Abstract
Purpose
To present an overview of current machine learning methods and their use in medical research, focusing on select machine learning techniques, best practices, and deep learning.
Methods
A systematic literature search in PubMed was performed for articles pertinent to the topic of artificial intelligence methods used in medicine with an emphasis on ophthalmology.
Results
A review of machine learning and deep learning methodology for the audience without an extensive technical computer programming background.
Conclusions
Artificial intelligence has a promising future in medicine; however, many challenges remain.
Translational Relevance
The aim of this review article is to provide the nontechnical readers a layman's explanation of the machine learning methods being used in medicine today. The goal is to provide the reader a better understanding of the potential and challenges of artificial intelligence within the field of medicine.
Keywords: deep learning, machine learning, artificial intelligence
Introduction
Over the past decade, artificial intelligence (AI) has become a popular subject both within and outside of the scientific community; an abundance of articles in technology and non-technology-based journals have covered the topics of machine learning (ML), deep learning (DL), and AI.1–6 Yet there still remains confusion around AI, ML, and DL. The terms are highly associated, but are not interchangeable. In this review, we (attempt to) forgo technical jargon to better explain these concepts to a clinical audience.
In 1956, a group of computer scientists proposed that computers could be programmed to think and reason, “that every aspect of learning or any other feature of intelligence [could], in principle, be so precisely described that a machine [could] be made to simulate it.”7 They described this principle as “artificial intelligence.”7 Simply put, AI is a field focused on automating intellectual tasks normally performed by humans, and ML and DL are specific methods of achieving this goal. That is, they are within the realm of AI (Fig. 1). However, AI includes approaches that do not involve any form of “learning.” For instance, the subfield known as symbolic AI focuses on hardcoding (i.e., explicitly writing) rules for every possible scenario in a particular domain of interest. These rules, written by humans, come from a priori knowledge of the particular subject and task to be completed. For example, if one were to program an algorithm to modulate room temperature of an office, he or she likely already know what temperatures are comfortable for humans to work in and would program the room to cool if temperatures rise above a specific threshold and heat if they drop below a lower threshold. Although symbolic AI is proficient at solving clearly defined logical problems, it often fails for tasks that require higher-level pattern recognition, such as speech recognition or image classification. These more complicated tasks are where ML and DL methods perform well. This review summarizes machine learning and deep learning methodology for the audience without an extensive technical computer programming background.
Figure 1.
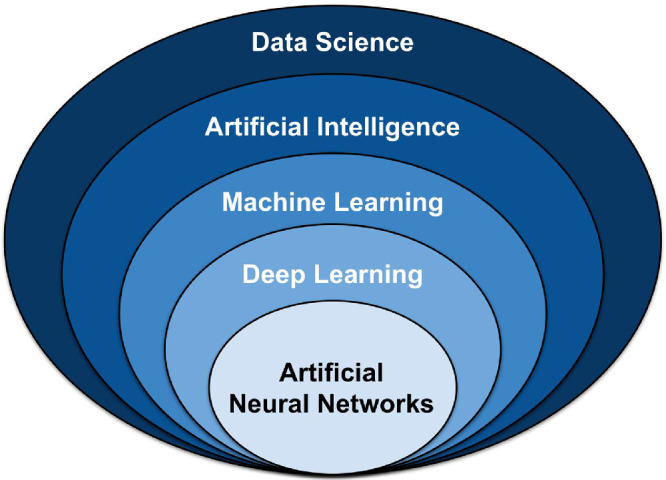
Umbrella of select data science techniques. Artificial intelligence (AI) falls within the realm of data science, and includes classical programming and machine learning (ML). ML contains many models and methods, including deep learning (DL) and artificial neural networks (ANN).
Methods
We conducted a literature search in PubMed for articles that were pertinent to leading artificial intelligence methods being utilized in medical research. Selection of articles was at the sole discretion of the authors. The goal of our literature search was to provide the nontechnical readers a layman's explanation of the machine learning methods being used in medicine today.
Results
We found 33 articles that were pertinent to the main AI methods being used in medicine today.
Discussion
Introduction to Machine Learning
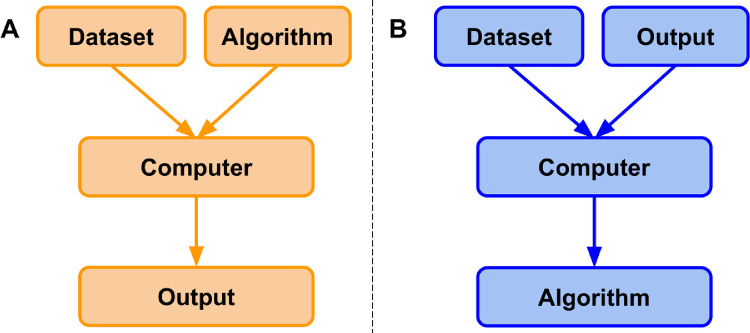
ML is a field that focuses on the learning aspect of AI by developing algorithms that best represent a set of data. In contrast to classical programming (Fig. 2A), in which an algorithm can be explicitly coded using known features, ML uses subsets of data to generate an algorithm that may use novel or different combinations of features and weights than can be derived from first principles (Fig. 2B).8,9 In ML, there are four commonly used learning methods, each useful for solving different tasks: supervised, unsupervised, semisupervised, and reinforcement learning.8–10 To better understand these methods, they will be defined via an example of a hypothetical real estate company that specializes in predicting housing prices and features associated with those houses.
Figure 2.
Classical programming versus machine learning paradigm. (A) In classical programming, a computer is supplied with a dataset and an algorithm. The algorithm informs the computer how to operate upon the dataset to create outputs. (B) In machine learning, a computer is supplied with a dataset and associated outputs. The computer learns and generates an algorithm that describes the relationship between the two. This algorithm can be used for inference on future datasets.
