+ cells infected with lentivirus-encoding miR-1165-3p. In contrast, upon the down-regulation of miR-1165-3p, type 2 cytokines (IL-4, IL-5 and IL-13) and the Th2 master transcription factor GATA-3 were increased. In the HDM-induced allergic airway inflammation model, miR-1165-3p lentivirus treatment relived airway hyper-responsiveness, total IgE and pulmonary inflammation. Moreover, miR-1165-3p suppressed Th2 differentiation in vivo. MiR-1165-3p was shown to target IL-13 and PPM1A, which regulated Th2 polarization via STAT1, STAT5 and AKT signaling pathways. Ectopic expression of PPM1A in CD4+ T cells over-expressing miR-1165-3p promoted Th2 differentiation. In addition, PPM1A was significantly elevated in asthmatic patients. Combined with our previous observation that miR-1165-3p was a diagnostic marker for asthma,12 these results suggested that miR-1165-3p targeted IL-13 and PPM1A to control Th2 differentiation and pulmonary inflammation in asthma.
The prevalence of asthma has increased considerably in the past 3 decades.30 Though heterogeneity in asthma phenotypes has been recognized, allergic asthma is the most common type.31 For almost 3 decades, the Th1 and Th2 paradigm has provided a productive model for exploring the pathogenesis of allergic diseases.32 Though Th9,33 Th17,34 Th22,35 Tfh,36 Treg37 and other non-Th2 CD4+ cells may contribute to asthma development, Th2 cells still orchestrate the type 2 immune response, leading to IgE-primed sensitization, airway hyper-reactivity and eosinophilia.38 Therefore, a better understanding of the mechanisms behind the differentiation of Th2 in asthma is a prerequisite for Th2-based therapy for allergic asthma and other diseases.
In a previous study, we reported that miR-1165-3p was lower in Th2 cells.11 Therefore, we transfected Th2 cells with lentivirus-encoding miR-1165-3p. With elevated miR-1165-3p, type 2 cytokines (IL-4, IL-5 and IL-13) were decreased in Th2 cells; furthermore, the Th2 master transcription factor GATA-3 was also suppressed, suggesting that miR-1165-3p down-regulated GATA-3-dependent Th2 differentiation. Moreover, in the HMD-induced allergic airway inflammation model, ectopic expression of miR-1165-3p suppressed Th2 pulmonary inflammation, suggesting that miR-1165-3p may be a potential candidate for therapeutic intervention against allergic airway inflammation.
To explore the direct targets of miR-1165-3p, we used biotin-miRNA pull-down and RNA-seq, in which 159 mRNAs were enriched in the Bio-miR-1165-3p pull-down compared to the RNA pull-down with a biotinylated negative control. Using the combination of bioinformatics analysis and a luciferase reporter assay, we further demonstrated that IL-13 and PPM1A were direct targets of miR-1165-3p. As a fundamental mediator of allergic asthma, IL-13 mediates eosinophilic infiltration, mucus secretion and airway hyper-responsiveness.39 In addition, IL-13 mediates tissue fibrosis40; therefore, miR-1165-3p-dependent degradation of IL-13 may alleviate chronic asthma as well as acute asthma.
In genome-wide association studies (GWAS), PPM1A was a risk factor for allergic rhinitis.41 In resting CD4+ T cells, PPM1A is relatively high.42 Compared to Th1 cells, PPM1A transcripts were higher in Th2 cells,43 suggesting that PPM1A may regulate Th2 differentiation. In fact, PPM1A is a negative threshold regulator of type 1 macrophage differentiation through AKT or STAT1, establishing it as a key phosphatase that orchestrates cell polarization.29 The direct association between PPM1A and IL-13 remains elusive in the literature. However, PP1β augmented IL-5 and IL-13 expression in IL-2 stimulated T cells.44 In contrast, protein tyrosine phosphatase Shp-1 is a negative modulator of IL-13,45 implying that protein phosphatase may be involved in IL-13 and Th2 differentiation. Collectively, our results showed that PPM1A activated STAT5, and inhibited AKT and STAT1, which cooperatively promoted Th2 differentiation.
