Abstract
The abundance of lipopolysaccharide (LPS) in house dust mite (HDM) preparations is broad and mirrors the variability seen in the homes of people with asthma. LPS in commercially available stocks ranges from 31 to 5,2000 endotoxin units. The influence of vastly different LPS loads on the mechanisms that define the immune and inflammatory phenotype of HDM-challenged mice has not been defined. This aim of the study was to understand the lung phenotype of mice challenged with HDM extract containing high or low levels of LPS. Female BALB/c mice were sensitized for 2 wk with commercial HDM extract containing either high (36,000 endotoxin units; HHDM) or low (615 endotoxin units; LHDM) levels of LPS. Lung phenotype was characterized by measuring lung function, total and differential cell counts, cytokine abundance, and the lung transcriptome by RNA-sequencing. LPS levels in HDM stocks used for preclinical asthma research in mice remain poorly reported. In 2019, only 14% of papers specified LPS concentration in HDM lots. Specific differences existed in airway responsiveness between mice challenged with HHDM or LHDM. HHDM- and LHDM-induced cytokine profiles of bronchial lavage were significantly different and the lung transcriptome was differentially enriched for genes involved in DNA damage repair or cilium movement, following HHDM or LHDM challenge, respectively. The abundance of LPS in commercially available HDM influences the phenotype of allergic airways inflammation in mice. Failure to report the level of LPS in HDM extracts used in animal models of airway disease will lead to inconsistency in reproducibility and reliability of published data.
Keywords: asthma, endotoxin, house dust mite, lipopolysaccharide, lung, RNA-seq
INTRODUCTION
Asthma is a chronic inflammatory lung disease that is characterized by variable airflow limitation, airway hyperresponsiveness, and airway remodeling (3). To recapitulate the complexities of asthma pathobiology, researchers use allergen-challenged animal models to unravel mechanisms and pathophysiological features in preclinical studies that are needed for the development of novel therapeutics (27). Several allergens are commonly used to induce airway inflammation and responsiveness in mice including ovalbumin (OVA), house dust mite (HDM) extract, and cockroach antigens. HDM (Dermatophagoides pteronyssinus or D. farina) is favored for preclinical mouse models of asthma due to its relevance as a human aeroallergen for asthma [~90% of people with asthma are sensitized to HDM (25)]. Moreover, mice readily develop allergic airway inflammation to inhaled or nasally instilled HDM without the need for adjuvants.
In mice, acute, repeated challenge with HDM induces significant eosinophilic and neutrophilic airway inflammation, with increased airway responsiveness to methacholine (6, 18). Chronic HDM challenge (5–8 wk) leads to inflammation and significant airway remodeling that mimics the human asthma (15). The composition of the HDM extract used for challenge is important, as proteases contribute to epithelial barrier dysfunction that leads to increased sensitivity of airways to spasmogens (19). Endotoxin or bacterial LPS is a component of HDM known to mediate proinflammatory responses via the Toll-like receptor innate immune signaling pathway, and correlated with asthma risk and severity (7, 22). Higher household LPS loads are associated with lower forced expiratory volume in 1 s (FEV1)/forced vital capacity (FVC) in asthmatics (5). For OVA sensitization mouse models of allergic asthma, spiking OVA with 1µg of LPS induces greater eosinophilic inflammation, but spiking with 10µg of LPS induces neutrophilic inflammation and greater IL-12 expression (13). Additionally, LPS instillation in an OVA mouse model of asthma leads to increased IL-5, IL-4, and TNF-α in bronchoalveolar lavage fluid and greater IgE levels in the serum (14). These studies not only indicate that LPS can function as an adjuvant to enhance allergen-mediated responses but also demonstrate that the amount of LPS in the allergen results in different inflammatory profiles and immune phenotypes in the lungs of mice.
Paradoxically, the significant variability in the abundance of LPS present in commercially available HDM extracts is either not reported or not considered in many studies. LPS concentration in commercial HDM can vary up to 1,000-fold. For example, HDM stocks can have as low as 31 EU to as high as 5,2000 EU. In the absence of commercially available standardized stocks, it is critical to consider the abundance of LPS in HDM extracts to fully understand the implications of data reported using HDM-challenged murine models, in particular for preclinical and drug development studies for asthma and allergic inflammation. In this study, we performed a comparative analysis of two lots of commercially available HDM extract with significantly different LPS concentrations to assess the impact on murine lung function, inflammation, and the lung transcriptome. The aim of this study was to better understand the impact of divergent abundance of LPS in HDM, and highlight the importance of reporting LPS abundance HDM extract used for individual studies. This information is important as it can promote reproducibility in preclinical studies of allergic asthma.
METHODS
Animal allergen exposure and lung function.