Supervised Learning
Suppose the real estate company would like to predict the price of a house based on specific features of the house. To begin, the company would first gather a dataset that contains many instances.8,9,11 Each instance represents a singular observation of a house and associated features. Features are the recorded properties of a house that might be useful for predicting prices (e.g., total square-footage, number of floors, the presence of a yard).8,9,11 The target is the feature to be predicted, in this case the housing price.8,9,11 Datasets are generally split into training, validation, and testing datasets (models will always perform optimally on the data they are trained on).8,9 Supervised learning uses patterns in the training dataset to map features to the target so that an algorithm can make housing price predictions on future datasets. This approach is supervised because the model infers an algorithm from feature-target pairs and is informed, by the target, whether it has predicted correctly.8–10 That is, features, x, are mapped to the target, Y, by learning the mapping function, f, so that future housing prices may be approximated using the algorithm Y = f(x). The performance of the algorithm is evaluated on the test dataset, data that the algorithm has never seen before.8,9 The basic steps of supervised machine learning are (1) acquire a dataset and split it into separate training, validation, and test datasets; (2) use the training and validation datasets to inform a model of the relationship between features and target; and (3) evaluate the model via the test dataset to determine how well it predicts housing prices for unseen instances. In each iteration, the performance of the algorithm on the training data is compared with the performance on the validation dataset. In this way, the algorithm is tuned by the validation set. Insofar as the validation set may differ from the test set, the performance of the algorithm may or may not generalize. This concept will be discussed further in the section on performance evaluation.
The most common supervised learning tasks are regression and classification.8–10 Regression involves predicting numeric data, such as test scores, laboratory values, or prices of an item, much like the housing price example.8–10 Classification, on the other hand, entails predicting to which category an example belongs.8–10 Sticking with the previous example, imagine that rather than predicting exact housing prices in a fluctuating market, the real estate company would now like to predict a range of prices for which a house will likely sell, such as (0, 125K), (125K, 250K), (250K, 375K), and (375K, ∞). To accomplish this, data scientists would transform the numeric target variable into a categorical variable by binning housing prices into separate classes. These classes would be ordinal, meaning that there is a natural order associated with the categories.9 However, if their task was to determine whether houses had wood, plastic, or metal siding, classes would be nominal; they are independent of one another and have no natural order.9
Unsupervised Learning
In contrast to supervised learning, unsupervised learning aims to detect patterns in a dataset and categorize individual instances in the dataset to said categories.8–10 These algorithms are unsupervised because the patterns that may or may not exist in a dataset are not informed by a target and are left to be determined by the algorithm. Some of the most common unsupervised learning tasks are clustering, association, and anomaly detection.8–10 Clustering, as the name suggests, groups instances in a dataset into separate clusters based upon specific combinations of their features.8–10 Say the real estate company now uses a clustering algorithm on its dataset and it finds three distinct clusters. Upon further investigation, it might find that the clusters represent the three separate architects responsible for designing the homes in their dataset, which is a feature that was not present in the training dataset.
Semisupervised Learning
Semisupervised learning can be thought of as the “happy medium” between supervised and unsupervised learning and is particularly useful for datasets that contain both labeled and unlabeled data (i.e., all features are present, but not all features have associated targets).10 This situation typically arises when labeling images become time-intensive or cost-prohibitive. Semisupervised learning is often used for medical images, where a physician might label a small subset of images and use them to train a model. This model is then used to classify the rest of the unlabeled images in the dataset. The resultant labeled dataset is then used to train a working model that should, in theory, outperform unsupervised models.10
Reinforcement Learning
Finally, reinforcement learning is the technique of training an algorithm for a specific task where no single answer is correct, but an overall outcome is desired.9,10 It is arguably the closest attempt at modeling the human learning experience because it also learns from trial and error rather than data alone.9,10 Although reinforcement learning is a powerful technique, its applications in medicine are currently limited and thus will be presented with a new example. Imagine one would like to train an algorithm to play the video game Super Mario Bros, where the purpose of the game is to move the character Mario from the left side of the screen to the right side in order to reach the flag pole at the end of each level while avoiding hazards such as enemies and pits. There is no correct sequence of controller inputs; there are sequences that lead to a win and those that do not. In reinforcement learning, an algorithm would be allowed to “play” on its own. It would attempt many different controller inputs and when it finally moves Mario forward (without receiving damage), the algorithm is “rewarded” (i.e., the behavior is reinforced). Through this process, the algorithm begins to learn what behavior is desired (e.g., moving forward is better than moving backward, jumping over enemies is better than running into them). Eventually, the algorithm learns how to move from start to finish. Although reinforcement has its place in the field of computer science and machine learning, it has yet to make a substantial impact in clinical medicine.
