Abstract
Widespread adoption of electronic health records (EHRs) has resulted in the collection of massive amounts of clinical data. In ophthalmology in particular, the volume range of data captured in EHR systems has been growing rapidly. Yet making effective secondary use of this EHR data for improving patient care and facilitating clinical decision-making has remained challenging due to the complexity and heterogeneity of these data. Artificial intelligence (AI) techniques present a promising way to analyze these multimodal data sets. While AI techniques have been extensively applied to imaging data, there are a limited number of studies employing AI techniques with clinical data from the EHR. The objective of this review is to provide an overview of different AI methods applied to EHR data in the field of ophthalmology. This literature review highlights that the secondary use of EHR data has focused on glaucoma, diabetic retinopathy, age-related macular degeneration, and cataracts with the use of AI techniques. These techniques have been used to improve ocular disease diagnosis, risk assessment, and progression prediction. Techniques such as supervised machine learning, deep learning, and natural language processing were most commonly used in the articles reviewed.
Keywords: artificial intelligence, electronic health record, machine learning, ophthalmology
Introduction
The rapid adoption of electronic health records (EHRs) in recent decades has generated large volumes of clinical data with potential to support secondary use in research.1–3 Indeed, a recurring justification for EHR adoption has been to support the collection and analysis of “big data” to gain meaningful insights.4,5 The clinical research community has expressed growing interest in developing effective techniques to reuse clinical data from EHRs, in part because of the benefits of secondary data reuse over primary data collection.6,7 Researchers reusing EHR data may not need to recruit patients or collect new data, potentially reducing cost compared with traditional clinical research. Moreover, EHR data often contain valuable longitudinal data regarding a patient's status, medical care, and disease progression, which have been previously shown to support clinical decision support,8 medical concept extraction,9 diagnosis,10 and risk assessment.11
However, there are challenges associated with reusing EHR data, particularly because of its complexity and heterogeneity. For example, in ophthalmology, patient data contained in EHRs may include fields as diverse as demographic information, diagnoses, laboratory tests, prescriptions, eye examinations, imaging, and surgical records. Interpreting these heterogeneous data requires strategies such as information extraction, dimension reduction, and predictive modeling typical of machine learning and, more broadly, artificial intelligence (AI) techniques. Applying AI to EHR data has been productive in a variety of domains. For instance, studies in cardiology have broadly used AI techniques with EHR data for the early detection of heart failure,12 to predict the onset of congestive heart failure,13 and to improve risk assessment in patients with suspected coronary artery disease.14 Likewise in ophthalmology, machine learning classifiers with EHR data have been used to predict risks of cataract surgery complications, improve diagnosis of glaucoma and age-related macular degeneration (AMD), and perform risk assessment of diabetic retinopathy (DR).15–18
Although the application of AI to EHR data related to ocular diseases has increased during the past decade, there have been no published reviews of this literature. One literature review of machine learning techniques applied in ophthalmology was published in 201719; however, the included studies mainly focused on the application of machine learning techniques to imaging data, rather than EHR data. This manuscript addresses this knowledge gap by reviewing the literature applying AI techniques to EHR data for ocular disease diagnosis and monitoring. With this review, we explore the type of AI techniques used, the performance of these techniques, and how AI has been applied to specific ocular diseases, providing future directions to clinical practice and research.
Methods
An exhaustive search was performed in the PubMed database using search terms related to “Artificial intelligence”, “Electronic health records,” and “Eye” in any field of articles. See the Appendix for the full query. The results were then examined and narrowed according to the following criteria:
-
1.
Duplicates were removed.
-
2.
Studies were eliminated for lack of relevance after review of the title and abstract; studies that used only imaging data without any EHR data were excluded.
-
3.
Studies without direct clinical application or not related to the topic were excluded.
The review process is summarized in Figure 1. One author (WL) identified articles for inclusion through manual title, abstract, and content review. Two authors (WL and JSC) extracted data for each study: the aim, disease, algorithm, specific techniques, performance assessment, and conclusion of the articles that met the inclusion criteria, as summarized in the Table.15–18,20–28
Figure 1.
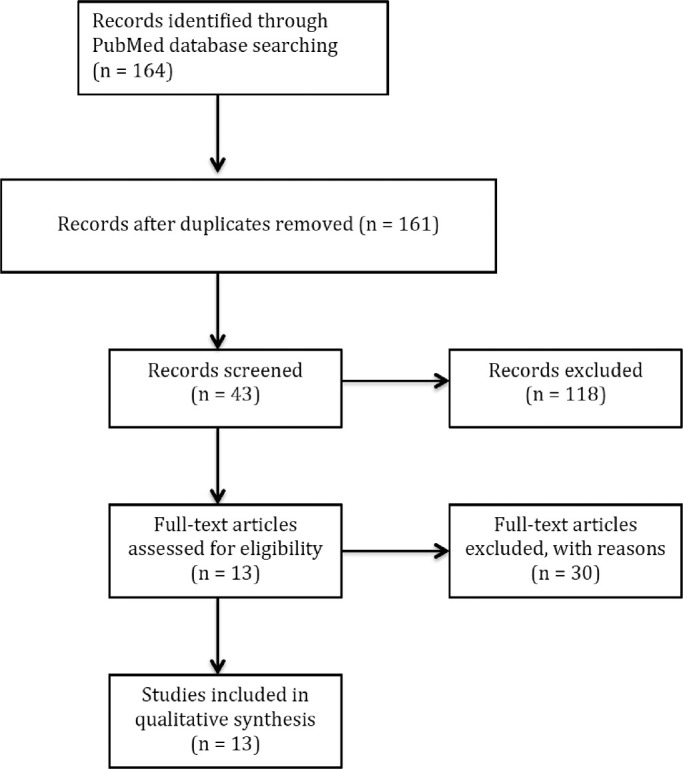
Flow diagram for the literatures selection.
Table.
