Abstract
Background
The number of cases from the coronavirus disease 2019 (COVID-19) global pandemic has overwhelmed existing medical facilities and forced clinicians, patients, and families to make pivotal decisions with limited time and information.
Main body
While machine learning (ML) methods have been previously used to augment clinical decisions, there is now a demand for “Emergency ML.” Throughout the patient care pathway, there are opportunities for ML-supported decisions based on collected vitals, laboratory results, medication orders, and comorbidities. With rapidly growing datasets, there also remain important considerations when developing and validating ML models.
Conclusion
This perspective highlights the utility of evidence-based prediction tools in a number of clinical settings, and how similar models can be deployed during the COVID-19 pandemic to guide hospital frontlines and healthcare administrators to make informed decisions about patient care and managing hospital volume.
Keywords: Artificial intelligence (AI), Clinical decision-making, Coronavirus disease 19 (COVID-19), Healthcare, Machine learning (ML)
Background
Coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV2), has spread to the level of a global pandemic (World Health Organization 2020) with, at the time of writing, over 1.6 million reported cases in the United States and 5.6 million worldwide. In the United States, New York City was the epicenter of the disease with over 199,000 confirmed cases and over 16,400 deaths (Center for Systems Science and Engineering (CSSE) at Johns Hopkins University 2020). At the pandemic’s peak, existing medical facilities were overwhelmed, with emergency departments (EDs), floor units, and Intensive Care Units (ICUs) stretched beyond capacity and resources (Evans and Armour 2020). Due to the challenges of this novel disease, healthcare providers, patients, and their families have been required to rapidly make crucial and difficult decisions with limited information. The phenotypes of COVID-19 range from no or relatively mild symptoms and uneventful recovery to rapid deterioration, acute respiratory distress syndrome (ARDS), multi-organ system failure, and death. The trajectory for patients most likely to decompensate is being investigated but remains elusive at present; lack of standardized care is forcing unprecedented workflow for physicians and nurses. Given the gravity of these circumstances and increase in the number of cases, there is a pressing need for tools that can augment current healthcare resources. Machine learning (ML) and artificial intelligence (AI) methods can be applied to understand subgroups of patients, guide clinical decision-making, and improve both operation- and patient-centered outcomes. This perspective highlights the benefits of these tools observed at various clinical settings and describes how the value of ML and AI algorithms, when conscientiously built, may be augmented during the COVID-19 pandemic.
Main text
Identifying underlying clinical patterns is already an area of active investigation in the field of ML/AI in healthcare; programs range from modulating single parameters to advanced predictive modeling to forecast decompensation, among other clinical outcomes, and augment medical decisions (Churpek and Edelson 2016; Kipnis et al. 2016; Brekke et al. 2019). The ongoing COVID-19 health crisis has transformed the aspiration for enhanced clinical decision-making tools into a demand for “Emergency ML.” To support the medical response to COVID-19, researchers are compressing traditional timelines of retrospective training and testing, prospective validation, incremental launches, and deployment of algorithms. Clinical outcomes that an ML algorithm could predict resemble a perpetually moving target. The clinical data, including multiple patient cohorts and a variety of outcomes, are updated daily or more frequently with evolving statistics and ever-increasing observations (Santosh 2020). Since the phenotype of COVID-19 deviates from typical ARDS or other acute organ dysfunctions, existing models of decompensation, risk of mortality, or clinical trajectory prediction created for non-COVID-19 cohorts are not guaranteed to maintain previously reported performance (Wang et al. 2019; Zampieri et al. 2019; Tomasev et al. 2019; Mousavizadeh and Ghasemi 2020). Some currently proposed diagnostic models to detect COVID-19 infection in symptomatic patients excluded proportions of patients with the disease or favored certain predictors, therefore losing key information and potentially limiting performance in widespread screenings (Meng et al. 2020; Song et al. 2020a; Lopez-Rincon et al. 2020; Batista et al. 2020). Thus, adapting and training algorithms on this unique patient population is essential to construct effective prediction models.
As patients move between a range of clinical settings (outpatient clinics, EDs, floor units, ICUs), they generate unique personal datasets that reflect their phenotypes and require varying clinical resources and participate in multiple decisions, ranging from trivial to potentially life altering. In these settings and during each transition, ML/AI can help clinicians, patients, and their families efficiently process all available data to generate informed, evidence-based recommendations and participate in shared decision-making to identify the optimal course of action. In the age of COVID-19, this can be incorporated into several opportunities across the spectrum of care (Fig. 1a).
Fig. 1.
