Abstract
Nuclear genome architecture relies on interactions between the genome and various nuclear scaffolds. One such nuclear scaffold is the nuclear pore complex (NPC), which in addition to its nuclear transport function, can interact with underlying chromatin. In particular, NPCs have been recently reported to associate with a number of enhancers and super-enhancers in metazoan genomes, and select NPC components have been shown to promote the formation of specific genomic loops. Here we provide a brief overview of current models of enhancer function, and discuss recent evidence that NPCs bind enhancers and contribute to topological genome organization. We also examine possible models of how gene and enhancer targeting to NPCs may contribute to tissue-specific genome architecture and expression programs, including the possibility that NPCs may promote phase separation of transcriptional compartments.
Introduction
The complexity of multicellular organisms relies on specific and highly regulated transcriptional programs that control activation of genes in a cell and developmental time dependent manner. To achieve such specificity, gene expression is controlled by the orchestrated action of cis-regulatory elements, such as enhancers, that are located at large distances away from the transcription start sites (TSSs) [1]. Gene expression is also influenced by the nuclear architecture of the genome, or how the genome is folded and arranged inside the nuclear space [2–5]. Nuclear architecture includes both long-range interactions between distant genomic loci and the interactions of loci with nuclear scaffolds and structures. One of the most pronounced nuclear scaffolds is the nuclear envelope (NE), which includes nuclear lamina and nuclear pore complexes (NPCs). NPCs are large protein assemblies, whose classically defined cellular function is to allow and regulate selective nuclear-cytoplasmic transport [6]. NPCs consist of approximately 30 conserved components, termed Nucleoporins (Nups). A susbset of Nups, classified as stable, comprise the NE-embedded NPC core structure, while other Nups, classified as dynamic, associate with the NPC core in an on-and-off manner [7,8]. A recurring protein domain found in the majority of dynamic Nups is the extended array of phenylalanine-glycine (FG) repeats, which is known to mediate the selective permeability transport barrier of NPCs [9–12].
In addition to transport, multiple studies have implicated NPCs in gene regulation via chromatin binding, and thus in functioning as nuclear scaffolds for genome organization [13–15]. As discussed below, recent evidence has also shown that nuclear pore complexes target a number of enhancers and super-enhancers in metazoan genomes [16–18]. These findings have highlighted the notion that NPC-genome interactions contribute to long-range folding of the genome, to enhancer functions and to tissue-specific transcriptional programs. Here we provide a brief overview of the current models of enhancer function, and of the recent evidence that NPCs bind enhancers and participate in nuclear architecture of the genome. We then discuss possible models as to what functional purpose is carried out by gene and enhancer targeting to NPCs.
Mechanisms of genome organization
Enhancers consist of short DNA segments that are found some distance away from promoters and that can direct tissue-specific gene expression via recruitment of sequence-specific transcription factors (TFs) (refs) [19]. In addition to their functional definition, enhancers can often be recognized through a set of chromatin characteristics, such as enrichment of H3K27 acetylation, H3K4 mono-methylation, binding of the p300/CBP histone acetyl transferase and non-promoter binding of RNA Pol II (refs) [1,20,21]. In some cases, multiple enhancers are found in close genomic proximity, resulting in a cluster of closely spaced enhancers that has been termed locus control region (LCR) or super-enhancer (SE). Besides the chromatin characteristics found in enhancers, SEs also show strong enrichment for coactivators, specifically BRD4 and the Mediator complex, and exhibit particularly high levels of H3K27 acetylation (refs) [20,22]. Generally, SEs and LCRs are believed to bring a coalition of transcription factor (TFs) and cofactors to the target promoter, delivering a unique regulatory signature that will determine the appropriate functional outcome for the cell type and the time in development.
Enhancer-promoter communication is encompassed in a higher-order layer of regulation, determined by the organization of the genome in nuclear space. Data from microscopy studies and the development of molecular methods for capturing the spatial organization of the genome, such as genome architecture mapping (GAM) [23] or experiments based on DNA-ligation approaches (chromosome conformation capture (3C) technique and its subsequent maturation) [24], have revealed that the genome is spatially compartmentalized into topologically associated domains (TADs) [25–27]. One of the functional purposes of TADs is believed to be the limitation of enhancer activity to a particular genomic region – such that the activity of enhancers is constrained to cognate promoters within the same TAD, while being insulated from regulatory activity in neighboring domains [28,29]. In this manner, enhancer-promoter loops and other smaller scale loops represent sub-TAD structures, contained within a larger self-interacting TAD. The boundaries of TADs are stabilized by the binding of architectural proteins and by the presence of highly transcribed genes and repetitive elements [30]. The interruption of these boundaries can lead to TAD disorganization and spurious contacts between previously isolated domains, which has been linked to irregular gene expression, developmental pathologies and disease [31,32].