This protocol was approved by the University of Manitoba Animal Care Ethics Board. Seven-week-old female BALB/c mice (Charles River Laboratories, MA) were anesthetized under isoflurane (4%) and challenged with HDM Dermatophagoides pteronyssinus (Greer Laboratories) at 25 µg of protein in 35 µL of sterile saline intranasally five times a week for 2 wk, as previously reported (18). Forty-eight hours after the final HDM dose, mice were anesthetized with pentobarbital and a catheter was inserted into the trachea to measure lung function using the FlexiVent small animal ventilator (Scireq, Montreal Canada), using increasing dose of nebulized methacholine. Two HDM stocks with different endotoxin concentrations were used; high-endotoxin HDM (HHDM; XPB82D3A2.5, lot no. 253983) refers to 3,6000 endotoxin units (EU) per vial or 7,877 EU/mg of protein, and low-endotoxin HDM (LHDM; XPB70D3A2.5, lot no. 259585) refers to 615 EU per vial or 115 EU/mg of protein. The two stocks had similar levels of Der p 1 protein (4.9% HHDM vs. 5.5% LHDM of total protein).
Sample collection.
Lung lavage (BAL) was collected by filling the lungs with two 1-mL aliquots of sterile saline. The cell pellet was collected by spinning the samples at ×100 g for 10 min. BAL was aliquoted and stored at −20°C for future analysis. Cell pellet was resuspended in 1 mL sterile saline for quantification of total and differential cell influx into the airways using Hema 3 Stat Pack (Fisher Scientific). Lungs were removed and the left lung was placed into cold RNAlater (Qiagen) for RNA stabilization and isolation for RNA-sequencing.
Cytokine profiling.
The Mouse Cytokine 31-plex and Mouse TGF-β 3-plex Luminex array from Eve Technologies (Calgary, Alberta) was used to analyze the BAL samples. Samples were corrected by volume of BAL recovered from the lung.
RNA isolation and transcriptomic analysis.
Total RNA was extracted from lung tissue samples using the RNeasy Mini Kit (Qiagen) following manufacturer’s protocol. Quality assessment of the extracted RNA was performed using the Eukaryotic Total RNA Nano Chip with the 2100 Bioanalyzer (Agilent Technologies). All samples had an RNA integrity number >6. RNA-Seq library construction was performed by enriching for poly-A-tailed RNA using the Magnetic mRNA Isolation Kit (New England Biolabs), followed by complementary DNA (cDNA) library preparation using the Kapa Stranded Total RNA Kit (Kapa Biosystems). DNA libraries were PCR-amplified followed by cleaning and size selection using the AMPure XP kit (Agencourt). DNA samples were quantified using the Quant-iT dsDNA Assay Kit (Invitrogen) and normalized to 4 nM. Samples were then barcode labeled, multiplexed, and sequenced on the HiSeq 2500 sequencer (Illumina) using the High Output single read run of 100-bp-long sequence reads (+ adapter/index sequences) at the University of British Columbia Sequencing Centre. Sequence quality was assessed using FastQC v0.11.5 (2) and MultiQC v0.8.dev0 (10). Reads were not trimmed in this workflow. The FASTQ sequence reads were aligned to the mouse genome (Ensembl GRCm38) using STAR v2.5 and mapped to Ensembl GRCm38 transcripts. Read counts were generated using htseq-count [HTSeq 0.6.1p1 (1)]. Genes with <10 counts across all treatment groups were prefiltered and removed. Differential genes were identified using the Wald statistics test (16) and filtering for any genes that had an adjusted P < 0.05 as the threshold. Based on clustering by principle component analysis (PCA), one naive mouse lung sample was identified as an outlier. It clustered away from the other naive samples and in the opposite direction of changes from the HDM-treated samples. Thus data were removed from future transcriptomic analyses.
Data analysis.
Statistical analysis was done in R (version 3.5.1) with the DESeq2 package, version 1.14.1 (16). Functional discovery of pathway enrichment and network analyses was performed using InnateDB (4), and NetworkAnalyst (26) respectively. Lung function and cell count data were analyzed with a repeated measures two-way ANOVA with multiple comparisons between treatment group at each methacholine dose (P value adjusted using Benjamini-Hochberg method). Cytokine array was analyzed using a MANOVA in R with an adjusted P < 0.05 considered significant. Graphs were produced in R using ggplot2 (version 3.0.0).
RESULTS
Literature search.
