Abstract
Severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) has resulted in a pandemic and continues to spread around the globe at an unprecedented rate. To date, no effective therapeutic is available to fight its associated disease, COVID-19. Our discovery of a novel insertion of glycosaminoglycan (GAG)-binding motif at S1/S2 proteolytic cleavage site (681–686 (PRRARS)) and two other GAG-binding-like motifs within SARS-CoV-2 spike glycoprotein (SGP) led us to hypothesize that host cell surface GAGs may interact SARS-CoV-2 SGPs to facilitate host cell entry. Using a surface plasmon resonance direct binding assay, we found that both monomeric and trimeric SARS-CoV-2 SGP bind more tightly to immobilized heparin (KD = 40 pM and 73 pM, respectively) than the SARS-CoV and MERS-CoV SGPs (500 nM and 1 nM, respectively). In competitive binding studies, the IC50 of heparin, tri-sulfated non-anticoagulant heparan sulfate, and non-anticoagulant low molecular weight heparin against SARS-CoV-2 SGP binding to immobilized heparin were 0.056 μM, 0.12 μM, and 26.4 μM, respectively. Finally, unbiased computational ligand docking indicates that heparan sulfate interacts with the GAG-binding motif at the S1/S2 site on each monomer interface in the trimeric SARS-CoV-2 SGP, and at another site (453–459 (YRLFRKS)) when the receptor-binding domain is in an open conformation. The current study serves a foundation to further investigate biological roles of GAGs in SARS-CoV-2 pathogenesis. Furthermore, our findings may provide additional basis for further heparin-based interventions for COVID-19 patients exhibiting thrombotic complications.
Keywords: SARS-CoV-2, COVID-19, Glycosaminoglycans, Heparin, Spike glycoprotein, Binding interactions
Abbreviations
- ACE2
angiotensin-converting enzyme 2
- CoV
coronavirus
- COVID-19
coronavirus disease 2019
- CSA
chondroitin sulfate A
- CSC
chondroitin sulfate C
- CSD
chondroitin sulfate D
- CSE
chondroitin sulfate E
- dp
degree of polymerization
- DS
dermatan sulfate
- DPP4
dipeptidyl peptidase 4
- ECM
extracellular matrix
- GAG
glycosaminoglycan
- HA
hyaluronan
- HIV-1
human immunodeficiency virus type 1
- HS
heparan sulfate
- HP
heparin
- IBV
infectious bronchitis virus
- MERS-CoV
Middle East respiratory syndrome-related coronavirus
- MHV
mouse hepatitis virus
- NACH
non-anticoagulant heparin
- N-DeS HP
N-desulfated heparin
- PG
proteoglycan
- RBD
receptor binding domain
- RU
resonance unit
- SGP
spike glycoprotein
- S1
subunit 1
- S2
subunit 2
- SARS-CoV
severe acute respiratory syndrome-related coronavirus
- SARS-CoV-2
severe acute respiratory syndrome-related coronavirus 2
- SDS
sodium dodecyl sulfonate
- SPR
surface plasmon resonance
- TMPRSS2
transmembrane serine protease 2
- TriS
trisulfated
- 2-DeS HP
2-O-desulfated heparin
- 6-DeS HP
6-O-desulfated heparin derivative
- 2OST/6OST
2-/6-O-sulfotransferase
1. Introduction
In March 2020, the World Health Organization (WHO) declared severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) a pandemic less than three months after its initial emergence in Wuhan, China (Chan et al., 2020b; World Health Organization, 2020a). SARS-CoV-2 is a zoonotic Betacoronavirus transmitted through person-person contact through airborne and fecal-oral routes, and has caused over 10,021,401 confirmed coronavirus disease 2019 (COVID-19) cases and 499,913 associated deaths worldwide as of June 29th, 2020 (Chan et al., 2020b; van Doremalen et al., 2020; World Health Organization, 2020b; Yeo et al., 2020). While there is limited understanding of SARS-CoV-2 pathogenesis, extensive studies have been performed on how its closely related cousins, SARS-CoV and MERS-CoV (Middle East respiratory syndrome-related coronavirus), invade host cell. Upon initially contacting the surface of a host cell, SARS-CoV and MERS-CoV exploit host cell proteases to prime their surface spike glycoproteins (SGPs) for fusion activation, which is achieved by receptor binding, low pH, or both (Belouzard et al., 2009; Matsuyama et al., 2018). The receptor binding domain (RBD) resides within subunit 1 (S1) while subunit 2 (S2) facilitates viral-host cell membrane fusion (Belouzard et al., 2009). Activated SGP undergoes a conformational change followed by an initiated fusion reaction with the host cell membrane (Belouzard et al., 2009). Endocytosed virions are further processed by the endosomal protease cathepsin L in the late endosome (Huang et al., 2006; Matsuyama et al., 2018). Both MERS-CoV and SARS-CoV require proteolytic cleavage at their S2’ site, but not at their S1–S2 junction, for successful membrane fusion and host cell entry (Belouzard et al., 2009; Matsuyama et al., 2018). Additionally, receptors involved in fusion activation of SARS-CoV and MERS-CoV include heparan sulfate (HS) and angiotensin-converting enzyme 2 (ACE2), and dipeptidyl peptidase 4 (DPP4), respectively (Lang et al., 2011; Li et al., 2005; Raj et al., 2013). Recent progress confirms that SARS-CoV-2 also utilizes ACE2 and host cell proteases during host cell entry (Hoffmann et al., 2020).
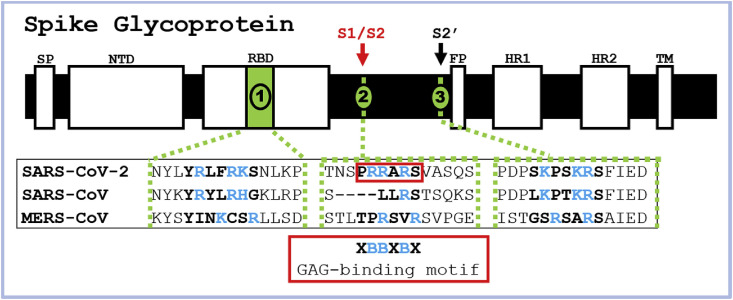
SARS-CoV and other pathogens arrive at a host cell surface by clinging, through their surface proteins, to linear, sulfated polysaccharides called glycosaminoglycans (GAGs) (Belouzard et al., 2012; Kamhi et al., 2013; Kim et al., 2017). The repeating disaccharide units of GAGs, comprised of a hexosamine and a uronic acid or a galactose residue, are often sulfated (Fig. S1) (Lindahl et al., 2015). GAGs are generally found covalently linked to core proteins as proteoglycans (PGs) and reside inside the cell, at the cell surface, and in the extracellular matrix (ECM) (Lindahl et al., 2015). GAGs facilitate various biological processes, including cellular signaling, pathogenesis, and immunity, and possess diverse therapeutic applications (Lindahl et al., 2015). For example, an FDA approved anticoagulant heparin (HP) is a secretory GAG released from granules of mast cells during infection (Kim et al., 2018; Lindahl et al., 2015). Some GAG binding proteins can be identified by amino acid sequences known as Cardin-Weintraub motifs corresponding to ‘XBBXBX’ and ‘XBBBXXBX’, where X is a hydropathic residue and B is a basic residue, such as arginine and lysine, responsible for interacting with the sulfate groups present in GAGs (Cardin and Weintraub, 1989; Hileman et al., 1998a). Examination of the SARS-CoV-2 SGP sequence revealed that the GAG-binding motif resides within S1/S2 proteolytic cleavage motif (furin cleavage motif BBXBB) that is not present in SARS-CoV or MERS-CoV SGPs (Fig. 1, S2, and S3) (Coutard et al., 2020). Additionally, we discovered GAG-binding-like motifs within RBD and S2′ proteolytic cleavage site in SARS-CoV-2 SGP (Fig. 1, S2, and S3). This discovery prompted us to hypothesize that host cell surface GAGs may interact SARS-CoV-2 SGPs to potentially facilitate host cell entry. We performed surface plasmon resonance (SPR)-based binding assays to determine binding kinetics of the interactions between various GAGs and SARS-CoV-2 SGP in comparison with SARS-CoV, and MERS-CoV SGP to address this question. Lastly, we performed blind docking on the trimeric SARS-CoV-2 SGP model to objectively identify the preferred binding GAG-binding sites on the SGP.
