Abstract
Indoleamine 2,3-dioxygenase 1 (IDO1) catalyzes the first and rate-limiting reaction of l-tryptophan (Trp) conversion into l-kynurenine (Kyn). The depletion of Trp, and the accumulation of Kyn have been proposed as mechanisms that contribute to the suppression of the immune response—primarily evidenced by in vitro study. IDO1 is therefore considered to be an immunosuppressive modulator and quantification of IDO1 metabolism may be critical to understanding its role in select immunopathologies, including autoimmune- and oncological-conditions, as well as for determining the potency of IDO1 enzyme inhibitors. Because tryptophan 2,3-dioxygenase (TDO), and to a significantly lesser extent, IDO2, also catabolize Trp into Kyn, it’s important to differentiate the contribution of each enzyme to Trp catabolism and Kyn generation. Moreover, a great variety of detection methods have been developed for the quantification of Trp metabolites, but choosing the suitable protocol remains challenging. Here, we review the differential expression of IDO1/TDO/IDO2 in normal and malignant tissues, followed by a comprehensive analysis of methodologies for quantifying Trp and Kyn in vitro and in vivo, with an emphasis on the advantages/disadvantages for each application.
1. Introduction
A fundamental biological event of all living organisms is their sensation and response to changes in nutrient availability, which is a major determinant along with other environmental cues that shape cellular behavior. Amino acid metabolism has long been recognized as a crucial regulatory mechanism that underlies cellular responses during select pathophysiological conditions. Among the nine “essential” amino acids in higher vertebrates that are required for dietary intake, tryptophan (Trp) requires more energy than any other amino acid during post–transcriptional modification, and is the least abundant amino acid found in mammalian proteins. Moreover, less than 1% of dietary Trp is used for protein synthesis, while the remainder is degraded through decarboxylation, transamination, hydroxylation, or oxidation (Badawy, 2014), leading to generation of physiologically active compounds including neuroactive tryptamine, neuroprotective melatonin, and/or immunomodulatory kynurenine (Kyn). Approximately 80–90% of dietary Trp is metabolized through the Kyn pathway (Fig. 1) and generation of nicotinamide adenine dinucleotide (NAD) is an important enzyme co-factor that is ultimately created during full pathway biosynthesis. In addition, the methoxyindole pathway utilizes about 1–2% of l-Trp and provides the neuroactive compounds, serotonin and melatonin (Henykova et al., 2016).
Fig. 1.
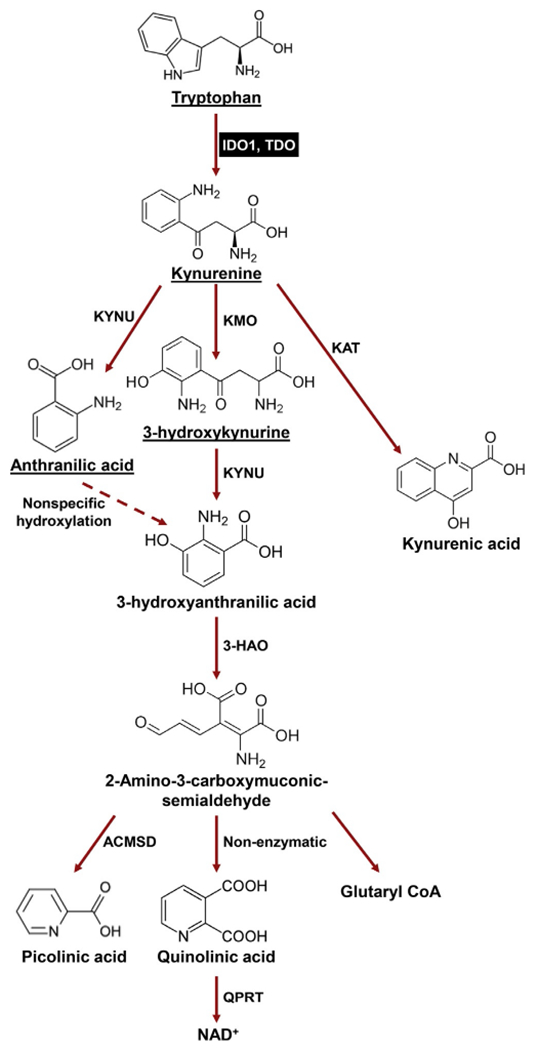
The metabolic kynurenine (Kyn) conversion pathway. In addition to being used as building block for protein synthesis, the majority of dietary tryptophan (95%) is catabolized via the Trp→Kyn pathway (red arrows). Within the Kyn pathway, underlined metabolites can cross the blood brain barrier (BBB). IDO1 (and TDO) are highlighted in the black box. 3-HAO, 3-hydroxyanthranilate 3,4-dioxygenase; ACMSD, 2-amino-3-carboxymuconate semialdehyde carboxylase; KAT, kynurenine aminotransferase (I, II, III); KAT, kynurenine aminotransferase (I, II, III); KMO, kynurenine 3-monooxygenase; KYNU, kynureninase; QPRT, quinolinic-acid phosphoribosyl transferase.
Both IDO1 and TDO catalyze the rate-limiting cleavage of the Trp indole ring 2,3-double bond and incorporate molecular oxygen. The product of this reaction is N-formylkynurenine, which is rapidly and spontaneously converted into l-Kyn. The latter catabolite is further converted into downstream intermediates, including 3-hydroxy-l-kynurenine (3-HK), 3-hydroxyanthranilate (3-HAA) and quinolinic acid (Quin), which can also affect the immune response (Zhai et al., 2015). In addition to IDO1 and TDO, the IDO1 paralog, referred to as IDO2, has been identified and proposed to be a third enzyme that is capable of catalyzing Trp degradation through the Kyn pathway (Ball et al., 2007; Metz et al., 2007; Yuasa et al., 2007).
However, the Km of human IDO1 and IDO2 for l-Trp, is 20.90 ± 3.95 μmol/L and 6809 ± 917 μmol/L, respectively, indicating a dramatic decrease in the enzymic potential for IDO2 to convert Trp into Kyn (Pantouris, Serys, Yuasa, Ball, & Mowat, 2014). Strikingly, the transcriptome analysis of 129 human tumor samples and 25 human tumor cell lines demonstrated very limited IDO2 expression (van Baren & Van den Eynde, 2015). In combination with the finding that human IDO2 possesses negligible Trp catabolic activity, the role of IDO2 in tumor-induced immunosuppression is still elusive. Also notable is that mice constitutionally deficient for IDO2 do not show decreased systemic Kyn levels as compared to wild-type counterparts (Metz et al., 2014), which is a significant contrast to the decreased Kyn levels in animal subjects constitutionally deficient for IDO1(Nagano et al., 2013).
