Abstract
Porcine epidemic diarrhea virus (PEDV) is responsible for the acute infectious swine disease porcine epidemic diarrhea (PED). PED causes damage to the intestine, including villus atrophy and shedding, leading to serious economic losses to the pig industry worldwide. We carried out an in vitro study to investigate cell apoptosis and the cell cycle in a PEDV-infected host using transcriptomic shotgun sequencing (RNA-Seq) to study gene responses to PEDV infection. Results revealed that the PEDV infection reduced proliferation activity, blocked the cell cycle at S-phase and induced apoptosis in IPEC-J2 cells. The expression of gene levels related to ribosome proteins and oxidative phosphorylation were significantly up-regulated post-PEDV infection. Although the significantly down-regulated on PI3K/Akt signaling pathway post-PEDV infection, the regulator–related genes of mTOR signaling pathway exerted significantly up-regulated or down-regulated in IPEC-J2 cells. These results indicated that ribosome proteins and oxidative phosphorylation process were widely involved in the pathological changes and regulation of host cells caused by PEDV infection, and PI3K/AKT and mTOR signaling pathways played a vital role in antiviral regulation in IPEC-J2 cells. These data might provide new insights into the specific pathogenesis of PEDV infection and pave the way for the development of effective therapeutic strategies.
Keywords: PEDV, IPEC-J2 cell, RNA-Seq, Cell cycle, Apoptosis
Highlights
-
•
PEDV infection reduced proliferation activity, blocked cell cycle and induced apoptosis in IPEC-J2.
-
•
PEDV infection significantly up-regulated the expression of ribosome proteins and oxidative phosphorylation process.
-
•
PI3K/Akt and mTOR signaling pathways played a vital role in modulating the effects of PEDV infection.
1. Introduction
Porcine epidemic diarrhea (PED) is an acute intestinal infectious disease caused by porcine epidemic diarrhea virus (PEDV) [1]. PEDV infection, which is characterized watery diarrhea, vomiting and dehydration, can affect pigs of all ages and may be fatal in newborn piglets [2]. PEDV, family Coronaviridae, was first reported in 1978 and subsequently erupted in Europe and Asia, with the first case of PEDV infection in the United States emerging in 2013 [3,4]. PED has thus spread worldwide leading to significant financial concerns in the global pork industry [5].
PEDV is an enveloped positive-strand RNA virus. Its genome contains seven open reading frames (ORFs) encoding three non-structural proteins (ORF1a, ORF1b, and ORF3) and four structural proteins (spike glycoprotein, envelope protein, membrane glycoprotein, and nucleocapsid protein) [6]. PEDV is mainly transmitted via the fecal-oral route and numerous virions are found in the intestines of infected pigs. Inactivated and attenuated vaccines can provide some protection against PEDV; however, the emergence of immunodeficiency or new variant strains may lead to the failure of traditional attenuated vaccines, especially in Asian countries after 2010 [7,8]. The intestinal epithelial cell monolayer plays a key role in preventing infection by pathogenic microorganisms [8]. However, intestinal epithelial cells are the main target cells of PEDV infection, and the virus then replicates in the small intestine villi. PEDV-infected pigs thus show reduced intestinal villus height and decreased transepithelial resistance [9].
Cell perception of various extracellular stimuli, such as viral infection, leads to the activation of specific intracellular signaling networks that then change the biological properties of the cells, such as cell proliferation, apoptosis, and the cell cycle. Kim et al. found that a caspase-independent mitochondrial programmed cell death protein 8 (apoptosis-inducing factor)-mediated pathway played a central role in PEDV-induced apoptosis, facilitating viral replication and pathogenesis [10]. Lee et al. showed that the JNK and p38 mitogen-activated protein kinase (MAPK) pathways also contributed to PEDV infection, while another study showed that activated p53 and the accumulation of reactive oxygen species participated in PEDV-induced apoptosis, and that p53 could be regulated by reactive oxygen species during PEDV infection [11]. Activated p38 MAPK and SAPK/JNK had no effect on PEDV-induced apoptosis [12]. Virus infection destroys the cell cycle by inducing a series of signal transduction pathways, which in turn play an important role in promoting viral replication. PEDV infection causes cell cycle arrest in the G0/G1 phase via a p53-dependent pathway. Other studies have also shown that the PEDV ORF3, envelope protein, and membrane glycoprotein can prolong and block the cell cycle in S phase [13,14].
Although the understanding of the molecular virology, epidemiology, and vaccinology of PEDV has improved greatly since the reemergence of these strains, the molecular mechanism of the host cell response to PEDV infection remains largely unclear, especially in terms of the pathological changes in the biological characteristics of the host cells. Furthermore, most studies of the pathogenesis and molecular mechanism of PEDV infection of host cells to date have been based on African green monkey kidney cells (Vero cells). However, several recent studies have identified the IPEC-J2 non-transformed porcine jejunal epithelial cell line as a suitable model system for PEDV infection [[15], [16], [17]].
In the present study, we determined the effect of PEDV infection on apoptosis and the cell cycle in porcine IPEC-J2 cells, and investigated the mechanisms involved in the cell-virus interactions by RNA-Seq transcriptomic assay.