Models of enhancer-promoter communication
How distally located enhancers can convey regulatory information to its promoters has become a topic of intense study. The most established and supported model of enhancer-promoter communication proposes that chromatin loops bring enhancers and promoters into close physical proximity, and that such enhancer looping drives transcriptional activation [2,5,33]. Although put forward decades ago, most of the molecular evidence for this model has come from locus-specific and genome-wide 3C methods. One of many examples of enhancer looping is the mouse β-globin SE, also known as locus control region (LCR), which consists of four regions with enhancer activity [34]. The LCR is located 50 kb away from the β-globin gene cluster and can loop over to contact its target promoters in erythroid cells (where the β-globin gene is active) but shows no interaction with the promoters in cells from other lineages [35]. Importantly, loop formation appears to be sufficient to activate transcription, as demonstrated by force-tethering the self-association domain of the transcriptional cofactor Ldb1 to the β-globin promoter, which induced both looping of LCR and transcriptional activation in pro-erythroblast cells [36]. Recent studies have also shown that individual elements of the β-globin SE can aggregate to form a hub that can accommodate multiple loops or target promoters simultaneously [37]. Enhancer-promoter looping has also been observed for a variety of active genes controlling a wide aspect of biological processes, such as the CFTR gene, which codes for cystic fibrosis transmembrane protein, [38] or the proto-oncogene c-Myc locus [39], among many others.
On the other hand, recent evidence has challenged the looping model as the only model to explain communication between enhancers and promoters. This evidence has come primarily from imaging of in situ labeled gene loci, using fixed or live microscopy. For example, induction of the sonic hedgehog morphogen (Shh) gene in neural progenitor cells is controlled by the action of distal brain enhancers that appear to move further away instead of closer to the activated Shh gene [40]. The authors propose that instead of looping over, the Shh enhancers may drive regional chromatin decompaction to create an active environment. Interestingly, this regulatory method seems to be cell type specific, since expression of Shh in the limb buds is regulated by a distant limb enhancer that does appear to loop out the intervening chromatin to contact its promoter [41]. Similarly, the spatial organization of the key pluripotency regulator Sox2 and its essential Sox2 Control Region (SCR) SE was investigated using live-cell microscopy in mouse embryonic stem cells (mESCs) [42]. In this study, Sox2 and SCR also show no evidence of increased spatial proximity upon activation, suggesting that newly formed enhancer-promoter contacts do not drive Sox2 transcription. Furthermore, a genome-wide detection of distal promoter-interacting regions in multiple cell lines revealed that while some enhancer-promoter loops are very dynamic in response to lineage commitment signals, other enhancer contacts remain stable during cell differentiation [43]. In agreement with this observation, 4C experiments in Drosophila embryos suggested that transcriptional changes during Drosophila development mainly occur in the context of pre-formed chromatin loops [44]. These data indicate that the enhancer-promoter looping event does not always result in concurrent gene activation, but instead may be formed beforehand to allow subsequent transcription or as the Shh study suggests [40], may not be a continuous requirement for transcription.
To reconcile these conflicting conclusions on the significance of enhancer looping, one intriguing scenario that has been proposed is a situation where enhancers such as SCR establish a large isolated compartment for transcription [33,40,42]. Within such compartments, enhancers and promoters are engaged in preferential communication but do not have to be in constant physical contact. This model is supported by recent studies, which propose that coactivators form phase-separated condensates at SEs of key cell-identity genes [45–48]. Such phase-separated regulatory clusters are thought to be sustained by cooperative interactions between enhancers’ bound factors and the transcriptional machinery. These clusters exhibit properties of selectively permeable liquid droplets, resulting in spatial compartmentalization that assures the concentration of the transcriptional apparatus, thus providing robust transcription. Supporting the idea of transcriptional phase separation, these studies demonstrate that the low complexity domains (LCDs) of TFs can phase separate with co-activators frequently found at SEs, such as the Mediator complex and BRD4 [45,47,48]. It has been suggested that multiple properties of SEs can be successfully explained by the phase separation model [49].
Enhancers at NPCs and the roles of Nups in nuclear architecture
Multiple studies over the last two decades have uncovered functional roles of Nups in gene regulation via chromatin binding [50]. Recently, several reports have demonstrated that enhancers and SEs are frequently targeted to nuclear pores [16–18], and that NPC components contribute to formation of long-range genomic contacts [16,51–53]. In Drosophila, comparison of Nup ChIP-seq datasets to distribution of the H3K27acetyl mark and to genome-wide enhancer maps [54] have revealed the targeting of Nups to a subset of promoters and enhancers in multiple cell types [16]. In particular, a dynamic NPC component Nup98 was found to bind promoters and enhancers of Hox genes and of genes induced by a steroid hormone ecdysone [16]. In mammalian cells, two separate reports have similarly identified association of SEs and LCRs with NPCs and specific Nups [17,18]. First, DamID mapping of stable Nup93 and dynamic Nup153 revealed extensive binding of these Nups to tissue-specific SEs in human cells [18]. For instance, in U2OS cells, nearly half of the classified SEs were found to be targeted by Nup153. Depletion of Nup153 resulted in gene expression changes that were particularly severe for SE-regulated targets, providing evidence for the potential functionality of this binding [18].