2. Materials and methods
2.1. Materials
Monomeric SARS-CoV-2, SARS-CoV, and MERS-CoV SGPs were purchased from Sino Biological Inc. Trimeric SARS-CoV-2 SGP was kindly provided by Prof. Jason McLellan from University of Texas at Austin (Wrapp et al., 2020). The GAGs used in this study were porcine intestinal HP (average molecular weight, Mw = 16 kDa) and porcine intestinal heparan sulfate (HS) (Mw = 14 kDa) from Celsus Laboratories (Cincinnati, OH); chondroitin sulfate A (CSA, Mw = 20 kDa) from porcine rib cartilage (Sigma, St. Louis, MO), dermatan sulfate (DS, Mw = 30 kDa) from porcine intestine (Sigma), chondroitin sulfate C (CSC, Mw = 20 kDa) from shark cartilage (Sigma), chondroitin sulfate D (CSD, Mw = 20 kDa) from whale cartilage (Seikagaku, Tokyo, Japan) and chondroitin sulfate E (CSE, Mw = 20 kDa) from squid cartilage (Seikagaku). N-desulfated HP (N-DeS HP, Mw = 14 kDa) and 2-O-desulfated HP (2-DeS HP, MW = 13 kDa) were prepared in house based on previously established protocols (Yates et al., 1996). The 6-O-desulfated HP derivative, 6-DeS HP, Mw = 13 kDa, was generously provided by Prof. Lianchun Wang from University of South Florida. Non-anticoagulant low molecular weight HP (NACH) was synthesized from dalteparin, a nitrous acid depolymerization product of porcine intestinal HP, followed by periodate oxidation as described in our previous work (Islam et al., 2002). TriS HS (NS2S6S) was synthesized from N-sulfo heparosan with subsequent modification with C5-epimerase and 2-O- and 6-O-sulfotransferases (2OST and 6OST1/6OST3) (Linhardt et al., 2007). HP oligosaccharides included tetrasaccharide (dp4), hexasaccharide (dp6), octasaccharide (dp8), decasaccharide (dp10), dodecasaccharide (dp12), tetradecasaccharide (dp14), hexadecasaccharide (dp16) and octadecasaccharide (dp18) and were prepared from porcine intestinal HP controlled partial heparin lyase 1 treatment followed by size fractionation. The chemical structures of the GAGs are shown in Fig. S1. The SA sensor chips were from GE Healthcare (Uppsala, Sweden). SPR measurements were performed on a BIAcore 3000 operated using BIAcore 3000 control and BIAevaluation software (version 4.0.1).
2.2. Preparation of HP biochip
Biotinylated HP was prepared by conjugating its reducing end to amine-PEG3-Biotin (Pierce, Rockford, IL). In brief, HP (2 mg) and amine-PEG3-Biotin (2 mg, Pierce, Rockford, IL) were dissolved in 200 μL H2O, 10 mg NaCNBH3 was added. The reaction mixture was heated at 70 °C for 24 h, after that a further 10 mg NaCNBH3 was added and the reaction was heated at 70 °C for another 24 h. After cooling to room temperature, the mixture was desalted with the spin column (3000 MWCO). Biotinylated HP was collected, freeze-dried and used for SA chip preparation. The biotinylated HP was immobilized to streptavidin (SA) chip based on the manufacturer's protocol. The successful immobilization of HP was confirmed by the observation of a 200-resonance unit (RU) increase on the sensor chip. The control flow cell (FC1) was prepared by 2 min injection with saturated biotin.
2.3. Measurement of interaction between HP and CoV SGP using BIAcore
SGP samples were diluted in buffer (0.01 M HEPES, 0.15 M NaCl, 3 mM EDTA, 0.005% surfactant P20, pH 7.4). Different dilutions of protein samples were injected at a flow rate of 30 μL/min. At the end of the sample injection, the same buffer was flowed over the sensor surface to facilitate dissociation. After a 3 min dissociation time, the sensor surface was regenerated by injecting with 30 μL of 0.25% sodium dodecyl sulfonate (SDS) to get fully regenerated surface. The response was monitored as a function of time (sensorgram) at 25 °C.
2.4. Solution competition study between HP on chip surface and HP, HP-derived oligosaccharides, chemically modified HP or GAGs in solution using SPR
SARS-CoV-2 SGP (50 nM) mixed with 1 μM HP, HP-derived oligosaccharides, chemically modified HP or GAGs in SPR buffer were injected over HP chip at a flow rate of 30 μL/min, respectively. After each run, the dissociation and the regeneration were performed as described above.
2.5. SPR solution competition IC50 measurement of glycans (HP, TriS HS and NACH) inhibition on SARS-CoV-2 SGP-HP interaction
Solution competition studies between a surface HP and soluble glycans (HP, TriS HS and NACH) to measure IC50 were performed using SPR (Fu et al., 2013). In brief, SARS-CoV-2 S-protein (50 nM) samples alone or mixed with different concentrations of glycans in SPR buffer were injected over the HP chip at a flow rate of 30 μl/min, respectively. After each run, dissociation and regeneration were performed as described above. For each set of competition experiments, a control experiment (only protein without glycan) was performed to ensure the surface was completely regenerated.
2.6. Protein modeling
The 3D coordinates for the SGP trimer (NCBI reference sequence YP_009724390.1) were downloaded from the SWISS-MODEL homology modeling server (Waterhouse et al., 2018). The selected model was generated with the Cryo-EM structure PDB ID 6VSB as a template, which has a 99.26% sequence identity and 95% coverage for amino acids 27 to 1146. The template and resulting model is the “prefusion” structure with one of the three receptor binding domains (Chain A) in the “up” or “open” conformation (Wrapp et al., 2020). Cryo-EM studies have revealed that the SARS-CoV-2 SGP trimer exists in two conformational states in approximately equal abundance (Walls et al., 2020). In one state, all SGP monomers have their hACE2-binding domain closed, and in the other, one monomer has its hACE2-binding domain open, where it is positioned away from the interior of the protein.