2. Materials and methods
2.1. Cells and viruses
The porcine small intestinal epithelial cell line IPEC-J2 was obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA) and cultured in Dulbecco's Modified Eagle's Medium (Gibco BRL, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT, USA) and 1% penicillin-streptomycin. Cells were incubated at 37 °C with 5% CO2. The classical PEDV CV777 strain (GenBank: KT323979.1) was kindly provided by the Jiangsu Academy of Agricultural Sciences.
2.2. Indirect immunofluorescence assay for detection of PEDV
IPEC-J2 cells were grown in 12-well plates and infected with CV777 at a multiplicity of infection (MOI) of 1.0. The cells were washed with phosphate-buffered saline (PBS) at 24, 48, and 72 h post infection (hpi), fixed with 4% paraformaldehyde for 15 min, and then permeabilized with 0.2% Triton X-100 for 10 min. The cells were then incubated with 1:200 anti-PEDV N protein mouse monoclonal antibody (prepared and stored in our laboratory) for 12 h at 4 °C, washed, and incubated with fluorescein isothiocyanate-conjugated goat-anti-mouse IgG (1:500) (Boster, China) secondary antibody for 1 h at 37 °C. The cells were then stained with DAPI (1 μg/mL) (Beyotime Biotechnology, China), the sample was covered for 15min, and then washed three times with PBS for 3 min each time. Cell staining was observed by fluorescence microscopy.
2.3. Cell viability assay
IPEC-J2 cells were grown in 96-well plates (1 × 104 cells/well) and infected with CV777 at an MOI of 1.0. Control cells were treated with the same dose of culture medium. MTT (20 μL, 5 mg/mL) was added to each well at 24, 48, and 72 hpi. The culture medium was removed after 4 h of incubation, and 50 μL dimethyl sulfoxide was added. After mixing, the absorbance was measured at 490 nm using a 550 Microplate Reader (Bio-Rad, CA, USA). Experiments were repeated three times, with twelve samples taken at each time point.
2.4. Cell cycle assessment by flow cytometry
IPEC-J2 cells were grown in 6-well plates (1 × 106 cells/well) and infected with CV777 at an MOI of 1.0. Control cells were treated with the same dose of culture medium. Cells were trypsinized at 24, 48, and 72 hpi, harvested, and fixed with 1 mL 80% pre-cooled ethanol in test tubes and incubated for 2 days at 4 °C. The cells were then centrifuged at 500×g for 5 min and the cell pellets were resuspended in PBS containing 5 U RNase and 50 mg/mL of propidium iodide (PI). The cells were then incubated on ice for 30 min in the dark. Cell cycle distribution was calculated from 10,000 cells and determined using a Coulter Epics XL flow cytometer (Beckman Coulter, CA, USA).
2.5. Cell apoptosis assessment by CCK8 assay and fluorescent staining
IPEC-J2 cells were grown in 96-well plates (1 × 104 cells/well) in 100 μL culture medium and infected with CV777 at an MOI of 1.0. Control cells were treated with the same dose of culture medium. After washing with PBS, the cells were further cultured in serum-free medium. CCK-8 solution (Beyotime Biotechnology) (10 μL) was added to each well at 24, 48, and 72 hpi and incubated 37 °C for 2 h in a cell culture incubator. The absorbance was measured at 450 nm using a 550 Microplate Reader (Bio-Rad). Experiments were repeated three times, with twelve samples taken at each time point.
2.6. Cell apoptosis detection by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay
IPEC-J2 cells were grown on microscope cover slips placed in 6-well tissue culture plates and mock-infected or infected with PEDV at an MOI of 1.0.The virus-infected cells were fixed at 48 hpi with 4% paraformaldehyde for 25 min at 4 °C and permeabilized with 0.2% TritonX-100 in PBS at room temperature for 5 min. Cell samples were rinsed twice with PBS, and the TUNEL reaction mixture (Beyotime Biotechnology) was added and incubated for 60 min at 37 °C, followed by three washes with PBS. TUNEL-labeled cells were subjected to immunofluorescence assay using anti-PEDV N protein mouse monoclonal antibody and goat anti-mouse antibody, as described above. After sealing with an anti-fluorescence quenching liquid, the samples were mounted on a fluorescence microscope and examined at an excitation wavelength of 550 nm and emission wavelength of 570 nm (red fluorescence).
2.7. Cell apoptosis detection by annexin V and PI staining assay
IPEC-J2 cells were grown in 6-well tissue culture plates and mock-infected or infected with PEDV at an MOI of 1.0.Mock- and PEDV-infected cells were then harvested and washed with cold PBS at 24, 48, and 72 hpi. Cell apoptosis was determined using an AlexaFluor488 AnnexinV/Dead Cell Apoptosis kit (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions. The cells were suspended in 100 μL annexin-binding buffer, and then incubated with AlexaFluor488-conjugated AnnexinV and PI at room temperature for 15 min in the dark. After incubation, 400 μL annexin-binding buffer was added to each sample and mixed gently on ice. The samples were analyzed using a Coulter Epics XL flow cytometer (Beckman Coulter) and Kaluza software.