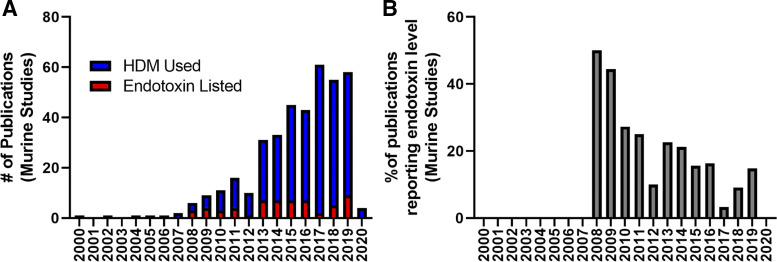
Searching PubMed for “HDM”[Title/Abstract]) AND Mice[Title/Abstract] AND Asthma[Title/Abstract], we found a total of 389 publications dating back to 2000. Of these, only 15.2% (59 papers), explicitly stated the specific concentration of LPS in HDM stocks. Figure 1A shows that while the number of papers using the HDM-challenged animal model studies has increased in recent years, the number of papers indicating how much LPS was in stock HDM supplies has remained at 15% or lower. In fact, the percentage of publications per year reporting LPS specific concentration in HDM extracts used has decreased from a peak in 2009 (Fig. 1B). This peak in 2009 can be explained by a relatively low number of publications (6) with three reporting LPS.
Fig. 1.
Literature using the house dust mite (HDM) model of asthma in mice underreports the LPS abundance in their extracts. A: number of publications per year using the HDM mouse model of asthma (blue) and number of publications per year indicating the specific concentration of endotoxin (LPS) in the HDM extract (red). B: percentage of publications per year reporting LPS level in HDM extract.
Airway dysfunction.
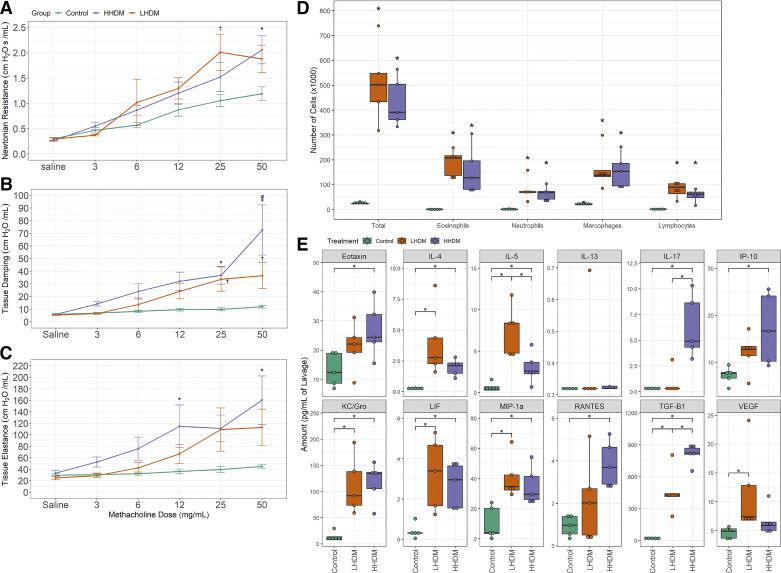
HHDM and LHDM both induced a significant increase in methacholine responsiveness for Newtonian resistance and tissue damping (Fig. 2, A and B, P < 0.05) compared with the allergen-naive mice. Notably, HHDM induced a significantly greater change in tissue damping when compared with LHDM (Fig. 2B, P < 0.05). Furthermore, HHDM, but not LHDM, induced significant changes in tissue elastance (Fig. 2C, P < 0.05).
Fig. 2.
House dust mite (HDM) endotoxin abundance alters lung function and inflammatory parameters in the female murine lung. A–E: Newtonian resistance (A), tissue damping (B), tissue elastance (C), lavage differential cell counts (D), and inflammatory cytokines (E) in lavage (normalized to lavage volume). For lung function data, maximum values in response to methacholine were collected. Lung function (A–C): *P < 0.05, high-endotoxin HDM (HHDM) vs. control mice; †P < 0.05 LHDM vs. control mice; #P < 0.05 vs. LHDM challenged mice (repeated measures two-way ANOVA with multiple comparisons between groups). Differential cell counts (D): *P < 0.05 vs. control mice (two-way ANOVA with Tukey comparison between groups). Cytokine abundance (E): *P < 0.05 (MANOVA); n = 5 female mice per treatment group. Green: control; orange: low-endotoxin HDM; purple: high-endotoxin HDM.
Airway inflammation.