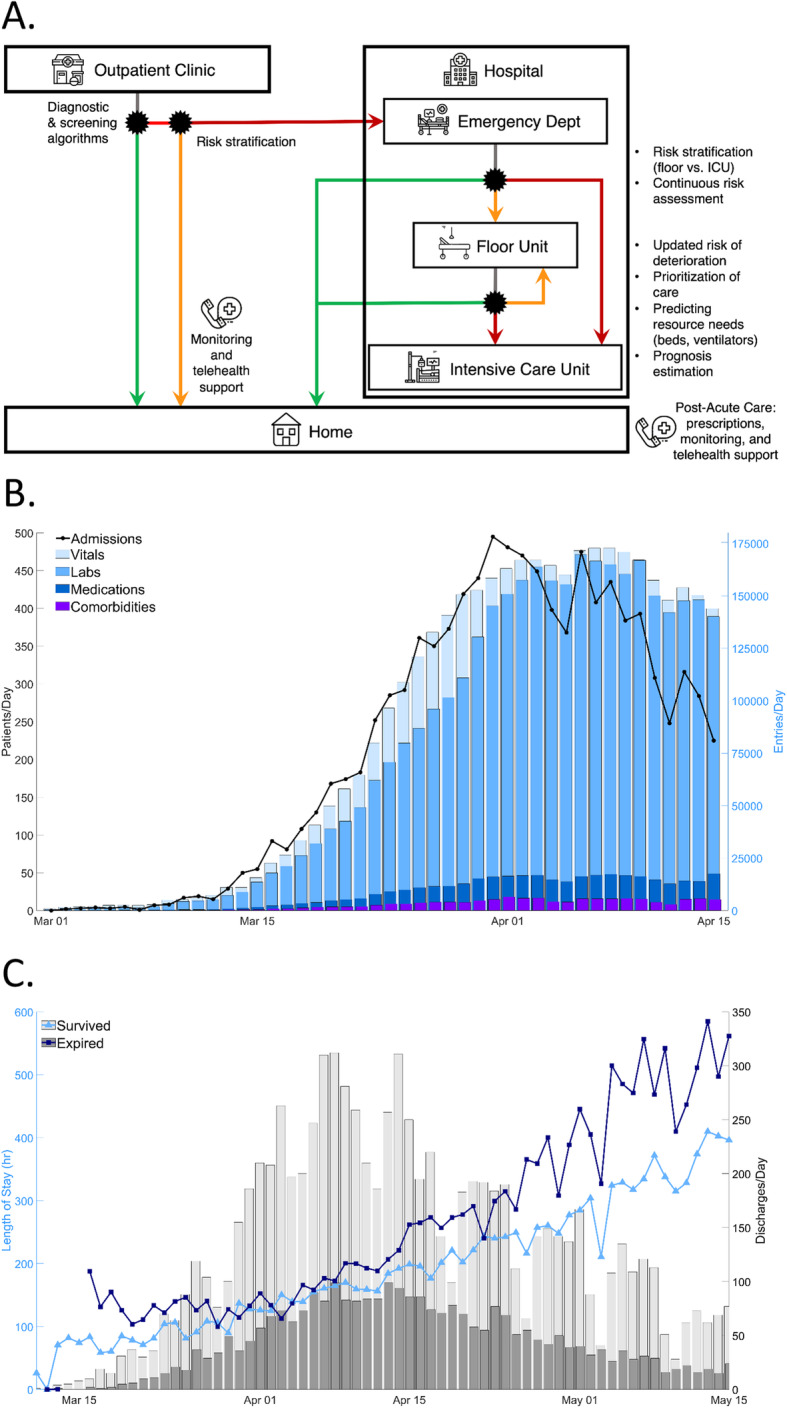
a ML/AI in the patient care pathway. The black asterisks represent multiple decision points during the patient care pathway that could be augmented by ML/AI tools. The green traces represent a COVID-19 negative diagnosis or recovery while the orange and red traces represent risk stratification of patients by lower and higher risks of deterioration, respectively, as determined by a potential ML/AI model. Additional decisions in the hospital include prioritization of care, allocation of resources, and estimation of prognosis. b An expanding COVID-19 database. Since March 1, 2020, there has been an increasing amount of COVID-19 patient data, shown here by new admissions and new medical data entries at Northwell Health, New York’s largest health system, facilities. Given increasing hospital admissions (black trace, left y-axis), there have been hundreds of thousands of new data entries per day (colored bars, right y-axis), including vitals, laboratory results, medication orders, and patient comorbidities. This vast data allows a unique opportunity to implement ML/AI to support medical frontlines and healthcare administrators in the fight against COVID-19. c Evolving patient profiles and discharge rates. Basic characteristics of the patient population changed during the progression of the wave of new cases, which can affect performance of a predictive model. For example, the average length of stay for expired patients and those discharged alive (dark blue with square markers and light blue with triangle markers, respectively, left y-axis) diverged in mid-April. Because of these changes, a predictive model with good early performance may decline because of differences between patients hospitalized for three weeks compared to less than a week. Also, an individual’s patient profile may have evolved significantly from hospital admission to those timepoints later during hospitalization. Discharges per day (grey bars, right y-axis) increased with the pandemic’s peak and declined with reduced numbers of new cases
The first opportunity lies during initial screening and evaluation of symptomatic people in outpatient facilities. Development of ML models to risk-stratify these individuals and separate low-risk patients from those at higher risk for deterioration can enable focus on the needs of approximately 15% that require more resource-intensive care (Center for Disease Control and Prevention (CDC) COVID-19 Response Team 2020). Identification of low-risk patients can lead to increased utilization of telehealth and virtual care to avoid unnecessary hospital admissions (Dorsey and Topol 2016). Previous ML models have already been developed to reduce avoidable initial admissions (Ngo et al. 2019), predict risk of 30-day readmissions (Frizzell et al. 2017; Golas et al. 2018), and improve pharmaceutical prescriptions (Ribers and Ullrich 2019). Even disease diagnosis can be possible using emerging ML/AI technology; Parkinson’s Disease can be successfully detected by a smartphone-based monitoring platform that extracts features from voice, gait, and reaction time data (Zhan et al. 2018). These approaches suggest the possibility of improving COVID-19-screening phone calls and follow-up survey information (Rao and Vazquez 2020). A model could predict the probability of a confirmed COVID-19 diagnosis and its severity by taking answers from symptomatic individuals and amplifying it with clinical information from electronic health records (EHRs), including comorbidities (DeCaprio et al. 2020), complaints, and demographics (including geography). An ML model could even predict levels of dyspnea over the phone with estimations of emotional affect and cough sounds from speech already possible (Fayek et al. 2017; Porter et al. 2019). The severity prediction could also indicate the level of necessary care: self-monitoring, outpatient doctor visit, or ED visit (Greenhalgh et al. 2020). Prediction models can reduce the number of patients that are admitted to already-overloaded hospitals. However, this type of model may be limited by lack of vital clinical information such as oxygen saturation or severity of dyspnea, which is highly subjective via self-report. Additionally, variability in the exact time that patients develop symptoms after exposure imposes additional challenges (Lauer et al. 2020). Finally, specific patient behavior could influence the quality of data labels used for building ML algorithms. For example, individuals may not call because they have no or mild upper respiratory symptoms; studies estimate that over half of COVID-19 infections are undocumented (Li et al. 2020a). Conversely, individuals may be advised to self-monitor but seek treatment, regardless. These groups could lead to inaccurate labeling for training and testing ML algorithms; more widespread testing and big data could reduce the effects of this hurdle.
The ED represents a second opportunity to apply ML/AI to a data-rich, time-pressured environment where clinicians are called upon to collect, assimilate, and analyze large amounts of data. This begins at the point of triage, where severity of symptoms and the need for urgent intervention are first assessed, to the continued course of patient evaluation through vitals measurements and laboratory results. The changing status of the patient’s health culminates in the decision to either discharge the patient or admit them to the hospital. If escalation of care in the hospital is necessary, determining the most appropriate environment (i.e., floor or critical care unit) is intimately linked to assessing future risk of deterioration. ML/AI models have been developed to be applied at each of these phases within ED care; in triage, vitals representing illness severity can predict resource needs (Levin et al. 2018; Raita et al. 2019) while risk profiles can be determined as new data is obtained (Janke et al. 2016). Lastly, patient condition while in the ED can be used to calculate risk of deterioration or death (Brajer et al. 2020). For patients with COVID-19, ML/AI can provide decision support at each stage by calculating likelihood of admission at triage, refining risk estimates with real data from clinical evaluation, and predicting the patient’s trajectory as well as the effects of prompt ventilator use. A possible input to these models may be chest X-rays and computed tomography (CT) images from diagnosis and disease progression; CT severity scores have been shown to identify patients with severe cases of COVID-19 (Yang et al. 2020), and diagnostic models based on CT images have been proposed to support diagnosis and monitor progression (Jin et al. 2020a; Song et al. 2020b; Xu et al. 2020; Shan et al. 2020; Wang et al. 2020; Ozturk et al. 2020;Jin et al. 2020, 2020a; Li et al. 2020b; Shi et al. 2020a). Within the ED, assessing the possibility of respiratory failure can help institutions and clinicians prioritize and allocate scarce resources as demand outpaces supply.