2.8. RNA-Seq transcriptomic assay
IPEC-J2 cells were grown in 25 cm2 cell culture flasks and infected with CV777 at an MOI of 1.0. Control cells were treated with the same dose of culture medium. Three replicates were used for each group. Total RNA was extracted from the cells using TRIzol reagent according the manufacturer's instructions (Invitrogen) and genomic DNA was removed using DNase I (TaKaRa, Japan). cDNA was synthesized using a Super Script double-stranded cDNA synthesis kit (Invitrogen) and sequenced by Shanghai Major bioBiopharm Technology Co. Ltd. (Shanghai, China) using an Illumina HiSeq2500 system (Illumina, CA, USA). Differential expression analysis was carried out using EdgeR software [18]. Differences in expression levels between groups were considered significant after adjusting for multiple testing based on a q value < 0.05. We first filtered the genes based on q < 0.05 and an absolute difference > ±2-fold (i.e., log2 change > ±1.0). DEG enrichment was analyzed using Gene Ontology (GO) tools (https://github.com/tanghaibao/GOatools), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was carried out using KOBAS (http://kobas.cbi.pku.edu.cn/home.do) [19].
2.9. Real-time quantitative PCR (qPCR) validation
Total RNA was extracted from infected and control IPEC-J2 cells (8 repetitions per group) using TRIzol reagent (Invitrogen) and cDNA was synthesized using a Revert Aid kit (Thermo Scientific, USA) according to the manufacturer's instructions. Thirteen of the most significantly differentially expressed genes between the PEDV infection and control groups according to RNA sequencing data (log2-fold change; P < 0.05, false discovery rate > 95%) were selected for real-time qPCR. The primer pairs for qPCR were designed using the assembled RNA-Seq sequence data (Table 1 ). qPCR was carried out with a SYBR Green PCR Master Mix Kit (Toyobo, Osaka, Japan) using an ABI7500 Detection System (Applied Biosystems, USA). Two replicates per sample were performed for each qPCR assay and β-actin was used as the reference gene for normalization. The relative expression of each gene was calculated according to the comparative threshold cycle (Ct) method by 2−ΔΔCt: ΔΔCt = (Ctij−Ctβ-actin j)−(Cti1−Ctβ-actin1), where Ctij and Ctβ-actin j are the Ct for gene i and for β-actin in a sample (named j), and Cti1 and Ctβ-actin1 are the Ct in sample 1, expressed as the standard.
Table 1.
Sequences of the PCR primers used in this study.
| Gene | Forward primer | Reverse primer | Product (bp) |
|---|---|---|---|
| β-actin | CTCTTCCAGCCCTCCTTCC | GGTCCTTGCGGATGTCG | 97 |
| TNFAIP8L2 | CATGACGGTGCTCAGCTTTA | GGTGAAGTCAGGCCCATAGA | 202 |
| TMEM74B | TTGTTTCTGCCCTGGTTTTC | GCACAGAGAGACCATCAGCA | 233 |
| TMEM40 | CAAGACCACAGTCAGGTCCA | AATGTCAGGCTCTCCATGCT | 189 |
| TTC5 | TCACCCATTGCAAGAACAAA | GGGTTCTGGCCAGTATTGAA | 211 |
| RGCC | GCCACTTCCACTACGAGGAG | TGTCTCCGAGTTTGGCTTTC | 213 |
| KCNU1-5 | GAAGGCAGAGTTCCCGTCTT | TTCTTCCCTTTGACCACTTCA | 250 |
| MYPN | TGGTCATTGCCGAGGTATTT | TGGAGTTGGCTGAGTGAAGA | 150 |
| GPX8 | GGCTTTTCCATGCAATCAGT | CCCTCAGGGTTGACCAGATA | 213 |
| GJB4 | GCATCCTGCTCAACCTCAGT | GAAGGCAGAGTTCCCGTCTT | 167 |
| COL23A1 | TTGACTACGATGGCAGGATCT | TCTCCTTTGGGTCCGATTG | 193 |
| COL16A1 | CGTGACAGGCTTCAATCTCA | TCCATCGGTCACTTGAAACA | 235 |
| CRBN | TGTGTTGCCTTCAACCATGT | CATCCCATTCACGTAGCTGTT | 224 |
| ADGRD1 | GCACACACCTCACCAACTTC | GACAGAGAGCAGCCGATGTA | 109 |
2.10. Statistical analysis
Statistical analyses were carried out using Prism software and expressed as the mean ± standard deviation. Differences among multiple groups were analyzed by one-way analysis of variance followed by the Student-Newman-Keuls test. Differences were considered significant at P < 0.05.
2.11. Accession number
The sequence was deposited in the GenBank database (Accession number PRJNA554982).
3. Results
3.1. Influence of PEDV infection on proliferation of IPEC-J2 cells
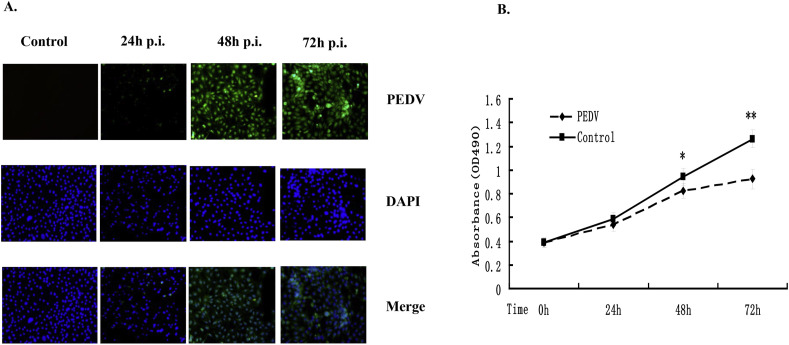
Cell proliferation activity gradually reduced in cell infected with PEDV at 1 MOI compared with the control group (Fig. 1 A and B). Cell viability decreased with increasing infection time, and cell growth was significantly inhibited at 48 and 72 hpi (Fig. 1A and B), indicating that PEDV inhibited IPEC-J2 cell growth in a time-dependent manner.