The clinical observation that plasma PPM1A was higher in asthmatic patients further supported the importance of PPM1A in Th2 allergic asthma. Previously, we have showed that miR-1165-3p was lower in Th2 cells or CD4+ T cells from asthmatic mice11 and that miR-1165-3p was paradoxically increased in the sera from clinical asthmatic patients.12 Extracellular miRNAs may not necessarily reflect the abundance of miRNAs in the cell of origin. For example, miR-141 was up-regulated in the plasma of prostate cancer patients and was down-regulated in prostate cancer cell lines.46 Patients with asthma were given medications to control symptoms, which may influence the release of miR-1165-3p from cells into blood. Moreover, expression levels of miRNAs may be not associated with their regulatory activities.47 For example, miR-34c-5p and its target STAB2 were both down-regulated in CRC tissues,48 implicating that interactions between miRNAs and their targets may be more intricate. The mechanisms for the discrepancy of miR-1165-3p and the target PPM1A in asthma warrant further research. Indeed, our study was hampered by the inadequate clarifications in the expression and the regulation of miR-1165-3p.
In summary, we investigated the role of miR-1165-3p in the progression of allergic airway inflammation and its potential underlying mechanism to regulate Th2 differentiation. Our results provided direct evidence that miR-1165-3p inhibits the Th2 response of allergy through the STAT and AKT signaling pathways by targeted inhibition of PPM1A, shedding the light that miR-1165-3p and PPM1A may be effective targets for the prevention and treatment of allergic asthma and associated diseases.
ACKNOWLEDGMENTS
This research was supported by the Precision Medicine Research of The National Key Research and Development Plan of China (2016YFC0905800), National Natural Science Foundation of China (81671563, 81770031, 81700028), Natural Science Foundation of Jiangsu Province (BK20171501, SBK2017080, BK20181497), Jiangsu Province's Young Medical Talent Program, China (QNRC2016600) and Jiangsu Provincial Health and Family Planning Commission Foundation (Q2017001).
Footnotes
Disclosure: There are no financial or other issues that might lead to conflict of interest.
SUPPLEMENTARY MATERIALS
Primers in the study
The transfection efficiency of lentiviral on CD4+ T cells. CD4+ T cells were cultured in T-helper type 2 cell conditions for 5 days and were transfected with LentimiRa-GFP-miR-1165-3p viral on day 1. GFP was used as a marker for miR-1165-3p overexpression.
The total IgE in the plasma of the asthmatic and control groups. The IgE levels in the plasma was measured using ELISA kit (n = 18–20).
References
- 1.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18:716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- 2.Malmhäll C, Alawieh S, Lu Y, Sjöstrand M, Bossios A, Eldh M, et al. MicroRNA-155 is essential for T(H)2-mediated allergen-induced eosinophilic inflammation in the lung. J Allergy Clin Immunol. 2014;133:1429–1438. 1438.e1–1438.e7. doi: 10.1016/j.jaci.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Barnes PJ. The cytokine network in asthma and chronic obstructive pulmonary disease. J Clin Invest. 2008;118:3546–3556. doi: 10.1172/JCI36130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenner RG, Townsend MJ, Jackson I, Sun K, Bouwman RD, Young RA, et al. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proc Natl Acad Sci U S A. 2009;106:17876–17881. doi: 10.1073/pnas.0909357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, et al. T-bet is a STAT1-induced regulator of IL-12R expression in naïve CD4+ T cells. Nat Immunol. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- 6.Zhu J, Cote-Sierra J, Guo L, Paul WE. Stat5 activation plays a critical role in Th2 differentiation. Immunity. 2003;19:739–748. doi: 10.1016/s1074-7613(03)00292-9. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 8.Maneechotesuwan K. Role of microRNA in severe asthma. Respir Investig. 2019;57:9–19. doi: 10.1016/j.resinv.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Simpson LJ, Patel S, Bhakta NR, Choy DF, Brightbill HD, Ren X, et al. A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nat Immunol. 2014;15:1162–1170. doi: 10.1038/ni.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pua HH, Steiner DF, Patel S, Gonzalez JR, Ortiz-Carpena JF, Kageyama R, et al. MicroRNAs 24 and 27 suppress allergic inflammation and target a network of regulators of T helper 2 cell-associated cytokine production. Immunity. 2016;44:821–832. doi: 10.1016/j.immuni.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Chen Z, Xu K, Wang Z, Wu C, Sun Z, et al. Next generation sequencing for miRNA profile of spleen CD4+ T cells in the murine model of acute asthma. Epigenomics. 2018;10:1071–1083. doi: 10.2217/epi-2018-0043. [DOI] [PubMed] [Google Scholar]