A third point of intervention lies in the transition from the ED to the inpatient setting and throughout the patient’s stay. The staggering wave of admissions in this pandemic is overwhelming even the most efficient of hospital frontlines. Under normal conditions, hospitals with a large influx of patients tend to struggle to manage them in a timely manner (Morley et al. 2018). Studies have shown that as the time to see admitted patients increases, so does the risk for adverse events (Eriksson et al. 2017). Any time delay in this crisis can lead to a missed opportunity or limited time to save a life. ML/AI techniques can augment the acumen of healthcare providers and aid hospitals in the Herculean task of managing patient volume by calculating measures that can prioritize patients and potentially decrease adverse events. As patient numbers keep increasing, ML/AI tools, based on ever-increasing ongoing vitals, labs, medications, and orders (Fig. 1b), can be applied to calculate risk scores for multiple time points. Short-term predictions (4–8 h) can be used for nurses and physicians to prioritize care. Mid-term predictions (12–24 h) can help units identify patients with the least likelihood of decompensation; this measure of stability can support decisions by clinicians to adapt care on the path to discharge. Lastly, long-term models (more than 24 h) can help administrators allocate precious resources, such as ventilators, beds, and staffing. Clinically predictive tools already exist to predict mortality risks by calculating a score from Multilobular infiltration, hypo-Lymphocytosis, Bacterial coinfection, Smoking history, hyper-Tension, and Age (MuLBSTA) to separate pneumonia patients into relevant categories of care and guide clinical decisions (Guo et al. 2019), and early work with small patient cohorts of COVID-19 has led to models that identify key clinical characteristics that can predict severe cases (Yan et al. 2020; Jiang et al. 2020; Xie et al. 2020; Bai et al. 2020; Caramelo et al. 2020; Lu et al. 2020; Gong et al. 2020; Shi et al. 2020b). One particular model—the Northwell COVID-19 Survival (“NOCOS”) calculator – was built from demographic, laboratory, clinical, and treatment data of over 5200 inpatients to predict survival probability; seven variables from patient EHRs were identified as early predictors of survival, and the easily comprehensible output of the calculator is being used by clinicians to provide critical decision support (Levy et al. 2020). Finally, ML can assist healthcare providers with the most emotionally difficult conversations with patients and their families: goals of care. By providing an objective measure of mortality risk, ML offers clinicians, patients, and their families essential information in this shared decision process.
While researchers in the field of digital medicine and healthcare ML have built predictive models for decades, the urgency to clinically operationalize them is novel. Practices established to tackle COVID-19 can offer a future path for clinical translation of ML/AI tools. The ultimate goal of any model is to maximize patient outcomes for the greatest number of patients within an acceptable ethical framework. However, the inputs to these models can vary based on availability of equipment or staff; heterogeneity in standards of care; responses to anecdotes, personal communications, and preprinted scientific manuscripts; and evolving government policies and professional society guidelines. Such extreme variability in inputs makes setting a fixed operating point, usually based on clinicians’ acceptability of risk, very difficult. For these reasons, models should be operationalized with frequent and automated re-training and re-validation.
Another extremely important consideration is ensuring that all development studies are scientifically and ethically sound. Firstly, available data only represents patients that are admitted and treated. Data-driven models may magnify current disparities in healthcare because there is already less data to represent those with diminished access to care. This issue of equity can limit generalizability of ML/AI models. Secondly, in an outbreak setting, access to any therapy is pressing but still requires strong proof of efficacy (Lane et al. 2016). In this public health emergency, rapid responses are critical, and testing of COVID-19 vaccines has been fast-tracked to phase I trials for early spring of 2020 (Ahmed et al. 2020; Fauci et al. 2020). This provides a compelling precedent to accelerate the deployment of accurate ML models. If an algorithm is proven on retrospective data, it should be implemented into clinical translation for validation and use as quickly as possible. It is crucial to note, however, that an expedited process for model creation and validation can lead to inaccurate results, as seen by many of the early projection models for the progression of COVID-19. To prevent errors and strengthen the accuracy of models over time, active learning is required, with continued emphases on increased frequency for re-training and re-validation as more data becomes available. Moreover, different types of validations can strengthen the stakeholders’ trust in a model’s predictive performance. Prospective validation can address the model’s stability over time, while external validation can address the model’s clinical portability to various hospitals and geographic locations. Whenever possible, both types of validation should be carried out. This concern is not only specific to COVID-19 related models but any application of healthcare ML/AI.
Specifically, during the COVID-19 pandemic, basic characteristics of the patient population can change during the evolving stages of the patient wave, affecting the performance of a predictive model (Fig. 1c). Throughout April, both expired and discharged surviving patients followed similar rates in discharges per day. However, the average length of stay for these two cohorts increased daily, but also diverged significantly by early April, with the patients that expire after that date presenting a five-fold increase in hospitalization duration, compared to a four-fold increase for discharged surviving patients. Such a massive difference in an integral aspect of the dataset, like length of stay, can have a profound effect in prognostic algorithms of arbitrary horizons; while early on in the pandemic, these algorithms would perform very well, they would naturally degrade later on. This decline is due to both the differences in profile of patients that stayed on average 3 weeks in the hospital compared to those that stay less than a week and internal comparisons per patient, when analyzing their medical profile at the beginning of their hospitalization relative to the end. Figure 1c showcases the unique characteristics of a COVID-19 dataset, as we have captured it from our database. As shown, discharge numbers increased and remained high during the pandemic’s peak.
