Abstract
Artificial intelligence (AI)-based automated classification and segmentation of optical coherence tomography (OCT) features have become increasingly popular. However, its 3-dimensional volumetric nature has made developing an algorithm that generalizes across all patient populations and OCT devices challenging. Several recent studies have reported high diagnostic performances of AI models; however, significant methodological challenges still exist in applying these models in real-world clinical practice. Lack of large-image datasets from multiple OCT devices, nonstandardized imaging or post-processing protocols between devices, limited graphics processing unit capabilities for exploiting 3-dimensional features, and inconsistency in the reporting metrics are major hurdles in enabling AI for OCT analyses. We discuss these issues and present possible solutions.
Keywords: optical coherence tomography, artificial intelligence, deep learning
Introduction
The rapid growth of artificial intelligence (AI) capabilities and widespread applications continues to expand technological frontiers. AI was first described in 1956 as a machine capable of independent thinking and human-like behavior after training.1 Machine learning, a subfield of AI, was subsequently introduced in 1959, as an algorithm that can automatically modify its behavior after exposure to multiple inputs.2 Recent technological breakthroughs in computing power has led to deep learning, a relatively new subfield of machine learning that involves convolutional neural networks (CNNs).3 Convolutional layers, the basis of CNNs, use weights in filter kernels that are applied to each pixel position in the image. Instead of ingesting the whole image as a high-dimensional tensor as in a multilayer perceptron network, CNNs learn to extract features by learning the convolutional filters.4 Deep learning has contributed to transformative changes in AI and computer vision, resulting in driverless cars, language translation, and facial recognition technologies.5–7 Within ophthalmology, deep learning has been applied to automated diagnosis, segmentation, big data analysis, and outcome predictions.8 Many recent studies have used deep learning to diagnose and segment features of diabetic retinopathy,9,10 age-related macular degeneration (AMD),11,12 and glaucoma,13,14 performing comparably or superiorly to human experts.
One of the important AI-based applications in ophthalmology is OCT image analysis. The advent of OCT has revolutionized the clinical management of many retinal diseases, including AMD,15,16 diabetic macular edema,17–19 and retinal vein occlusions.20–22 OCT is the most commonly obtained imaging modality in ophthalmology with 6.74 million performed in the US Medicare population alone in 2017.23 Given its ubiquitous availability, increasing attention has been directed toward implementing a fully automated disease detection system.24 Studies have shown robust diagnostic performance in using deep learning and OCTs to detect retinal diseases and triage the urgency of referrals for potentially sight-threatening ocular conditions.10,25,26
Despite these reported high diagnostic performances, numerous methodological challenges have resulted in difficulties of translating these algorithms into the clinical practice. These include (1) lack of large-image datasets from multiple OCT devices, (2) nonstandardized imaging and/or postprocessing protocols between devices, (3) limited graphics processing unit (GPU) capabilities, and (4) inconsistency in the reporting metrics. This review article will delve into the above-mentioned difficulties with potential solutions.
Limited Number of Datasets
The lack of publicly available volumetric OCT datasets is a major barrier to deep learning–based OCT image analysis. Having numerous input examples is required, particularly in deep learning, to optimize training and reduce overfitting. Many currently existing datasets are limited in the number of scans of normal and pathologic features, are not publicly available, or contain only images acquired from 1 OCT manufacturer. The Medical Image Computing and Computer Assisted Intervention (MICCAI) Society has created benchmark studies to address the limitations of small datasets. In 2017, the MICCAI RETOUCH Challenge assembled a segmentation and detection benchmark study using a large dataset with 112 manually segmented spectral-domain (SD)-OCT volumes from 3 different OCT vendors.27 This dataset was significantly larger than the dataset provided in the previous 2015 MICCAI OPTIMA Cyst Segmentation Challenge that was composed of only 30 SD-OCT volumes from 4 different OCT vendors.28 Robust algorithms were produced within the challenge, and this motivated the need for algorithms that generalize widely across diverse patient populations and different OCT vendors. However, even larger, publicly available multivendor OCT datasets with validated manual annotations are required to meet the need to advance deep learning.