Performance Evaluation
To maximize the chance of generalizability to the performance of the algorithm on unseen data, the training dataset is usually split into a slightly smaller training dataset and a separate validation dataset.8,9 Metrics used for evaluation of a model depend upon the model itself and whether it is in the training or testing phase. The validation dataset is meant to mimic the test dataset and helps data scientists tune an algorithm by identifying when a model may generalize well and work in a new population. Because the validation dataset is a small sample of the true (larger) population, it may not accurately represent the population itself due to an unknown sampling bias. Therefore, model performance and generalizability should not be assessed via validation set performance. It is conceivable that a data scientist could create a validation dataset with an unknown bias and use it to tune a model. Although the model might perform well on the validation dataset, it would likely not perform well on the much larger test dataset (i.e., it would not be a generalizable model)
Typically, model performance is monitored via some form of accuracy on the training and validation datasets during this phase. So long as the accuracy of the model on the training set (X%) and validation set (Y%) are increasing and converging after each training iteration, the model is considered to be learning. If both converge, but do not increase (e.g., X converges on Y at 50%), the model is not learning and may be underfit to the data, that is, it may not have learned enough of the relationship between features and targets in a way that it would be expected to work in another population. Finally, if training performance increases far more than validation set performance (e.g., the model has an accuracy of 99% on the data it was trained on, but only 80% on the validation data), the model is overfit. That is, it has learned features specific to the training dataset population at the expense of generalizability to another population. Although the validation dataset is not specifically used to train the algorithm, it is used to iteratively tune the algorithm. Therefore, the validation dataset is not necessarily a reliable indicator of model performance on unseen data.8,9
Upon completion of the training phase, a data scientist has, ideally, trained a highly generalizable model; however, this must be confirmed via a separate test dataset. In the case of supervised learning, which will be the focus of this review from here on, the performance of a learned model can be evaluated in a number of ways, but is most commonly evaluated based on prediction accuracy (classification) or error and residuals (regression).8,9 As previously mentioned, the test dataset contains instances of the original dataset that have not been seen by the algorithm during the training phase. If the predictive power of a model is strong on the training dataset, but poor on the test dataset, then the model is too specific to the patterns from the training data and is considered to be overfit to the training dataset.8,9 That is, it has memorized patterns rather than learned a generalizable model. An underfit model, on the other hand, is one that performs poorly on both training and test datasets and has neither learned nor memorized the training dataset and still is not generalizable.8,9 An ideally fitted model is one that performs strongly on both datasets, suggesting it is generalizable (i.e., it will perform well on other similar datasets).8,9
With regression models, the average mean squared error (MSE) can be an indicator of model performance.8,9 MSE measures how close a predicted value is to the intended target value. MSE is calculated by summing the differences between predicted values and target values, squaring the results, and dividing by the total number of instances .8,9 There are many other measures of performance for regression models that are out of the scope of this review.
For binary classification, the output of the model is a class. However, before the class designation, the probability of an instance belonging to class A or class B is determined.8,9 Normally, this probability threshold is set at 0.5. A receiver operating characteristic curve evaluates a model's true positive rate (TPR; i.e., sensitivity, recall), the number of samples correctly identified as positive divided by the total number of positive samples, versus its false-positive rate (FPR; i.e., 1 - specificity), the number of samples incorrectly identified as positive divided by the total number of negative samples (Fig. 3, Fig. 4A).8,9 Similarly, the precision-recall curve evaluates a model's positive predictive value (PPV; i.e., precision), the number of samples correctly identified as positive divided by the total number of samples identified as positive, versus its recall (Fig. 3, Fig. 4B).8,9 Each curve is evaluated across the range of model probability thresholds from 1 to 0, left to right. A receiver operating characteristic curve starts at the point (FPR = 0, TPR = 0), which corresponds to a decision threshold of 1 (every sample is classified as negative, and thus there are no false or true positives). It ends at the point (FPR = 1, TPR = 1), which corresponds to a decision threshold of 0 (where every sample is classified as positive, and thus all points are either truly or falsely labeled positive). The points in between, which create the curve, are obtained by calculating the TPR and FPR for different decision thresholds between 1 and 0, trading off sensitivity (minimizing false negatives) with specificity (minimizing false positives). The area under the curve (AUC) of the receiver operating characteristics curve (AUROC) can be calculated and used as a metric for evaluating the overall performance of a classifier, assuming the classes of the dataset are balanced. If classes are not balanced, the area under the precision-recall curve (AUPR) may be a better metric of model performance because the threshold (set at 0.5 in Fig. 4B) may be adjusted. For example, if a dataset comprised 75% of class A and 25% of class B, the ratio between the two would be computed as the threshold (0.75). In practice, an AUROC value of 0.50 indicates a model that performs no better than chance, and an AUC of 1.00 indicates that the model performs perfectly; the higher the value of the AUC, the stronger the performance of the ML model.8,9 Similarly, an AUPR value at the preset threshold indicates a model that performs no better than chance, and an AUPR value of 1.00 indicates a perfect model.8,9
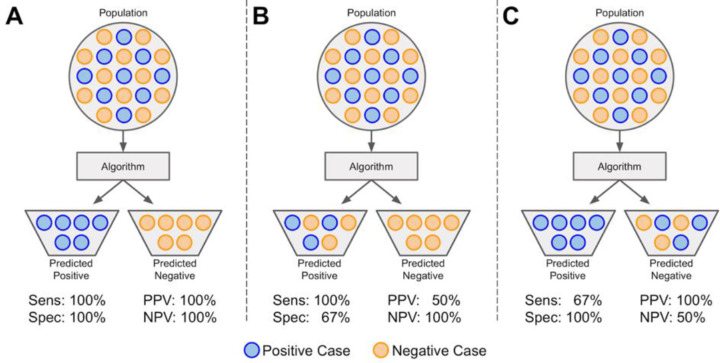
Figure 3.
Sensitivity, specificity, positive predictive value, and negative predictive value. A population (dataset) is represented as circles colored blue if positive or orange if negative. The dataset is input to an algorithm that predicts each instance's class association. If an instance is correctly predicted as positive or negative, it is a true positive (TP) or true negative (TN), respectively. If an instance is incorrectly labeled positive or negative, it is a false positive (FP) or false negative (FN), respectively. (A) A model with perfect sensitivity ( ) and specificity (). (B) A model with perfect sensitivity (ability to correctly classify all positive cases), but poor specificity (ability to correctly classify all negative cases) and (C) a model with perfect specificity, but poor sensitivity. Although a model might have perfect sensitivity (B), it can have many false positives. Similarly, a model with perfect specificity (C) might have many false negatives. Therefore, it is also useful to evaluate the positive predictive value (PPV; ) and the negative predictive value (NPV; ). PPV and NPV are also thus dependent on the prevalence of disease in a population.