Other potential challenges regard curating high fidelity inputs that can produce reliable outputs for clinician use. It will be necessary to perform automated data scrubbing to all new data entries and establish efficient deployment of updated algorithms. This goal must be achieved by essential integration into standardized pipelines of EHR platforms. The COVID-19 pandemic has mobilized efforts for increasing patient data availability, but these large amounts of new data require continuous quality checks to ensure that desired outputs are achieved. Without high quality data inputs, any model’s output, especially those that rely on evolving databases to increase accuracy, will suffer from garbage in, garbage out (GIGO). In addition, outputs of any model must be easy to evaluate and factor into traditional medical care. An indexed value could represent a patient priority based on probability of outcome. Priority scores can guide allocation of care, ventilators, and beds as well as designate patients to the appropriate medical departments (White and Lo 2020). Finally, amid imposed lockdowns, maintaining communication between the ML engineers, data scientists, and stakeholders, including hospital frontlines and healthcare administrators, can be challenging. Sustaining increased productivity and morale is necessary for the success of all such endeavors.
Conclusions
While caring for thousands of COVID-19 patients, hospital staff, nurses, physicians, administrators, scientists, and engineers have also been pursuing ways to optimize care to face the onslaught of daily new cases. ML and AI is becoming more prevalent in healthcare and medicine, and the worldwide COVID-19 crisis presents a critical situation that demands implementation of ML approaches, whether applications are for medical treatment research, patient care, allocating resources, or managing hospital volume. Medical personnel in all clinical settings, including doctors and nurses, need a support system in a shared decision-making process that includes patients and their families. Expanding datasets provide an example of available information in this unique opportunity for deployment of an unprecedented “emergency ML” effort. Northwell Health, New York’s largest health system, serves multiple communities at the world’s current epicenter of the COVID-19 pandemic and maintains an example of a sizeable COVID-19 patient database—featuring vitals, laboratory results, ordered medications, and continuously captured and updated demographic information, and it has already been used to present characteristics and early clinical outcomes of 5700 hospitalized COVID-19 patients in the New York City area (Richardson et al. 2020). With increasing health data, evidence-based prediction tools trained and validated properly and often can guide overwhelmed hospital frontlines and administrators to make informed decisions in a challenging time. With the best possible data and analytics, the field of ML/AI can be a key ally in the fight to limit the devastating consequences of COVID-19.
Acknowledgments
We acknowledge and honor all of our Northwell team members who consistently put themselves in harm’s way during the COVID-19 pandemic. We dedicate this article to them, as their vital contribution to knowledge about COVID-19 and sacrifices on the behalf of patients made it possible.
Abbreviations
- AI
Artificial intelligence
- ARDS
Acute respiratory distress syndrome
- COVID-19
Coronavirus disease 2019
- CT
Computed tomography
- ED
Emergency department
- EHR
Electronic health record
- ICU
Intensive care unit
- ML
Machine learning
- MuLBSTA
-
Multilobular infiltration, hypo-Lymphocytosis, Bacterial coinfection, Smoking history
hyper-Tension, and Age
- NOCOS
Northwell COVID-19 Survival
- SARS-CoV2
Severe acute respiratory syndrome coronavirus 2
Authors’ contributions
SD and TPZ conceptualized and designed the study. JSH and TPZ had full access to all data in the study and take responsibility for the integrity of the data. KC performed data extraction and cleaning. DPB, AM, EK, SC, VT, TL, SLC, and MdP contributed to many discussions at different stages of manuscript development and supported literature review. SD, DPB, AM, EJ, and TPZ contributed to drafts of the manuscript. SD, DPB, and SC designed and created the Fig. SC, MdP, and JSH critically reviewed the paper. The Northwell COVID-19 Research Consortium prioritized this manuscript, organized meetings between contributing authors, and provided support in finalizing the manuscript for submission. The authors read and approved the final submitted manuscript.
Funding
This work was supported by grants R24AG064191 from the National Institute on Aging of the National Institutes of Health and R01LM012836 from the National Library of Medicine of the National Institutes of Health. Neither source of funding had any role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
Availability of data and materials
The data that support the findings of this study are available on request from COVID19@northwell.edu. The data are not publicly available due to restrictions as it could compromise the privacy of research participants.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Theodoros P. Zanos, Email: tzanos@northwell.edu
the Northwell COVID-19 Research Consortium:
Lance B. Becker, Jennifer Cookingham, Karina W. Davidson, Andrew J. Dominello, Louise Falzon, Thomas McGinn, Jazmin N. Mogavero, and Gabrielle A. Osorio
References
- Ahmed SF, Quadeer AA, McKay MR. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses 2020; 12:doi: 210.3390/v12030254. [DOI] [PMC free article] [PubMed]
- Bai X, Fang C, Zhou Y, Bai S, Liu Z, Chen Q, Xu Y, Xia T, Gong S, Xie X, et al. Predicting COVID-19 malignant progression with AI techniques. medRxiv. 2020.03.20.20037325.
- Batista AFM, Miraglia JL, Donato THR, Chiavegatto Filho ADP. COVID-19 diagnosis prediction in emergency care patients: a machine learning approach. medRxiv. 2020.04.04.20052092.