2.7. Ligand docking
Initial coordinates for a hexasaccharide fragment of HS (GlcA(2S)-GlcNS(6S))3 were generated using the GAG-Builder tool (Singh et al., 2019) at GLYCAM-Web (glycam.org) and used for unbiased (blind) docking. A hexasaccharide was chosen as being sufficiently long to represent a typical GAG length found in protein co-complexes (Singh et al., 2019) and to avoid introducing so many degrees of internal flexibility that the efficiency of the docking conformational search algorithm was impaired. Docking was performed using a version of Vina-Carb (Nivedha et al., 2016) that has been modified to improve its performance for GAGs. A grid box with dimensions (x = 190, y = 223, z = 184 Å) was placed at the geometric center the protein enclosing its entire surface. Docking was performed with default values, with the following exceptions: exhaustiveness = 80, chi_cutoff = 2, and chi_coeff = 0.5. All sulfate and hydroxyl groups and glycosidic torsion angles were treated as flexible, resulting in 83 ligand poses.
3. Results
3.1. Kinetic measurements of CoV SGP-HP interactions
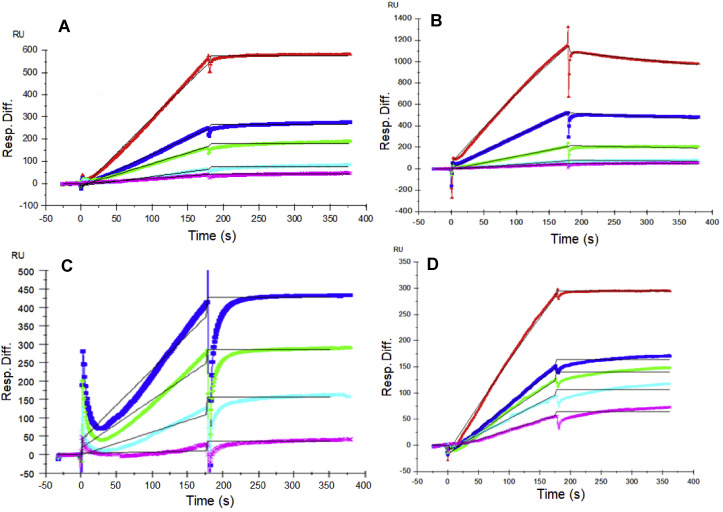
Previous reports showed that various CoVs bind GAGs through their SGPs to invade host cells (Belouzard et al., 2012). In the current study, we utilized SPR to measure the binding kinetics and interaction affinity of monomeric and trimeric SARS-CoV-2, monomeric SARS-CoV and MERS-CoV with SGP-HP using a sensor chip with immobilized HP. Sensorgrams of CoV SGP-HP interactions are shown in Fig. 2 . The sensorgrams were fit globally to obtain association rate constant (k a), dissociation rate constant (k d) and equilibrium dissociation constant (KD) (Table 1 ) using the BiaEvaluation software and assuming a 1:1 Langmuir model. SARS-CoV-2 and MERS CoV SGP exhibited a markedly low dissociation rate constant (k d ~10−7 1/s) suggesting excellent binding strength. The HP binding properties of monomeric SARS-CoV-2 SGP were comparable to that of the trimeric form (KD of monomer and trimer were 40 pM and 73 pM, respectively). In comparison, previously known HP binding SARS-CoV SGP showed nearly 10-fold lower affinity, 500 nM. The length of monomeric SARS-CoV-2 SGP (Sino Biological Inc.) spans subunit 1 containing proposed GAG-binding motifs 1 and 2 while trimeric SARS-CoV-2 SGP was expressed and characterized by Prof. Jason McLellan (Wrapp et al., 2020). Comparable KD values suggest that the folding of monomeric SGP is in the same correct conformation as the trimer. Trimeric SGP, surprisingly, does not possess greater binding affinity than that of monomer, this may be partly due to the monomer having both GAG-binding motif sites 1 and 2 while trimer has three of the GAG-binding site 1 and mutated site 2 from RRAR to GSAS (Wrapp et al., 2020).
Fig. 2.
SPR sensorgrams for binding kinetics/affinity measurements for SGP-HP interactions. (A) SARS-CoV-2 SGP (monomer), concentration of SGP (from top to bottom): 100, 50, 25, 12.5 and 6.25 nM. (B) SARS-CoV SGP, concentrations of SARS-CoV SGP (from top to bottom): 100, 50, 25, 12.5 and 6.25 nM. (C) MERS CoV SGP, concentrations of MERS CoV SGP (from top to bottom): 100, 50, 25, 12.5 and 6.25 nM. (D) SARS-CoV-2 SGP (trimer), concentration of SGP (from top to bottom): 800, 400, 200, 100 and 50 nM. The black curves are the fits using a 1:1 Langmuir model from BIAevaluate 4.0.1.
Table 1.
Summary of kinetic data of CoV SGP-HP interactionsa.
| Interaction | ka(M−1s1) | kd(1/s) | KD(M) |
|---|---|---|---|
| SARS-CoV-2 SGP (monomer) | 2.5 × 103 (±62.7) | 1.0 × 10−7 (±7.9 × 10−8) | 4.0 × 10−11 |
| SARS-CoV-2 SGP (trimer) | 1.6 × 103 (±127) | 1.2 × 10−7 (±5.5 × 10−8) | 7.3 × 10−11 |
| SARS-CoV SGP (monomer) | 4.12 × 104 (±136) | 4.01 × 10−4 (±6.49 × 10−6) | 5.0 × 10−7 |
| MERS-CoV SGP (monomer) | 339 (±27) | 3.5 × 10−7 (±2.6 × 10−6) | 1.0 × 10−9 |
The data with (±) in parentheses are the standard deviations (SD) from global fitting of five injections.
The extremely high binding affinity of SARS-CoV-2 SGP to HP was supported by the chip surface regeneration conditions. The immobilized HP surface could only be regenerated using a harsh regeneration reagent, 0.25% SDS, instead of the standard 2M NaCl solution used for removing HP-binding proteins. One reason for SARS-CoV-2 SGP's extremely high affinity to immobilized heparin is the high density of surface bound ligands might promote polyvalent interactions. The difference of binding kinetics and affinity of CoV SGPs to HP may also be due in part to the difference in protein sequence of the CoV SGPs. Based on amino acid alignment analysis using the Basic Local Alignment Search Tool (BLAST), SARS-CoV and SARS-CoV-2 SGPs share 76% similarity. Association rate constants (ka) for MERS-CoV SGP (339 (±27) 1/M−1s1) was the lowest, followed by monomeric and trimeric SARS-CoV-2 SGPs (2.5 × 103 (±62.7) M−1s−1 and 1.6 × 103 (±127) M−1s−1, respectively) (Table 1). SARS-CoV SGP had the highest Ka, which was 4.12 × 104 (±136) M−1s−1. The differences in ka values suggest a different mechanism when each SGP binds HP in addition to differences in binding strengths.