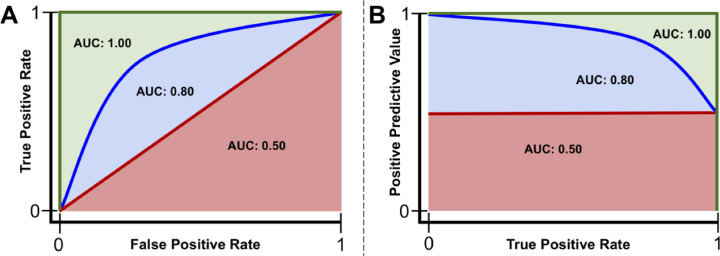
Figure 4.
Example receiver operating characteristics and precision-recall curves. Red line: a model that performs no better than chance has an area under the curve (AUC) of the receiver operating characteristics curve (AUROC) of 0.50 or area under the precision-recall curve (AUPR) at the class ratio (red shaded area). Blue line: a model that performs better than chance, but not perfectly, will have an AUC between 0.50 and 1.00 (blue + red shaded areas). Green line: a model that performs perfectly has an AUC of 1.00 (red + blue + green shaded areas).
Classic Machine Learning Methods
There are many machine learning algorithms used in medicine. Described next are some of the most popular to date.
Linear Regression
Linear regression is arguably the simplest ML algorithm. The main idea behind regression analysis is to specify a relationship between one or more numeric features and a single numeric target.8,9 Linear regression is an analysis technique used to solve a regression problem by using a straight line to describe a dataset. Univariate linear regression, a regression problem where only a single feature is used for predicting a target value, can be represented in a slope-intercept form: y = ax + b.8,9 Here, a is a weight describing the slope, which describes how much a line increases on the y-axis for each increase in x. The intercept, b, describes the point where the line intercepts the y-axis. Linear regression models a dataset using this slope-intercept form, where the machine's task is to identify values of a and b such that the determined line is best able to relate the supplied values of x values to the values of y. Multivariate linear regression is similar; however, there are multiple weights in the algorithm, each describing to what degree each feature influences the target.8,9
In practice, there is rarely a single function that fits a dataset perfectly. To measure the error associated with a fit, the residuals are measured. Conceptually, residuals are the vertical distances between predicted values, , and actual values, y. In machine learning, the cost function is a calculus derived term that aims to minimize errors associated with a model.8,9 The process of minimizing the cost function involves an iterative optimization algorithm known as gradient descent, of which the mathematical calculations involved are outside the scope of this article.8,9,12 In linear regression, the cost function is the previously described MSE. Minimizing this function often obtains estimates of a and b that best model a dataset. All model-based learning algorithms have a cost function, and the goal is to minimize this function to find the best-fit model.8,9
Logistic Regression
Logistic regression is a classification algorithm where the goal is to find a relationship between features and the probability of a particular outcome. Rather than using the straight line produced by linear regression to estimate class probability, logistic regression uses a sigmoidal curve to estimate class probability (Fig. 5). This curve is determined by the sigmoid function, , which produces an S-shaped curve that converts discrete or continuous numeric features (x) into a single numerical value (y) between 0 and 1.8,9 The major advantage of this method is that probabilities are bounded between 0 and 1 (i.e., probabilities cannot be negative or greater than 1). It can be either binomial, where there are only two possible outcomes, or multinomial, where there can be three or more possible outcomes.8,9
Figure 5.
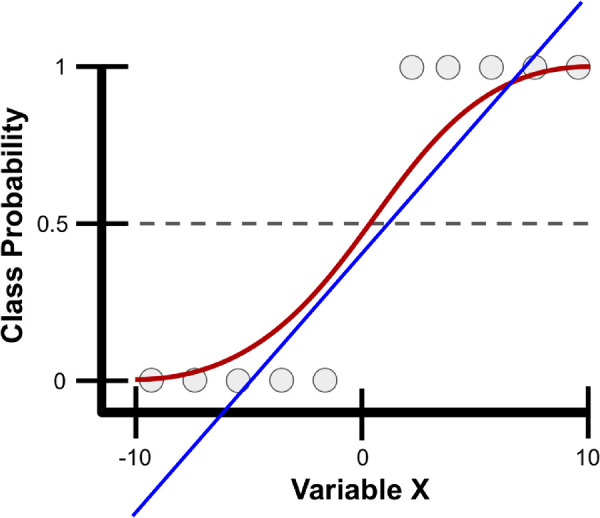
Example class probability prediction using linear and logistic regression. Presented are linear (blue line) and logistic (red line) regression models for predicting the probability of various samples (gray circles) as belonging to a particular class using a single variable, variable X, which ranges from -10 to 10. With logistic regression, variable X is transformed into class probabilities that are bounded between 0 and 1 using the sigmoid function. Simple linear regression attempts to estimate class probabilities, but is not bounded between 0 and 1; thus, it breaks a fundamental law of probability that does not allow for negative probabilities or those greater than 1.