Second, a dCas9-based proteomics approach, which aimed to identify protein composition of regulatory elements via sequence-specific guide RNAs, has revealed multiple components of the NPC as highly abundant at LCR enhancers of the human β-globin gene cluster [17]. This approach yielded several known regulators of enhancer function, such as GATA1 and chromatin remodeling complexes, but also the dynamic Nups discussed above, Nup98 and Nup153. Targeting of Nup153 and Nup98 to LCR enhancers was further confirmed by ChIP-seq analysis of both Nups, and depletion of either Nup lead to a down-regulation of globin genes in primary erythroid cells. Nup98 and Nup153 were also found to be highly enriched at multiple erythroid SEs [17], providing further evidence for the frequent association between SEs and Nups. Together, these studies suggest that complex enhancer landscapes such as those in LCRs, SEs and Hox gene clusters, are preferentially associated with nuclear pores.
Although Nup153 and Nup98 can interact with chromatin at the NPC or in the nuclear interior [55–58], the Nup-enhancer contacts were found to preferentially occur at the NE-embedded NPCs [16,18]. For both mammalian Nup153-SE contacts and Drosophila Nup98-enhancer contacts, this was demonstrated by FISH and supported by chromatin binding of a stable NE-embedded Nup, Nup93. Interestingly, Nup98 targeting to promoters and enhancers of ecdysone-inducible genes occurred at the NPC regardless of the transcriptional state of these genes [16]. Similarly, the mammalian HoxA cluster was found to exhibit ChIP signal of Nup93 in cells where HoxA is normally silenced, and this localization was found to depend on Nup93 [59,60]. These findings suggest that NPCs can target silent or poised genes and enhancers, and highlight the role of NPCs as stable binding scaffolds for developmentally regulated targets.
The recurrent detection of enhancer targeting has suggested architectural roles for Nups, and multiple lines of evidence have now demonstrated a functional relationship between Nups and topological looping, particularly at the more local sub-TAD level. Depletion of Drosophila Nup98 was found to destabilize enhancer-promoter loops that are induced by addition of ecdysone [16]. This functional role of Nup98 in loop formation parallels similar findings in yeast, where transcriptional 5’−3’ looping of galactose-inducible genes was found to depend on another NPC component Mlp1/2 [53,61]. Moreover, inter-chromosomal clustering and long-range interactions between distant co-induced genes were found to involve binding to the NPCs [51,52]. Interestingly, in the fly system, Nup98 and NPC targeting have also been repeatedly linked to boundary elements. Genome-wide studies have revealed enrichment of DNA binding motifs of insulator proteins among NPC-genome contacts [62] and a high level of ChIP-seq overlap as well as a physical interactions between Nup98 and insulator proteins such as CTCF and Su(Hw) [16]. Genome-wide comparison of various ChIP-seq datasets to Hi-C data has identified Nup98 as one of the factors enriched at the bases of sub-TAD architectural loops containing enhances in fly cells [63]. Furthermore, Nup98 binding was also observed at a subset of TAD boundary regions, suggesting that association with nuclear pore proteins may result in stronger boundaries [64]. These reports support the notion that NPCs function in architectural folding of the genome, perhaps especially in the establishment of sub-TAD structures or certain TAD boundaries, and contribute to formation and stabilization of enhancer-promoter loops.
Nuclear pores as hubs of transcriptional regulation – possible models
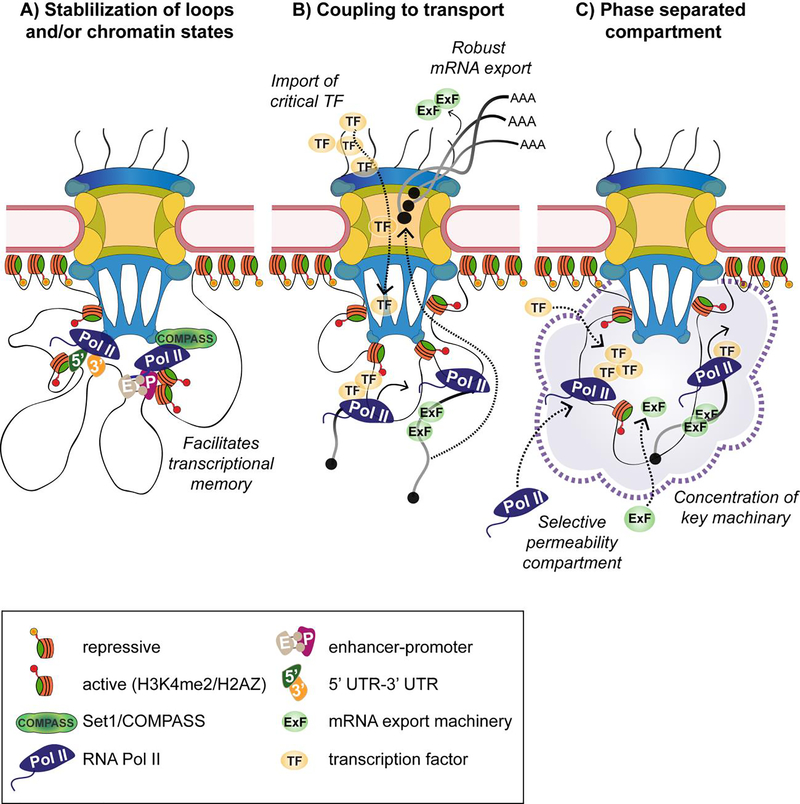
The findings described above demonstrate that nuclear pore proteins are frequently found at critical enhancer elements, and that select Nups contribute to expression and architectural states of their target genes. Based on this, we hypothesize 3 possible, non-mutually exclusive models for the chromatin-binding function of NPCs (Figure 1). These models offer potential cellular reasons for the involvement of nuclear pores in enhancer regulation, genome architecture and epigenetic maintenance of gene expression programs.