The large amount of data required to train a deep learning algorithm poses a significant challenge. Bigger datasets result in adequate training of the model's parameters and further improve generalizability. Ideally, these training datasets are composed of numerous, manually annotated real data samples. However, these datasets are costly and, therefore, scarce. Although a few large datasets exist, training deep learning models using small datasets is still possible through 3 strategies. The first technique exploits B-scans adjacent to the training image within a volume. These neighboring B-scans can be used as additional examples in the training dataset because of their similar, but slightly different, anatomical structures. Similarly in computed tomography (CT) scanning, Ben-Cohen et al.29 used unlabeled liver CT scan slices adjacent to the labeled training image as additional images. Second, transfer learning and/or fine-tuning where a network is pretrained on a larger unrelated dataset such as ImageNet can be used to start closer to a local minimum because the lower-level filters are already “learned.” Finally, applying data augmentations to individual scans can expand and diversify datasets without the need to acquire new images. Common transformations, including flipping, shearing, rotation, and outward/inward scaling, are generalizable because they are representative of the true variance captured in OCT imaging. Lee et al.25 used a 432 × 32 window to OCT images and varied its position throughout the scan to significantly increase their training dataset size. Morley et al.30 performed rotation and a unique myopic warping transformation to increase the RETOUCH dataset size by 45 times the original amount. Kuwayama et al.31 improved their training dataset from 1,100 B-scans to 59,400 through horizontal flipping, rotation, and translation, producing an algorithm that correctly classified rare diseases such as Vogt-Koyanagi-Harada disease. Similarly, Kihara et al.32 applied data augmentation (transformation, rotation, horizontal reflection) to increase their training dataset from 67,899 OCT B-scans to 103,053. Gao et al.33 used a mirroring operation for data augmentation. Devalla et al.13 compared the performance of a deep learning model with and without data augmentation (rotation, horizontal flipping, shifting, additive white noise, multiplicative speckle noise, elastic deformation, and occluding patches). Superior performance was reported in the model trained using both real and synthetic OCT data compared with the model trained using only real images. These results were attributed to less overfitting and improved generalizability because of additional synthetic training inputs. Although data augmentation can generate large and diverse training datasets, it is important to emphasize that numerous real data in the form of validated datasets are superior to synthetic images.34 In addition to these techniques, newer methods have recently demonstrated potential in expanding datasets.
Generative adversarial networks (GANs) are a novel, unsupervised machine learning technique that has been used to augment datasets.35 These GANs are composed of a generator subnetwork and a discriminator subnetwork that work in concert to yield an image indistinguishable from real-world data. Consequently, stunningly realistic images can be produced and used to produce large, diverse, and high-quality datasets while avoiding costly data acquisition, manual annotation, and data augmentation steps.36 Even datasets that have already experienced data transformations can further benefit from GAN-based data augmentation.37 Within ophthalmology, GANs have been applied to synthesize realistic retinal OCT images.38
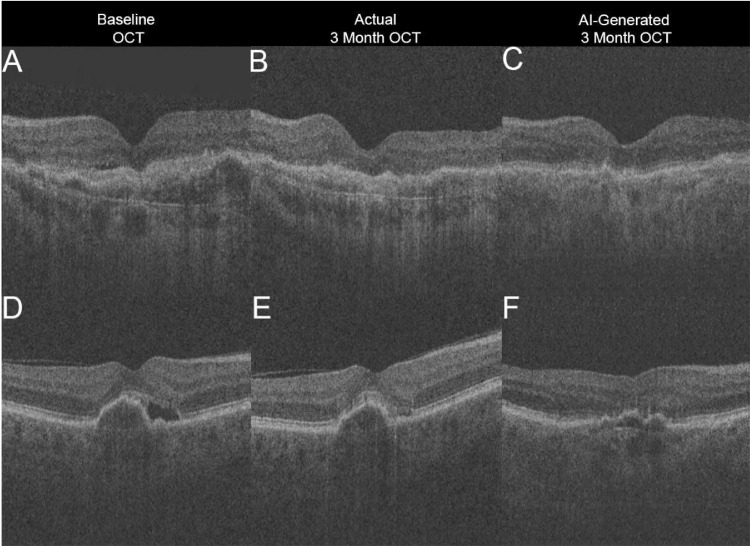
Similarly, we recently used a GAN to create an OCT image that predicted changes in subretinal fluid (SRF) after anti–vascular endothelial growth factor (VEGF) therapy in treatment-naive neovascular AMD patients. Based on the baseline B-scan, this GAN was trained to generate a corresponding OCT image depicting treatment response at 3 months after monthly intravitreal anti-VEGF injections (postloading dose). A total of 60,895 macula-centered OCT volumes at 512 × 885 × 129 were extracted from a Topcon 3D OCT-2000 (Topcon Inc, Tokyo, Japan). Of these, 625 OCT volumes (6875 paired B-scans) of patients with both baseline and postloading dose images were used to train, validate, and test a conditional GAN.39 Images were allocated to training, validation, and testing OCT volumes in 60%, 20%, and 20%, respectively, with patient-level partitioning. The GAN occasionally predicted resolution of SRF correctly (Figs. A–C), but in the majority of images (Figs. D–F), the model overestimated SRF resolution. The GAN was found to “over optimistically” generate the best possible OCT image by assuming complete resolution of neovascular AMD features after loading dose anti-VEGF therapy. We concluded that the poor performance was likely the result of overfitting and that much more training data and direct contextual information are needed to improve the GAN. Although GANs represent a promising field of deep learning research and can impressively reproduce biomedical imaging features, they require thorough clinical validation before they can be medically applied.
