SUMMARY
Biological aging involves an interplay of conserved and targetable molecular mechanisms, summarized as the hallmarks of aging. Metformin- a biguanide that combats age-related disorders and improves healthspan, is the first drug to be tested for its age-targeting effects in the large clinical trial- TAME (Targeting Aging by MEtformin). This review focuses on metformin’s mechanisms in attenuating hallmarks of aging and their interconnectivity, by improving nutrient-sensing, enhancing autophagy and intercellular communication, protecting against macromolecular damage, delaying stem-cell aging, modulating mitochondrial function, regulating transcription, and lowering telomere attrition and senescence. These characteristics make metformin an attractive gerotherapeutic to translate to human trials.
Graphical Abstract
Metformin is the first drug to be tested for its age-targeting effects in a large clinical trial. In this Perspective, Kulkarni et al. review how metformin acts on its primary and secondary targets to attenuate the hallmarks of aging, highlighting its utility as an effective gerotherapeutic intervention.
Introduction
Aging is characterized by a progressive loss of physiological function, which drives the development of chronic morbidities including metabolic, cardiovascular, neoplastic and neurodegenerative disorders as well as geriatric symptoms like frailty and immobility. Aging is accompanied by an inherent biological mechanism that is malleable and can be targeted using therapeutic interventions. Indeed, over the past few decades, scientists have achieved remarkable progress in extending healthspan and lifespan of model organisms using several genetic, dietary and pharmacological interventions. These advancements have urged the geroscience research community to initiate clinical trials to investigate the efficacy of interventions in targeting human aging, starting with the TAME (Targeting Aging with MEtformin) study (Barzilai 2017, Campisi et al. 2019). The TAME study, soon to be launched in the near future, aims to prove the concept that human aging can be targeted while simultaneously preventing a multitude of major age-related outcomes. Furthermore, TAME is a potential tool to facilitate the FDA to approve “aging” as a target for drug discovery and development. Thus, TAME will pave the way for the development of novel interventions that could target and delay the aging process and improve human healthspan, by modulating the conserved mechanistic pathways involved in aging.
To systematically dissect the biological aging process, Lopez-Otin et al characterized nine major hallmarks of aging, widely accepted by the geroscience research community, namely 1) Genomic Instability, 2) Epigenetic Alterations, 3) Loss of Proteostasis, 4) Deregulated Nutrient-Sensing, 5) Mitochondrial Dysfunction, 6) Cellular Senescence, 7) Stem Cell Exhaustion, 8) Altered Intercellular Communication and 9) Telomere Attrition (López-Otín et al. 2013). These are divided as primary, antagonistic and integrative hallmarks depending on their functional characteristics as causes of damage, responses to damage, and end results of the first two categories, respectively (López-Otín et al. 2013). The hallmarks and their interconnectivity can serve as an evaluation tool to assess and prioritize interventions that can be deemed effective in targeting aging. Although the contribution of each of these hallmarks towards the progression of biological aging is not yet fully elucidated, interventions that can modulate several of these hallmarks, at least in part, need to be studied extensively to provide newer insights into the druggable targets of biological aging.
In humans, metformin has been in clinical use for over 60 years, studied extensively, has a high safety profile and is uniquely positioned to intervene several crucial pathways responsible for aging and age-related diseases (Barzilai et al. 2016). As recommended by the American Diabetes Association, due to its glucose-lowering effects, metformin monotherapy is the preferred first-line pharmacological action against type-2 diabetes (2019). Epidemiological studies have revealed metformin’s gerotherapeutic effect in lowering the incidence of multiple age-related diseases as well as all-cause mortality, in both diabetics and non-diabetics (Campbell et al. 2017, Valencia et al. 2017). Clinical studies including the Diabetes Prevention Program (DPP) in non-diabetics and U.K. Prospective Diabetes Study (UKPDS) in diabetics, support metformin’s role as an effective intervention against diabetes and cardiovascular disease. Association studies suggest a decrease in the incidence of most age-related cancers, Alzheimer’s Disease while clinical studies support metformin’s role in a decrease in cognitive decline and reduced mortality in diabetics taking metformin compared to non-diabetics (Barzilai et al. 2016).
Studies in multiple model organisms and human cell lines have elucidated metformin’s role in targeting multiple mechanisms of aging. In mice and C. elegans, metformin extends lifespan and improves several indicators of healthspan (Anisimov et al. 2008, Martin-Montalvo et al. 2013, De Haes et al. 2014, Chen et al. 2017). Intermittent metformin treatment, when initiated in mice, even later in life and administered every other week, has also shown to provide health benefits such as a reduction in hepatic steatosis through regulation of the liver transcriptome and metabolome (Alfaras et al. 2017). Recently, metformin’s extraordinary ability as a gerotherapeutic is established through several experimental, clinical and observational evidence (Novelle et al. 2016, Piskovatska et al. 2018, Glossmann and Lutz 2019, Soukas et al. 2019). Moreover, in older human adults, we recently demonstrated metformin’s efficacy in targeting multiple age-associated metabolic and non-metabolic pathways and reverting derangements, using cardiometabolic and transcriptomic outcomes (Kulkarni et al. 2018).
In this review, we aim to determine the influence of metformin’s mechanisms of action on the hallmarks of biological aging. We demonstrate that each hallmark of aging, as well as their interconnectivity is attenuated by metformin’s direct and/or downstream effects (Figure 1).
Figure 1-. Mechanisms of metformin action in attenuating hallmarks of biological aging.
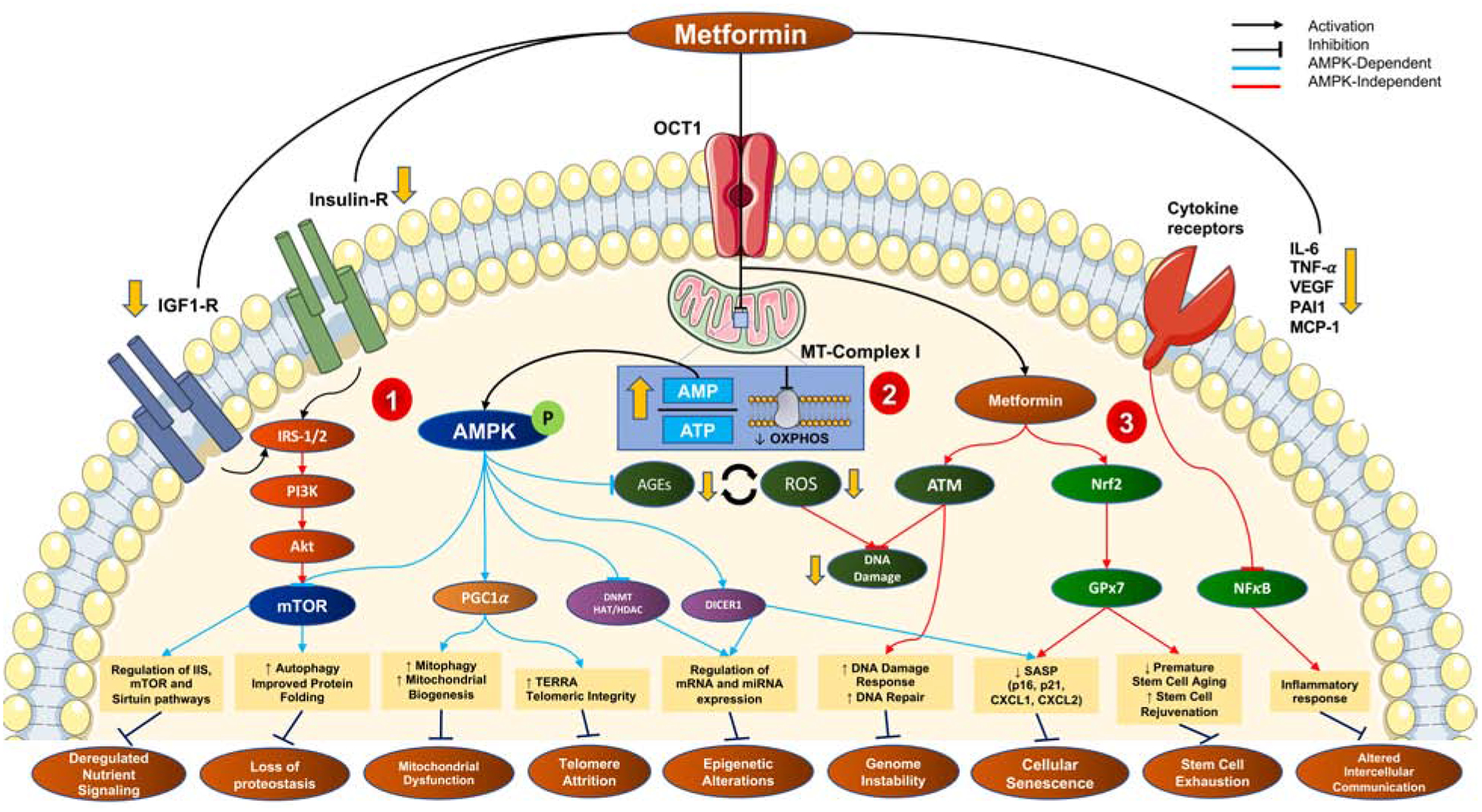
The cellular uptake of metformin is via the organic cationic transporter 1 (OCT1), after which it exerts three arms of action- 1) metabolic; 2) oxidative and 3) inflammatory.
1) Metformin inhibits mitochondrial complex I and thereby oxidative phosphorylation leading to an increased AMP:ATP ratio, causing a direct activation of AMPK. AMPK-dependent mechanisms (blue) contribute to the downstream inhibition of mTORC1 (improved nutrient-sensing and autophagy), activation of PGC-1α (improved mitochondrial biogenesis), transcriptional regulation via DNA/histone modifications and miRNAs. Extracellularly, metformin downregulated Insulin/IGF1 signaling, also leading to mTORC1 inhibition.
2) The inhibition of mitochondrial ETC also leads to AMPK-independent effects (red) including reduced reactive oxygen species (ROS), reduced advanced glycation end-products (AGEs) and thereby reduced macromolecular damage.
3) The AMPK-independent (red) anti-inflammatory and senotherapeutic effects of metformin are evident via the downregulation of pro-inflammatory cytokines, NF-κB signaling, and activation of Nrf2-Gpx7 and ATM-signaling, respectively.
These three arms work to mitigate the aging-induced dysregulation in cells, thereby attenuating hallmarks of aging.
Mechanisms of metformin in targeting biological aging
Metformin was introduced to the world in 1957, as an antihyperglycemic agent by the French physician Jean Sterne, and today, it has become one of the most commonly used pharmaceutical interventions and the most prescribed glucose-lowering medication, worldwide (Bailey 2017). Indeed, it is important to note that metformin does not cause hypoglycemia per se but rather reduces hepatic glucose production through improved hepatic insulin sensitivity, which results in the reduction of fasting plasma glucose levels (Jackson et al. 1987, DeFronzo et al. 1991). Furthermore, with the epidemiological, preclinical and clinical evidence of metformin exhibiting beneficial effects beyond glycemic control in diabetics, it has been suggested to be repurposed against cancer and its recurrence (Heckman-Stoddard et al. 2017), cardiovascular disease (Rena and Lang 2018), neurodegenerative diseases (Rotermund et al. 2018), autoimmune diseases (Ursini et al. 2018) and most recently, systemic aging as a whole (Barzilai et al. 2016, Barzilai 2017). Despite such widespread use and efficacy, the mechanisms by which metformin regulates fundamental pathways in aging and diseases are not fully elucidated. Metformin’s antihyperglycemic role is attributed to its action on glucose metabolism, specifically as a suppressor of hepatic gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase, thereby modifying the hepatocellular redox state to reduce glucose formation from lactate and glycerol (Madiraju et al. 2014). On the other hand, metformin’s suppression of hepatic glucose production is shown to be the result of AMP-induced inhibition of fructose-1,6-bisphosphatase-1, a rate-controlling enzyme in gluconeogenesis (Hunter et al. 2018). In addition to these mechanisms, metformin’s metabolic action includes a reduction in glucose absorption in the intestine (Wu et al. 2017), restoring insulin secretion in pancreatic beta-cells (Patane et al. 2000) and to a lesser extent increasing insulin-mediated glucose uptake in the peripheral tissues of muscle and adipose (Galuska et al. 1991).
Being a hydrophilic compound charged positively at physiological pH, metformin enters and leaves the cells mainly via organic cationic transporters (OCTs) and multidrug and toxin extrusion transporters (MATEs) (Gong et al. 2012). The hepatic, intestinal and adipocytic uptake of metformin is primarily mediated through the OCT1 (Wang et al. 2002, Moreno-Navarrete et al. 2011) (Figure 1). Furthermore, it has been shown to accumulate in mitochondria due to the membrane potential across the mitochondrial inner membrane (Owen et al. 2000). One of the main actions of metformin is the inhibition of mitochondrial complex I (NADH:ubiquinone oxidoreductase), that leads to multiple downstream effects both on metabolic and non-metabolic pathways responsible in the aging process (El-Mir et al. 2000, Foretz et al. 2014, Barzilai et al. 2016). The mechanisms by which metformin inhibits mitochondrial complex I remain unsolved. Although, very high concentrations of metformin are needed to directly inhibit complex I activity in isolated mitochondria, micromolar concentrations of the drug are effective in achieving a dose- and time-dependent weak, reversible and selective complex I inhibition (Vial et al. 2019). As a result, the effects of metformin diverge on metabolic and oxidative pathways and their downstream targets. However, some actions of metformin, including its antiproliferative effect can be demonstrated regardless of its effect on mitochondria, especially in Rho0 cells deficient in mitochondrial DNA (Liu et al. 2014).