Figure 1. Possible non-mutually exclusive models for the genome-binding functions of NPCs.
A) Binding of inducible genes and their promoter and enhancer elements to the NPCs facilitates epigenetic maintenance of the activated state by stabilizing histone modifications, binding of the COMPASS complex and enhancer-promoter loops. In this manner, the NPC may function as a nuclear compartment/complex that aids in the interplay between these processes to promote transcriptional memory. B) Targeting active genes and enhancer units to NPCs may be coupled to transport-related functions of NPCs. Such transport-related functions may include the connection between transcription and mRNA export, to streamline the generation of mature mRNA, or the connection between the import of critical transcription factors and their targeting to NPC-bound genes, to promote the efficiency of gene targeting. C) Phase-separating properties of FG Nups at NPCs may promote phase separated transcriptional compartments at enhancers and super-enhancers and their target genes. Such compartments may help concentrate key chromatin and transcriptional regulators, and/or create isolated selectively permeable hubs, using known functions of FG Nups in regulation of selective permeability.
Epigenetic stabilization of chromatin loops and states by Nups
NPC targeting and Nup98 in particular have been repeatedly linked to the phenomenon of transcriptional memory. Transcriptional memory or priming describes the enhanced response of previously activated genes to later rounds of activation, after a period of repression [65,66]. This ability of genes to respond more rapidly or robustly upon repeated exposure can be transmitted through cell divisions, i.e. epigenetically. Mechanistically, transcriptional memory has been shown to depend on a number of cellular players, but Nup98 has emerged as an evolutionary conserved factor required for transcriptional memory, necessary for priming of diverse inducible genes in yeast, flies and mammals [16,67]. As in other systems, Nup98 was found to be required for transcriptional memory of ecdysone-inducible genes in Drosophila culture cells [16]. Namely, depletion of Nup98 did not affect the transcriptional activation dynamics during initial exposure to ecdysone, but resulted in a less robust transcriptional response upon re-exposure. At the same time, Nup98 depletion also resulted in the loss of enhancer-promoter loops that were induced by initial ecdysone exposure [16]. This suggests that enhancer-promoter loops at ecdysone-inducible genes are not linked to concurrent transcriptional output, but instead may form as part of a memory complex to mark genes as recently activated. In support of this notion, enhancer-promoter loops and enhanced interactions of Nup98 with architectural proteins, induced by initial ecdysone exposure, were found to persist through the period of transcriptional repression [16].
These findings are also in line with previous reports, discussed above, which show that some enhancer-promoter loops do not correlate with transcriptional output. Thus, one possibility is that Nup98 and NPC targeting function in stabilization of enhancer-promoter loops, and that this stabilization is part of the overall process of epigenetic transcriptional memory that allows for more robust or more coordinated transcriptional responses in the future (Figure 1A). Another key pathway implicated in transcriptional memory is deposition of H3K4 methylation, particularly di-methylation (H3K4Me2), through the action of the conserved histone methylase COMPASS [67,68]. Transcriptional memory has been shown to rely on the deposition of H3K4Me2 mark at primed genes [67] and metazoan Nup98 homologues have been found to interact with H3K4 methylases Set1, MLL1 and Trithorax (Trx) [69–71]. It is currently unclear how H3K4 methylation and enhancer looping are integrated with each other, but both have been shown to depend on Nup98 and to function as components of epigenetic memory. In this manner, cells may utilize genome targeting to the NPC via Nup98 for assembly of a transcriptional memory complex, which includes a specialized chromatin and architectural state (Figure 1A).