Studies on Ocular Diseases Using Artificial Intelligence Techniques With EHR Data
| Authors | Aim | Disease | Algorithm Type | Specific Techniques | Performance | Conclusions |
|---|---|---|---|---|---|---|
| Lin et al.20 | Disease detection | Myopia | Supervised machine learning | Random forest | 95% CI for predicting onset of high myopia. 3 years onset prediction (AUC: 94%–98.5%), 5 years (85.6%–90.1%), 8 years (80.1%–83.7%) | Machine learning with EHR data can accurately predict myopia onset |
| Lee et al.21 | Improve diagnostic accuracy | AMD | Deep learning | Convolutional neural networks | For each patient, AUC (97.45%), accuracy (93.54%), sensitivity (92.64%), and specificity (93.69%) | Linked OCT images to EMR data can improve the accuracy of a deep learning model when used to distinguish AMD from normal OCT images |
| Baxter et al.22 | Risk assessment | Open-angle glaucoma | Supervised machine learningDeep learning | Logistic regression, random forests,ANNs | AUC of logistic model (67%), random forest (65%), ANNs (65%) | Existing systemic data in the EHR can identify POAG patients at risk of progression to surgical intervention |
| Chaganti et al.16 | Identify risk factors and improve diagnostic accuracy | Glaucoma, intrinsic optic nerve disease, optic nerve edema, orbital inflammation, and thyroid eye disease | Supervised machine learning | Random forest | AUC of classifiers: glaucoma (88%), intrinsic optic neuritis (76%), optic nerve edema (78%), orbital inflammation (77%), thyroid eye disease (85%) | EMR phenotype (from pyPheWAS) can improve the predictive performance of a random forest classifier with imaging biomarkers |
| Apostolova et al.23 | Patient identification | Open globe injury | Supervised machine learning & Text-mining | SVM NLP–Word embeddings |
Text classification: precision (92.50%), recall (89.83%) | Free-form text with machine learning methods can used to identify open globe injury |
| Saleh et al.18 | Risk assessment | DR | Supervised machine learning | FRF, DRSA | Performance of FRF: Accuracy (80.29%), sensitivity (80.67%), specificity (80.18%) Performance of DRSA: Accuracy (77.32 %), sensitivity (76.89 %), specificity (77.43%) of DRSA. |
Ensemble classifiers (RFR and DRSA) can be applied for diabetic retinopathy risk assessment. The 2-step aggregation procedure is recommended |
| Rohm et al.24 | Predict progression | AMD | Supervised machine learning | AdaBoost, Gradient Boosting, Random Forests, Extremely Randomized trees, LASSO | Accuracy of logMAR VA prediction after VEGF injections.3 months: MAE (0.14), RMSE (0.18)12 months: MAE (0.16), RMSE (0.2) | EHR data of patients with neovascular AMD can be used to predict visual acuity by using machine learning models |
| Yoo and Park25 | Risk assessment | DR | Supervised machine learning | Ridge, elastic net, and LASSO | In external validation, LASSO predicted DR: AUC (82%), accuracy (75.2%), sensitivity (72.1%), and specificity (76.0%) | LASSO with EHR data can be used to predict DR risk among diabetic patients |
| Fraccaro et al.17 | Improve diagnostic accuracy | AMD | Supervised machine learning | Logistic regression, decision trees, SVM, random forests, and AdaBoost | AUC of random forest, logistic regression, and AdaBoost (92%); SVM, decision trees (90%) | Machine learning algorithms using clinical EHR data can be used to improve diagnostic accuracy of AMD |
| Sramka et al.26 | Improve surgical outcome | Cataracts | Supervised machine learningDeep learning | SVM-RMMLNN-EM | Both SVM-RM and MLNN-EM achieved significantly better results than the Barrett Universal II formula in the ±0.50 D PE category | SVM-RM and MLNN-EM with EHR data can be used to improve clinical IOL calculations and improve cataract surgery refractive outcomes |
| Peissig et al.27 | Patient identification | Cataracts | Text-mining | NLP | The multimodal model shows results including sensitivity (84.6%), specificity (98.7%), PPV (95.6%), and NPV (95.1%) | A multimodal strategy incorporating optical character recognition and natural language processing can increase the number of cataracts cases identified |
| Gaskin et al.15 | Identify and predict risks of cataract surgery complications | Cataract | Supervised machine learning Text-mining |
Bootstrapped LASSO, random forest NLP |
Based on the LASSO model, younger age (<60 years old), prior anterior vitrectomy or refractive surgery, history of AMD, and complex cataract surgery were risk factors associated with postoperative complicationsThe random forest model shows high NPV > 95% and moderate sensitivity (67%) and AUC (65%) | Bootstrapped LASSO can be used to identify risk factors of postoperative complications of cataract surgeryRandom forest shows good reliability for predicting cataract surgery complications |
| Skevofilakas et al.28 | Risk assessment | DR | Deep learningSupervised machine learning | FNN and iHWNNCART | AUC of hybrid DSS (98%), iHWNN (97%), FNN (88%), and CART (86%). | Hybrid DSS trained on imaging and related EHR data can estimate the risk of a type 1 diabetic patient developing diabetic retinopathy |
AMD, age-related macular degeneration; ANN, artificial neural network; AUC, area under the curve; CART, classification and regression tree; CI, confidence interval; DR, diabetic retinopathy; DRSA, dominance-based rough set approach; DSS, decision support system; EHR, electronic medical record; EMR, electronic medical record; FNN, feed forward neural network; FRF, fuzzy random forest; iHWNN, improved hybrid wavelet neural network; IOL, intraocular lens; LogMAR, logarithm of the minimum angle of resolution; LASSO, least absolute shrinkage and selection operator; MAE, mean absolute error; MLNN-EM, multilayer neural network ensemble model; NLP, natural language processing; NPV, negative predictive value; OCT, optical coherence tomography; POAG, primary open-angle glaucoma; RFR, random forest regression; RMSE, root mean squared error; SVM, support vector machine; SVM-RM, support vector machine regression model; VA, visual acuity; VEGF, vascular endothelial growth factor.