In parallel to complex I inhibition and its action on the mitochondrial electron transport chain (ETC), metformin’s mechanism of action is attributed both to its 5’ adenosine monophosphate -activated protein kinase (AMPK)-dependent and AMPK-independent roles (Figure 1). The downstream effect of mitochondrial complex I inhibition is directly evident from the increase in cytoplasmic AMP:ATP and ADP:ATP ratios, which in turn leads to phosphorylation and activation of AMPK (Foretz et al. 2014). The direct activation of AMPK and inhibition of mTORC1 is a result of metformin’s action on the lysosomal pathway, requiring v-ATP-ase-AXIN/LKB1, proposed to be a mechanism to increase the lifespan of C. elegans (Zhang et al. 2016, Chen et al. 2017). Phosphorylation and activation of AMPK leads to further inhibition of mTORC1, activation of peroxisome proliferator-activated receptor-gamma coactivator-1 alpha (PGC1-α) and mitochondrial biogenesis, activation of SIRT1 and other nutrient-sensing pathways, inhibition of advanced-glycation end products partly by inhibiting Nuclear Factor kappa-light-chain-enhancer of activated B cells (NF-κB) and pro-inflammatory cytokines, activation of Ulk1 and regulation of autophagy, among others (Kim et al. 2011, Salminen and Kaarniranta 2012, Barzilai et al. 2016, Zhou et al. 2016, Herzig and Shaw 2018) (Figure 1). Metformin’s AMPK-independent mechanisms also contribute to the direct activation of SIRT1, direct inhibition of mTORC1 via Rag-GTPases, suppression of adipogenesis through inhibition of p70S6K pathway, activation of DNA damage-like response via the activation of ATM/Chk2 pathway, and activation of Nuclear factor erythroid 2-Related Factor 2 (Nrf2), all of which result in downregulation of inflammatory responses (Kalender et al. 2010, Vazquez-Martin et al. 2011, Chen et al. 2017, Prasad et al. 2017, Cuyas et al. 2018) (Figure 1). A novel role of metformin was elucidated in modulating the gut microbiota and the availability of branched-chain amino acids to the gut microbiota, which is directly attributable to its effect on nutrient-sensing and aging. This function is further discussed below in metformin’s capability to improve intercellular signaling and attenuate inflammation. More recently, metformin’s beneficial effects on maintaining energy balance and body weight were shown to be regulated via growth/differentiation factor 15 (GDF15), which further warrants more understanding of metformin’s GDF15-mediated targeting of biological aging (Coll et al. 2020).
Although the mechanisms of metformin in targeting fundamental pathways in biological aging are far from completely understood, here we attempt to link its mode of action by highlighting its role on individual hallmarks of biological aging as evidenced in cell lines and model organisms (Table 1, Figure 1).
Table 1-.
Summary of key targets and pathways impacted by metformin as evidenced by their modulation in cell lines, C. elegans, Drosophila, and rodents.
| Attenuation of Hallmarks of Aging | Effects of Metformin on key targets and pathways involved in regulating each hallmark of aging | |||
|---|---|---|---|---|
 |
 |
 |
||
| Improved Nutrient Signaling |
|
|
|
|
| Enhanced Intercellular Communication |
|
|
– |
|
| Ameliorated Proteostasis |
|
|
– |
|
| Protection against Genomic Instability |
|
– |
|
|
| Regulated Mitochondrial Function |
|
|
– |
|
| Increased Stem Cell rejuvenation capacity |
|
– |
|
|
| Regulated Epigenetic Alterations |
|
|
– |
|
| Minimized Telomere Attrition |
|
– | – | – |
| Attenuated Cellular Senescence |
|
– | – |
|
Effects of metformin on attenuating individual hallmarks of aging
Primary targets of metformin among aging hallmarks
1. Deregulated nutrient-sensing
Nutrient availability and sensing are major regulators of cell-signaling pathways that take cues from environmental stimuli and intracellular activity to maintain energy homeostasis within a cell. These pathways determine cellular energy levels and communicate and coordinate with hormonal and nutrient signaling cascades to induce a positive feedback loop whereby an optimal level is achieved. However, with aging, there is a decline in the ability of the cell to maintain metabolic homeostasis, further contributing to the organismal aging phenotype (Bettedi and Foukas 2017). The most important nutrient sensors and signaling cascades that regulate aging and determine longevity are identified from genetic polymorphism studies of long-lived individuals and genetic manipulation screens in model organisms. These include highly conserved nutrient-sensing systems- 1) Somatotropic axis (GH/IGF-1) that elicits a response similar to insulin, regarding the cellular availability of glucose; 2) mTOR signaling responsible for sensing high amino acid concentrations; 3) AMPK that is involved in sensing low energy states via high AMP levels; 4) Sirtuins that complement in sensing low energy states by detecting high NAD+ levels (López-Otín et al. 2013). Although there are inconsistencies in the disease- and age-associated changes in the directionality of the GH/IGF-1 axis, a number of human studies suggest that its attenuation may protect from age-related conditions (Milman et al. 2016). Impaired insulin/IGF-1 signaling (IIS) extends lifespan by 2-fold in C. elegans daf-2 mutants, by promoting L-proline catabolism and endogenous stress defense response via a transient ROS signal (Zarse et al. 2012). On the other hand, although the effects are tissue- and sex-specific, unequivocal evidence lies in showing that aging and age-related diseases upregulate mTORC1, while AMPK and sirtuins act in the opposite direction to mTOR signaling (López-Otín et al. 2013).
Metformin activates AMPK and SIRT1 and downregulates IIS and mTORC1
Metformin’s beneficial effects on energy metabolism and aging are partly a consequence of directly targeting these key energy sensors. Metformin is shown to decrease insulin and IGF-1 levels and downregulate IIS in cancer cells (Sarfstein et al. 2013, Xie et al. 2014, Klement and Fink 2016). Additionally, metformin activates AMPK via LKB1-dependent mechanism, mediating its effect on lifespan and healthspan extension in mice and C. elegans (Onken and Driscoll 2010, Martin-Montalvo et al. 2013). AMPK-activation by metformin is also indicative of its suppressive effect on advanced glycation end products (AGEs), thereby mitigating the expression of inflammatory cytokines (Chung et al. 2017) (Figure 1). Metformin is shown to directly inhibit mTORC1 via Rag-GTPase inhibition and indirectly via REDD1 upregulation promoting TSC2 activity and AMPK activation-mediated phosphorylation of S6K1 and 4E-BP1- mTORC1 substrates decreasing its translation (Dowling et al. 2007, Kalender et al. 2010, Ben Sahra et al. 2011, Amin et al. 2019). It is also revealed to be a direct activator of SIRT1, especially in low NAD+ concentrations (Cuyas et al. 2018).
As an established AMPK activator, metformin’s role has been very well studied in modulating nutrient-sensing pathways. However, recent evidence in its direct inhibition of mTORC1 as well as sirtuins can have potential implications downstream of nutrient depletion, mimicking the benefits of dietary restriction. The tissue-specificity and dose-dependency in mediating these nutrient-signaling pathways in the context of human aging need to be further elucidated.
2. Altered intercellular communication
Endocrine, neuronal and neuroendocrine pathways provide cues to the cells to respond effectively to environmental changes, pathogens, tissue disruptors and mechanical stressors (López-Otín et al. 2013). Aging leads to a systemic dysregulation of effective cell-cell connectivity and its associated response disrupting the maintenance of intercellular communication. A key consequence of this dysregulation is age-associated chronic, sterile, low-grade inflammatory state called inflammaging, that is usually accompanied by a consistent increase in pro-inflammatory cytokine secretion (TNF, IL-6, and IL-1β), activation of NF-κB and interferon signaling, increased burden of senescent cells and their secretome as well as altered autophagic response (López-Otín et al. 2013, Franceschi et al. 2018). Importantly, inflammation is shown to trigger or respond to multiple other hallmarks including epigenetic alterations, stem cell dysfunction and loss of proteostasis, thereby contributing to the pathogenesis of age-related diseases and accelerated aging (Franceschi et al. 2018, Sanada et al. 2018, Vinatier et al. 2018, Benayoun et al. 2019, Josephson et al. 2019).
Metformin suppresses pro-inflammatory cytokines and inhibits NF-κB pathway
Several studies suggest metformin’s role in modulating several key immunoregulatory mechanisms, often with improvement in metabolic parameters (Saisho 2015). In older diabetic individuals, metformin monotherapy lowered the levels of circulating pro-inflammatory cytokines and the associated mortality risk in a recent five-year follow-up study (Tizazu et al. 2019). In primary hepatocytes, using a distinct mechanism to its anti-hyperglycemic effect, metformin suppressed TNF-α dependent NF-κB signaling and expression of IL-6 and IL-1β (Cameron et al. 2016). This suppression of proinflammatory cytokine markers along with senescence-associated secretory phenotype (SASP) is also evident through metformin’s inhibition of IKK/NF-κB activation (Moiseeva et al. 2013). A working model of metformin’s anti-inflammatory properties also suggests that metformin, in an AMPK-dependent inhibition of STAT3, prevents the differentiation of monocytes to macrophages (Vasamsetti et al. 2015). In addition to direct inhibition of inflammation, metformin’s action in lowering body weight and improving insulin metabolism and sensitivity has indirect implications on lowering systemic inflammation as well (Saisho 2015).
Metformin modulates the gut microbiota further improving metabolism and reducing inflammation
There is increasing evidence on the role of gut microbiota in effectively mediating inflammatory responses. Age-associated microbial dysbiosis (alterations in gut microbiota) is shown to promote inflammation, gut permeability and proinflammatory cytokine release (Thevaranjan et al. 2017). Healthy gut microbiome has been shown to be associated with optimal healthy immune function (Seidel and Valenzano 2018). Early evidence of metformin’s role in delaying aging and increasing lifespan, via modulating the microbial folate and inducing methionine restriction was demonstrated in C. elegans (Cabreiro et al. 2013). Similarly, recent evidence suggests that metformin extends Drosophila lifespan in a dose-dependent manner, only in control OP50 E.coli colonized strains, and not germ-free strains, indicating the role of microbiota in mediating metformin-induced lifespan extension across species (Pryor et al. 2019). Later, animal studies revealed that metformin’s glucoregulatory role was an effect of increased Lactobacilli abundance in the upper small intestine, while its anti-inflammatory role was associated with increased abundance of Akkermansia, Bacteroides, Butyricimonas, and Parabacteroides genera (Bauer et al. 2018, Lee et al. 2018). In human diabetic individuals, metformin was shown to shift gut microbiota composition to predominantly short-chain fatty-acid producing microbes (de la Cuesta-Zuluaga et al. 2017). Furthermore, metformin’s role in improving metabolic dysfunction in diabetics was linked to an increase in levels of bile acid glycoursodeoxycholic acid (GUDCA) by inhibiting the growth of B. fragilis, thereby altering intestinal Farnesoid X Receptor (FXR) signaling (Sun et al. 2018).
Metformin’s role as an anti-inflammatory is directly responsive and consequential to regulating cellular senescence, stem cell function, autophagy, and macromolecular damage. Cell-cell communication and response are maintained, thereby leading to a decrease in the abundance of pro-inflammatory cytokines. It is essential to understand how metformin mitigates inflammatory responses against various stressors. Moreover, understanding the role of gut microbiota in regulating metformin’s action on inflammation and intercellular communication is crucial for establishing this drug as a candidate to target aging.
3. Genomic instability
Genomic instability refers to the accumulation of genetic damage throughout one’s lifespan, due to a multitude of DNA alterations like mutations, indels and chromosomal rearrangements (Vijg and Suh 2013). Attributed to both endogenous (mitochondrial ROS, DNA replication errors, etc.) and exogenous (environmental and iatrogenic) agents, genomic instability often accompanies biological aging, while its artificial induction can often lead to an accelerated aging phenotype (Niedernhofer et al. 2018). Such genetic damage involves oxidation of constituent DNA bases, crosslinking of DNA and protein as well as accumulation of DNA double-stranded breaks. Although the extent of lesions occurring daily in each somatic cell can be quite high, most of them are countervailed by the vast network of DNA repair mechanisms. However, as evidenced by increased DNA repair deficits in mouse models of accelerated aging and human progeroid syndromes as well as increased nuclear and mitochondrial DNA damage, genomic instability is a pivotal hallmark of biological aging (Vijg and Suh 2013).
Metformin’s genome protective effects are a result of increased DNA-damage like response and DNA repair
Several mechanisms have been associated with metformin’s response to mitigate genomic instability, in the context of aging and various types of cancers. Studies have shown metformin’s genome protective effects via reduction of oxidative stress, DNA damage and DNA damage response, regulation of Ataxia Telangiectasia Mutated (ATM) protein kinase and epigenetic effects, however, there is no consensus regarding the underlying mechanism of the drug in regulating components of genome stability (Najafi et al. 2018). In mouse embryonic fibroblasts (MEFs), metformin is shown to reduce paraquat-associated endogenous ROS levels and associated DNA damage by preventing ROS toxicity (Algire et al. 2012). Furthermore, in a dose-dependent manner, metformin prevents activation of ATM-protein kinase, thereby lowering ROS and γH2AX-a sensitive marker of DNA-double stranded breaks and genomic instability (Halicka et al. 2011). Similarly, it takes 10-fold lower levels of metformin to scavenge mitochondrial ROS than required to inhibit mitochondrial complex I, without directly impacting respiratory capacity as evidenced in rats (Kane et al. 2010). The genotoxicity-protective effects of metformin are demonstrated in human lymphocytes, normal and diabetic rats, rat and mouse bone marrow cells, mouse kidney cells as evidenced by a reduction in micronuclei and chromosomal aberrations (Sant’Anna et al. 2013, Cheki et al. 2016, Othman et al. 2016, Najafi et al. 2018, Cheki et al. 2019). In T2D patients, metformin induces an antioxidant response further leading to DNA Base Excision Repair system (Dogan Turacli et al. 2018). In the presence of DNA damage, metformin also activates ATM and Checkpoint kinases-2 thereby phosphorylating serine 132 on histone H2AX → γH2AX, and recruiting DNA repair complexes at the DNA double-strand breaks (Vazquez-Martin et al. 2011). Additionally, we have previously shown that short-term metformin treatment in older adults induced BRCA-mediated DNA damage response and DNA repair in skeletal muscle (Kulkarni et al. 2018).