Fig. 1.
Influence of PEDV infection on proliferation of IPEC-J2 cells. (A) IPEC-J2 cells were grown in a 12-well plates and infected with CV777 at an MOI of 1.0, and maintained for 24, 48, and 72 h. For immunostaining, the infected cells were fixed at 24, 48, and 72 hpi and incubated with anti-PEDV N protein mouse monoclonal antibody followed by Alexa green-conjugated goat anti-mouse secondary antibody (first row). The cells were then counterstained with DAPI (second row) and examined using a fluorescence microscope at 100× magnification. Third row is a merged image. (B) IPEC-J2 cells at 1 × 104 cells/well in 96-well plates were infected with PEDV at an MOI of 1.0 or mock-infected and MTT assays were performed at various times post-infection. Data presented as absorbance at 490 nm in mock- and PEDV-infected cells at 24, 48, and 72 hpi. Data based on three repeated experiments. *P < 0.05, **P < 0.01 versus mock infection.
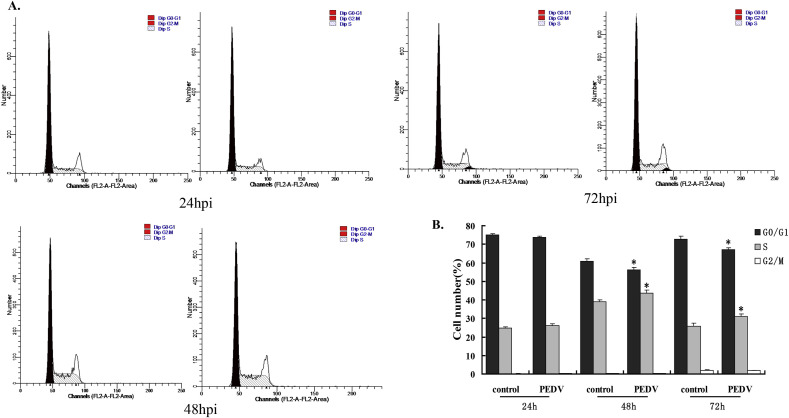
3.2. Influence of PEDV infection on IPEC-J2 cell cycle
Cell cycle disorders are known to be a common response to many infections. We performed flow cytometry analysis based on the DNA content in nuclei stained with PI to determine if the effect of PEDV infection on the growth of IPEC-J2 cells was due to cell cycle arrest at a certain phase of the cell cycle (Fig. 2 A). There were no significant differences in the proportions of G0/G1, S, and G2/M phases between PEDV-infected and control cells at 24 h. The proportions of cells in G0/G1 in the PEDV-infected group at 48 and 72 hpi were significantly lower than in the control group (56.05% versus 60.85% and 67.01% versus 72.63%, respectively), while the proportions in S-phase were significantly higher (43.67% versus 38.91% and 31.09% versus 25.87%, respectively). There was no significant difference in the proportions of G2/M phase cells between the groups at any time point. Further quantitative analysis of the histograms was performed to determine the percentages of cells in each phase (Fig. 2B), where G0/G1 phase cells had diploid DNA content and G2/M phase cells showed tetraploid DNA content. PEDV infection caused a significant increase in the proportion of cells in S phase accompanied by a decrease in G0/G1 phase. Overall, these results revealed that PEDV infection induced cell cycle block in the S phase of the cell cycle.
Fig. 2.
Cell cycle distribution in control and PEDV-infected cells by flow cytometry. (A) Flow cytometry analysis of cells following PI staining. (B) Percentage of cells in each phase of the cell cycle based on flow cytometry data. Results given as mean ± standard deviation of three independent experiments. *P < 0.05 S phase in PEDV-infected vs control cells.
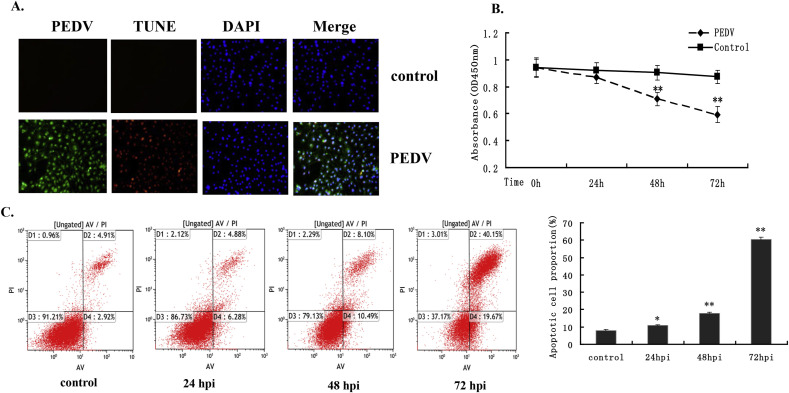
3.3. PEDV infection induced IPEC-J2 cell apoptosis
We investigated the effect of PEDV infection on apoptotic cell death by TUNEL assay. TUNEL-labeled cells were only observed among virus-infected cells and not among control cells (Fig. 3 A). PEDV-induced cell apoptosis was further confirmed by CCK8 assay. The absorbance of PEDV-infected cells was significantly decreased at 48 and 72 hpi compared with mock-infected cells (Fig. 3B). The process of PEDV-induced apoptosis was then assessed using annexinV/PI flow cytometry. Virus-or mock-infected cells were stained with annexinV and PI and then examined using fluorescence-activated cell sorting (FACS) flow cytometry to quantitatively determine the percentage of apoptotic cells. PEDV infection significantly increased cell apoptosis at 24, 48 and 72 hpi, and the percentage of apoptotic cells increased with infection time, reaching a maximum of 60.43% at 72 hpi (Fig. 3C). Taken together, these data demonstrate that PEDV infection induced IPEC-J2 cell apoptosis.