Figure.
Artificial intelligence (AI)-generated prediction optical coherence tomography (OCT) image overestimates treatment response in treatment-naive neovascular age-related macular degeneration patients 3 months after monthly injections (loading dose) of anti–vascular endothelial growth factor. In case 1, subretinal fluid (SRF) at baseline (A) resolves after treatment at month 3 per ground truth OCT image (B), which the AI-generated OCT image correctly predicts (C). In case 2, SRF at baseline (D) improves but persists at postinjection month 3 (E); however, AI-generated prediction (F) incorrectly assumes complete resolution of SRF.
Lack of a Standardized Acquisition, Image Registration, and Postprocessing Protocol
OCT acquires a series of cross-sectional B-scans of the retina, and these 2-dimensional images are often compared with ultrasound. However, 3-dimensional (3D) OCT volumes more closely resemble magnetic resonance images and CT scans. Nevertheless, in contrast to scanning the entire body, OCT captures 49 to 128 B-scans over a 6 × 6 mm region of the retina, resulting in much denser volumes.40 OCT scans acquire significantly finer detail and higher resolution, ranging from 1 to 10 µm, compared with 300 µm on high-resolution CT and 1 mm on magnetic resonance imaging.41 Despite high-resolution scanning, image quality can vary substantially because of noise and motion artifacts.42 Therefore, analyzing dense, volumetric OCT scans through deep learning presents a unique challenge.
A major challenge of AI-based OCT volumetric analysis is the lack of a standardized acquisition protocol within a single or multiple OCT device. Without standardization, images of different sizes, contrast levels, and textures that are not generalizable to a single AI algorithm are obtained.43,44 Raster pattern dimensions are not consistent between devices, ranging from 128 × 256 to 256 × 768.40 Different acquisition times and signal-to-noise ratios among devices result in variable image quality. Some devices achieve a high signal-to-noise ratio through averaging multiple images but sacrifice B-scan volume density.27 Variations in B-scan quality and density can affect volumetric analysis because of inconsistent voxel intensities and distributions. With these differences, many deep learning algorithms are restricted to 1 OCT imaging device and 1 scan pattern, thereby limiting generalizability.
To overcome this lack of standardization, studies have attempted to develop a single model that is generalizable to OCT images from multiple imaging platforms. de Sisternes et al.45 created 4 models, each trained using voxel features extracted at 4 different resolutions that correspond to the resolutions of each OCT imaging device. Venhuizen et al.44 combined predictions generated from 3 CNNs at different image scales across entire OCT volumes to segment intraretinal fluid. Lu et al.36 produced individual CNNs for each of the 3 OCT devices separately. Each model performed pixelwise segmentation on each B-scan volume, followed by random forest classification of each pixel to determine the probability of fluid. The mean of the 10 highest probabilities was calculated to determine the probability of fluid within the volume. Other groups standardized image size and intensities for all the scans across OCT devices.30,46,47 Another study used neighboring B-scans to provide 3D contextual information.48 De Fauw et al.10 proposed a 2-step framework that involved segmentation followed by a device vendor-independent classification second step. The segmentation network was retrained separately on different OCT imaging devices while the classification network remained untouched, allowing for an easily adaptable model to new OCT protocols. These benchmark studies have motivated research necessary for multivendor volumetric analysis allowing for further generalizability among OCT imaging devices.