Although there is no consensus on how metformin regulates oxidative damage and attenuates genome instability, the evidence is stronger for metformin stimulating DNA damage responses and protective mechanisms against genotoxic stress. It is essential to further elucidate metformin’s role in preventing or reverting age-induced dysregulation in both nuclear and mitochondrial genome stability, chromosomal structure and its interplay with key processes like senescence and gene regulatory changes.
4. Loss of proteostasis
A highly regulated network of > 2000 proteins comprising of molecular chaperones, proteolytic systems and regulators, is crucial to maintain a stable proteome or protein homeostasis (proteostasis) (Hipp et al. 2019). Aging, age-associated pathologies and neurodegenerative diseases including Alzheimer’s and Parkinson’s diseases, exhibit an impaired network of coordinated proteostasis, with the accumulation of intracellular damage (Kaushik and Cuervo 2015). Three key aspects of the proteostasis network, namely- protein synthesis and folding, maintenance of conformational stability and autophagy-mediated protein degradation are deteriorated with aging, leading to an imbalance in protein abundance (Hipp et al. 2019). Model organism studies have revealed lifespan extensions through alterations within pathways enhancing proteostasis, including Insulin/IGF-1 signaling, mitochondrial ETC, enhanced autophagy, and nutrient-sensing.
Metformin augments autophagy and rescues protein misfolding
Metformin’s stabilizing effect on proteostasis is mainly the result of augmented autophagy and inhibited protein synthesis via direct and indirect inhibition of mTOR signaling (Shi et al. 2012). In a Clk1 mutant Parkinson’s Disease mouse models, with increased activity of neurotoxin MPP, metformin treatment reversed behavioral impairments, reduced α-synuclein accumulation and enhanced LC3-II-mediated autophagy in dopaminergic neurons and midbrain, in an AMPK-dependent mechanism (Lu et al. 2016, Yan et al. 2017). Autophagy-related proteins LAMP-1 and Beclin-1 are upregulated by metformin treatment, which have been shown to attenuate hyperglycemia-induced apoptosis in cardiomyocytes (Wang et al. 2017). Accumulating evidence also suggests that metformin induces enhanced autophagic responses in δ-Sarcoglycan deficiency-induced dilated cardiomyopathic mouse hearts and enhanced mitophagy in ob/ob mice (Song et al. 2016, Kanamori et al. 2019). Furthermore, metformin is also shown to rescue misfolding and trafficking of rhodopsin, downstream of AMPK-activation (Athanasiou et al. 2017).
These studies in model systems reveal metformin’s role against impairments in proteostasis. But whether the prevention of protein misfolding and trafficking translates to attenuation of aging in target organs is yet to be elucidated. However, further studies are warranted to understand the drug’s effect on rescuing age-associated imbalance in protein abundance and misfolding. An ongoing clinical trial examining metformin’s pro-autophagy effects in prediabetic individuals may help elucidate the drug’s role in humans (NCT03309007).
Secondary targets of metformin among aging hallmarks
5. Mitochondrial dysfunction
There is longstanding evidence that the decline in mitochondrial function with aging, may contribute to age-associated dysregulation in energy homeostasis and increased predisposition to age-related diseases (Sun et al. 2016). Aging induced disruptions in mitochondrial function is a result of several intra- and extracellular stresses including mtDNA mutations, nuclear genomic instability, reduced mitochondrial biogenesis, defective mitophagy, and altered mitochondrial dynamics and ETC regulation, among others (López-Otín et al. 2013). With an increased understanding of the role of mitochondria in addition to serving as the powerhouse of the cell, its dysfunction with aging can have implications on inflammation, senescence, autophagy and retrograde nuclear signaling (Jang et al. 2018).
Metformin lowers oxidative stress via mitochondrial complex I inhibition and improves mitochondrial biogenesis via PGC-1α
Due to their key role in regulating these vital cellular processes along with energy homeostasis and oxidative stress via ROS, mitochondria are deemed to be an attractive therapeutic target against aging and age-related diseases and pathologies (Madreiter-Sokolowski et al. 2018, Murphy and Hartley 2018). Metformin inhibits complex I (NADH:ubiquinone oxidoreductase) of the mitochondrial ETC, thereby directly impacting mitochondria-induced oxidative stress via lowering ROS or by indirect scavenging mechanism (Kelly et al. 2015, Fontaine 2018, Vial et al. 2019). This inhibition also induced an anti-inflammatory response by specifically inhibiting pro-IL1β production in macrophages (Kelly et al. 2015). Similarly, complex I inhibition by metformin also reduces ROS mediated IL-6 release in alveolar macrophages, thereby preventing arterial thrombosis in mice (Soberanes et al. 2019). Decreased hepatic gluconeogenesis is also demonstrated to be associated with metformin’s direct inhibition of mitochondrial glycerophosphate dehydrogenase (mGPDH)- a flavin-linked respiratory chain-linked dehydrogenase, which in turn modulates cytosolic redox state (Madiraju et al. 2014). Metformin increases the expression and protein activity of hepatic and skeletal muscle associated PGC-1α, a co-transcriptional regulator that induces mitochondrial biogenesis (Suwa et al. 2006, Aatsinki et al. 2014). Interestingly, in vitro studies have demonstrated the ability of metformin to revert mitochondrial dysfunction associated with Down syndrome, a human model of accelerated aging, via restoration of mitochondrial gene expression network and rescue of mitochondrial morphology (Izzo et al. 2017). Linking its effects on histone methylation and sirtuin activation, metformin was also shown to induce mitochondrial biogenesis and delay cellular senescence by acting through SIRT3 upregulation (Karnewar et al. 2018). Metformin’s direct activation of SIRT1 may help combat age-related mitochondrial dysfunction which is attributed to a reduction in SIRT1 activity leading to downregulation of a nuclear-encoded mitochondrial transcription factor A (TFAM) and declines in NAD+ levels (Gomes et al. 2013, Cuyas et al. 2018). Recently, metformin’s cardioprotective role in human arterial appendage tissue was elucidated through its dose-dependent mild inhibitory effect on the activity of complexes I, IV and V with a reduced superoxide production and attenuation of mitochondrial permeability transition pore (Emelyanova et al. 2019).
Although there is substantial evidence of the role of metformin in inhibiting mitochondrial complex I and on improving mitochondrial biogenesis by increasing PGC-1α expression, there is no consensus on whether this effect is tissue-specific. For instance, in skeletal muscle, metformin is shown to inhibit aerobic exercise-induced mitochondrial adaptations in older adults (Konopka et al. 2019). But, in mice with AMPK-kinase-dead mutation, even short-term metformin treatment enhances mitochondrial function and biogenesis in skeletal muscle (Kristensen et al. 2013). Hence, it is important to further expand the understanding of metformin’s species- and tissue-specific mitochondrial regulation as a monotherapy and in combination with other interventions.
6. Stem cell exhaustion
With aging, there is a systemic decrease in the regenerative capacity of tissues. It is imperative to maintain homeostasis of stem and progenitor cells and their regeneration potential since both a reduced number of stem cells and excessive proliferation can induce aging phenotypes (López-Otín et al. 2013). Age-associated decrease in stem cell function is observed in several stem cell populations, including those of hematopoietic cells, intestinal stem cells, satellite cells, neuronal stem cells, hair follicle cells, melanocytes, and germline cells (Schultz and Sinclair 2016). Currently, there is no consensus regarding the association between the change in stem cell number of various tissue sub-types and aging. However, some studies have demonstrated that hematopoietic and intestinal stem cells increase with age in humans as well as animal models (mice, Drosophila spp.) (Schultz and Sinclair 2016). The exact mechanism for decreased stem and progenitor cell capacity to rejuvenate is not known, but the common mechanisms of stem cell aging can be explained by some cell-intrinsic and extrinsic factors including telomere attrition, molecular damage, epigenetic drift and deregulation of developmental pathways (Ermolaeva et al. 2018).
Metformin induces stem cell rejuvenation capacity and delays stem cell aging
Fortunately, metformin targets several pathways that cause stem cell exhaustion, thereby reinforcing the rationale to understand the role of metformin in addressing age-associated stem cell exhaustion. The previously mentioned gerotherapeutic effect of metformin in activating Gpx7 via Nrf2 is shown to delay cellular attrition and prevent premature aging thereby increasing the lifespan of human mesenchymal stem cells (Fang et al. 2018). The early evidence of metformin delaying stem cell aging was demonstrated in Drosophila midgut intestinal stem cells (ISCs), where it inhibited DNA-damage and downstream centrosome amplification via the AKT/TOR pathway, later found out to be also mediated by Atg6- an autophagy-related factor (Na et al. 2013, Na et al. 2015, Na et al. 2018). The anti-aging effect of metformin in stem cell function is not limited to ISCs and can be extended to other stem cell populations. Recently, in female mice, metformin was found to expand the neural stem cell pool, while still promoting neurogenesis and cognitive recovery in both sexes (Ruddy et al. 2019). Interestingly, metformin also reversed age-associated dysregulation of rejuvenating and differentiating capacity of oligodendrocyte progenitor cells, further improving remyelination (Neumann et al. 2019). Metformin-induced retention of self-renewing capacity and neurogenesis are mediated through two distinct mechanisms- the former via a transcription factor TAp73, and the latter via AMPK-activated αPKC-CBP pathway (Fatt et al. 2015). In muscle, aged satellite cells fail to maintain quiescence and the capacity to self-renew once activated. Metformin treatment is shown to delay satellite cell activation and maintain a quiescent, low metabolic state thereby maintaining stem cell numbers (Pavlidou et al. 2019).
Maintenance of stem cell number and function can help combat age-associated loss of rejuvenation potential and thereby promote tissue repair. In several instances, metformin has been effective in targeting stem cell exhaustion, delaying stem cell aging and maintaining stem cell function. It is very important to delineate metformin’s role as a gerotherapeutic agent preserving stem cell function, while also preventing abnormal differentiation and leading to tumorigenesis. Furthermore, this role of metformin needs to be investigated further in studies and trials in regenerative medicine.
7. Epigenetic alterations
Loss of histones, imbalance in histone modifications, changes in chromatin architecture, breakdown of the nuclear lamina, as well as DNA and histone methylation changes are characteristics of biological aging (Sen et al. 2016). Histone modifications such as acetylation and phosphorylation can lead to active transcription, loss of cellular homeostasis and age-associated metabolic decline while post-translational modifications like ubiquitination, sumoylation, O-GlcNAcylation, and ADP-ribosylation, can have varying influences on transcriptional dysregulation (Pal and Tyler 2016, Peleg et al. 2016). Furthermore, DNA and histone methylation landscapes change during aging, generally leading to global hypomethylation and promoter-specific hypermethylation (Michalak et al. 2019). These changes are determinants of conserved, tissue-specific, age-associated transcriptional changes, that contribute to metabolic and inflammatory phenotypes of aging (Benayoun et al. 2015, Benayoun et al. 2019).
Metformin regulates transcriptional activity via histone modifications, DNA methylation, and miRNAs
Due to their modifiable nature, epigenetic and transcriptional changes are generally indicative of the effect of therapeutic interventions on age-related diseases and promoting healthy aging. Metformin’s AMPK-dependent and independent mechanisms have been observed to influence histone modifications through phosphorylation of histone acetyltransferases (HATs), inhibition of Class II histone deacetylases (HDACs) and activation of SIRT1 (Bridgeman et al. 2018). In non-diabetic HER2-positive breast cancer patients as well as human breast cancer xenografts injected into nude mice, the drug augmented global H3K27me3 levels, directly targeted demethylase KDM6A/UTX and moreover, reverted age-associated loss of global H3K27me3 in fibroblasts from both healthy older donors and Werner’s syndrome patients (Cuyàs et al. 2017). This effect was irrespective of metformin’s mitochondrial complex I inhibition, as evidenced in Rho0 cells devoid of mitochondria. Additionally, metformin acts as a metaboloepigenetic regulator by promoting global DNA methylation via the mitochondrial one-carbon metabolism pathway and in cancer cells via the H19/S-adenosylhomocysteine hydrolase (SAHH) axis (Cuyàs et al. 2017, Zhong et al. 2017). Metformin’s direct upregulation of global DNA methylation and its downstream tissue-specific (skeletal muscle, subcutaneous adipose tissue, peripheral blood mononuclear cells) transcriptional profiles in healthy and diabetic individuals as well as cancer cell lines (breast and colorectal), reflect its age-targeting and anti-tumoral role (de Kreutzenberg et al. 2015, Yu et al. 2017, Bridgeman et al. 2018, Elbere et al. 2018, Kulkarni et al. 2018, Sabit et al. 2018, Schulten and Bakhashab 2019). Apart from its effect on DNA and histone methylation, metformin also upregulates DICER1 and downstream miRNAs, several of which are reduced with cellular senescence, suggesting metformin’s multiple mechanisms to regulate transcription and post-transcription (Noren Hooten et al. 2016). Most recently, metformin, in combination with recombinant human growth hormone. (rhGH) and dehydroepiandrosterone (DHEA) was shown to revert the mean “epigenetic age”- a DNA-methylation based marker of biological age, by 1.5 years after a year of treatment in healthy human adults (Fahy et al. 2019).