Fig. 3.
Cell apoptosis analysis by CCK8 assay. (A) TUNEL labeling of mock- and PEDV-infected cells. Mock- and PEDV-infected cells were fixed at 48 hpi and subjected to TUNEL assay (red), followed by staining with anti-PEDV-N antibody (green). The cells were then counterstained with DAPI and observed under a confocal microscope at 400× magnification. In merged images, all TUNEL-positive cells were localized within the nuclei of the corresponding PEDV-infected cells. (B) IPEC-J2 cells at 1 × 104 cells/well in 96-well plates were mock-infected of infected with PEDV at an MOI of 1.0. CCK8 assays were performed at various times post-infection. Data presented as absorbance at 450 nm in mock- and PEDV-infected cells at 24, 48, and 72 hpi. Data based on three repeated experiments. *P < 0.05, **P < 0.01 versus mock infection. (C) Cell apoptosis analysis by flow cytometry with dual annexinV-PI cell labeling. PEDV-infected cells collected at different time periods were dual-labeled with annexinV and PI and analyzed by FACS. Lower left quadrants represent intact cells (annexinV−/PI−); lower right quadrants represent early apoptotic cells (annexinV+/PI−); upper right quadrants indicate late apoptotic cells (annexinV+/PI−); upper left quadrants indicate necrotic cells (annexinV−/PI+). Figure representative of three independent experiments. Graph on the right represents the percentage of apoptotic cells. *P < 0.05, **P < 0.01 versus mock infection.
3.4. Comparing significantly differential gene expressions (DEGs) of infection stages with the control group
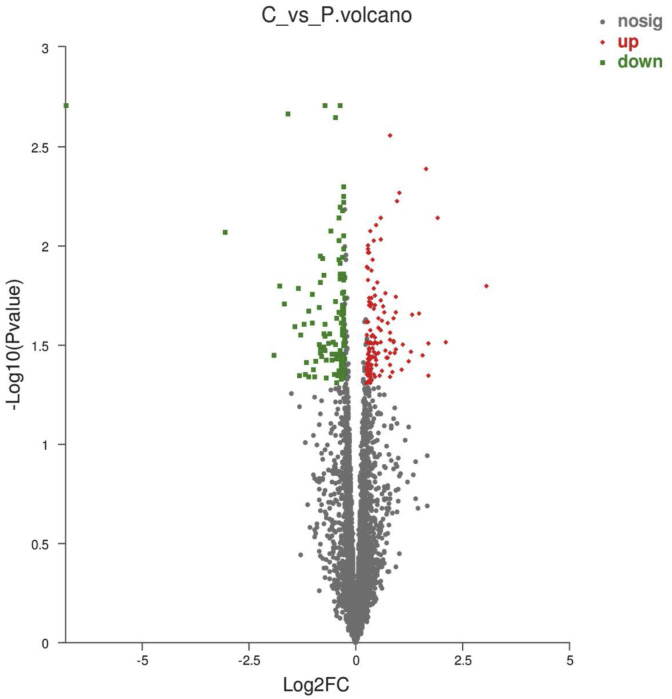
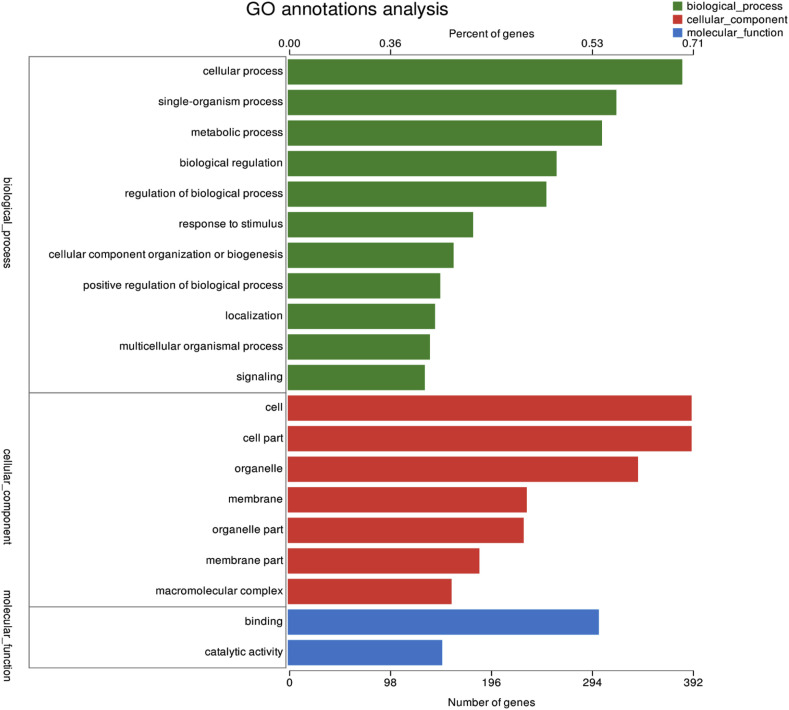
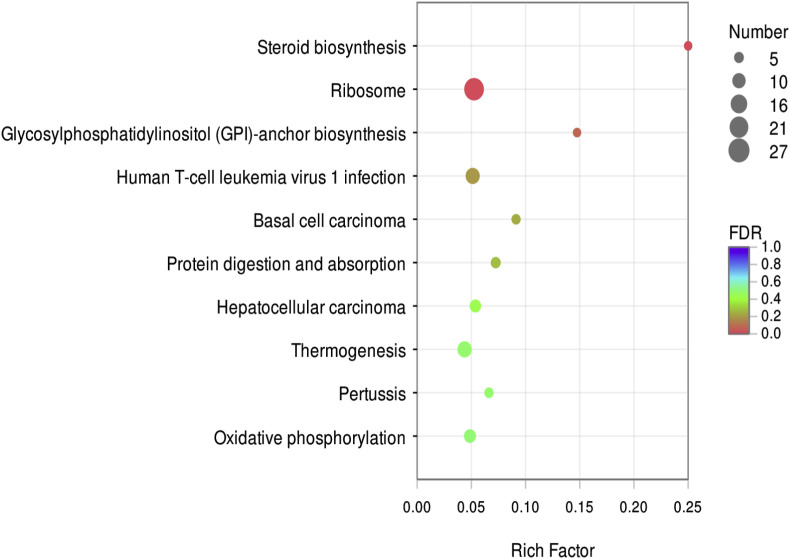
There 25,880 DEGs were normalized and 251 significant DEGs were detected between the PEDV-infected and control groups, including 133 up-regulated and 118 down-regulated genes (Fig. 4 ). GO functional cluster analysis divided the DEGs into three categories involving biological process, cellular component and molecular function (Fig. 5 ). Three categories include 20 sub-categories and 55 term. There are the most gene related to cellular process and single-organism process in biological process category. Cell and cell part -related genes are most in cellular component category, and the most genes related to binding and catalytically activity in molecular function. KEGG pathway analysis was performed to evaluate the biological and ontological significance of DEGs. The results showed that the DEGs were involved in a variety of life activities, and more genes were annotated