The variability of the image registration and postprocessing protocols across devices poses an additional challenge for clinical deployment. Analyzing OCT volumes using 3D convolutions would harness additional structural data in another dimension and possibly improve segmentation accuracy across a volume. However, some OCT devices acquire B-scans that are widely spaced and image registration (ie, the alignment of consecutive images in an OCT volume) is not performed. Large interscan distances and misaligned images can cause high variability between adjacent scans because of eye movement (in the x, y, and z directions), thus rendering 3D convolutions meaningless.49 Although image registration provides important benefits, its inconsistent use among OCT manufacturers also has drawbacks. During the process of image registration, volumes are subjected to various postprocessing transformations, such as scaling and rotation, in order to optimize alignment between images. Such postprocessing procedures are inconsistent among OCT vendors, further contributing to discrepancies between devices and limiting the use of 3D convolutions in OCT.50 Because of these technical shortcomings, vast amounts of data harbored within 3D OCT volumes are underexploited.
Inconsistent Metrics
Another important consideration is determining the appropriate metrics for classification and segmentation performance. Classification is most commonly measured using area under the receiver operating characteristics curve (AUROC) and area under the precision-recall curve (AUPR). AUROC and AUPR allow direct comparison of models at different thresholds and summarize the algorithm's ability to correctly predict positive classes. Of the 2, AUROC is most commonly used and is most appropriate when using balanced datasets. However, when class distributions are skewed, AUROC is overly optimistic, even though the model's true performance remains constant.51,52 AUPR, which measures the true positives among positive predictions, does not consider negative predictions as AUROC does. When distributions are imbalanced (ie, more negative classifications exist than positive classifications, or vice versa), AUROC will become skewed, but AUPR will not. Therefore, AUPR is the more accurate metric when using imbalanced datasets to evaluate classification performance.53
A variety of metrics are used to measure segmentation performance, such as Dice, F1 score, Jaccard coefficient (Jaccard), and intersection over union.54,55 All of these measures are overlap indexes that range from 0 (no overlap) to 1 (complete overlap) when comparing the segmentation performance of an algorithm against human experts.56 These metrics are closely related as Dice is equivalent to F1 score and Jaccard equates to intersection over union, but Dice and Jaccard are mathematically distinct, given by the equations:
Therefore, Dice is always larger than Jaccard except when there is complete overlap (true positive = 1) or complete discordance (true positive = 0). This is problematic because Dice scores may unfairly overinflate results when comparing studies that exclusively use Jaccard. Because both metrics are positively correlated with each other, reporting both Dice and Jaccard does not offer any additional information.55 Many of these metrics do not perform well when there are no features of interest present in the image because the true negatives are not in the denominator and smoothed variants of these metrics are often applied.
Computational Restrictions
Computational constraints are another key concern. Insufficient dynamic random access memory (DRAM) in GPUs is pervasive in deep learning. Inadequate computational capacities can often limit minibatch size, depth of convolutions, or choice of algorithms to a less robust one.57 Rhu et al.58 reported that VGG-16 architectures, which contain 16 convolutional layers and 3 fully connected layers, require 28 GB of memory using a batch size of 256. Because a single GPU with 7 GB of DRAM can only accommodate a batch size of 64, the VGG-16 architecture requires multiple GPUs for a large batch size or reducing the batch size significantly during training. Applying these deep learning algorithms to entire OCT volumes requires large GPU DRAM capacities.59 Maetschke et al.60 used a 3D CNN to classify healthy and glaucomatous eyes using raw, unsegmented OCT volumes of the optic nerve head. However, because of limited GPU space, the authors restricted their work to only 5 convolutional layers. Li et al.61 used a U-Net architecture with 3D convolutions to perform large 4 × 496 × 512 × 19 3D convolutions using only two 8-GB GPUs. However, advances in GPUs are forthcoming, including application-specific integrated circuits and the recently released NVIDIA DGX-2, which has 256 GB of GPU DRAM. Until then, these GPU limitations impede the optimal use of deep learning for training OCT algorithms.
Future Directions
Moving forward, many hurdles, such as the practicality and real-world implementation of AI, must also be overcome. Despite recent US Food and Drug Administration approval of multiple AI-based devices,9,62 it remains unclear how and where they will be incorporated into real clinical settings and how much patients will benefit from the device.63 In addition, because of the complex nature of AI, particularly deep learning, patients and physicians alike may not trust the clinical utility of AI due to the black-box phenomenon.64 Elucidating the decision-making process of deep learning algorithms is largely unknown and is a significant hurdle. This is potentiated by minorly modified, visually imperceptible (adversarialized) images that can fool CNNs to incorrectly predict diseases with high confidence.65 This brings security and safety measures of such systems into questions. Shah et al.66 demonstrated a significant decline in performance when image-based CNNs were presented adversarialized color fundus photographs of referable DR. Using 50 original color fundus photographs and 50 corresponding adversarialized images, 98% of the original images were classified correctly, but only 53% of the adversarialized images were correctly identified. In contrast, humans correctly classified all original and corresponding adversarialized images as referable DR. Clinically, adversarial attacks could cause a system to misdiagnose a patient as healthy when they actually have vision-threatening DR, or vice versa. Therefore, these systems could cause many patients to have inappropriate overtreatment or undertreatment, worsen outcomes, and further diminish trust.67 Much work is needed to validate deep learning networks, strengthen security, and improve the reliability of these networks. Novel techniques such as class activation maps68 and occlusion testing69,70 could identify areas of importance to AI systems and explain the system's “thought process.”