- Brajer N, Cozzi B, Gao M, Nichols M, Revoir M, Balu S, et al. Prospective and external evaluation of a machine learning model to predict in-hospital mortality of adults at time of admission. JAMA Netw Open. 2020;3(2):e1920733. doi: 10.1001/jamanetworkopen.2019.20733. [DOI] [PubMed] [Google Scholar]
- Brekke IJ, Puntervoll LH, Pedersen PB, Kellett J, Brabrand M. The value of vital sign trends in predicting and monitoring clinical deterioration: a systematic review. PLoS One. 2019;14(1):e0210875. doi: 10.1371/journal.pone.0210875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramelo F, Ferreira N, Oliveiros B. Estimation of risk factors for COVID-19 mortality - preliminary results. medRxiv. 2020.02.24.20027268.
- Center for Disease Control and Prevention (CDC). CDC COVID-19 Response Team. Severe outcomes among patients with coronavirus disease 2019 (COVID-19) — United States, February 12–march 16, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:343-6. [DOI] [PMC free article] [PubMed]
- Center for Systems Science and Engineering (CSSE) at Johns Hopkins University . Coronavirus disease 2019 (COVID-19): cases in U.S. (U.S) Atlanta: Department of Health & Human Services; 2020. [Google Scholar]
- Churpek MM, Edelson DP. Moving beyond single-parameter early warning scores for rapid response system activation. Crit Care Med. 2016;44:2283–2285. doi: 10.1097/CCM.0000000000002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaprio D, Gartner JA, Burgess T, Kothari S, Sayed S, McCall CJ. Building a COVID-19 vulnerability index. medRxiv. 2020.03.16.20036723.
- Dorsey ER, Topol EJ. State of telehealth. N Engl J Med. 2016;375:154–161. doi: 10.1056/NEJMra1601705. [DOI] [PubMed] [Google Scholar]
- Eriksson CO, Stoner RC, Eden KB, Newgard CD, Guise JM. The association between hospital capacity strain and inpatient outcomes in highly developed countries: a systematic review. J Gen Intern Med. 2017;32(6):686–696. doi: 10.1007/s11606-016-3936-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M, Armour S. Hospital capacity crosses tipping point in U.S. coronavirus hot spots. In: the wall street journal. New York: Dow Jones & Company, Inc.; 2020. [Google Scholar]
- Fauci AS, Lane HC, Redfield RR. Covid-19 - navigating the uncharted. N Engl J Med. 2020;382(13):1268–1269. doi: 10.1056/NEJMe2002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayek HM, Lech M, Cavedon L. Evaluating deep learning architectures for speech emotion recognition. Neural Netw. 2017;92:60–68. doi: 10.1016/j.neunet.2017.02.013. [DOI] [PubMed] [Google Scholar]
- Frizzell JD, Liang L, Schulte PJ, Yancy CW, Heidenreich PA, Hernandez AF, et al. Prediction of 30-day all-cause readmissions in patients hospitalized for heart failure: comparison of machine learning and other statistical approaches. JAMA Cardiol. 2017;2(2):204–209. doi: 10.1001/jamacardio.2016.3956. [DOI] [PubMed] [Google Scholar]
- Golas SB, Shibahara T, Agboola S, Otaki H, Sato J, Nakae T, et al. A machine learning model to predict the risk of 30-day readmissions in patients with heart failure: a retrospective analysis of electronic medical records data. BMC Med Inform Decis Mak. 2018;18(1):44. doi: 10.1186/s12911-018-0620-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Ou J, Qiu X, Jie Y, Chen Y, Yuan L, et al. A tool to early predict severe 2019-novel coronavirus pneumonia (COVID-19): a multicenter study using the risk nomogram in Wuhan and Guangdong, China. medRxiv. 2020.03.17.20037515.