into ribosome, oxidative phosphorylation and signal transduction of viral infection (Fig. 6 ). The ribosome is a large assembly of proteins and ribosomal RNAs (rRNAs) that functions to translate messenger RNAs (mRNAs) into proteins. In our results, compared with the control group, the ribosome gene expression significant up-regulated in PEDV-infected IPEC-J2 cells, such as 60S ribosomal protein (RPL10A, RPL14, RPL17, RPL22 and RPL23), 40S ribosomal protein (PPS18, RPS3A, RPS26 and RPS27A) and mitochondrial ribosomal protein (MRPL12, MRPS14, MRPL1 and MRPL27). Compared with the control, ATP synthase (ATP5PO, ATP6V1G1 and ATP5F1C), NADH dehydrogenase (NDUFA8, NDUFS4 and NDUFB2, NDUFB6) and cytochrome C oxidase (COX5B, UQCRC2) had significant up-regulated in PEDV-infected cell that these genes related with oxidative phosphorylation process. A significant change of DEGs was also observed in multiple cellular signaling pathways. The phosphatidylinositol 3′-kinase(PI3K)-Akt signaling pathway is activated by many types of cellular stimuli or toxic insults and regulates fundamental cellular functions such as proliferation, growth, and survival. In this study, We found 8 significant DEGs in the PI3K/Akt signaling pathway compared with the control. Most of the positive regulators (EPHA2, ITGB4, EFNA1, IL2RG, TNC, CSF3 and MYC) were down-regulated and the negative regulator PTEN was up-regulated. These results indicated that IPEC-J2 cells infected with PEDV could negatively regulate PI3K/Akt signaling pathway. The mammalian target of rapamycin (mTOR) signaling pathway serves as a master regulator of cell metabolism, growth, proliferation and survival. In the study, 8 significant DEGs were filtered out in the mTOR signaling pathway. Compared with the control group, the mTOR upstream regulators (ATP6V1G1, FZD2 and LAMTOR2) were significantly up-regulated and the negative regulator PTEN was up-regulated. However, some of the upstream regulators(SLC3A2, SLC7A5, ITGA9 and LRP5) exerted significantly down-regulated. These signaling pathways significant change provide possible mechanisms responsible for the effects of PEDV on cell proliferation, cell cycle and apoptosis in IPEC-J2 cells. Differentially expressed genes related to cell apoptosis and the cell cycle determined by RNA-Seq analysis showed in Table 2 .
Fig. 4.
Volcano chart showing distribution of DEGs. The X–Y axis represents the log10 transformed gene expression level, red represents up-regulated genes, blue-green represents down-regulated genes, and gray indicates non-DEGs.