- 12.Wu C, Xu K, Wang Z, Chen Z, Sun Z, Yu W, et al. A novel microRNA miR-1165-3p as a potential diagnostic biomarker for allergic asthma. Biomarkers. 2019;24:56–63. doi: 10.1080/1354750X.2018.1501762. [DOI] [PubMed] [Google Scholar]
- 13.Draijer C, Robbe P, Boorsma CE, Hylkema MN, Melgert BN. Dual role of YM1+ M2 macrophages in allergic lung inflammation. Sci Rep. 2018;8:5105. doi: 10.1038/s41598-018-23269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takyar S, Vasavada H, Zhang JG, Ahangari F, Niu N, Liu Q, et al. VEGF controls lung Th2 inflammation via the miR-1-Mpl (myeloproliferative leukemia virus oncogene)-P-selectin axis. J Exp Med. 2013;210:1993–2010. doi: 10.1084/jem.20121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boldogh I, Bacsi A, Choudhury BK, Dharajiya N, Alam R, Hazra TK, et al. ROS generated by pollen NADPH oxidase provide a signal that augments antigen-induced allergic airway inflammation. J Clin Invest. 2005;115:2169–2179. doi: 10.1172/JCI24422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myou S, Leff AR, Myo S, Boetticher E, Tong J, Meliton AY, et al. Blockade of inflammation and airway hyperresponsiveness in immune-sensitized mice by dominant-negative phosphoinositide 3-kinase-TAT. J Exp Med. 2003;198:1573–1582. doi: 10.1084/jem.20030298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tumes DJ, Onodera A, Suzuki A, Shinoda K, Endo Y, Iwamura C, et al. The polycomb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells. Immunity. 2013;39:819–832. doi: 10.1016/j.immuni.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 18.Sekiya T, Yoshimura A. In vitro Th differentiation protocol. Methods Mol Biol. 2016;1344:183–191. doi: 10.1007/978-1-4939-2966-5_10. [DOI] [PubMed] [Google Scholar]
- 19.Chae WJ, Ehrlich AK, Chan PY, Teixeira AM, Henegariu O, Hao L, et al. The Wnt antagonist dickkopf-1 promotes pathological type 2 cell-mediated inflammation. Immunity. 2016;44:246–258. doi: 10.1016/j.immuni.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tussiwand R, Everts B, Grajales-Reyes GE, Kretzer NM, Iwata A, Bagaitkar J, et al. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity. 2015;42:916–928. doi: 10.1016/j.immuni.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Awan HM, Shah A, Rashid F, Wei S, Chen L, Shan G. Comparing two approaches of miR-34a target identification, biotinylated-miRNA pulldown vs miRNA overexpression. RNA Biol. 2018;15:55–61. doi: 10.1080/15476286.2017.1391441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 25.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Liu Y, Shi G. Gene microarray analysis of expression profiles in Suberoyllanilide hyroxamic acid-treated dendritic cells. Biochem Biophys Res Commun. 2019;508:392–397. doi: 10.1016/j.bbrc.2018.11.143. [DOI] [PubMed] [Google Scholar]
- 27.Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CD, et al. MicroRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity. 2011;35:169–181. doi: 10.1016/j.immuni.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.d'Avenia M, Citro R, De Marco M, Veronese A, Rosati A, Visone R, et al. A novel miR-371a-5p-mediated pathway, leading to BAG3 upregulation in cardiomyocytes in response to epinephrine, is lost in Takotsubo cardiomyopathy. Cell Death Dis. 2015;6:e1948. doi: 10.1038/cddis.2015.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith SR, Schaaf K, Rajabalee N, Wagner F, Duverger A, Kutsch O, et al. The phosphatase PPM1A controls monocyte-to-macrophage differentiation. Sci Rep. 2018;8:902. doi: 10.1038/s41598-017-18832-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HY, DeKruyff RH, Umetsu DT. The many paths to asthma: phenotype shaped by innate and adaptive immunity. Nat Immunol. 2010;11:577–584. doi: 10.1038/ni.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schatz M, Rosenwasser L. The allergic asthma phenotype. J Allergy Clin Immunol Pract. 2014;2:645–648. doi: 10.1016/j.jaip.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Maggi E. The TH1/TH2 paradigm in allergy. Immunotechnology. 1998;3:233–244. doi: 10.1016/s1380-2933(97)10005-7. [DOI] [PubMed] [Google Scholar]