- Greenhalgh T, Koh GCH, Car J. Covid-19: a remote assessment in primary care. BMJ. 2020;368:m1182. doi: 10.1136/bmj.m1182. [DOI] [PubMed] [Google Scholar]
- Guo L, Wei D, Zhang X, Wu Y, Li Q, Zhou M, et al. Clinical features predicting mortality risk in patients with viral pneumonia: the MuLBSTA score. Front Microbiol. 2019;10:2752. doi: 10.3389/fmicb.2019.02752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke AT, Overbeek DL, Kocher KE, Levy PD. Exploring the potential of predictive analytics and big data in emergency care. Ann Emerg Med. 2016;67(2):227–236. doi: 10.1016/j.annemergmed.2015.06.024. [DOI] [PubMed] [Google Scholar]
- Jiang X, Coffee M, Bari A, Wang J, Jiang X, Huang J, et al. Towards an artificial intelligence framework for data-driven prediction of coronavirus clinical severity. Computers, Materials & Continua. 2020;63(1):537–551. doi: 10.32604/cmc.2020.010691. [DOI] [Google Scholar]
- Jin C, Chen W, Cao Y, Xu Z, Zhang X, Deng L, Zheng C, Zhou J, Shi H, Feng J. Development and evaluation of an AI system for COVID-19 diagnosis. medRxiv. 2020a.03.20.20039834. [DOI] [PMC free article] [PubMed]
- Jin S, Wang B, Xu H, Luo C, Wei L, Zhao W, Hou X, Ma W, Xu Z, Zheng Z, et al. AI-assisted CT imaging analysis for COVID-19 screening: building and deploying a medical AI system in four weeks. medRxiv 2020b.03.19.20039354. [DOI] [PMC free article] [PubMed]
- Kipnis P, Turk BJ, Wulf DA, LaGuardia JC, Liu V, Churpek MM, et al. Development and validation of an electronic medical record-based alert score for detection of inpatient deterioration outside the ICU. J Biomed Inform. 2016;64:10–19. doi: 10.1016/j.jbi.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane HC, Marston HD, Fauci AS. Conducting clinical trials in outbreak settings: points to consider. Clin Trials. 2016;13(1):92–95. doi: 10.1177/1740774515618198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer SA, Grantz KH, Bi Q, Jones, FK, Zheng Q, Meredith HR, Azman AS, Reich NG, Lessler J. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172(9):577–82. [DOI] [PMC free article] [PubMed]
- Levin S, Toerper M, Hamrock E, Hinson JS, Barnes S, Gardner H, et al. Machine-learning-based electronic triage more accurately differentiates patients with respect to clinical outcomes compared with the emergency severity index. Ann Emerg Med. 2018;71(5):565–e562. doi: 10.1016/j.annemergmed.2017.08.005. [DOI] [PubMed] [Google Scholar]
- Levy TJ, Richardson S, Coppa K, Barnaby DP, McGinn T, Becker LB, Davidson KW, Hirsch JS, Zanos T. Estimating survival of hospitalized COVID-19 patients from admission information. medRxiv. 2020.04.22.20075416.
- Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q, et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020b:200905. [DOI] [PMC free article] [PubMed]
- Li R, Pei S, Chen B, Song Y, Zhang T, Yang W, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2). Science. 2020;367(6490): 489-93. [DOI] [PMC free article] [PubMed]
- Lopez-Rincon A, Tonda A, Mendoza-Maldonado L, Claassen E, Garssen J, Kraneveld AD. Accurate identification of SARS-CoV-2 from viral genome sequences using deep learning. bioRxiv. 2020.03.13.990242.
- Lu J, Hu S, Fan R, Liu Z, Yin X, Wang Q, Lv Q, Cai Z, Li H, Hu Y, et al. ACP risk grade: a simple mortality index for patients with confirmed or suspected severe acute respiratory syndrome coronavirus 2 disease (COVID-19) during the early stage of outbreak in Wuhan, China. medRxiv. 2020.02.20.20025510.
- Meng Z, Wang M, Song H, Guo S, Zhou Y, Li W, Zhou Y, Li M, Song X, Zhou Y, et al. Development and utilization of an intelligent application for aiding COVID-19 diagnosis. medRxiv. 2020.03.18.20035816.
- Morley C, Unwin M, Peterson GM, Stankovich J, Kinsman L. Emergency department crowding: a systematic review of causes, consequences and solutions. PLoS One. 2018;13(8):e0203316. doi: 10.1371/journal.pone.0203316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavizadeh L, Ghasemi S. Genotype and phenotype of COVID-19: their roles in pathogenesis. J Microbiol Immunol Infect. 2020. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- Ngo T, Georgescu V, Gervet C, Laurent A, Libourel T, Mercier G. Machine learning application to the reduction of ambulatory care sensitive admissions (ACSA) Eur J Public Health. 2019;29(Suppl 4):ckz187.061. doi: 10.1093/eurpub/ckz187.061. [DOI] [Google Scholar]
- Ozturk S, Ozkaya U, Barstugan M. Classification of coronavirus images using shrunken features. medRxiv. 2020.04.03.20048868. [DOI] [PMC free article] [PubMed]
- Porter P, Abeyratne U, Swarnkar V, Tan J, Ng TW, Brisbane JM, et al. A prospective multicentre study testing the diagnostic accuracy of an automated cough sound centred analytic system for the identification of common respiratory disorders in children. Respir Res. 2019;20(1):81. doi: 10.1186/s12931-019-1046-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raita Y, Goto T, Faridi MK, Brown DFM, Camargo CA, Jr, Hasegawa K. Emergency department triage prediction of clinical outcomes using machine learning models. Crit Care. 2019;23(1):64. doi: 10.1186/s13054-019-2351-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A, Vazquez JA. Identification of COVID-19 can be quicker through artificial intelligence framework using a Mobile phone-based survey in the populations when cities/towns are under quarantine. Infect Control Hosp Epidemiol. 2020. p. 1–5. [DOI] [PMC free article] [PubMed]
- Ribers MA, Ullrich H. Battling antibiotic resistance: can machine learning improve prescribing? CESifo working paper series. Munich: CESifo group; 2019.