Because of its role as a molecular switch for nutrient availability and its inducible expression upon inflammatory stimulation, IDO1 was originally thought of as an innate immune modulator that restricts Trp and essential for inhibiting microbial growth and neoplastic cell proliferation (Taylor & Feng, 1991). This initial notion was revised by Munn and colleagues, demonstrating that the in vivo administration of an IDO1 pathway inhibitor, 1-methyl tryptophan (1-MT), leads to T-cell-dependent fetal allograft rejection (Munn et al., 1998). Subsequent work demonstrated that IDO1-expressing macrophages, dendritic cells (DC), and tumor cells mediate the inhibition of T-cell proliferation (Frumento et al., 2002; Hwu et al., 2000; Munn et al., 1999, 2002). Further mechanistic studies indicated that the T cell inhibition is caused by Trp depletion-induced GCN2 and/or mTOR signaling pathways. Moreover, in vitro studies demonstrated that exogenously-added Kyn acts as an immunosuppressive metabolite when further combined with transforming growth factor beta, resulting in the promotion of naïve T cell differentiation into FoxP3+ regulatory T cells (Mezrich et al., 2010). Additionally, the exogenous addition of other Kyn pathway metabolites, including 3-HK, 3-HAA, and quinolinic acid, also suppress T cell proliferation in vitro (Zhai et al., 2015). Since TDO, and to a significantly lesser extent, IDO2, catabolize Trp into Kyn, it’s possible that the combined contributions of IDO1, TDO, and IDO2 collectively contribute to the Trp depletion/Kyn accumulation hypothesis of metabolism-induced immunosuppression. Furthermore, given the broad expression of IDO1 in many types of cancer (Theate et al., 2015), it’s not surprising that there is a high level of interest in studying IDO1 in tumor-induced immunotolerance, as well as the multitude of clinical trials evaluating IDO1 pathway and enzyme inhibitors in patients with cancer (Zhai et al., 2015). In contrast, fewer studies have reported an immunosuppressive role for TDO in cancer (Platten, von Knebel Doeberitz, Oezen, Wick, & Ochs, 2015).
The current dogma for how IDO1 mediates immunosuppression still primarily focuses on the depletion of Trp and/or accumulation of Kyn. It is therefore important to be equipped with the proper tools for accurately and sensitively measuring IDO1 enzyme activity. It’s important to note, however, that there is no available methodology for distinguishing between the competing activities of IDO1, TDO and IDO2. Tissue expression profiling, in addition to analysis of enzyme activity, is therefore necessary to fully understand the unique contributions of different Trp catabolic enzymes. Furthermore, the availability of reliable techniques for quantifying Trp and Kyn in biological samples with complex composition (bone, muscle, brain, etc.) is another challenge for measuring IDO1 enzyme activity. The detection of trace amounts of Kyn, tissue contaminants that interfere with sample analysis, and the instability of some Trp metabolites, require the careful refinement and preparation of samples prior to conducting downstream analytic methods. Next, we summarize current knowledge for how IDO1 and TDO expression is distributed and regulated among normal and malignant tissues, followed by a discussion of their relationship with the Kyn pathway. Finally, methodologies for detection Trp and downstream Kyn metabolites are discussed in detail.
2. Differential expression of IDO1 and TDO in normal and malignant tissues
Normally, TDO is primarily expressed in the liver and is responsible for regulation of serum Trp homeostasis. TDO expression is affected by dietary Trp intake, whereby oral consumption of Trp-containing factors leads to a significant increase in TDO expression and/or activity when Trp is present in the blood at supraphysiological concentrations (Knox, 1951). As compared to liver, the expression of TDO in other normal tissues is negligible. In contrast, a number of human tumors express increased TDO levels including bladder urothelial carcinoma, cholangiocarcinoma, and hepatocellular carcinoma. Strikingly, many cancer types express a basal levels of TDO, although there is significant variability between individual patients (van Baren & Van den Eynde, 2015).
Normally, IDO1 is constitutively expressed in the epithelia and endothelial cells of several organs in both human and/or mouse placenta, as well as the lung and female genital tract (Blaschitz et al., 2011; Dai & Zhu, 2010; Sedlmayr et al., 2002; Smith et al., 2012). IDO1 is also highly expressed in mouse epididymis (Theate et al., 2015). Expression of IDO1 is induced in select immune cells that are primarily myeloid in nature and include dendritic cells and macrophages, during periods of cytokine stimulation including interferon-gamma (IFN-γ), interleukins-1 beta and −6 (IL-β, IL-6), tumor necrosis factor-alpha (TNF-α), prostagladin-E2 (PGE2), and lipopoly-saccharide (LPS). Upregulation of IDO1 expression has also been detected in a large variety of transformed cells and cancerous tissues (Theate et al., 2015).
The co-expression pattern of IDO1 and TDO in many cancers requires the expression level analysis for these two enzymes when also attempting to assess each enzyme’s individual contribution to Kyn pathway-associated tumor-induced immunosuppression. It should be noted, however, that the mRNA and protein levels for IDO1 do not always correlate with one another (Theate et al., 2015; Zhai et al., 2017a). This may be due to different folding conformations of the protein or to post-translation modifications (PTMs) that interfere with antibody binding (Fujigaki, Seishima, & Saito, 2012).
3. Kyn pathway and IDO1 enzyme activity
During the first step of the Kyn pathway, l-Trp is oxidized by IDO or TDO into N′-formylkynurenine, followed by the rapid and spontaneous conversion into Kyn (Hwu et al., 2000) (Fig. 1). Because dietary intake of Trp can affect serum amino acid levels, investigators often refer to the Kyn/Trp ratio as a proxy for IDO1 enzyme activity (Pertovaara et al., 2005). Different tissues possess variable Kyn levels, and levels in the blood plasma, serum, and brain are typically relatively low in the micromolar range (Baran, Jellinger, & Deecke, 1999; Pertovaara et al., 2005; Zhao, Gao, & Zhu, 2010). Kynurenine aminotransferases (KAT I and KAT II) facilitate the generation of kynurenic acid (KYNA) by irreversible transamination of Kyn (Han, Robinson, & Li, 2008; Rossi, Han, Li, Li, & Rizzi, 2004). In the brain, KYNA is generated by astrocytes (Guidetti, Hoffman, Melendez-Ferro, Albuquerque, & Schwarcz, 2007) and present at micromolar concentrations, while its level in cerebrospinal fluids is much lower in the nanomolar range (Baran et al., 1999). Kyn is also a precursor for 3-hydroxykynurenine (3-HK) which is converted by kynurenine hydroxylase (KMO), or for anthranilic acid (AA) which is converted by kynureninase (KYNU). Kynurenine hydroxylase is expressed by several cell types including microglia (Parrott & O’Connor, 2015), decidual or placental cells (Ligam, Manuelpillai, Wallace, & Walker, 2005) and is upregulated by pro-inflammatory cytokines (Parrott & O’Connor, 2015). Therefore, despite the generation of Kyn by IDO1 and TDO, its level at homeostasis is also affected by the expression and activity of several other enzymes including KAT, KMO, and KYNU. Notably, the interactions and regulatory mechanisms between these Kyn pathway enzymes under most pathogenic conditions have yet to be elucidated.
4. Methods for quantification of IDO1 enzyme activity
Since identifying the metabolic potential of IDO1, different methods have been developed to measure the levels of Trp and Kyn in biological samples, including: (i) absorbance-; (ii) radioactivity-; (iii) fluorescence-; (iv) cell-; (v) high-performance liquid chromatography (HPLC)-based assays. The first four approaches are generally utilized for the analysis of in vitro samples collected from cell free or cell culture systems, while the latter technique can be used for the analysis of more complex samples collected from in vivo modeling.