There is inherent variability in metformin’s transcriptional and epigenetic regulation via histone and DNA modifications and its effects on miRNA. Most of the studies so far have investigated this regulation in the cell lines at suprapharmacological doses that may limit their generalizability to clinical studies. It is essential to elucidate these responses further, in the context of pharmacologically relevant doses in tissue and cell-specific manner as well as at systemic levels.
8. Telomere attrition
Telomeres are highly conserved ribonucleoprotein complexes on the termini of eukaryotic chromosomes that play a crucial role in protecting chromosomal ends from degradation caused by an incomplete DNA replication (Casagrande and Hau 2019). With aging and stress exposure, these regions are particularly vulnerable to deterioration, and due to insufficient telomerase in adult cells to replenish this loss, they undergo progressive attrition with cell proliferation (Blackburn et al. 2015). Progressive telomere shortening is found to be associated with biological aging and age-related morbidities, frailty, and mortality (Armanios et al. 2009, Araujo Carvalho et al. 2019, Whittemore et al. 2019, Zhu et al. 2019). Furthermore, telomere shortening is also shown to trigger cellular senescence, inflammation, and mitochondrial dysfunction via p53-mediated DNA damage response pathways (Zhu et al. 2019).
Metformin activates TERRA and is shown to reduce telomere shortening
There is little evidence on the action of interventions including metformin on telomere length and especially on their consequences on healthy aging and increased lifespan. However, Diman et al. identified downstream targets of AMPK activated by metformin-nuclear respiratory factor 1 and PGC1-α as regulators of human telomere transcription via telomeric repeat-containing RNA (TERRA) (Diman et al. 2016). In diabetic individuals, metformin is suggested to reduce telomere shortening, when compared to those not taking metformin (Robb-MacKay 2018). This is similar to other studies where metformin monotherapy to improve glucose tolerance or insulin sensitivity prevented shortening of leukocyte telomere length in diabetic individuals (de Zegher et al. 2015, Rosa et al. 2018, Liu et al. 2019). In mothers with gestational diabetes, metformin treatment prevented telomere shortening in their male offspring (Garcia-Martin et al. 2018).
Further studies are warranted to understand how metformin may prevent telomere attrition and maintain telomeric integrity. Additionally, metformin’s effect on mitochondrial function and cellular senescence may induce protective feedback mechanisms on telomere maintenance. It is also important to delineate metformin’s effects on telomere length and integrity in aging versus cancer.
9. Cellular senescence
Senescence, defined as a stable cell cycle arrest when cells reach their replicative potential or are exposed to internal or external stressors, is claimed to be a major cause of aging (Gil 2019). Seminal discoveries, first by Hayflick and Moorehead, and lately by Baker et al., concretized cellular senescence as a major driver of aging with the age-associated accumulation of senescent cells, and attenuation of age-related tissue dysfunction after their removal, even late in life (Hayflick and Moorhead 1961, Baker et al. 2011). Although senescence acts as a protective mechanism against malignancies, serving as a tumor-suppressive mechanism, recent evidence suggests that senescent cells can morph their phenotype to SASP, which can paradoxically cause tumor growth (Coppé et al. 2010). Furthermore, senescent cell accumulation via aging-induced epigenetic, proteotoxic and genotoxic stresses usually exhibits a causal role in many age-related diseases, including glaucoma, cataracts, diabetes mellitus, osteoarthritis, among others (Childs et al. 2015). More recently, senolytics- drugs that selectively induce apoptosis in senescent cells have shown tremendous potential and clinical utility to alleviate aging and age-related phenotypes (Kirkland et al. 2017, Xu et al. 2018, Justice et al. 2019).
Metformin downregulates SASP and lowers senescent cell burden
Metformin- although not exhibiting senolytic properties, has been effective in suppressing cellular senescence and SASP in multiple age-associated dysfunctions. Chronic metformin administration at low doses delays senescence in human diploid fibroblasts and human mesenchymal stem cells, as evidenced by reduced SA-β-Gal staining, via Nrf2-mediated upregulation of Glutathione peroxidase 7 (GPx7) (Fang et al. 2018). Metformin’s senotherapeutic role is also mediated by its anti-inflammatory effect by preventing NF-κB translocation into the nucleus thereby not phosphorylating IκB and its kinase and inhibiting NF-κB pathway altogether (Moiseeva et al. 2013). Inhibition of NF-κB by metformin in ex-vivo cultures of murine olfactory ensheathing cells is also associated with decreased SA-β-Gal activity along with a decreased expression of pro-inflammatory cytokines and oxidative stress markers (Śmieszek et al. 2017). In a DICER1-dependent mechanism, metformin is shown to lower the protein levels of p16 and p21, and RNA levels of SASP hallmarks including IL-6 and IL-8 in human fibroblasts (Noren Hooten et al. 2016). Recently, in human periodontal ligament cells, the protective effect against senescence and oxidative stress was linked to metformin-induced stimulation of autophagy, further highlighting the role of metformin in targeting the interconnectedness of these two hallmarks of aging (Kuang et al. 2019). Similarly, in in vitro and in vivo models of intervertebral disc degeneration, metformin treatment reduced senescence in nucleus pulposus cells by upregulating AMPK-mediated autophagy (Chen et al. 2016).
It is evident that metformin attenuates the increased burden of senescent cells and upregulation of SASP with aging. Its role as a SASP modulator via Nrf2-Gpx7 activation in mediating oxidative stress, and via NF-κB inhibition in mediating inflammatory response provides a better understanding of its senotherapeutic mechanism. However, in the context of breast cancer and hepatoma, low dose metformin has been previously shown to induce SASP gene expression signature and p53-dependent senescence, respectively (Williams et al. 2013, Yi et al. 2013). Thus, metformin’s effect on cellular senescence and SASP is context-dependent (anti-cancer by inducing apoptosis and anti-aging by inhibiting inflammation), probing for further understanding of what stimulates metformin to induce an anti-senescence response in aging tissues as opposed to pro-senescence in cancerous tissues. With the advent of clinical trials with senolytics, it is also crucial to understand how metformin can be best utilized in combination with other drugs to optimize the combinatorial effect on senescence and aging.
Side Effects of Metformin
Clearly, the unexpected positive side effects of metformin include increased healthspan and decreased mortality. Recently, it was evident that diabetic patients with liver, kidney and heart diseases, who did not receive metformin could have benefitted from its use with reduced hospitalization and mortality (Crowley et al. 2017). Hence, the ‘black box’ warning for metformin has been steadily disappearing, as evidenced by less strict indication for use in patients with reduced kidney function (Bakris and Molitch 2016). As in most drugs, minor side effects occur with metformin, but it is important to understand whether they reveal anything about the mechanism of action of this drug. Most of these minor side effects include gastrointestinal discomfort such as abdominal or stomach pain, early satiety, decreased appetite, and diarrhea, which usually subside after one or two weeks of use (McCreight et al. 2016). If diarrhea persists beyond a week (in ~3% of users), the drug is discontinued. These side effects could be the result of an acute change in the gut microbiome, as evidenced in nematodes and other smaller organisms (Cabreiro et al. 2013). Microbiome changes may also be secondary to reduced vitamin B12 levels with chronic metformin use, although the mechanism is still unclear and few patients need vitamin B12 replacement (Aroda et al. 2016). A condition termed Metformin Associated Lactic Acidosis (MALA) is associated with metformin use in patients who already have high lactic acid levels as in severe kidney, liver and heart diseases, while also taking metformin (DeFronzo et al. 2016). Few patients may complain about anxiety, sleeplessness, fast breathing, and other symptoms soon after taking metformin and will usually stop the drugs on their own. Metformin increases lactate levels, while maintaining the normal range, but it is possible that those with severe side effects including MALA subjects may be more sensitive to the inhibition of mitochondrial complex-1, attributable to genetic mechanisms.
The TAME trial aims to examine the subjects who stop taking metformin for all the aforementioned reasons and determine if they are prone to different outcomes, with their biological samples available for further mechanistic studies. It is also important to understand that, consistent with its biological effects since metformin is not a hypoglycemic agent it is not likely to cause hypoglycemia unless used in combination with other glucose-lowering drugs in diabetic individuals. Thus, it is very crucial to note that outside of its indication for diabetes, the use of metformin is recommended only in the context of clinical trials, under physician supervision.
Conclusions and Perspectives
Metformin’s efficacy in attenuating hallmarks of biological aging is reflective of its strength and potential as a therapeutic that can target crucial mechanistic pathways involved in aging. The metabolic effects of metformin are mainly through its metabolic arm via activation of AMPK and oxidative arm through inhibition of complex I of the mitochondrial ETC. There are additional direct effects on mTORC1, PGC1-α, Insulin-IGF1 signaling, SIRT1, NF-κB signaling, and pro-inflammatory cytokines thereby allowing us to classify the four hallmarks (deregulated nutrient-sensing, altered intercellular communication, genomic instability and loss of proteostasis) as the primary targets of metformin. The effects on mitochondrial function, DNA and histone modifications, stem cell rejuvenation, preventing telomere shortening and downregulation of senescence and SASP are downstream of the primary targets (Figure 2).
Figure 2-. The primary and secondary targets of metformin among the hallmarks of aging.
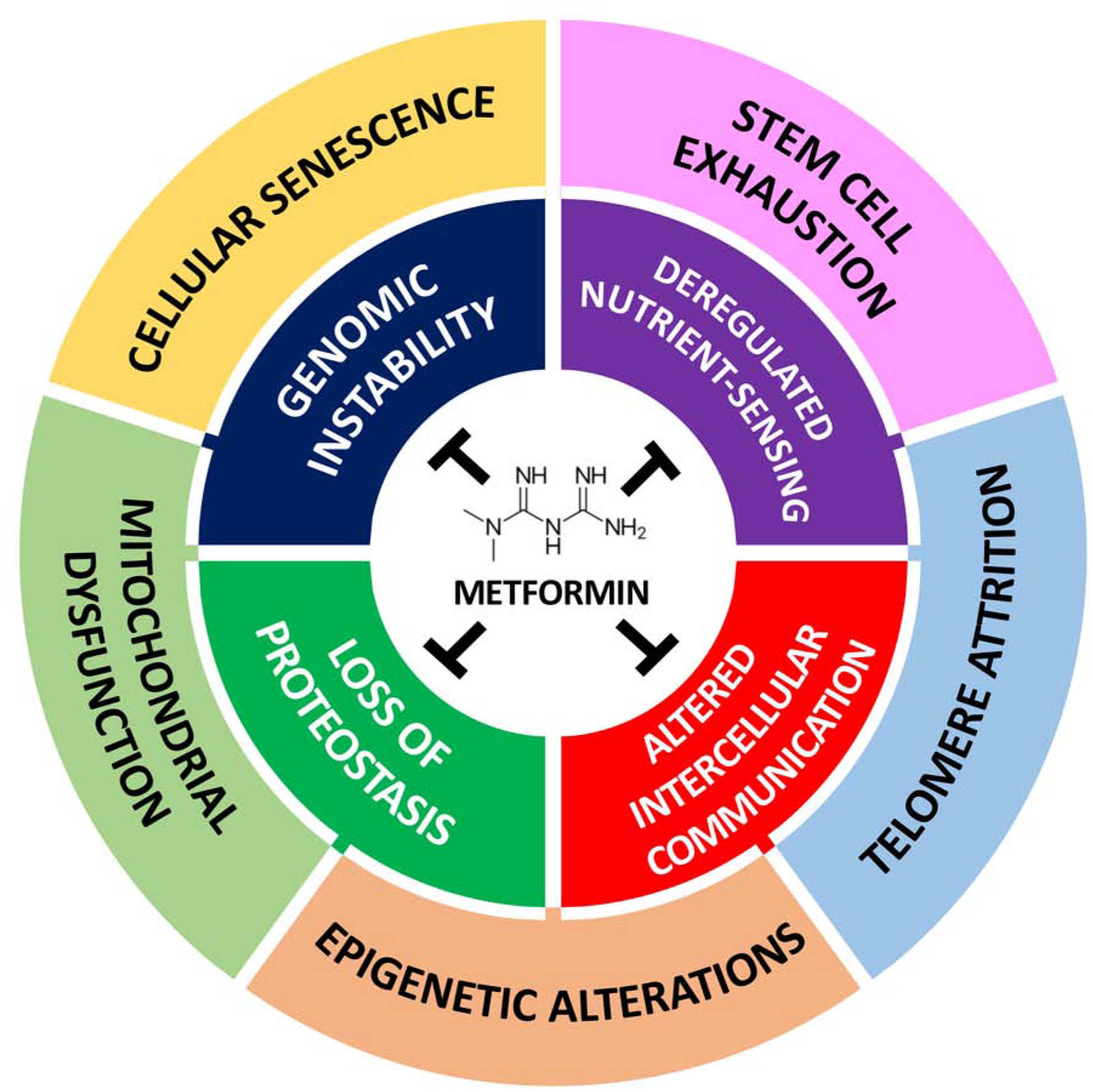
Metformin attenuates hallmarks of aging to varying degrees and in turn, contributes to its gerotherapeutic effects. We classified the hallmarks in the inner circle (deregulated nutrient-sensing, genomic instability, loss of proteostasis and altered intercellular communication) as primary, reflective of them being direct targets of metformin via its action on AMPK, SIRT1, mTORC1, IIS pathways, protection against macromolecular damage, improved autophagy response and reduced inflammation, respectively. The hallmarks in the outer circle (stem cell exhaustion, cellular senescence, mitochondrial dysfunction, epigenetic alterations, telomere attrition) are secondary targets, due to their attenuation being mediated by metformin’s role on a primary target. Due to the high degree of interconnectedness between the hallmarks of aging, metformin’s attenuation of any single hallmark has a major influence on several others, thereby leading to a widespread response against aging.