Fig. 5.
GO classification and functional annotation of molecular biological function, cellular components, and biological process.
Fig. 6.
KEGG pathway classification and functional enrichment of predicted DEGs.
Table 2.
Summary of differentially expressed genes related to cell apoptosis and the cell cycle determined by RNA-Seq analysis.
| KEGG enrichment | Type of genes/regulators | Genes | Gene change |
|---|---|---|---|
| Ribosome | 60S ribosomal protein | RPL10A, RPL14, RPL17, RP L22, RPL23 | up |
| 40S ribosomal protein | PPS18, RPS3A, RPS26, RPS27A | up | |
| mitochondrial ribosomal protein | MRPL12, MRPS14, MRPL1, MRPL27 | up | |
| Oxidative phosphorylation | ATP synthase | ATP5PO, ATP6V1G1, ATP5F1C | up |
| NADH dehydrogenase | NDUFA8, NDUFS4, NDUFB2, NDUFB6 | up | |
| cytochrome C oxidase | COX5B, UQCRC2 | up | |
| PI3K/Akt signaling pathway | up-regulator | EPHA2, ITGB4, EFNA1, IL2RG, TNC, CSF3,MYC | down |
| down-regulator | PTEN | up | |
| mTOR signaling pathway | up-regulator | ATP6V1G1, FZD2, LAMTOR2 | up |
| SLC3A2, SLC7A5, ITGA9, LRP5 | down | ||
| down-regulator | PTEN | up |
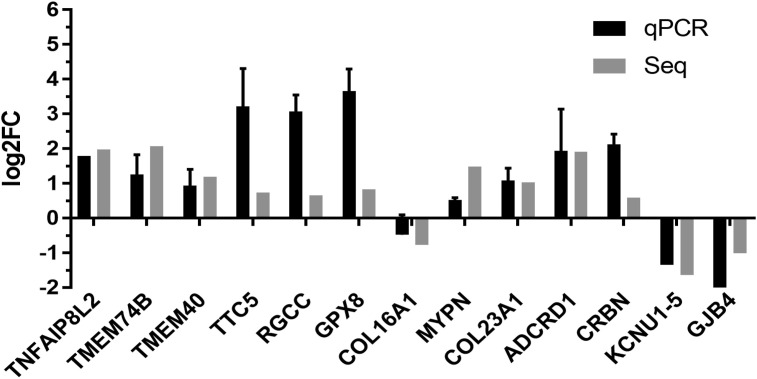
3.5. The significant DEGs were confirmed by real-time qPCR
Thirteen of the significantly differentially expressed genes between the PEDV infection and control groups were selected for real-time qPCR. β-actin as a reference gene for qPCR. The results showed that the expression level of thirteen genes were consistent with the results from RNA-Seq (Fig. 7 ).
Fig. 7.
RNA-Seq results were confirmed by real-time qPCR.
4. Discussion
PEDV suppresses the host protein synthesis machinery to promote cell apoptosis and block the cell cycle [20,21]. Detailed information on the interaction between PEDV and these cell processes will thus contribute to a comprehensive understanding of such antiviral mechanisms. PEDV infection remains one of the most important swine diseases in terms of vaccination and immunology [22]. Moreover, PEDV has a strong tendency to infect specific cells, particularly intestinal epithelial cells, where it can replicate efficiently. In this study, we therefore selected porcine IPEC-J2 cells to study the mechanisms by which PEDV affects apoptosis and the cell cycle at different times post-infection.
The current results showed that PEDV infection inhibited the growth of IPEC-J2 cells by blocking the cell cycle in S phase. In addition, PEDV CV777 increased apoptosis in IPEC-J2 cells. We also used RNA-Seq technology to analyze the differential expression profiles of intestinal epithelial cell genes in response to PEDV infection, and verified the accuracy and reliability of the transcriptome sequencing data by mRNA relative expression. Some DEGs involved in critical processes of apoptosis and the cell cycle in PEDV-infected pig cells were significantly up-regulated or down-regulated, including genes involved in signal transduction and related signaling pathways.
Apoptosis is an important process regulating the pathogenesis of many infectious diseases [10]. During the process of virus infection, apoptosis inhibition can prevent premature cell death and thus maximize viral replication, while apoptosis induction can facilitate virus release and dissemination from infected cells [23]. Furthermore, viruses manipulate progression through the cell cycle and alter checkpoint signaling to provide a favorable environment for their own replication [24]. The severe acute respiratory syndrome-related coronavirus N protein was recently reported to induce cell cycle arrest in S phase by down-regulating the expression of S phase gene products. S phase is a critical phase during cell cycle progression because it provides a cellular environment that is beneficial for viral replication. In the current study, we verified that PEDV infection blocked the cell cycle in S phase, and used RNA-Seq technology to analyze the functions and pathways of the annotated genes [25].