4.1. Colorimetric absorbance assay
The absorbance-based assay was first designed by Takikawa et al. to determine the IDO1 enzyme activity in a cell-free system (Takikawa, Kuroiwa, Yamazaki, & Kido, 1988). In this method, purified recombinant IDO1 protein or cell extract is added into the assay mixture containing 50mM potassium phosphate buffer (pH 6.5), 20mM ascorbate, 10μM methylene blue, 100μg/mL catalase, 400μM l-tryptophan. The reaction is carried out at 37 °C for 30 or 60min, terminated by adding 20μL of 30% (w/v) trichloroacetic acid (TCA), and further incubated at 50°C for 30min to hydrolyze N-formylkynurenine (NFK) produced by IDO1 into Kyn. After centrifugation at 2500 × g for 15 min at 20°C, Kyn present in the supernatant is measured by a Jasco HPLC system with a reverse phase column (Ø, 4.5 mm × 15 cm). The mobile phase is held at 10mM ammonium acetate containing 10% (w/v) methanol and Kyn is detected at an absorbance of 360 nm. This method was later simplified by Tamantha et al. to remove the HPLC step (Littlejohn et al., 2000). Specifically, after the hydrolyzation of NFK and centrifugation, the supernatant is mixed with 2% (w/v) p-dimethylaminobenzaldehyde (p-DMAB) in acetic acid (Ehrlich’s reagent). The accumulation of yellow pigment derived from Kyn can then be quantified by measuring the 480 nm wavelength.
The primary advantage of the absorbance-based assay is its easy operation and low cost. However, the reaction mixture must always be freshly prepared with special attention to the ascorbic acid and catalase. Because the absorbance method relies on an absorption of the imine produced by reaction of the aromatic amino group of Kyn with p-DMAB at λmax 480 nm, it is incompatible with compounds containing select functional groups; most notably related to primary amines that react with Ehrlich’s reagent. Moreover, colored compounds with an absorption of similar wavelength can also interfere with assay results. Furthermore, because IDO1 enzyme activity requires the reduction of heme iron from the ferric- to the ferrous-state, characterization of IDO1 activity often begins by a reaction containing methylene blue (MB) as an electron source. The physiological reductant of IDO1 was originally proposed to be a superoxide anion, but more recently that designation was passed along to cytochrome b5. Human IDO1 shows reduced activity in a reaction containing cytochrome b5 as compared with the MB assay, indicating the influence of co-factors on measuring enzyme activity with the absorbance-based methodology.
4.2. Radioactivity assay
The radioactivity-based IDO1 assay relies on the same reaction constituents as those in the absorbance-based system, except that radiolabeled l-[ring-2-14C] tryptophan is used as a substrate. The radioactivity of the supernatant generated from the [14C]-formic acid through NFK hydrolysis is detected by liquid scintillation spectroscopy and indicates the degree of Kyn formation (Ozaki, Nichol, & Duch, 1987). This method is more sensitive and reliable than the absorbance-based assay but is less convenient due to the hazard of working with radioactive products.
4.3. Fluorescence assay
Since Kyn fluoresces at λex 365 nm and λem 480nm, Matin et al. developed a method for quantifying IDO1 enzyme activity utilizing the quantification of fluorescence with the maximal fluorescence intensity of Kyn produced from hydrolysis of NFK under basic conditions (Matin, Streete, Jamie, Truscott, & Jamie, 2006). This technique has limits of detection similar to the standard absorbance assay, but with a disadvantage of low signal to background (S/B) ratio and a weak signal that decays over a short period of time. A more sensitive fluorescence-based assay was developed by Tomek, Palmer, Kendall, Flanagan, and Ching (2015), utilizing a novel NFK-derived fluorophore, NFK-piperidine (NFKPIP). For this modified fluorescence assay, and unlike the absorbance assay in which the enzymatic reaction is terminated by 30% TCA, the reaction is instead terminated by adding PIP. The reaction plates are then covered with a sealing film and heated at 65 °C for 20min. After 0.5–4 h, the fluorescence intensity in each well is read using a multimode plate reader with an excitation wavelength of 400nm and an emission wavelength of 500nm at 25°C (3mm measurement height, 100 flashes). This newer fluorescence assay is also suitable for high through-put screening of compounds for new IDO1 enzyme inhibitors; indicated by its superior S/B ratio as compared to the standard absorbance assay.
Similar to the NFKPIP fluorescence assay, Seegers et al. developed a high-throughput screening assay for both IDO1 and TDO based on a novel chemical probe, NFK Green, that reacts specifically with NFK to form a green fluorescent molecule with an excitation wavelength of 400nm and an emission wavelength of 510nm (Seegers et al., 2014). In this fluorescence assay, after mixing compounds and IDO1, the reaction is started by addition of Trp. After 10–30min, the reaction is stopped by the addition of NFK Green at 37°C for 4h. The system can be easily read on commonly used fluorescence or multimodal plate readers. Compared to the previous fluorescence assays, the NFK Green assay works at ambient temperature and neutral pH, which is more suitable for high throughput screens. Moreover, this assay doesn’t require heat and is performed without the addition of acidic or toxic reagents.
4.4. Cell-based assay
Since IDO1 is induced by various inflammatory stimuli, including IFN-γ, TNF-α, and LPS, Peng et al. used the conditioned medium from IFN-γ stimulated HeLa cells as the source of IDO1 activity and developed a cell-based assay for specific inhibitor screening (Peng et al., 2016). HeLa cells are first seeded into a 96-well culture plate at a density of 1 × 104 per well. The following day, human IFN-γ (10ng/mL) and compounds are added in a total volume of 200μL culture medium containing 15μg/mL of l-tryptophan. After an additional 24h incubation, 140 μL of the supernatant is mixed with 10 μL of 6.1 N TCA and the mixture is incubated for 30 min at 50 °C. The reaction mixture is then centrifuged for 10min at 2500 rpm to remove sediment. 100μL of the supernatant is further mixed with 100 μL of 2% (w/v) p-DMAB in acetic acid and the wavelength is measured at 480 nm. Because of its simplicity and low cost, this cell-based IDO1 activity assay is widely used in the screening for potential IDO1 enzyme inhibitors. One major advantage of this method is that it utilizes the intracellular Kyn pathway enzyme machinery instead of the cell-free reaction mixtures, providing ex vivo data that reflect the inhibitory potential of screened compounds.
4.5. HPLC assay
To date, chromatography remains as one of the most accurate and sensitive approaches for quantifying both Trp and Kyn from complex biological matrices. However, the methodology does require costly and sophisticated equipment. The standardization of sample extraction protocols also results in a rigorous validation of the results. Chromatographic analyses can be classified into two major groups: (i) liquid chromatography-mass spectrometry (LC-MS) or LC-MS/MS and gas chromatography (GC)-MS or GC-MS/MS; (ii) HPLC-based detection. The former approaches represent the most sensitive and accurate methodologies for the simultaneous detection of multiple Kyn metabolites, but requires meticulous sample preparation [including solid phase extraction (SPE) or derivatization], as well as costly isotope-labeled internal standards. The HPLC systems equipped with the UV, fluorescence, or electrochemical detectors are more accessible for Trp and Kyn analysis in clinical and simple laboratory settings.