Several clinical trials are currently underway to assess the response of metformin as a monotherapy and in combination with lifestyle interventions like exercise, on clinical and molecular outcomes of individual hallmarks of aging. These are particularly involved in measuring metformin’s effect on frailty and associated inflammatory and SASP candidate biomarkers (NCT03451006), pro-autophagy effects by measuring LC3 levels (NCT03309007), changes in muscle size (NCT03107884), improvements in immune response via the gut microbiota (NCT03713801), and effective influenza vaccine responses (NCT03996538).
An important limitation to studying metformin’s action on aging is its tissue-specificity, dose-dependency and the role of mitochondrial inhibition in contributing to any of its effects. No doubt, further studies will potentially shed more light on each of these limitations. But as the accumulation of evidence links metformin to all hallmarks of aging, it is very important to remember that metformin’s impact on any of these hallmarks has direct consequences on systemic attenuation across several other hallmarks of aging (Figure 2). For example, as highlighted above, evidence suggests that targeting autophagy impacts mitochondrial function, nutrient-sensing, and macromolecular damage. With this line of thought, we suggest that targeting aging at any of the primary or secondary levels, will ultimately make the cells, tissues, and systems more youthful, with a direct effect on the systemic biology of aging (Figure 2). Thus, some effects that are observed are a result of achieving systemic youthfulness and not direct targets of the drug. In fact, reviewing the effects of other interventions like rapalogs and sirtuins also shows that they may be impacting many of the similar hallmarks of aging (Lamming et al. 2013, Grabowska et al. 2017).
However, for any biotechnology or pharmaceutical firm that wants to design metformin-like drugs, it is also possible that all effects of metformin need to be combined for the optimal effect. For instance, targeting complex I is reasonable for the development of anti-cancer drugs and a drug that does not depend on tissue selectivity and gets to the mitochondria in all tissues (that is also relevant for aging) is warranted. But to show that it is superior to metformin in preventing age-related diseases may be risky, not to mention-expensive for patients.
In conclusion, there is extensive epidemiological, basic science, and clinical data highlighting the effectiveness of metformin in targeting several age-related morbidities in humans. Studies in model organisms and cell lines provide compelling evidence on metformin’s beneficial effects against crucial pathways in aging. In addition to its known safety profile and long-term use in humans, metformin-induced attenuation of the major hallmarks of biological aging and their interconnectivity makes it a very attractive candidate against aging, which the TAME study is set to prove and change the landscape of healthspan around the world.
Acknowledgments
This work is supported by the grants from the National Institutes of Health (P01AG021654) (N.B.), Nathan Shock Center of Excellence in the Basic Biology of Aging (P30AG038072) (N.B.), Paul F. Glenn Center for the Biology of Aging Research at the Albert Einstein College of Medicine (N.B.). A.S.K. is supported by the Albert Einstein College of Medicine’s Ph.D. in Clinical Investigation program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests
References
- (2019). “9. Pharmacologic Approaches to Glycemic Treatment: Standards of Medical Care in Diabetes—2019.” Diabetes Care 42(Supplement 1): S90. [DOI] [PubMed] [Google Scholar]
- Aatsinki SM, Buler M, Salomaki H, Koulu M, Pavek P and Hakkola J (2014). “Metformin induces PGC-1alpha expression and selectively affects hepatic PGC-1alpha functions.” Br J Pharmacol 171(9): 2351–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaras I, Mitchell SJ, Mora H, Lugo DR, Warren A, Navas-Enamorado I, Hoffmann V, Hine C, Mitchell JR, Le Couteur DG, Cogger VC, Bernier M and de Cabo R (2017). “Health benefits of late-onset metformin treatment every other week in mice.” NPJ aging and mechanisms of disease 3: 16–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algire C, Moiseeva O, Deschenes-Simard X, Amrein L, Petruccelli L, Birman E, Viollet B, Ferbeyre G and Pollak MN (2012). “Metformin reduces endogenous reactive oxygen species and associated DNA damage.” Cancer Prev Res (Phila) 5(4): 536–543. [DOI] [PubMed] [Google Scholar]
- Amin S, Lux A and O’Callaghan F (2019). “The journey of metformin from glycaemic control to mTOR inhibition and the suppression of tumour growth.” Br J Clin Pharmacol 85(1): 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimov VN, Berstein LM, Egormin PA, Piskunova TS, Popovich IG, Zabezhinski MA, Tyndyk ML, Yurova MV, Kovalenko IG, Poroshina TE and Semenchenko AV (2008). “Metformin slows down aging and extends life span of female SHR mice.” Cell Cycle 7(17): 2769–2773. [DOI] [PubMed] [Google Scholar]
- Araujo Carvalho AC, Tavares Mendes ML, da Silva Reis MC, Santos VS, Tanajura DM and Martins-Filho PRS (2019). “Telomere length and frailty in older adults-A systematic review and meta-analysis.” Ageing Res Rev 54: 100914. [DOI] [PubMed] [Google Scholar]
- Armanios M, Alder JK, Parry EM, Karim B, Strong MA and Greider CW (2009). “Short telomeres are sufficient to cause the degenerative defects associated with aging.” Am J Hum Genet 85(6): 823–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroda VR, Edelstein SL, Goldberg RB, Knowler WC, Marcovina SM, Orchard TJ, Bray GA, Schade DS, Temprosa MG, White NH, Crandall JP and Diabetes G Prevention Program Research (2016). “Long-term Metformin Use and Vitamin B12 Deficiency in the Diabetes Prevention Program Outcomes Study.” The Journal of clinical endocrinology and metabolism 101(4): 1754–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou D, Aguila M, Opefi CA, South K, Bellingham J, Bevilacqua D, Munro PM, Kanuga N, Mackenzie FE, Dubis AM, Georgiadis A, Graca AB, Pearson RA, Ali RR, Sakami S, Palczewski K, Sherman MY, Reeves PJ and Cheetham ME (2017). “Rescue of mutant rhodopsin traffic by metformin-induced AMPK activation accelerates photoreceptor degeneration.” Hum Mol Genet 26(2): 305–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CJ (2017). “Metformin: historical overview.” Diabetologia 60(9): 1566–1576. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL and van Deursen JM (2011). “Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders.” Nature 479(7372): 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakris GL and Molitch ME (2016). “Should Restrictions Be Relaxed for Metformin Use in Chronic Kidney Disease? Yes, They Should Be Relaxed! What’s the Fuss?” Diabetes Care 39(7): 1287. [DOI] [PubMed] [Google Scholar]
- Barzilai N, Crandall JP, Kritchevsky SB and Espeland MA (2016). “Metformin as a Tool to Target Aging.” Cell Metab 23(6): 1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzilai NR (2017). “TARGETING AGING WITH METFORMIN (TAME).” Innovation in Aging: 743–743. [Google Scholar]
- Bauer PV, Duca FA, Waise TMZ, Rasmussen BA, Abraham MA, Dranse HJ, Puri A, O’Brien CA and Lam TKT (2018). “Metformin Alters Upper Small Intestinal Microbiota that Impact a Glucose-SGLT1-Sensing Glucoregulatory Pathway.” Cell Metab 27(1): 101–117.e105. [DOI] [PubMed] [Google Scholar]
- Bauer PV, Duca FA, Waise TMZ, Rasmussen BA, Abraham MA, Dranse HJ, Puri A, O’Brien CA and Lam TKT (2018). “Metformin Alters Upper Small Intestinal Microbiota that Impact a Glucose-SGLT1-Sensing Glucoregulatory Pathway.” Cell Metabolism 27(1): 101–117.e105. [DOI] [PubMed] [Google Scholar]
- Ben Sahra I, Regazzetti C, Robert G, Laurent K, Le Marchand-Brustel Y, Auberger P, Tanti JF, Giorgetti-Peraldi S and Bost F (2011). “Metformin, independent of AMPK, induces mTOR inhibition and cell-cycle arrest through REDD1.” Cancer Res 71(13): 4366–4372. [DOI] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA and Brunet A (2015). “Epigenetic regulation of ageing: linking environmental inputs to genomic stability.” Nat Rev Mol Cell Biol 16(10): 593–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Singh PP, Mahmoudi S, Harel I, Casey KM, Dulken BW, Kundaje A and Brunet A (2019). “Remodeling of epigenome and transcriptome landscapes with aging in mice reveals widespread induction of inflammatory responses.” Genome Research 29(4): 697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettedi L and Foukas LC (2017). “Growth factor, energy and nutrient sensing signalling pathways in metabolic ageing.” Biogerontology 18(6): 913–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Epel ES and Lin J (2015). “Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection.” Science 350(6265): 1193–1198. [DOI] [PubMed] [Google Scholar]
- Bridgeman SC, Ellison GC, Melton PE, Newsholme P and Mamotte CDS (2018). “Epigenetic effects of metformin: From molecular mechanisms to clinical implications.” Diabetes Obes Metab 20(7): 1553–1562. [DOI] [PubMed] [Google Scholar]
- Cabreiro F, Au C, Leung KY, Vergara-Irigaray N, Cocheme HM, Noori T, Weinkove D, Schuster E, Greene ND and Gems D (2013). “Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism.” Cell 153(1): 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron AR, Morrison VL, Levin D, Mohan M, Forteath C, Beall C, McNeilly AD, Balfour DJ, Savinko T, Wong AK, Viollet B, Sakamoto K, Fagerholm SC, Foretz M, Lang CC and Rena G (2016). “Anti-Inflammatory Effects of Metformin Irrespective of Diabetes Status.” Circ Res 119(5): 652–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JM, Bellman SM, Stephenson MD and Lisy K (2017). “Metformin reduces all-cause mortality and diseases of ageing independent of its effect on diabetes control: A systematic review and meta-analysis.” Ageing Res Rev 40: 31–44. [DOI] [PubMed] [Google Scholar]
- Campisi J, Kapahi P, Lithgow GJ, Melov S, Newman JC and Verdin E (2019). “From discoveries in ageing research to therapeutics for healthy ageing.” Nature 571(7764): 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casagrande S and Hau M (2019). “Telomere attrition: metabolic regulation and signalling function?” Biol Lett 15(3): 20180885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheki M, Ghasemi MS, Rezaei Rashnoudi A and Erfani Majd N (2019). “Metformin attenuates cisplatin-induced genotoxicity and apoptosis in rat bone marrow cells.” Drug Chem Toxicol: 1–8. [DOI] [PubMed] [Google Scholar]
- Cheki M, Shirazi A, Mahmoudzadeh A, Bazzaz JT and Hosseinimehr SJ (2016). “The radioprotective effect of metformin against cytotoxicity and genotoxicity induced by ionizing radiation in cultured human blood lymphocytes.” Mutat Res 809: 24–32. [DOI] [PubMed] [Google Scholar]
- Chen D, Xia D, Pan Z, Xu D, Zhou Y, Wu Y, Cai N, Tang Q, Wang C, Yan M, Zhang JJ, Zhou K, Wang Q, Feng Y, Wang X, Xu H, Zhang X and Tian N (2016). “Metformin protects against apoptosis and senescence in nucleus pulposus cells and ameliorates disc degeneration in vivo.” Cell Death & Disease 7(10): e2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ou Y, Li Y, Hu S, Shao LW and Liu Y (2017). “Metformin extends C. elegans lifespan through lysosomal pathway.” Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SC, Brooks R, Houskeeper J, Bremner SK, Dunlop J, Viollet B, Logan PJ, Salt IP, Ahmed SF and Yarwood SJ (2017). “Metformin suppresses adipogenesis through both AMP-activated protein kinase (AMPK)-dependent and AMPK-independent mechanisms.” Mol Cell Endocrinol 440: 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs BG, Durik M, Baker DJ and van Deursen JM (2015). “Cellular senescence in aging and age-related disease: from mechanisms to therapy.” Nature medicine 21(12): 1424–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MM, Nicol CJ, Cheng YC, Lin KH, Chen YL, Pei D, Lin CH, Shih YN, Yen CH, Chen SJ, Huang RN and Chiang MC (2017). “Metformin activation of AMPK suppresses AGE-induced inflammatory response in hNSCs.” Exp Cell Res 352(1): 75–83. [DOI] [PubMed] [Google Scholar]
