Abstract
The COVID-19 pandemic raised by SARS-CoV-2 is a public health emergency. However, lack of antiviral drugs and vaccine against human coronaviruses demands a concerted approach to challenge the SARS-CoV-2 infection. Under limited resource and urgency, combinatorial computational approaches to identify the potential inhibitor from known drugs could be applied against risen COVID-19 pandemic. Thereof, this study attempted to purpose the potent inhibitors from the approved drug pool against SARS-CoV-2 main protease (Mpro). To circumvent the issue of lead compound from available drugs as antivirals, antibiotics with broad spectrum of viral activity, i.e. doxycycline, tetracycline, demeclocycline, and minocycline were chosen for molecular simulation analysis against native ligand N3 inhibitor in SARS-CoV-2 Mpro crystal structure. Molecular docking simulation predicted the docking score >−7 kcal/mol with significant intermolecular interaction at the catalytic dyad (His41 and Cys145) and other essential substrate binding residues of SARS-CoV-2 Mpro. The best ligand conformations were further studied for complex stability and intermolecular interaction profiling with respect to time under 100 ns classical molecular dynamics simulation, established the significant stability and interactions of selected antibiotics by comparison to N3 inhibitor. Based on combinatorial molecular simulation analysis, doxycycline and minocycline were selected as potent inhibitor against SARS-CoV-2 Mpro which can used in combinational therapy against SARS-CoV-2 infection.
Keywords: COVID-19, Tetracyclines, Drug repurposing, SARS-CoV-2 Mpro, Molecular simulation calculations
Graphical abstract
1. Introduction
Human coronaviruses (HCoVs), such as middle-east respiratory syndrome coronavirus (MERS-CoV), severe acute respiratory syndrome coronavirus (SARS-CoV), and 2019 novel coronavirus (2019-nCoV, also called as SARS-CoV-2) that emerged from animal reservoirs, have aggravate the pandemic with high mortality and morbidity. Coronaviruses (CoVs) are positive-sense, single-stranded RNA (ssRNA) virus which contain the largest RNA genome of 27–32 kb with six to ten open reading frames (ORFs) enclosed in an enveloped protein to form spherical particles of 100–160 nm size [1,2]. The genetic material of HCoVs is highly susceptible to frequent recombination process which has been suggested to direct the establishment of new HCoV strains with altered virulence [3]. As a result, a total of seven main HCoVs have been recorded, viz. HCoV-229E, HCoV-NL63, HCoV-OC43, HCoV-HKU1, MERS-CoV, SARS-CoV, and SARS-CoV-2; these viruses are held accountable for fatal infections in the respiratory system and other system symptoms, including diarrhea and nausea [4,5].
HCoVs belong to the Coronaviridae family of the order Nidovirales and classified into four genera (α, β, γ, and δ) [6]. Recently discovered SARS-CoV-2 belongs to the β genus which shares 79.5% sequence similarity to SARS-related coronavirus and is 96% identical at the whole-genome level to a bat coronavirus [7,8]. The genome analysis of coronaviruses, including SARS-CoV-2 genome (GenBank:MN908947.3), revealed four non-structural proteins, i.e. papain-like protease (PLpro), 3-chymotrypsin-like (3CLpro), helicase, and RNA polymerase; four structural proteins, viz. nucleocapsid (N) protein, membrane (M) protein, envelope (E) protein, Spike (S) protein, and accessory proteins [1,[9], [10], [11]]. Both 3CLpro and PLpro are reported in transcription and replication of the virus [6]. Remarkably, in vitro inhibition of these proteases by the selected inhibitors has shown antiviral activity against coronaviruses (CoVs) [12,13]. These observations concluded the 3CLpro as the main coronavirus protease (Mpro) [14], which is essentially required to conduct the replication cycle of the virus [6,15]. Like in other HCoVs, SARS-CoV-2 Mpro digests the polyprotein at conserved sites; it starts with autolytic cleavage from pp1a and pp1ab site encoded by the virus ORF1a/b which shares 96% sequence similarity with Mpro of SARS-CoV strain [16,17]. To conquer coronavirus infections, the inhibition of viral Mpro, which is functionally important in the viral life cycle and absence of its human homologues, has been approved as conventional antiviral therapeutic strategy [12].
In current scenario, despite numerous efforts, no drug or vaccine is yet approved against the HCoVs and expected to take further months to years for the new interventions [11]. Recently, various Mpro inhibitors were demonstrated for broad-spectrum in vitro activities against CoVs via interacting with conserved key residues required for the substrate recognition at respective Mpro enzymes [18,19]. Moreover, with the urgency of SARS-CoV-2 outbreak, drug repurposing is represented as an effective drug discovery approach from existing drugs to suggestively shorten the time and condense the cost by comparison to de novodrug discovery strategy and randomized clinical trials [20]. To facilitate the rapid drug discovery, computational drug repurposing approaches have been established as an effective method to reveal the new indications from already known drugs with the aid of computational algorithms such as molecular docking simulation and molecular dynamics simulation to predict the drug-target interactions and binding stability, respectively [21,22]. Previously, computational drug repurposing approaches were employed to identify the potential drug candidates against viral infections, including Ebola, ZIKA, dengue, and influenza infections [23], and for HCoVs such as SARS-CoV and MERS-CoV [[24], [25], [26]]. Also, nonpeptidic active-site-directed inhibition of SARS-CoV Mpro was discovered by a combination of screening and docking methods [27]. Likewise, bifunctional aryl boronic acid compounds were discovered to inhibit the SARS-CoV Mpro using combination of experimental and computational methods [28]. After SARS-CoV-2 outbreak, computational drug repurposing was also applied to find the potential drugs and some of the investigations are already reported [[29], [30], [31]]. In area of drug repurposing, antibiotics from tetracyclines and its derivatives have been repurposed against viral infections [32,33]. These antibiotics are well designated as broad-spectrum bacteriostatic inhibitors which selectively inhibit the microbial protein synthesis only, including naturally occurring polyketide compounds (chlortetracycline, oxytetracycline, and tetracycline) and their semisynthetic derivatives (minocycline, doxycycline, and methacycline) [32]. Notably, tetracycline derivatives, i.e. doxycycline, was reported to exert antiviral effects against antimurine retrovirus [34], Chikungunya infection [33], and blockage of dengue virus replication in infected cell lines [35,36]. Besides, demeclocycline is known to exert anti-viral activity on west Nile virus (WNV) [37] and included in the list of antiviral compounds [38]. Another, broad spectrum tetracycline derivative minocycline was initially reported against human immunodeficiency virus (HIV) type 1 infection [39], and later studied for other viruses such as WNV, Japanese encephalitis virus (JEV), Simian Immunodeficiency virus (SIV), Sindbis, human T-lymphocytic virus type-1 (HTLV-1), Rabies, and Reoviruses [40]. Moreover, tetracyclines (e.g. tetracycline, doxycycline, and minocycline) which are known as zinc chelating agents were postulated to inhibit COVID-19 infection by restricting viral replication in the host [41]. Additionally, synthetic halogenated tetracycline eravacycline was demonstrated with inhibitory effect against SARS-CoV, SARS-CoV-2, MERS-CoV, and hepatitis C virus (HCV) using computational studies [42].