4.5.1. Sample preparation for HPLC
It can be challenging to measure Trp and its metabolites from biological samples due to their lability, low physiological concentration, and/or the potential presence of interfering compounds. Some precautions should be undertaken when performing the analysis. Work with the samples quickly, at low temperature, and protect them from light to reduce degradation of Trp and its downstream metabolites. To remove the proteinaceous materials from biological samples including plasma, serum, and tissues, a protein precipitation step is required. Proteins in plasma are frequently removed through pretreatment of samples with acids and by SPE. Deproteinization with perchloric acid (PCA) and TCA can be used for this purpose. However, indole derivatives are sensitive to acidic conditions (Amirkhani, Heldin, Markides, & Bergquist, 2002; Moller, Du Preez, & Harvey, 2012) and the precipitation with TCA lowers the Kyn signal (Amirkhani et al., 2002). Other acids have been tested for the purpose of deproteination including sulfosalicylic acid (Pinhati et al., 2012), hydrochloric acid (Baran, Gramer, Honack, & Loscher, 1995; Baran et al., 1999), mixture of ascorbic acid, and PCA (Forrest, Mackay, Stoy, Stone, & Darlington, 2007) or TCA with addition of hydrochloric acid (Notarangelo, Wang, Horning, & Schwarcz, 2016) or acetonitrile (Zagajewski et al., 2012).
In addition to acid precipitation, sample pretreatment by SPE is the other useful approach for removing trace amounts of interfering compounds. It is based on the nonpolar, polar, ion exchange (cation and anion), and mixed mode interactions of sorbent with an analyte dissolved in liquid phase and subsequent elution with an appropriate solvent. This purification method is frequently used in more sensitive chromatographic approaches such as LS-MS/MS.
To date, the most commonly analyzed tumor tissue samples are plasma from cancer patients because of the easy availability. Analyses on solid tumors for Trp-Kyn pathway metabolites are less common compared to those of plasma samples. Many cancer studies performed metabolomic analyses on solid tumors using LC-MS/MS or GC-MS/MS for more sensitive and non-targeted screening, which were not specifically optimized for Trp-Kyn metabolites detection (Reznik et al., 2018). Interestingly, studies that focus on Trp-Kyn pathway metabolites in non-tumor diseases have generated many protocols for solid tissue processing. Table 1 provides a brief summary on previously reported processing methods for various tissue types using HPLC or LC-MS detection.
Table 1.
Tumor and non-tumor tissue sample processing methods for HPLC detection.
| Tissue type | Processing method | Detection method | Reference |
|---|---|---|---|
| CT26 subcutaneous tumor, plasma | Tumor homogenized in H2O, both supernatants from tumor homogenate and plasma were separated using SPE, elute in methanol-acetonitrile (5:5 v/v), twofold dilution in H2O for analysis | LC-MS/MS | Wang et al. (2018a) |
| LLC subcutaneous tumor, serum | Serum protein was precipitated by mixing with 3 vol 3% PCA at 4 °C for 1 h followed by centrifugation; tumor samples were homogenized in 2 vol KP buffer, then supernatants were mixed with 1 vol of 3% PCA | HPLC-UV, HPLC-FD |
Watcharanurak et al. (2014) |
| CSF, serum | Protein was precipitated by mixing with 1 vol 5% TCA, supernatants filtered through 0.2μm filters | HPLC-FD | Yan et al. (2015) |
| Maternal brain, fetal brain, placenta | Tissue thawed and sonicated in ultrapure H2O, supernatants diluted in 0.1% ascorbic acid, then mixed with ½ vol acetone to remove protein followed by methanol: chloroform extraction and derivatization | GC-MS | Notarangelo and Schwarcz (2016) |
| Forebrain, cerebellum, liver | Homogenized by sonication in ultrapure H2O, protein was precipitated in 1.2% PCA and removed by centrifugation | HPLC-FD | Notarangelo et al. (2016) |
| Liver | Tissue homogenate diluted in 5mM ammonium formate containing 0.1% TCA (2:5v/v), protein precipitated with 100% TCA |
HPLC-MS | Mole et al. (2016) |
| Cervical cancer punch biopsy | Snap frozen tissue homogenized in 9% (w/v) PCA, protein was precipitated in 6% (w/v) PCA | HPLC-UV | Hascitha et al. (2016) |
| Endometriotic tissue | Frozen tissue homogenized using TissueLyzer in a methanol: H2O mixture (1:1). Supernatants were vacuum dried and re-dissolved in H2O, protein was precipitated by methanol | LC/ESI–MS | Mariuzzi et al. (2016) |
| Brain | Frozen tissue homogenized in DPBS, supernatants were mixed with 20% w/v ice-cold TCA in a 4:1 v/v ratio on ice. Supernatants after centrifugation were neutralized with 1vol of 1mol/L phosphate buffer (pH 7.3) before analysis | HPLC-FD, HPLC-UV | Too et al. (2016) |
| Plasma, brain | Plasmatic proteins were removed by acetonitrile (0.2 mL ACN /0.1 mL plasma). Supernatants from the homogenate were further filtered through nylon filters; brain tissues were homogenized and sonicated in ACN, hydrochloric acid (100 mM) and EDTA (27 mM solution) mixture at 5:4:1 (v/v/v) ratio, followed by supernatants filtration | HPLC-ESI-MS/MS | Vondrousova et al. (2019) |
| Forebrain, liver | Frozen tissues were homogenized (1:20, w/v) in an aqueous solution containing 0.1% ascorbic acid and internal standards. Proteins were denatured using acetone followed by centrifugation. 50μL of methanol: chloroform (20:50, v/v) were added to the supernatant and centrifuged | GC-MS | Larkin et al. (2016) |
| Plasma, liver, lung, kidney | Plasma samples were deproteinated by 2M HClO4 and centrifuged. The supernatant was filtered using 0.45μm syringe filters; solid tissues homogenized in ice by thrice the volume of 20% TCA followed by centrifugation filtration using 0.45 mm filters | HPLC-FD | Bartosiewicz et al. (2017) |
| Liver, kidney, brain, lung, and spleen | Tissue was homogenized in 3% PCA and the homogenate then kept at 4°C overnight followed by centrifugation at 4°C (7000 g, 20 min) and filtration through 0.45mm filters | HPLC-UV | Ohta et al. (2017) |
| Aorta | Tissues were lyzed, sonicated and deproteinized in 7.5% PCA, followed by centrifugation. Supernatants were directly used for analysis | HPLC-ECD | Metghalchi et al. (2018) |
| Pancreatic tumor | Frozen tissues were pulverized by agitation with stainless steel balls at liquid nitrogen temperature, then mixed by vortexing with –80°C 80:20 methanol:water, soluble extract from the mixture were isolated by centrifugation and repeat the extraction twice | HPLC-MS | Kamphorst et al. (2015) |
| Human feces | Homogenized cold feces (1 g) was mixed with 1mL of H2O (4°C), vortexed for 30s, ultrasonicated for 2min (4°C), and vortexed for 30 s prior to ultracentrifugation (171,500 g) for 30min at 4 °C | HPLC-ECD | Laurans et al. (2018) |
| Serum, urine, kidney | 50μL of serum, urine and tissue homogenate were mixed with an equal volume of PBS (pH 7.4), followed by an addition of 10μL of 60% PCA and vortexed. After incubation on ice, samples were centrifuged at 4 °C for 10min and the supernatant collected and filtered with a Millipore MSRLN0410 filter plate | HPLC-UV | Wang et al. (2018b) |
| Human placenta | Tissues were homogenized using Bertin Minilys® bead tubes containing 10% TCA. After centrifugation, supernatants were filtered through a 0.22μm polytetrafluoroethylene filter before analysis | HPLC-UV | Keaton et al. (2019) |
| Glioblastoma | GBM was homogenized in a 0.1 N hyperchloric acid/10μM ascorbate solution, followed by the Costar Spin-X filtration | HPLC-ECD | Zhai et al. (2017b) |
4.5.2. Detection of Trp and Kyn by HPLC
The most popular method for Trp and Kyn quantification is with an HPLC system coupled to different detection modules. Among these, the HPLC-UV method is most frequently used. However, the sensitivity (Xiao, Luo, Pi, & Tang, 2008) and selectivity (Ma, Xu, Wang, Deng, & Ding, 2009) of HPLC-UV-based techniques may be insufficient for detecting Kyn present at micromolar concentrations in biological fluids or tissues. Kyn quantification using the UV detector is generally performed at wavelength of 225 and 360±5nm. Due to prolific detection of endogenous compounds observed at 225 nm, the clinical analysis is favored at a detection of 360±5nm (Vignau, Jacquemont, Lefort, Imbenotte, & Lhermitte, 2004; Zhen et al., 2011).