Results
The PubMed query returned 164 articles published through August 2019. In total, 161 articles were reviewed after removing 3 duplicates. Then 118 articles were excluded because of lack of relevance based on the title and abstract. A total of 13 articles were considered that met inclusion criteria (Fig. 1).
AI Techniques
Three major techniques were used in these studies: 11 studies used supervised machine learning, of which 3 studies specifically used a deep learning technique; 2 studies also used natural language processing (NLP) to generate structured data suitable for analysis from unstructured text. Only 1 study used deep learning by itself, and another study used NLP independent of other techniques (Table). Figure 2 illustrates a simplified machine learning process and the relationship among these 3 techniques. In short, NLP can be used to extract useful information from text-based data and process it into a format suitable for machine learning. Supervised machine learning techniques, some of which use deep learning algorithms, can then be applied to these and other structured data sets to develop predictive models or classifiers.
Figure 2.
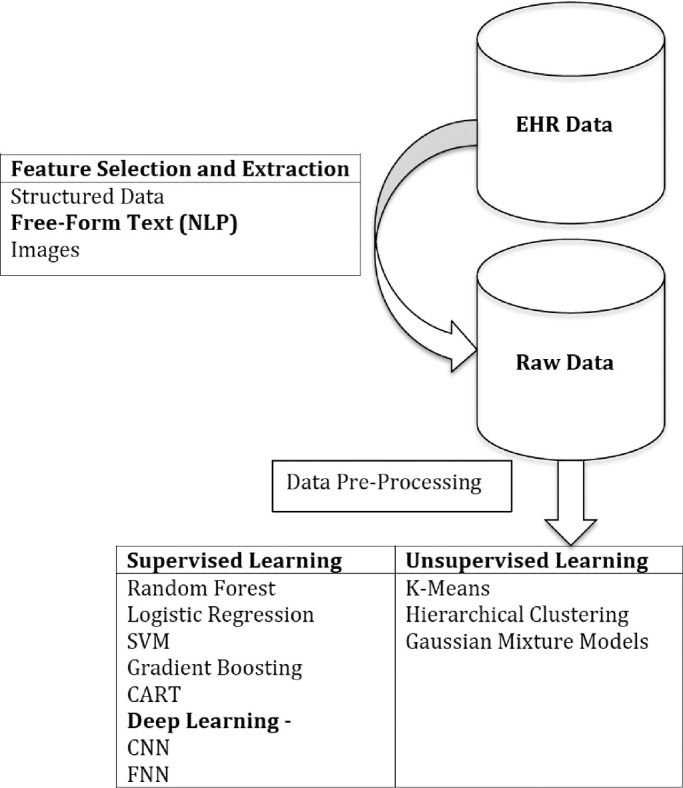
Schematic of the steps of machine learning application. NLP, natural language processing; SVM, support vector machine; CART, classification and regression tree; CNN, convolutional neural network; FNN, feed forward neural network.
Machine Learning
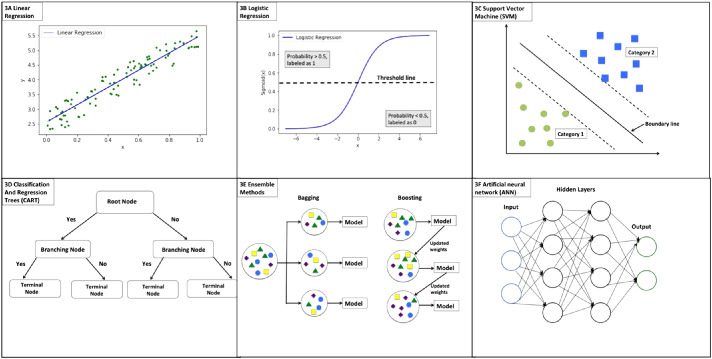
Machine learning techniques are computational methods that learn patterns or classifications within data without being explicitly programmed to do so.29 Machine learning can be divided into 2 methods based on the use of “ground truth” data: supervised learning and unsupervised learning. In supervised learning, a model learns from “ground truth” data in a training data set that contains labeled output data and then can predict the output for new cases. The algorithm is typically a classifier with categorical output or a regression algorithm with continuous output. In unsupervised learning, the model learns from a training data set without labeled output and identifies underlying patterns or structures within its input data. In medicine, machine learning has been widely used in several specialties such as radiology, cardiology, oncology, and ophthalmology to improve diagnostic accuracy and early disease detection.30 In this review, most studies used supervised machine learning techniques such as random forest,31 logistic regression,32 support vector machines (SVMs),33 gradient boosting,34 least absolute shrinkage and selection operator (LASSO),35 AdaBoost,36 and classification and regression tree (CART).37
As shown in Figure 3B, logistic regression is an extension of linear regression (Fig. 3A). In linear regression, the data is modeled as a linear relationship that can be used to predict a value for a given input. In logistic regression, a non-linear function, called the logistic function, converts prediction values into binary categories based on a threshold. Some methods can be used to improve the prediction accuracy of logistic regression, such as least absolute shrinkage and selection operator (LASSO).35 LASSO is a statistical method that selects a smaller subset of predictor variables most related to the outcome variable and shrinks regression coefficients to improve accuracy and generalizability. SVM is another popular machine learning model used for classification analysis. As shown in Figure 3C, a boundary is created to split input data into two distinct groups and can be used to classify new data into similar distinct categories.
Figure 3.
Illustrations of machine learning models. 3A. Linear regression; 3B. Logistic regression; 3C. Support vector machine; 3D. Classification and regression trees (CART); 3E. Ensemble methods; 3F. Artificial neural network (ANN).
A decision tree is an important supervised machine learning algorithm. Figure 3D illustrates a decision tree with a root node as a start followed by the branched nodes and terminal nodes. The root node is the first decision node representing the best predictor variable. Each branched node represents the output of a given input variable. As more input variables are added to subsequent branching nodes, the decision tree becomes more sophisticated in predicting the outcome variable at the terminal nodes.
