Abstract
Prolonged obesity is associated with blunted feeding and thermogenic autonomic responses to leptin, but cardiovascular responses to leptin are maintained. This state of “selective leptin resistance” (SLR) is therefore proposed to contribute to the pathogenesis and maintenance of obesity-associated hypertension. Cells of the arcuate nucleus of the hypothalamus (ARC) detect leptin, and although the cellular and molecular mechanisms remain unclear, altered ARC biology is hypothesized to contribute to SLR. Male C57BL/6J mice were fed a high fat diet (HFD) or chow from 8–18 weeks of age, as this paradigm models SLR. Nuclei were then isolated from ARC for single-nucleus RNA sequencing (snRNA-seq). HFD caused expected gains in adiposity and circulating leptin. Twenty-three unique cell-type clusters were identified, and Ingenuity Pathway Analysis (IPA) was used to explore changes in gene expression patterns due to chronic HFD within each cluster. Notably, gene expression signatures related to leptin signaling exhibited suppression predominantly in neurons identified as the Agouti-related peptide (Agrp) subtype. IPA results were also consistent with alterations in cAMP response element-binding protein (CREB) signaling in Agrp neurons after HFD, and reduced phosphorylated CREB was confirmed in ARC after prolonged HFD by capillary electrophoresis-based Western blotting. These findings support the concept that prolonged HFD-induced obesity is associated with selective changes in Agrp neuron biology, possibly secondary to altered CREB signaling.
Keywords: Obesity, Metabolism, Leptin, Arcuate Nucleus, Hypothalamus
Graphical Abstract
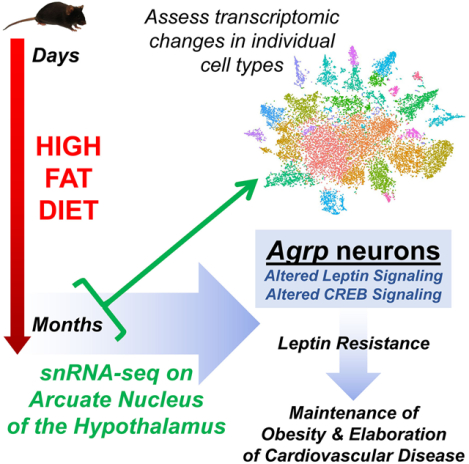
Introduction
Obesity represents a major risk factor for the development of hypertension, and the leptin and melanocortin systems are implicated in this process.1 Although leptin normally acts to inhibit energy intake and increase energy output, obesity is associated with hyperleptinemia and attenuated energy balance effects of leptin. Simultaneously, the stimulatory effects of leptin upon autonomic projections innervating cardiovascular tissues is maintained during obesity. This resistance to the metabolic effects and maintenance of cardiovascular effects of leptin (i.e. selective leptin resistance, SLR) has been proposed to contribute to the development and maintenance of obesity and obesity-associated cardiovascular disease.2
Leptin is known to act at its receptor (LEPR) in the arcuate nucleus of the hypothalamus (ARC) to stimulate an array of second-messenger signaling cascades; these mechanisms result in the subsequent activation of multiple neural networks throughout the hypothalamus and beyond to ultimately promote melanocortin signaling and autonomic activity to many effector tissues (recently reviewed3). Importantly, although many mechanisms of LEPR signaling that contribute to the control of energy balance have been mapped, the changes in these networks that mediate leptin resistance during prolonged obesity remain unclear. We hypothesize that changes in second-messenger signaling cascades within specific cell types of the ARC, such as proopiomelanocortin (Pomc)- or Agouti-related peptide (Agrp)-expressing neurons, underlie the development of SLR.
Previously, we demonstrated that ten weeks of a moderate high fat diet (HFD; OpenSource D12451; 45% kcal from fat / 35% kcal from carbohydrate) feeding in C57BL/6J male mice caused diet-induced obesity (DIO) and SLR.4 Mice fed HFD from 8 to 18 weeks of age exhibited attenuated autonomic activation and feeding behaviors in response to leptin compared to mice maintained on standard chow (Teklad 7913; 18% kcal from fat / 59% kcal from carbohydrate) for the same timeframe. In the current study, we utilized single nucleus (sn)RNA-seq to dissect the unique transcriptional changes within individual cell types of the ARC that are induced in this model of prolonged HFD feeding, to identify molecular perturbations that may underlie changes in leptin sensitivity.
Methods
All methods are included in the associated supplemental methods document, and all data and materials that support the findings of this study are available from the corresponding authors upon reasonable request. All studies were approved by the Institutional Animal Care and Use Committee at the University of Iowa. Briefly, male C57BL/6J mice were maintained on a chow diet (Teklad 7913) or fed HFD or from 8–18 weeks of age before nuclei from the ARC were isolated and analyzed by snRNA-seq using the 10X Genomics platform. Sequencing libraries were constructed using the Chromium Single Cell 3’ GEM, Library & Gel Bead Kit v3 and sequenced on an Illumina HiSeq 4000 with 150 basepair (bp) paired-end reads. Resulting FASTQ sequencing files are publicly available at PRJNA604055. Data were analyzed using Cell Ranger v3.0.1 (10X Genomics) and the R package Seurat v3.0. Differentially expressed genes within resulting nuclei clusters were used for canonical pathway and upstream pathway analyses using the Ingenuity Pathway Analysis (IPA) software package.5 In a separate cohort of mice, CREB phosphorylation was assessed in ARC punches using the Simple Wes system (Protein Simple, San Jose, CA) as previously described.6
Results
HFD increases adiposity and circulating leptin
Male C57BL/6J mice (Jackson Laboratories, #000664) were randomly assigned to diet treatment. Body masses and adiposity were confirmed to not differ between groups at 6 weeks of age (Fig 1A). Mice were switched to 45% HFD (OpenSource D12451) at 8 weeks of age or maintained on standard chow (Teklad 7913) for the remainder of the study. As expected, HFD caused increased body mass, adiposity, and plasma leptin levels compared to chow-fed mice at 18 weeks of age (Fig 1B). At 10 weeks of age and again at 18 weeks of age, food intake was assessed by daily measures of food hopper masses. Caloric intake was significantly increased at 10 weeks of age, but this increase was attenuated at 18 weeks of age, despite the increased body mass (Fig 1C). Whereas feeding efficiency (a rough, inverse metric of energy expenditure) was not different between HFD- and chow-fed mice at 10 weeks of age, it was significantly increased in the HFD-fed group at 18 weeks of age. Such results support the concept that total energy expenditure relative to weight gain is reduced in the HFD-fed group despite increased plasma leptin levels. Thus, energy output mechanisms appear to exhibit resistance to the effects of increased endogenous plasma leptin levels after ten weeks of exposure to HFD; these results are consistent with our previous demonstration that thermogenic autonomic and feeding behavior responses to acute exogenous leptin administration are attenuated after ten weeks of HFD.4
Figure 1. Prolonged HFD feeding increases body mass and plasma leptin.