Conclusions
Many important methodological and technical challenges exist in analyzing OCT with deep learning. A standardized framework for OCT scans is necessary to increase generalizability. Recent studies have inspired models that standardize OCT images from various devices. The performance of these models can only be accurately compared when the appropriate metrics are used consistently. Large, manually annotated datasets using real patient data are also required to optimize the performance of these models to improve generalizability and is superior to data augmentation and adversarialized images. Although deep learning and OCT have individually revolutionized ophthalmology, optimizing the combined technologies will be integral to accelerating progress in the field.
Acknowledgments
Supported by grants from the NEI, Bethesda, MD (CSL; K23EY02492), (AYL; K23EY029246); Research to Prevent Blindness, Inc. New York, NY (CSL, AYL); and Lowy Medical Research Institute (CSL, AYL). The sponsors or funding organizations had no role in the preparation or approval of the manuscript.
Disclosure: R.T. Yanagihara, None; C.S. Lee, None; D.S.W. Ting, Singapore Eye Lesion Analyzer (SELENA+) (P); A.Y. Lee, NVIDIA (F), Microsoft (F), Novartis (F), Carl Zeiss Meditec (F), Topcon (R), Verana Health (C), Genetech/Roche (C)
References
- 1. McCarthy J, Minsky ML, Rochester N, Shannon CE. A proposal for the Dartmouth Summer Research Project on Artificial Intelligence, August 31, 1955. AIMagazine. 2006; 27: 12–12. [Google Scholar]
- 2. Samuel AL. Some studies in machine learning using the game of checkers. IBM JResDev. 1959; 3: 210–229. 10.1147/rd.33.0210 [DOI] [Google Scholar]
- 3. Lee A, Taylor P, Kalpathy-Cramer J, Tufail A. Machine learning has arrived! Ophthalmology. 2017; 124: 1726–1728. 10.1016/j.ophtha.2017.08.046 [DOI] [PubMed] [Google Scholar]
- 4. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015; 521: 436–444. 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 5. Parkhi OM, Vedaldi A, Zisserman A, Others. Deep face recognition. In: Proceedings of the British Machine Vision Conference. 2015; Vol. 1: 6. [Google Scholar]
- 6. Waldrop MM. Autonomous vehicles: no drivers required. Nature. 2015; 518: 20–23. [DOI] [PubMed] [Google Scholar]
- 7. Jean S, Cho K, Memisevic R, Bengio Y. On using very large target vocabulary for neural machine translation. arXiv [csCL] December 2014, http://arxiv.org/abs/1412.2007.
- 8. Schmidt-Erfurth U, Sadeghipour A, Gerendas BS, Waldstein SM, Bogunović H. Artificial intelligence in retina. Prog Retin Eye Res. 2018; 67: 1–29. [DOI] [PubMed] [Google Scholar]
- 9. Abràmoff MD, Lavin PT, Birch M, Shah N, Folk JC. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. npj Digital Medicine. 2018; 1: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De Fauw J, Ledsam JR, Romera-Paredes B, et al.. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018; 24: 1342–1350. [DOI] [PubMed] [Google Scholar]
- 11. Schlegl T, Waldstein SM, Bogunovic H, et al.. Fully automated detection and quantification of macular fluid in OCT using deep learning. Ophthalmology. 2018; 125: 549–558. [DOI] [PubMed] [Google Scholar]
- 12. Li F, Chen H, Liu Z, Zhang X, Wu Z. Fully automated detection of retinal disorders by image-based deep learning. Graefes Arch Clin Exp Ophthalmol. 2019; 257: 495–505. [DOI] [PubMed] [Google Scholar]
- 13. Devalla SK, Renukanand PK, Sreedhar BK, et al.. DRUNET: a dilated-residual U-Net deep learning network to segment optic nerve head tissues in optical coherence tomography images. Biomed Opt Express. 2018; 9: 3244–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fu H, Baskaran M, Xu Y, et al.. A deep learning system for automated angle-closure detection in anterior segment optical coherence tomography images. Am J Ophthalmol. 2019; 203: 37–45. [DOI] [PubMed] [Google Scholar]
