To the Editor,
The coronavirus disease (COVID-19), which has created the recent pandemic, caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and has spread over to 227 countries globally. This virus has taken a heavy toll on human beings: as on date (3rd July 2020), an estimated 11.03 million people are infected and about 0.52 million have died from COVID-19. The infection and mortality rate in different countries are seemingly different. The genetic makeup of the subject might be playing a role in susceptibility to COVID-19 disease or poor prognosis. Recently, Delanghe et al. [1] highlighted the association of angiotensin-converting enzyme 1 (ACE-1) genetic variant with susceptibility to SARS-CoV-2 infection and related mortality in some of the European countries. Thus, the role of biochemical receptors in inducing susceptibility to SARS-CoV-2 infection and death cannot be ruled out. C-C Chemokine Receptor 5 (CCR5) is an essential member of the G protein-coupled receptor family abundantly present on the surface of monocytes, T cell, and macrophages. CCR5 is known to be responsible for the induction of inflammation in a wide range of infectious diseases and recruit leukocytes towards inflammation sites [2]. The critical role of CCR5 has been elegantly described in a wide range of viral infections: Human Immunodeficiency Virus (HIV), Hepatitis C Virus (HCV) and Hepatitis B Virus (HBV), West Nile Virus (WNV), Tick-Borne Encephalitis Virus (TBEV) [2]. Differential surface expression of CCR5has been linked with susceptibility/resistance against viral diseases. CCR5 gene is located at the short arm (p.21) of chromosome 3. A common 32 bp deletion variant at the coding region leads to the creation of a premature stop codon and produce 215 amino acids length instead of a full length of 352 amino acids. CCR5 Δ32 variant produces truncated protein and significantly diminish surface expression of the receptor [3]. The CCR5Δ32 polymorphism has been reported worldwide (https://www.ncbi.nlm.nih.gov/snp/rs333#frequency_tab, accessed on 29th June 2020).
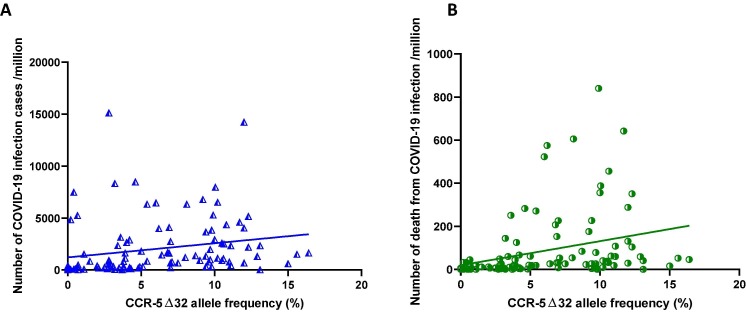
As the CCR5 plays an essential role in the pathogenesis of various viral infections and the Δ32 variants regulate the surface expression, we attempted to address a preliminary question, i.e. “Is the differential infection and mortality rate with COVID-19 worldwide correlated with the distribution of CCR5 Δ32 mutant?”. Accordingly, data of COVID-19 disease and mortality rate per million of inhabitants were obtained from the website (https://www.worldometers.info/coronavirus/, accessed on 29th June 2020). The prevalence of CCR5 Δ32 allele in healthy controls from 107 countries was obtained from an earlier publication [4] and PubMed search. The data (CCR-5 Δ32 allele frequency, rate of COVID-19 infections and deaths per million) were subjected to Spearman Rank Correlation test at α = 0.0001 level. A significant positive correlation was observed between COVID-19 infection rate/million (Spearman r = 0.4628, p < 0.0001, n = 107) and mortality rate/million of inhabitants (Spearman r = 0.5517, p < 0.0001, n = 107) with the frequency of CCR5 Δ32 allele (Fig. 1 ). Further, CCR5 Δ32 minor allele frequency and COVID-19 mortality rate in an African population revealed a positive correlation (Spearman r = 0.6210, p = 0.0045). These data and findings are indicative of an association of CCR5 Δ32 with susceptibility to SARS-CoV-2 infection and mortality. However, the mechanism of CCR5 Δ32 allele offering predisposition to SARS-CoV-2 infection susceptibility and death of the patient is not known. An earlier investigation on CCR5 deficient mice demonstrated the suppression of Th1 immune response and susceptibility to viral and bacterial infections [5], [6]. Higher incidence of HCV [7] and WNV [8] have been associated with the deletion allele of the CCR5 gene, further corroborating our observations. On the contrary, 32 bp deletion allele of the CCR5 gene is known to offer protection against HIV infection by hindering the entry of viruses inside the immune cells [9]. Earlier reports have demonstrated significant correlation of ACE-1 (D allele: Spearman r = -0.510 and p = 0.01) [1] and C3 (S allele:r2 = 0.480, p < 0.001) [10] with mortality due to SARS-CoV-2 infection. However, in the present study we observed a positive correlation on 55% of included population, projecting CCR5 Δ32 allele as an important genetic marker of SARS-CoV-2 related death.