Both HHDM and LHDM induced robust infiltration of immune cells into the airways, as determined by total and differential cell counts in the BAL fluid (Fig. 2D). There were no significant differences in the profile of leukocytes in BAL fluid between HHDM and LHDM challenged mice (Fig. 2D). We detected 11 cytokines in BAL fluid that were significantly elevated by either LHDM or HHDM challenge, compared with allergen-naive animals (Fig. 2E). Of these, IL-17, eotaxin, interferon gamma-induced protein 10 (IP-10) and RANTES were significantly induced only by HHDM, but VEGF was significantly induced only by LHDM challenge (P < 0.05). The concentration of IL-17 and TGF-β were 10 and 1.8 times higher, respectively, in BAL fluid from HHDM-challenged mice compared with LHDM-challenged mice (P < 0.05). In contrast, the abundance of IL-5 was 2.5 times higher in the BAL samples from LHDM-challenged mice (P < 0.05).
Lung transcriptome.
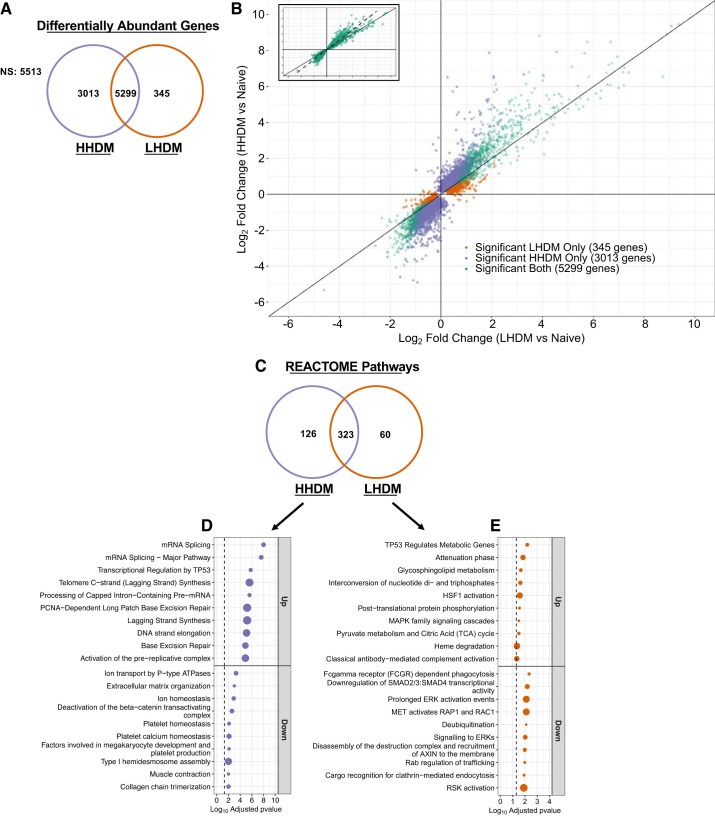
Whole lung transcriptome analysis was performed on mice from allergen-naive, HHDM- and LHDM-challenged groups. These data were submitted to the Gene Expression Omnibus (GEO) database under the accession number GSE116967. We compared changes in the differentially abundant gene profile in lung lysates following HHDM or LHDM challenge, relative to the levels in allergen-naive mice. Figure 3, A and B, highlights the relationship between LHDM-induced effects (fold changes, x-axis) and HHDM-induced effects (fold changes, y-axis). Genes that are commonly altered in both data sets (green, 5,299 genes), or are uniquely altered by HHDM (purple, 3,013 genes), or uniquely altered by LHDM (orange, 345 genes) are shown in Fig. 3A. All genes shown in Fig. 3B reached an adjusted P value threshold of <0.05 (compared with naive animals); however, 5,513 genes were not significantly altered by either HDM exposure. Figure 3B, inset, shows the linear relationship between fold change in commonly shared genes in LHDM and HHDM exposures and reveals a deviation from the normal line toward the HHDM axis indicating that this exposure generally induced greater change. Figure 3C summarizes the pathways enriched by HHDM and LHDM exposure. There were 323 pathways commonly enriched by both HHDM and LHDM, 126 pathways were uniquely enriched by HHDM challenge, and 60 pathways were uniquely enriched after LHDM challenge. Figure 3, D and E, highlights some of the most significantly altered (ranked by adjusted P value) pathways unique to each exposure. In HHDM, pathways involved in DNA damage repair and extracellular matrix organization were significantly altered. In LHDM, pathways involved in sphingolipid metabolism and extracellular signal-regulated kinase (ERK) signaling were altered.
Fig. 3.
House dust mite (HDM) endotoxin abundance significantly alters lung transcriptomic profile. A: Venn diagram showing common and unique genes whose abundance is altered by HDM relative to control mice. NS, not significant. B: scatter plot comparing log2-fold change (FC) in low-endotoxin HDM (LHDM)-challenged animals (x-axis) vs. log2FC in high-endotoxin HDM (HHDM) challenged animals (y-axis) to identify unique and common genes in each data set. Diagonal line indicates line of equity (y = x). Inset: plots line of best fit for common genes (dashed line). C: Venn diagram showing overlap of REACTOME pathways between the 2 HDM lots. D and E: HHDM (D) and LHDM (E) unique altered REACTOME pathways. Point size indicates %enrichment for the pathways. Dashed line indicates adjusted. P value cutoff of 0.05; n = 4 female mice in control group; n = 5 in LHDM and HHDM group. Green: significant both HDM lots; orange: significant LHDM; purple: significant HHDM.