- Coll AP, Chen M, Taskar P, Rimmington D, Patel S, Tadross JA, Cimino I, Yang M, Welsh P, Virtue S, Goldspink DA, Miedzybrodzka EL, Konopka AR, Esponda RR, Huang JTJ, Tung YCL, Rodriguez-Cuenca S, Tomaz RA, Harding HP, Melvin A, Yeo GSH, Preiss D, Vidal-Puig A, Vallier L, Nair KS, Wareham NJ, Ron D, Gribble FM, Reimann F, Sattar N, Savage DB, Allan BB and O’Rahilly S (2020). “GDF15 mediates the effects of metformin on body weight and energy balance.” Nature 578(7795): 444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppé J-P, Desprez P-Y, Krtolica A and Campisi J (2010). “The senescence-associated secretory phenotype: the dark side of tumor suppression.” Annual review of pathology 5: 99–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley MJ, Diamantidis CJ, McDuffie JR, Cameron CB, Stanifer JW, Mock CK, Wang X, Tang S, Nagi A, Kosinski AS and Williams JW Jr. (2017). “Clinical Outcomes of Metformin Use in Populations With Chronic Kidney Disease, Congestive Heart Failure, or Chronic Liver Disease: A Systematic Review.” Annals of internal medicine 166(3): 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuyàs E, Fernández-Arroyo S, Verdura S, García RÁ-F, Stursa J, Werner L, Blanco-González E, Montes-Bayón M, Joven J, Viollet B, Neuzil J and Menendez JA (2017). “Metformin regulates global DNA methylation via mitochondrial one-carbon metabolism.” Oncogene. [DOI] [PubMed] [Google Scholar]
- Cuyas E, Verdura S, Llorach-Pares L, Fernandez-Arroyo S, Joven J, Martin-Castillo B, Bosch-Barrera J, Brunet J, Nonell-Canals A, Sanchez-Martinez M and Menendez JA (2018). “Metformin Is a Direct SIRT1-Activating Compound: Computational Modeling and Experimental Validation.” Front Endocrinol (Lausanne) 9: 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Haes W, Frooninckx L, Van Assche R, Smolders A, Depuydt G, Billen J, Braeckman BP, Schoofs L and Temmerman L (2014). “Metformin promotes lifespan through mitohormesis via the peroxiredoxin PRDX-2.” Proc Natl Acad Sci U S A 111(24): E2501–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kreutzenberg SV, Ceolotto G, Cattelan A, Pagnin E, Mazzucato M, Garagnani P, Borelli V, Bacalini MG, Franceschi C, Fadini GP and Avogaro A (2015). “Metformin improves putative longevity effectors in peripheral mononuclear cells from subjects with prediabetes. A randomized controlled trial.” Nutr Metab Cardiovasc Dis 25(7): 686–693. [DOI] [PubMed] [Google Scholar]
- de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, Velasquez-Mejia EP, Carmona JA, Abad JM and Escobar JS (2017). “Metformin Is Associated With Higher Relative Abundance of Mucin-Degrading Akkermansia muciniphila and Several Short-Chain Fatty Acid-Producing Microbiota in the Gut.” Diabetes Care 40(1): 54–62. [DOI] [PubMed] [Google Scholar]
- de Zegher F, Díaz M and Ibáñez L (2015). “Association Between Long Telomere Length and Insulin Sensitization in Adolescent Girls With Hyperinsulinemic Androgen Excess.” JAMA Pediatrics 169(8): 787–788. [DOI] [PubMed] [Google Scholar]
- DeFronzo R, Fleming GA, Chen K and Bicsak TA (2016). “Metformin-associated lactic acidosis: Current perspectives on causes and risk.” Metabolism 65(2): 20–29. [DOI] [PubMed] [Google Scholar]
- DeFronzo RA, Barzilai N and Simonson DC (1991). “Mechanism of metformin action in obese and lean noninsulin-dependent diabetic subjects.” J Clin Endocrinol Metab 73(6): 1294–1301. [DOI] [PubMed] [Google Scholar]
- Diman A, Boros J, Poulain F, Rodriguez J, Purnelle M, Episkopou H, Bertrand L, Francaux M, Deldicque L and Decottignies A (2016). “Nuclear respiratory factor 1 and endurance exercise promote human telomere transcription.” Science Advances 2(7): e1600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan Turacli I, Candar T, Yuksel EB, Kalay S, Oguz AK and Demirtas S (2018). “Potential effects of metformin in DNA BER system based on oxidative status in type 2 diabetes.” Biochimie 154: 62–68. [DOI] [PubMed] [Google Scholar]
- Dowling RJ, Zakikhani M, Fantus IG, Pollak M and Sonenberg N (2007). “Metformin inhibits mammalian target of rapamycin-dependent translation initiation in breast cancer cells.” Cancer Res 67(22): 10804–10812. [DOI] [PubMed] [Google Scholar]
- El-Mir M-Y, Nogueira V, Fontaine E, Avéret N, Rigoulet M and Leverve X (2000). “Dimethylbiguanide Inhibits Cell Respiration via an Indirect Effect Targeted on the Respiratory Chain Complex I.” Journal of Biological Chemistry 275(1): 223–228. [DOI] [PubMed] [Google Scholar]
- Elbere I, Silamikelis I, Ustinova M, Kalnina I, Zaharenko L, Peculis R, Konrade I, Ciuculete DM, Zhukovsky C, Gudra D, Radovica-Spalvina I, Fridmanis D, Pirags V, Schioth HB and Klovins J (2018). “Significantly altered peripheral blood cell DNA methylation profile as a result of immediate effect of metformin use in healthy individuals.” Clin Epigenetics 10(1): 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelyanova L, Rudic T, Ross GR, Rizvi F, Kress DC and Jahangir A (2019). “Metformin Has Direct Protective Effect on Human Cardiac Mitochondria.” The FASEB Journal 33(1_supplement): 794.794–794.794. [Google Scholar]
- Ermolaeva M, Neri F, Ori A and Rudolph KL (2018). “Cellular and epigenetic drivers of stem cell ageing.” Nat Rev Mol Cell Biol 19(9): 594–610. [DOI] [PubMed] [Google Scholar]
- Fahy GM, Brooke RT, Watson JP, Good Z, Vasanawala SS, Maecker H, Leipold MD, Lin DTS, Kobor MS and Horvath S (2019). “Reversal of epigenetic aging and immunosenescent trends in humans.” Aging Cell: e13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Yang J, Wu X, Zhang G, Li T, Wang X, Zhang H, Wang CC, Liu GH and Wang L (2018). “Metformin alleviates human cellular aging by upregulating the endoplasmic reticulum glutathione peroxidase 7.” Aging Cell: e12765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatt M, Hsu K, He L, Wondisford F, Miller FD, Kaplan DR and Wang J (2015). “Metformin Acts on Two Different Molecular Pathways to Enhance Adult Neural Precursor Proliferation/Self-Renewal and Differentiation.” Stem cell reports 5(6): 988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine E (2018). “Metformin-Induced Mitochondrial Complex I Inhibition: Facts, Uncertainties, and Consequences.” Front Endocrinol (Lausanne) 9: 753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foretz M, Guigas B, Bertrand L, Pollak M and Viollet B (2014). “Metformin: from mechanisms of action to therapies.” Cell Metab 20(6): 953–966. [DOI] [PubMed] [Google Scholar]
- Foretz M, Hébrard S, Leclerc J, Zarrinpashneh E, Soty M, Mithieux G, Sakamoto K, Andreelli F and Viollet B (2010). “Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state.” The Journal of clinical investigation 120(7): 2355–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C, Garagnani P, Parini P, Giuliani C and Santoro A (2018). “Inflammaging: a new immune–metabolic viewpoint for age-related diseases.” Nature Reviews Endocrinology 14(10): 576–590. [DOI] [PubMed] [Google Scholar]
- Galuska D, Zierath J, Thorne A, Sonnenfeld T and Wallberg-Henriksson H (1991). “Metformin increases insulin-stimulated glucose transport in insulin-resistant human skeletal muscle.” Diabete Metab 17(1 Pt 2): 159–163. [PubMed] [Google Scholar]
- Garcia-Martin I, Penketh RJA, Janssen AB, Jones RE, Grimstead J, Baird DM and John RM (2018). “Metformin and insulin treatment prevent placental telomere attrition in boys exposed to maternal diabetes.” PloS one 13(12): e0208533–e0208533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J (2019). “Cellular senescence causes ageing.” Nature Reviews Molecular Cell Biology 20(7): 388–388. [DOI] [PubMed] [Google Scholar]
- Gillespie ZE, Wang C, Vadan F, Yu TY, Ausio J, Kusalik A and Eskiw CH (2019). “Metformin induces the AP-1 transcription factor network in normal dermal fibroblasts.” Sci Rep 9(1): 5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glossmann HH and Lutz OMD (2019). “Metformin and Aging: A Review.” Gerontology: 1–10. [DOI] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJY, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, Mercken EM, Palmeira CM, de Cabo R, Rolo AP, Turner N, Bell EL and Sinclair DA (2013). “Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging.” Cell 155(7): 1624–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L, Goswami S, Giacomini KM, Altman RB and Klein TE (2012). “Metformin pathways: pharmacokinetics and pharmacodynamics.” Pharmacogenet Genomics 22(11): 820–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowska W, Sikora E and Bielak-Zmijewska A (2017). “Sirtuins, a promising target in slowing down the ageing process.” Biogerontology 18(4): 447–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halicka HD, Zhao H, Li J, Traganos F, Zhang S, Lee M and Darzynkiewicz Z (2011). “Genome protective effect of metformin as revealed by reduced level of constitutive DNA damage signaling.” Aging (Albany NY) 3(10): 1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Gadalla AE, Olsen GS and Hardie DG (2002). “The antidiabetic drug metformin activates the AMP-activated protein kinase cascade via an adenine nucleotide-independent mechanism.” Diabetes 51(8): 2420–2425. [DOI] [PubMed] [Google Scholar]
- Hayflick L and Moorhead PS (1961). “The serial cultivation of human diploid cell strains.” Exp Cell Res 25: 585–621. [DOI] [PubMed] [Google Scholar]
- Heckman-Stoddard BM, DeCensi A, Sahasrabuddhe VV and Ford LG (2017). “Repurposing metformin for the prevention of cancer and cancer recurrence.” Diabetologia 60(9): 1639–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S and Shaw RJ (2018). “AMPK: guardian of metabolism and mitochondrial homeostasis.” Nature reviews. Molecular cell biology 19(2): 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp MS, Kasturi P and Hartl FU (2019). “The proteostasis network and its decline in ageing.” Nat Rev Mol Cell Biol 20(7): 421–435. [DOI] [PubMed] [Google Scholar]
- Howell JJ, Hellberg K, Turner M, Talbott G, Kolar MJ, Ross DS, Hoxhaj G, Saghatelian A, Shaw RJ and Manning BD (2017). “Metformin Inhibits Hepatic mTORC1 Signaling via Dose-Dependent Mechanisms Involving AMPK and the TSC Complex.” Cell Metabolism 25(2): 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter RW, Hughey CC, Lantier L, Sundelin EI, Peggie M, Zeqiraj E, Sicheri F, Jessen N, Wasserman DH and Sakamoto K (2018). “Metformin reduces liver glucose production by inhibition of fructose-1–6-bisphosphatase.” Nat Med 24(9): 1395–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo A, Nitti M, Mollo N, Paladino S, Procaccini C, Faicchia D, Cali G, Genesio R, Bonfiglio F, Cicatiello R, Polishchuk E, Polishchuk R, Pinton P, Matarese G, Conti A and Nitsch L (2017). “Metformin restores the mitochondrial network and reverses mitochondrial dysfunction in Down syndrome cells.” Hum Mol Genet 26(6): 1056–1069. [DOI] [PubMed] [Google Scholar]
- Jackson RA, Hawa MI, Jaspan JB, Sim BM, Disilvio L, Featherbe D and Kurtz AB (1987). “Mechanism of metformin action in non-insulin-dependent diabetes.” Diabetes 36(5): 632–640. [DOI] [PubMed] [Google Scholar]
- Jang JY, Blum A, Liu J and Finkel T (2018). “The role of mitochondria in aging.” The Journal of Clinical Investigation 128(9): 3662–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephson AM, Bradaschia-Correa V, Lee S, Leclerc K, Patel KS, Muinos Lopez E, Litwa HP, Neibart SS, Kadiyala M, Wong MZ, Mizrahi MM, Yim NL, Ramme AJ, Egol KA and Leucht P (2019). “Age-related inflammation triggers skeletal stem/progenitor cell dysfunction.” Proceedings of the National Academy of Sciences 116(14): 6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice JN, Nambiar AM, Tchkonia T, LeBrasseur NK, Pascual R, Hashmi SK, Prata L, Masternak MM, Kritchevsky SB, Musi N and Kirkland JL (2019). “Senolytics in idiopathic pulmonary fibrosis: Results from a first-in-human, open-label, pilot study.” EBioMedicine 40: 554–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalender A, Selvaraj A, Kim SY, Gulati P, Brule S, Viollet B, Kemp BE, Bardeesy N, Dennis P, Schlager JJ, Marette A, Kozma SC and Thomas G (2010). “Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner.” Cell Metab 11(5): 390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori H, Naruse G, Yoshida A, Minatoguchi S, Watanabe T, Kawaguchi T, Yamada Y, Mikami A, Kawasaki M, Takemura G and Minatoguchi S (2019). “Metformin Enhances Autophagy and Provides Cardioprotection in delta-Sarcoglycan Deficiency-Induced Dilated Cardiomyopathy.” Circ Heart Fail 12(4): e005418. [DOI] [PubMed] [Google Scholar]