Hence, under the impression of tetracyclines and their derivatives inhibits viral infections, we hypothesized that antibiotics reported for antiviral activities may be helpful in the SARS-CoV-2 inhibition by blockage of its main protease, viz. Mpro. Thereof, we employed structure based molecular docking for selected four tetracyclines, i.e. doxycycline, tetracycline, demeclocycline, and minocycline known for antiviral activities against other viruses, with Mpro from SARS-CoV-2. Similar virtual screening approach was recently applied to identify the tetracycline derivatives as Flavivirus inhibitors [36]. Following, docked protein-antibiotics complexes were evaluated for the complex stability via classical molecular dynamics simulation. The binding affinity and stability of selected viral protease-antibiotics complexes were also compared to the docked complexes of protease with native ligand N3 inhibitor to generate a comparative study as mentioned in Fig. 1 .
Fig. 1.
Repurposing of antibiotics tetracyclines as potential inhibitors of SARS-CoV-2 main protease (Mpro) via combinatorial molecular simulation calculations.
2. Methodology
2.1. Receptor and ligand collection
Crystal structure of the novel SARS-CoV-2 main protease (SARS-CoV-2 Mpro) was downloaded from PDB database (https://www.rcsb.org/) with PDB ID:6lu7 [43]. The viral Mpro was elucidated at 2.16 Å resolution as dimer, where each monomer contained three domains; domain I (residues 8–101), domain II (residues 102–184), and domain III (residues 201–303) connected by long interconnecting loop (residues 185–200) between the domains II and III. The catalytic dyad (His41 and Cys145) and substrate binding sites were also defined between the domains I and II in the crystal structure with N3 inhibitor [43]. Besides, 3D structures of selected antibiotics, i.e. doxycycline (CID: 54671203), tetracycline (CID: 54675776), demeclocycline (CID: 54680690), and minocycline (CID: 54675783), were also downloaded from PubChem database (https://pubchem.ncbi.nlm.nih.gov/).
2.2. Molecular docking simulation
Molecular docking simulation between the SARS-CoV-2 Mpro and selected antibiotics was conducted in Chimera-AutoDock Vina plugin setup to determine the interacting residues of the protein with respective ligands. Briefly, protein and ligand structures were minimized using the default parameters in structure minimization tool in USCF Chimera-1.14 [44]. Following, receptor and ligands were prepared using Dock prep tool in Chimera under default parameters, where native ligand from the crystal structure was removed, polar hydrogen atoms and charges were added. Finally, molecular docking simulations were performed using AutoDock Vina [45] plugin with default setting at the native ligand binding pocket by adopting the docking grid size of 60 × 40 × 40 Å along both three (X, Y, and Z) axes, covering all the essential residues center at −8.918, 17.918, 62.905 Å region, to provide copious space for the ligand conformations. By docking, at least 10 conformers were generated and conformer with lowest binding energy and RMSD were chosen for further analysis in free academic Maestro v12.3 (Schrödinger Release 2020-1: Maestro, Schrödinger, LLC, New York, NY, 2020). Herein, noncovalent interactions, viz. hydrogen bonding, hydrophobic, π-π interaction, π-cation interaction, positive (basic or positive amino acids), negative (acidic or negative amino acids), polar (polar amino acids), glycine (non-polar interaction), and salt bridges interactions, were calculated at cutoff radius of 4 Å under default conditions, and both 3D and 2D interaction images were generated. Similar docking methodology was also employed for the crystal structure ligand, i.e. Michael acceptor inhibitor, known as N3 inhibitor [43], to validate the docking procedure and for comparative analysis with the selected antibiotics.
2.3. Molecular dynamics simulation
The best scored poses for receptor-ligand complexes were evaluated for complex stability by 100 ns molecular dynamics (MD) simulations using free academic Desmond v5.6 module of Schrödinger-Maestro v11.8 [46] under Linux environment on HP Z2 workstation. All the MD systems for protein-ligand complexes were fabricated as an orthorhombic grid box (10 Å × 10 Å × 10 Å buffer) followed by addition of TIP4P (transferable intermolecular potential 4 point) water molecules for system minimization using System Builder tools in Desmond-Maestro interface. Later, salt and ion placement were omitted at 20 Å from the ligand and whole system was neutralized by addition of counter ions. Besides, 0.002 ps time interval was set for the anisotropic diagonal position scaling to reserve the constant pressure during MD simulation. Additionally, temperature of the system was set at 300 K coupled with 20 ps NPT reassembly at 1 atm pressure. Furthermore, system density was preserved near 1 g/cm3 and all the calculations were conducted under default parameters. Finally, MD simulation for each complex was conducted for 100 ns interval under similar conditions. Subsequently, the simulation trajectories were analyzed by simulation interaction diagram tool in Desmond v5.6 module of Schrödinger-Maestro v11.8 [46].