3.2. SPR solution competition study on the interaction between surface-bound HP and SARS-CoV-2 SGP to HP-derived oligosaccharides in solution
Solution/surface competition experiments were performed by SPR to examine the effect of the saccharide chain length of HP on the SARS-CoV-2 SGP-HP interaction. HP-derived oligosaccharides of different lengths, from tetrasaccharide (dp4) to octadecasaccharide (dp18), were used in these competition studies. The same concentration (1000 nM) of HP oligosaccharides were mixed in the SARS-CoV-2 SGP protein (50 nM)/HP interaction solution. Negligible competition was observed (Fig. S4) when 1000 nM of oligosaccharides (from dp4 to dp18) were present in the protein solution suggesting that the SARS-CoV-2 SGP-HP interaction is chain-length dependent and it prefers to bind full chain (~dp30) HP.
3.3. SPR solution competition study of different chemically modified HP derivatives and GAGs
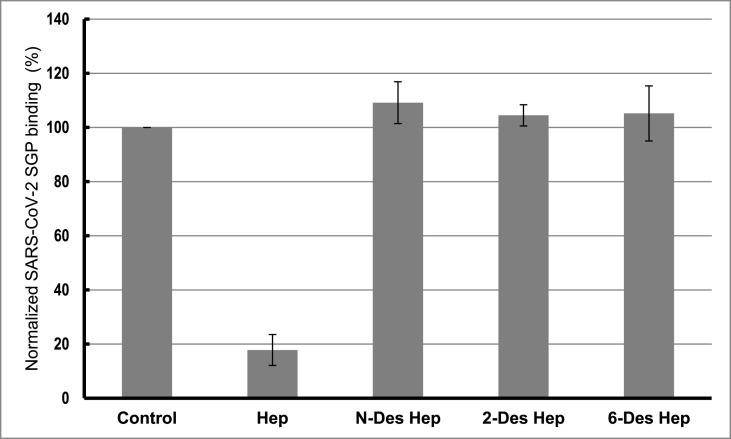
Competition levels measured by SPR for chemically modified HP derivatives are shown in Fig. 3 . The results of these studies demonstrate that all the chemically modified HPs, N-desulfated HP, 2-O-desulfated HP and 6-O-desulfated HP, were unable to compete with immobilized HP for binding to SARS-CoV-2 SGP-HP suggesting all the sulfate groups within HP have critical impact on this interaction.
Fig. 3.
Bar graphs of normalized SARS-CoV-2 SGP binding preference to surface HP by competing with different chemical modified HP in solution. Concentration was 50 nM for SARS-CoV-2 SGP and 1000 nM for different chemical modified HP. All bar graphs based on triplicate experiments.
SPR competition assay was also used to test the binding preference of SARS-CoV-2 SGP for various GAGs (Fig. S1), including various chondroitin sulfates, dermatan and keratan sulfates, and the results are shown in Fig. S5. Weak or no inhibitory activities were observed for all GAGs tested, suggesting that the binding of SARS-CoV-2 SGP protein to GAGs appears to be HP specific and greatly influenced by the level of sulfation within the GAG.
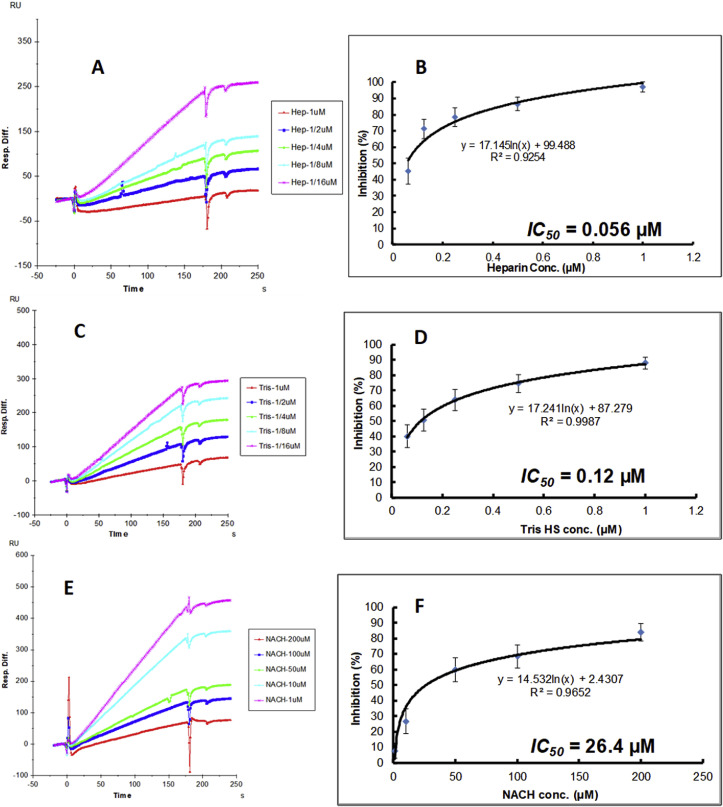
3.4. SPR solution competition dose response analysis of HP, tri-sulfated HS and NACH
Solution competition dose response analysis between surface immobilized HP and various soluble glycans (HP, non-anticoagulant trisulfated (TriS) HS, and non-anticoagulant low molecular weight HP (NACH)) was performed to calculate their IC50 values (Fig. 4 A–E). SARS-CoV-2 SGP protein (50 nM) samples were pre-mixed with different concentrations of glycans before injection into the HP chip. The sensorgrams (Fig. 4A, C, and 4E) show that once the active binding sites on SARS-CoV-2 SGP were occupied by glycans in solution, the binding of SARS-CoV-2 SGP to the surface-immobilized HP decreased resulting in a reduction of signal in a concentration dependent fashion. The IC50 values (concentration of competing analyte resulting in a 50% decrease in RU) were calculated from the plots SARS-CoV-2 SGP binding signal (normalized) vs. glycans concentration in solution (Fig. 4B, D, and 4F). The IC50 values of HP, TriS HS and NACH were 0.056 μM, 012 μM and 26.4 μM, respectively.
Fig. 4.
Inhibition analysis of glycans on the interactions between SARS-CoV-2 SGP and HP using SPR; SARS-CoV-2 SGP concentration was 50 nM. (A) Competition SPR sensorgrams of SARS-CoV-2 SGP-HP interaction inhibiting by different concentration of heparin. (B) Dose response curves for IC50 calculation of heparin using SARS-CoV-2 SGP inhibition data from surface competition SPR. (C) Competition SPR sensorgrams of SARS-CoV-2 SGP-HP interaction inhibiting by different concentration of TriS HS. (D) Dose response curves for IC50 calculation of TriS HS using SARS-CoV-2 SGP inhibition data from surface competition SPR. (E) Competition SPR sensorgrams of SARS-CoV-2 SGP-HP interaction inhibiting by different concentration of NACH. (F) Dose response curves for IC50 calculation of NACH using SARS-CoV-2 SGP inhibition data from surface competition SPR.