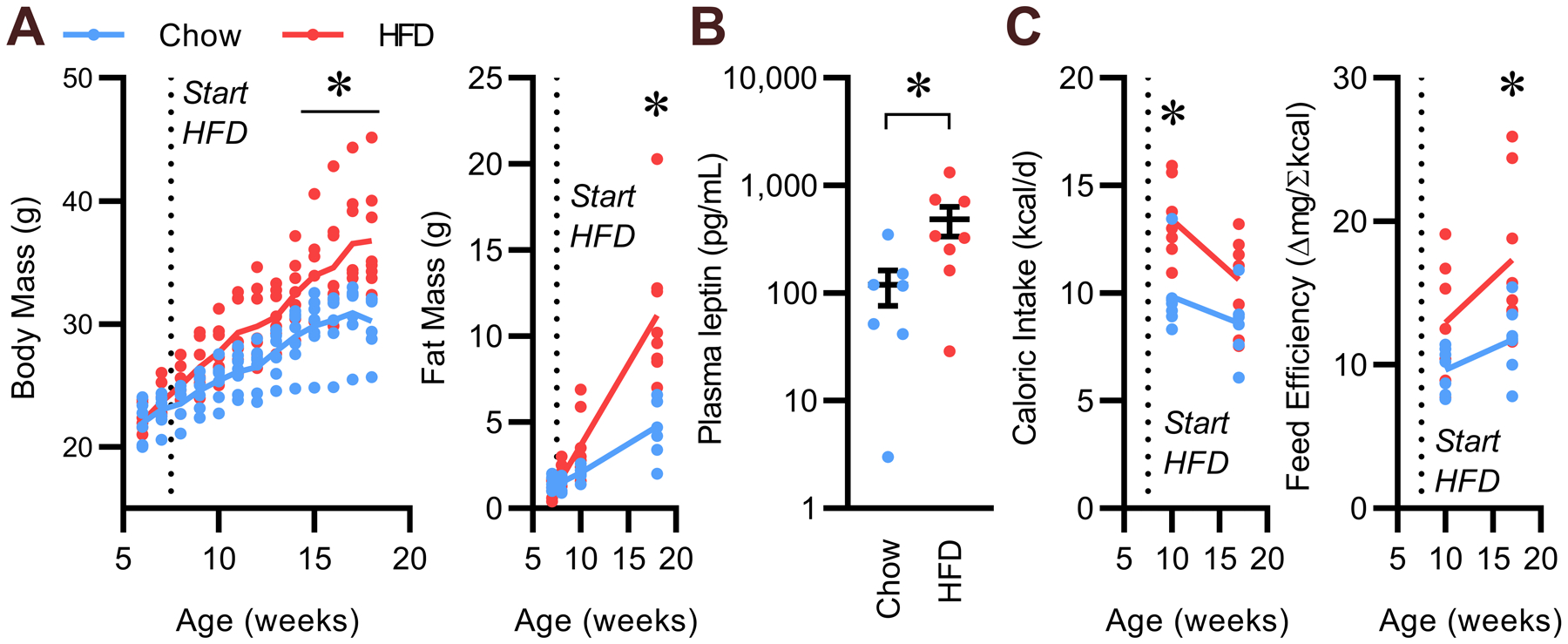
(A) Body masses and fat masses determined by time-domain NMR, versus age in mice fed HFD (45% kcal from fat) versus chow from 8 – 18 weeks of age. (B) Plasma leptin at 18 weeks of age. (C) Caloric intake and feeding efficiency after 2 and 10 weeks of dietary intervention. n=7 chow and n=8 HFD; trendlines represent mean; summary data presented as mean ± SEM; dots represent individual animals; *p<0.05 by Tukey multiple-comparisons procedure (A, C) or two-tailed Student’s t-test after Log10 transform (B).
snRNA-seq identifies 23 cell types in ARC
At 18 weeks of age, each mouse was euthanized, the hypothalamus was dissected, and nuclei from the ARC were isolated and utilized for snRNA-seq (Tables S1 & S2). Sequence alignment, quality control (QC) assessment and feature-barcode generation were performed using the Cell Ranger software suite (10x Genomics). The QC metrics of this dataset were of similar quality to other recent snRNA-seq studies7, 8 (Table S3, and Figures S1 & S2). After filtering out low-quality nuclei, followed by combining nuclei from all six samples, unbiased clustering analyses via the R package Seurat9 identified twenty-three unique clusters of nuclei (Fig 2A). Each cluster was populated by nuclei from both chow- and HFD-fed animals (Fig 2B) and defined by the top uniquely expressed gene(s) in that cluster (Fig 2C). The cell types represented by each cluster were then identified based on comparison to three previous studies examining the transcriptomes of individual cell types within the ARC of mice.10–12 Clusters without a previously-defined cell type were identified by the two most abundantly expressed genes associated specifically with that cluster. As expected, major cell types that are sensitive to leptin and known to be present within the ARC were identified, including cells characterized by high levels of expression of Pomc (cluster 5) or Agrp (cluster 15).
Figure 2. Prolonged HFD feeding causes differential expression of various genes in individual cell types of the ARC.
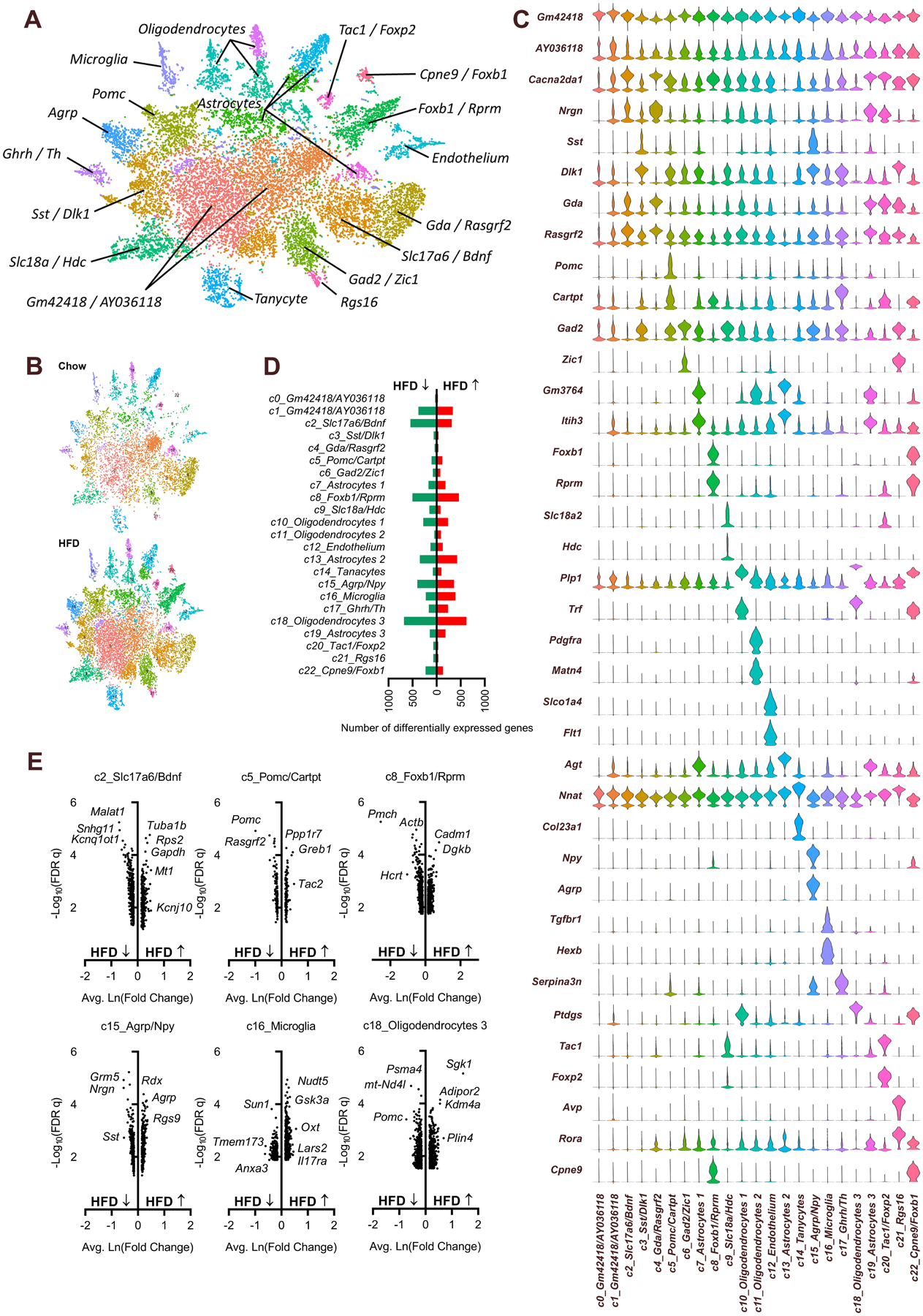
(A) t-distributed stochastic neighbor embedding (TSNE) plot of cell clusters, identified by Seurat. (B) TSNE plot of cell clusters, split by diet. (C) Expression of top genes that uniquely identify individual clusters. (D) Differential gene expression counts, HFD vs Chow, by cluster. (E) Volcano plots of differentially-expressed genes, HFD vs Chow, in selected clusters. Related information also included in Tables S1–S3 and Figures S1–S2.
Cluster-specific alterations in gene expression with HFD
The impact of chronic HFD on the transcriptome of each nuclear cluster was determined by performing differential gene expression analyses (FDR q < 0.1, |lnFC| ≥ 0.15, HFD v Chow) using Seurat (Supplemental Data File 2). With these cutoff values, HFD was associated with profound changes in gene expression ranging from 60 differentially expressed genes in cluster 0, to 1300 genes in cluster 18 (median across all clusters = 328, mean across all clusters = 418) (Fig 2D). Volcano plots were constructed to visualize gene expression changes within clusters with the greatest number of differentially expressed genes (clusters 2, 8, and 18), clusters of canonically leptin-sensitive cell types including cluster 5 (Pomc neurons) and cluster 15 (Agrp/Npy neurons), and inflammatory cells such as cluster 16 (microglia) (Fig 2E). Notably, despite elevated endogenous plasma leptin levels within the HFD group (Fig 1B), several well-recognized leptin targets were differentially expressed in these clusters in directions opposite that which is expected in response to leptin. For example, within cluster 5 (Pomc neurons), Pomc expression was significantly suppressed by prolonged HFD feeding, and within cluster 15 (Agrp/Npy neurons) the expression of Agrp was significantly increased by prolonged HFD feeding (Fig 2E).