Coupling of genome binding to nuclear transport
One of the initial ideas for the function of NPC-gene interactions was the “gene gating hypothesis”, which postulated that genes may be targeted to nuclear pores to couple the production and export of mRNAs [72]. This hypothesis has received biochemical support when the NPC-associated mRNA export complex TREX-2 was discovered to share a component, Sus1, with the transcriptional activator SAGA complex [73], and to physically interact with the Mediator complex [74]. NPC components are also involved in multiple steps of mRNA maturation, and in addition to export factors, interact with the quality control machinery that ensures retention of intron-containing transcripts [75]. On the other hand, it is difficult to explain how targeting enhancers, SEs and boundary elements to NPCs would integrate with mRNA export. Nonetheless, one function of gene targeting to the NPCs may be to streamline the mRNA biogenesis process by coupling multiple steps in transcription, chromatin regulation, mRNA processing and eventually, export (Figure 1B). In this manner, the reported interaction between Nups and the Mediator complex may play a role in both the stabilization of enhancer-promoter loops and the export of resulting transcripts.
Another possible transport-related role for NPC-genome contacts is the connection to import of critical transcription factors (Figure 1B). Multiple transcription factors in yeast and metazoan systems have been shown to physically interact with NPC components or to mediate the targeting of specific genes to the NPC [76–78]. The ecdysone receptor (EcR), for instance, is thought to undergo nuclear translocation upon activation [79]. Reported binding of poised EcR-regulated genes at the NPC [16] may promote rapid targeting of EcR to its target genes upon nuclear entry. Another NPC component, the transmembrane Nup210, was recently shown to associate with a key transcriptional regulator of myoblast differentiation, Mef2C, and to physically bind its gene targets [77]. As Nup210 is required for muscle differentiation, this mechanism suggests that NPC-bound Nup210 functions to promote recruitment of Mef2C to its target genes to drive differentiation. And although no direct evidence exists for the functional coupling of import and gene binding at the NPC, it remains an intriguing possibility.
NPCs as phase-separated compartments for gene expression
One of the critical functions of FG domain-containing Nups (FG Nups) is to selectively regulate transport between the nucleus and the cytoplasm. The FG domains are LCD domains as they are highly repetitive, intrinsically disordered and show high conformational flexibility [12,80]. Extensive FG repeats are found in most dynamics Nups, including those located at the cytoplasmic fibrils, nuclear basket, and the nuclear pore channel. Selective transport is mediated through dynamic contacts between nuclear transport receptors and FG motifs [9,80]. Consistently with the intrinsically disordered nature of FG domains, higher densities of FG domains can phase separate in vitro, and the formation of FG hydrogels can recapitulate some of the selective transport properties of NPCs [9,80,81]. Both Nup98 and Nup153, discussed above, contain lengthy FG domains, and the FG domains of Nup98 homologs have been shown to phase separate from relatively dilute solutions [82].
Given these biochemical properties of FG Nups, and the recent evidence for the existence of SEs in phase separated compartments, discussed above, it is tempting to speculate that NPCs may help create a phase-separated compartment for SEs and other enhancer clusters (Figure 1C). NPCs can provide a high local density of FG domains that can seed or promote the formation of phase-separated environments around bound SEs and gene-enhancer clusters. This may be facilitated by the conserved interactions between FG-containing Nups and key regulators such as the Mediator complex, architectural proteins and Set1/Trx homologues. Such Nup-generated compartments may provide a way to concentrate regulatory molecules, promote an isolated environment for tighter transcriptional control and delimit a space for enhancer-promoter interactions. This phase-separating function of Nups is perhaps easiest to envision occurring at the actual NPCs, which offer a high local concentration of FG domains, but could theoretically also occur at nucleoplasmic binding sites of FG Nups such as Nup98 and Nup153. Moreover, such local environments can benefit from the function of Nups in regulation of selective permeability. It is conceivable that much like at the NPC transport channel, FG Nups may help set up a selectively permeable barrier to the entry of transcriptional and chromatin regulators. Currently, there is no direct evidence that phase-separating or permeability-regulating properties of Nups contribute to their transcriptional roles. But future experiments can address this question as well as the other models both in vivo and in vitro, elucidating the mechanistic role of the NPC in genome control.