Ensemble methods combine multiple machine learning models and are commonly used to improve the performance of prediction models. The two most common methods: bootstrapping aggregation (bagging) and boosting were shown in Figure 3E. In a bagging method, multiple subsets of data are randomly selected from the original dataset and each subset data are used to train a separate prediction model. The final predictions will be aggregated from all prediction models. Random forest algorithms are examples of an ensemble machine learning method that combine bagging and decision trees. Boosting is another technique that combines multiple models to create a more accurate one. Adaboost and gradient boosting are widely used boosting machine learning algorithms.
As shown in the Table, random forest was used by Lin et al.20 to predict myopia onset and by Chaganti et al.16 to improve the diagnostic accuracy of glaucoma. In addition, Baxter et al.22 used random forest and logistic regression to identify patients with open-angle glaucoma who had a risk of progression to surgical intervention. Fraccaro et al.17 used logistic regression, decision trees, SVMs, random forests, and AdaBoost to improve diagnostic accuracy of AMD. In addition, fuzzy random forest (FRF) and dominance-based rough set approach (DRSA) were used by Saleh et al.18 for DR risk assessment. And Gaskin et al.15 used random forest and bootstrapped LASSO to identify and predict risks of cataract surgery complications. Moreover, Yoo and Park25 used elastic net and LASSO to predict DR risk among diabetic patients.
Deep Learning
Deep learning is a subset of machine learning techniques based on artificial neural networks (ANNs) that mimic human brain processing. As shown in Figure 3F, multiple layers of computation are constructed in a deep learning model, and each layer is used to perform computations on data from the previous layer. The layers between the input layer and the output layer are called hidden layers. While the information may flow from the input to subsequent output layers (feedforward), information can also flow backward from hidden layers to input layers (backpropagation). The inputs and outputs of hidden layers are not reported; deep learning algorithms present only the final outcome of the output layer.38 Deep learning does not use structured features for input as machine learning does; therefore, deep learning is useful for raw images because they do not have to be prefiltered as they do for machine learning algorithms. After processing raw input through multiple layers within deep neural networks, the algorithms find appropriate features for classifying output. In this review, several articles used deep learning algorithms such as ANNs,39 convolutional neural networks (CNNs),40 multilayer neural network ensemble models (MLNN-EMs),41 and feed forward neural networks (FNNs).42 CNN is a subtype of deep neural network commonly used in image classification. In a CNN model, special convolution and pooling layers are used to reduce a raw image to essential features necessary for the model to classify or label the image. In other words, these techniques use machine learning to determine model input features from the raw image data, rather than a human or a separate image processing program. MLNN-EM is a learning technique that integrates several neural networks to aggregated outcome. In addition, FNN is another subtype of neural network where the information moves forward in (one direction) from root nodes; information never moves backwards. The nodes between input and out layers do not form a cycle of information.
As shown in the Table, Lee et al.21 used CNNs to distinguish AMD from normal OCT images, Baxter et al.22 used ANNs to identify open-angle glaucoma patients at risk of progression to surgery. Also, Sramka et al.26 used models MLNN-EMs and support vector machine regression models (SVM-RM) to improve clinical intraocular lens (IOL) calculations, and Skevofilakas et al.28 used feed forward neural network (FNN) and improved hybrid wavelet neural networks to develop hybrid decision support system for predicting DR risk among diabetic patients.
NLP
NLP is a branch of AI in which computers attempt to interpret human language in written or spoken form. By using NLP, researchers can extract information from text; some uses in medicine include separating progress notes into sections, determining diagnoses from notes, and identifying the documentation of adverse events.43 As shown in the Table, Apostolova et al.,23 Peissig et al.,27 and Gaskin et al.15 describe the use of NLP in extracting cataract information from free-form text clinical notes.
Outcome Metrics for Evaluation of Performance of AI Techniques
Performance evaluation of different AI techniques depends on the chosen algorithm, the purpose of the study, and the input data set. In supervised machine learning algorithms, classifiers are evaluated based on a comparison between the known categorical output and the predicted categorical output. For outputs with 2 categories, the accuracy, sensitivity, specificity, positive predictive value, and negative predictive value can be computed. Another important evaluation metric is the AUC-ROC (area under the curve–receiving operating characteristic), which is used to evaluate the performance of classifiers based on different thresholds. ROC is a probability curve that visualizes the true positive rate (sensitivity) change with respect to false positive rate (1–specificity) for different threshold values used in the model. The AUC represents the ability of a model to distinguish between different outcome values.44 An AUC equal to 1 is ideal and represents the model's ability to perfectly distinguish between two outcomes. On the other hand, an AUC of approximately 0.5 is the worst case because it means that the model is not better than chance for distinguishing between two outcomes.
As shown in the Table, 8 studies used AUC-ROC to evaluate the performance of classifiers.15–17,20–22,25,28 The range of AUC-ROC was from 65% to 98.5%, and the median AUC in all included studies was 90%. In addition, precision and recall were used to evaluate the performance of text-mining algorithms.45 Apostolova et al.23 and Peissig et al.27 used precision and recall to evaluate the performance of text classification.
For regression models, 2 evaluation metrics—mean absolute error (MAE) and root mean squared error (RMSE)—are commonly used to measure accuracy for continuous variables. They measure the average difference between actual observations and predictions. MAE shows the absolute differences with equal weight for each difference. In contrast, RMSE penalized larger errors by taking the square of the difference before averaging. In the study by Rohm et al.,24 MAE and RMSE were used to evaluate visual acuity prediction.
Application of AI to Clinical Ophthalmology
AI techniques have been applied clinically to improving ocular disease diagnosis, predicting disease progression, and risk assessment (Table). Several diseases were studied in articles included in this review including glaucoma, cataracts, AMD, and DR. We will present the benefits of AI techniques with EHR data in these diseases.