To improve sensitivity, the fluorescence detectors coupled with HPLC (HPLC–FD) are incorporated into the UV detection system for Kyn analysis. Vignau et al. used the 150×4.6mm I.D. analytical column packed with Platinum EPS C18 100 Å (Vignau et al., 2004). The sample is analyzed in a mobile phase of 5 mmol/L aqueous zinc acetate solution in acetonitrile (92:8, v/v; pH 4.9) with an injection volume of 20 μL and a flow rate at 1.0mL/min using a Waters LC-600 coupled with two detectors: a Waters 2487 Dual Absorbance for UV detection at 365nm for Kyn, and a Waters 474 Scanning Fluorescence for Trp detection with excitation at 254 nm and emission at 404 nm. For the internal standard, fluorescence conditions are modified to an excitation of 220nm with an emission at 354nm. The chromatogram of serum samples shows a good separation efficiency for Kyn (retention time 3.29min) and Trp (retention time 3.98min).
In addition to the HPLC-UV and HPLC-FD methods, HPLC methods employing electrochemical detection (HPLC-ED) (Sinz et al., 1998) and mass spectrometric detection (HPLC-MS) (Curto et al., 2015; Heyes, Saito, Milstien, & Schiff, 1995) have been developed to further increase the sensitivity of detection. Dr. Dantzer’s group developed a protocol using HPLC-ED to measure the levels of Trp and Kyn from plasma and brain tissue samples (O’Connor et al., 2008). Specifically, plasma samples are first deproteinated by mixing with 10% sulfosalicyclic acid solution on ice for at least 30 min. After centrifugation, the supernatant is further purified through a Costar Spin-X centrifuge filter. For the brain samples, tissue is homogenized in a 0.1 N hyperchloric acid/10μM ascorbate solution, followed by the Costar Spin-X filtration. The processed samples are analyzed in a mobile phase (pH 4.6) consisting of 75 mM NaH2PO4, 25mM EDTA (disodium salt) and 100μLL−1 triethylamine in acetonitrile/water (6:94, v/v) through an HPLC system equipped with an ESA Coulochem II detector and a 5041 Enhanced Analytical cell containing a glassy carbon electrode (+600mV). Our laboratory adapted this protocol using a UHPLC-ECD system and obtained an optimal detection for chromatography (Fig. 2): separation within 7min (at a flow rate of 0.2mL/min) with Kyn retention at 3.4min and Trp retention at 5.6min; limit of detection is 0.02μM (Zhai et al., 2017b).
Fig. 2.
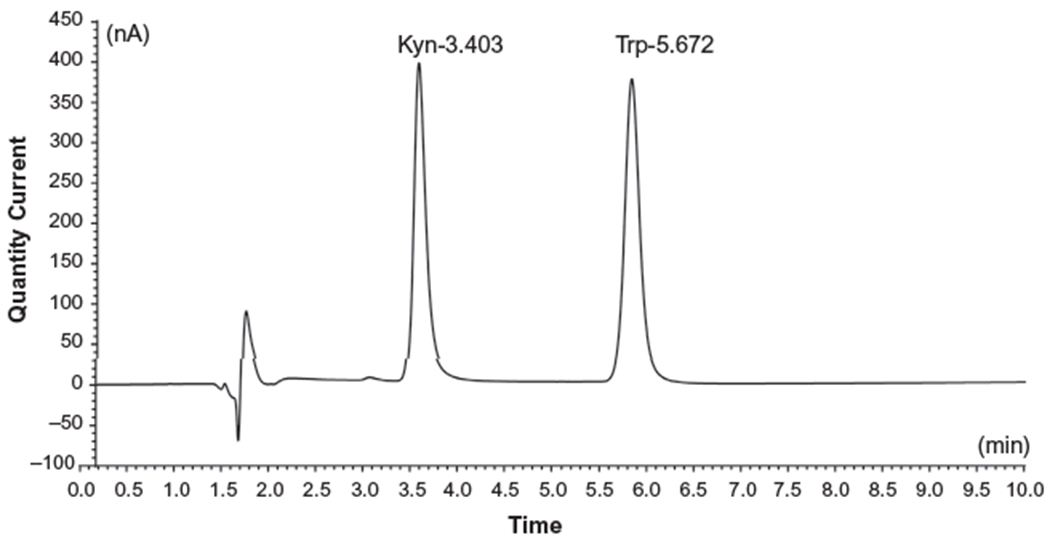
Quantification of Trp and Kyn levels in mouse plasma using a Coulechem III high performance liquid chromatography electrochemical detector system. The retention times and peaks for Trp and Kyn indicated as labeled.
The HPLC-MS method is the most sensitive approach for Trp metabolite measurements and is suitable for detecting multiple analytes simultaneously. Hényková et al. have proposed a UHPLC–MS/MS protocol for quantifying the profile of Trp, Kyn, kynurenic acid, 3-HK, 3-HAA, AA and 11 other Trp-related neuroactive compounds in human serum and cerebrospinal fluid within 10min (Henykova et al., 2016). Despite impressive developments, the main drawback of the LC–MS approaches is a low ionization response of Trp and the Kyns as compared to less polar compounds, and a significant impact of sample impurities during the ionization process (Moller et al., 2012). Thus, the LC–MS method requires very careful sample preparation (i.e., by SPE) that reduces matrix interference and improves extraction efficiency (Moller et al., 2012). The addition of internal standards (especially isotope-labeled analogues) minimizes assay variation. The derivatization step likely helps in the analysis to improve sensitivity and specificity of the method since it increases mass of the target compounds and eliminates the interference of matrix components occurring in the low-m/z region (Zhang et al., 2015).
