Abstract
Tsetse fly exhibit species-specific olfactory uniqueness potentially underpinned by differences in their chemosensory protein repertoire. We assessed 1) expansions of chemosensory protein orthologs in Glossina morsitans morsitans, Glossina pallidipes, Glossina austeni, Glossina palpalis gambiensis, Glossina fuscipes fuscipes and Glossina brevipalpis tsetse fly species using Café analysis (to identify species-specific expansions) and 2) differential expressions of the orthologs and associated proteins in male G. m. morsitans antennae and head tissues using RNA-Seq approaches (to establish associated functional molecular pathways). We established accelerated and significant (P<0.05, λ = 2.60452e-7) expansions of gene families in G. m. morsitans Odorant receptor (Or)71a, Or46a, Ir75a,d, Ionotropic receptor (Ir) 31a, Ir84a, Ir64a and Odorant binding protein (Obp) 83a-b), G. pallidipes Or67a,c, Or49a, Or92a, Or85b-c,f and Obp73a, G. f. fuscipes Ir21a, Gustatory receptor (Gr) 21a and Gr63a), G. p. gambiensis clumsy, Ir25a and Ir8a, and G. brevipalpis Ir68a and missing orthologs in each tsetse fly species. Most abundantly expressed transcripts in male G. m. morsitans included specific Or (Orco, Or56a, 65a-c, Or47b, Or67b, GMOY012254, GMOY009475, and GMOY006265), Gr (Gr21a, Gr63a, GMOY013297 and GMOY013298), Ir (Ir8a, Ir25a and Ir41a) and Obp (Obp19a, lush, Obp28a, Obp83a-b Obp44a, GMOY012275 and GMOY013254) orthologs. Most enriched biological processes in the head were associated with vision, muscle activity and neuropeptide regulations, amino acid/nucleotide metabolism and circulatory system processes. Antennal enrichments (>90% of chemosensory transcripts) included cilium-associated mechanoreceptors, chemo-sensation, neuronal controlled growth/differentiation and regeneration/responses to stress. The expanded and tsetse fly species specific orthologs includes those associated with known tsetse fly responsive ligands (4-methyl phenol, 4-propyl phenol, acetic acid, butanol and carbon dioxide) and potential tsetse fly species-specific responsive ligands (2-oxopentanoic acid, phenylacetaldehyde, hydroxycinnamic acid, 2-heptanone, caffeine, geosmin, DEET and (cVA) pheromone). Some of the orthologs can potentially modulate several tsetse fly species-specific behavioral (male-male courtship, hunger/host seeking, cool avoidance, hygrosensory and feeding) phenotypes. The putative tsetse fly specific chemosensory gene orthologs and their respective ligands provide candidate gene targets and kairomones for respective downstream functional genomic and field evaluations that can effectively expand toolbox of species-specific tsetse fly attractants, repellents and other tsetse fly behavioral modulators.
Author summary
Tsetse flies are insect vectors of sleeping sickness in humans and nagana in livestock in sub-Sahara Africa. Tsetse flies identify their hosts (preferred and non-preferred) by detecting and processing odor cues emitted by the hosts in their environment. Tsetse flies use chemosensory proteins and associated pathways in their antennae to identify these cues. In this study, we identified expansions of these chemosensory protein in six tsetse fly species (Glossina morsitans morsitans, Glossina pallidipes, Glossina austeni, Glossina palpalis gambiensis, Glossina fuscipes fuscipes and Glossina brevipalpis) with different known hosts. We also identified potential ligands to these proteins based on fruit fly (Drosophila melanogaster) orthologs. With G. m. morsitans as an example, we identified the proteins and associated molecular pathways preferentially expressed in tsetse fly antennae. These proteins may be responsible for the tsetse fly species-specific host discrimination, with the ligands eliciting species-specific behavioral responses in the flies. The expressed orthologs may be functionally important in odor detection in tsetse fly and lay down useful groundwork for downstream functional genomics R&D for more effective tsetse fly species-specific odor attractants and repellents for routine tsetse fly control operations.
Introduction
Human African Trypanosomiasis (HAT) constitutes one of the most neglected tropical diseases (NTDs) with devastating health and economic consequences in sub-Sahara Africa [1,2]. On the other hand, African Animal Trypanosomiasis (AAT) is rampant in livestock inhabiting tsetse-infested areas throughout the continent. The AAT cause death of about three million cattle each year [3], and in terms of agricultural Gross Domestic Product (GDP), loss of about US$ 4.75 billion per year [3]. The HAT and AAT causative trypanosomes are transmitted by different groups of tsetse species. Tsetse control is considered an effective approach and constitutes the corner stone in trypanosomiasis suppression [4,5]. Tsetse fly species belong to Glossina genus and are generally restricted to sub-Saharan Africa. Twenty-three species and eight sub-species of tsetse flies are recognized [6,7]. These species are divided into Morsitans, Palpalis and Fusca clade sub-genera, described by respective savanna, riverine/lacustrine and forest ecological niches they occupy. The Morsitans group consists of five species that include Glossina morsitans morsitans and Glossina pallidipes restricted to savannah grassland and Glossina austeni occupying coastal woodlands [8]. This group is adapted to drier habitats than Palpalis and Fusca [9] and preferentially feeds on livestock and wildlife. They are thus important vectors of African Animal Trypanosomiasis (AAT) also known as nagana. On the other hand, Palpalis group consists of five species, including Glossina palpalis gambiensis and Glossina fuscipes fuscipes in West, Central and East Africa. These species are predominant vectors of Human African Trypanosomosis (HAT), also known as sleeping sickness, despite their preferential predilection to feeding on reptiles and ungulates. Fusca group consist of 13 species largely inhabiting damp evergreen forests of West Africa (except Glossina brevipalpis) and are mainly associated with livestock. Glossina brevipalpis is of limited medical and agricultural significance and occurs discontinuously in other parts of sub-Saharan Africa [6].
These tsetse fly species exhibit different olfactory uniqueness, which partly accounts for their gradation of preferences for their particular hosts. This olfactory uniqueness (and visual responses) has been exploited in designing effective tsetse fly bait technologies that consist of synthetic blends of attractants and repellents that mimic those of their natural hosts and non-hosts respectively [10–13]. These technologies are especially applicable for G. m. morsitans and G. pallidipes but not G. austeni (among savanna species) [14] and palpalis group. For example, G. pallldipes, G. m. morsitans and to some extent G. brevipalpis are attracted to traps baited with POCA (3-n-propylphenol, 1-octen-3-ol, 4-cresol and acetone) and to which G. austeni poorly responds [15–17]. Molecular bases of these natural differential responses are poorly understood but may be underpinned by differences in their chemosensory apparatus. The chemosensory apparatus facilitate reception of odorants and tastants, and consist of Odorant-binding proteins (Obps), Odorant-degrading enzymes (Odes), Odorant receptors (Ors), Ionotropic receptors (Irs), Gustatory receptors (Grs), Chemosensory proteins (Csps), Sensory neuron membrane proteins (Snmps) and CD36-like pheromone sensors [18–24]. These chemosensory proteins mediate decoding of ecological odors and odorant specific behavioral responses in insect hosts. These responses include seeking for hosts, location of oviposition sites, searching for mates, and detecting and escaping from potential predators. The Obp transport pheromone molecules and general odorants to Ors [25]. The Ors are odorant-gated ion channels composed of an odorant-binding subunit and olfactory co-receptor Orco [26,27]. The Irs have higher specificity to volatiles than Ors, detecting specific variety of odors, such as acids, aldehydes, amines and humidity [20,28]. The Ir25a and Ir8a are putative conserved Ir co-receptors [23]. The Grs discern odor tastes and contact pheromones [29]. Only two Snmp subfamilies (Snmp 1 and Snmp 2) have been identified in insects, where Snmp1 is expressed in pheromone-sensitive Olfactory Receptor Neurons (ORNs) while Snmp 2 is expressed in supporting cells [30–32]. Some of these chemosensory proteins are present in non-canonical chemosensory organs, such as legs [33,34], wings [35,36] and pheromone glands [37], where only a subset of Irs are specifically expressed in D. melanogaster antennae [20]. Among tsetse flies, genomes of G. pallidipes, G. m. morsitans, G. austeni, G. p. gambiensis, G. f. fuscipes and G. brevipalpis (representative of the different clades/sub-general) have been sequenced [38], and their respective chemosensory proteins annotated [39–41].
Here we report on 1) expansions of chemosensory protein orthologs in six tsetse fly species/subspecies (G. pallidipes, G. m. morsitans, G. austeni, G. p. gambiensis, G. f. fuscipes and G. brevipalpis) to identify species-specific expansions and 2) differential expressions of these and associated proteins in antennae and head tissues G. m. morsitans to establish probable functional pathways influencing host seeking behaviors in this specie.
Materials and methods
Differential expansions of D. melanogaster chemosensory gene orthologs among tsetse flies
We obtained complete D. melanogaster gene set release 79 (Drosophila_melanogaster. BDGP6.pep.all.fa) from Ensembl project [42] in fasta format. We then isolated D. melanogaster chemosensory genes from the gene set by searching and retrieving flybase [43] chemosensory gene IDs in the gene set using “Odorant receptor”, “Gustatory receptor”, “Ionotropic receptor”, “Odorant-binding protein”, “Sensory neuron membrane protein” and “Glutamate receptor” Linux bash regular expressions. For Csp orthologs, we extracted D. melanogaster IDs from Macharia et al., (2016) [40]. We separately obtained VectorBase Release VB-2019-02 homologs (gene trees) of disease vectors from VectorBase database [44] in OrthoXML formats. The gene trees were pre-computed by Gene Orthology/Paralogy prediction pipeline in VectorBase [44] that identified gene duplications within species and speciation events. We probed the VectorBase homologs for ortholog groups (gene families) with the D. melanogaster chemonsensory genes (flybase IDs) to identify their respective tsetse flies (G. austeni, G. f. fuscipes, G. p. gambiensis, G. brevipalpis, G. pallidipes and G. m. morsitans) orthologs. We identified presence of the individual genes in each gene family (ortholog group) and species. Gene families with accelerated gene expansions were pre-computed through Computational Analysis of gene Family Evolution (CAFE) [45] in VectorBase [44]. We considered the VectorBase [44] pre-computed gene expansions/contractions reliable since they are 1) community reviewed and adopted and with stable ortholog IDs and 2) regularly updated (with new gene-sets and genomes). We also conducted Principal Component Analysis (PCA) in R using FactoMineR and Factoextra packages with species-specific gene counts as input data to establish relationship between the expanded/contracted chemosensory genes (Ors, Irs, Grs and Obps) and tsetse species.