Decision Trees and Random Forests
A decision tree is a supervised learning technique, primarily used for classification tasks, but can also be used for regression.8,9 A decision tree begins with a root node, the first decision point for splitting the dataset, and contains a single feature that best splits the data into their respective classes (Fig. 6).8,9 Each split has an edge that connects either to a new decision node that contains another feature to further split the data into homogenous groups or to a terminal node that predicts the class. This process of separating data into two binary partitions is known as recursive partitioning.8,9 A random forest is an extension of this method, known as an ensemble method, that produces multiple decision trees.8,9 Rather than using every feature to create every decision tree in a random forest, a subsample of features are used to create each decision tree. Trees then predict a class outcome, and the majority vote among trees is used as the model's final class prediction.8,9
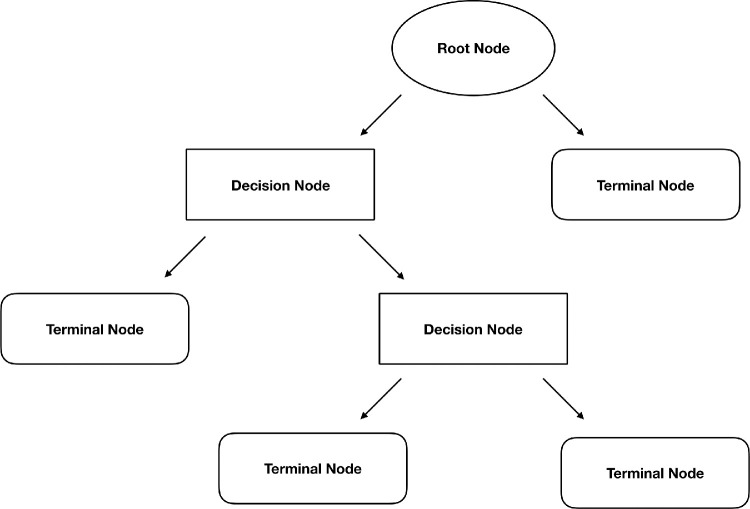
Figure 6.
Structure of a decision tree. Splitting of the dataset begins at the root node. Each split connects to either another decision node, which results in further splitting of the data, or a terminal node that predicts the class of the data.
Classic Machine Learning in Ophthalmology
Although DL has become a highly popular technique in ophthalmology, there are a multitude of examples of classic ML algorithms being used in the field. Simple linear models have been used to predict patients who would develop advanced age-related macular degeneration and to discern which factors separate patients into who will respond to anti-vascular endothelial growth factor treatment versus those who will not.13–16 Random forest algorithms have been used to discover features that are most predictive of progression to geographic atrophy in age-related macular degeneration and find prognostic features for visual acuity outcomes of intravitreal anti-vascular endothelial growth factor treatment.17,18 Random forest classifiers have also been applied to diagnose and grade cataracts from ultrasound images, as well as identify patients with glaucoma based on retinal nerve fiber layer and visual field data.19,20
Neural Networks and Deep Learning
An artificial neural network (ANN) is a machine learning algorithm inspired by biological neural networks.8,9,21 Each ANN contains nodes (analogous to cell bodies) that communicate with other nodes via connections (analogous to axons and dendrites). Much in the way synapses between neurons are strengthened when their neurons have correlated outputs in a biological neural network (the Hebbian theory postulates that “nerves that fire together, wire together”), connections between nodes in an ANN are weighted based upon their ability to provide a desired outcome.8,9,21
Feedforward Neural Networks
A perceptron is a machine learning algorithm that takes in a series of features and their targets as input and attempts to find a line, plane, or hyperplane that separates the classes in a two-, three-, or hyper-dimensional space, respectively.9,22,23 These features are transformed using the sigmoid function (Fig. 7A). Thus, this method is similar to logistic regression; however, it only provides class associations, and not the probability of an instance belonging to a class.
Figure 7.
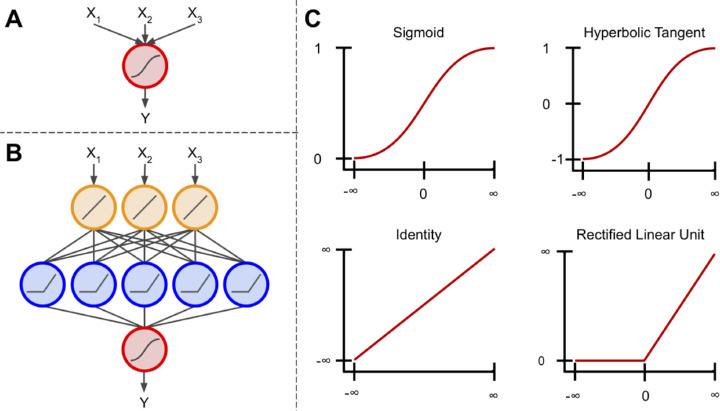
Components of a neural network. (A) The basis of an artificial neural network, the perceptron. This algorithm uses the sigmoid function to scale and transform multiple inputs into a single output ranging from 0 to 1. (B) An artificial neural network connects multiple perceptron units, so that the output of one unit is used as input to another. Additionally, these units are not limited to using the sigmoid activation function. (C) Examples of four different activation functions: sigmoid, hyperbolic tangent, identity, and rectified linear unit. The sigmoid scales inputs between 0 and 1 using an S-shaped curved. Similarly, the hyperbolic tangent function uses an S-shaped curve, but scales inputs between -1 and 1. The identity function can multiply its input by any number to produce a linear output. The rectified linear unit is similar to the identity function, however all inputs < 0 are given an output value of 0. There are other activation functions outside of these, but these are arguably.