4.5.3. Comparison of different HPLC detection systems
Choosing the most suitable chromatographic method for measurement of Trp and Kyns can be challenging, which is determined by several factors: (1) the estimated concentration of substrate(s) in the sample; (2) whether simultaneous analysis of multiple compounds is required; (3) affordability of expensive detection equipment. The HPLC-UV detector is mainly used for Kyn quantification, although these methods suffer from low sensitivity and selectivity due to an interference of endogenous compounds present in biological samples. HPLC–FD methods address the low S/B ratio issue, but are limited since not all Kyns fluoresce or their native fluorescence is too low for accurate quantification. For HPLC-ED detection, the selectivity and repeatability are compromised due to interference of sample components and the electrode clogging. The HPLC-MS methods requires expensive equipment and precise sample preparation including a purification step and derivatization of target analytes, but is advantageous due to its high sensitivity and small requirement for sample volume.
5. Concluding remarks
IDO1 is widely accepted as a critical immunosuppressive mediator involved in many pathological conditions including autoimmunity, infectious diseases, and cancer. Based on a competing hypothesis, IDO1 suppresses the immune response through its intrinsic capacity to deplete Trp and/or cause accumulation of Kyn locally. Determination of Trp and Kyn levels is therefore critical for evaluating its related immunosuppressive contribution(s) and provides a rapid and direct readout while investigating potentially new IDO1 enzyme inhibitors. Yet, challenges still exist in the field and include a difficulty distinguishing between the metabolic contribution of IDO1 versus other Trp degrading enzymes, as well as the choice of optimal methodology for quantifying the Kyn pathway metabolites. Expression profiling of IDO1/TDO/IDO2 is necessary for the analyzed specimens and correlation of expression profiling with Trp/Kyn ratio changes may provide a more accurate interpretation of IDO1-specific activity. Issues of sample concentration, sample complexity, rate of analyte decomposition, and differences in distribution of Kyn across different tissues with or without treatments, should be considered when choosing the chromatographic method. To provide an equivocal comparison between data obtained from different groups, it is important to set up standard protocols for each IDO1 activity quantification method and for providing raw data in the supplementary figures of published results. Ultimately, quantification of Trp and Kyn through the methods described here will inform future researchers as to determining the importance of targeting IDO1 enzyme activities, and its effects in the model systems or patients under study.
References
- Amirkhani A, Heldin E, Markides KE, & Bergquist J (2002). Quantitation of tryptophan, kynurenine and kynurenic acid in human plasma by capillary liquid chromatography-electrospray ionization tandem mass spectrometry. Journal of Chromatography B, Analytical Technologies in the Biomedical and Life Sciences, 780, 381–387. [DOI] [PubMed] [Google Scholar]
- Badawy AA (2014). The tryptophan utilization concept in pregnancy. Obstetrics & Gynecology Science, 57, 249–259. 10.5468/ogs.2014.57.4.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball HJ, et al. (2007). Characterization of an indoleamine 2,3-dioxygenase-like protein found in humans and mice. Gene, 396, 203–213. 10.1016/j.gene.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Baran H, Gramer M, Honack D, & Loscher W (1995). Systemic administration of kainate induces marked increases of endogenous kynurenic acid in various brain regions and plasma of rats. European Journal of Pharmacology, 286, 167–175. [DOI] [PubMed] [Google Scholar]
- Baran H,Jellinger K,& Deecke L (1999). Kynurenine metabolism in Alzheimer’s disease. Journal of Neural Transmission (Vienna, Austria: 1996), 106, 165–181. 10.1007/s007020050149. [DOI] [PubMed] [Google Scholar]
- Bartosiewicz J, et al. (2017). The activation of the kynurenine pathway in a rat model with renovascular hypertension. Experimental Biology and Medicine (Maywood, N.J.), 242, 750–761. 10.1177/1535370217693114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschitz A, et al. (2011). Vascular endothelial expression of indoleamine 2,3-dioxygenase 1 forms a positive gradient towards the feto-maternal interface. PLoS One, 6, e21774 10.1371/journal.pone.0021774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curto M, et al. (2015). Altered kynurenine pathway metabolites in serum of chronic migraine patients. The Journal of Headache and Pain, 17, 47 10.1186/s10194-016-0638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, & Zhu BT (2010). Indoleamine 2,3-dioxygenase tissue distribution and cellular localization in mice: Implications for its biological functions. The Journal of Histochemistry and Cytochemistry, 58, 17–28. 10.1369/jhc.2009.953604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest CM, Mackay GM, Stoy N, Stone TW, & Darlington LG (2007). Inflammatory status and kynurenine metabolism in rheumatoid arthritis treated with melatonin. British Journal of Clinical Pharmacology, 64, 517–526. 10.1111/j.1365-2125.2007.02911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frumento G, et al. (2002). Tryptophan-derived catabolites are responsible for inhibition of T and natural killer cell proliferation induced by indoleamine 2,3-dioxygenase. The Journal of Experimental Medicine, 196, 459–468. 10.1084/jem.20020121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujigaki H, Seishima M, & Saito K (2012). Posttranslational modification of indoleamine 2,3-dioxygenase. Analytical and Bioanalytical Chemistry, 403, 1777–1782. 10.1007/s00216-012-5946-2. [DOI] [PubMed] [Google Scholar]
- Guidetti P, Hoffman GE, Melendez-Ferro M, Albuquerque EX, & Schwarcz R (2007). Astrocytic localization of kynurenine aminotransferase II in the rat brain visualized by immunocytochemistry. Glia, 55, 78–92. 10.1002/glia.20432. [DOI] [PubMed] [Google Scholar]
- Han Q, Robinson H, & Li J (2008). Crystal structure of human kynurenine aminotransferase II. The Journal of Biological Chemistry, 283, 3567–3573. 10.1074/jbc.M708358200. [DOI] [PubMed] [Google Scholar]
- Hascitha J, et al. (2016). Analysis of kynurenine/tryptophan ratio and expression of IDO1 and 2 mRNA in tumour tissue of cervical cancer patients. Clinical Biochemistry, 49, 919–924. 10.1016/jclinbiochem.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Henykova E, et al. (2016). Stable isotope dilution ultra-high performance liquid chromatography-tandem mass spectrometry quantitative profiling of tryptophan-related neuroactive substances in human serum and cerebrospinal fluid. Journal of Chromatography. A, 1437, 145–157. 10.1016/jchroma.2016.02.009. [DOI] [PubMed] [Google Scholar]
- Heyes MP, Saito K, Milstien S, & Schiff SJ (1995). Quinolinic acid in tumors, hemorrhage and bacterial infections of the central nervous system in children. Journal of the Neurological Sciences, 133, 112–118. [DOI] [PubMed] [Google Scholar]
- Hwu P, et al. (2000). Indoleamine 2,3-dioxygenase production by human dendritic cells results in the inhibition of T cell proliferation. The Journal of Immunology, 164, 3596–3599. 10.4049/jimmunol.164.7.3596. [DOI] [PubMed] [Google Scholar]