Transcriptional expression of D. melanogaster chemosensory gene orthologs in male G. m. morsitans
We employed high throughput Illumina based RNA-Seq approach to establish expression profiles of the D. melanogaster chemosensory gene orthologs in male G. m. morsitans. We established expression levels of the orthologs in the antennae and in relation to the head libraries. We isolated and sequenced RNA from antennae or head tissues from colony reared G. m. morsitans as described previously [46]. Briefly, we fed teneral male G. m. morsitans (1–3 days old) on defibrinated bovine blood meal (their initial blood meal post-eclosion) (commercially supplied by Hemostat Laboratories, Dixon, CA, USA) to putatively prime their chemosensory system. We then extracted their antennae in two independent biological replicates (from 50 flies each) using liquid nitrogen-based method of Menuz et al. (2014) [47] 72 hrs post-feeding. We envisaged that the 72 hrs deprivation of blood meal (food) would biologically prime potential host seeking chemosensory apparatus in the flies and enhance RNA-seq detection of chemosensory gene expressions, specifically those associated with hunger/host seeking.
The G. m. morsitans show marked die1changes in their biting activity in the field, with their peak activity in the morning and afternoon [48,49]. We thus snap froze individual tsetse flies in liquid nitrogen in the morning (09:30 hrs) and carefully hand-dissected their antennae from the head into 1.5 ml microfuge tubes kept cold in liquid nitrogen. We then isolated RNA by mechanically crushing the antennae with disposable RNAseq-free plastic pestles in TRIzol reagent (Invitrogen, Carlsbad, USA) following the manufacturer’s protocol. We removed traces of potential carry over DNA (that could potentially confound our RNA-Seq analysis) by digesting possible contaminating genomic DNAs (gDNA) in the total RNA using TURBO DNase (Ambion life technologies, TX, USA) following manufacturer’s instructions. We confirmed removal of the gDNA from total RNA by qualitative assessment of PCR amplicons from final RNA samples using tsetse fly specific beta-tubulin gene primers as documented in Bateta et al. (2017) [46]. We verified quality and integrity of RNA samples using Agilent Bioanalyzer 2100 (Agilent, Palo Alto, CA, USA) following manufacturer’s instructions. cDNA was then generated from the RNA using Illumina TruSeq RNA Sample Preparation Kit (Illumina, Hayward, CA, USA) and the cDNA (75 bp single-end read) and sequenced on Illumina HiSeq 2500 at Yale University Center of Genome Analysis (YCGA), New Haven, CT, USA. We similarly prepared head transcriptomes from two independent biological replicates (50 flies each) from 72 hrs starved 40 days old males. We deposited all transcriptome sequences at the Sequence Read Archive (SRA) under study accession numbers PRJNA343267 and PRJNA343269 for the antennae and head libraries respectively.
Expression profiles of D. melanogaster chemosensory gene orthologs in male G. m. morsitans antennae and head libraries
We established quality of the reads in each individual transcriptome library using FastQC (Babraham Bioinformatics) software package (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/)). We then used the FastQC results to clean (trimm) the reads using CLC genomic workbench version 10 software (CLC Bio, Aarhus, Denmark) through settings that permitted 1) removal of low quality sequences (limit = 0.05), 2) removal of ambiguous nucleotides (maximum 2 nucleotides allowed), 3) removal of terminal nucleotides (10 nucleotides from the 5’ end and 1 nucleotide from the 3’ end) and 4) removal of sequences on length (minimum length 15 nucleotides, maximum length 1000 nucleotides). We then mapped the cleaned reads on to G. m. morsitans transcripts gene-set version 1.9 from Vectorbase [44] using CLC genomic workbench version 10 software (CLC Bio, Aarhus, Denmark) thorough settings that permitted 1) mismatch cost of 2, 2) insertion/deletion cost of 3, 3)length fraction of 0.8, 4) similarity fraction of 0.8, 5) maximum number of reads per hit of 10, and 6) strand specificity set as both strands.
From the mappings, we established reads mapping per transcript and reads per kilobase of transcripts per Million mapped reads (RPKM), a normalized index of relative gene expression associated with each transcript (including chemosensory genes) in the gene-set for individual transcriptomes [50]. We then established differentially expressed transcripts between the antennae and the head transcriptomes by comparing the reads mapped in the genes sets from respective transcriptomes using edgeR software [51,52]. We considered transcripts validly differentially expressed if they had at least two-fold changes, p-value corrected False Detection Rate (FDR) < 0.05 and one Counts Per Million (CPM) coverage to mitigate against type I statistical errors. We then determined antennae or head enriched molecular processes using canonical Gene Set Enrichment Analysis (GSEA) using WEB-based GEne SeT AnaLysis Toolkit (WebGestalt) [53]. Since WebGestalt database did not include tsetse flies, but D. melanogaster gene set, we obtained homologs of the entire G. m. morsitans gene-set in D. melanogaster through Basic Alignment Search Tool (BLAST) analysis of protein sequences (Blastp) [54] of the G. m. morsitans gene-set against those of D. melanogaster and accepted hits with e-value < 0.001 as significantly homologous. We then used these D. melanogaster homologs as proxy in WebGestalt to assess enrichment of their associated G. m. morsitans homologs. We used the FDR corrected p-value ranked D. melanogaster homolog gene-sets of differentially expressed G. m. morsitans transcripts as input for the analysis [55]. We considered selection of 5–2000 Entrez Gene IDs, FDR < 0.05, 1000 permutations and 20 categories with the outputted leading-edge genes default parameters for the analysis. Through GSEA, we separated and identified significantly enriched non-redundant biological processes, cellular components and molecular function Gene Ontology (GO) terms, Kyoto Encyclopedia of Genes and Genomes, KEGG, PANTHER, Reactome, pathways and Database of Protein, Chemical and Genetic Interactions (BioGRID) network [56–61]. Next, we identified antennae or head (tissue) specific chemosensory genes by mapping the global most differentially (based on fold change) and abundantly (based on CPM) or significantly expressed (based on p-value) transcripts in MA or volcano plots respectively using edgeR software package [52,62] in R software [63]. We considered chemosensory genes with fold changes (FC) ≥ 1.25 as of chemosensory biological significance as previously documented [64].
Results
Expansions of chemosensory gene orthologs among tsetse fly species
We identified 60 each of Ors, Irs or Grs, 51 Obps, seven GluR and two Snmps (excluding isoforms) in D. melanogaster [43] and four Csps [40], with 58, 34, 13, 22, 2 and 3 orthologs (VectorBase gene trees, Release VB-2019-02) [44] respectively among the tsetse fly species (S1 Table). Café gene expansion analysis [45] revealed significant (P<0.05, λ = 2.60452e-7) accelerated expansions of several gene families/clusters including VBGT00190000010263 (Or71a and Or46a), VBGT00190000009736 (Ir75a,d, Ir31a, Ir84a and Ir64a) and VBGT00190000009994 (Obp83a-b) in G. m. morsitans, VBGT00840000047907 (Or67a,c, Or49a, Or92a, Or85b-c,f) and VBGT00190000013627 (Obp73a) in G. pallidipes, VBGT00190000012412 (Ir21a) and VBGT00190000010879 (Gr21a and Gr63a) carbon dioxide receptors orthologs [65] in G. f. fuscipes, VBGT00820000046003 (clumsy, Ir25a and Ir8a) in G. p. gambiensis and VBGT00190000013104 (Ir68a) in G. brevipalpis (S1 Table). No gene families were significantly expanded in G. austeni. We also identified several orthologs that were missing/absent in specific tsetse fly species (S1 Table). The Ir76b ortholog was absent in four tsetse fly species (G. p. gambiensis, G. m. morsitans, G. pallidipes and G. brevipalpis) while Gr33a was missing in G. brevipalpis. Both Gr32a and Gr68a were missing in G. brevipalpis and G. m. morsitans. The Gr64a-f, Gr5a, Gr43a, Obp56a/d/e and Or71a orthologs were absent in all tsetse fly species. The Snmp1, Or67d and Obp19a and Orco ortholog appeared to be conserved across all tsetse fly species. Our PCA analysis revealed a general positive correlation between tsetse species across four chemosensory groups (Ors, Irs, Grs or Obps). Additionally, Gr and Ir orthologs appeared to be positively correlated (S1 Fig panels B2 and B3) in relation to a unique G. m. morsitans cluster (S1 Fig panels A2 and A3).