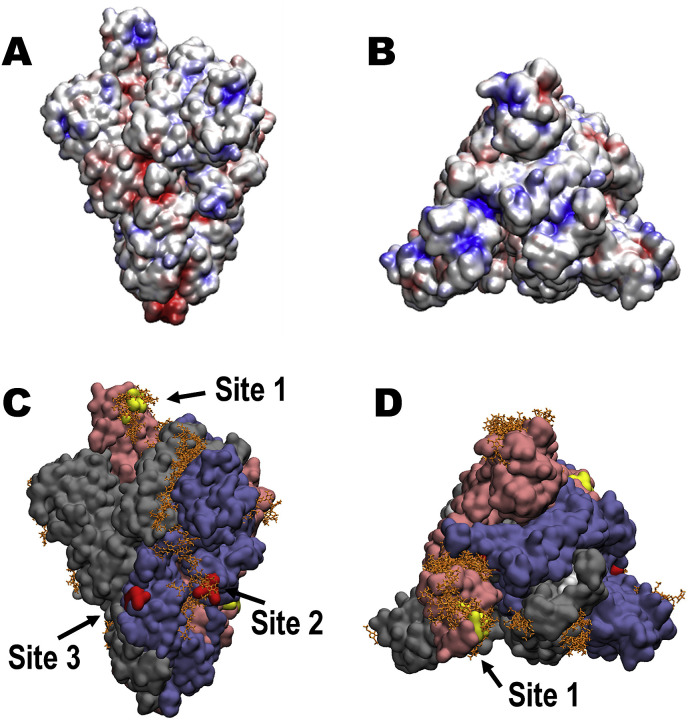
3.5. Identification of GAG-binding motifs by blinding docking analysis
Using a modified version of Autodock Vina tuned for use with carbohydrates (Vina-Carb) (Nivedha et al., 2016; Trott and Olson, 2009), we performed blind docking on the trimeric SARS-CoV-2 SGP model to discover objectively the preferred binding GAG-binding sites on the SGP protein surface. The SGP contains three putative GAG-binding motifs with the following sequences: 453–459 (YRLFRKS), 681–686 (PRRARS), and 810–816 (SKPSKRS), which we define as sites 1, 2, and 3, respectively (Fig. 1, S2, and S3). An HS hexasaccharide fragment (GlcA(2S)-GlcNS(6S)) binds site 2 in each monomer chain in the trimeric SGP (Fig. 5C and S3). The docking results also indicates that HS may bind to site 1 when the apex of the S1 monomer is in an open conformation, as this allows basic residues to be more accessible to ligand binding. The site 1 residues are less accessible for GAG binding when the domain is in a closed conformation (Fig. 5 D). The electrostatic potential surface representation of the trimeric SGP confirms that the GAG-binding poses generally prefer regions of positive charge, as expected, and illustrates that basic residues within site 3 are not exposed for binding to HS on any of the chains (Fig. 5A). Finally, our blind docking analysis reveals that a longer HS polymer may span an inter-domain channel that contains site 2.
Fig. 5.
Structure of trimeric SARS-CoV-2 SGP and proposed GAG-binding motifs. (A) Electrostatic potential surface (-ve charge (red) to + ve charge (blue)) computed with Chimera. (B) Electrostatic potential surface showing a top view of the SGP trimer. (C) Solvent accessible surface of the SARS-CoV-2 SGP trimer (pink (Chain A), grey (Chain B), blue (Chain C)) showing the predicted poses of HS hexasaccharides (orange) obtained from unbiased docking, and the three GAG-binding motifs (yellow (Chain A), white (Chain B), red (Chain C)), image generated with VMD (Humphrey et al., 1996). (D) Solvent accessible surface showing a top view of the SGP trimer. Amino acid sequences for GAG-binding motifs site 1, 2, and 3 are YRLFRKS, PRRARS, and SKPSKRS.
4. Discussion
The original SARS-CoV and numerous pathogens exploit host cell surface GAGs during the initial step of host cell entry (Agelidis and Shukla, 2020; Belouzard et al., 2012; Kamhi et al., 2013; Kim et al., 2017). Based on our initial discovery of GAG-binding and GAG-binding-like motifs at site 1 (within the RBD, Y453–S459), site 2 (at the proteolytic cleavage site at S1/S2 junction, P681–S686), and site 3 (at the S2’ proteolytic cleavage site, S810–S816), we hypothesized that SARS-CoV-2 may also interact with host cell surface GAGs through its SGPs to invade host cell (Fig. 1, S2, and S3). The predominant GAG in normal human lung is HS followed by CS (Frevert and Sannes, 2005) and it is noteworthy that lung tissue is rich in mast cells and has been a source of commercial HP (Guan et al., 2016). Using unbiased docking, we found that TriS HS hexasaccharide (GlcA(2S)-GlcNS(6S)) binds site 2 in each monomer chain in the trimeric SARS-CoV-2 SGP (Fig. 5C). TriS HS hexasaccharide may additionally bind site 1 when the SGP monomer is in an open conformation, but not site 3 where the basic residues are not accessible to the surface (Fig. 5C and D). Docking results indicated that the HS hexasaccharides could span an inter-domain channel that includes site 2, suggesting a mechanism for the binding of a longer HS sequence (Fig. 5C).
Next, we experimentally determined binding kinetics for the interactions between HP (rich (60–80%) in TriS domains) and monomeric SARS-CoV-2, trimeric SARS-CoV-2, monomeric SARS-CoV, and monomeric MERS-CoV SGPs using SPR binding assays (Fig. 2 and Table 1). GAG-protein interactions are mainly electrostatically driven (Hileman et al., 1998b), thus, HS-binding proteins generally bind HP due to its higher degree of sulfation (Lindahl et al., 2015). We discovered that HP binds both monomeric and trimeric SARS-CoV-2 SGP with remarkable affinity (KD = 40 pM and 73 pM, respectively) (Fig. 2 and Table 1). This was unexpectedly tight binding for a GAG-protein interaction as even one of one of the prototypical HP-binding proteins, fibroblast growth factor 2 (FGF2), has a KD of 39 nM (Ibrahimi et al., 2004). Additionally, high affinity may be partly due to polyvalent interactions that the high density of surface bound HP provides. In comparison, SARS-Cov and MERS-CoV SGPs also bind HP, however, much more weakly with binding strengths of KD = 500 nM and 1 nM, respectively (Fig. 2 and Table 1). While HS facilitates SARS-CoV host cell entry and is an essential host cell surface receptor, its involvement in MERS-CoV host cell entry or binding kinetics for SARS-Cov and MERS-CoV SGPs had not previously been reported (Lang et al., 2011).