- Kamphorst JJ, et al. (2015). Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Research, 75, 544–553. 10.1158/0008-5472.Can-14-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keaton SA, et al. (2019). Altered tryptophan catabolism in placentas from women with pre-eclampsia. International Journal of Tryptophan Research: IJTR, 12, 1178646919840321. 10.1177/1178646919840321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox WE (1951). Two mechanisms which increase in vivo the liver tryptophan peroxidase activity: Specific enzyme adaptation and stimulation of the pituitary adrenal system. British Journal of Experimental Pathology, 32, 462–469. [PMC free article] [PubMed] [Google Scholar]
- Larkin PB, et al. (2016). Tryptophan 2,3-dioxygenase and indoleamine 2,3-dioxygenase 1 make separate, tissue-specific contributions to basal and inflammation-induced kynurenine pathway metabolism in mice. Biochimica et Biophysica Acta, 1860, 2345–2354. 10.1016/j.bbagen.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurans L, et al. (2018). Genetic deficiency of indoleamine 2,3-dioxygenase promotes gut microbiota-mediated metabolic health. Nature Medicine, 24,1113–1120. 10.1038/s41591-018-0060-4. [DOI] [PubMed] [Google Scholar]
- Ligam P, Manuelpillai U, Wallace EM, & Walker D (2005). Localisation of indoleamine 2,3-dioxygenase and kynurenine hydroxylase in the human placenta and decidua: Implications for role of the kynurenine pathway in pregnancy. Placenta, 26, 498–504. 10.1016/j.placenta.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Littlejohn TK, et al. (2000). Expression and purification of recombinant human indoleamine 2, 3-dioxygenase. Protein Expression and Purification, 19, 22–29. 10.1006/prep.2000.1214. [DOI] [PubMed] [Google Scholar]
- Ma L, Xu B, Wang W, Deng W, &Ding M (2009). Analysis of tryptophan catabolism in HBV patients by HPLC with programmed wavelength ultraviolet detection. Clinica Chimica Acta: International Journal of Clinical Chemistry, 405, 94–96. 10.1016/j.cca.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Mariuzzi L, et al. (2016). Functional expression of aryl hydrocarbon receptor on mast cells populating human endometriotic tissues. Laboratory Investigation: A Journal of Technical Methods and Pathology, 96, 959–971. 10.1038/labinvest.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matin A, Streete IM, Jamie IM, Truscott RJ, &Jamie JF. (2006). Afluorescence- based assay for indoleamine 2,3-dioxygenase. Analytical Biochemistry, 349, 96–102. 10.1016/j.ab.2005.10.039. [DOI] [PubMed] [Google Scholar]
- Metghalchi S, et al. (2018). Indoleamine 2 3-dioxygenase knockout limits angiotensin II-induced aneurysm in low density lipoprotein receptor-deficient mice fed with high fat diet. PLoS One, 13, e0193737 10.1371/journal.pone.0193737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz R, et al. (2007). Novel tryptophan catabolic enzyme IDO2 is the preferred biochemical target of the antitumor indoleamine 2,3-dioxygenase inhibitory compound d-1-methyl-tryptophan. Cancer Research, 67, 7082–7087. 10.1158/0008-5472.can-07-1872. [DOI] [PubMed] [Google Scholar]
- Metz R, et al. (2014). IDO2 is critical for IDO1-mediated T-cell regulation and exerts a non-redundant function in inflammation. International Immunology, 26, 357–367. 10.1093/intimm/dxt073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezrich JD, et al. (2010). An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. Journal of Immunology, 185, 3190–3198. 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mole DJ, et al. (2016). Kynurenine-3-monooxygenase inhibition prevents multiple organ failure in rodent models of acute pancreatitis. Nature Medicine, 22, 202–209. 10.1038/nm.4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller M, Du Preez JL, & Harvey BH (2012). Development and validation of a single analytical method for the determination of tryptophan, and its kynurenine metabolites in rat plasma. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences, 898 121–129. 10.1016/j.jchromb.2012.04.030. [DOI] [PubMed] [Google Scholar]
- Munn DH,et al. (1998). Prevention of allogeneic fetal rejection by tryptophan catabolism. Science (New York, N.Y.), 281, 1191–1193. [DOI] [PubMed] [Google Scholar]
- Munn DH, et al. (1999). Inhibition of T cell proliferation by macrophage tryptophan catabolism. The Journal of Experimental Medicine, 189, 1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn DH, et al. (2002). Potential regulatory function of human dendritic cells expressing indoleamine 2,3-dioxygenase. Science (New York, N.Y.), 297, 1867–1870. 10.1126/science.1073514. [DOI] [PubMed] [Google Scholar]
- Nagano J, et al. (2013). Effects of indoleamine 2,3-dioxygenase deficiency on high-fat diet- induced hepatic inflammation. PLoS One, 8, e73404 10.1371/journal.pone.0073404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notarangelo FM, & Schwarcz R (2016). Restraint stress during pregnancy rapidly raises kynurenic acid levels in mouse placenta and fetal brain. Developmental Neuroscience, 38, 458–468. 10.1159/000455228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notarangelo FM, Wang XD, Horning KJ, & Schwarcz R (2016). Role of d-amino acid oxidase in the production of kynurenine pathway metabolites from d-tryptophan in mice. Journal of Neurochemistry, 136, 804–814. 10.1111/jnc.13455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor JC, et al. (2008). Lipopolysaccharide-induced depressive-like behavior is mediated by indoleamine 2,3-dioxygenase activation in mice. Molecular Psychiatry, 14, 511–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y, et al. (2017). Effect of water-immersion restraint stress on tryptophan catabolism through the kynurenine pathway in rat tissues. The Journal of Physiological Sciences, 67, 361–372. 10.1007/s12576-016-0467-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki Y, Nichol CA, & Duch DS (1987). Utilization of dihydroflavin mononucleotide and superoxide anion for the decyclization of L-tryptophan by murine epididymal indoleamine 2,3-dioxygenase. Archives of Biochemistry and Biophysics, 257, 207–216. [DOI] [PubMed] [Google Scholar]
- Pantouris G, Serys M, Yuasa HJ, Ball HJ, & Mowat CG (2014). Human indoleamine 2,3-dioxygenase-2 has substrate specificity and inhibition characteristics distinct from those of indoleamine 2,3-dioxygenase-1. Amino Acids, 46, 2155–2163. 10.1007/s00726-014-1766-3. [DOI] [PubMed] [Google Scholar]
- Parrott JM, & O’Connor JC (2015). Kynurenine 3-monooxygenase: An influential mediator of neuropathology. Frontiers in Psychiatry, 6, 116 10.3389/fpsyt.2015.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YH, et al. (2016). Important hydrogen bond networks in indoleamine 2,3-dioxygenase 1 (IDO1) inhibitor design revealed by crystal structures of imidazoleisoindole derivatives with IDO1. Journal of Medicinal Chemistry, 59, 282–293. 10.1021/acs.jmedchem.5b01390. [DOI] [PubMed] [Google Scholar]
- Pertovaara M, et al. (2005). Mechanisms dependent on tryptophan catabolism regulate immune responses in primary Sjogren’s syndrome. Clinical and Experimental Immunology, 142, 155–161. 10.1111/j.1365-2249.2005.02889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinhati RR, et al. (2012). Quantification of tryptophan in plasma by high performance liquid chromatography. Quimica Nova, 35, 623–626. [Google Scholar]