3. Results and discussion
3.1. Intermolecular interaction analysis
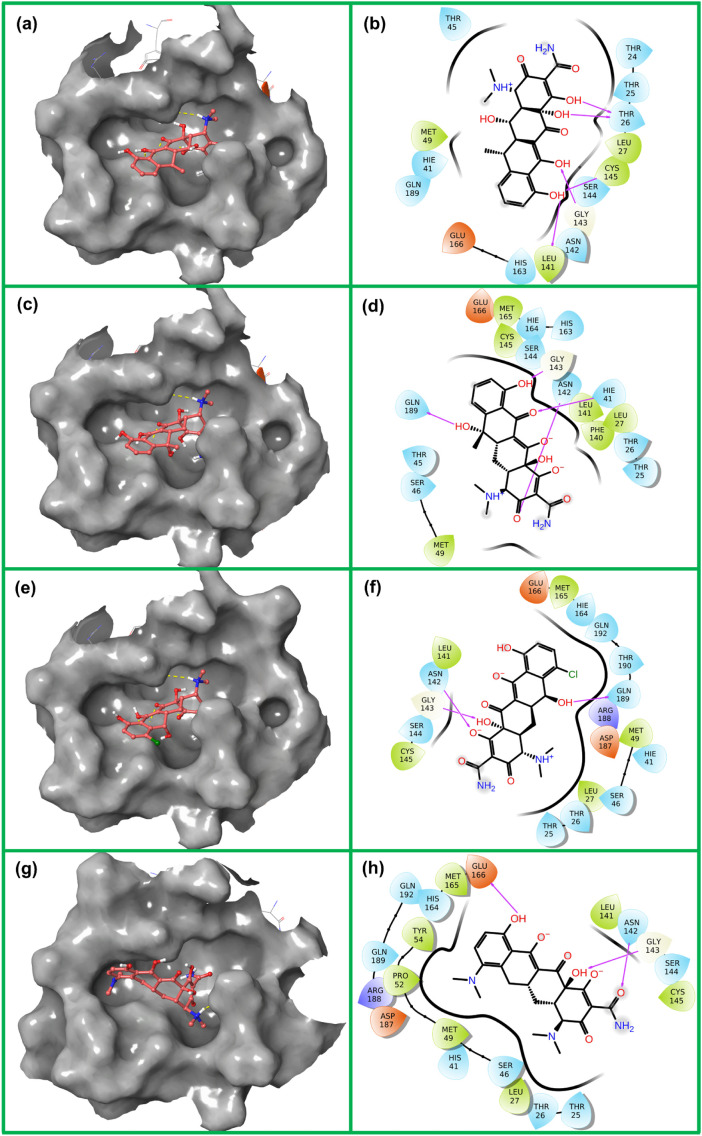
The molecular docking algorithms have been widely used to predict the bioactive compounds or drug repurposing against various drug targetable proteins in diseases and infections [36,47]. Likewise, this study also employed the molecular docking simulation approach for the selected antibiotic tetracyclines, i.e. doxycycline, tetracycline, demeclocycline, and minocycline, to predict their instinctively potential against SARS-CoV-2 infection by its Mpro inhibition. All the docked drugs and N3 inhibitor showed significant docking confirmation with binding affinity energies >−7 kcal/mol at least RMSD (0 value by default in AutoDock Vina under default parameters) in the active pocket of SARS-CoV-2 Mpro with respect to N3 inhibitor (Figs. 2 , S1). The docked SARS-CoV-2 Mpro-doxycycline complex was recorded with significant −7.6 kcal/mol docking score and formed five hydrogen bonds with the viral protease by active residues Thr26 (2.8 Å and 2.33 Å), Leu141 (2.33 Å), Gly143 (2.86 Å), and Cys145 (3.23 Å); additional interactions, i.e. hydrophobic (Leu27, Met49, Leu141, Cys145, Met165), polar (Thr24, Thr25, Thr26, Hie41, Thr45, Asn142, Ser144, His163, and Gln189), negative (Glu166), and glycine (Gly143), were logged in SARS-CoV-2 Mpro-doxycycline complex (Fig. 2a–b). Whereas, SARS-CoV-2 Mpro docked complex with tetracycline was logged for docking energy of −7.5 kcal/mol and formation of four hydrogen bonds with the active residues His41 (2.84 Å), Asn142 (3.33 Å), Gly143 (2.84 Å), and Gln189 (3.86 Å) was also recorded. Moreover, hydrophobic interactions (Leu27, Met49, Phe140, Leu141, Cys145, and Met165), polar interactions (Thr25, Thr26, His41, Thr45, Ser46, Asn142, Ser144, His163, His164, and Gln189), negative interactions (Glu166), and glycine interactions (Gly143) were also observed between the residues of SARS-CoV-2 Mpro and atoms of tetracycline (Fig. 2c–d). Similarly, SARS-CoV-2 Mpro-demeclocycline complex exhibited significant docking score of −7.4 kcal/mol along with formation of three hydrogen bonds at essential residues Asn142 (3.03 Å), Gly143 (2.80 Å), and Gln189 (3.74 Å) of SARS-CoV-2 Mpro. Additionally, the respective complex was noted for hydrophobic interactions (Leu27, Met49, Leu141, Cys145, and Met165), polar interactions (Thr25, Thr26, Hie41, Ser46, Asn142, Ser144, His164, Gln192, Thr190, and Gln189), negative interactions (Glu166 and Asp187), positive interactions (Arg188), and glycine interactions (Gly143) (Fig. 2e–f). While, SARS-CoV-2 Mpro-minocycline complex showed substantial binding energy of −7.1 kcal/mol followed by formation of three hydrogen bonds with the residues Asn142 (2.64 Å), Gly143 (2.54 Å), and Glu166 (2.84 Å) in the active pocket of viral protease. Other intermolecular interactions, viz. hydrophobic interactions (Leu27, Met49, Pro52, Tyr54, Leu141, Cys145, and Met165), polar interactions (Thr25, Thr26, His41, Ser46, Asn142, Ser144, His164, and Gln189), negative interactions (Glu166 and Asp187), positive interactions (Arg188), and glycine interactions (Gly143), were also observed for SARS-CoV-2 Mpro-minocycline docked complex (Fig. 2g–h). The substantial docking energy and formation of strong hydrogen bonds between the atoms of respective antibiotics with active residues of SARS-CoV-2 Mpro indicate the substantial stability of docked protease-antibiotics complexes.