Acknowledgements
We thank members of the Capelson lab, as well as labs of Raj Jain and Shawn Little, for helpful scientific discussions. We apologize to colleagues in the field whose work could not be cited due to space limitations. M.C. is supported by the Research Scholar Grant RSG-15-159-01-CSM from the American Cancer Society, and by the NIH R01GM124143 grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
*of special interest
**of outstanding interest
- 1.Ong CT, Corces VG: Enhancers: emerging roles in cell fate specification. EMBO Rep 2012, 13:423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vermunt MW, Zhang D, Blobel GA: The interdependence of gene-regulatory elements and the 3D genome. J Cell Biol 2019, 218:12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dekker J, Mirny L: The 3D Genome as Moderator of Chromosomal Communication. Cell 2016, 164:1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonev B, Cavalli G: Organization and function of the 3D genome. Nat Rev Genet 2016, 17:772. [DOI] [PubMed] [Google Scholar]
- 5.Dekker J, Misteli T: Long-Range Chromatin Interactions. Cold Spring Harb Perspect Biol 2015, 7:a019356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Angelo MA, Hetzer MW: Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol 2008, 18:456–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capelson M, Doucet C, Hetzer MW: Nuclear pore complexes: guardians of the nuclear genome. Cold Spring Harb Symp Quant Biol 2011, 75:585–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rabut G, Doye V, Ellenberg J: Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol 2004, 6:1114–1121. [DOI] [PubMed] [Google Scholar]
- 9.Xu S, Powers MA: In vivo analysis of human nucleoporin repeat domain interactions. Mol Biol Cell 2013, 24:1222–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frey S, Gorlich D: A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 2007, 130:512–523. [DOI] [PubMed] [Google Scholar]
- 11.Strawn LA, Shen T, Shulga N, Goldfarb DS, Wente SR: Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat Cell Biol 2004, 6:197–206. [DOI] [PubMed] [Google Scholar]
- 12.Denning DP, Patel SS, Uversky V, Fink AL, Rexach M: Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc Natl Acad Sci U S A 2003, 100:2450–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raices M, D’Angelo MA: Nuclear pore complexes and regulation of gene expression. Curr Opin Cell Biol 2017, 46:26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sood V, Brickner JH: Nuclear pore interactions with the genome. Curr Opin Genet Dev 2014, 25:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ptak C, Aitchison JD, Wozniak RW: The multifunctional nuclear pore complex: a platform for controlling gene expression. Curr Opin Cell Biol 2014, 28:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pascual-Garcia P, Debo B, Aleman JR, Talamas JA, Lan Y, Nguyen NH, Won KJ, Capelson M: Metazoan Nuclear Pores Provide a Scaffold for Poised Genes and Mediate Induced Enhancer-Promoter Contacts. Mol Cell 2017, 66:63–76 e66* Here we report the prevalence of Nups, particularly Nup98, at promoters and enhancers of the Drosophila genome, and demonstrate a functional requirement of Nup98 in enhancer-promoter loops of ecdysone-inducible genes. Significantly, the Nup98-stabilized enhancer-promoter loops occur independently of concurrent transcription and instead contribute to epigenetic memory of activating events.
- 17.Liu X, Zhang Y, Chen Y, Li M, Zhou F, Li K, Cao H, Ni M, Liu Y, Gu Z, et al. : In Situ Capture of Chromatin Interactions by Biotinylated dCas9. Cell 2017, 170:1028–1043 e1019.** This study reports an unbiased identfication of chromatin factors associated with enhancers, using guide RNAs to specific regulatory elements and dCas9, coupled to proteomics, to capture these interactions. Using this approach, the authors identify several Nups, including Nup98 and Nup153, as highly abundant at LCR enhancers and super-enhancers in erythroid cells.
- 18.Ibarra A, Benner C, Tyagi S, Cool J, Hetzer MW: Nucleoporin-mediated regulation of cell identity genes. Genes Dev 2016, 30:2253–2258.* The authors analyze genome-wide binding of Nups in several human cell types, and report recurrent binding of Nup153 to a large number of cell type specific super-enhancers (SEs) in humal cells. These interactions preferentially occur at the NPCs in the nuclear periphery, and Nup 153 is required for expression of SEs-directed genes.
- 19.Spitz F, Furlong EE: Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 2012, 13:613–626. [DOI] [PubMed] [Google Scholar]
- 20.Heinz S, Romanoski CE, Benner C, Glass CK: The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 2015, 16:144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calo E, Wysocka J: Modification of enhancer chromatin: what, how, and why? Mol Cell 2013, 49:825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA: Super-enhancers in the control of cell identity and disease. Cell 2013, 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beagrie RA, Scialdone A, Schueler M, Kraemer DC, Chotalia M, Xie SQ, Barbieri M, de Santiago I, Lavitas LM, Branco MR, et al. : Complex multi-enhancer contacts captured by genome architecture mapping. Nature 2017, 543:519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. : Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G: Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 2012, 148:458–472. [DOI] [PubMed] [Google Scholar]
- 26.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. : Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 2012, 485:381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B: Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowley MJ, Corces VG: Organizational principles of 3D genome architecture. Nat Rev Genet 2018, 19:789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hnisz D, Day DS, Young RA: Insulated Neighborhoods: Structural and Functional Units of Mammalian Gene Control. Cell 2016, 167:1188–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou C, Li L, Qin ZS, Corces VG: Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell 2012, 48:471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun JH, Zhou L, Emerson DJ, Phyo SA, Titus KR, Gong W, Gilgenast TG, Beagan JA, Davidson BL, Tassone F, et al. : Disease-Associated Short Tandem Repeats Co-localize with Chromatin Domain Boundaries. Cell 2018, 175:224–238 e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spielmann M, Lupianez DG, Mundlos S: Structural variation in the 3D genome. Nat Rev Genet 2018, 19:453–467. [DOI] [PubMed] [Google Scholar]
- 33.Furlong EEM, Levine M: Developmental enhancers and chromosome topology. Science 2018, 361:1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dean A: On a chromosome far, far away: LCRs and gene expression. Trends Genet 2006, 22:38–45. [DOI] [PubMed] [Google Scholar]
- 35.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W: Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell 2002, 10:1453–1465. [DOI] [PubMed] [Google Scholar]
- 36.Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA: Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 2012, 149:1233–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allahyar A, Vermeulen C, Bouwman BAM, Krijger PHL, Verstegen M, Geeven G, van Kranenburg M, Pieterse M, Straver R, Haarhuis JHI, et al. : Enhancer hubs and loop collisions identified from single-allele topologies. Nat Genet 2018, 50:1151–1160* Here the authors report the multi-contact 4C (MC-4C) method, which is able to detect multi-way DNA contacts. Using this approach, they show that the β-globin super-enhancer can accommodate more than one gene simultaneously, uncovering a more detailed picture of the architectural properties of super-enhancers.