Leptin pathway analysis identifies robust changes in Agrp neuron after prolonged HFD
To more comprehensively examine cell type-specific changes in response to leptin signaling after prolonged HFD, we used Ingenuity Pathway Analysis (IPA) to probe each cluster for enrichment of the “Leptin Signaling in Obesity” gene set. This analysis method reports both a p-value and a z-value. The p-value is calculated by Fisher’s exact test and reflects confidence that genes in the selected gene set (i.e. “Leptin Signaling in Obesity”) from the selected cluster are responding to the stimulus (i.e. HFD) as a group, regardless of the directionality of the response of each gene. In contrast, the z-value is an assessment of the match between the expected direction vs the observed gene expression responses, integrated across all genes in the gene set. It reflects the overall direction of response to the stimulus, such that a positive value (z>0) indicates that the gene set is responding in the predicted direction, which may be up- or down-regulated depending on the expected responses of individual genes within the set. A negative z-value (z<0) instead indicates that the genes in the set are generally responding in the opposite direction than would be predicted. A zero z-value (z=0) indicates that although the expression levels of individual genes may be significantly changed within the set, that cumulatively there is no pattern in the directionality of response of the group of genes; some genes are responding as predicted and others are responding in the opposite direction as would be predicted.
Surprisingly, despite increased plasma leptin levels in the HFD-fed group (Fig 1B), no cluster exhibited a statistically significant (p<0.05) positive (z>0) response for this set of leptin-responsive genes (Table 1, and Supplemental File 3). More surprisingly, of the five clusters assigned a significant (p<0.05) p-value, only three were awarded a non-zero z-score; indeed, the three clusters exhibiting significant p-values were awarded negative z-scores. Cluster 8, whose cell type was not clearly identified but was characterized by strong expression of Foxb1 and Rprm, has significant enrichment of leptin-responsive genes but there was little pattern in the directionality of the response of the genes as a group. Cluster 15, the Agrp/Npy neuronal cluster, and cluster 18, one of three oligodendrocyte clusters, were each awarded a more impressive z-score of −1.633. Of these two, however, enrichment was only statistically significant in the Agrp/Npy cluster (cluster 15). The determination that canonical leptin-responsive genes exhibit expression patterns opposite of what is expected given elevated plasma leptin, combined with the selective negative z-score for the leptin-responsive gene set specifically in cluster 15 but not cluster 5, support multiple interim conclusions. First, molecular evidence for leptin resistance is present within the ARC after 10 weeks of HFD feeding, thereby confirming alterations in ARC biology during prolonged obesity. Second, the most pronounced reduction or reversal in responsiveness to leptin after this prolonged HFD feeding is observed in the Agrp/Npy neuron, which strongly contrasts with the traditional field-wide focus upon the Pomc neuron subtype but agrees with a handful of recent high-profile studies.13, 14
Table 1.