Expression profiles of chemosensory ortholog transcripts in male G. m. morsitans antennae
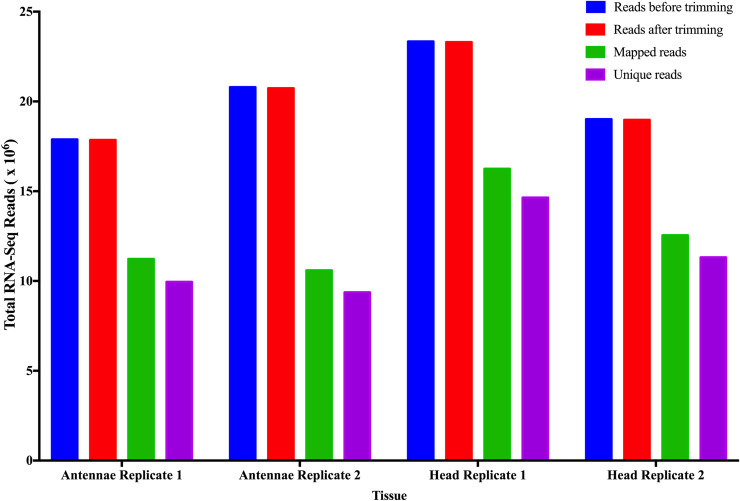
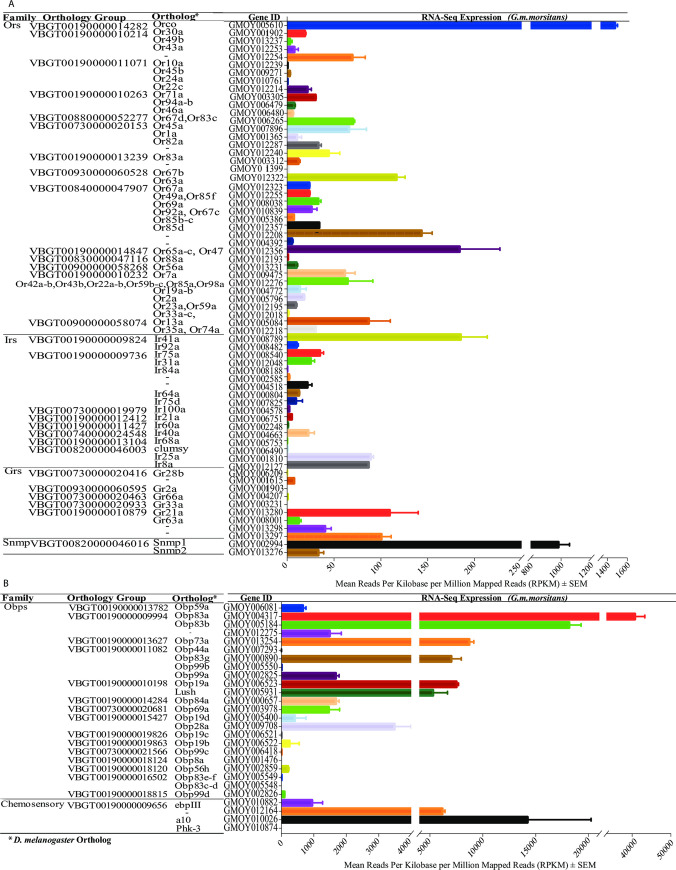
The RNA-Seq of the antennae and head libraries yielded 23.3 to 17.9 million reads from respective libraries. We successfully mapped 51.0 to 69.6% of these reads onto G. m. morsitans transcripts where we established about 88.4% unique mappings of the reads to specific transcripts (Fig 1). We have summarized expressions profiles of the chemosensory orthologs in Fig 2. Orco, Or56a, 65a-c, Or47b and Or67b, and three G. m. morsitans specific orthologs (GMOY012254, GMOY009475, and GMOY006265) were among most abundantly expressed transcripts with Or33a-c orthologs exhibiting the least expression. Expressions of the members of the significantly expanded Ors gene families were marginal. Only six Gr orthologs were expressed among which Gr21a and Gr63a orthologs (carbon dioxide receptors) [65] and related two G. m. morsitans specific (GMOY013297 and GMOY013298) orthologs were abundantly expressed. The putative conserved core-receptors (Ir8a and Ir25a) and Ir41a were among the most abundantly expressed Irs orthologs. All but Ir75a-c expanded Ir orthologs were expressed. Most abundantly expressed Obp orthologs include Obp19a, lush, Obp28a, Obp83a-b Obp44a and two G. m. morsitans specific (GMOY012275 and GMOY013254) orthologs. Among these, Obp83a-b were among the significantly expanded Obp families. Both Snmps (Snmp 1 and Snmp 2) and Csp2 were also abundantly expressed.
Fig 1. Summary of processing and mapping statistics of RNA-Seq reads from male G. m. morsitans antennae and head transcriptomes.
Fig 2. Expression profiles of D. melanogaster chemosensory gene orthologs in male G. m. morsitans antennae 72 hrs post feeding.
Enriched pathways between male G. m. morsitans antennae and head libraries
Our Gene Set Enrichment Analysis (GSEA) of transcripts between the antennae and head libraries revealed several enriched pathways and processes between these tissues (Table 1, S2 Table). Our GoSlim GO analysis component of the GSEA assigned 85.4% of our transcripts to biological process, cellular components and molecular function ontologies (S2 Table). The most predominantly enriched biological processes between the antennae and head include metabolic processes, biological regulations, multicellular organismal processes, developmental processes and responses to stimuli. Most of these biological processes appeared to be localized in the membrane, macromolecular complex and nucleus cellular components, and were predominantly involved in protein binding, nucleic acid binding, ion binding and hydrolase activity molecular functions (S2 Table). More specifically, most enriched biological processes in the head were associated with vision, muscle activity and associated structural proteins and neuropeptide regulations, amino acid/nucleotide metabolism and circulatory system processes. The enriched cellular components were predominantly associated with vision and muscular functions. On the other hand, most enriched antennal biological processes were cilium-associated mechanoreceptors, chemo-sensation, neuronal controlled growth and differentiation, and regeneration/responses to stress, while enriched cellular components were associated with chemo-sensation, mechano-reception and muscular activities. Most enriched molecular functions in the head and antennae were associated with vision/muscular activities and chemo-sensation, respectively. The KEGG pathway analysis revealed enrichment of vision-associated pathways. Similarly, PANTHER pathway analysis also identified vision, in addition to neuropeptide signaling and muscular associated activities among the most enriched pathways in the head. We identified similar outcomes from our protein-protein interactions BIOGRID analysis in the head library. The Reactome pathway analysis identified vision and amino acids and derivative metabolism pathways predominating in the head transcriptome. We did not identify pathways or networks significantly enriched in the antennae library.
Table 1. Summary of Canonical Gene-set Enrichment Analysis (GSEA) of differentially expressed transcripts between male G. m. morsitans tsetse fly antennae and head transcriptomes.
| Functional Database | Tissue | Annotation | Statistics | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Class | Tissue | Process ID | Description | General Function | Size | L | ES | NES | P Value | FDR |
| Gene | Biological | Head | GO:0050953 | Sensory perception of light stimulus | Vision | 59 | 22 | 0.898 | 1.883 | 0.000 | 0.000 |
| Ontology | Process | GO:0007186 | G-protein coupled receptor signaling pathway | Vision | 162 | 56 | 0.796 | 1.869 | 0.000 | 0.000 | |
| GO:0032101 | Regulation of response to external stimulus | Vision | 101 | 8 | 0.801 | 1.833 | 0.000 | 0.000 | |||
| GO:0010927 | Cellular component assembly involved in morphogenesis | Muscle activity | 108 | 18 | 0.773 | 1.765 | 0.000 | 0.001 | |||
| GO:0042440 | Pigment metabolic process | Vision | 115 | 18 | 0.735 | 1.692 | 0.000 | 0.004 | |||
| GO:0009628 | Response to abiotic stimulus | Vision | 360 | 36 | 0.682 | 1.689 | 0.000 | 0.003 | |||
| GO:0003012 | Muscle system process | Muscle activity | 27 | 12 | 0.879 | 1.676 | 0.000 | 0.005 | |||
| GO:0044057 | Regulation of system process | Neuropeptide muscle regulations | 48 | 12 | 0.795 | 1.661 | 0.000 | 0.007 | |||
| GO:0043473 | Pigmentation | Vision | 103 | 18 | 0.706 | 1.610 | 0.003 | 0.025 | |||
| GO:0006730 | One-carbon metabolic process | Vision | 15 | 5 | 0.910 | 1.607 | 0.000 | 0.024 | |||
| GO:0003013 | Circulatory system process | Neuropeptide regulations | 40 | 12 | 0.784 | 1.590 | 0.005 | 0.032 | |||
| Antennae | GO:0044782 | Cilium organization | Mechanoreception | 62 | 22 | -0.839 | 2.170 | 0.000 | 0.000 | ||
| GO:0031503 | Protein complex localization | Mechanoreception | 30 | 12 | -0.849 | 1.903 | 0.000 | 0.001 | |||
| GO:0007606 | Sensory perception of chemical stimulus | Chemo-sensation | 124 | 57 | -0.628 | 1.806 | 0.000 | 0.006 | |||
| GO:0035218 | Leg disc development | Growth/differentiation | 87 | 15 | -0.665 | 1.781 | 0.000 | 0.007 | |||
| GO:0030705 | Cytoskeleton-dependent intracellular transport | Mechanoreception | 66 | 11 | -0.676 | 1.751 | 0.000 | 0.012 | |||
| GO:0030031 | Cell projection assembly | Mechanoreception | 112 | 35 | -0.624 | 1.742 | 0.005 | 0.011 | |||
| GO:0031099 | Regeneration | Repair/response to stress | 18 | 4 | -0.828 | 1.711 | 0.000 | 0.015 | |||