Fig. 1.
Correlation of CCR-5 Δ32 minor allele prevalence with the number of COVID-19 infection cases/million and the number of patients death due to COVID-19/million worldwide. Prevalence of CCR-5 Δ32 mutant allele in healthy controls was searched from earlier published reports and correlated with COVID-19 infection (A) and death rate (B) per million throughout the globe. Each dot represented a country. A positive correlation was observed in the distribution of CCR-5 Δ32 minor allele with the number of cases (Spearman r = 0.4628, p < 0.0001, n = 107) and death (Spearman r = 0.5517, p < 0.0001, n = 107) from COVID-19. A total of one hundred seven countries were considered for the analysis based on availability of data. The list of countries are follows “Afghanistan, Albania, Algeria, Argentina, Armenia, Australia, Austria, Azerbaijan, Bahrain, Bangladesh, Belarus, Belgium, Bosnia and Herzegovina, Brazil, Bulgaria, Burkina Faso, Cameroon, Canada, Chile, China, Colombia, Croatia, Cuba, Cyprus, Czech Republic, DR Congo, Denmark, Dominican Republic, Ecuador, Egypt, El Salvador, Eritrea, Estonia, Ethiopia, Faeroe Islands, Finland, France, Georgia, Germany, Ghana, Greece, Guinea, Hong Kong, Hungary, India, Indonesia, Iran, Iraq, Ireland, Israel, Italy, Ivory Coast, Jamaica, Japan, Jordan, Kazakhstan, Kenya, Kosovo, Kyrgyzstan, Latvia, Lebanon, Lithuania, Luxembourg, Malawi, Malta, Mexico, Moldova, Mongolia, Montenegro, Morocco, Netherlands, New Zealand, Nigeria, Norway, Oman, Pakistan, Papua New Guinea, Peru, Philippines, Poland, Portugal, Romania, Russia, Rwanda, Saudi Arabia, Senegal, Serbia, Slovakia, Slovenia, Somalia, South Africa, South Korea, Spain, Sri Lanka, Sweden, Switzerland, Syria, Taiwan, Thailand, Tunisia, Turkey, Ukraine, United Arab Emirates, United Kingdom, United State, Venezuela, Vietnam”.
Although the present report highlighted a significant association of CCR5 Δ32 variant with susceptibility and mortality from SARS-CoV-2 infection, it has set the stage for in-depth analysis by factoring in various other aspects. Inclusion of other genetic polymorphisms of the CCR5 gene can further highlight the role of CCR5 in COVID-19 pathogenesis. Additionally, other parameters such as testing capacity, inter-country movement frequency, health policy of the regional government, the density of the population, demographic age profile of the infected cases, various co-morbidities phenotypes [11] can place crucial role in enhancing the understanding and strengthening the present analysis. In conclusion, COVID-19 infection and mortality are associated with CCR5 Δ32 allele, and population-based genetic association studies in different cohorts are required to validate our findings.