When multiple perceptrons are connected, the model is referred to as a multilayer perceptron algorithm or an ANN. Commonly, ANNs contain a layer of input nodes, a layer of output nodes, and a number of “hidden layers” between the two.9 In simple ANNs, there exists an input layer between zero and three hidden layers and an output layer, whereas deep neural networks contain tens or even hundreds of hidden layers.9,24 For most tasks, ANNs feed information forward. This is known as a feedforward neural network, meaning information from each node in the previous layer is passed to each node in the next layer, transformed, and passed forward to each node in the next layer (Fig. 7B).9 In recurrent neural networks, which are out of the scope of this paper, information can be passed between nodes within a layer or to previous layers, where their output is operated on and fed forward once again.22
Each layer in an ANN can contain any number of nodes; however, the number of nodes in the output layer typically corresponds to the number of classes being predicted if the goal is multiclass classification, a single node with a sigmoidal activation for binary classification, or a linear activation function if the goal is regression.9,24 These activation functions simply transform a node's input into a desired output (Fig. 7C). Each node in an ANN contains an activation function (not just the output layer; Fig. 7B). These activation functions, although not always linear, do not have to be complex. For instance, the rectified linear unit applies a linear transformation to inputs ≥ 0, and sets inputs < 0 to 0.25 It follows that as inputs proceed through an ANN, they are progressively modified at each layer so that at the final layer they no longer resemble their original state. However, this final representation of the input is, in theory, more predictive of the specified outcome.
Convolutional Neural Networks
For image recognition tasks, each input into a feedforward ANN corresponds to a pixel in the image. However, this is not ideal because there are no connections between nodes in a layer. In practice, this means that the spatial context of features in the image are lost.24,26,27 In other words, pixels that are close to one another in an image are likely more correlated than pixels on opposite sides of the image, but a feedforward ANN does not take this into account.
A convolutional neural network (CNN) is a special case of the ANN that overcomes this issue by preserving the spatial relationship between pixels in an image.24,26,27 Rather than using single pixels as input, a CNN feeds patches of an image to specific nodes in the next layer of nodes (rather than all nodes), thereby preserving the spatial context from which a feature was extracted.9,24,26,27 These patches of nodes learn to extract specific features and are known as convolutional filters.
Convolutions are widely used in the realm of image processing, and are often used to blur or sharpen images, or for other tasks such as edge detection.28 A visible-light digital image is simply a single matrix if the image is grayscale or three stacked matrices if the image is color (red, green, and blue color channels).28 These matrices contain values, typically between 0 and 255, that represent pixels in the image and the intensity of each color channel at each pixel.28 A convolutional filter is a much smaller matrix that is typically square and range in size from 2 × 2 to 9 × 9.28 This filter is passed over the original image and, at each position, element-wise matrix multiplication is performed (Fig. 8).28 The output of this convolution is mapped to a new matrix (a feature map) that contains values corresponding to whether or not the convolutional filter detected a feature of interest.24,26–29
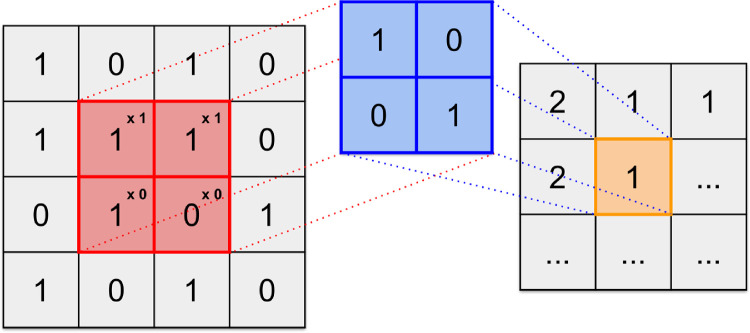
Figure 8.
Example of a digital image convolved with a filter. The image (left) is transformed into the feature map (right) via a convolutional filter (center). The convolutional filter is designed to locate diagonal lines running from top left to bottom right of the image. The filter passes over the image in a specified manner and each element in the image (red) is multiplied by the corresponding element in the convolutional filter (blue). The summation of these elements (orange) is output into a new matrix that reports the presence of a diagonal line. The feature map indicates 2 when the specified diagonal line is found, 1 if a portion of it is found, and 0 if none of it is found.
In CNNs, filters are trained to extract specific features from images (e.g., vertical lines, U-shaped objects,) and mark their location on the feature map.26,27 A deep CNN then uses the feature map as input for the next layer, which uses new filters to create another new feature map.24,26,27 This can continue for many layers and, as it continues, the extracted features become abstract, but highly useful for prediction. The final features maps are then compressed from their square representations and input to a feedforward ANN, where classification of the image based on the extracted features and textures can occur.24,26,27 This process is referred to as DL.24
Aside from image classification tasks, DL has shown promise for image segmentation tasks.1,30,31 Rather than classifying images as a whole, this method aims to identify objects within an image. To accomplish this task, DL classifies individual pixels given surrounding pixel information. For example, in diabetic retinopathy, a segmentation algorithm might segment (outline) the retinal vasculature by assigning probabilities to individual pixels as belonging to a retinal blood vessel or not belonging to a retinal blood vessel. A similar method for breast cancer detection could mark pixels as belonging to a mass or not belonging to a mass, and the output image could be provided to a radiologist for further review.