| Cellular | Head | GO:0019898 | Extrinsic component of membrane | Vision | 72 | 11 | 0.870 | 1.891 | 0.000 | 0.000 | |
| Component | GO:0016028 | rhabdomere | Vision | 34 | 17 | 0.955 | 1.886 | 0.000 | 0.000 | ||
| GO:0043292 | Contractile fiber | Muscle activity | 50 | 20 | 0.871 | 1.822 | 0.000 | 0.000 | |||
| GO:0015629 | Actin cytoskeleton | Vision/Muscle activity | 99 | 18 | 0.794 | 1.807 | 0.000 | 0.000 | |||
| GO:0098796 | Membrane protein complex | Vision | 233 | 10 | 0.690 | 1.689 | 0.000 | 0.001 | |||
| GO:0098858 | Actin-based cell projection | Vision | 22 | 4 | 0.861 | 1.600 | 0.002 | 0.012 | |||
| GO:0031984 | Organelle sub-compartment | Vision | 86 | 9 | 0.684 | 1.515 | 0.007 | 0.046 | |||
| Antennae | GO:0005929 | Cilium | Chemo-sensation/ Mechanoreception | 80 | 30 | -0.846 | 2.256 | 0.000 | 0.000 | ||
| GO:0031252 | Cell leading edge | Chemo-sensation | 52 | 28 | -0.811 | 2.005 | 0.000 | 0.000 | |||
| GO:0005815 | Microtubule organizing center | Mechanoreception/Muscle activity | 111 | 20 | -0.666 | 1.849 | 0.000 | 0.001 | |||
| Molecular | Head | GO:0005516 | Calmodulin binding | Vision/Muscle activity | 43 | 6 | 0.834 | 1.706 | 0.000 | 0.009 | |
| Function | Antennae | GO:0005549 | Odorant binding | Chemo-sensation | 49 | 35 | -0.843 | 2.170 | 0.000 | 0.000 | |
| Pathway | KEGG | Head | dme04745 | Phototransduction—fly—Drosophila melanogaster (fruit fly) | Vision | 25 | 14 | 0.954 | 1.772 | 0.000 | 0.000 |
| Analysis | Panther | Head | P00057 | Wnt signaling pathway | Vision | 62 | 8 | 0.827 | 1.749 | 0.000 | 0.000 |
| P00031 | Inflammation mediated by chemokine and cytokine signaling pathway | Vision/Muscle activity | 26 | 5 | 0.895 | 1.716 | 0.000 | 0.002 | |||
| P00044 | Nicotinic acetylcholine receptor signaling pathway | Vision/Muscle activity | 38 | 9 | 0.845 | 1.705 | 0.000 | 0.002 | |||
| P00016 | Cytoskeletal regulation by Rho GTPase | Vision/Muscle activity | 21 | 4 | 0.911 | 1.680 | 0.000 | 0.004 | |||
| P00004 | Alzheimer disease-presenilin pathway | Vision/Muscle activity | 25 | 5 | 0.848 | 1.593 | 0.005 | 0.026 | |||
| P00012 | Cadherin signaling pathway | Muscle activity | 25 | 3 | 0.839 | 1.585 | 0.003 | 0.025 | |||
| P00042 | Muscarinic acetylcholine receptor 1 and 3 signaling pathway | Vision/Neuropeptide regulations | 20 | 7 | 0.836 | 1.568 | 0.011 | 0.033 | |||
| P04374 | 5HT2 type receptor mediated signaling pathway | Vision | 18 | 7 | 0.841 | 1.567 | 0.014 | 0.030 | |||
| P00028 | Heterotrimeric G-protein signaling pathway-rod outer segment phototransduction | Vision | 5 | 3 | 0.991 | 1.563 | 0.000 | 0.031 | |||
| Reactome | Head | R-DME-1852241 | Organelle biogenesis and maintenance | Vision | 38 | 4 | 0.904 | 1.778 | 0.000 | 0.000 | |
| R-DME-2514856 | The phototransduction cascade | Vision | 12 | 6 | 0.961 | 1.687 | 0.000 | 0.025 | |||
| R-DME-5620920 | Cargo trafficking to the periciliary membrane | Vision | 15 | 4 | 0.956 | 1.683 | 0.000 | 0.018 | |||
| R-DME-5617833 | Cilium Assembly | Vision | 15 | 4 | 0.956 | 1.674 | 0.000 | 0.018 | |||
| R-DME-5620916 | VxPx cargo-targeting to cilium | Vision | 12 | 4 | 0.965 | 1.655 | 0.000 | 0.026 | |||
| R-DME-2514859 | Inactivation, recovery and regulation of the phototransduction cascade | Vision | 12 | 6 | 0.961 | 1.644 | 0.000 | 0.029 | |||
| R-DME-2187338 | Visual phototransduction | Vision | 14 | 6 | 0.957 | 1.644 | 0.000 | 0.025 | |||
| R-DME-76002 | Platelet activation, signaling and aggregation | Vision/Muscle activity | 47 | 9 | 0.784 | 1.640 | 0.000 | 0.024 | |||
| R-DME-71291 | Metabolism of amino acids and derivatives | Metabolism | 57 | 19 | 0.761 | 1.634 | 0.000 | 0.027 | |||
| R-DME-2672351 | Stimuli-sensing channels | Vision | 9 | 3 | 0.954 | 1.622 | 0.000 | 0.034 | |||
| R-DME-500792 | GPCR ligand binding | Vision | 14 | 4 | 0.920 | 1.618 | 0.002 | 0.034 | |||
| Network Analysis | PPI_BIOGRID | Head | PPI_BIOGRID M119 | Muscle activity | 33 | 16 | 0.872 | 1.711 | 0.000 | 0.004 | |
| PPI_BIOGRID M37 | Muscle activity | 71 | 20 | 0.767 | 1.683 | 0.000 | 0.004 | ||||
| PPI_BIOGRID M80 | Vision | 12 | 8 | 0.957 | 1.643 | 0.000 | 0.017 | ||||
*Non-Redundant
Differentially expressed transcripts between male G. m. morsitans antennae and head libraries
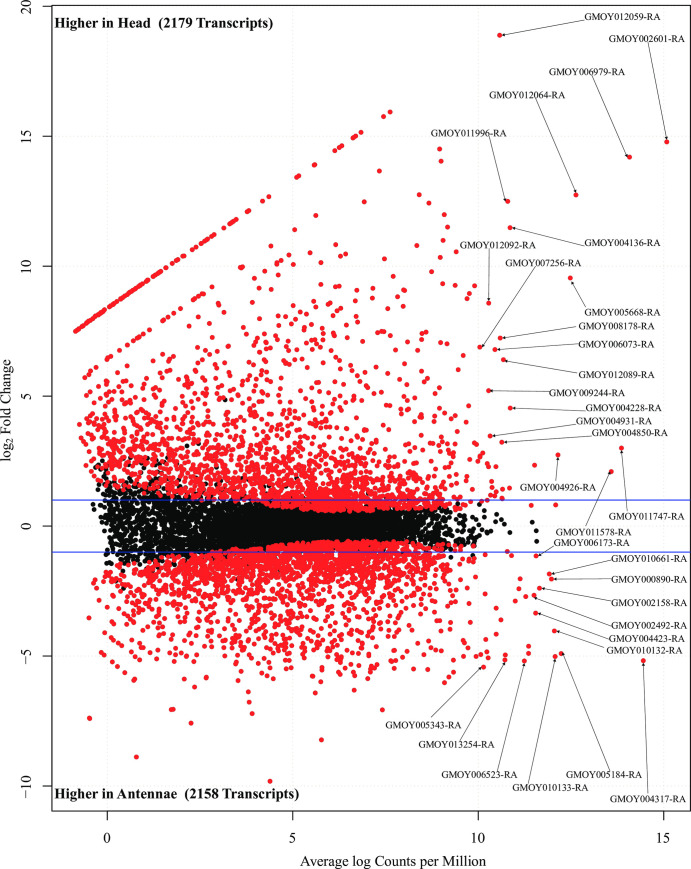
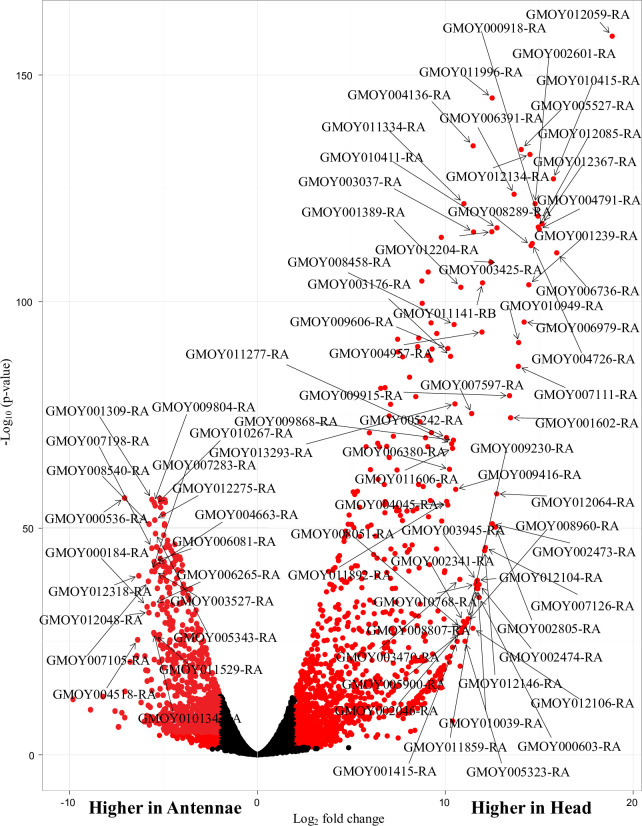
Our search for both differentially (FC > 2) and abundantly expressed (CPM > 1) transcripts between the head and antennae libraries identified 2179 and 2158 transcripts respectively differentially expressed (FDR corrected p value < 0.05) between each library as summarized in our MA plot (Fig 3). Among these transcripts, at least 52 transcripts were most differentially and abundantly expressed (log FC > 2 and Average log CPM > 10) in both libraries. These transcripts were predominantly associated with vision, iron transport, metabolism and signal transduction in the head. In the antennae, the transcripts were involved in odor sensing and clearing, fatty acid synthesis and regulation of feeding behavior and locomotor activity (S3 Table). Analysis of both differentially (FC) and significantly expressed (p-value) transcripts between the head and antennae libraries identified 49 and 61 transcripts as most significantly expressed (FC >10 or <-5, and–log10 p-value > 25) in the head and antennae libraries respectively as summarized in our volcano plot (Fig 4). Overall, about 40 and 52 percent of the transcripts were associated with vision (head) and chemo-sensation (antennae) respectively. Most significantly expressed transcripts in the head library were functionally associated with energy mobilization, feeding, immunity, cytoskeleton integrity, amino acid metabolism, endocrine signaling and neuronal development and support. In the antennae, most significantly expressed transcripts were functionally associated chemo-sensation, metabolism, and cell proliferation, regulation of gene expression, signal transduction, anatomical integrity, neuron integrity/development and mechanoreception (S3 Table).
Fig 3. MA plot showing abundantly and differentially expressed transcripts between the male G. m. morsitans head and antennae transcriptomes.
Dots indicate points-of-interest that display individual transcript abundance (x axis) and fold-change (y axis). Red dots indicate transcripts with fold-changes of two or more (log2 ≥ 1) and False Detection Rate (FDR) corrected p values of less than 0.05 (significant) between the head and antennae transcriptomes. Black dots indicate transcripts with non-significant changes between the transcriptomes.