- 15. Rosenfeld PJ, Moshfeghi AA, Puliafito CA. Optical coherence tomography findings after an intravitreal injection of bevacizumab (Avastin) for neovascular age-related macular degeneration. Ophthalmic Surgery, Lasers and Imaging Retina. 2005; 36: 331–335. 10.3928/1542-8877-20050701-14 [DOI] [PubMed] [Google Scholar]
- 16. Lalwani GA, Rosenfeld PJ, Fung AE, et al.. A variable-dosing regimen with intravitreal ranibizumab for neovascular age-related macular degeneration: year 2 of the PrONTO Study. Am J Ophthalmol. 2009; 148: 43–58.e1. [DOI] [PubMed] [Google Scholar]
- 17. Martidis A, Duker JS, Greenberg PB, et al.. Intravitreal triamcinolone for refractory diabetic macular edema. Ophthalmology. 2002; 109: 920–927. 10.1016/s0161-6420(02)00975-2 [DOI] [PubMed] [Google Scholar]
- 18. Korobelnik J-F, Do DV, Schmidt-Erfurth U, et al.. Intravitreal aflibercept for diabetic macular edema. Ophthalmology. 2014; 121: 2247–2254. [DOI] [PubMed] [Google Scholar]
- 19. Virgili G, Menchini F, Casazza G, et al.. Optical coherence tomography (OCT) for detection of macular oedema in patients with diabetic retinopathy. Cochrane Database Syst Rev. 2015; 1: CD008081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Park CH, Jaffe GJ, Fekrat S. Intravitreal triamcinolone acetonide in eyes with cystoid macular edema associated with central retinal vein occlusion. Am J Ophthalmol. 2003; 136: 419–425. [DOI] [PubMed] [Google Scholar]
- 21. Costa RA, Jorge R, Calucci D, Melo LA Jr, Cardillo JA, Scott IU. Intravitreal bevacizumab (Avastin) for central and hemicentral retinal vein occlusions: IBeVO study. Retina. 2007; 27: 141–149. [DOI] [PubMed] [Google Scholar]
- 22. Prager F, Michels S, Kriechbaum K, et al.. Intravitreal bevacizumab (Avastin) for macular oedema secondary to retinal vein occlusion: 12-month results of a prospective clinical trial. Br J Ophthalmol. 2009; 93: 452–456. [DOI] [PubMed] [Google Scholar]
- 23. CMS.gov. Medicare provider utilization and payment data: physician and other supplier. July 2019. https://www.cms.gov/Research-Statistics-Data-and-Systems/Statistics-Trends-and-Reports/Medicare-Provider-Charge-Data/Physician-and-Other-Supplier.html Accessed July 17, 2019.
- 24. Huang D, Swanson EA, Lin CP, et al.. Optical coherence tomography. Science. 1991; 254: 1178–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee CS, Tyring AJ, Deruyter NP, Wu Y, Rokem A, Lee AY. Deep-learning based, automated segmentation of macular edema in optical coherence tomography. Biomed Opt Express. 2017; 8: 3440–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kermany DS, Goldbaum M, Cai W, et al.. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018; 172: 1122–1131.e9. [DOI] [PubMed] [Google Scholar]
- 27. Bogunovic H, Venhuizen F, Klimscha S, et al.. RETOUCH–the retinal OCT fluid detection and segmentation benchmark and challenge. IEEE Transactions on Medical Imaging. 2019: 1–1. 10.1109/tmi.2019.2901398 [DOI] [PubMed] [Google Scholar]
- 28. Wu J, Philip A-M, Podkowinski D, et al.. Multivendor spectral-domain optical coherence tomography dataset, observer annotation performance evaluation, and standardized evaluation framework for intraretinal cystoid fluid segmentation. J Ophthalmol. 2016; 2016: 3898750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ben-Cohen A, Klang E, Amitai MM, Goldberger J, Greenspan H. Anatomical data augmentation for CNN based pixel-wise classification. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). Washington, DC: IEEE; 2018: 1096–1099. [Google Scholar]
- 30. Morley D, Foroosh H, Shaikh S, Bagci U. Simultaneous detection and quantification of retinal fluid with deep learning. arXiv [csCV] August 2017, http://arxiv.org/abs/1708.05464.