- 38.Ott CJ, Blackledge NP, Kerschner JL, Leir SH, Crawford GE, Cotton CU, Harris A: Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc Natl Acad Sci U S A 2009, 106:19934–19939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe JS, Minder JL, Mercan F, Wang E, Eckersley-Maslin MA, et al. : Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev 2013, 27:2648–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benabdallah NS, Williamson I, Illingworth RS, Boyle S, Grimes GR, Therizols P, Bickmore WA: PARP mediated chromatin unfolding is coupled to long-range enhancer activation. bioRxiv 2017. doi: 10.1101/155325. [DOI] [Google Scholar]
- 41.Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T: Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev Cell 2009, 16:47–57. [DOI] [PubMed] [Google Scholar]
- 42.Alexander JM, Guan J, Huang B, Lomvardas S, Weiner OD: Live-Cell Imaging Reveals Enhancer-dependent Sox2 Transcription in the Absence of Enhancer Proximity. bioRxiv 2018. doi: 10.1101/409672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rubin AJ, Barajas BC, Furlan-Magaril M, Lopez-Pajares V, Mumbach MR, Howard I, Kim DS, Boxer LD, Cairns J, Spivakov M, et al. : Lineage-specific dynamic and pre-established enhancer-promoter contacts cooperate in terminal differentiation. Nat Genet 2017, 49:1522–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghavi-Helm Y, Klein FA, Pakozdi T, Ciglar L, Noordermeer D, Huber W, Furlong EE: Enhancer loops appear stable during development and are associated with paused polymerase. Nature 2014, 512:96–100. [DOI] [PubMed] [Google Scholar]
- 45.Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. : Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361* This study, together with two other studies published concurrently (46 and 47), describe in vivo presence of phase-separated transcriptional condensates that are driven by the interactions of low complexity protein domains (LCD) of transcriptional regulators. Here, the authors characterize MED1 and BRD4 LCDs that can form liquid-like droplets in vitro and form discrete nuclear bodies that appear to form around super-enhancers. The authors suggest that these phase-separated condensates around super-enhacers facilitate compartimentalization of transcriptional hubs.
- 46.Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, Cattoglio C, Heckert A, Banala S, Lavis L, Darzacq X, et al. : Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 2018, 361. * [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V, Cisse, II: Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 2018, 361:412–415.* [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, et al. : Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 2018, 175:1842–1855 e1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA: A Phase Separation Model for Transcriptional Control. Cell 2017, 169:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buchwalter A, Kaneshiro JM, Hetzer MW: Coaching from the sidelines: the nuclear periphery in genome regulation. Nat Rev Genet 2019, 20:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brickner DG, Sood V, Tutucci E, Coukos R, Viets K, Singer RH, Brickner JH: Subnuclear positioning and interchromosomal clustering of the GAL1–10 locus are controlled by separable, interdependent mechanisms. Mol Biol Cell 2016, 27:2980–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brickner DG, Brickner JH: Interchromosomal clustering of active genes at the nuclear pore complex. Nucleus 2012, 3:487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan-Wong Sm, Wijayatilake HD, Proudfoot NJ: Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev 2009, 23:2610–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shlyueva D, Stelzer C, Gerlach D, Yanez-Cuna JO, Rath M, Boryn LM, Arnold CD, Stark A: Hormone-responsive enhancer-activity maps reveal predictive motifs, indirect repression, and targeting of closed chromatin. Mol Cell 2014, 54:180–192. [DOI] [PubMed] [Google Scholar]
- 55.Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW: Dynamic association of NUP98 with the human genome. PLoS Genet 2013, 9:e1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A: Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet 2010, 6:e1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kalverda B, Pickersgill H, Shloma VV, Fornerod M: Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 2010, 140:360–371. [DOI] [PubMed] [Google Scholar]
- 58.Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW: Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 2010, 140:372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Labade AS, Karmodiya K, Sengupta K: HOXA repression is mediated by nucleoporin Nup93 assisted by its interactors Nup188 and Nup205. Epigenetics Chromatin 2016, 9:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA: Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev 2008, 22:627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hampsey M, Singh BN, Ansari A, Laine JP, Krishnamurthy S: Control of eukaryotic gene expression: gene loops and transcriptional memory. Adv Enzyme Regul 2011, 51:118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalverda B, Fornerod M: Characterization of genome-nucleoporin interactions in Drosophila links chromatin insulators to the nuclear pore complex. Cell Cycle 2010, 9:4812–4817. [DOI] [PubMed] [Google Scholar]