- Richardson S, Hirsch JS, Narasimhan M, Crawford JM, McGinn T, Davidson KW, and the Northwell Covid-Research Consortium, Barnaby DP, Becker LB, Chelico JD, et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the new York City area. JAMA. 2020; [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- Santosh KC. AI-driven tools for coronavirus outbreak: need of active learning and cross-population train/test models on Multitudinal/multimodal data. J Med Syst. 2020;44(5):93. doi: 10.1007/s10916-020-01562-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan F, Gao Y, Wang J, Shi W, Shi N, Han M, Xue Z, Shen D, Shi Y. Lung infection quantification of COVID-19 in CT images with deep learning. In: arXiv e-prints. 2020: arXiv:2003.04655.
- Shi F, Xia L, Shan F, Wu D, Wei Y, Yuan H, Jiang H, Gao Y, Sui H, Shen D. Large-scale screening of COVID-19 from community acquired pneumonia using infection size-aware classification. In: arXiv e-prints. 2020a: arXiv:2003.09860. [DOI] [PubMed]
- Shi Y, Yu X, Zhao H, Wang H, Zhao R, Sheng J. Host susceptibility to severe COVID-19 and establishment of a host risk score: findings of 487 cases outside Wuhan. Crit Care. 2020;24(1):108. doi: 10.1186/s13054-020-2833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C-Y, Xu J, He J-Q, Lu Y-Q. COVID-19 early warning score: a multi-parameter screening tool to identify highly suspected patients. medRxiv. 2020a.03.05.20031906.
- Song Y, Zheng S, Li L, Zhang X, Zhang X, Huang Z, Chen J, Zhao H, Jie Y, Wang R, et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. medRxiv. 2020b.02.23.20026930. [DOI] [PMC free article] [PubMed]
- Tomasev N, Glorot X, Rae JW, Zielinski M, Askham H, Saraiva A, et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature. 2019;572(7767):116–119. doi: 10.1038/s41586-019-1390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, Cai M, Yang J, Li Y, Meng X, et al. A deep learning algorithm using CT images to screen for Corona virus disease (COVID-19). medRxiv. 2020.02.14.20023028. [DOI] [PMC free article] [PubMed]
- Wang T, Tschampel T, Apostolova E, Velez T. Using latent class analysis to identify ARDS sub-phenotypes for enhanced machine learning predictive performance. In: arXiv e-prints. 2019: arXiv:1903.12127.
- White DB, Lo B. A framework for rationing ventilators and critical care beds during the COVID-19 pandemic. JAMA. 2020. p. 27. [Epub ahead of print]. [DOI] [PubMed]
- World Health Organization . WHO director-General's opening remarks at the media briefing on COVID-19 - 20 march 2020. Geneva: World Health Organization; 2020. [Google Scholar]
- Xie J, Hungerford D, Chen H, Abrams ST, Li S, Wang G, Wang Y, Kang H, Bonnett L, Zheng R, et al. Development and external validation of a prognostic multivariable model on admission for hospitalized patients with COVID-19. medRxiv. 2020.03.28.20045997.
- Xu X, Jiang X, Ma C, Du P, Li X, Lv S, Yu L, Chen Y, Su J, Lang G, et al. Deep learning system to screen coronavirus disease 2019 pneumonia. In: arXiv e-prints. 2020: arXiv:2002.09334. [DOI] [PMC free article] [PubMed]
- Yan L, Zhang H-T, Goncalves J, Xiao Y, Wang M, Guo Y, Sun C, Tang X, Jin L, Zhang M, et al. A machine learning-based model for survival prediction in patients with severe COVID-19 infection. medRxiv. 2020.02.27.20028027.
- Yang R, Li X, Liu H, Zhen Y, Zhang X, Xiong Q, et al. Chest CT severity score: an imaging tool for assessing severe COVID-19. Radiology: Cardiothoracic Imaging. 2020;2(2):e200047. doi: 10.1148/ryct.2020200047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampieri FG, Costa EL, Iwashyna TJ, Carvalho CRR, Damiani LP, Taniguchi LU, et al. Heterogeneous effects of alveolar recruitment in acute respiratory distress syndrome: a machine learning reanalysis of the alveolar recruitment for acute respiratory distress syndrome trial. Br J Anaesth. 2019;123(1):88–95. doi: 10.1016/j.bja.2019.02.026. [DOI] [PubMed] [Google Scholar]
- Zhan A, Mohan S, Tarolli C, Schneider RB, Adams JL, Sharma S, et al. Using smartphones and machine learning to quantify Parkinson disease severity: the Mobile Parkinson disease score. JAMA Neurol. 2018;75(7):876–880. doi: 10.1001/jamaneurol.2018.0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from COVID19@northwell.edu. The data are not publicly available due to restrictions as it could compromise the privacy of research participants.