Comparison of IPA “Leptin Signaling in Obesity” gene expression signature enrichment between cells assigned to each cluster, from chow- vs HFD-fed animals.
| Cluster | Cell type | Genes within cluster that match “Leptin Signaling in Obesity” gene expression signature & exhibit significant changes in expression with HFD | Z-score | P-value |
|---|---|---|---|---|
| 0 | Gm42418/AY036118 | Pomc | (none) | 0.197 |
| 1 | Gm42418/AY036118 | Plcb4, Pik3c2a, Map2k2, Akt3 | (none) | 0.288 |
| 2 | Slc17a6/Bdnf | Pik3c2a, Fgfr1, Pomc | (none) | 1.000 |
| 3 | Sst/Dlk1 | Npy, Agrp | (none) | 0.048 |
| 4 | Gda/Rasgrf2 | Pomc | (none) | 0.238 |
| 5 | Pomc/Cartpt | Agrp, Pomc | (none) | 0.201 |
| 6 | Gad2/Zic1 | (none) | (none) | (none) |
| 7 | Astrocytes 1 | Fgfr3, Prkacb, Plce1, Pomc | (none) | 0.044 |
| 8 | Foxb1/Rprm | Npy, Plcb4, Mapk1, Fgfr1, Plcb1, Pomc, Stat3, Adcy8 | −0.447 | 0.031 |
| 9 | unkSlc18a/Hdc | (none) | (none) | (none) |
| 10 | Oligodendrocytes 1 | Pik3r3, Gab1, Fgfr1, Fgfr2, Plcb1, Agrp, Pomc | 0.000 | 0.003 |
| 11 | Oligodendrocytes 2 | Pik3ca | (none) | 0.413 |
| 12 | Endothelial Cells | Pde3a, Pomc | (none) | 0.252 |
| 13 | Astrocytes 2 | Mapk3, Pomc | (none) | 1.000 |
| 14 | Tanycytes | (none) | (none) | (none) |
| 15 | Agrp/Npy | Prkar2b, Ptpn11, Fgfr1, Plcb1, Agrp, Pomc, Stat3, Map2k1 | −1.633 | 0.009 |
| 16 | Microglia | Prkar2b, Pik3r2 | (none) | 1.000 |
| 17 | Ghrh/Th | Plcb4, Prkar2b, Adcy1 | (none) | 0.188 |
| 18 | Oligodendrocytes 3 | Gab2, Akt1, Prkar2b, Map2k2, Pik3r1, Pomc, Jak2, Pik3r2 | −1.633 | 0.122 |
| 19 | Astrocytes 3 | Prkar2a | (none) | 1.000 |
| 20 | Tac1/Foxp2 | Lepr | (none) | 0.316 |
| 21 | Rgs16 | Mapk1 | (none) | 0.324 |
| 22 | Cpne9/Foxb1 | Lepr, Plcg1, Pik3r2 | (none) | 0.154 |
Additional information included in Supplemental File 2.
Long-term HFD feeding interrupts CREB signaling in the Agrp neuron
A previous study examined the effects of short-term (1 week) feeding of an ultra-HFD / low-carbohydrate (D12492: 60% kcal from fat / 20% kcal from carbohydrate) versus low-fat / high-carbohydrate (D12450B, 10% kcal from fat / 70% kcal from carbohydrate) diet upon the transcriptomes of cells within the ARC.11 To dissociate the effects of short-term versus prolonged exposure to HFD, we compared the list of differentially expressed genes within the current Agrp/Npy neuronal cluster (cluster 15) to genes that were differentially expressed in analogous Agrp/Npy clusters from the dataset from Campbell (Fig 3A, and Tables S4–S7). We determined that 529 genes were differentially expressed in this cell type with only short-term HFD feeding, and 47 genes were differentially expressed in similar ways with both short-term and prolonged HFD feeding. In contrast, 683 genes were differentially expressed only after prolonged HFD feeding, and 34 genes that were differentially expressed during short-term HFD feeding exhibited the opposite expression pattern after long-term HFD feeding. The resulting 717 genes that exhibited unique patterns of expression with prolonged HFD feeding were then analyzed by IPA to identify canonical pathways that are uniquely altered within Agrp/Npy neurons after prolonged HFD feeding. Given our biased focus upon this cell cluster, it was unsurprising to note significant negative z-scores for pathways associated with leptin signaling (such as Leptin Signaling in Obesity, z-score = −1.633, and JAK/STAT Signaling, z-score = −1.890). Interestingly, the cAMP Response Element-Binding Protein (CREB) Signaling in Neurons pathway was identified as having the most consistent and robust alteration in expression pattern, and this pattern was negative (z-score = −2.714) (Table 2, and Supplemental File 3). Similarly, the identification of candidate upstream regulators by the IPA algorithm supports likely inhibition of Protein Kinase A (PKA) in cluster 15 (z-score = −2.208) (Supplemental File 4).
Figure 3. Comparison of differentially expressed gene lists within Agrp/Npy neuron clusters between short-term and prolonged HFD exposures, and altered CREB signaling in the ARC after prolonged HFD feeding.
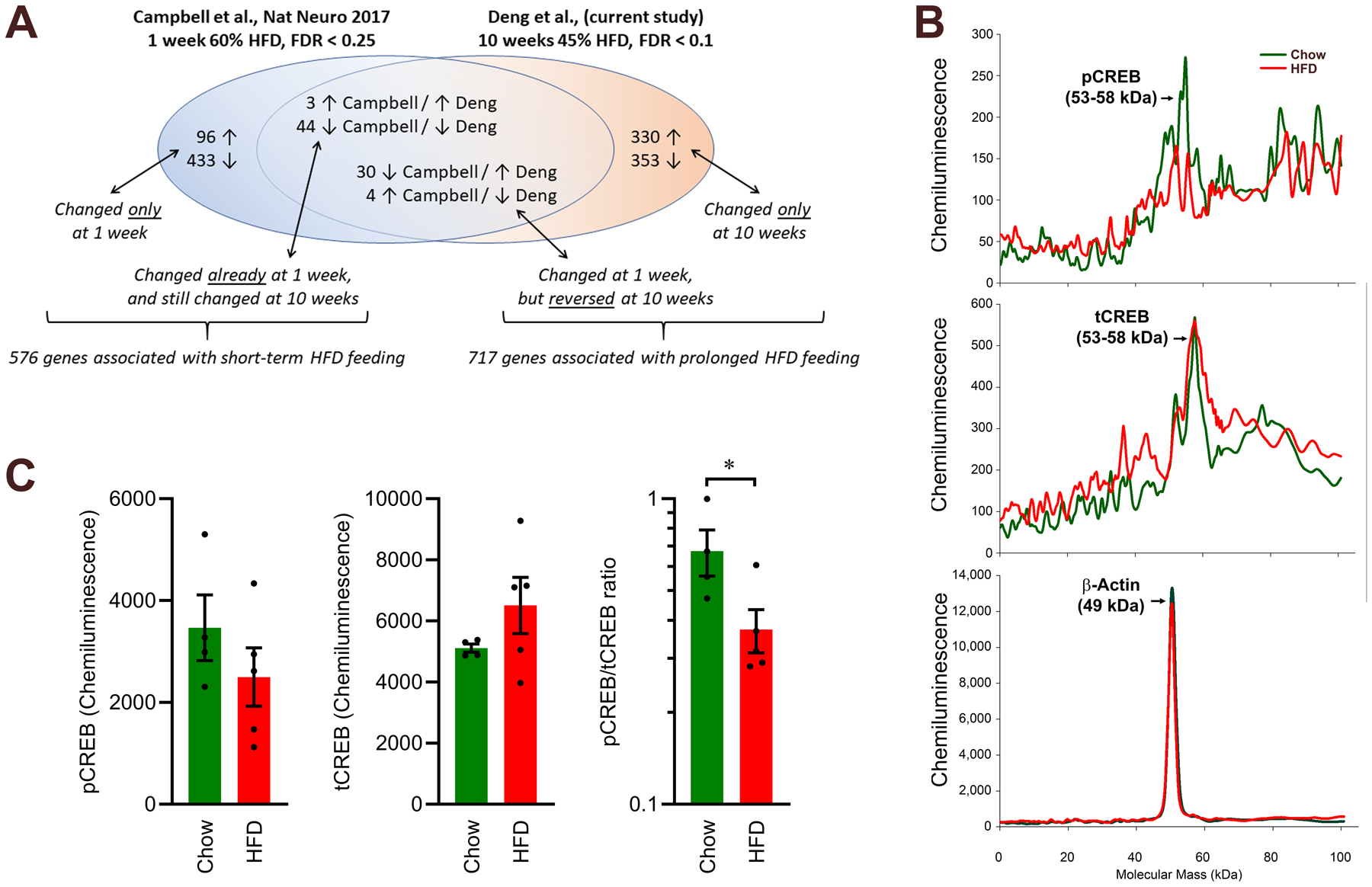
(A) Genes that were differentially expressed in Agrp/Npy cells from Campbell et al. (1 week ultra-HFD vs low-HFD) and the current study (10 weeks HFD vs chow) were compared to identify patterns of gene expression that are unique to short-term versus prolonged HFD exposures. (B) Example chemiluminescent profile of capillary electrophoresis-based Western blots for pCREB, tCREB, and β-actin in samples from ARC tissue. (C) Quantification of area under the curve from chemiluminescence profiles. Chow n=4, HFD n=5. *p<0.05 by two-tailed Student’s t-test after Log10 transform (C). Related information also included in Tables S4–S7.