Glaucoma
Two studies in this review focused on the field of glaucoma and used supervised machine learning techniques to improve diagnosis and predict progression.16,22 In the study by Chaganti et al.,16 a good performance was obtained (AUC of glaucoma diagnosis 88%), and results showed that the addition of an EMR phenotype could improve the classification accuracy of a random forest classifier with imaging biomarkers.16 On the other hand, Baxter et al.22 reported a moderate performance (AUC 67%) in a study that used EHR data alone to predict risk of progression to surgical intervention in patients with open-angle glaucoma. In addition to model performance, it is important to know which factors can be used to improve disease diagnosis. The work performed by Chaganti et al.16 began to explore this problem by comparing the performance of classifiers using EMR phenotypes, visual disability scores, and imaging metrics.
Cataracts
Three studies applying different AI techniques to cataract diagnosis and management were reviewed. In the study by Peissig et al.,27 NLP was used to extract cataract information from free-text documents. An EHR-based cataract phenotyping algorithm, which consisted of structured data, information from free-text notes, and optical character recognition on scanned clinical images, was developed to identify cataract subjects. The result of the study showed good performance (predictive positive value >95%).27 Additionally, Gaskin et al.15 used supervised machine learning algorithms to identify risk factors and to predict intraoperative and postoperative complications of cataract surgery. The investigators used data mining via NLP to extract cataract information from the EHR system.15 The predictive model showed moderate performance (AUC 65%), and the risk factors associated with surgical complications included younger patients, refractive surgery history, AMD history, and complex cataract surgery. These risk factors were associated with postoperative complications, and the predictive model showed moderate performance (AUC 65%). Supervised machine learning (SVM-RM) and deep learning (MLNN-EM) algorithms were used to improve the IOL power calculation by Sramka et al.26 Both SVM-RM and MLNN-EM model provided better IOL calculations than the Barrett Universal II formula.
AMD
Three studies used AI in AMD. Lee et al.21 used deep learning techniques to improve the diagnosis of AMD. Optical coherence tomography (OCT) images of each patient were linked to EMR clinical end points extracted from EPIC (Verona, WI) for each patient to predict a diagnosis of AMD. The model had high accuracy with an AUC 97% in distinguishing AMD from normal OCT images.21 Another study conducted by Rohm et al.24 used supervised regression models to accurately predict visual acuity in response to anti–vascular endothelial growth factor injections in patients with neovascular AMD. Models predicting treatment response may have implications in encouraging patients adhering to intravitreal therapy. Also, as demonstrated by Fraccaro et al.,17 supervised machine learning techniques can be incorporated into EHR systems providing real-time support for AMD diagnosis.
DR
DR is one of the most common comorbidities of diabetes, and frequent screening examinations for diabetic patients are resource consuming. Three studies explore this problem by using AI techniques with EHR data to determine patient risk for the development of DR. Saleh et al.18 used 2 kinds of ensemble classifiers—FRF and DRSA—to predict DR risk using EHRs. Good performance (accuracy 80%) of the FRF model was shown in this study. Similarly, Yoo and Park25 proposed a comparison between the learning models—ridge, elastic net, and LASSO—using the traditional indicators of DR. They showed that the performance of LASSO (AUC 81%) was significantly better than the traditional indicators (AUC of glycated hemoglobin 69%; AUC of fasting plasma glucose 54%) in diagnosing DR. In addition, a hybrid DSS was developed by Skevofilakas et al.28 to estimate the risk of a patient with type 1 diabetes to develop DR. The hybrid DSS showed an excellent performance with an AUC of 98%. Overall, these studies show that integrating these techniques with an EHR system has promise in improving early detection of diabetic patients at risk of DR progression.
Discussion
This article reviews the literature applying AI techniques to EHR data to aid in ocular disease diagnosis and risk assessment. We focus the discussion on 3 areas: AI techniques used to analyze EHR data, the performance of techniques, and the ocular diseases most commonly analyzed.
First, secondary use of EHR data via AI techniques can be used to improve ocular disease diagnosis, risk assessment, and disease progression. The predictive models across the 8 classifiers showed good performance with a median AUC of 90%. One study, prediction of postoperative complications of cataract surgery, reported moderate accuracy with 65%,15 perhaps because of insufficient predictors, such as lack of surgeon-relevant information. Also, the prevalence of various complications may affect the reliability of prediction outcomes. For example, a rare prevalence complication may not be handled well with standard classification techniques because of imbalanced data.46,47 When a dataset contains a very few number of cases of disease or complications, there is not enough data about these cases for the model to accurately learn how to predict them. On the other hand, excellent performance of classifiers trained on combined EHR and image data were reported by Skevofilakas et al.28 and Lee et al.21 For future studies, a feasible direction might be to develop a hybrid model that uses both the routine EHR data and image data sets to have a more complete picture of patient variables associated with the outcome of interest.
Second, supervised machine learning was the most common technique used with EHR data to analyze ocular diseases. These studies focused on improving diagnosis, predicting progression, or assessing risk for early detection. The predictors defined were based on the risk factors of disease, demographic features found from literature review, and clinical experiences. None of the studies reviewed used unsupervised machine learning techniques where the desired output and the relationship between the outcome variable and the predictors are unknown. These methods are used to identify clusters of data that are similar and can help discover the hidden factors that are useful for improving the diagnosis. However, unsupervised learning has been successfully applied to other fields. For example, Marlin et al.48 demonstrated that the probabilistic clustering model for time-series data from real-world EHRs could be able to capture patterns of physiology and be used to construct mortality prediction models. For future studies, unsupervised machine learning techniques might be used to find hidden patterns from EHR data for improving clinical predictions of ocular diseases.
Finally, in this review, studies that analyzed EHR data with AI techniques mainly focused on 4 diseases: glaucoma, DR, AMD, and cataracts. The focus on these diseases (glaucoma, AMD, and DR) is likely due to their prevalence as the major causes of irreversible blindness in the world.49 Early detection or treatment can delay or halt the progression of such diseases, reduce visual morbidity, and preserve a patient's quality of life.50,51 These studies suggest that AI techniques can be used to achieve this goal. Furthermore, cataract surgery is the most common refractive surgical procedure and is one of the most common surgeries performed in ophthalmology.52 Risk assessment of the postoperative complications and decreasing the risk of reoperation are crucial to patient outcomes, and AI techniques can help approach these issues.