- Kane DA, Anderson EJ, Price JW 3rd, Woodlief TL, Lin C-T, Bikman BT, Cortright RN and Neufer PD (2010). “Metformin selectively attenuates mitochondrial H2O2 emission without affecting respiratory capacity in skeletal muscle of obese rats.” Free radical biology & medicine 49(6): 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnewar S, Neeli PK, Panuganti D, Kotagiri S, Mallappa S, Jain N, Jerald MK and Kotamraju S (2018). “Metformin regulates mitochondrial biogenesis and senescence through AMPK mediated H3K79 methylation: Relevance in age-associated vascular dysfunction.” Biochim Biophys Acta Mol Basis Dis 1864(4 Pt A): 1115–1128. [DOI] [PubMed] [Google Scholar]
- Kaushik S and Cuervo AM (2015). “Proteostasis and aging.” Nat Med 21(12): 1406–1415. [DOI] [PubMed] [Google Scholar]
- Kelly B, Tannahill GM, Murphy MP and O’Neill LA (2015). “Metformin Inhibits the Production of Reactive Oxygen Species from NADH:Ubiquinone Oxidoreductase to Limit Induction of Interleukin-1beta (IL-1beta) and Boosts Interleukin-10 (IL-10) in Lipopolysaccharide (LPS)-activated Macrophages.” J Biol Chem 290(33): 20348–20359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B and Guan KL (2011). “AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1.” Nat Cell Biol 13(2): 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland JL, Tchkonia T, Zhu Y, Niedernhofer LJ and Robbins PD (2017). “The Clinical Potential of Senolytic Drugs.” J Am Geriatr Soc 65(10): 2297–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klement RJ and Fink MK (2016). “Dietary and pharmacological modification of the insulin/IGF-1 system: exploiting the full repertoire against cancer.” Oncogenesis 5(2): e193–e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka AR, Laurin JL, Schoenberg HM, Reid JJ, Castor WM, Wolff CA, Musci RV, Safairad OD, Linden MA, Biela LM, Bailey SM, Hamilton KL and Miller BF (2019). “Metformin inhibits mitochondrial adaptations to aerobic exercise training in older adults.” Aging Cell 18(1): e12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen JM, Larsen S, Helge JW, Dela F and Wojtaszewski JF (2013). “Two weeks of metformin treatment enhances mitochondrial respiration in skeletal muscle of AMPK kinase dead but not wild type mice.” PLoS One 8(1): e53533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Y, Hu B, Feng G, Xiang M, Deng Y, Tan M, Li J and Song J (2019). “Metformin prevents against oxidative stress-induced senescence in human periodontal ligament cells.” Biogerontology. [DOI] [PubMed] [Google Scholar]
- Kulkarni AS, Brutsaert EF, Anghel V, Zhang K, Bloomgarden N, Pollak M, Mar JC, Hawkins M, Crandall JP and Barzilai N (2018). “Metformin regulates metabolic and nonmetabolic pathways in skeletal muscle and subcutaneous adipose tissues of older adults.” Aging Cell 17(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Ye L, Sabatini DM and Baur JA (2013). “Rapalogs and mTOR inhibitors as anti-aging therapeutics.” The Journal of clinical investigation 123(3): 980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Lee Y, Kim J, An J, Lee S, Kong H, Song Y, Lee CK and Kim K (2018). “Modulation of the gut microbiota by metformin improves metabolic profiles in aged obese mice.” Gut Microbes 9(2): 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Doonan BB, Wu JM and Hsieh TC (2016). “Combined metformin and resveratrol confers protection against UVC-induced DNA damage in A549 lung cancer cells via modulation of cell cycle checkpoints and DNA repair.” Oncol Rep 35(6): 3735–3741. [DOI] [PubMed] [Google Scholar]
- Liu J, Ge Y, Wu S, Ma D, Xu W, Zhang Y and Yang Y (2019). “Association between antidiabetic agents use and leukocyte telomere shortening rates in patients with type 2 diabetes.” Aging 11(2): 741–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chhipa RR, Pooya S, Wortman M, Yachyshin S, Chow LML, Kumar A, Zhou X, Sun Y, Quinn B, McPherson C, Warnick RE, Kendler A, Giri S, Poels J, Norga K, Viollet B, Grabowski GA and Dasgupta B (2014). “Discrete mechanisms of mTOR and cell cycle regulation by AMPK agonists independent of AMPK.” Proceedings of the National Academy of Sciences 111(4): E435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Blasco MA, Partridge L, Serrano M and Kroemer G (2013). “The Hallmarks of Aging.” Cell 153(6): 1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Su C, Qiao C, Bian Y, Ding J and Hu G (2016). “Metformin Prevents Dopaminergic Neuron Death in MPTP/P-Induced Mouse Model of Parkinson’s Disease via Autophagy and Mitochondrial ROS Clearance.” Int J Neuropsychopharmacol 19(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madiraju AK, Erion DM, Rahimi Y, Zhang XM, Braddock DT, Albright RA, Prigaro BJ, Wood JL, Bhanot S, MacDonald MJ, Jurczak MJ, Camporez JP, Lee HY, Cline GW, Samuel VT, Kibbey RG and Shulman GI (2014). “Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase.” Nature 510(7506): 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madreiter-Sokolowski CT, Sokolowski AA, Waldeck-Weiermair M, Malli R and Graier WF (2018). “Targeting Mitochondria to Counteract Age-Related Cellular Dysfunction.” Genes 9(3): 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Montalvo A, Mercken EM, Mitchell SJ, Palacios HH, Mote PL, Scheibye-Knudsen M, Gomes AP, Ward TM, Minor RK, Blouin MJ, Schwab M, Pollak M, Zhang Y, Yu Y, Becker KG, Bohr VA, Ingram DK, Sinclair DA, Wolf NS, Spindler SR, Bernier M and de Cabo R (2013). “Metformin improves healthspan and lifespan in mice.” Nat Commun 4: 2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCreight LJ, Bailey CJ and Pearson ER (2016). “Metformin and the gastrointestinal tract.” Diabetologia 59(3): 426–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak EM, Burr ML, Bannister AJ and Dawson MA (2019). “The roles of DNA, RNA and histone methylation in ageing and cancer.” Nature Reviews Molecular Cell Biology 20(10): 573–589. [DOI] [PubMed] [Google Scholar]
- Milman S, Huffman DM and Barzilai N (2016). “The Somatotropic Axis in Human Aging: Framework for the Current State of Knowledge and Future Research.” Cell metabolism 23(6): 980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseeva O, Deschenes-Simard X, St-Germain E, Igelmann S, Huot G, Cadar AE, Bourdeau V, Pollak MN and Ferbeyre G (2013). “Metformin inhibits the senescence-associated secretory phenotype by interfering with IKK/NF-kappaB activation.” Aging Cell 12(3): 489–498. [DOI] [PubMed] [Google Scholar]
- Moreno-Navarrete JM, Ortega FJ, Rodriguez-Hermosa JI, Sabater M, Pardo G, Ricart W and Fernandez-Real JM (2011). “OCT1 Expression in adipocytes could contribute to increased metformin action in obese subjects.” Diabetes 60(1): 168–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP and Hartley RC (2018). “Mitochondria as a therapeutic target for common pathologies.” Nat Rev Drug Discov 17(12): 865–886. [DOI] [PubMed] [Google Scholar]
- Na H-J, Park J-S, Pyo J-H, Lee S-H, Jeon H-J, Kim Y-S and Yoo M-A (2013). “Mechanism of metformin: Inhibition of DNA damage and proliferative activity in Drosophila midgut stem cell.” Mechanisms of Ageing and Development 134(9): 381–390. [DOI] [PubMed] [Google Scholar]
- Na HJ, Park JS, Pyo JH, Jeon HJ, Kim YS, Arking R and Yoo MA (2015). “Metformin inhibits age-related centrosome amplification in Drosophila midgut stem cells through AKT/TOR pathway.” Mech Ageing Dev 149: 8–18. [DOI] [PubMed] [Google Scholar]
- Na HJ, Pyo JH, Jeon HJ, Park JS, Chung HY and Yoo MA (2018). “Deficiency of Atg6 impairs beneficial effect of metformin on intestinal stem cell aging in Drosophila.” Biochem Biophys Res Commun 498(1): 18–24. [DOI] [PubMed] [Google Scholar]
- Najafi M, Cheki M, Rezapoor S, Geraily G, Motevaseli E, Carnovale C, Clementi E and Shirazi A (2018). “Metformin: Prevention of genomic instability and cancer: A review.” Mutat Res Genet Toxicol Environ Mutagen 827: 1–8. [DOI] [PubMed] [Google Scholar]
- Neumann B, Baror R, Zhao C, Segel M, Dietmann S, Rawji KS, Foerster S, McClain CR, Chalut K, van Wijngaarden P and Franklin RJM (2019). “Metformin Restores CNS Remyelination Capacity by Rejuvenating Aged Stem Cells.” Cell Stem Cell 25(4): 473–485.e478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Gurkar AU, Wang Y, Vijg J, Hoeijmakers JHJ and Robbins PD (2018). “Nuclear Genomic Instability and Aging.” Annu Rev Biochem 87: 295–322. [DOI] [PubMed] [Google Scholar]
- Noren Hooten N, Martin-Montalvo A, Dluzen DF, Zhang Y, Bernier M, Zonderman AB, Becker KG, Gorospe M, de Cabo R and Evans MK (2016). “Metformin-mediated increase in DICER1 regulates microRNA expression and cellular senescence.” Aging Cell 15(3): 572–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novelle MG, Ali A, Diéguez C, Bernier M and de Cabo R (2016). “Metformin: A Hopeful Promise in Aging Research.” Cold Spring Harbor Perspectives in Medicine 6(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onken B and Driscoll M (2010). “Metformin Induces a Dietary Restriction–Like State and the Oxidative Stress Response to Extend C. elegans Healthspan via AMPK, LKB1, and SKN-1.” PLOS ONE 5(1): e8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othman EM, Oli RG, Arias-Loza PA, Kreissl MC and Stopper H (2016). “Metformin Protects Kidney Cells From Insulin-Mediated Genotoxicity In Vitro and in Male Zucker Diabetic Fatty Rats.” Endocrinology 157(2): 548–559. [DOI] [PubMed] [Google Scholar]
- Owen MR, Doran E and Halestrap AP (2000). “Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain.” Biochem J 348 Pt 3: 607–614. [PMC free article] [PubMed] [Google Scholar]
- Pal S and Tyler JK (2016). “Epigenetics and aging.” Sci Adv 2(7): e1600584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patane G, Piro S, Rabuazzo AM, Anello M, Vigneri R and Purrello F (2000). “Metformin restores insulin secretion altered by chronic exposure to free fatty acids or high glucose: a direct metformin effect on pancreatic beta-cells.” Diabetes 49(5): 735–740. [DOI] [PubMed] [Google Scholar]
- Pavlidou T, Marinkovic M, Rosina M, Fuoco C, Vumbaca S, Gargioli C, Castagnoli L and Cesareni G (2019). “Metformin Delays Satellite Cell Activation and Maintains Quiescence.” Stem Cells Int 2019: 5980465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg S, Feller C, Ladurner AG and Imhof A (2016). “The Metabolic Impact on Histone Acetylation and Transcription in Ageing.” Trends Biochem Sci 41(8): 700–711. [DOI] [PubMed] [Google Scholar]
- Piskovatska V, Stefanyshyn N, Storey KB, Vaiserman AM and Lushchak O (2018). “Metformin as a geroprotector: experimental and clinical evidence.” Biogerontology. [DOI] [PubMed] [Google Scholar]
- Prasad S, Sajja RK, Kaisar MA, Park JH, Villalba H, Liles T, Abbruscato T and Cucullo L (2017). “Role of Nrf2 and protective effects of Metformin against tobacco smoke-induced cerebrovascular toxicity.” Redox Biol 12: 58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor R, Norvaisas P, Marinos G, Best L, Thingholm LB, Quintaneiro LM, De Haes W, Esser D, Waschina S, Lujan C, Smith RL, Scott TA, Martinez-Martinez D, Woodward O, Bryson K, Laudes M, Lieb W, Houtkooper RH, Franke A, Temmerman L, Bjedov I, Cocheme HM, Kaleta C and Cabreiro F (2019). “Host-Microbe-Drug-Nutrient Screen Identifies Bacterial Effectors of Metformin Therapy.” Cell 178(6): 1299–1312.e1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rena G and Lang CC (2018). “Repurposing Metformin for Cardiovascular Disease.” Circulation 137(5): 422–424. [DOI] [PubMed] [Google Scholar]
- Robb-MacKay C (2018). The Effect of Metformin on Absolute Telomere Length. Master of Science- Biology, Lakehead University. [Google Scholar]
- Rosa E, Dos Santos RRC, Fernandes LFA, Neves FAR, Coelho MS and Amato AA (2018). “Leukocyte telomere length correlates with glucose control in adults with recently diagnosed type 2 diabetes.” Diabetes Res Clin Pract 135: 30–36. [DOI] [PubMed] [Google Scholar]
- Rotermund C, Machetanz G and Fitzgerald JC (2018). “The Therapeutic Potential of Metformin in Neurodegenerative Diseases.” Frontiers in endocrinology 9: 400–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddy RM, Adams KV and Morshead CM (2019). “Age- and sex-dependent effects of metformin on neural precursor cells and cognitive recovery in a model of neonatal stroke.” Sci Adv 5(9): eaax1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabit H, Abdel-Ghany SE, OA MS, Mostafa MA and El-Zawahry M (2018). “Metformin Reshapes the Methylation Profile in Breast and Colorectal Cancer Cells.” Asian Pac J Cancer Prev 19(10): 2991–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saisho Y (2015). “Metformin and Inflammation: Its Potential Beyond Glucose-lowering Effect.” Endocr Metab Immune Disord Drug Targets 15(3): 196–205. [DOI] [PubMed] [Google Scholar]