- 63.Cubenas-Potts C, Rowley MJ, Lyu X, Li G, Lei EP, Corces VG: Different enhancer classes in Drosophila bind distinct architectural proteins and mediate unique chromatin interactions and 3D architecture. Nucleic Acids Res 2017, 45:1714–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramirez F, Bhardwaj V, Arrigoni L, Lam KC, Gruning BA, Villaveces J, Habermann B, Akhtar A, Manke T: High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun 2018, 9:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.D’Urso A, Brickner JH: Epigenetic transcriptional memory. Curr Genet 2017, 63:435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brickner JH: Transcriptional memory at the nuclear periphery. Curr Opin Cell Biol 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Light WH, Freaney J, Sood V, Thompson A, D’Urso A, Horvath CM, Brickner JH: A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol 2013, 11:e1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.D’Urso A, Takahashi YH, Xiong B, Marone J, Coukos R, Randise-Hinchliff C, Wang JP, Shilatifard A, Brickner JH: Set1/COMPASS and Mediator are repurposed to promote epigenetic transcriptional memory. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Franks TM, McCloskey A, Shokirev MN, Benner C, Rathore A, Hetzer MW: Nup98 recruits the Wdr82-Set1A/COMPASS complex to promoters to regulate H3K4 trimethylation in hematopoietic progenitor cells. Genes Dev 2017, 31:2222–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu H, Valerio DG, Eisold ME, Sinha A, Koche RP, Hu W, Chen CW, Chu SH, Brien GL, Park CY, et al. : NUP98 Fusion Proteins Interact with the NSL and MLL1 Complexes to Drive Leukemogenesis. Cancer Cell 2016, 30:863–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pascual-Garcia P, Jeong J, Capelson M: Nucleoporin Nup98 associates with Trx/MLL and NSL histone-modifying complexes and regulates Hox gene expression. Cell Rep 2014, 9:433–442. [DOI] [PubMed] [Google Scholar]
- 72.Blobel G: Gene gating: a hypothesis. Proc Natl Acad Sci U S A 1985, 82:8527–8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E: Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 2004, 116:75–86. [DOI] [PubMed] [Google Scholar]
- 74.Schneider M, Hellerschmied D, Schubert T, Amlacher S, Vinayachandran V, Reja R, Pugh BF, Clausen T, Kohler A: The Nuclear Pore-Associated TREX-2 Complex Employs Mediator to Regulate Gene Expression. Cell 2015, 162:1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bonnet A, Palancade B: Intron or no intron: a matter for nuclear pore complexes. Nucleus 2015, 6:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Su Y, Pelz C, Huang T, Torkenczy K, Wang X, Cherry A, Daniel CJ, Liang J, Nan X, Dai MS, et al. : Post-translational modification localizes MYC to the nuclear pore basket to regulate a subset of target genes involved in cellular responses to environmental signals. Genes Dev 2018, 32:1398–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Raices M, Bukata L, Sakuma S, Borlido J, Hernandez LS, Hart DO, D’Angelo MA: Nuclear Pores Regulate Muscle Development and Maintenance by Assembling a Localized Mef2C Complex. Dev Cell 2017, 41:540–554 e547.* This study provides a mechanistic analyis for the role of Nup210 in muscle differentiation in mammalian myoblast differentiation system and in the zebrafish model. Importantly, the authors find a requirement of Nup210 in the NPC-coupled assembly of the Mef2C transcriptional complex at the Mef2C target genes, which is necessary for the muscle-specific gene expression program.
- 78.Randise-Hinchliff C, Coukos R, Sood V, Sumner MC, Zdraljevic S, Meldi Sholl L, Garvey Brickner D, Ahmed S, Watchmaker L, Brickner JH: Strategies to regulate transcription factor-mediated gene positioning and interchromosomal clustering at the nuclear periphery. J Cell Biol 2016, 212:633–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johnston DM, Sedkov Y, Petruk S, Riley KM, Fujioka M, Jaynes JB, Mazo A: Ecdysone- and NO-mediated gene regulation by competing EcR/Usp and E75A nuclear receptors during Drosophila development. Mol Cell 2011, 44:51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Milles S, Huy Bui K, Koehler C, Eltsov M, Beck M, Lemke EA: Facilitated aggregation of FG nucleoporins under molecular crowding conditions. EMBO Rep 2012, 14:178–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frey S, Rees R, Schunemann J, Ng SC, Funfgeld K, Huyton T, Gorlich D: Surface Properties Determining Passage Rates of Proteins through Nuclear Pores. Cell 2018, 174:202–217 e209. [DOI] [PubMed] [Google Scholar]
- 82.Schmidt HB, Gorlich D: Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity. Elife 2015, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]