Table 2.
Ingenuity Pathway Analysis pathway enrichment in Agrp/Npy cell cluster (cluster 15) after prolonged (10 wk) but not short-term (1 wk) HFD (p<0.05, |z|>1.5).
| IPA Pathway | Z-score |
|---|---|
| CREB Signaling in Neurons | −2.714 |
| Actin Cytoskeleton Signaling | −2.496 |
| Insulin Receptor Signaling | −2.333 |
| Huntington’s Disease Signaling | −2.236 |
| 14-3-3-mediated Signaling | −2.121 |
| FcγRIIB Signaling in B Lymphocytes | −2.000 |
| Amyotrophic Lateral Sclerosis Signaling | −1.890 |
| Prolactin Signaling | −1.890 |
| JAK/Stat Signaling | −1.890 |
| Synaptic Long Term Potentiation | −1.667 |
| Leptin Signaling in Obesity | −1.633 |
| ATM Signaling | 1.633 |
| Androgen Signaling | 1.633 |
| Cardiac β-adrenergic Signaling | 2.121 |
Additional information included in Supplemental Files 3 and 4.
CREB phosphorylation is reduced in ARC after prolonged HFD
Finally, we confirmed reductions in CREB phosphorylation within the ARC using capillary electrophoresis-based Western blotting. ARC tissue from chow- and HFD-fed animals was isolated, homogenized, and analyzed for phosphorylated (pCREB) and total (tCREB) cAMP Response Element Binding Protein and β-actin content (Fig 3B and Figure S3). As anticipated given the above sequencing data, ARC samples from HFD-fed mice were characterized by a reduced pCREB/tCREB ratio (Fig 3C). Together these data support the working hypothesis that prolonged HFD feeding results in disrupted PKA/CREB signaling within the ARC, and more specifically, within Agrp neurons.
Discussion
Our group previously established that 10 weeks of HFD feeding was sufficient to model SLR in mice, as this simple intervention resulted in blunted thermogenic brown adipose sympathetic nervous activity responses to leptin while renal sympathetic nervous responses to leptin were maintained.4 While disparate data from our group and others supports the involvement of various second-messenger systems within the hypothalamus in SLR, snRNAseq-based dissection of the transcriptomic changes induced within cell types of the ARC in this diet-induced model provides a simple and unbiased method to tease apart the cellular and molecular mechanisms that contribute to SLR. Given the overwhelming focus of the field upon the biology of the Pomc neuron in mediating the physiological and pathophysiological responses to leptin, it was surprising to discover a much more robust signature for leptin dysfunction in Agrp neurons after prolonged obesity. These results lead to the conclusion that molecular evidence of SLR is present at the level of the ARC, independent of whether other synergistic changes are discovered in second-order regions of the hypothalamus or within the brainstem. Further, these results support the iconoclastic conclusion that the Agrp neuron may contribute a much more important role in the development of SLR than other cell types of the ARC. Further, data presented herein provide new, independent, unbiased identification of altered CREB signaling – specifically within the Agrp neuron – as a potential mediator of SLR.
Other experimental approaches have supported the concept that changes in Agrp neuron biology contribute to the pathogenesis of SLR, and identified potential genetic mediators. For example, leptin modulates the electrical activity of Agrp neurons, and diet-induced obesity is associated with persistent activation of these cells.15 Activation of c-Jun N-terminal kinase 1 in Agrp neurons is sufficient to induce leptin resistance,16 and inhibition of PKA activity17 or ablation of Forkhead Box O118 in these cells prevents leptin resistance. Conditional deletion of Lepr from Agrp neurons also attenuates sympathetic nerve activity to interscapular brown and subcutaneous white tissues, and accelerates weight gain.19 Thus, identification of altered CREB, 14-3-3, JAK/STAT, and leptin signaling in Agrp neurons after prolonged HFD in the current study (Table 2) provides independent support for these molecular mechanisms, specifically within Agrp neurons, in the pathogenesis of SLR.
Multiple studies have also demonstrated that leptin resistance is correlated with changes in the control of Agrp expression within the ARC of C57BL/6J mice. Short-term HFD (1- to 7-weeks) causes suppression of Agrp mRNA despite 4- to 6-fold elevations in endogenous plasma leptin.20, 21 In contrast, longer-term HFD exposure (8- to 18-weeks) is associated with loss of this suppression despite maintenance of elevated plasma leptin.21, 22 Further, 20-weeks HFD feeding has been shown to abolish ARC Agrp expression, food intake, and body mass responses to acute exogenous leptin in diet-sensitive C57BL/6J mice.23 Importantly, all of these studies analyzed Agrp expression in the complete ARC, and did not analyze expression in individual cell types. Future studies to discern changes in responses of Agrp (and other genes) to acute leptin delivery within individual cell types after prolonged diet-induced obesity are logical, given the results of the current study.