This review presents the AI techniques used in vision sciences based on EHR data. However, several problems still need to be addressed for future studies. One of the major problems is data quality. EHR data required for research are essentially different from data collected during a traditional clinical research study. EHR data collected from clinical practice may have incomplete information due to incorrect data entry, nonanswers, and recording errors. Consequently, the performance of machine learning models will be dependent on data quality and is an issue when using AI techniques with EHR data.53–56 Additionally, except for the work reported by Lin et al.,20 all reviewed studies were single-center studies. Thus, the results of studies may not be generalizable to other healthcare systems.
Although imaging data do not suffer from the data quality issues of other clinical data, there is no well-established gold standard for many imaging techniques. For instance, Garvin et al.57 presented an automated 3-dimensional intraretinal layer segmentation algorithm using OCT image data. The gold standard was determined by 2 retinal experts’ recommendations. This requires more time and resources to analyze and cross-validate the outcomes. Also, different preprocessing and postprocessing algorithms, hardware configurations, and image processing steps are intended to improve image quality for easier automated diagnosis. However, these factors often make models difficult to replicate. In addition, using imaging analysis without other prior information, such as medical history information, may affect the model performance and lead to biased results. Therefore, integration of imaging data and routine EHR data allows us to obtain prior information to input to the predictive model.
Conclusion
AI techniques are rapidly being adopted in ophthalmology and have the potential to improve the quality and delivery of ophthalmic care. Moreover, secondary use of EHR data is an emerging approach for clinical research involving AI, particularly given the availability of large-scale data sets and analytic methods.58–60 In this review, we describe applications of AI methods to ocular diseases and problems such as diagnostic accuracy, disease progression, and risk assessment and find that the number of published studies in this area has been relatively limited due to challenges with the current quality of EHR data. In the future, we expect that AI using EHR data will be applied more widely in ophthalmic care, particularly as techniques improve and EHR data quality issues are resolved.
Acknowledgments
Supported by grants P30EY10572 (MFC), R00LM012238 (MRH), and T15LM007088 (WL) from the National Institutes of Health, Bethesda, MD; unrestricted departmental funding from Research to Prevent Blindness, New York, NY (MFC); and Research to Prevent Blindness Medical Student Fellowship (JSC). The funding organizations had no role in the design or conduct of this research.
Disclosure: W.-C. Lin, None; J.S. Chen, None; M.F. Chiang, Novartis (C), and Inteleretina, LLC (E); M.R. Hribar, None
Appendix: Appendix: PubMed Search Query
(
( ("electronic"[All Fields] AND "health"[All Fields] AND record[All Fields])
OR (“electronic"[All Fields] AND "medical"[All Fields] AND record[All Fields])
OR (("computerised"[All Fields] OR "computerized"[All Fields]) AND "medical"[All Fields] AND record[All Fields])
OR ("electronic health records"[MeSH Terms])
OR (“medical records systems, computerized"[MeSH Terms])
OR (“electronic health data” [All Fields])
OR (“personal health data” [All Fields])
OR (“personal health record” [All Fields])
OR (“personal health records” [All Fields])
OR (“Health Record” [All Fields])
OR (“computerized patient medical records” [All Fields])
OR (“computerized medical record” [All Fields])
OR (“computerized medical records” [All Fields])
OR (“computerized patient records” [All Fields])
OR (“computerized patient record” [All Fields])
OR (“computerized patient medical record” [All Fields])
OR (“electronic health records” [All Fields])
OR (“electronic patient record” [All Fields])
OR (“electronic healthcare record” [All Fields])
OR (“ehr” [All Fields])
OR (“ehrs” [All Fields])
OR (“emr” [All Fields])
OR (“emrs” [All Fields])
OR (“phr” [All Fields])
OR (“phrs” [All Fields])
OR (“patient record” [All Fields])
OR (“patient health record” [All Fields])
OR (“healthcare record” [All Fields])
)
AND
(
("Machine"[All Fields] AND "Learning"[All Fields])
OR (“Artificial"[All Fields] AND "intelligence"[All Fields])
OR ("deep learning"[All Fields])
OR (“Machine intelligence"[All Fields])
OR (“Natural language processing"[All Fields])
OR (“NLP"[All Fields])
OR (“ML"[All Fields])
OR (“DL"[All Fields])
OR (“vector machine "[All Fields])
OR (“random forest"[All Fields])
OR (“neural network "[All Fields])
)
AND
English[lang]
AND
(
("Ophthalmology"[All Fields])
OR (“Eye"[All Fields])
OR (“Vision"[All Fields])
OR (“visual"[All Fields])
OR (“Diabetic retinopathy"[All Fields])
OR (“Cataract"[All Fields])
OR (“Glaucoma” [All Fields])
OR (“Cornea” [All Fields])
OR (“Pediatric Ophthalmology and Strabismus” [All Fields])
OR (“Retina” [All Fields])
OR (“Retinal disease” [All Fields])
OR (“Uveitis” [All Fields])
OR (“Neuro-ophthalmology” [All Fields])
OR (“Ophthalmic genetics” [All Fields])
OR (“Inherited retina diseases” [All Fields])
OR (“Oculoplastics” [All Fields])
OR (“Ocular Oncology” [All Fields])
OR (“Cataract surgery” [All Fields])
OR (“Comprehensive eye care” [All Fields])
OR (“Oculofacial plastics and reconstructive surgery” [All Fields])
OR (“Vision rehabilitation” [All Fields])
OR (“Contact Lenses” [All Fields])
OR (“Myopia” [All Fields])
OR (“Age-related macular degeneration” [All Fields])
OR (“ROP” [All Fields])
OR (“Congenital cataract” [All Fields])
OR (“Low vision” [All Fields])
OR (“Pediatric ophthalmology” [All Fields])
OR (“Strabismus” [All Fields])
OR (“Oculoplastic surgery” [All Fields])
OR (“Comprehensive ophthalmology” [All Fields])
OR (“Refractive error” [All Fields])
OR (“Refractive surgery” [All Fields])
)
References
- 1. Safran C, Bloomrosen M, Hammond WE, et al.. Toward a national framework for the secondary use of health data: an American Medical Informatics Association White Paper. J Am Med Inform Assoc. 2007; 14: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Charles D, Gabriel M, Searcy T. Adoption of electronic health record systems among US non-federal acute care hospitals: 2008-2012. J ONC data brief. 2013; 9: 1–9. [Google Scholar]
- 3. Coorevits P, Sundgren M, Klein GO, et al.. Electronic health records: new opportunities for clinical research. J Intern Med. 2013; 274: 547–560. [DOI] [PubMed] [Google Scholar]