Deep Learning in Ophthalmology
The popularity for DL has especially risen in the field of ophthalmology for image-based diagnostic systems. On the simpler end of visual interpretation tasks, Coyner et al. devised a DL system for automated assessment of retinal fundus image quality with an output of “acceptable” or “not acceptable” based on multiple graded expert labels.3 Presumably, the network learned that the retinal vasculature must be easily distinguishable for an image to be deemed acceptable. In a more complex task, Gulshan et al. demonstrated that DL could classify diabetic retinopathy, in agreement with the Early Treatment for Diabetic Retinopathy Study scale, using only retinal fundus images as input and the consensus diagnoses of multiple clinicians as the “class labels.”2 The presence of features such as microaneurysms, intraretinal hemorrhages, or neovascularization were not supplied to the DL method as signs of diabetic retinopathy. Rather, the DL model either learned these features or learned novel features that aid in the diagnosis of diabetic retinopathy. Further, Brown et al. trained a similar DL network for the diagnosis of plus disease in retinopathy of prematurity. First, an algorithm was trained to segment retinal vasculature into binary vessel maps. Then another DL algorithm was trained to examine the vessel maps and conclude whether the vasculature appeared normal or abnormal.1 This network, too, performs on par or better than most experts in the field. One of the most impressive examples of DL in ophthalmology was conducted by De Fauw et al. Using three-dimensional optical coherence tomography images, a DL framework was trained to not only detect a single disease, but more than 50 common retinal diseases.6
Challenges with DL Models
In recent years, DL has become a hot topic within the field of medicine given the digital availability of information; however, many challenges still exist. DL is limited by the quantity and quality of data used to train the model. It is difficult to estimate how much data are necessary to sufficiently and reliably train DL systems because it depends both on the quality of the input training data as well as the complexity of the task. Typically, thousands of training examples are required to create a model that is both accurate and generalizable. Thus, developing models for identification of rare diseases, where large datasets may not be readily available, is especially challenging. On the other hand, although one might assume that more data will always lead to better models, if the quality of the training data is imprecise, mislabeled, or somehow systematically different than the test population, training on very large datasets may result in models that do not perform well in real-world scenarios. Furthermore, there is an implicit assumption that datasets are accurately labeled by human graders. Unfortunately, this is often not the case, and noisy and/or missing labels are often a bane for data scientists.
DL methods also suffer from the “black box” problem: input is supplied to the algorithm and an output emerges, but it is not exactly clear what features were identified or how they informed the model output.29,32,33 In contrast, simple linear algorithms, although not always as powerful as DL, are easily interpretable. The computed weights for each feature are supplied upon completion of the training process, which allow for one to interrogate exactly how the model works and possibly discover important predictors that may be useful for prevention of a disease. With deep learning, a complex series of matrix multiplication and abstract filters makes interpretability significantly more challenging.29,32,33 Activation maps, or heatmaps, are methods that attempt to address the “black box” issue by highlighting areas of images that highlight regions of an image that “fire together” with the output classification label.29,32,33 Unfortunately, these methods still require human interpretation, as they are often not examined critically (examples are cherry picked for publication, highly subject to confirmation bias, etc.), and thus this remains an active area of research. For instance, if a DL model classifies a fundus image as having proliferative diabetic retinopathy, a heatmap will highlight feature areas on that fundus image that contributed to the decision of being classified as having proliferative diabetic retinopathy. It is up to the physician to interpret whether these DL model identified features are the same features the physician would use to diagnose the disease, and the implications of such findings.
Conclusion
AI methods have shown to be a promising tool in the field of medicine. Recent work has demonstrated that these methods can develop effective diagnostic and predictive tools to identify various diseases. In the future, AI-based programs may become an integral part of patients’ clinic visits with their ability to assist in diagnosis and management of various diseases. Physicians should take an active approach to understand the theories behind AI and its utility in medicine with the goal of providing optimal patient care.
Acknowledgments
This project was supported by grants R01EY19474, K12 EY027720, and P30EY10572 from the National Institutes of Health; SCH-1622679, SCH-1622542, and SCH-1622536 from the National Science Foundation; and by unrestricted departmental funding and a Career Development Award (JPC) from Research to Prevent Blindness.
Disclosure: R.Y. Choi, None; A.S. Coyner, None; J. Kalpathy-Cramer, None; M.F. Chiang, None; J.P. Campbell, None
References
- 1. Brown JM, Campbell JP, Beers A, et al.. Automated diagnosis of plus disease in retinopathy of prematurity using deep convolutional neural networks. JAMA Ophthalmol. 2018; 136: 803–810. 10.1001/jamaophthalmol.2018.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gulshan V, Peng L, Coram M, et al.. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016; 316: 2402–2410. 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 3. Coyner AS, Swan R, Campbell JP, et al.. Automated fundus image quality assessment in retinopathy of prematurity using deep convolutional neural networks. Ophthalmol Retina. 2019; 3: 444–450. 10.1016/j.oret.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rajpurkar P, Irvin J, Zhu K, et al.. CheXNet: radiologist-level pneumonia detection on chest X-rays with deep learning. ArXiv171105225 Cs Stat. November 2017. http://arxiv.org/abs/1711.05225. Accessed October 23, 2019.
- 5. Jones LD, Golan D, Hanna SA, Ramachandran M. Artificial intelligence, machine learning and the evolution of healthcare: a bright future or cause for concern? Bone Jt Res. 2018; 7: 223–225. 10.1302/2046-3758.73.BJR-2017-0147.R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De Fauw J, Ledsam JR, Romera-Paredes B, et al.. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018; 24: 1342–1350. 10.1038/s41591-018-0107-6. [DOI] [PubMed] [Google Scholar]
- 7. Moor J. The Dartmouth College Artificial Intelligence Conference: the next fifty years. AI Mag. 2006; 27: 87–87. 10.1609/aimag.v27i4.1911. [DOI] [Google Scholar]
- 8. James G, Witten D, Hastie T, Tibshirani R, An Introduction to Statistical Learning: With Applications in R. New York: Springer; 2013. [Google Scholar]
- 9. Hastie T, Tibshirani R, Friedman JH. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2nd ed. New York, NY: Springer; 2009. [Google Scholar]
- 10. NVIDIA Blog: Supervised Vs. Unsupervised Learning. The Official NVIDIA Blog. https://blogs.nvidia.com/blog/2018/08/02/supervised-unsupervised-learning/ Published August 2, 2018. Accessed October 24, 2019.