A large body of evidence supports a major role for the Pomc-expressing neuron in the control of sympathetic nervous system activity to both cardiovascular and metabolic targets, and the signaling cascades and neural projections that mediate these effects are largely defined (recently reviewed in detail24, 25). In contrast, mechanisms contributing to the control of Agrp expression and the activity of Agrp-expressing neurons are not deeply characterized at this time, however, a role for CREB has been suggested. For example, manipulation of phosphodiesterase-3B resulted in reciprocal changes in Agrp expression within mouse hypothalamic mHypoE-46 cells.26 Further, mice lacking the RIIβ regulatory subunit of PKA exhibit altered leptin signaling and resistance to diet-induced obesity, and inhibition of PKA activity within Agrp neurons partially recapitulated this effect.17 CREB signaling has also been implicated in the induction of NOR1 in response to leptin, which additionally acts to modulate the expression of Agrp within the ARC.27 Studies of mouse hypothalamic GT1–7 cells have demonstrated a complex interaction between CREB and ERK signaling in the transcriptional control of Agrp,28 and insulin has also been shown to act via Extracellular signaling Related Kinase (ERK) within mHypoE-46 cells to control Agrp expression.29 Importantly, we previously demonstrated that intracerebroventricular delivery of ERK inhibitors PD98059 or U0126, but not the phosphoinositide 3-kinase (PI3K) inhibitor LY294002, attenuated leptin-induced sympathetic nervous activity responses to thermogenic brown adipose while having no effect on renal, lumbar, or adrenal sympathetic nerve responses to leptin.30 Interestingly, in other cell types it has been demonstrated that the PD98059 and U0126 ERK inhibitors are also known to indirectly interfere with PKA/CREB signaling,31, 32 while the PI3K inhibitor LY294002 stimulates CREB signaling.33 Finally, angiotensin II type 1A receptors (AT1A) are known to activate second-messenger signaling cascades involving both CREB and ERK, and we previously demonstrated a critical role for AT1A, localized to a subset of Agrp-expressing neurons of the ARC, in the control of Agrp gene expression, brown adipose sympathetic nerve activity, and resting metabolic rate in response to various stimuli.34, 35 These findings reinforce the working hypothesis that a complex mechanism involving CREB and ERK (but apparently not PI3K) contributes to the control of Agrp. Future work to comprehensively characterize the transcriptional control of Agrp and the functional control of Agrp neuron activity is clearly required to understand the development of SLR.
Finally, increasing evidence supports the concept that glial (astrocytes, microglia, oligodendrocyte, etc.) cell dysfunctions and hypothalamic inflammation may contribute to obesity and leptin resistance.36–38 While no significant changes in canonical leptin signaling after prolonged HFD were observed in glial cell types in the current study (Table 1), further exploration of IPA outputs for these various cell types (Supplemental Files 2–4) may inform these mechanisms. For example, interleukin-3, −6, −17A, and −23 signaling pathways were significantly changed (p<0.05, z≤−1) in Cluster 18, and such cytokines have been implicated in the control of leptin sensitivity and Agrp expression.39, 40 Thus, a role for diet-induced changes in the activities of other (non-Agrp) cell types in the development of SLR may additionally be supported by the current study.
Limitations
This study must be interpreted in light of its limitations. First, this study is limited by the inherent sequencing depth issues that plague all single nucleus and single cell RNA-seq studies; changes in expression of low-abundance transcripts are not detectable. This complication limits our ability to analyze the distinct transcriptomes of subtypes of each major cell type (eg – subtypes of Agrp neurons), and also limits analysis of changes in low-abundance transcripts (eg – G protein coupled receptors, and leptin receptor). Second, the study is limited by a relatively small number of replicates. Consensus on methods to calculate power and sample size has not been reached with regard to single nucleus RNA-seq studies, but we suspect that the current study is underpowered to detect smaller changes in gene expression within and across clusters. The current sample size was selected for pragmatic reasons including cost and the goal of restricting sequencing to a small (6 to 8) number of samples to permit simultaneous sequencing of all samples in a single lane, which reduces experimental error. Third, and again for pragmatic reasons, only one sex was examined in the current study. Males were chosen for study because of their greater propensity for weight gain on a 45% HFD, plus our previous demonstration that ten weeks of 45% HFD is sufficient to model leptin resistance in male C57BL/6J mice.4 It has been clearly established that sex differences exist with regard to Agrp neuron biology,41–43 and thus the identification of changes specifically within Agrp neurons in the current study provides a critical rationale to pursue future studies of gene expression changes in the ARC after prolonged HFD in both sexes. Fourth, our comparison of the effects of long-term versus short-term HFD feeding took advantage of the previous study from Campbell et al.11 In their study, the authors examined gene expression in cells of the ARC between mice that were fed an ultra-HFD (60% kcal from fat) compared to a low fat / high carbohydrate (10% kcal from fat, 70% from carbohydrates) diet for just 1-week. In the current study, mice were fed a HFD (45% kcal from fat) diet for ten weeks, or maintained on a standard chow. It is likely that the difference in diets (both “HFD” and “control” diets) between studies will therefore account for some of the variability between datasets, and thus contribute to the relatively low overlap of the lists of differentially expressed genes at 1- vs 10-weeks (Fig 2A). As the cost of snRNA-seq studies decreases, it will be appropriate to replicate time-course studies in a much more controlled manner to more comprehensively identify differences in the transcriptomes of individual cell types of the ARC after short- vs long-term HFD feeding and diet-induced obesity. Fifth, in the current study we utilized the single nucleus (snRNA-seq) instead of a single cell (scRNA-seq) approach. Because scRNA-seq requires a large fraction of viable cells, snRNA-seq has become the preferred method to analyze transcriptomes of banked or frozen tissues. Most importantly, several studies have compared the outcomes of simultaneous analyses by each method and a high congruence of results has been observed in various tissue types including mouse visual cortex,44 and mouse kidney,45 and human induced pluripotent stem cells.46 Nonetheless, future validation of the snRNA-seq approach specifically within the ARC is warranted. Sixth, we utilized the C57BL/6J mouse for these studies, and it is known that these animals exhibit spontaneous individual sensitivity or resistance to weight gain and other cardiometabolic phenotypes in response to HFD.23 Thus, some within-group variability in the current study may result from our grouping of animals that individually may or may not have exhibited sensitivity to the HFD stimulus. Finally, we utilized an analysis approach that focused on gene network analysis, which reduces reliance upon analysis of low-abundance transcripts. As a result, some biologically-significant changes in expression of individual genes within each cell type are likely to be missed. It is important to therefore appreciate that the current study provides positive implication of alterations in generalized leptin and CREB signaling within the Agrp neuron of the ARC during prolonged diet-induced obesity – but should not be taken as evidence against the importance of other mechanisms that are likely active in other cell types such as the Pomc neuron.
Perspectives
The findings of this study lead to several exciting conclusions. First, prolonged HFD feeding is associated with unique gene expression patterns that are qualitatively and quantitatively distinct from the patterns observed with short-term HFD exposure. Second, we conclude that long-term HFD feeding is associated with molecular evidence of leptin resistance at the level of the ARC, the primary central nervous system detection site for circulating leptin. Third, this decoupling of leptin signaling is most pronounced in specific cell types of the ARC, such as the Agrp/Npy neuron – but not the substantially more well-characterized Pomc neuron. Fourth, unbiased exploration of the dysfunctions within the Agrp/Npy neuron after prolonged HFD feeding point to dysfunction of CREB signaling pathways. Thus, it follows that selective leptin resistance may involve dysfunctional CREB-mediated control of Agrp neuronal function, and subsequent disruption of the portion of the melanocortin signaling system that specifically contributes to the control of energy balance. Clarification of the mechanisms by which CREB contributes to Agrp/Npy neuronal function in health and disease is warranted.