- 4. Cano I, Tenyi A, Vela E, Miralles F, Roca J. Perspectives on Big Data applications of health information. Curr Opin Syst Biol. 2017; 3: 36–42. [Google Scholar]
- 5. Khennou F, Khamlichi YI, Chaoui NEH. Improving the use of big data analytics within electronic health records: a case study based OpenEHR. Procedia Comput Sci. 2018; 127: 60–68. [Google Scholar]
- 6. Meystre S, Lovis C, Bürkle T, Tognola G, Budrionis A, Lehmann C. Clinical data reuse or secondary use: current status and potential future progress. Yearb Med Inform. 2017; 26: 38–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cowie MR, Blomster JI, Curtis LH, et al.. Electronic health records to facilitate clinical research. Clin Res Cardiol. 2017; 106: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuperman GJ, Bobb A, Payne TH, et al.. Medication-related clinical decision support in computerized provider order entry systems: a review. J Am Med Inform Assoc. 2007; 14: 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jiang M, Chen Y, Liu M, et al.. A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. J Am Med Inform Assoc. 2011; 18: 601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kononenko I. Machine learning for medical diagnosis: history, state of the art and perspective. Artif Intell Med. 2001; 23: 89–109. [DOI] [PubMed] [Google Scholar]
- 11. Persell SD, Dunne AP, Lloyd-Jones DM, Baker DW. Electronic health record-based cardiac risk assessment and identification of unmet preventive needs. Med Care. 2009; 47: 418–424. [DOI] [PubMed] [Google Scholar]
- 12. Choi E, Schuetz A, Stewart WF, Sun J. Using recurrent neural network models for early detection of heart failure onset. J Am Med Inform Assoc. 2016; 24: 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheng Y, Wang F, Zhang P, Hu J. Risk prediction with electronic health records: A deep learning approach. Paper presented at: Proceedings of the 2016 SIAM International Conference on Data Mining 2016.
- 14. Motwani M, Dey D, Berman DS, et al.. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: a 5-year multicentre prospective registry analysis. Eur Heart J. 2016; 38: 500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gaskin GL, Pershing S, Cole TS, Shah NH. Predictive modeling of risk factors and complications of cataract surgery. Eur J Ophthalmol. 2016; 26: 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chaganti S, Nabar KP, Nelson KM, Mawn LA, Landman BA. Phenotype analysis of early risk factors from electronic medical records improves image-derived diagnostic classifiers for optic nerve pathology. Paper presented at: Medical Imaging 2017: Imaging Informatics for Healthcare, Research, and Applications 2017. Orlando, FL, USA, Feb 15–16, 2017. [DOI] [PMC free article] [PubMed]
- 17. Fraccaro P, Nicolo M, Bonetto M, et al.. Combining macula clinical signs and patient characteristics for age-related macular degeneration diagnosis: a machine learning approach. BMC Ophthalmol. 2015; 15: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saleh E, Błaszczyński J, Moreno A, et al.. Learning ensemble classifiers for diabetic retinopathy assessment. Artif Intell Med. 2018; 85: 50–63. [DOI] [PubMed] [Google Scholar]
- 19. Caixinha M, Nunes S.. Machine learning techniques in clinical vision sciences. Curr Eye Res. 2017; 42: 1–15. [DOI] [PubMed] [Google Scholar]
- 20. Lin H, Long E, Ding X, et al.. Prediction of myopia development among Chinese school-aged children using refraction data from electronic medical records: a retrospective, multicentre machine learning study. PLoS Med. 2018; 15: e1002674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee CS, Baughman DM, Lee AY. Deep learning is effective for classifying normal versus age-related macular degeneration OCT images. Ophthalmol Retina. 2017; 1: 322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baxter SL, Marks C, Kuo T-T, Ohno-Machado L, Weinreb RN. Machine learning-based predictive modeling of surgical intervention in glaucoma using systemic data from electronic health records. Am J Ophthalmol. 2019; 208: 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Apostolova E, White HA, Morris PA, Eliason DA, Velez T. Open globe injury patient identification in warfare clinical notes. AMIA AnnuSympProc. 2017; 2017: 403–410. [PMC free article] [PubMed] [Google Scholar]
- 24. Rohm M, Tresp V, Muller M, et al.. Predicting visual acuity by using machine learning in patients treated for neovascular age-related macular degeneration. Ophthalmol. 2018; 125: 1028–1036. [DOI] [PubMed] [Google Scholar]
- 25. Yoo TK, Park E-C.. Diabetic retinopathy risk prediction for fundus examination using sparse learning: a cross-sectional study. BMC Med Inform Decis Mak. 2013; 13: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sramka M, Slovak M, Tuckova J, Stodulka P. Improving clinical refractive results of cataract surgery by machine learning. PeerJ. 2019; 7: e7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peissig PL, Rasmussen LV, Berg RL, et al.. Importance of multi-modal approaches to effectively identify cataract cases from electronic health records. J Am Med Inform Assoc. 2012; 19: 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Skevofilakas M, Zarkogianni K, Karamanos BG, Nikita KS. A hybrid decision support system for the risk assessment of retinopathy development as a long term complication of type 1 diabetes mellitus. Paper presented at: 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology; Buenos Aires, Argentina, Aug 31 - Sep 4, 2010. [DOI] [PubMed]
- 29. Anzai Y. PatternRecognitionand Machine Learning. Burlington, MA: Morgan Kaufmann; 2012. [Google Scholar]
- 30. Ramesh A, Kambhampati C, Monson JR, Drew P. Artificial intelligence in medicine. Ann R Coll Surg Engl. 2004; 86: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liaw A, Wiener M.. Classification and regression by randomForest. RNews. 2002; 2: 18–22. [Google Scholar]
- 32. Kleinbaum DG, Dietz K, Gail M, Klein M, Klein M. LogisticRegression. New York: : Springer-Verlag, 2002. [Google Scholar]
- 33. Burges CJ. A tutorial on support vector machines for pattern recognition. Data Min Knowl Discov. 1998; 2: 121–167. [Google Scholar]
- 34. Friedman JH. Stochastic gradient boosting. Comput Stat Data Anal. 2002; 38: 367–378. [Google Scholar]
- 35. Kukreja SL, Löfberg J, Brenner M. A least absolute shrinkage and selection operator (LASSO) for nonlinear system identification. IFACProcVol. 2006; 39: 814–819. [Google Scholar]
- 36. Schapire RE. Explaining adaboost. In: EmpiricalInference. Berlin: : Springer; 2013: 37–52. [Google Scholar]
- 37. Lewis RJ. An introduction to classification and regression tree (CART) analysis. Paper presented at: Annual Meeting of the Society for Academic Emergency Medicine; San Francisco, CA; 2000. May 22–25.