- 11. Molnar C. Interpretable Machine Learning: A Guide for Making Black Box Models Explainable. Morrisville, North Carolina: Lulu Press, Inc.; 2019. https://christophm.github.io/interpretable-ml-book/. [Google Scholar]
- 12. Mei S, Montanari A, Nguyen P-M. A mean field view of the landscape of two-layer neural networks. Proc Natl Acad Sci U S A. 2018; 115: E7665–E7671. 10.1073/pnas.1806579115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bogunovic H, Waldstein SM, Schlegl T, et al.. Prediction of Anti-VEGF Treatment Requirements in Neovascular AMD Using a Machine Learning Approach. Invest Ophthalmol Vis Sci. 2017; 58: 3240–3248. 10.1167/iovs.16-21053. [DOI] [PubMed] [Google Scholar]
- 14. de Sisternes L, Simon N, Tibshirani R, Leng T, Rubin DL. Quantitative SD-OCT imaging biomarkers as indicators of age-related macular degeneration progression. Invest Ophthalmol Vis Sci. 2014; 55: 7093–7103. 10.1167/iovs.14-14918. [DOI] [PubMed] [Google Scholar]
- 15. Schmidt-Erfurth U, Waldstein SM, Klimscha S, et al.. Prediction of individual disease conversion in early AMD using artificial intelligence. Invest Ophthalmol Vis Sci. 2018; 59: 3199–3208. 10.1167/iovs.18-24106. [DOI] [PubMed] [Google Scholar]
- 16. Vogl W-D, Waldstein SM, Gerendas BS, Schlegl T, Langs G, Schmidt-Erfurth U. Analyzing and predicting visual acuity outcomes of anti-VEGF therapy by a longitudinal mixed effects model of imaging and clinical data. Invest Ophthalmol Vis Sci. 2017; 58: 4173–4181. 10.1167/iovs.17-21878. [DOI] [PubMed] [Google Scholar]
- 17. Bogunovic H, Montuoro A, Baratsits M, et al.. Machine learning of the progression of intermediate age-related macular degeneration based on OCT imaging. Invest Ophthalmol Vis Sci. 2017; 58: BIO141–BIO150. 10.1167/iovs.17-21789. [DOI] [PubMed] [Google Scholar]
- 18. Niu S, de Sisternes L, Chen Q, Rubin DL, Leng T. Fully automated prediction of geographic atrophy growth using quantitative spectral-domain optical coherence tomography biomarkers. Ophthalmology. 2016; 123: 1737–1750. 10.1016/j.ophtha.2016.04.042. [DOI] [PubMed] [Google Scholar]
- 19. Caixinha M, Amaro J, Santos M, Perdigao F, Gomes M, Santos J. In-vivo automatic nuclear cataract detection and classification in an animal model by ultrasounds. IEEE Trans Biomed Eng. 2016; 63: 2326–2335. 10.1109/TBME.2016.2527787. [DOI] [PubMed] [Google Scholar]
- 20. Kim SJ, Cho KJ, Oh S. Development of machine learning models for diagnosis of glaucoma. PloS One. 2017; 12: e0177726 10.1371/journal.pone.0177726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hebb DO. Animal and physiological psychology. Annu Rev Psychol. 1950; 1: 173–188. 10.1146/annurev.ps.01.020150.001133. [DOI] [PubMed] [Google Scholar]
- 22. Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature. 1986; 323: 533–536. 10.1038/323533a0. [DOI] [Google Scholar]
- 23. Orbach J. Principles of neurodynamics. Perceptrons and the theory of brain mechanisms. Arch Gen Psychiatry. 1962; 7: 218–219. 10.1001/archpsyc.1962.01720030064010. [DOI] [Google Scholar]
- 24. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015; 521: 436–444. 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 25. Hahnloser RHR, Sarpeshkar R, Mahowald MA, Douglas RJ, Seung HS. Digital selection and analogue amplification coexist in a cortex-inspired silicon circuit. Nature. 2000; 405: 947–951. 10.1038/35016072. [DOI] [PubMed] [Google Scholar]
- 26. Krizhevsky A, Sutskever I, Hinton GE. ImageNet Classification with Deep Convolutional Neural Networks. In: Pereira F, Burges CJC, Bottou L, Weinberger KQ, Advances in Neural Information Processing Systems 25. Red Hook, NY: Curran Associates, Inc.; 2012: 1097–1105. http://papers.nips.cc/paper/4824-imagenet-classification-with-deep-convolutional-neural-networks.pdf. [Google Scholar]
- 27. Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science. 2006; 313: 504–507. 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- 28. Kaehler A, Bradski GR. Learning OpenCV 3: Computer Vision in C++ with the OpenCV Library. Sebastopol, CA: O'Reilly Media; 2017. [Google Scholar]
- 29. Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Learning deep features for discriminative localization. ArXiv151204150 Cs December 2015. http://arxiv.org/abs/1512.04150 Accessed October 23, 2019.
- 30. Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. ArXiv150504597 Cs May 2015. http://arxiv.org/abs/1505.04597 Accessed October 28, 2019.
- 31. Ghosh S, Das N, Das I, Maulik U. Understanding deep learning techniques for image segmentation. ArXiv190706119 Cs July 2019. http://arxiv.org/abs/1907.06119 Accessed October 28, 2019.
- 32. Zhang Q, Zhu S-C. Visual interpretability for deep learning: a survey. ArXiv180200614 Cs February 2018. http://arxiv.org/abs/1802.00614 Accessed October 23, 2019.
- 33. Zeiler MD, Fergus R. Visualizing and understanding convolutional networks. ArXiv13112901 Cs November 2013. http://arxiv.org/abs/1311.2901 Accessed October 23, 2019.