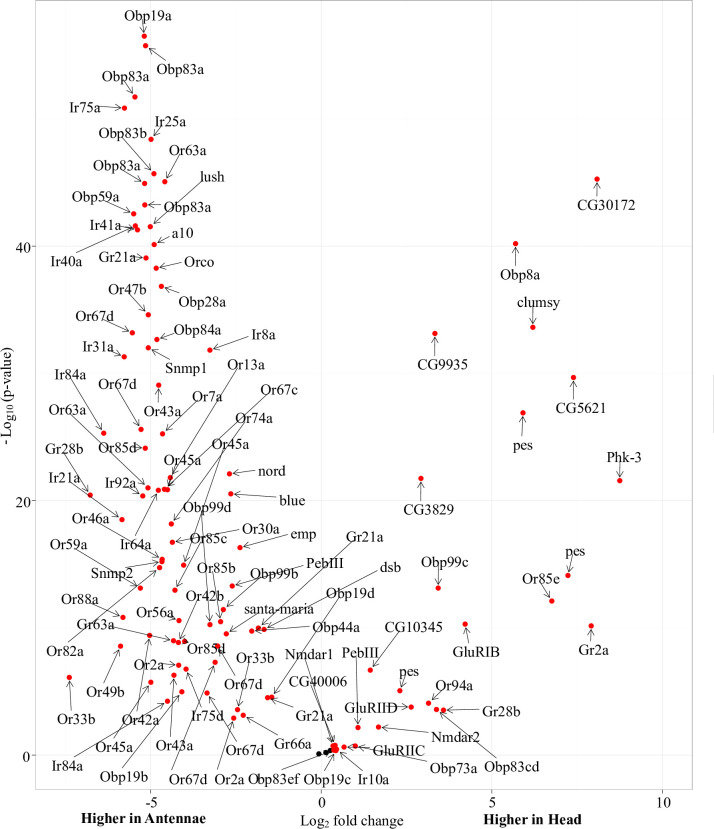
Fig 4. Volcano plot showing abundantly and significantly expressed transcripts between the male G. m. morsitans head and antennae transcriptomes.
Dots indicate points-of-interest that display fold-changes (x axis) and statistical significance (-log10 of p value, y axis) in transcripts between the head and antennae transcriptomes. Red dots indicate transcripts with fold-changes of two or more (log2 ≥ 1) and False Detection Rate (FDR) corrected p values of less than 0.05 and are indicate transcripts with significant changes between the transcriptomes. Black dots represent transcripts with non-significant changes between the transcriptomes.
Differential expression of chemosensory gene transcripts between male G. m. morsitans antennae and head libraries
When we considered fold change greater than 1.25 as of biological chemosensory significance [64], most (> 90%) chemosensory transcripts showed significantly higher expressions in the antennae than in the head (Fig 5). Among these, significantly expressed chemosensory transcripts (p-value < 1e-20) in the antennae include several Obp (Lush, Obp19a, Obp28a, Obp59a, Obp83a/b and Obp84a), Ir (Ir25a, Ir31a, Ir40a, Ir41a, Ir64a, Ir75a, Ir76b, Ir84a, Ir8a and Ir92a), Or (Orco, Or7a, Or13a, Or43a, Or45a, Or47b, Or63a/c/d and Or85d), Gr (Gr21a), Csp [Csp2 (a10) and Csp4 (Phk-3)] and Snmp1 orthologs. Specifically, most significantly expressed transcripts were predominantly Obp orthologs. On the other hand, we identified a subset of obp (Obp8a, Clumsy, Obp99c Obp83cd), Or (Or85e, Or71a), Grs (Gr2a, Gr28b) and Csp4 (Phk-3) orthologs with significantly higher expression in the head than in the antennae libraries.
Fig 5. Volcano plot showing abundantly and significantly expressed chemosensory gene orthologs between the male G. m. morsitans head and antennae transcriptomes.
Dots indicate points-of-interest that display fold-changes (x axis) and statistical significance (-log10 of p value, y axis) in transcripts between the head and antennae transcriptomes. Red dots indicate transcripts with fold-changes of two or more (log2 ≥ 1) and False Detection Rate (FDR) corrected p values of less than 0.05 and are indicate transcripts with significant changes between the transcriptomes. Black dots represent transcripts with non-significant changes between the transcriptomes.
Discussion
In this study, we profiled expansions of chemosensory gene orthologs among six tsetse fly species/subspecies (G. pallidipes, G. m. morsitans, G. austeni, G. p. gambiensis, G. f. fuscipes and G. brevipalpis) and employed RNA-seq to discern differential expressions of the orthologs and associated proteins in antennae and head tissues male G. m. morsitans. Our café analysis for gene expansion revealed significant accelerated expansion of 4-methyl phenol and 4-propyl phenol responsive Or71a [66] in G. m. morsitans. The 4-methyl phenol and 4-propyl phenol are known G. m. morsitans and G. pallidipes attractants present in natural ox odor [17,67]. These findings probably account for the observed differential responses of these species to synthetic blends of these odors [68]. On the other hand, expansions of Ir75a,d, Ir31a, Ir84a and Ir64a orthologs in G. m. morsitans suggest differential odor-tuning and responses to acetic acid and 2-oxopentanoic acid in this species [69–73]. Acetic acid component of the vertebrate breath is an attractant of most hematophagous vectors while 2‐oxopentanoic acid elicit a landing response from Anopheles gambiae [74]. Whether there is enhanced attraction and landing behavior in G. m. morsitans in the presence of these kairomones remains to be determined. Expansion of Ir84a in G. m. morsitans may also indicate enhanced response to phenylacetaldehyde and male-male courtship [75] in this tsetse fly specie relative to the other species. Expansion of hunger responsive Obp83a ortholog [76] in G. m. morsitans suggest enhanced host seeking persistence in this specie relative to the other species. The G. pallidipes appears to be characterized by potentially muted responses to feeding stimulating hydroxycinnamic acids linked to missing Or71a [77], but enhanced responses to butanol, 2-heptanone and ketones lactones and phenolic compounds associated with the expanded Or49a [78,79], Or67a [80], Or85f [81] and Or85c [82] orthologs. The responses to butanol, lactones, ketones and phenolic compounds have been evaluated in development of baits used routinely in field control of G. pallidipes. Carbon dioxide receptors Gr21a and Gr63a orthologs [65] were expanded in G. f. fuscipes and most abundantly expressed in male G. m. morsitans antennae. These findings are indicative of the heavier investment by G. f. fuscipes than other tsetse flies in carbon dioxide detection and consequently host location [83]. The potential impact of the expansion (in G. f. fuscipes) of the Ir21a required for cool avoidance behavior [84] is not clear, but may be tied to the humid and warm habitat preference in the G. f. fuscipes lacustrine habitats. The Gr64a-f, Gr5a and Gr43a sugar receptor orthologs [85,86] were conspicuously absent in tsetse flies, consistent with our previous finding [40], a phenomenon attributable to exclusive sugar deficient blood diet in tsetse flies. The G. brevipaplis specific expansions of hygrosensory behavior mediating Ir68a ortholog [87] suggest potential behavioral responses to these and related odor cues specific to this tsetse fly. We did not identify expansion of Or67d in tsetse flies, contrary to previous reports [39,40].
We identified several missing/absent or conserved tsetse fly species specific orthologs with potential implications on respective tsetse species phenotypes. Absent Gr33a ortholog responsive to nonvolatile repulsive chemicals, including N,N-diethyl-meta-toluamide (DEET) [88,89] in G. brevipalpis and marginal expression of Gr66a ortholog in male G. m. morsitans antennae, suggest diminished responses in these species to some repellents. This phenomenon is further supported by absence of another caffeine and DEET responsive Gr32a ortholog [88,89] and courtship pheromone associated Gr68a ortholog [90] in G. brevipalpis and G. m. morsitans. The missing Ir76b ortholog in four tsetse fly species (G. p. gambiensis, G. m. morsitans, G. pallidipes and G. brevipalpis) suggests that these tsetse species may have reduced responses to Ir76b ortholog mediated feeding preferences for amino acids [73] relative to remaining tsetse fly species. The conspicuous absence of Obp56a,d,e orthologs in tsetse flies, point to possible reduction in their responses to the associated pheromones [91]. Geosmin responsive Or56a ortholog [92] was most abundantly expressed Or after Orco in the G. m. morsitans antennae. Since Geosmin is a microbial odorant that alerts flies of presence of harmful microbes and induces avoidance behavior [92], the findings suggest potential repellence of tsetse flies by Geosmin and associated compounds, which can form a basis for a search for tsetse fly specific repellents. Conserved Gr2a, Gr28b and Gr66a orthologs across most species supports a notion of general aversion of salts [93], caffeine, DEET and some amino acids (theophylline, threonine and valine) [88,94–97] among the vectors. The Snmp1 ortholog associated with detection of pheromones appears to be conserved across all the tsetse fly species, which in concert with similarly conserved Or67d and Orco orthologs, are functionally associated with detection of lipid-derived pheromones [98,99]. Other conserved pheromone responsive orthologs, include male-specific pheromone 11-cis-vaccenyl acetate (cVA) responsive lush and Obp19a [100] (absent in G. austeni) and l-carvone, 2-heptanone and acetophenone responsive Obp83a [101]. Lush, Or67d, Or83c and Obp83a were predominantly expressed in male G. m. morsitans antennae. We identified Ir93a ortholog in G. austeni contrary to previous findings [40]. Overall, we identified potential tsetse fly specific receptors and semiochemicals/ligands for downstream functional validations that can be employed to expand the toolbox of tsetse fly attractants, repellents and regulators.