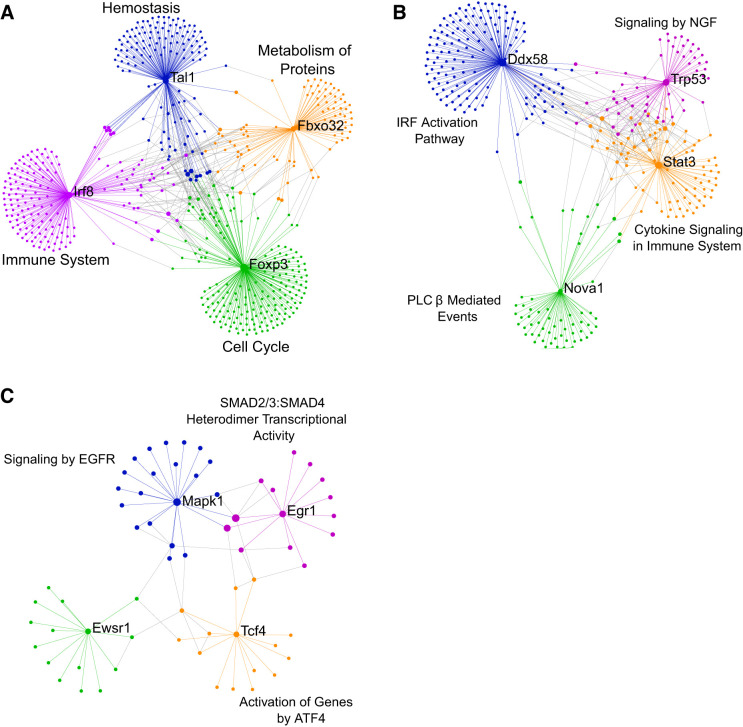
Protein-protein interaction networks were created from the HHDM- and LHDM-challenged transcriptome data using NetworkAnalyst. The top four hubs that were common to both HHDM and LHDM or that were unique to either challenge are shown in Fig. 4, A–C. The top four nodes that were common between HHDM and LHDM establish a network (Fig. 4A), that includes T cell acute lymphocytic leukemia 1 (TAL1), interferon regulatory factor 8 (IRF8), Forkhead box P3 (FOXP3), and F-box protein 32 (FBXO32). Pathway analysis on each node network revealed enrichment for hemostasis (blue, TAL1 network), immune system (purple, IRF8), cell cycle (green, FOXP3), and metabolism of proteins (orange, FBXO32). The top four nodes that comprise a HHDM-unique network (Fig. 4B) included DExD/H-Box Helicase 58 (DDX58), NOVA alternative splicing regulator 1 (NOVA1), signal transducer and activator of transcription 3 (STAT3), and tumor protein P53 (TRP53 or TP53). Pathway analysis of these nodes revealed enrichment in IRF activation (blue, DDX58), PLCβ-mediated events (green, NOVA1), cytokine signaling in immune system (orange, STAT3), and signaling by NGF (purple, TRP53). Finally, the top four nodes of a LHDM-unique network (Fig. 4C) were mitogen-activated protein kinase 1 (MAPK1), EWS RNA binding protein 1 (EWSR1), transcription factor 4 (TCF4), and early growth response 1 (EGR1). Pathway analysis of these nodes revealed enrichment in signaling by epidermal growth factor receptor (blue, MAPK1), activation of genes by ATF4 (orange, TCF4), and SMAD2/3:SMAD4 heterodimer transcriptional activity (purple, EGR1).
Fig. 4.
Network analysis identified hub genes common to either house dust mite (HDM) and unique to high-endotoxin HDM (HHDM) and low-endotoxin HDM (LHDM). Networks showing keys hubs and the associated most significant (by P value) REACTOME pathways enriched at those hubs for common HDM genes (A), HHDM unique genes (B), and LHDM unique genes (C). Main hubs labeled with text. Different colors show networks connected to the main hubs. Network and pathway analysis done using NetworkAnalyst.