Fig. 2.
3D and 2D interaction profiles for SARS-CoV-2 Mpro-antibiotics; (a–b) doxycycline, (c–d), tetracycline, (e–f) demeclocycline, and (g–h) minocycline, depicting active residues around the ligand at 4 Å area in the active pocket of SARS-CoV-2 Mpro. In 2D interaction maps, hydrogen bond formation (pink arrows), hydrophobic (green), polar (blue), red (negative), violet (positive) and glycine (grey) interaction are also logged between the docked SARS-CoV-2 Mpro and selected antibiotics. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Furthermore, the docked poses of the antibiotics with SARS-CoV-2 Mpro were also compared to the SARS-CoV-2 Mpro-N3 inhibitor docked complex (Fig. S2). The comparative docking analysis includes docking energy and intermolecular interaction profiling between the selected complexes. From molecular docking simulation analysis, N3 inhibitor (−7.6 kcal/mol) showed relatively similar docking energy by comparison to selected antibiotics (maximum docking score −7.6 kcal/mol logged for SARS-CoV-2 Mpro-doxycycline complex). Moreover, SARS-CoV-2 Mpro-N3 inhibitor complex exhibited formation of four hydrogen bonds with residues Cys145 (2.01 Å), Glu166 (2.16 Å), and Gln189 (2.30 and 3.06 Å) along with other intermolecular interactions (Fig. S2). Interestingly, in SARS-CoV-2 Mpro-antibiotic complexes, doxycycline also generated five hydrogen bonds and minimum of three hydrogen bonds were noted for the minocycline docked at the active pocket of viral protease. Besides, both antibiotics and N3 inhibitor exhibited significant hydrophobic, polar, negative, positive, and glycine interactions with common residues of the SARS-CoV-2 Mpro (Figs. 2, S2). Interestingly, selected antibiotics docked with SARS-CoV-2 Mpro exhibited one or more than type of interaction with the catalytic dyad (Cys145 and His41), substrate binding residues (Phe140, Leu14, Gly143, Ser144, His163, Met165, and Glu166) and other essential residues required for the substrate binding pocket (Thr24, Thr25, Met49, Phe140, Asn142, His163, Met165, Asp187, and Gln189) (Figs. 2, S2) as reported in the viral protease crystal structure with N3 inhibitor [43]. Additionally, Gln189 residue, which is absolute requirement for S1 subsite at the P1 position was logged in the interaction profile with the selected antibiotics with SARS-CoV-2 Mpro inhibitors [43]. As, the antibiotics were logged for intermolecular interaction with residues of active site and substrate-binding pocket located between the clefts of domain I and domain II of vial main protease, suggested to inhibit the proteolytic function of SARS-CoV-2 Mpro, which is required for the viral replication and pathogenesis [43]. Moreover, interacted residues of SARS-CoV-2 Mpro in the antibiotics docked complexes are highly conserved as substrate-recognition pocket of the CoVs, suggested the selected screened antibiotics can be employed for the development of broad spectrum of antiviral drugs against HCoVs. Hence, molecular docking simulation analysis for the screened antibiotics with respect to N3 inhibitor suggested that the selected drug molecules have the potential to inhibit SARS-CoV-2 Mpro by interacting with its catalytic dyad and substrate binding residues.
3.2. Molecular dynamics simulation analysis
Molecular dynamics simulation can be employed to the docked complexes to study their stability and intermolecular interaction profiling with respect to time [47,48]. In this study, docked complexes of antibiotics and N3 inhibitor with viral protease were subjected to 100 ns molecular dynamics (MD) simulation to predict the respective complex confirmation change, stability, and intermolecular interactions between the active residues of the receptor and atoms of the ligand molecules. Initially, last pose from simulation trajectory was extracted and studied for the intermolecular interaction for the respective complexes. A maximum of three hydrogen bonds were recorded for SARS-CoV-2 Mpro-demeclocycline, two hydrogen bonds for SARS-CoV-2 Mpro-tetracycline, only one hydrogen bond in each SARS-CoV-2 Mpro-doxycycline and SARS-CoV-2 Mpro-minocycline complex against SARS-CoV-2 Mpro-N3 inhibitor (no hydrogen bonding) (Figs. S3, S4). These observations predicted that selected antibiotics have substantial affinity with the active pocket of SARS-CoV-2 Mpro by comparison to N3 inhibitor. To further verified the selected receptor-ligand complex stabilities, four properties were calculated from respective simulation trajectories, i.e. (a) root mean square deviation (RMSD), (b) root mean square fluctuation (RMSF), (c) protein secondary structure element (SSE) changes to nullify abnormal structural changes and conformational modification of docked ligands with respect to simulation time, and (d) protein-ligand contacts mapping, was performed to analyze the type of interaction between the ligand atoms and residues of the protein during MD simulation.