Our gene and pathway enrichment analyses suggest that male G. m. morsitans head and antennae are predominately involved with vision and olfaction (odor sensing and clearing) respectively. In addition to the classical and canonical olfaction pathways, we also established fatty acid synthesis and associated xenobiotic responsive cytochrome P450 (Cyp6g1/2, Cyp304a1) and Glutathione S transferase pathways preferentially enriched in the antennae. Similar observations have been made in cutworm moth (Agrotis ipsilon) antennae [102] and may indicate significant investment in odor/pheromone clearing [103], probably as a strategy for faster desensitization of antennae responses in the absence or disengagement with relevant cues. Other enriched pathways and transcripts included lush, lush-like Obp19a, Obp28a and Obp83a/b, Obp84a, Or7a and Snmp1 that are associated with responses to pheromones [91,104]. The antennae transcriptome appears to be dominated with abundant, differentially expressed Ir75a-c, Ir31a, Ir84a, Ir41a, Ir92a and Gr21a orthologs, functionally associated with responses to various odor cues including acetic acid, 2-oxopentanoic acid [70–72], pyridine, 1,4-diaminobutane, cadaverine, spermidine, pyrrolidine [72], phenylacetyaldehyde [26], ammonia [20] and carbon dioxide [65]. Some of the cues, such as butanol, carbon dioxide and acetic acid are documented odor cues in the breath of the tsetse fly vertebrate hosts and are actively employed by tsetse fly in host location [10,15], suggesting that the rest might perform similar functions in nature.
The antennae were also enriched with transcripts associated with cilium mechanoreceptors/locomotor activity, indicating possible significant role of antennae in the detection of kinetic energy (energy of movement, e.g. touch, sound, vibration, changing pressure) or potential energy (e.g. gravity) and hence guiding physical orientation of the fly. Stress induced neuronal controlled growth and differentiation and regeneration pathways were also enriched in the antennae, suggesting important role of the antennae in modulating responses of the fly to fluctuations in oxygen levels, temperature and redox state [105]. In addition to vision gene, the head was enriched with muscle and associated structural proteins, and energy mobilization potentially associated with feeding, as well as neuropeptide regulations associated with modification of nervous and endocrine systems. Most differential and abundantly expressed head specific chemosensory transcripts were also functionally associated with feeding. These included Obp8a involved in food perception [106] and host location [107], and Gr28a/b and Gr2a linked to regulation of aversion to high-salt associated diet [93]. Phenotypic roles of other head-specific chemosensory transcripts, such as Csp2 (a10) and Csp4 (Phk-3), Clumsy, Obp99c Obp83cd, Or85e, Or71a and Csp4 (Phk-3), remain to be elucidated. Other than vision, olfaction and associated molecular processes, other processes appear to dominate physiological and molecular functions in the head and antennae libraries, respectively, indicating other functional roles of these tissues. Since these tissues (antennae and head) where extracted in the morning, the transcriptional responses coincided with the peak activity of the tsetse flies and hence reflect chemosensory and visual processes associated with host finding behavior predominant in that duration. Since our gene analyses were focused on antennae from male G. m. morsitans, our gene expression results were potentially biased toward male tsetse flies and G. m. morsitans subspecies. It would therefore be prudent to further assess for similar response in the remaining five tsetse fly species/subspecies, both gender and at different physiological states that influence their olfactory responses.
Conclusions
We identified tsetse fly specific chemosensory gene orthologs and their putative ligands, as potential candidates for downstream functional genomic and field validations. The validations could yield new tsetse fly attractants, repellents and pheromones with potential in incremental improvements of current tsetse fly control strategies. We also identified major sensory pathways and processes potentially active in the tsetse fly antennae and head that can be exploited in modulating tsetse fly behavior.
Supporting information
(A) Clustering of chemosensory orthologs between tsetse species (B) Clustering of individual orthologs within chemosensory gene families.
(TIF)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We are thankful to Ms. Yineng Wu for technical assistance during this study.
Data Availability
All RNA-Seq data files are available from the NCBI database (accession number(s) PRJNA343267 and PRJNA343269).
Funding Statement
None of the authors received direct funding for the research but were supported by Fogarty International Center of the National Institutes of Health (FIC-NIH) grants R03TW009444 and, D43TW007391, and NIH/NIAID supported grant U01AI115648 to Professor Serap Aksoy, Yale School of Public Health, Yale University, NH,CT, USA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Brun R, Blum J, Chappuis F, Burri C. Human African trypanosomiasis. Lancet. Elsevier BV; 2010;375: 148–159. 10.1016/S0140-6736(09)60829-1 [DOI] [PubMed] [Google Scholar]
- 2.Hotez PJ, Kamath A. Neglected Tropical Diseases in Sub-Saharan Africa: Review of Their Prevalence, Distribution, and Disease Burden. PLoS Negl Trop Dis. Public Library of Science; 2009;3: e412 Available: 10.1371/journal.pntd.0000412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.FAO. Impacts of trypanosomosis on African agriculture. PAAT Technical and Scientific Series No. 2. Rome; 2000.
- 4.Kettle DS. Medical and veterinary entomology. Wallingford, Oxon, UK: CAB International, in association with the International Livestock Research Institute, Nairobi, Kenya; 1990. [Google Scholar]
- 5.Elliott M. Properties and Applications of Pyrethroids. Environ Health Perspect. [National Institute of Environmental Health Sciences, Brogan & Partners]; 1976;14: 3–13. 10.2307/3428357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leak S. Tsetse Biology and Ecology: Their Role in the Epidemiology and Control of Trypanosomosis. Wallingford: CAB International, in association with the International Livestock Research Institute, Nairobi, Kenya; 1998. [Google Scholar]
- 7.Krafsur ES. Tsetse flies: Genetics, evolution, and role as vectors. Infect Genet Evol. 2009;9: 124–141. 10.1016/j.meegid.2008.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colvin J, Gibson G. Host-Seeking Behavior and Management of Tsetse. Annu Rev Entomol. Annual Reviews; 1992;37: 21–40. 10.1146/annurev.en.37.010192.000321 [DOI] [PubMed] [Google Scholar]
- 9.Rogers DJ, Robinson TP, Maudlin I, Holmes P, Miles M, Rogers ME, et al. Tsetse distribution. InEds Trypanos Oxford pp Antennal SNMPs Sens neuron Membr proteins Lepid Defin a unique Fam Invertebr CD36like proteins J Neurobiol 49 47. 2004;61: 139–179. [Google Scholar]
- 10.Vale GA. Development of baits for tsetse flies (Diptera: Glossinidae) in Zimbabwe. J Med Entomol. 1993;30: 831–842. 10.1093/jmedent/30.5.831 [DOI] [PubMed] [Google Scholar]
- 11.Mangwiro TNC, Torr SJ, Cox JR, Holloway MTP. The efficacy of various pyrethroid insecticides for use on odour-baited targets to control tsetse. Med Vet Entomol. 1999;13: 315–323. 10.1046/j.1365-2915.1999.00165.x [DOI] [PubMed] [Google Scholar]
- 12.Gikonyo NK, Hassanali A, Njagi PGN, Gitu PM, Midiwo JO. Odor composition of preferred (buffalo and ox) and nonpreferred (waterbuck) hosts of some savanna tsetse flies. J Chem Ecol. Springer Nature; 2002;28: 969–981. 10.1023/a:1015205716921 [DOI] [PubMed] [Google Scholar]
- 13.Gikonyo NK, Hassanali A, Njagi PGN, Saini RK. Responses of Glossina morsitans morsitans to blends of electroantennographically active compounds in the odors of its preferred (buffalo and ox) and nonpreferred (waterbuck) hosts. J Chem Ecol. Springer Nature; 2003;29: 2331–2345. 10.1023/a:1026230615877 [DOI] [PubMed] [Google Scholar]
- 14.Gibson G, Torr SJ. Visual and olfactory responses of haematophagous Diptera to host stimuli. Med Vet Entomol. Wiley-Blackwell; 1999;13: 2–23. 10.1046/j.1365-2915.1999.00163.x [DOI] [PubMed] [Google Scholar]
- 15.Torr SJ, Mangwiro TNC, Hall DR. Responses of Glossina pallidipes to synthetic repellents in the field. Bull Entomol Res. Cambridge University Press (CUP); 1996;86: 609–616. 10.1017/S0007485300039419 [DOI] [Google Scholar]
- 16.Dransfield RD, Brightwell R, Chaudhury MF, Golder TK, Tarimo SAR. The use of odour attractants for sampling Glossina pallidipes Austen(Diptera: Glossinidae) at Nguruman, Kenya. Bull Entomol Res. Cambridge University Press (CUP); 1986;76: 607 10.1017/s000748530001511x [DOI] [Google Scholar]
- 17.Hassanali A, McDowell PG, Owaga MLA, Saini RK. Identification of tsetse attractants from excretory products of a wild host animal, Syncerus caffer. Int J Trop Insect Sci. Cambridge University Press (CUP); 1986;7: 5–9. 10.1017/s1742758400003027 [DOI] [Google Scholar]
- 18.Carey AF, Carlson JR. Insect olfaction from model systems to disease control. Proc Natl Acad Sci. Proceedings of the National Academy of Sciences; 2011;108: 12987–12995. 10.1073/pnas.1103472108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benton R, Vannice KS, Vosshall LB. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature. Springer Nature; 2007;450: 289–293. 10.1038/nature06328 [DOI] [PubMed] [Google Scholar]