- 38. Goodfellow I, Bengio Y, Courville A. DeepLearning. Cambridge, MA: : MIT Press; 2016. [Google Scholar]
- 39. Jain AK, Mao J, Mohiuddin KM. Artificial neural networks: a tutorial. Computer. 1996; 29: 31–44. [Google Scholar]
- 40. Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Paper presented at: Advances in Neural Information Processing Systems; Lake Tahoe, NV, USA, Dec 3–8, 2012.
- 41. Kourentzes N, Barrow DK, Crone SF. Neural network ensemble operators for time series forecasting. Expert Syst Appl. 2014; 41: 4235–4244. [Google Scholar]
- 42. Svozil D, Kvasnicka V, Pospichal J. Introduction to multi-layer feed-forward neural networks. Chemometr Intell Lab Syst. 1997; 39: 43–62. [Google Scholar]
- 43. Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc. 2011; 18: 544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hanley JA, McNeil B.. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982; 143: 29–36. [DOI] [PubMed] [Google Scholar]
- 45. Buckland M, Gey F.. The relationship between recall and precision. J Am Med Inform Assoc. 1994; 45: 12–19. [Google Scholar]
- 46. Chawla NV, Japkowicz N, Kotcz A. Special issue on learning from imbalanced data sets. ACM Sigkdd Explorations Newsletter. 2004; 6: 1–6. [Google Scholar]
- 47. Van Hulse J, Khoshgoftaar TM, Napolitano A. Experimental perspectives on learning from imbalanced data. Paper presented at: Proceedings of the 24th International Conference on Machine Learning; Corvallis, OR, USA, June 20–24, 2007.
- 48. Marlin BM, Kale DC, Khemani RG, Wetzel RC. Unsupervised pattern discovery in electronic health care data using probabilistic clustering models. Paper presented at: Proceedings of the 2nd ACM SIGHIT International Health Informatics Symposium; Miami, FL, USA, Jan 28–30, 2012.
- 49. Organization WH. Global Data on Visual Impairments 2010. Lausanne, Switzerland: : WHO Press; 2012. [Google Scholar]
- 50. Alghamdi HF. Causes of irreversible unilateral or bilateral blindness in the Al Baha region of the Kingdom of Saudi Arabia. Saudi J Ophthalmol. 2016; 30: 189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Weinreb RN, Khaw PT. Primary open-angle glaucoma. Lancet. 2004; 363: 1711–1720. [DOI] [PubMed] [Google Scholar]
- 52. Wang W, Yan W, Fotis K, et al.. Cataract surgical rate and socioeconomics: a global study. Invest Ophthalmol Vis Sci. 2016; 57: 5872–5881. [DOI] [PubMed] [Google Scholar]
- 53. Hersh WR. Adding value to the electronic health record through secondary use of data for quality assurance, research, and surveillance. Clin Pharmacol Ther. 2007; 81: 126–128. [PubMed] [Google Scholar]
- 54. Botsis T, Hartvigsen G, Chen F, Weng C. Secondary use of EHR: data quality issues and informatics opportunities. Summit Transl Bioinform. 2010; 2010: 1. [PMC free article] [PubMed] [Google Scholar]
- 55. Ashfaq HA, Lester CA, Ballouz D, Errickson J, Woodward MA. Medication accuracy in electronic health records for microbial keratitis. JAMAOphthalmol. 2019; 137: 929–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Berdahl CT, Moran GJ, McBride O, Santini AM, Verzhbinsky IA, Schriger DL. Concordance between electronic clinical documentation and physicians' observed behavior. JAMA Netw Open. 2019; 2: e1911390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Garvin MK, Abramoff MD, Wu X, Russell SR, Burns TL, Sonka M. Automated 3-D intraretinal layer segmentation of macular spectral-domain optical coherence tomography images. IEEE Trans Med Imaging. 2009; 28: 1436–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Parke DW, Rich WL, Sommer A, Lum F. The American Academy of Ophthalmology's IRIS Registry (Intelligent Research in Sight Clinical Data): a look back and a look to the future. Ophthalmol. 2017; 124: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 59. Clark A, Ng JQ, Morlet N, Semmens JB. Big data and ophthalmic research. Surv Ophthalmol. 2016; 61: 443–465. [DOI] [PubMed] [Google Scholar]
- 60. Coleman AL. How big data informs us about cataract surgery: the LXXII Edward Jackson Memorial Lecture. Am J Ophthalmol. 2015; 160: 1091–1103. e1093. [DOI] [PubMed] [Google Scholar]