3.2.1. RMSD and RMSF
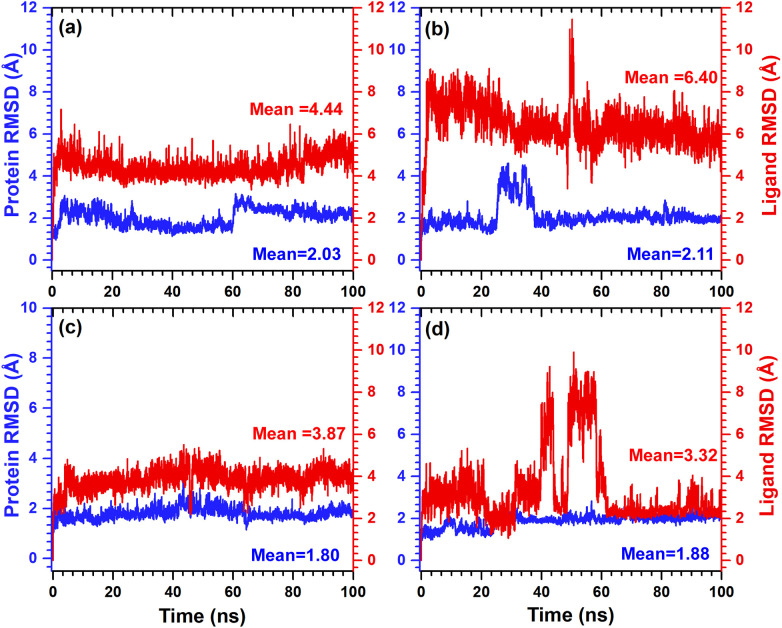
To quantify the degree of conformational changes in protein and ligand of the docked complexes, initially RMSD for protein (Cα, backbone, side-chain, and heavy) and fit ligand on protein were calculated from 100 ns simulation trajectories for each complex. The Cα atoms of SARS-CoV-2 Mpro docked with selected antibiotics and N3 inhibitor showed acceptable fluctuations (mean = 1.80 to 2.11 Å) with equilibration till end of simulation (Figs. 3 , S5). Likewise, backbone, side-chain, and heavy atoms of SARS-CoV-2 Mpro from respective complexes were noted for stable deviations (<3 Å), except for receptor complex with tetracycline (<4.5 Å) showed fluctuation around 20–40 ns followed by state of equilibrium till end of 100 ns simulation (Fig. S6). Besides, RMSD of the four antibiotics fit on protein exhibited acceptable deviations (Fig. 3). Notably, tetracycline and minocycline showed maximum variation (<10.5 Å) between 40 and 60 ns interval followed by equilibrium at 5.8 Å and 2.2 Å, respectively at the end of 100 ns simulation (Fig. 3). While RMSD for the N3 inhibitor fit on protein showed variations (~6 Å) between 40 and 60 ns and then higher deviations (~9-18 Å) after 80 ns till end of 100 ns interval (Fig. S5). These results indicate that antibiotics docked protease complexes have attained the equilibrium during 100 ns simulation while N3 inhibitor docked complex required longer simulation interval to achieve the complex stability. Thereof, based on mean of the RMSD trajectories, the selected antibiotics were concluded with significant complex stability by comparison to N3 inhibitor docked with SARS-CoV-2 Mpro.
Fig. 3.
Calculated RMSD values for α-carbon atoms (blue curves) of SARS-CoV-2 Mpro and selected ligands (red curves), viz. (a) doxycycline, (b), tetracycline, (c) demeclocycline, and (d) minocycline plotted with respect to 100 ns MD simulation time. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Furthermore, flexibility of SARS-CoV-2 Mpro docked with selected compounds was analyzed to monitor the deviations in the protein residues and ligand atoms during simulation (Fig. S7). In all the complexes, acceptable RMSF values for the protein residues were recorded with respect to time where residues on the N-terminal (<2 Å) exhibited low fluctuation compared to the C-terminal (last 10 residues with variation of >4.5 Å). Besides, ligands were also monitored to have significant contacts with the residues in β-strands (containing active pocket of the protein), supports the integration of the docked ligands with residues on the active region of SARS-CoV-2 Mpro (Fig. S7). Moreover, intermolecular interactions of the ligands with secondary structure elements (α-helices and β-strands) of the protein indicate induction of slight rigidity in the protein structure. Hence, these acceptable RMSF values for protein structure and residue-ligand contact mapping suggested the structural stability of SARS-CoV-2 Mpro complexes with antibiotics by comparison to native ligand N3 inhibitor of crystal structure during 100 ns simulation.
Likewise, RMSF calculated for the selected antibiotics fit on SARS-CoV-2 Mpro with respect to simulation time suggested the acceptable deviations (<3.5 Å) in mean structure of the ligand, expect for tetracycline (atoms at position 19 to 28) and minocycline (atoms at position 19 to 28) (Fig. S8). These variations in heteroatoms of the ligands were also concluded to contribute in relatively higher ligand fit on protein RMSD against other antibiotics during simulation. Furthermore, RMSF calculated for the N3 inhibitor fit on SARS-CoV-2 Mpro exhibited higher deviations (5–10 Å) (Fig. S8), which were suggested to contribute higher RMSD in the fit ligand on protein during MD simulation (Fig. S5). In conclusion, RMSD and RMSF indicate the acceptable changes in the antibiotics docked SARS-CoV-2 Mpro complexes by comparison to SARS-CoV-2 Mpro-N3 inhibitor during 100 ns MD simulations.