After discovering the high binding affinity between HP and SARS-CoV-2 SGP, we next found that the degree and position of sulfation within HP was important for its successful binding to monomeric SARS-CoV-2 SGP (Fig. 3, Fig. 4). N-, 2-O, and 6-O-sulfation were all required for binding to SARS-CoV-2 SGP (Fig. 3). This was additionally demonstrated when competitive binding studies gave IC50 values of HP (0.056 μM), TriS HS (NS2S6S) (0.12 μM), and NACH (26.4 μM) for the inhibition of SARS-CoV-2 SGP binding to immobilized HP (Fig. 4). Both HP and TriS HS are sulfated at N-, 2-O-, and 6-O- positions and have approximately the same molecular weight (and chain length) but HP has additional 3-O-sulfation, responsible for its anticoagulant activity (Fig. S1). NACH lacks an intact antithrombin binding site, has a lower average molecular weight of 5 kDa than HP, and a higher content (>90%) of TriS (Islam et al., 2002).
The low IC50 of these GAGs suggest that the FDA approved anticoagulant HP, or its non-anticoagulant derivatives, might have therapeutic potential against SARS-CoV-2 infection as competitive inhibitors. The location of proposed GAG-binding sites is also of interest. Unlike SARS-CoV and MERS-CoV SGPs, SARS-CoV-2 SGP has a novel insert in the amino acid sequence (681–686 (PRRARS)) that fully follows GAG-binding Cardin-Weintraub motif (XBBXBX) and a furin-cleavage motif (BBXBB) at the S1/S2 junction (Fig. 1). This site was also shown to be a preferred GAG-binding motif by our unbiased docking study (Fig. 5). Proteolytic cleavage at S1/S2 is not required for successful viral-host cellular membrane fusion in SARS-CoV and MERS-CoV SGPs (Belouzard et al., 2009; Matsuyama et al., 2018). Transmembrane serine protease 2 (TMPRSS2) and cathepsin B/L have been recently confirmed as host cell proteases that SARS-CoV-2 exploits to prime for fusion activation and membrane fusion (Hoffmann et al., 2020). Proteolytic cleavage primes the SGP for fusion activation and may additionally influence cell-cell fusion, host cell entry, and/or the infectivity of the virus (Belouzard et al., 2012; Follis et al., 2006). Additionally, it is notable that the proteolytic cleavage motif PRRARS is almost identical to the HP binding sequence in fibronectin, PRRARV. The nucleocapsid protein of SARS-CoV induces apoptosis and actin reorganization partly through downregulation of fibronectin in mammalian cells under stressed conditions (Surjit et al., 2004). It also has been proposed that SARS-COV-2 SGP has acquired the general binding motif for integrin, one of fibronectin binding proteins for ECM maintenance, which facilitates virus transmission efficiency (Tresoldi et al., 2020).
Fig. 1.
Identification of GAG-binding motif within SARS-CoV-2, SARS-CoV, and MERS-CoV SGPs. Domains in SGP include signal peptide (SP), N-terminal domain (NTD), receptor-binding domain (RBD), fusion peptide (FP), heptad repeat 1/2 (HR 1/2).
Some CoVs possess both GAG-binding and furin cleavage motifs at their S1/S2 junction in their SGPs (Belouzard et al., 2012). In the case of MHV A59 SGP, a single amino acid mutation near the GAG-binding and furin cleavage motifs, resulted from a cell culture adaptation (known as MHV/BHK), determines whether a virion binds GAGs or exploits host cell surface protease, but not both (de Haan et al., 2008, 2005). While not within the CoV family, human immunodeficiency virus type 1 (HIV-1) requires HS-binding to achieve optimal furin processing because HS binding allows selective exposure of furin cleavage site on the envelope glycoprotein (Pasquato et al., 2007). It is of interest to further investigate potential crosstalk between host cell surface GAGs and cellular proteases during SARS-CoV-2 host cell entry.
COVID-19 hospitalized patients appear to be at increased risk of thrombotic complications and WHO recommends HP to reduce incidence of venous thromboembolism (Giannis et al., 2020; World Health Organization, 2020c). Additional studies report that soluble unfractionated heparin and its derivatives inhibit SARS-CoV-2 host cell entry in vitro (Partridge et al., 2020; Tandon et al., 2020). Nebulized Heparin is currently being evaluated in COVID-19 patients (Dixon et al., 2020) and a separate clinical study demonstrates the association between heparin and lower mortality in COVID-19 patients (Ayerbe et al., 2020). With additional randomized controlled trials in various therapeutic regimes (Ayerbe et al., 2020), HP may be an excellent candidate to be repurposed as prophylactic COVID-19 therapeutic.
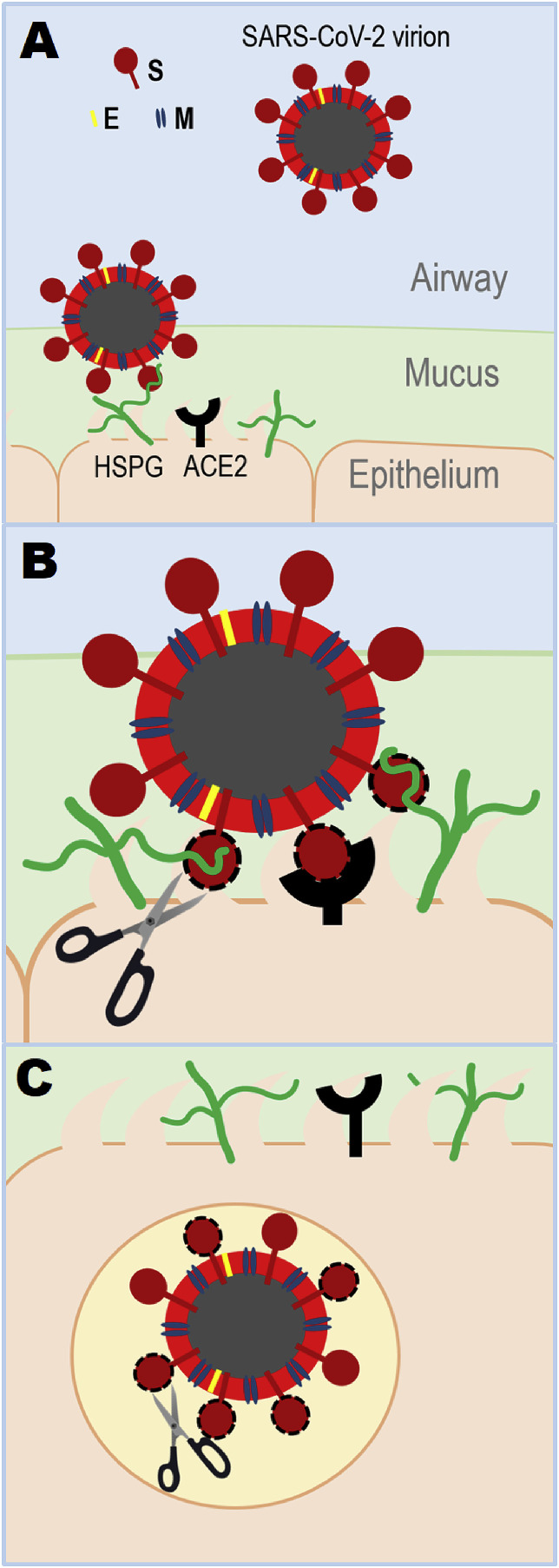
Based on our findings, we propose a model on how GAGs may facilitate host cell entry of SARS-CoV-2 (Fig. 6 ). First, virions land on the epithelial surface in the airway by binding to HS through their SGPs (Fig. 6A). Host cell surface proteoglycans utilize their long HS chains to securely wrap around the trimeric SGP (Fig. 6A). During this step, heavily sulfated HS chains span inter-domain channel containing GAG-binding site 2 on each monomer in the trimeric SGP and binds site 1 within the RBD in an open conformation (Fig. 5). Host cell surface and extracellular proteases, including TMPRSS2 (Hoffmann et al., 2020), may process site 2 (S1/S2 junction) and/or 3 (S2’) and GAG chains come off from site 2 upon cleavage (Fig. 6B). HS and an established host cell surface receptor ACE2 binding to more readily accessible RBD containing site 1 may drive conformational change of SGP and activate viral-cellular membrane fusion (Wrapp et al., 2020). A separate study reports that HP binding results in conformational change in RBD demonstrated by circular dichroism (Mycroft-west et al., 2020). Finally, SGP on the endocytosed virion may utilize an endosomal host cell protease, such as cathepsin B/L, to further execute viral-cellular membrane fusion (Hoffmann et al., 2020) (Fig. 6C). If taken an additional route of viral entry, receptor-dependent endocytosis, reported for SARS-CoV (Wang et al., 2008), concurrently endocytosed and recycled HSPGs may be further exploited by SARS-CoV-2.