- Salminen A and Kaarniranta K (2012). “AMP-activated protein kinase (AMPK) controls the aging process via an integrated signaling network.” Ageing Res Rev 11(2): 230–241. [DOI] [PubMed] [Google Scholar]
- Sanada F, Taniyama Y, Muratsu J, Otsu R, Shimizu H, Rakugi H and Morishita R (2018). “Source of Chronic Inflammation in Aging.” Frontiers in cardiovascular medicine 5: 12–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant’Anna JR, Yajima JP, Rosada LJ, Franco CC, Prioli AJ, Della-Rosa VA, Mathias PC and Castro-Prado MA (2013). “Metformin’s performance in in vitro and in vivo genetic toxicology studies.” Exp Biol Med (Maywood) 238(7): 803–810. [DOI] [PubMed] [Google Scholar]
- Sarfstein R, Friedman Y, Attias-Geva Z, Fishman A, Bruchim I and Werner H (2013). “Metformin Downregulates the Insulin/IGF-I Signaling Pathway and Inhibits Different Uterine Serous Carcinoma (USC) Cells Proliferation and Migration in p53-Dependent or -Independent Manners.” PLOS ONE 8(4): e61537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulten HJ and Bakhashab S (2019). “Meta-Analysis of Microarray Expression Studies on Metformin in Cancer Cell Lines.” Int J Mol Sci 20(13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MB and Sinclair DA (2016). “When stem cells grow old: phenotypes and mechanisms of stem cell aging.” Development 143(1): 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel J and Valenzano DR (2018). “The role of the gut microbiome during host ageing.” F1000Research 7: F1000 Faculty Rev-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen P, Shah PP, Nativio R and Berger SL (2016). “Epigenetic Mechanisms of Longevity and Aging.” Cell 166(4): 822–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi WY, Xiao D, Wang L, Dong LH, Yan ZX, Shen ZX, Chen SJ, Chen Y and Zhao WL (2012). “Therapeutic metformin/AMPK activation blocked lymphoma cell growth via inhibition of mTOR pathway and induction of autophagy.” Cell Death Dis 3: e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack C, Foley A and Partridge L (2012). “Activation of AMPK by the Putative Dietary Restriction Mimetic Metformin Is Insufficient to Extend Lifespan in Drosophila.” PLOS ONE 7(10): e47699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Śmieszek A, Stręk Z, Kornicka K, Grzesiak J, Weiss C and Marycz K (2017). “Antioxidant and Anti-Senescence Effect of Metformin on Mouse Olfactory Ensheathing Cells (mOECs) May Be Associated with Increased Brain-Derived Neurotrophic Factor Levels-An Ex Vivo Study.” International journal of molecular sciences 18(4): 872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Śmieszek A, Stręk Z, Kornicka K, Grzesiak J, Weiss C and Marycz K (2017). “Antioxidant and Anti-Senescence Effect of Metformin on Mouse Olfactory Ensheathing Cells (mOECs) May Be Associated with Increased Brain-Derived Neurotrophic Factor Levels—An Ex Vivo Study.” International Journal of Molecular Sciences 18(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberanes S, Misharin AV, Jairaman A, Morales-Nebreda L, McQuattie-Pimentel AC, Cho T, Hamanaka RB, Meliton AY, Reyfman PA, Walter JM, Chen CI, Chi M, Chiu S, Gonzalez-Gonzalez FJ, Antalek M, Abdala-Valencia H, Chiarella SE, Sun KA, Woods PS, Ghio AJ, Jain M, Perlman H, Ridge KM, Morimoto RI, Sznajder JI, Balch WE, Bhorade SM, Bharat A, Prakriya M, Chandel NS, Mutlu GM and Budinger GRS (2019). “Metformin Targets Mitochondrial Electron Transport to Reduce Air-Pollution-Induced Thrombosis.” Cell Metab 29(2): 335–347.e335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YM, Lee WK, Lee Y-H, Kang ES, Cha B-S and Lee B-W (2016). “Metformin Restores Parkin-Mediated Mitophagy, Suppressed by Cytosolic p53.” International journal of molecular sciences 17(1): 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukas AA, Hao H and Wu L (2019). “Metformin as Anti-Aging Therapy: Is It for Everyone?” Trends Endocrinol Metab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Xie C, Wang G, Wu Y, Wu Q, Wang X, Liu J, Deng Y, Xia J, Chen B, Zhang S, Yun C, Lian G, Zhang X, Zhang H, Bisson WH, Shi J, Gao X, Ge P, Liu C, Krausz KW, Nichols RG, Cai J, Rimal B, Patterson AD, Wang X, Gonzalez FJ and Jiang C (2018). “Gut microbiota and intestinal FXR mediate the clinical benefits of metformin.” Nature Medicine 24(12): 1919–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Youle RJ and Finkel T (2016). “The Mitochondrial Basis of Aging.” Molecular cell 61(5): 654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwa M, Egashira T, Nakano H, Sasaki H and Kumagai S (2006). “Metformin increases the PGC-1alpha protein and oxidative enzyme activities possibly via AMPK phosphorylation in skeletal muscle in vivo.” J Appl Physiol (1985) 101(6): 1685–1692. [DOI] [PubMed] [Google Scholar]
- Thevaranjan N, Puchta A, Schulz C, Naidoo A, Szamosi JC, Verschoor CP, Loukov D, Schenck LP, Jury J, Foley KP, Schertzer JD, Larche MJ, Davidson DJ, Verdu EF, Surette MG and Bowdish DME (2017). “Age-Associated Microbial Dysbiosis Promotes Intestinal Permeability, Systemic Inflammation, and Macrophage Dysfunction.” Cell Host Microbe 21(4): 455–466.e454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tizazu AM, Nyunt MSZ, Cexus O, Suku K, Mok E, Xian CH, Chong J, Tan C, How W, Hubert S, Combet E, Fulop T, Ng TP and Larbi A (2019). “Metformin Monotherapy Downregulates Diabetes-Associated Inflammatory Status and Impacts on Mortality.” Frontiers in Physiology 10(572). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H-H, Lai H-Y, Chen Y-C, Li C-F, Huang H-S, Liu H-S, Tsai Y-S and Wang J-M (2017). “Metformin promotes apoptosis in hepatocellular carcinoma through the CEBPD-induced autophagy pathway.” Oncotarget 8(8): 13832–13845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursini F, Russo E, Pellino G, D’Angelo S, Chiaravalloti A, De Sarro G, Manfredini R and De Giorgio R (2018). “Metformin and Autoimmunity: A “New Deal” of an Old Drug.” Front Immunol 9: 1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia WM, Palacio A, Tamariz L and Florez H (2017). “Metformin and ageing: improving ageing outcomes beyond glycaemic control.” Diabetologia 60(9): 1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasamsetti SB, Karnewar S, Kanugula AK, Thatipalli AR, Kumar JM and Kotamraju S (2015). “Metformin inhibits monocyte-to-macrophage differentiation via AMPK-mediated inhibition of STAT3 activation: potential role in atherosclerosis.” Diabetes 64(6): 2028–2041. [DOI] [PubMed] [Google Scholar]
- Vazquez-Martin A, Oliveras-Ferraros C, Cufi S, Martin-Castillo B and Menendez JA (2011). “Metformin activates an ataxia telangiectasia mutated (ATM)/Chk2-regulated DNA damage-like response.” Cell Cycle 10(9): 1499–1501. [DOI] [PubMed] [Google Scholar]
- Vial G, Detaille D and Guigas B (2019). “Role of Mitochondria in the Mechanism(s) of Action of Metformin.” Front Endocrinol (Lausanne) 10: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijg J and Suh Y (2013). “Genome instability and aging.” Annu Rev Physiol 75: 645–668. [DOI] [PubMed] [Google Scholar]
- Vinatier C, Domínguez E, Guicheux J and Caramés B (2018). “Role of the Inflammation-Autophagy-Senescence Integrative Network in Osteoarthritis.” Frontiers in physiology 9: 706–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DS, Jonker JW, Kato Y, Kusuhara H, Schinkel AH and Sugiyama Y (2002). “Involvement of organic cation transporter 1 in hepatic and intestinal distribution of metformin.” J Pharmacol Exp Ther 302(2): 510–515. [DOI] [PubMed] [Google Scholar]
- Wang GY, Bi YG, Liu XD, Zhao Y, Han JF, Wei M and Zhang QY (2017). “Autophagy was involved in the protective effect of metformin on hyperglycemia-induced cardiomyocyte apoptosis and Connexin43 downregulation in H9c2 cells.” Int J Med Sci 14(7): 698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu B, Yang Y, Wang Y, Zhao Z, Miao Z and Zhu J (2019). “Metformin exerts antidepressant effects by regulated DNA hydroxymethylation.” Epigenomics. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xu W, Yan Z, Zhao W, Mi J, Li J and Yan H (2018). “Metformin induces autophagy and G0/G1 phase cell cycle arrest in myeloma by targeting the AMPK/mTORC1 and mTORC2 pathways.” Journal of Experimental & Clinical Cancer Research 37(1): 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheaton WW, Weinberg SE, Hamanaka RB, Soberanes S, Sullivan LB, Anso E, Glasauer A, Dufour E, Mutlu GM, Budigner GS and Chandel NS (2014). “Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis.” eLife 3: e02242–e02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittemore K, Vera E, Martínez-Nevado E, Sanpera C and Blasco MA (2019). “Telomere shortening rate predicts species life span.” Proceedings of the National Academy of Sciences 116(30): 15122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CC, Singleton BA, Llopis SD and Skripnikova EV (2013). “Metformin induces a senescence-associated gene signature in breast cancer cells.” Journal of health care for the poor and underserved 24(1 Suppl): 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhou B, Oshiro-Rapley N, Li M, Paulo JA, Webster CM, Mou F, Kacergis MC, Talkowski ME, Carr CE, Gygi SP, Zheng B and Soukas AA (2016). “An Ancient, Unified Mechanism for Metformin Growth Inhibition in C. elegans and Cancer.” Cell 167(7): 1705–1718.e1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Xie C, Wu H, Jones KL, Horowitz M and Rayner CK (2017). “Metformin reduces the rate of small intestinal glucose absorption in type 2 diabetes.” Diabetes Obes Metab 19(2): 290–293. [DOI] [PubMed] [Google Scholar]
- Xie Y, Wang J-L, Ji M, Yuan Z-F, Peng Z, Zhang Y, Wen J-G and Shi H-R (2014). “Regulation of insulin-like growth factor signaling by metformin in endometrial cancer cells.” Oncology letters 8(5): 1993–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Pirtskhalava T, Farr JN, Weigand BM, Palmer AK, Weivoda MM, Inman CL, Ogrodnik MB, Hachfeld CM, Fraser DG, Onken JL, Johnson KO, Verzosa GC, Langhi LGP, Weigl M, Giorgadze N, LeBrasseur NK, Miller JD, Jurk D, Singh RJ, Allison DB, Ejima K, Hubbard GB, Ikeno Y, Cubro H, Garovic VD, Hou X, Weroha SJ, Robbins PD, Niedernhofer LJ, Khosla S, Tchkonia T and Kirkland JL (2018). “Senolytics improve physical function and increase lifespan in old age.” Nat Med 24(8): 1246–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Han C, Wang G, Waddington JL, Zheng L and Zhen X (2017). “Activation of AMPK/mTORC1-Mediated Autophagy by Metformin Reverses Clk1 Deficiency-Sensitized Dopaminergic Neuronal Death.” Mol Pharmacol 92(6): 640–652. [DOI] [PubMed] [Google Scholar]
- Yi G, He Z, Zhou X, Xian L, Yuan T, Jia X, Hong J, He L and Liu J (2013). “Low concentration of metformin induces a p53-dependent senescence in hepatoma cells via activation of the AMPK pathway.” Int J Oncol 43(5): 1503–1510. [DOI] [PubMed] [Google Scholar]
- Yu X, Mao W, Zhai Y, Tong C, Liu M, Ma L, Yu X and Li S (2017). “Anti-tumor activity of metformin: from metabolic and epigenetic perspectives.” Oncotarget 8(3): 5619–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarse K, Schmeisser S, Groth M, Priebe S, Beuster G, Kuhlow D, Guthke R, Platzer M, Kahn CR and Ristow M (2012). “Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal.” Cell Metab 15(4): 451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CS, Li M, Ma T, Zong Y, Cui J, Feng JW, Wu YQ, Lin SY and Lin SC (2016). “Metformin Activates AMPK through the Lysosomal Pathway.” Cell Metab 24(4): 521–522. [DOI] [PubMed] [Google Scholar]
- Zhong T, Men Y, Lu L, Geng T, Zhou J, Mitsuhashi A, Shozu M, Maihle NJ, Carmichael GG, Taylor HS and Huang Y (2017). “Metformin alters DNA methylation genome-wide via the H19/SAHH axis.” Oncogene 36(17): 2345–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z. e., Tang Y, Jin X, Chen C, Lu Y, Liu L and Shen C (2016). “Metformin Inhibits Advanced Glycation End Products-Induced Inflammatory Response in Murine Macrophages Partly through AMPK Activation and RAGE/NFκB Pathway Suppression.” Journal of diabetes research 2016: 4847812–4847812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Liu X, Ding X, Wang F and Geng X (2019). “Telomere and its role in the aging pathways: telomere shortening, cell senescence and mitochondria dysfunction.” Biogerontology 20(1): 1–16. [DOI] [PubMed] [Google Scholar]