3.2.2. Protein secondary structure elements (SSE)
The protein SSE (α-helices and β-strands) were monitored for SSE distribution and SSE composition analysis for the respective residue index during the simulation to observed changes in the domains of SARS-CoV-2 Mpro docked with respective ligands (Fig. S9). The selected complexes of SARS-CoV-2 Mpro with antibiotics and N3 inhibitor exhibited only α-helix structures in the C-terminal region (201–303) which formed the domain III of the viral protease. While, N-terminal region showed both α-helices and β-strands (1–200) which constitute the domain I and domain III for the viral protease, defined in the crystal structure for the formation of active pocket [43]. These observations concluded the secondary structure of SARS-CoV-2 Mpro complexed with all the selected ligands (%total SSE = 45 ± 1.2). Interestingly, β-strand (%total 25 ± 1.1) secondary structures in the active pocket of SARS-CoV-2 Mpro were monitored to be significantly occupied by selected antibiotics and N3 inhibitor (Fig. S9). Furthermore, torsion angle potential plot was also studied for all the selected ligands which exhibits the relation between the torsion angle of the respective ligands to their respective potential energy [49]. The torsion angle provides the necessary information for the number of rotatable bonds in the ligand molecule (Fig. S10). Here, doxycycline, tetracycline, demeclocycline, minocycline, and N3 inhibitor were predicted with 6, 6, 7, 6, and 22 rotatable bonds in their structures. The torsion potential and histogram relationships provide a standard method to get an insight into the conformational strain introduced in the ligands during simulation, which assisted to understand the protein-bound ligand conformation. These results indicate that ligand with a smaller number of rotatable bonds showed stronger rigidity and vice-versa may contribute to generate optimum geometry for the ligand in the active pocket to attained higher stability. This can be correlated with the docking score and number for intermolecular interactions in the respective docked complexes (Figs. 2, S2). Moreover, modifications introduced in the fit ligand on protein during MD simulation were also calculated in terms of ligand RMSD, radius of gyration, intra-molecular hydrogen bonds, molecular surface area (MolSA), solvent accessible surface area (SASA), and polar surface area (PSA) (Fig. S11). These results suggested the stability of selected antibiotics at the active pocket of SARS-CoV-2 Mpro by comparison to N3 inhibitor.
3.2.3. Protein-ligand interaction mapping
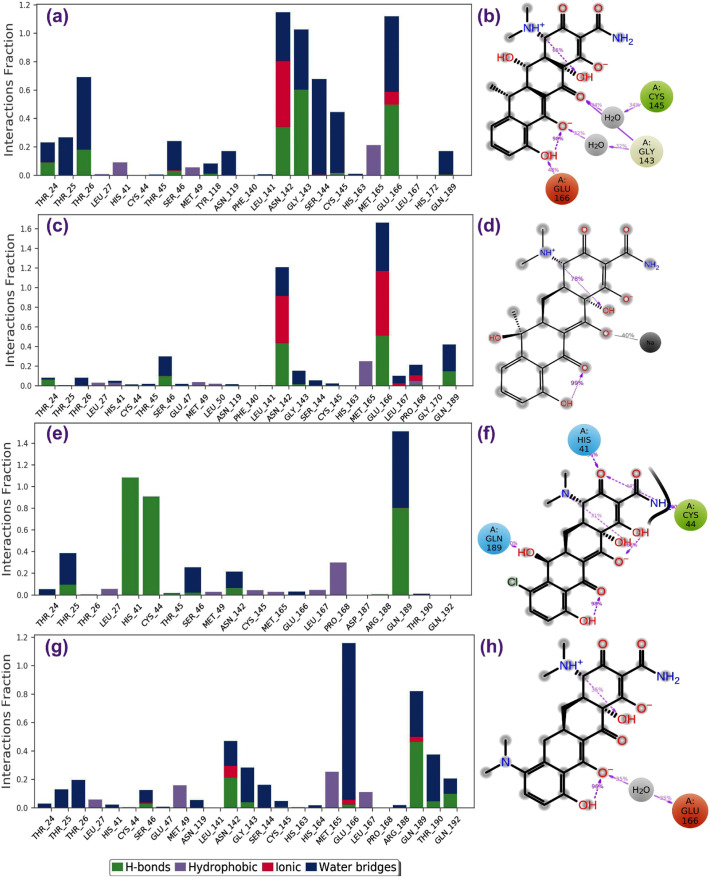
The binding mode of ligand with the active pocket of SARS-CoV-2 Mpro can be further analyzed for the atomic level interactions which includes hydrogen bond formation, hydrophobic contact, ionic interaction, and salt bridge formation during MD simulation. Hence, these protein-ligand interactions were extracted from 100 ns simulation trajectories to support the respective complex stability during MD simulation. All the antibiotics by comparison to N3 inhibitor showed significant interactions with the essential catalytic residues, i.e. His41 and Cys145, in addition to other substrate binding residues in the active pocket of SARS-CoV-2 Mpro (Figs. 4 , S12); these residues were also recorded for the intermolecular interaction with the ligands during molecular docking simulation (Figs. 2. S2). Interestingly, all the antibiotics were observed for higher number of interactions with residues in protein-ligand mapping and formation of hydrogen bond with active residues of the protein for 30% of the simulation interval with an exception in SARS-CoV-2 Mpro-tetracycline complex (Fig. 4). However, only Asn142 and Glu166 were predicted with substantial hydrogen boning and water bridges formation in SARS-CoV-2 Mpro-N3 inhibitor complex from protein-ligand interaction mapping data analysis (Fig. S12). Furthermore, total number of contacts and their density with respect to time were also calculated (Fig. S13). Again, these plots suggested the multiple interactions of the selected antibiotics by comparison to N3 inhibitor with the essential residues (His41, Asn142, Gly143, Cys145, Glu166, and Gln189) in the active pocket of SARS-CoV-2 Mpro. These observations suggested the substantial protein-ligand intermolecular interactions and stability of four antibiotics by comparison to N3 inhibitor throughout the 100 ns MD simulations.
Fig. 4.