Fig. 6.
Proposed model of SARS-CoV-2 host cell entry. SARS-CoV-2 surface is decorated with envelop (E), membrane (M), and SGP (Chan et al., 2020a). (A) Virion lands on host cell surface by binding to heparan sulfate proteoglycan (HSPG). (B) SGP goes through proteolytic digestion by host cell surface protease, which initiates viral-host cell membrane fusion by conformational change caused by host cell receptor binding (HSPG and ACE2). ACE2 is an established host cell surface receptor in SARS-CoV-2 host cell entry (Hoffmann et al., 2020; Wrapp et al., 2020). (C) Virion enters the host cell and may further experience proteolytic processing by endosomal host cell protease. In the case of receptor-dependent endocytic viral entry (Wang et al., 2008), virions may further utilize endocytosed and recycled HSPGs for their advantage.
In conclusion, we have characterized binding kinetics between GAGs and SARS-CoV-2 SGP and determined potential GAG-binding motifs within SARS-CoV-2 SGP in the current work. SPR studies demonstrate that both monomeric and trimeric SARS-CoV-2 SGP bind HP with remarkably high affinity and it prefers long, heavily sulfated (TriS rich) structures. Additionally, we reported low IC50 of HP and derivatives against HP and SARS-CoV-2 SGP interactions suggesting therapeutic potential of HP as COVID-19 competitive inhibitors. Lastly, unbiased computational ligand docking indicated that a TriS HS oligosaccharide preferably interacts with GAG-binding motifs at the S1/S2 junction and within receptor binding domain and hinted at mechanism of binding. This study provides groundwork for biological evaluation of GAGs as host cell surface receptors and facilitate designing of GAG-based COVID-19 therapeutics.
Acknowledgements
We appreciate Prof. Jason McLellan from University of Texas Austin for providing trimeric SARS-CoV-2 SGP. Additionally, we thank professor Lianchun Wang from University of South Florida for providing 6-O-desulfated HP derivative. This work was supported by the National Heart, Lung, and Blood Institute Grant K12 5K12HL141956-02 to S.Y.K. and M.M.F.; the National Institutes of Health Grants U01 CA221216 and U01 CA207824 to R.J.W.; and the National Institutes of Health Grants DK111958, CA231074, NS088496, and AG062344 to R.J.L.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2020.104873.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Agelidis A., Shukla D. Heparanase, heparan sulfate and viral infection. Adv. Exp. Med. Biol. 2020 doi: 10.1007/978-3-030-34521-1_32. [DOI] [PubMed] [Google Scholar]
- Ayerbe L., Risco C., Ayis S. The association between treatment with heparin and survival in patients with Covid-19. J. Thromb. Thrombolysis. 2020 doi: 10.1007/s11239-020-02162-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardin A.D., Weintraub H.J.R. Molecular modeling of protein-glycosaminoglycan interactions. Arterioscler. Thromb. Vasc. Biol. 1989;9:21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., Yuan S., Kok K.H., To K.K.W., Chu H., Yang J., Xing F., Liu J., Yip C.C.Y., Poon R.W.S., Tsoi H.W., Lo S.K.F., Chan K.H., Poon V.K.M., Chan W.M., Ip J.D., Cai J.P., Cheng V.C.C., Chen H., Hui C.K.M., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A.M., Haijema B.J., Schellen P., Schreur P.W., te Lintelo E., Vennema H., Rottier P.J.M. Cleavage of group 1 coronavirus spike proteins: how furin cleavage is traded off against heparan sulfate binding upon cell culture adaptation. J. Virol. 2008;82:6078–6083. doi: 10.1128/jvi.00074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A.M., Li Z., te Lintelo E., Bosch B.J., Haijema B.J., Rottier P.J.M. Murine coronavirus with an extended host range uses heparan sulfate as an entry receptor. J. Virol. 2005;79:14451–14456. doi: 10.1128/jvi.79.22.14451-14456.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon B., Smith R.J., Artigas A., Laffey J., McNicholas B., Schmidt E., Nunes Q., Skidmore M., de Lima A., Moran J.L., VanHaren F., Doig G., Ghosh A., Said S., Santamaria J.D. 2020. Can Nebuilsed Heparin Reduce Time to Extubation in SARS-CoV-2 (CHARTER Study) - Protocol. medRxiv. [DOI] [Google Scholar]
- Follis K.E., York J., Nunberg J.H. Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell-cell fusion but does not affect virion entry. Virology. 2006;350:358–369. doi: 10.1016/j.virol.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frevert C.W., Sannes P.L. Matrix proteoglycans as effector molecules for epithelial cell function. Eur. Respir. Rev. 2005;14:137–144. doi: 10.1183/09059180.05.00009703. [DOI] [Google Scholar]
- Fu L., Li G., Yang B., Onishi A., Li L., Sun P., Zhang F., Linhardt R.J. Structural characterization of pharmaceutical heparins prepared from different animal tissues. J. Pharmaceut. Sci. 2013;102:1447–1457. doi: 10.1002/jps.23501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannis D., Ziogas I.A., Gianni P. Coagulation disorders in coronavirus infected patients: COVID-19, SARS-CoV-1, MERS-CoV and lessons from the past. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104362. 104363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Xu X., Liu X., Sheng A., Jin L., Linhardt R.J., Chi L. Comparison of low-molecular-weight heparins prepared from bovine lung heparin and porcine intestine heparin. J. Pharmaceut. Sci. 2016;105:1843–1850. doi: 10.1016/j.xphs.2016.03.037. [DOI] [PubMed] [Google Scholar]
- Hileman R.E., Fromm J.R., Weiler J.M., Linhardt R.J. Glycosaminoglycan-protein interactions: definition of consensus sites in glycosaminoglycan binding proteins. Bioessays. 1998;20:156–167. doi: 10.1002/(SICI)1521-1878(199802)20:2<156::AID-BIES8>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Hileman R.E., Jennings R.N., Linhardt R.J. Thermodynamic analysis of the heparin interaction with a basic cyclic peptide using isothermal titration calorimetry. Biochemistry. 1998;37:15231–15237. doi: 10.1021/bi980212x. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:1–10. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I.C., Bosch B.J., Li F., Li W., Kyoung H.L., Ghiran S., Vasilieva N., Dermody T.S., Harrison S.C., Dormitzer P.R., Farzan M., Rottier P.J.M., Choe H. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 2006;281:3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Ibrahimi O.A., Zhang F., Hrstka S.C.L., Mohammadi M., Linhardt R.J. Kinetic model for FGF, FGFR, and proteoglycan signal transduction complex assembly. Biochemistry. 2004;43:4724–4730. doi: 10.1021/bi0352320. [DOI] [PubMed] [Google Scholar]