- 33.Tong R, Xu L, Liang L, Huang H, Wang R, Zhang Y. Analysis of the levels of Th9 cells and cytokines in the peripheral blood of mice with bronchial asthma. Exp Ther Med. 2018;15:2480–2484. doi: 10.3892/etm.2018.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choy DF, Hart KM, Borthwick LA, Shikotra A, Nagarkar DR, Siddiqui S, et al. TH2 and TH17 inflammatory pathways are reciprocally regulated in asthma. Sci Transl Med. 2015;7:301ra129. doi: 10.1126/scitranslmed.aab3142. [DOI] [PubMed] [Google Scholar]
- 35.Leyva-Castillo JM, Yoon J, Geha RS. IL-22 promotes allergic airway inflammation in epicutaneously sensitized mice. J Allergy Clin Immunol. 2019;143:619–630.e7. doi: 10.1016/j.jaci.2018.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varricchi G, Harker J, Borriello F, Marone G, Durham SR, Shamji MH. T follicular helper (Tfh) cells in normal immune responses and in allergic disorders. Allergy. 2016;71:1086–1094. doi: 10.1111/all.12878. [DOI] [PubMed] [Google Scholar]
- 37.Aron JL, Akbari O. Regulatory T cells and type 2 innate lymphoid cell-dependent asthma. Allergy. 2017;72:1148–1155. doi: 10.1111/all.13139. [DOI] [PubMed] [Google Scholar]
- 38.Muehling LM, Lawrence MG, Woodfolk JA. Pathogenic CD4+ T cells in patients with asthma. J Allergy Clin Immunol. 2017;140:1523–1540. doi: 10.1016/j.jaci.2017.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grünig G, Corry DB, Reibman J, Wills-Karp M. Interleukin 13 and the evolution of asthma therapy. Am J Clin Exp Immunol. 2012;1:20–27. [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Z, Homer RJ, Wang Z, Chen Q, Geba GP, Wang J, et al. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest. 1999;103:779–788. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ortiz RA, Barnes KC. Genetics of allergic diseases. Immunol Allergy Clin North Am. 2015;35:19–44. doi: 10.1016/j.iac.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Budhiraja S, Ramakrishnan R, Rice AP. Phosphatase PPM1A negatively regulates P-TEFb function in resting CD4(+) T cells and inhibits HIV-1 gene expression. Retrovirology. 2012;9:52. doi: 10.1186/1742-4690-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lund R, Ahlfors H, Kainonen E, Lahesmaa AM, Dixon C, Lahesmaa R. Identification of genes involved in the initiation of human Th1 or Th2 cell commitment. Eur J Immunol. 2005;35:3307–3319. doi: 10.1002/eji.200526079. [DOI] [PubMed] [Google Scholar]
- 44.Yamaoka K, Kaminuma O, Kitamura N, Mori A, Tatsumi H, Nemoto S, et al. Protein phosphatase 1 is involved in IL-2-induced IL-5 and IL-13 expression in human T cells. Genes Cells. 2012;17:611–618. doi: 10.1111/j.1365-2443.2012.01610.x. [DOI] [PubMed] [Google Scholar]
- 45.Haque SJ, Harbor P, Tabrizi M, Yi T, Williams BR. Protein-tyrosine phosphatase Shp-1 is a negative regulator of IL-4- and IL-13-dependent signal transduction. J Biol Chem. 1998;273:33893–33896. doi: 10.1074/jbc.273.51.33893. [DOI] [PubMed] [Google Scholar]
- 46.Russo F, Di Bella S, Bonnici V, Laganà A, Rainaldi G, Pellegrini M, et al. A knowledge base for the discovery of function, diagnostic potential and drug effects on cellular and extracellular miRNAs. BMC Genomics. 2014;15(Suppl 3):S4. doi: 10.1186/1471-2164-15-S3-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang Z, Zhou H, Zheng H, Wu J. Expression levels of microRNAs are not associated with their regulatory activities. Biol Direct. 2011;6:43. doi: 10.1186/1745-6150-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen QY, Des Marais T, Costa M. Deregulation of SATB2 in carcinogenesis with emphasis on miRNA-mediated control. Carcinogenesis. 2019;40:393–402. doi: 10.1093/carcin/bgz020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers in the study
The transfection efficiency of lentiviral on CD4+ T cells. CD4+ T cells were cultured in T-helper type 2 cell conditions for 5 days and were transfected with LentimiRa-GFP-miR-1165-3p viral on day 1. GFP was used as a marker for miR-1165-3p overexpression.
The total IgE in the plasma of the asthmatic and control groups. The IgE levels in the plasma was measured using ELISA kit (n = 18–20).