- 31. Kuwayama S, Ayatsuka Y, Yanagisono D, et al.. Automated detection of macular diseases by optical coherence tomography and artificial intelligence machine learning of optical coherence tomography images. J Ophthalmol. 2019; 2019: 6319581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kihara Y, Heeren TFC, Lee CS, et al.. Estimating retinal sensitivity using optical coherence tomography with deep-learning algorithms in macular telangiectasia type 2. JAMA Netw Open. 2019; 2: e188029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gao K, Niu S, Ji Z, et al.. Double-branched and area-constraint fully convolutional networks for automated serous retinal detachment segmentation in SD-OCT images. Comput Methods Programs Biomed. 2019; 176: 69–80. [DOI] [PubMed] [Google Scholar]
- 34. Wong SC, Gatt A, Stamatescu V, McDonnell MD. Understanding data augmentation for classification: when to warp? In: 2016 International Conference on Digital Image Computing: Techniques and Applications (DICTA). Gold Coast, Queensland: IEEE; 2016: 1–6. [Google Scholar]
- 35. Goodfellow I, Pouget-Abadie J, Mirza M, et al.. Generative adversarial nets. In: Ghahramani Z, Welling M, Cortes C, Lawrence ND, Weinberger KQ, eds. Advances in Neural Information Processing Systems 27. Red Hook, NY: Curran Associates, Inc; 2014: 2672–2680. [Google Scholar]
- 36. Liu YC, Yang HH, Yang CH, et al. Synthesizing new retinal symptom images by multiple generative models. Computer Vision–ACCV 2018 Workshops. 2019: 235–250. 10.1007/978-3-030-21074-8_19 [DOI]
- 37. Antoniou A, Storkey A, Edwards H. Data augmentation generative adversarial networks. arXiv[statML] November 2017, http://arxiv.org/abs/1711.04340.
- 38. Odaibo SG. Generative adversarial networks synthesize realistic OCT images of the retina. arXiv[csCV] February 2019, http://arxiv.org/abs/1902.06676.
- 39. Zhu J-Y, Park T, Isola P, Efros AA. Unpaired image-to-image translation using cycle-consistent adversarial networks. In: Proceedings of the IEEE International Conference on Computer Vision. Venice: IEEE; 2017: 2223–2232. [Google Scholar]
- 40. Olson J, Sharp P, Goatman K, et al.. Improving the economic value of photographic screening for optical coherence tomography-detectable macular oedema: a prospective, multicentre, UK study. Health Technol Assess. 2013; 17: 1–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Popescu DP, Choo-Smith L-P, Flueraru C, et al.. Optical coherence tomography: fundamental principles, instrumental designs and biomedical applications. Biophys Rev. 2011; 3: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abràmoff MD, Garvin MK, Sonka M. Retinal imaging and image analysis. IEEE Rev Biomed Eng. 2010; 3: 169–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Girish GN, Thakur B, Chowdhury SR, Kothari AR, Rajan J. Segmentation of intra-retinal cysts from optical coherence tomography images using a fully convolutional neural network model. IEEE J Biomed Health Inform. 2019; 23: 296–304. [DOI] [PubMed] [Google Scholar]
- 44. Lu D, Heisler M, Lee S, et al.. Deep-learning based multiclass retinal fluid segmentation and detection in optical coherence tomography images using a fully convolutional neural network. Medical Image Analysis. 2019; 54: 100–110. 10.1016/j.media.2019.02.011 [DOI] [PubMed] [Google Scholar]
- 45. de Sisternes L, Hong J, Leng T, Rubin DL. A machine learning approach for device-independent automated segmentation of retinal cysts in spectral domain optical coherence tomography images. Proceeding Optima Challenge-MICCAI. 2015. https://optima.meduniwien.ac.at/fileadmin/Challenge2015/Sisternes-CystChallenge15.pdf.
- 46. Apostolopoulos S, Ciller C, Sznitman R, De Zanet S. Simultaneous classification and segmentation of cysts in retinal OCT. https://www.retinai.com/s/RETOUCH-RetinAI.pdf.
- 47. Tennakoon R, Gostar AK, Hoseinnezhad R, Bab-Hadiashar A. Retinal fluid segmentation in OCT images using adversarial loss based convolutional neural networks. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). Washington, DC: IEEE; 2018: 1436–1440. [Google Scholar]
- 48. Yadav S, Gopinath K, Sivaswamy J. A generalized motion pattern and FCN based approach for retinal fluid detection and segmentation. arXiv[csCV] December 2017, http://arxiv.org/abs/1712.01073.