Funding
AKP is supported by the DST-INSPIRE Faculty grant (IFA12/LSBM-46) from the Department of Science and Technology, Government of India, New Delhi.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Delanghe J.R., Speeckaert M.M., De Buyzere M.L. ACE Ins/Del genetic polymorphism and epidemiological findings in COVID-19. Clin. Chem. Lab. Med. 2020;58(7):1129–1130. doi: 10.1515/cclm-2020-0605. [DOI] [PubMed] [Google Scholar]
- 2.Klein R.S. A moving target: the multiple roles of CCR5 in infectious diseases. J. Infect. Dis. 2008;197(2):183–186. doi: 10.1086/524692. [DOI] [PubMed] [Google Scholar]
- 3.Mummidi S., Ahuja S.S., McDaniel B.L., Ahuja S.K. The human CC chemokine receptor 5 (CCR5) gene. Multiple transcripts with 5'-end heterogeneity, dual promoter usage, and evidence for polymorphisms within the regulatory regions and noncoding exons. J. Biol. Chem. 1997;272(49):30662–30671. doi: 10.1074/jbc.272.49.30662. [DOI] [PubMed] [Google Scholar]
- 4.Solloch U.V., Lang K., Lange V., Bohme I., Schmidt A.H., Sauter J. Frequencies of gene variant CCR5-Delta32 in 87 countries based on next-generation sequencing of 1.3 million individuals sampled from 3 national DKMS donor centers. Hum. Immunol. 2017;78(11–12):710–717. doi: 10.1016/j.humimm.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Nansen A., Christensen J.P., Andreasen S., Bartholdy C., Christensen J.E., Thomsen A.R. The role of CC chemokine receptor 5 in antiviral immunity. Blood. 2002;99(4):1237–1245. doi: 10.1182/blood.v99.4.1237. [DOI] [PubMed] [Google Scholar]
- 6.Dawson T.C., Beck M.A., Kuziel W.A., Henderson F., Maeda N. Contrasting effects of CCR5 and CCR2 deficiency in the pulmonary inflammatory response to influenza A virus. Am. J. Pathol. 2000;156(6):1951–1959. doi: 10.1016/S0002-9440(10)65068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahlenstiel G., Woitas R.P., Rockstroh J., Spengler U. CC-chemokine receptor 5 (CCR5) in hepatitis C–at the crossroads of the antiviral immune response? J. Antimicrob. Chemother. 2004;53(6):895–898. doi: 10.1093/jac/dkh239. [DOI] [PubMed] [Google Scholar]
- 8.Glass W.G., McDermott D.H., Lim J.K., Lekhong S., Yu S.F., Frank W.A., Pape J., Cheshier R.C., Murphy P.M. CCR5 deficiency increases risk of symptomatic West Nile virus infection. J. Exp. Med. 2006;203(1):35–40. doi: 10.1084/jem.20051970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNicholl J.M., Smith D.K., Qari S.H., Hodge T. Host genes and HIV: the role of the chemokine receptor gene CCR5 and its allele. Emerg. Infect. Dis. 1997;3(3):261–271. doi: 10.3201/eid0303.970302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delanghe J.R., De Buyzere M.L., Speeckaert M.M. C3 and ACE1 polymorphisms are more important confounders in the spread and outcome of COVID-19 in comparison with ABO polymorphism. Eur. J. Prevent. Cardiol. 2020 doi: 10.1177/2047487320931305. 2047487320931305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delanghe J.R., Speeckaert M.M., De Buyzere M.L. COVID-19 infections are also affected by human ACE1 D/I polymorphism. Clin. Chem. Lab. Med. 2020;58(7):1125–1126. doi: 10.1515/cclm-2020-0425. [DOI] [PubMed] [Google Scholar]