Protein-ligand interactions mapping for SARS-CoV-2 Mpro with selected antibiotics, i.e. (a–b) doxycycline, (c–d) tetracycline, (e–f) demeclocycline, and (g–h) minocycline, extracted from 100 ns MD simulations. In 2D interaction diagram, the residues cystine (green), glutamic acid (red), histidine and glutamine (blue), and glycine (grey) exhibit the hydrophobic, negative, polar and non-polar interactions, respectively along with hydrogen bonding (pink arrow) and ionic interaction with sodium ion (black line) with the receptor are extracted at 30% of the total MD simulation interaction interval. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Hence, based on docking score and simulation trajectory analysis, selected four antibiotics are predicted as potential SARS-CoV-2 Mpro inhibitor in the order, viz. doxycycline, minocycline, demeclocycline, and tetracycline, to challenge the SARS-CoV-2 infection by comparison to N3 inhibitor. Recently combinational therapy of lopinavir with the other potent compounds against SARS-CoV-2 virus has been suggested to increase synergy and decrease the effective concentration of lopinavir [50]. For instance, triple antiviral therapy composed of interferon β-1b, lopinavir–ritonavir, and ribavirin against SARS-CoV-2 Mpro was reported as superior and safe against lopinavir–ritonavir alone to reduce the virus shedding, lessening symptoms, and facilitating discharge of infected patients with mild to moderate degree of SARS-CoV-2 infection [51]. In this regard, based on our computational studies, the selected antibiotics, i.e. doxycycline, minocycline, demeclocycline, and tetracycline, may be helpful as combinational therapy with lopinavir-ritonavir and ribavirin against SARS-CoV-2 infection.
4. Conclusion
This study was aimed to repurpose the antibiotics with known anti-viral activity as potential inhibitor against SARS-CoV-2 Mpro with the aid of molecular simulation approaches. A total of four potent antibiotics, viz. doxycycline, tetracycline, demeclocycline, and minocycline were evaluated as SARS-CoV-2 Mpro inhibitor by comparison to native ligand of SARS-CoV-2 Mpro crystal structure (N3 inhibitor). Molecular docking simulation for the selected antibiotics showed docking score >−7 kcal/mol and interaction with catalytic dyad of SARS-CoV-2 Mpro, i.e. His41 and Cys145. These complexes were further analyzed for structural stability by 100 ns classical molecular dynamics simulation, suggested the considerable stability of SARS-CoV-2 Mpro docked complexes with antibiotics by comparison to N3 inhibitor of SARS-CoV-2 Mpro via strong multiple intermolecular interactions. From the binding mode analysis, before and after molecular dynamics simulation analysis followed by interaction profiles collected during the simulation interval supported the selected antibiotics for substantial stability on the active pocket of SARS-CoV-2 Mpro. In conclusion, based on docking score and molecular dynamics simulation trajectory analysis, selected four antibiotics can be used as potential SARS-CoV-2 Mpro inhibitor in the order, viz. doxycycline, minocycline, demeclocycline, and tetracycline to challenge the SARS-CoV-2 infection. Hence, the screened and computationally validated antibiotics can be further evaluated for in vitro SARS-CoV-2 Mpro inhibition and viral infection as single dose or in synergistic concentrations for the drug development and combinational therapy against SARS-CoV-2 infection.
Acknowledgments
Acknowledgment
This work was supported by the 2020 Yeungnam University Research Grant 220A380070.
Author contributions
Shiv Bharadwaj and Vivek Dhar Dwivedi designed the study. Shiv Bharadwaj performed all the computational experiments. Shiv Bharadwaj and Vivek Dhar Dwivedi analyzed the data. Kyung Eun Lee and Sang Gu Kang assisted in data analysis. Shiv Bharadwaj wrote the original and revised manuscript. Sang Gu Kang was responsible for the conduction and evaluation of the present work.
Data availability
The docking structures and molecular dynamics simulations trajectories are available upon request from the corresponding authors.
Declaration of competing interest
The author declares that there is no competing interest in this work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.lfs.2020.118080.
Appendix A. Supplementary data
Supplementary figures
References
- 1.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281(18):4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassetti M., Vena A., Giacobbe D.R. The novel Chinese coronavirus (2019-nCoV) infections: challenges for fighting the storm. Eur. J. Clin. Investig. 2020;50(3):e13209. doi: 10.1111/eci.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McIntosh K., Perlman S. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. vol. e2. 2015. Coronaviruses, including severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) pp. 1928–1936. [Google Scholar]
- 6.Wu C., Liu Y., Yang Y., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;579(7798):270–273. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.du Toit A. Outbreak of a novel coronavirus. Nat. Rev. Microbiol. 2020;18(3):123. doi: 10.1038/s41579-020-0332-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li G., de Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 12.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.-H. An overview of severe acute respiratory syndrome–coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.Y. Coronaviruses-drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15(5):327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morse J.S., Lalonde T., Xu S., Liu W.R. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. ChemBioChem. 2020;21:730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren Z., Yan L., Zhang N., Guo Y., Yang C., Lou Z., et al. The newly emerged SARS-like coronavirus HCoV-EMC also has an “Achilles’ heel”: current effective inhibitor targeting a 3C-like protease. Protein Cell. 2013;4:248–250. doi: 10.1007/s13238-013-2841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3(10):1742–1752. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng F. Cancer Bioinformatics. Springer; 2019. In silico oncology drug repositioning and polypharmacology; pp. 243–261. [DOI] [PubMed] [Google Scholar]
- 21.Karaman B., Sippl W. Computational drug repurposing: current trends. Curr. Med. Chem. 2019;26(28):5389–5409. doi: 10.2174/0929867325666180530100332. [DOI] [PubMed] [Google Scholar]