DISCUSSION
The objective of this report is to draw attention to the need to routinely report the composition, in particular LPS, of commercially available HDM products. While information from commercial sources on the exact composition of HDM extracts is lacking, LPS and Der p 1 levels are routinely provided and can be thus used for standardization when using HDM extracts in animal model studies of allergic airways disease. In our own review of the literature on murine models of asthma, only 15% of papers indicated the level of LPS in their extracts, a number that has been decreasing in the recent years (Fig. 1). We purchased two lots of lyophilized whole HDM stock that differed considerably in LPS abundance but were not different in Der p 1 abundance (based on information provided on data sheet from the commercial vendor). We compared responses in BALB/c mice using the two different HDM stocks to understand how difference in LPS content can influence the lung phenotype and molecular end points in mice.
We found that LPS abundance did not significantly alter immune cell infiltration, as these changes were significant for both HHDM and LHDM. However, we found LPS concentration-dependent differences in IL-5, IL-17, RANTES, and eotaxin, all important molecules in the pathogenesis and targets for treatment of asthma. Additionally, we noted that HHDM caused greater changes in tissue damping and elastance, which may be related to differences in fibrosis or secretions in the lower airways and parenchyma and could reflect the impact of LPS-Der p1 interactions, as has been reported (11, 12, 23). Using RNA-seq and bioinformatics, we uncovered significant differences in the underlying molecular phenotype that was related to the LPS amount in the HDM used for intranasal challenge. Key differences were evident in the lung transcriptome relating to DNA damage repair and ERK signaling. These data suggest that it is important to fully detail the composition of HDM extracts used in preclinical mouse models. We suggest that reporting LPS concentration will promote data reproducibility and enable appropriate comparison of studies that use the HDM-challenged in various murine models. While HDM extract is an intricate mixture of components, and the complex interactions between Der p 1 and LPS can alter biological response in a myriad of ways, providing the information from the manufacturer’s specification sheet on each lot is an easy starting point to address this issue.
HDM is a common aeroallergen affecting humans with asthma, which makes it a useful tool in mice to unravel the pathobiology of the disease. However, HDM is also a complex allergen composed of numerous proteins, enzymes, and bacterial products. A recent report identified 105 proteins in an extract from the Dermatophagoides farinae mite that could be classified into 7 distinct functional groups including the identification of novel allergens (8). This diversity of allergens means that, unlike OVA models of asthma, standardization of results across different HDM stocks can prove to be difficult. Extracts purchased from companies typically contain information on total protein, Der p 1, and LPS levels in each batch, which can be used to standardize dosing between experiments. Levels of Der p 1, a cysteine proteinase, can give an index of allergen potential as exposure to Der p 1 is associated with an increased risk for asthma (21) and disruption of the epithelial barrier (24). In our study, the levels of Der p 1 provided by the supplier were similar. LPS is also an important component of HDM extracts that is associated with asthma severity (17, 22) and a decrease in FEV1 (20). In mice, LPS can shift HDM-induced inflammation from eosinophilic to neutrophilic and promote the production of IL-17 and IL-33 in a dose-dependent manner (9). In OVA-sensitized mice, exposure to LPS increased levels of IgE, IL-4, IL-5, histamine, and oxidative stress response elements (14). Standardization against the other components of HDM extracts requires the use of external kits and reagents as their information is not provided by the HDM suppliers. Until this information is made available, LPS and Der p 1 levels can act as a way to standardize HDM extracts across experiments. It is important for researchers working with animal models to appreciate the association between endotoxin/LPS concentrations and airway inflammation in mouse models of asthma to better translate their findings to the human condition and better move preclinical therapeutics to clinical trials.
It is important to note that our study design was deliberate to compare commercially available HDM stocks, rather than using a single low-LPS stock that we might have spiked with LPS to simulate the HHDM scenario. This choice was made to ensure the data we generated represented real-world situations that researchers routinely encounter when purchasing HDM extract. Aside from the LPS levels in each HDM stock, we did not explore differences in proteases or chitinase activity between the two stocks. Previous work in epithelial cells and mice has shown that these two parameters are not crucial for the development of epithelial barrier dysfunction or airway sensitization (19). Finally, it is our suggestion that all papers using the HDM-challenge murine models, such as in preclinical studies of asthma, identify and state the endotoxin concentration in their stocks. This will directly facilitate to standardize the model and comparison of results across different publications. Unfortunately, providing the lot number in lieu of this information does not solve the problem of underreporting as there is currently no clear way to track lot number back to the endotoxin levels and composition of the HDM extract.
Conclusion.
Although the lung function phenotype between mice challenged with HDM with high or low endotoxin is largely similar, there exist substantial differences of responses at the molecular level that could strongly influence the development and success rate of novel therapeutics in preclinical studies. It is our recommendation that papers using the HDM model of airway inflammation state the endotoxin load of their stock in addition to the relative abundance of Der p 1 to facilitate reproducibility and comparison of data across publications using similar models.