- 20.Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant Ionotropic Glutamate Receptors as Chemosensory Receptors in Drosophila. Cell. Elsevier BV; 2009;136: 149–162. 10.1016/j.cell.2008.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su CY, Menuz K. Carlson perception: receptors, cells, and circuits. Transl from eng Cell eng. 2009;139: 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olivier V, Monsempes C, François M-CC, Poivet E, Jacquin-Joly E. Candidate chemosensory ionotropic receptors in a Lepidoptera. Insect Mol Biol. Wiley-Blackwell; 2011;20: 189–199. 10.1111/j.1365-2583.2010.01057.x [DOI] [PubMed] [Google Scholar]
- 23.Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. Public Library of Science (PLoS); 2010;6: e1001064 10.1371/journal.pgen.1001064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacquin-Joly E, Legeai F, Montagné N, Monsempes C, François MC, Poulain J, et al. Candidate chemosensory genes in female antennae of the noctuid moth spodoptera littoralis. Int J Biol Sci. Ivyspring International Publisher; 2012;8: 1036–1050. 10.7150/ijbs.4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogt RG. Biochemical diversity of odor detection-14: OBPs, ODEs and SNMPs. [Internet]. Insect Pheromone Biochemistry and Molecular Biology. 2003. 10.1016/B978-012107151-6/50016-5 [DOI] [Google Scholar]
- 26.Sato K, Pellegrino M, Nakagawa TTT, Nakagawa TTT, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. Springer Nature; 2008;452: 1002–1006. 10.1038/nature06850 [DOI] [PubMed] [Google Scholar]
- 27.Wicher D, Schäfer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, et al. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. Springer Nature; 2008;452: 1007–1011. 10.1038/nature06861 [DOI] [PubMed] [Google Scholar]
- 28.Yao CA. Chemosensory Coding by Neurons in the Coeloconic Sensilla of the Drosophila Antenna. J Neurosci. 2005;25: 8359–8367. 10.1523/JNEUROSCI.2432-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montell C. A taste of the Drosophila gustatory receptors. Curr Opin Neurobiol. 2009;19: 345–353. 10.1016/j.conb.2009.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogers ME, Krieger J, Vogt RG. Antennal SNMPs (sensory neuron membrane proteins) of lepidoptera define a unique family of invertebrate CD36-like proteins. J Neurobiol. Wiley-Blackwell; 2001;49: 47–61. 10.1002/neu.1065 [DOI] [PubMed] [Google Scholar]
- 31.Forstner M, Gohl T, Gondesen I, Raming K, Breer H, Krieger J. Differential Expression of SNMP-1 and SNMP-2 Proteins in Pheromone-Sensitive Hairs of Moths. Chem Senses. Oxford University Press (OUP); 2008;33: 291–299. 10.1093/chemse/bjm087 [DOI] [PubMed] [Google Scholar]
- 32.Gu S-H, Yang R-N, Guo M-B, Wang G-R, Wu K-M, Guo Y-Y, et al. Molecular identification and differential expression of sensory neuron membrane proteins in the antennae of the black cutworm moth Agrotis ipsilon. J Insect Physiol. Elsevier BV; 2013;59: 430–443. 10.1016/j.jinsphys.2013.02.003 [DOI] [PubMed] [Google Scholar]
- 33.Nomura A, Kawasaki K, Kubo T, Natori S. Purification and localization of p10, a novel protein that increases in nymphal regenerating legs of Periplaneta americana (American cockroach). Int J Dev Biol. 1992;36: 391–398. 10.1387/IJDB.1445782 [DOI] [PubMed] [Google Scholar]
- 34.Nomura Kitabayashi A, Arai T, Kubo T, Natori S. Molecular cloning of cDNA for p10, a novel protein that increases in the regenerating legs of Periplaneta americana (American cockroach). Insect Biochem Mol Biol. Elsevier BV; 1998;28: 785–790. 10.1016/s0965-1748(98)00058-7 [DOI] [PubMed] [Google Scholar]
- 35.Ban L, Scaloni A, Brandazza A, Angeli S, Zhang L, Yan Y, et al. Chemosensory proteins of Locusta migratoria. Insect Mol Biol. Wiley-Blackwell; 2003;12: 125–134. 10.1046/j.1365-2583.2003.00394.x [DOI] [PubMed] [Google Scholar]
- 36.Zhou SH, Zhang J, Zhang SG, Zhang L. Expression of chemosensory proteins in hairs on wings of Locusta migratoria (Orthoptera: Acrididae). J Appl Entomol. 2008;132: 439–450. 10.1111/j.1439-0418.2007.01255.x [DOI] [Google Scholar]
- 37.Jacquin-Joly E. Functional and Expression Pattern Analysis of Chemosensory Proteins Expressed in Antennae and Pheromonal Gland of Mamestra brassicae. Chem Senses. Oxford University Press (OUP); 2001;26: 833–844. 10.1093/chemse/26.7.833 [DOI] [PubMed] [Google Scholar]
- 38.IGGI. Genome sequence of the tsetse fly (Glossina morsitans): Vector of African trypanosomiasis. Science (80-). 2014;344: 380–386. 10.1126/science.1249656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obiero GFO, Mireji PO, Nyanjom SRG, Christoffels A, Robertson HM, Masiga DK. Odorant and Gustatory Receptors in the Tsetse Fly Glossina morsitans morsitans. PLoS Negl Trop Dis. 2014;8: e2663 10.1371/journal.pntd.0002663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macharia R, Mireji P, Murungi E, Murilla G, Christoffels A, Aksoy S, et al. Genome-Wide Comparative Analysis of Chemosensory Gene Families in Five Tsetse Fly Species. PLoS Negl Trop Dis. 2016;10: 1–30. 10.1371/journal.pntd.0004421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu R, He X, Lehane S, Lehane M, Hertz-Fowler C, Berriman M, et al. Expression of chemosensory proteins in the tsetse fly Glossina morsitans morsitans is related to female host-seeking behaviour Expression of chemosensory proteins in the tsetse fly Glossina morsitans morsitans is related to female host-seeking behaviour. Insect Mol Biol. 2011;21 (1): 41–48. 10.1111/j.1365-2583.2011.01114.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, et al. Ensembl 2016. Nucleic Acids Res. Oxford University Press (OUP); 2016;44: D710–6 SRC-GoogleScholar FG-0. 10.1093/nar/gkv1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, et al. FlyBase 2.0: The next generation. Nucleic Acids Res. 2019;47: D759–D765. 10.1093/nar/gky1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giraldo-Calderón GI, Emrich SJ, MacCallum RM, Maslen G, Dialynas E, Topalis P, et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. Oxford University Press (OUP); 2014;43: D707–D713. 10.1093/nar/gku1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: A computational tool for the study of gene family evolution. Bioinformatics. 2006;22: 1269–1271. 10.1093/bioinformatics/btl097 [DOI] [PubMed] [Google Scholar]
- 46.Bateta R, Wang J, Wu Y, Weiss BL, Warren WC, Murilla GA, et al. Tsetse fly (Glossina pallidipes) midgut responses to Trypanosoma brucei challenge. Parasit Vectors. BioMed Central; 2017;10: 614 10.1186/s13071-017-2569-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Menuz K, Larter NK, Park J, Carlson JR. An RNA-Seq Screen of the Drosophila Antenna Identifies a Transporter Necessary for Ammonia Detection. PLoS Genet. 2014;10 10.1371/journal.pgen.1004810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dean G, Dame D, Birkenmeyer D. Field cage evaluation of the competitiveness of male Glossina morsitans morsitans Vanderplanz sterilized with tepa or gamma radiation. Bull Entomol Res. 1969;59: 339–344. 10.1017/s0007485300003278 [DOI] [PubMed] [Google Scholar]
- 49.Pilson RD, Pilson BM. Behaviour studies of Glossina morsitans Westw. in the field. Bull Entomol Res. 2009/07/10. Cambridge University Press; 1967;57: 227–257. 10.1017/s0007485300049956 [DOI] [PubMed] [Google Scholar]
- 50.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5: 621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 51.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40: 4288–4297. 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017;45: W130–W137. 10.1093/nar/gkx356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. Elsevier BV; 1990;215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 55.Dębski KJ, Pitkanen A, Puhakka N, Bot AM, Khurana I, Harikrishnan KN, et al. Etiology matters–Genomic DNA Methylation Patterns in Three Rat Models of Acquired Epilepsy. Sci Rep. Springer Nature; 2016;6 10.1038/srep25668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. Oxford University Press (OUP); 2000;28: 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003;13: 2129–2141. 10.1101/gr.772403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. Oxford University Press (OUP); 2015;44: D481–D487. 10.1093/nar/gkv1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stark C. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34: D535–D539. 10.1093/nar/gkj109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.The Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. Oxford University Press (OUP); 2016;45: D331–D338. 10.1093/nar/gkw1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. Oxford University Press (OUP); 2010;26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.R Core Team. A Language and Environment for Statistical. Stat Comput Vienna Austria URL https://wwwRprojectorg. 2017; 900051. citeulike-article-id:2400517
- 64.Von Der Weid B, Rossier D, Lindup M, Tuberosa J, Widmer A, Col JD, et al. Large-scale transcriptional profiling of chemosensory neurons identifies receptor-ligand pairs in vivo. Nat Neurosci. Springer Nature; 2015;18: 1455–1463. 10.1038/nn.4100 [DOI] [PubMed] [Google Scholar]
- 65.Jones WD, Cayirlioglu P, Grunwald Kadow I, Vosshall LB. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. Springer Nature; 2007;445: 86–90. 10.1038/nature05466 [DOI] [PubMed] [Google Scholar]
- 66.Ray A, Van Der Goes Van Naters W, Carlson JR. A regulatory code for neuron-specific odor receptor expression. PLoS Biol. Public Library of Science; 2008;6: 1069–1083. 10.1371/journal.pbio.0060125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Owaga MLA, Hassanali A, McDowell PG. The role of 4-cresol and 3-n-propylphenol in the attraction of tsetse flies to buffalo urine. Insect Sci Appl. 1988th ed. 1988;9: 95–100. 10.1017/S1742758400010110 [DOI] [Google Scholar]
- 68.Torr SJ, Hall DR, Smith JL. Responses of tsetse flies (Diptera: Glossinidae) to natural and synthetic ox odours. Bull Entomol Res. 1995; 10.1017/S000748530005210X [DOI] [Google Scholar]