- 22.Vanhaelen Q., Mamoshina P., Aliper A.M., Artemov A., Lezhnina K., Ozerov I., et al. Design of efficient computational workflows for in silico drug repurposing. Drug Discov. Today. 2017;22(2):210–222. doi: 10.1016/j.drudis.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 23.Mani D., Wadhwani A., Krishnamurthy P.T. Drug repurposing in antiviral research: a current scenario. J Young Pharm. 2019;11(2):117–121. [Google Scholar]
- 24.Dyall J., Coleman C.M., Hart B.J., Venkataraman T., Holbrook M.R., Kindrachuk J., et al. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014;58(8):4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dyall J., Gross R., Kindrachuk J., Johnson R.F., Olinger G.G., Hensley L.E., et al. Middle East respiratory syndrome and severe acute respiratory syndrome: current therapeutic options and potential targets for novel therapies. Drugs. 2017;77(18):1935–1966. doi: 10.1007/s40265-017-0830-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abuhammad A., Al-Aqtash R.A., Anson B.J., Mesecar A.D., Taha M.O. Computational modeling of the bat HKU4 coronavirus 3CLpro inhibitors as a tool for the development of antivirals against the emerging Middle East respiratory syndrome (MERS) coronavirus. J. Mol. Recognit. 2017;30:e2644. doi: 10.1002/jmr.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaeppler U., Stiefl N., Schiller M., Vicik R., Breuning A., Schmitz W., et al. A new lead for nonpeptidic active-site-directed inhibitors of the severe acute respiratory syndrome coronavirus main protease discovered by a combination of screening and docking methods. J. Med. Chem. 2005;48(22):6832–6842. doi: 10.1021/jm0501782. [DOI] [PubMed] [Google Scholar]
- 28.Bacha U., Barrila J., Velazquez-Campoy A., Leavitt S.A., Freire E. Identification of novel inhibitors of the SARS coronavirus main protease 3CLpro. Biochemistry. 2004;43(17):4906–4912. doi: 10.1021/bi0361766. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6(1):1–18. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248 doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elfiky A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chukwudi C.U. rRNA binding sites and the molecular mechanism of action of the tetracyclines. Antimicrob. Agents Chemother. 2016;60(8):4433–4441. doi: 10.1128/AAC.00594-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothan H.A., Bahrani H., Mohamed Z., Teoh T.C., Shankar E.M., Rahman N.A., et al. A combination of doxycycline and ribavirin alleviated chikungunya infection. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0126360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sturtz F.G. Antimurine retroviral effect of doxycycline. Methods Find. Exp. Clin. Pharmacol. 1998;20(8):643–648. doi: 10.1358/mf.1998.20.8.487489. [DOI] [PubMed] [Google Scholar]
- 35.Rothan H.A., Mohamed Z., Paydar M., Rahman N.A., Yusof R. Inhibitory effect of doxycycline against dengue virus replication in vitro. Arch. Virol. 2014;159(4):711–718. doi: 10.1007/s00705-013-1880-7. [DOI] [PubMed] [Google Scholar]
- 36.Yang J.M., Chen Y.F., Tu Y.Y., Yen K.R., Yang Y.L. Combinatorial computational approaches to identify tetracycline derivatives as flavivirus inhibitors. PLoS One. 2007;2(5):e428. doi: 10.1371/journal.pone.0000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michaelis M., Kleinschmidt M.C., Doerr H.W., Cinatl J., Jr. Minocycline inhibits West Nile virus replication and apoptosis in human neuronal cells. J. Antimicrob. Chemother. 2007;60(5):981–986. doi: 10.1093/jac/dkm307. [DOI] [PubMed] [Google Scholar]
- 38.Aman M.J., Bavari S., Burnett J.C., Warfield K.L. 2011. Antiviral Compounds and Methods of Using Thereof U.S. Patent 7,999,001. 16 Aug. [Google Scholar]
- 39.Si Q., Cosenza M.A., Kim M.-O., Zhao M.-L., Brownlee M., Goldstein H., et al. A novel action of minocycline: inhibition of human immunodeficiency virus type 1 infection in microglia. J. Neuro-Oncol. 2004;10(5):284–292. doi: 10.1080/13550280490499533. [DOI] [PubMed] [Google Scholar]
- 40.Nagarakanti S., Bishburg E. Is minocycline an antiviral agent? A review of current literature. Basic Clin. Pharmacol. Toxicol. 2016;118(1):4–8. doi: 10.1111/bcpt.12444. [DOI] [PubMed] [Google Scholar]
- 41.Sodhi M., Etminan M. Therapeutic potential for tetracyclines in the treatment of COVID-19. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2020;40(5):487–488. doi: 10.1002/phar.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J. Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. J. Chem. Inf. Model. 2020;60(6):3277–3286. doi: 10.1021/acs.jcim.0c00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., et al. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 44.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., et al. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bowers K.J., Chow D.E., Xu H., Dror R.O., Eastwood M.P., Gregersen B.A., et al. SC'06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing. IEEE; 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters; p. 43. [Google Scholar]
- 47.Bharadwaj S., Lee K.E., Dwivedi D., Yadava U., Panwar A., Lucas S.J., et al. Discovery of Ganoderma lucidum triterpenoids as potential inhibitors against Dengue virus NS2B-NS3 protease. Sci. Rep. 2019;9(1):1–12. doi: 10.1038/s41598-019-55723-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bharadwaj S., Rao A.K., Dwivedi V.D., Mishra S.K., Yadava U. Structure-based screening and validation of bioactive compounds as Zika virus methyltransferase (MTase) inhibitors through first-principle density functional theory, classical molecular simulation and QM/MM affinity estimation. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1747545. [DOI] [PubMed] [Google Scholar]
- 49.Thangavel M., Chandramohan V., Shankaraiah L.H., Jayaraj R.L., Poomani K., Magudeeswaran S., et al. Design and molecular dynamic investigations of 7,8-dihydroxyflavone derivatives as potential neuroprotective agents against alpha-synuclein. Sci. Rep. 2020;10(1):1–10. doi: 10.1038/s41598-020-57417-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choy K.-T., Wong A.Y.-L., Kaewpreedee P., Sia S.-F., Chen D., Hui K.P.Y., et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hung I.F.N., Lung K.C., Tso E.Y.K., Liu R., Chung T.W.H., Chu M.Y., et al. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395(10238):1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Data Availability Statement
The docking structures and molecular dynamics simulations trajectories are available upon request from the corresponding authors.