GRANTS
This research was supported by funds to A. J. Halayko and N. M Mookherjee from the Canadian Institutes of Health Research (CIHR)-Canadian Respiratory Research Network and the Biology of Breathing Theme of the Children’s Hospital Research Institute of Manitoba and by CIHR Foundation Grant FDN-154287 (to R. E. Hancock). A. J. Halayko holds a Canada Research Chair in Lung Pathobiology and Treatment. R. E. Hancock holds a Canada Research Chair in Health and Genomics and a University of British Columbia Killam Professorship.
DISCLAIMERS
The content is solely the responsibility of the authors and does not necessarily represent the official views of the Canadian Institutes for Health Research.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
C.D.P. and A.J.H. conceived and designed research; C.D.P., S.B., A.L., and R.F. performed experiments; C.D.P., A.J., S.B., A.L., S.H., and R.E.W.H. analyzed data; C.D.P., A.J., T.H.M., N.M., and A.J.H. interpreted results of experiments; C.D.P. prepared figures; C.D.P. drafted manuscript; C.D.P., A.J., T.H.M., N.M., and A.J.H. edited and revised manuscript; C.D.P., N.M., and A.J.H. approved final version of manuscript.
Contributor Information
Collaborators: Canadian Respiratory Research Network
REFERENCES
- 1.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169, 2015. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews S. FastQC: A Quality Control Tool for High-Throughput Sequence Data. Cambridge, UK: The Babraham Institute; http://www.bioinformatics.babraham.ac.uk/projects/fastqc. 2010. [Google Scholar]
- 3.Bossé Y, Paré PD, Seow CY. Airway wall remodeling in asthma: from the epithelial layer to the adventitia. Curr Allergy Asthma Rep 8: 357–366, 2008. doi: 10.1007/s11882-008-0056-0. [DOI] [PubMed] [Google Scholar]
- 4.Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, Winsor GL, Hancock RE, Brinkman FS, Lynn DJ. InnateDB: systems biology of innate immunity and beyond–recent updates and continuing curation. Nucleic Acids Res 41, D1: D1228–D1233, 2013. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carnes MU, Hoppin JA, Metwali N, Wyss AB, Hankinson JL, O’Connell EL, Richards M, Long S, Freeman LEB, Sandler DP, Henneberger PK, Barker-Cummings C, Umbach DM, Thorne PS, London SJ. House dust endotoxin levels are associated with adult asthma in a U.S. farming population. Ann Am Thorac Soc 14: 324–331, 2017. doi: 10.1513/AnnalsATS.201611-861OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cates EC, Fattouh R, Wattie J, Inman MD, Goncharova S, Coyle AJ, Gutierrez-Ramos J-C, Jordana M. Intranasal exposure of mice to house dust mite elicits allergic airway inflammation via a GM-CSF-mediated mechanism. J Immunol 173: 6384–6392, 2004. doi: 10.4049/jimmunol.173.10.6384. [DOI] [PubMed] [Google Scholar]
- 7.Celedón JC, Milton DK, Ramsey CD, Litonjua AA, Ryan L, Platts-Mills TA, Gold DR. Exposure to dust mite allergen and endotoxin in early life and asthma and atopy in childhood. J Allergy Clin Immunol 120: 144–149, 2007. doi: 10.1016/j.jaci.2007.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choopong J, Reamtong O, Sookrung N, Seesuay W, Indrawattana N, Sakolvaree Y, Chaicumpa W, Tungtrongchitr A. Proteome, Allergenome, and Novel Allergens of House Dust Mite, Dermatophagoides farinae. J Proteome Res 15: 422–430, 2016. doi: 10.1021/acs.jproteome.5b00663. [DOI] [PubMed] [Google Scholar]
- 9.Daan de Boer J, Roelofs JJ, de Vos AF, de Beer R, Schouten M, Hommes TJ, Hoogendijk AJ, de Boer OJ, Stroo I, van der Zee JS, Veer CV, van der Poll T. Lipopolysaccharide inhibits Th2 lung inflammation induced by house dust mite allergens in mice. Am J Respir Cell Mol Biol 48: 382–389, 2013. doi: 10.1165/rcmb.2012-0331OC. [DOI] [PubMed] [Google Scholar]
- 10.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32: 3047–3048, 2016. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ichikawa S, Takai T, Yashiki T, Takahashi S, Okumura K, Ogawa H, Kohda D, Hatanaka H. Lipopolysaccharide binding of the mite allergen Der f 2. Genes Cells 14: 1055–1065, 2009. doi: 10.1111/j.1365-2443.2009.01334.x. [DOI] [PubMed] [Google Scholar]