Supplementary Material
Novelty and Significance.
What Is New?
Single nucleus RNAsequencing (snRNAseq) was used to examine transcriptomes of individual cell types of the mouse arcuate nucleus after prolonged diet-induced obesity
Unlike short-term dietary interventions, prolonged high fat diet caused selective changes in leptin signaling in the Agouti-related peptide (Agrp) neuron subtype
Changes in leptin signaling in the Agrp neuron after prolonged diet-induced obesity are paralleled by changes in CREB signaling
What Is Relevant?
Selective leptin resistance is proposed to contribute to the pathogenesis and maintenance of obesity-associated hypertension, but the cellular & molecular mechanisms remain unclear
The iconoclastic findings presented here support a dominant role for altered biology of the Agrp neuron, not the much more well-characterized proopiomelanocortin (Pomc) neuron, in selective leptin resistance
Summary
Understanding the cellular and molecular changes that occur with prolonged obesity is critical to understand and address selective leptin resistance and thereby obesity-associated cardiovascular disease.
Acknowledgments
The authors gratefully acknowledge the technical assistance of the Genomics Division of the Iowa Institute of Human Genetics.
Sources of Funding
This work and the authors were supported by grants from the NIH (HL134850, HL084207, HL007638, HL127673, GM008629) and the American Heart Association (18EIA33890055, 18PRE33960377, 19POST34380239). The authors were also supported through the MCW Clinical & Translational Science Institute “Obesity” Ensemble (UL1TR001436) and the Advancing a Healthier Wisconsin Endowment to MCW. Data presented herein were obtained at the Genomics Division of the Iowa Institute of Human Genetics which is supported, in part, by the University of Iowa Carver College of Medicine.
Footnotes
Disclosures
None.
Declarations of Interest: None
References
- 1.Hall JE, Carmo JMd, Silva AAd, Wang Z, Hall ME. Obesity-induced hypertension. Circulation research. 2015;116:991–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mark AL. Selective leptin resistance revisited. American journal of physiology. Regulatory, integrative and comparative physiology 2013;305:R566–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caron A, Richard D. Neuronal systems and circuits involved in the control of food intake and adaptive thermogenesis. Annals of the New York Academy of Sciences. 2017;1391:35–53 [DOI] [PubMed] [Google Scholar]
- 4.Rahmouni K, Morgan DA, Morgan GM, Mark AL, Haynes WG. Role of selective leptin resistance in diet-induced obesity hypertension. Diabetes. 2005;54:2012–2018 [DOI] [PubMed] [Google Scholar]
- 5.Kramer A, Green J, Pollard J Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics (Oxford, England). 2014;30:523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balapattabi K, Farmer GE, Knapp BA, Little JT, Bachelor M, Yuan JP, Cunningham JT. Effects of salt-loading on supraoptic vasopressin neurones assessed by clophensorn chloride imaging. Journal of neuroendocrinology. 2019;31:e12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, Choudhury SR, Aguet F, Gelfand E, Ardlie K, Weitz DA, Rozenblatt-Rosen O, Zhang F, Regev A. Massively parallel single-nucleus rna-seq with dronc-seq. Nature methods. 2017;14:955–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Velmeshev D, Schirmer L, Jung D, Haeussler M, Perez Y, Mayer S, Bhaduri A, Goyal N, Rowitch DH, Kriegstein AR. Single-cell genomics identifies cell type-specific molecular changes in autism. Science. 2019;364:685–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F, Huang D, Xu Y, Huang W, Jiang M, Jiang X, Mao J, Chen Y, Lu C, Xie J, Fang Q, Wang Y, Yue R, Li T, Huang H, Orkin SH, Yuan GC, Chen M, Guo G. Mapping the mouse cell atlas by microwell-seq. Cell. 2018;172:1091–1107.e1017 [DOI] [PubMed] [Google Scholar]
- 11.Campbell JN, Macosko EZ, Fenselau H, Pers TH, Lyubetskaya A, Tenen D, Goldman M, Verstegen AM, Resch JM, McCarroll SA, Rosen ED, Lowell BB, Tsai LT. A molecular census of arcuate hypothalamus and median eminence cell types. Nature neuroscience. 2017;20:484–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romanov RA, Zeisel A, Bakker J, Girach F, Hellysaz A, Tomer R, Alpar A, Mulder J, Clotman F, Keimpema E, Hsueh B, Crow AK, Martens H, Schwindling C, Calvigioni D, Bains JS, Mate Z, Szabo G, Yanagawa Y, Zhang MD, Rendeiro A, Farlik M, Uhlen M, Wulff P, Bock C, Broberger C, Deisseroth K, Hokfelt T, Linnarsson S, Horvath TL, Harkany T. Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nature neuroscience. 2017;20:176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J, Bartolome CL, Low CS, Yi X, Chien CH, Wang P, Kong D. Genetic identification of leptin neural circuits in energy and glucose homeostases. Nature. 2018;556:505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caron A, Dungan Lemko HM, Castorena CM, Fujikawa T, Lee S, Lord CC, Ahmed N, Lee CE, Holland WL, Liu C, Elmquist JK. Pomc neurons expressing leptin receptors coordinate metabolic responses to fasting via suppression of leptin levels. eLife. 2018;7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baver SB, Hope K, Guyot S, Bjorbaek C, Kaczorowski C, O’Connell KM. Leptin modulates the intrinsic excitability of agrp/npy neurons in the arcuate nucleus of the hypothalamus. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:5486–5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsaousidou E, Paeger L, Belgardt BF, Pal M, Wunderlich CM, Bronneke H, Collienne U, Hampel B, Wunderlich FT, Schmidt-Supprian M, Kloppenburg P, Bruning JC. Distinct roles for jnk and ikk activation in agouti-related peptide neurons in the development of obesity and insulin resistance. Cell reports. 2014;9:1495–1506 [DOI] [PubMed] [Google Scholar]
- 17.Yang L, McKnight GS. Hypothalamic pka regulates leptin sensitivity and adiposity. Nature communications. 2015;6:8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren H, Orozco IJ, Su Y, Suyama S, Gutierrez-Juarez R, Horvath TL, Wardlaw SL, Plum L, Arancio O, Accili D. Foxo1 target gpr17 activates agrp neurons to regulate food intake. Cell. 2012;149:1314–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell BB, Harlan SM, Morgan DA, Guo DF, Cui H, Rahmouni K. Differential contribution of pomc and agrp neurons to the regulation of regional autonomic nerve activity by leptin. Molecular metabolism. 2018;8:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Storlien LH, Huang XF. Effects of dietary fat types on body fatness, leptin, and arc leptin receptor, npy, and agrp mrna expression. American journal of physiology. Endocrinology and metabolism 2002;282:E1352–1359 [DOI] [PubMed] [Google Scholar]
- 21.Densmore VS, Morton NM, Mullins JJ, Seckl JR. 11 beta-hydroxysteroid dehydrogenase type 1 induction in the arcuate nucleus by high-fat feeding: A novel constraint to hyperphagia? Endocrinology. 2006;147:4486–4495 [DOI] [PubMed] [Google Scholar]