- 49. Xu J, Ishikawa H, Wollstein G, Kagemann L, Schuman JS. Alignment of 3-D optical coherence tomography scans to correct eye movement using a particle filtering. IEEE Trans Med Imaging. 2012; 31: 1337–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Baghaie A, Yu Z, D'Souza RM. State-of-the-art in retinal optical coherence tomography image analysis. Quant Imaging Med Surg. 2015; 5: 603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Davis J, Goadrich M.. The relationship between precision-recall and ROC curves. Proceedings of the 23rd International Conference on Machine Learning–ICML’06. AMC Press: New York; 2006. 10.1145/1143844.1143874 [DOI] [Google Scholar]
- 52. Fawcett T. ROC Graphs: Notes and Practical Considerations for Data Mining Researchers. Technical Report HPL-2003-4. HP Labs. Palo Alto: HP Laboratories; 2003. [Google Scholar]
- 53. Saito T, Rehmsmeier M.. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One. 2015; 10: e0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cárdenes R, de Luis-García R, Bach-Cuadra M. A multidimensional segmentation evaluation for medical image data. Comput Methods Programs Biomed. 2009; 96: 108–124. [DOI] [PubMed] [Google Scholar]
- 55. Taha AA, Hanbury A.. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imaging. 2015; 15: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zou KH, Warfield SK, Bharatha A, et al.. Statistical validation of image segmentation quality based on a spatial overlap index1. AcadRadiol. 2004; 11: 178–189. 10.1016/s1076-6332(03)00671-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ching T, Himmelstein DS, Beaulieu-Jones BK, et al. Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface. 2018; 15: 1–47. 10.1098/rsif.2017.0387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rhu M, Gimelshein N, Clemons J, Zulfiqar A, Keckler SW. vDNN: virtualized deep neural networks for scalable, memory-efficient neural network design. In: The 49th Annual IEEE/ACM International Symposium on Microarchitecture. MICRO-49. Piscataway, NJ, USA: IEEE Press; 2016:18: 1–18:13. [Google Scholar]
- 59. Smistad E, Falch TL, Bozorgi M, Elster AC, Lindseth F. Medical image segmentation on GPUs: a comprehensive review. Medical Image Analysis. 2015; 20: 1–18. 10.1016/j.media.2014.10.012 [DOI] [PubMed] [Google Scholar]
- 60. Maetschke S, Antony B, Ishikawa H, Wollstein G, Schuman J, Garnavi R. A feature agnostic approach for glaucoma detection in OCT volumes. PLoS One. 2019; 14: e0219126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li M-X, Yu S-Q, Zhang W, et al.. Segmentation of retinal fluid based on deep learning: application of three-dimensional fully convolutional neural networks in optical coherence tomography images. Int J Ophthalmol. 2019; 12: 1012–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Harrington SG, Johnson MK.. The FDA and artificial intelligence in radiology: defining new boundaries. J Am Coll Radiol. 2019; 16: 743–744. [DOI] [PubMed] [Google Scholar]
- 63. Keane PA, Topol EJ.. With an eye to AI and autonomous diagnosis. npj Digital Medicine. 2018; 1: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Castelvecchi D. Can we open the black box of AI? Nature. 2016; 538: 20–23. [DOI] [PubMed] [Google Scholar]
- 65. Su J, Vargas DV, Sakurai K. One pixel attack for fooling deep neural networks. IEEE Trans Evol Comput. 2019; 23: 828–841. [Google Scholar]
- 66. Shah A, Lynch S, Niemeijer M, et al.. Susceptibility to misdiagnosis of adversarial images by deep learning based retinal image analysis algorithms. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) . Washington, DC: IEEE; 2018: 1454–1457. [Google Scholar]
- 67. Ma X, Niu Y, Gu L, et al.. Understanding adversarial attacks on deep learning based medical image analysis systems. arXiv[csCV] July 2019; 5: 1421–1428. http://arxiv.org/abs/1907.10456. [Google Scholar]
- 68. Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Learning deep features for discriminative localization. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. Las Vegas, NV: IEEE; 2016: 2921–2929. [Google Scholar]
- 69. Zeiler MD, Fergus R.. Visualizing and understanding convolutional networks. In: Fleet D, Pajdla T, Schiele B, Tuytelaars T, eds. Computer Vision–ECCV 2014. Switzerland: Springer International Publishing; 2014: 818–833. [Google Scholar]
- 70. Lee CS, Baughman DM, Lee AY. Deep learning is effective for the classification of OCT images of normal versus age-related macular degeneration. Ophthalmol Retina. 2017; 1: 322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]