- 12.Irvin CG, Bates JH. Measuring the lung function in the mouse: the challenge of size. Respir Res 4: 1, 2003. doi: 10.1186/rr199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim YK, Oh SY, Jeon SG, Park HW, Lee SY, Chun EY, Bang B, Lee HS, Oh MH, Kim YS, Kim JH, Gho YS, Cho SH, Min KU, Kim YY, Zhu Z. Airway exposure levels of lipopolysaccharide determine type 1 versus type 2 experimental asthma. J Immunol 178: 5375–5382, 2007. doi: 10.4049/jimmunol.178.8.5375. [DOI] [PubMed] [Google Scholar]
- 14.Kumari A, Dash D, Singh R. Lipopolysaccharide (LPS) exposure differently affects allergic asthma exacerbations and its amelioration by intranasal curcumin in mice. Cytokine 76: 334–342, 2015. doi: 10.1016/j.cyto.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 15.Locke NR, Royce SG, Wainewright JS, Samuel CS, Tang ML. Comparison of airway remodeling in acute, subacute, and chronic models of allergic airways disease. Am J Respir Cell Mol Biol 36: 625–632, 2007. doi: 10.1165/rcmb.2006-0083OC. [DOI] [PubMed] [Google Scholar]
- 16.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550, 2014. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michel O, Kips J, Duchateau J, Vertongen F, Robert L, Collet H, Pauwels R, Sergysels R. Severity of asthma is related to endotoxin in house dust. Am J Respir Crit Care Med 154: 1641–1646, 1996. doi: 10.1164/ajrccm.154.6.8970348. [DOI] [PubMed] [Google Scholar]
- 18.Piyadasa H, Altieri A, Basu S, Schwartz J, Halayko AJ, Mookherjee N. Biosignature for airway inflammation in a house dust mite-challenged murine model of allergic asthma. Biol Open 5: 112–121, 2016. doi: 10.1242/bio.014464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Post S, Nawijn MC, Hackett TL, Baranowska M, Gras R, van Oosterhout AJ, Heijink IH. The composition of house dust mite is critical for mucosal barrier dysfunction and allergic sensitisation. Thorax 67: 488–495, 2012. doi: 10.1136/thoraxjnl-2011-200606. [DOI] [PubMed] [Google Scholar]
- 20.Rabinovitch N, Liu AH, Zhang L, Rodes CE, Foarde K, Dutton SJ, Murphy JR, Gelfand EW. Importance of the personal endotoxin cloud in school-age children with asthma. J Allergy Clin Immunol 116: 1053–1057, 2005. doi: 10.1016/j.jaci.2005.08.045. [DOI] [PubMed] [Google Scholar]
- 21.Sporik R, Holgate ST, Platts-Mills TA, Cogswell JJ. Exposure to house-dust mite allergen (Der p I) and the development of asthma in childhood. A prospective study. N Engl J Med 323: 502–507, 1990. doi: 10.1056/NEJM199008233230802. [DOI] [PubMed] [Google Scholar]
- 22.Thorne PS, Kulhánková K, Yin M, Cohn R, Arbes SJ Jr, Zeldin DC. Endotoxin exposure is a risk factor for asthma: the national survey of endotoxin in United States housing. Am J Respir Crit Care Med 172: 1371–1377, 2005. doi: 10.1164/rccm.200505-758OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanoirbeek JAJ, Rinaldi M, De Vooght V, Haenen S, Bobic S, Gayan-Ramirez G, Hoet PH, Verbeken E, Decramer M, Nemery B, Janssens W. Noninvasive and invasive pulmonary function in mouse models of obstructive and restrictive respiratory diseases. Am J Respir Cell Mol Biol 42: 96–104, 2010. doi: 10.1165/rcmb.2008-0487OC. [DOI] [PubMed] [Google Scholar]
- 24.Wan H, Winton HL, Soeller C, Tovey ER, Gruenert DC, Thompson PJ, Stewart GA, Taylor GW, Garrod DR, Cannell MB, Robinson C. Der p 1 facilitates transepithelial allergen delivery by disruption of tight junctions. J Clin Invest 104: 123–133, 1999. doi: 10.1172/JCI5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang JY, Chen WY. Inhalant allergens in asthmatic children in Taiwan: comparison evaluation of skin testing, radioallergosorbent test and multiple allergosorbent chemiluminescent assay for specific IgE. J Formos Med Assoc 91: 1127–1132, 1992. [PubMed] [Google Scholar]
- 26.Xia J, Gill EE, Hancock RE. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc 10: 823–844, 2015. doi: 10.1038/nprot.2015.052. [DOI] [PubMed] [Google Scholar]
- 27.Zosky GR, Sly PD. Animal models of asthma. Clin Exp Allergy 37: 973–988, 2007. doi: 10.1111/j.1365-2222.2007.02740.x. [DOI] [PubMed] [Google Scholar]