- Platten M, von Knebel Doeberitz N, Oezen I, Wick W, & Ochs K (2015). Cancer immunotherapy by targeting IDO1/TDO and their downstream effectors. Frontiers in Immunology, 5, 673 10.3389/fimmu.2014.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznik E, et al. (2018). A landscape of metabolic variation across tumor types. Cell Systems, 6, 301–313.e303. 10.1016/jxels.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi F, Han Q, Li J, Li J, & Rizzi M (2004). Crystal structure of human kynurenine aminotransferase I. The Journal of Biological Chemistry, 279, 50214–50220. 10.1074/jbc.M409291200. [DOI] [PubMed] [Google Scholar]
- Sedlmayr P, et al. (2002). Localization of indoleamine 2,3-dioxygenase in human female reproductive organs and the placenta. Molecular Human Reproduction, 8, 385–391. [DOI] [PubMed] [Google Scholar]
- Seegers N, et al. (2014). High-throughput fluorescence-based screening assays for tryptophan-catabolizing enzymes. Journal of Biomolecular Screening, 19, 1266–1274. 10.1177/1087057114536616. [DOI] [PubMed] [Google Scholar]
- Sinz EH, et al. (1998). Quinolinic acid is increased in CSF and associated with mortality after traumatic brain injury in humans. Journal of Cerebral Blood Flow and Metabolism: Official Journal of the International Society of Cerebral Blood Flow and Metabolism, 18, 610–615. 10.1097/00004647-199806000-00002. [DOI] [PubMed] [Google Scholar]
- Smith C, et al. (2012). IDO is a nodal pathogenic driver of lung cancer and metastasis development. Cancer Discovery, 2, 722–735. 10.1158/2159-8290.cd-12-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takikawa O, Kuroiwa T, Yamazaki F, & Kido R (1988). Mechanism of interferon- gamma action. Characterization ofindoleamine 2,3-dioxygenase in cultured human cells induced by interferon-gamma and evaluation of the enzyme-mediated tryptophan degradation in its anticellular activity. The Journal of Biological Chemistry, 263, 2041–2048. [PubMed] [Google Scholar]
- Taylor MW, & Feng GS (1991). Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. The FASEB Journal, 5, 2516–2522. [PubMed] [Google Scholar]
- Theate I, et al. (2015). Extensive profiling of the expression of the indoleamine 2,3- dioxygenase 1 protein in normal and tumoral human tissues. Cancer Immunology Research, 3, 161–172. 10.1158/2326-6066.cir-14-0137. [DOI] [PubMed] [Google Scholar]
- Tomek P, Palmer BD, Kendall JD, Flanagan JU, & Ching LM (2015). Formation of fluorophores from the kynurenine pathway metabolite N-formylkynurenine and cyclic amines involves transamidation and carbon-carbon bond formation at the 2-position of the amine. Biochimica et Biophysica Acta, 1850, 1772–1780. 10.1016/j.bbagen.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Too LK, et al. (2016). Deletion of TDO2, IDO-1 and IDO-2 differentially affects mouse behavior and cognitive function. Behavioural Brain Research, 312, 102–117. 10.1016/j.bbr.2016.06.018. [DOI] [PubMed] [Google Scholar]
- van Baren N, & Van den Eynde BJ (2015). Tryptophan-degrading enzymes in tumoral immune resistance. Frontiers in Immunology, 6, 34 10.3389/fimmu.2015.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignau J, Jacquemont MC, Lefort A, Imbenotte M, & Lhermitte M (2004). Simultaneous determination of tryptophan and kynurenine in serum by HPLC with UV and fluorescence detection. Biomedical Chromatography, 18, 872–874. 10.1002/bmc.445. [DOI] [PubMed] [Google Scholar]
- Vondrousova J, et al. (2019). Monitoring of kynurenine pathway metabolites, neurotransmitters and their metabolites in blood plasma and brain tissue of individuals with latent toxoplasmosis. Journal of Pharmaceutical and Biomedical Analysis, 170,139–152. 10.1016/j.jpba.2019.03.039. [DOI] [PubMed] [Google Scholar]
- Wang W, et al. (2018a). Determination of kynurnine and tryptophan, biomarkers of indoleamine 2,3-dioxygenase by LC-MS/MS in plasma and tumor. Bioanalysis, 10, 1335–1344. 10.4155/bio-2018-0041. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. (2018b). Regulation of indoleamine 2,3 dioxygenase and its role in a porcine model of acute kidney allograft rejection. Journal of Investigative Medicine: The Official Publication of the American Federation for Clinical Research, 66, 1109–1117. 10.1136/jim-2018-000742. [DOI] [PubMed] [Google Scholar]
- Watcharanurak K, et al. (2014). Effects of upregulated indoleamine 2, 3-dioxygenase 1 by interferon gamma gene transfer on interferon gamma-mediated antitumor activity. Gene Therapy, 21, 794–801. 10.1038/gt.2014.54. [DOI] [PubMed] [Google Scholar]
- Xiao LD, Luo XB, Pi LG, & Tang AG (2008). Simultaneous determination of kynurenine and kynurenic acid concentrations in human serum by HPLC with dual wavelengths fluorescence detection. Clinica Chimica Acta: International Journal of Clinical Chemistry, 395, 178–180. 10.1016/jxca.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Yan EB, et al. (2015). Activation of the kynurenine pathway and increased production of the excitotoxin quinolinic acid following traumatic brain injury in humans. Journal of Neuroinflammation, 12, 110 10.1186/s12974-015-0328-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa H, et al. (2007). Evolution of vertebrate indoleamine 2,3-dioxygenases. Journal of Molecular Evolution, 65, 705–714. 10.1007/s00239-007-9049-1. [DOI] [PubMed] [Google Scholar]
- Zagajewski J, et al. (2012). Conversion L-tryptophan to melatonin in the gastrointestinal tract: The new high performance liquid chromatography method enabling simultaneous determination of six metabolites of L-tryptophan by native fluorescence and UV-VIS detection. Journal of Physiology and Pharmacology: An Official Journal of the Polish Physiological Society, 63, 613–621. [PubMed] [Google Scholar]
- Zhai L, et al. (2015). Molecular pathways: Targeting IDO1 and other tryptophan dioxygenases for cancer immunotherapy. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research 21, 5427–5433. 10.1158/1078-0432.ccr-15-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L, et al. (2017a). Infiltrating T cells increase IDO1 expression in glioblastoma and contribute to decreased patient survival. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 23, 6650–6660. 10.1158/1078-0432.CCR-17-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L, et al. (2017b). Non-tumor cell IDO1 predominantly contributes to enzyme activity and response to CTLA-4/PD-L1 inhibition in mouse glioblastoma. Brain, Behavior, and Immunity, 62, 6 10.1016/j.bbi.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LH, et al. (2015). Simultaneous determination of multiple neurotransmitters and their metabolites in rat brain homogenates and microdialysates by LC-MS/MS. Analytical Methods, 7, 3929–3938. [Google Scholar]
- Zhao J, Gao P, & Zhu D (2010). Optimization of Zn2+-containing mobile phase for simultaneous determination of kynurenine, kynurenic acid and tryptophan in human plasma by high performance liquid chromatography. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences, 878, 603–608. 10.1016/j.jchromb.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Zhen Q, et al. (2011). Simultaneous determination of tryptophan, kynurenine and 5-hydroxytryptamine by HPLC: Application in uremic patients undergoing hemodialysis. Clinical Biochemistry, 44, 226–230. 10.1016/j.clinbiochem.2010.10.011. [DOI] [PubMed] [Google Scholar]