- Islam T., Butler M., Sikkander S.A., Toida T., Linhardt R.J. Further evidence that periodate cleavage of heparin occurs primarily through the antithrombin binding site. Carbohydr. Res. 2002;337:2239–2243. doi: 10.1016/S0008-6215(02)00229-X. [DOI] [PubMed] [Google Scholar]
- Kamhi E., Joo E.J., Dordick J.S., Linhardt R.J. Glycosaminoglycans in infectious disease. Biol. Rev. 2013;88:928–943. doi: 10.1111/brv.12034. [DOI] [PubMed] [Google Scholar]
- Kim S.Y., Zhang F., Gong W., Chen K., Xia K., Liu F., Gross R.A., Wang J.M., Linhardt R.J., Cotten M.L. Copper regulates the interactions of antimicrobial piscidin peptides from fish mast cells with formyl peptide receptors and heparin. J. Biol. Chem. 2018;293:15381–15396. doi: 10.1074/jbc.RA118.001904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., Zhao J., Liu X., Fraser K., Lin L., Zhang X., Zhang F., Dordick J.S., Linhardt R.J. Interaction of zika virus envelope protein with glycosaminoglycans. Biochemistry. 2017;56:1151–1162. doi: 10.1021/acs.biochem.6b01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang J., Yang N., Deng J., Liu K., Yang P., Zhang G., Jiang C. Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PloS One. 2011;6 doi: 10.1371/journal.pone.0023710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., Wong S.K., Huang I.C., Xu K., Vasilieva N., Murakami A., He Y., Marasco W.A., Guan Y., Choe H., Farzan M. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl U., Couchman J., Kimata K., Esko J.D. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; 2015. Proteoglycans and sulfated glycosaminoglycans. [DOI] [Google Scholar]
- Linhardt R.J., Dordick J.S., Deangelis P.L., Liu J. Enzymatic synthesis of glycosaminoglycan heparin. Semin. Thromb. Hemost. 2007;33:453–465. doi: 10.1055/s-2007-982076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Shirato K., Kawase M., Terada Y., Kawachi K., Fukushi S., Kamitani W. Middle East respiratory syndrome coronavirus spike protein is not activated directly by cellular furin during viral entry into target cells. J. Virol. 2018;92:1–12. doi: 10.1128/jvi.00683-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mycroft-west C.J., Su D., Pagani I., Rudd T.R., Elli S., Guimond S.E., Miller G., Meneghetti M.C.Z., Nader H.B., Li Y., Nunes Q.M., Procter P., Mancini N., Clementi M., Forsyth N.R., Turnbull J.E., Guerrini M., Fernig D.G., Vicenzi E., Yates E.A., Lima M.A., Skidmore M.A. Heparin inhibits cellular invasion by SARS-CoV-2: structural dependence of the interaction of the surface protein (spike) S1 receptor binding domain with heparin. 2020. bioRxiv Prepr. [DOI] [PMC free article] [PubMed]
- Nivedha A.K., Thieker D.F., Makeneni S., Hu H., Woods R.J. Vina-carb: improving glycosidic angles during carbohydrate docking. J. Chem. Theor. Comput. 2016;12:892–901. doi: 10.1021/acs.jctc.5b00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L.J., Green L.R., Monk P.N. 2020. Unfractionated Heparin Potently Inhibits the Binding of SARS-CoV-2 Spike Protein to a Human Cell Line. bioRxiv. [DOI] [Google Scholar]
- Pasquato A., Dettin M., Basak A., Gambaretto R., Tonin L., Seidah N.G., Di Bello C. Heparin enhances the furin cleavage of HIV-1 gp160 peptides. FEBS Lett. 2007;581:5807–5813. doi: 10.1016/j.febslet.2007.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki A., Fouchier R.A.M., Thiel V., Drosten C., Rottier P.J.M., Osterhaus A.D.M.E., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Montgomery D., Xue X., Foley B.L., Woods R.J. GAG Builder: a web-tool for modeling 3D structures of glycosaminoglycans. Glycobiology. 2019;29:515–518. doi: 10.1093/glycob/cwz027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Liu B., Jameel S., Chow V.T.K., Lal S.K. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochem. J. 2004;383:13–18. doi: 10.1042/BJ20040984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon R., Sharp J.S., Zhang F., Pomin V.H., Ashpole N.M., Mitra D., Jin W., Liu H., Sharma P., Linhardt R.J. 2020. Effective Inhibition of SARS-CoV-2 Entry by Heparin and Enoxaparin Derivatives. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresoldi I., Sangiuolo C.F., Manzari V., Modesti A. SARS-COV-2 and infectivity: possible increase in infectivity associated to integrin motif expression. J. Med. Virol. 2020;1–2 doi: 10.1002/jmv.25831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. Software news and updates AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd-Smith J.O., de Wit E., Munster V.J. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;1–3 doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;180:1–12. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., Jiang C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., De Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2020. WHO Director-General’s Opening Remarks at the Mission Briefing on COVID-19 -11 March 2020.https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-mission-briefing-on-covid-19---19-march-2020 [WWW Document]. URL. accessed 3.11.20. [Google Scholar]
- World Health Organization . 2020. Coronavirus Disease 2019 (COVID-19)https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports Situation Report. [WWW Document]. World Heal. Organ. URL. accessed 3.30.20. [Google Scholar]
- World Health Organization . 2020. Clinical Management of Severe Acute Respiratory Infection when Novel Coronavirus (2019-nCoV) Infection Is Suspected. [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 80. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates E.A., Santini F., Guerrini M., Naggi A., Torri G., Casu B. 1H and 13C NMR spectral assignments of the major sequences of twelve systematically modified heparin derivatives. Carbohydr. Res. 1996;294:15–27. doi: 10.1016/s0008-6215(96)90611-4. [DOI] [PubMed] [Google Scholar]
- Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020;5:335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.