- 69.Ai M, Min S, Grosjean Y, Leblanc C, Bell R, Benton R, et al. Acid sensing by the Drosophila olfactory system. Nature. 2010/11/17. 2010;468: 691–695. 10.1038/nature09537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Becher PG, Bengtsson M, Hansson BS, Witzgall P. Flying the Fly: Long-range Flight Behavior of Drosophila melanogaster to Attractive Odors. J Chem Ecol. Springer Nature; 2010;36: 599–607. 10.1007/s10886-010-9794-2 [DOI] [PubMed] [Google Scholar]
- 71.Prieto-Godino LL, Rytz R, Bargeton B, Abuin L, Arguello JR, Peraro MD, et al. Olfactory receptor pseudo-pseudogenes. Nature. 2016;539: 93–97. 10.1038/nature19824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Silbering AF, Rytz R, Grosjean Y, Abuin L, Ramdya P, Jefferis GSXE, et al. Complementary Function and Integrated Wiring of the Evolutionarily Distinct Drosophila Olfactory Subsystems. J Neurosci. 2011;31: 13357–13375. 10.1523/JNEUROSCI.2360-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ganguly A, Pang L, Duong V-K, Lee A, Schoniger H, Varady E, et al. A Molecular and Cellular Context-Dependent Role for Ir76b in Detection of Amino Acid Taste. Cell Rep. Elsevier BV; 2017;18: 737–750. 10.1016/j.celrep.2016.12.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Healy TP, Copland MJW. Human sweat and 2-oxopentanoic acid elicit a landing response from Anopheles gambiae. Med Vet Entomol. John Wiley & Sons, Ltd (10.1111); 2000;14: 195–200. 10.1046/j.1365-2915.2000.00238.x [DOI] [PubMed] [Google Scholar]
- 75.Grosjean Y, Farine J-P, Abuin L, Jefferis GSXE, Rytz R, Benton R, et al. An olfactory receptor for food-derived odours promotes male courtship in Drosophila. Nature. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved.; 2011;478: 236–240. 10.1038/nature10428 [DOI] [PubMed] [Google Scholar]
- 76.Liu R, Lehane S, He X, Lehane M, Hertz-Fowler C, Berriman M, et al. Characterisations of odorant-binding proteins in the tsetse fly _Glossina morsitans morsitans_. Cell Mol Life Sci. 2010;67: 919–929. 10.1007/s00018-009-0221-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dweck HKM, Ebrahim SAM, Farhan A, Hansson BS, Stensmyr MC. Olfactory Proxy Detection of Dietary Antioxidants in Drosophila. Curr Biol. Elsevier BV; 2015;25: 1111 10.1016/j.cub.2015.03.042 [DOI] [PubMed] [Google Scholar]
- 78.Montague SA, Mathew D, Carlson JR. Similar Odorants Elicit Different Behavioral and Physiological Responses, Some Supersustained. J Neurosci. 2011;31: 7891–7899. 10.1523/JNEUROSCI.6254-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoare DJ, Humble J, Jin D, Gilding N, Petersen R, Cobb M, et al. Modeling Peripheral Olfactory Coding in Drosophila Larvae. PLoS One. Public Library of Science (PLoS); 2011;6: e22996 10.1371/journal.pone.0022996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ramasamy S, Ometto L, Crava CM, Revadi S, Kaur R, Horner DS, et al. The Evolution of Olfactory Gene Families in Drosophila and the Genomic Basis of chemical-Ecological Adaptation in Drosophila suzukii. Genome Biol Evol. 2016;8: 2297–2311. 10.1093/gbe/evw160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hallem EA, Carlson JR. Coding of Odors by a Receptor Repertoire. Cell. Elsevier BV; 2006;125: 143–160. 10.1016/j.cell.2006.01.050 [DOI] [PubMed] [Google Scholar]
- 82.Kreher SA, Kwon JY, Carlson JR. The molecular basis of odor coding in the Drosophila larva. Neuron. Elsevier; 2005;46: 445–456. 10.1016/j.neuron.2005.04.007 [DOI] [PubMed] [Google Scholar]
- 83.Hall DR, Beevor PS, Cork A, Nesbitt BF, Vale GA. 1-Octen-3-ol. A potent olfactory stimulant and attractant for tsetse isolated from cattle odours. Int J Trop Insect Sci. Cambridge University Press (CUP); 1984;5: 335–339. 10.1017/s1742758400008626 [DOI] [Google Scholar]
- 84.Ni L, Klein M, Svec KV., Budelli G, Chang EC, Ferrer AJ, et al. The ionotropic receptors IR21a and IR25a mediate cool sensing in Drosophila. Calabrese RL, editor. Elife. eLife Sciences Publications, Ltd; 2016;5: e13254 10.7554/eLife.13254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thorne N, Chromey C, Bray S, Amrein H. Taste perception and coding in Drosophila. Curr Biol. Elsevier BV; 2004;14: 1065–1079. 10.1016/j.cub.2004.05.019 [DOI] [PubMed] [Google Scholar]
- 86.Wang Z, Singhvi A, Kong P, Scott K. Taste representations in the Drosophila brain. Cell. 2004;117: 981–991. 10.1016/j.cell.2004.06.011 [DOI] [PubMed] [Google Scholar]
- 87.Knecht ZA, Silbering AF, Cruz J, Yang L, Croset V, Benton R, et al. Ionotropic Receptor-dependent moist and dry cells control hygrosensation in Drosophila. Elife. eLife Sciences Organisation, Ltd.; 2017;6 10.7554/elife.26654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee Y, Kim SH, Montell C. Avoiding DEET through insect gustatory receptors. Neuron. Elsevier BV; 2010;67: 555–561. 10.1016/j.neuron.2010.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moon SJ, Lee Y, Jiao Y, Montell C. A Drosophila Gustatory Receptor Essential for Aversive Taste and Inhibiting Male-to-Male Courtship. Curr Biol. Elsevier BV; 2009;19: 1623–1627. 10.1016/j.cub.2009.07.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Isono. Molecular and cellular designs of insect taste receptor system. Front Cell Neurosci. Frontiers Media SA; 2010; 10.3389/fncel.2010.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Leal WS. Pheromone Reception. Top Curr Chem. 2004;240: 1–36. 10.1007/b98314 [DOI] [Google Scholar]
- 92.Stensmyr MC, Dweck HKM, Farhan A, Ibba I, Strutz A, Mukunda L, et al. A conserved dedicated olfactory circuit for detecting harmful microbes in drosophila. Cell. 2012;151: 1345–1357. 10.1016/j.cell.2012.09.046 [DOI] [PubMed] [Google Scholar]
- 93.Kim H, Jeong YT, Choi MS, Choi J, Moon SJ, Kwon JY. Involvement of a Gr2a-Expressing Drosophila Pharyngeal Gustatory Receptor Neuron in Regulation of Aversion to High-Salt Foods. Mol Cells. Korean Society for Molecular and Cellular Biology; 2017;40: 331–338. 10.14348/molcells.2017.0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stocker RF. Taste perception: Drosophila—A model of good taste. Curr Biol. 2004;14: R560–1. 10.1016/j.cub.2004.07.011 [DOI] [PubMed] [Google Scholar]
- 95.Marella S, Fischler W, Kong P, Asgarian S, Rueckert E, Scott K. Imaging taste responses in the fly brain reveals a functional map of taste category and behavior. Neuron. 2006;49: 285–295. 10.1016/j.neuron.2005.11.037 [DOI] [PubMed] [Google Scholar]
- 96.Lacaille F, Everaerts C, Ferveur J-F. Feminization and Alteration of Drosophila Taste Neurons Induce Reciprocal Effects on Male Avoidance Behavior. Behav Genet. Springer Nature; 2009;39: 554–563. 10.1007/s10519-009-9286-8 [DOI] [PubMed] [Google Scholar]
- 97.Lee Y, Kang MJ, Shim J, Cheong CU, Moon SJ, Montell C. Gustatory Receptors Required for Avoiding the Insecticide L-Canavanine. J Neurosci. 2012;32: 1429–1435. 10.1523/JNEUROSCI.4630-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pregitzer P, Greschista M, Breer H, Krieger J. The sensory neurone membrane protein SNMP1 contributes to the sensitivity of a pheromone detection system. Insect Mol Biol. 2014;23: 733–742. 10.1111/imb.12119 [DOI] [PubMed] [Google Scholar]
- 99.Gomez-Diaz C, Bargeton B, Abuin L, Bukar N, Reina JH, Bartoi T, et al. A CD36 ectodomain mediates insect pheromone detection via a putative tunnelling mechanism. Nat Commun. Springer Nature; 2016;7: 11866 10.1038/ncomms11866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Siciliano P, Scolari F, Gomulski GM, Falchetto M, Manni M, Gabrieli P, et al. Sniffing out chemosensory genes from the Mediterranean fruit fly, Ceratitis capitata. PLoS One. 2014;9: e85523 Available: 10.1371/journal.pone.0085523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Anholt RRH. Functional dissection of Odorant binding protein genes in Drosophila melanogaster. Genes Brain Behav Aug Gene Ontol Consort Expans Gene Ontol knowledgebase Resour Nucleic Acids Res Jan 445D1D331D338. 2011;10: 648–657. 10.1111/j.1601-183X.2011.00704.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gu SH, Sun L, Yang RN, Wu KM, Guo YY, Li XC, et al. Molecular characterization and differential expression of olfactory genes in the antennae of the black cutworm moth Agrotis ipsilon. PLoS One. Public Library of Science (PLoS); 2014;9: e103420 10.1371/journal.pone.0103420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Leal WS. Odorant Reception in Insects: Roles of Receptors, Binding Proteins, and Degrading Enzymes. Annu Rev Entomol. 2013;58: 373–391. 10.1146/annurev-ento-120811-153635 [DOI] [PubMed] [Google Scholar]
- 104.Lin C-CC, Prokop-Prigge KA, Preti G, Potter CJ. Food odors trigger Drosophila males to deposit a pheromone that guides aggregation and female oviposition decisions. Elife. eLife Sciences Organisation, Ltd.; 2015;4 10.7554/eLife.08688.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kagias K, Nehammer C, Pocock R. Neuronal Responses to Physiological Stress. Front Genet. Frontiers Media SA; 2012;3 10.3389/fgene.2012.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Arya GH, Weber AL, Wang P, Magwire MM, Negron YLS, Mackay TFC, et al. Natural Variation, Functional Pleiotropy and Transcriptional Contexts of Odorant Binding Protein Genes in Drosophila melanogaster. Genetics. Genetics Society of America; 2010;186: 1475–1485. 10.1534/genetics.110.123166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zheng W, Peng W, Zhu C, Zhang Q, Saccone G, Zhang H. Identification and expression profile analysis of odorant binding proteins in the oriental fruit fly Bactrocera dorsalis. Int J Mol Sci. 2013;14: 14936–14949. 10.3390/ijms140714936 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Clustering of chemosensory orthologs between tsetse species (B) Clustering of individual orthologs within chemosensory gene families.
(TIF)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All RNA-Seq data files are available from the NCBI database (accession number(s) PRJNA343267 and PRJNA343269).