- 22.Patterson CM, Villanueva EC, Greenwald-Yarnell M, Rajala M, Gonzalez IE, Saini N, Jones J, Myers MG Jr. Leptin action via lepr-b tyr1077 contributes to the control of energy balance and female reproduction. Molecular metabolism. 2012;1:61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enriori PJ, Evans AE, Sinnayah P, Jobst EE, Tonelli-Lemos L, Billes SK, Glavas MM, Grayson BE, Perello M, Nillni EA, Grove KL, Cowley MA. Diet-induced obesity causes severe but reversible leptin resistance in arcuate melanocortin neurons. Cell metabolism. 2007;5:181–194 [DOI] [PubMed] [Google Scholar]
- 24.Hall JE, do Carmo JM, da Silva AA, Wang Z, Hall ME. Obesity, kidney dysfunction and hypertension: Mechanistic links. Nat Rev Nephrol. 2019;15:367–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.da Silva AA, do Carmo JM, Wang Z, Hall JE. Melanocortin-4 receptors and sympathetic nervous system activation in hypertension. Current hypertension reports. 2019;21:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anamthathmakula P, Sahu M, Sahu A. Evidence suggesting phosphodiesterase-3b regulation of npy/agrp gene expression in mhypoe-46 hypothalamic neurons. Neuroscience letters. 2015;604:113–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim SG, Lee B, Kim DH, Kim J, Lee S, Lee SK, Lee JW. Control of energy balance by hypothalamic gene circuitry involving two nuclear receptors, neuron-derived orphan receptor 1 and glucocorticoid receptor. Molecular and cellular biology. 2013;33:3826–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park M, Oh H, York DA. Enterostatin affects cyclic amp and erk signaling pathways to regulate agouti-related protein (agrp) expression. Peptides. 2009;30:181–190 [DOI] [PubMed] [Google Scholar]
- 29.Mayer CM, Belsham DD. Insulin directly regulates npy and agrp gene expression via the mapk mek/erk signal transduction pathway in mhypoe-46 hypothalamic neurons. Molecular and cellular endocrinology. 2009;307:99–108 [DOI] [PubMed] [Google Scholar]
- 30.Rahmouni K, Sigmund CD, Haynes WG, Mark AL. Hypothalamic erk mediates the anorectic and thermogenic sympathetic effects of leptin. Diabetes. 2009;58:536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costes S, Broca C, Bertrand G, Lajoix AD, Bataille D, Bockaert J, Dalle S. Erk1/2 control phosphorylation and protein level of camp-responsive element-binding protein: A key role in glucose-mediated pancreatic beta-cell survival. Diabetes. 2006;55:2220–2230 [DOI] [PubMed] [Google Scholar]
- 32.Ashok C, Owais S, Srijyothi L, Selvam M, Ponne S, Baluchamy S. A feedback regulation of creb activation through the cul4a and erk signaling. Med Oncol. 2019;36:20. [DOI] [PubMed] [Google Scholar]
- 33.Xie Y, Perry BD, Espinoza D, Zhang P, Price SR. Glucocorticoid-induced creb activation and myostatin expression in c2c12 myotubes involves phosphodiesterase-3/4 signaling. Biochemical and biophysical research communications. 2018;503:1409–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Claflin KE, Sandgren JA, Lambertz AM, Weidemann BJ, Littlejohn NK, Burnett CM, Pearson NA, Morgan DA, Gibson-Corley KN, Rahmouni K, Grobe JL. Angiotensin at1a receptors on leptin receptor-expressing cells control resting metabolism. The Journal of clinical investigation. 2017;127:1414–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morselli LL, Claflin KE, Cui H, Grobe JL. Control of energy expenditure by agrp neurons of the arcuate nucleus: Neurocircuitry, signaling pathways, and angiotensin. Current hypertension reports. 2018;20:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Djogo T, Robins SC, Schneider S, Kryzskaya D, Liu X, Mingay A, Gillon CJ, Kim JH, Storch KF, Boehm U, Bourque CW, Stroh T, Dimou L, Kokoeva MV. Adult ng2-glia are required for median eminence-mediated leptin sensing and body weight control. Cell metabolism. 2016;23:797–810 [DOI] [PubMed] [Google Scholar]
- 37.de Git KC, Adan RA. Leptin resistance in diet-induced obesity: The role of hypothalamic inflammation. Obesity reviews : an official journal of the International Association for the Study of Obesity. 2015;16:207–224 [DOI] [PubMed] [Google Scholar]
- 38.Liu X, Zheng H. Leptin-mediated sympathoexcitation in obese rats: Role for neuron-astrocyte crosstalk in the arcuate nucleus. Front Neurosci. 2019;13:1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larsen L, Le Foll C, Dunn-Meynell AA, Levin BE. Il-6 ameliorates defective leptin sensitivity in dio ventromedial hypothalamic nucleus neurons. American journal of physiology. Regulatory, integrative and comparative physiology 2016;311:R764–r770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nogueira G, Solon C, Carraro RS, Engel DF, Ramalho AF, Sidarta-Oliveira D, Gaspar RS, Bombassaro B, Vasques AC, Geloneze B, Vinolo MA, Donato Junior J, Velloso LA. Interleukin-17 acts in the hypothalamus reducing food intake. Brain Behav Immun. 2019:S0889–1591(0819)30848–30847 [DOI] [PubMed] [Google Scholar]
- 41.Egan OK, Inglis MA, Anderson GM. Leptin signaling in agrp neurons modulates puberty onset and adult fertility in mice. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2017;37:3875–3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asarian L, Geary N. Sex differences in the physiology of eating. American journal of physiology. Regulatory, integrative and comparative physiology 2013;305:R1215–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheffer-Babila S, Sun Y, Israel DD, Liu SM, Neal-Perry G, Chua SC Jr. Agouti-related peptide plays a critical role in leptin’s effects on female puberty and reproduction. American journal of physiology. Endocrinology and metabolism 2013;305:E1512–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bakken TE, Hodge RD, Miller JA, Yao Z, Nguyen TN, Aevermann B, Barkan E, Bertagnolli D, Casper T, Dee N, Garren E, Goldy J, Graybuck LT, Kroll M, Lasken RS, Lathia K, Parry S, Rimorin C, Scheuermann RH, Schork NJ, Shehata SI, Tieu M, Phillips JW, Bernard A, Smith KA, Zeng H, Lein ES, Tasic B. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PloS one. 2018;13:e0209648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu H, Kirita Y, Donnelly EL, Humphreys BD. Advantages of single-nucleus over single-cell rna sequencing of adult kidney: Rare cell types and novel cell states revealed in fibrosis. Journal of the American Society of Nephrology : JASN. 2019;30:23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selewa A, Dohn R, Eckart H, Lozano S, Xie B, Gauchat E, Elorbany R, Rhodes K, Burnett J, Gilad Y, Pott S, Basu A. Systematic comparison of high-throughput single-cell and single-nucleus transcriptomes during cardiomyocyte differentiation. Sci Rep. 2020;10:1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