Host signaling pathways can be subverted by viruses to synthesize viral proteins and stifle the host's innate defense to facilitate its propagation [26]. The ribosome has traditionally been viewed as the cell's molecular machine, automatically chugging along, synthesizing proteins the cell needs to carry out the functions of life [27]. Researchers believed that ribosome appears to take a more active role, regulating the translation of specific proteins and ultimately how some viruses replicate, and further proved that blocking ribosomal protein rpL40 would disable vesicular stomatitis virus (VSV) while leaving normal cells largely unaffected [28] In our findings, the ribosome-related DEGs significant up-regulated in PEDV-infected IPEC-J2 cells, including 40s, 60s and mitochondrial ribosomal proteins. There some ribosomal proteins (RPL10A, RPS3A, RPS27A and RPL23) are closely related to the viral transcription by GO function annotation [29]. Because viral translation must occur in the face of translation compromising cellular defense mechanisms and in competition with abundant host transcripts [28]. These results suggested that PEDV may stressing IPEC-J2 cells to increase viral transcription through up-regulated the expression of ribosome proteins [30]. Interestingly, RPL23, RPS27A and RPS27L were also involved in the regulation of cell cycle and apoptotic processes [31]. Two research reports found that PEDV N protein [32] and E protein [33] induces endoplasmic reticulum (ER) stress through significantly up-regulated glucose regulated protein78 (GRP78). As we all know, a large number of ribosomes are attached to the ER [34], and thus we believe that the stress of the endoplasmic reticulum may be positively related to the up-regulation of ribosomes in host cell infected with PEDV. However, it has not been previously reported on the involvement of cellular ribosomes in intestinal epithelial cells infected with PEDV, and its role and mechanism in the process of infection are still unknown.
Oxidative phosphorylation is the process whereby the energy released from the oxidation reactions of the electron transfer chain is used for the synthesis of ATP. The overall process of oxidative phosphorylation is tightly controlled by transcriptional regulation at the level of DNA, translational effects via RNA levels and stability, by substrate feedback inhibition [35]. In this study, PEDV infection caused up-regulated of key enzymes (ATP synthase, NADH dehydrogenase and cytochrome C oxidase) involved in oxidative phosphorylation in IPEC-J2 cells. The enhancement of the oxidative phosphorylation process could produce more ATP, which may be related to the regulation of energy metabolism after the PEDV infects cells, and its role may be to meet the energy requirements of virus assembly [36]. While up-regulated oxidative phosphorylation caused cell damage and apoptosis through the production high levels of reactive oxygen species (ROS) [37]. Xu et al. found that PEDV infections induce apoptosis in vero cells via a reactive oxygen species (ROS)/p53. Therefore, These results should be considered that the upregulation of oxidative phosphorylation is related to apoptosis in IPEC-J2 cells infected with PEDV.
The signaling pathways of the interaction between virus and cells will greatly contribute to a comprehensive understanding of such antiviral mechanisms. In this study, the results indicated that PEDV infection could negatively regulate PI3K/Akt signaling pathway in IPEC-J2 cells. The positive regulators TNC(tenascin precursor) [38], CSF3(colony-stimulating factor 3) [39] and MYC (myc proto-oncogene protein) [40] are key molecules in PI3K/Akt signaling pathways, significantly down-regulation after PEDV infection compared with the control group, and they played important role in regulating cell proliferation and cell cycle arrest. The negative regulator PTEN (phosphatase and tensin homology deleted on chromosome 10) was up-regulated compared with the control group, and it is a key phosphatase that reduces AKT activation and prevents all downstream signaling events [41]. mTOR signaling pathway is a key downstream effector of the PI3K/Akt signaling pathway and is commonly targeted by viral proteins because of its role in cell apoptosis and cell cycle [42]. The up-regulated of negative regulator PTEN can inhibit mTOR signaling pathway activation. Similar to our results, Guo et al. [43] and Lin et al. [42] also found that the PEDV suppressed protein synthesis of host cells through down-regulation of the PI3K-AKT/mTOR signaling pathways. However, mTOR signaling can be manipulated by many genes [44]. In the current study, ITGA9 (integrin subunit alpha 9) [45] and LRP5 (Low-density lipoprotein receptor-related protein 5) [46] was down-regulated and FZD2 (frizzled class receptor 2) [47] was up-regulated, affecting mTOR activity. It is of note that FZD2 also involves the Wnt signaling pathway and Hippo signaling pathway [48]. Therefore, mTOR signaling pathway contributing to the cytopathology, apoptosis, and cell cycle changes through multiple regulation following PEDV infection [49]. These results may provide the basis for further analyses of the mechanism by which PEDV affects porcine intestinal epithelial cells apoptosis and the cell cycle.
5. Conclusion
In this study, we used RNA-Seq to profile the signature genes affected by PEDV infection in porcine intestinal epithelial cells. The results indicate that PEDV infection reduced proliferation activity, blocked cell cycle at the S phase and induced apoptosis in IPEC-J2 Cells. A novel discovery that the ribosome proteins and oxidative phosphorylation process were widely involved in the pathological changes and regulation of host cells caused by PEDV infection. Further findings that the mTOR and PI3K/Akt signaling pathways played a vital role in antiviral regulation in IPEC-J2 cells. These findings enhanced our understanding of the mechanism of PEDV infection in piglets, and will aid the development of clinical intervention strategies against PEDV infections.
Authors' contributions
XP conceived and designed the experiments; XS and LY performed the experiments; RZ analyzed the data; DZ contributed reagents/materials/Únalysis tools, XS and LY wrote the paper. All authors critically read and contributed to the manuscript and approved the final version.
Compliance with ethical standards
The authors declare that they have no conflicts of interest.
Author statement
We declare that the work described was original research that has not been published previously, and not under consideration for publication elsewhere, in whole or in part. All the authors listed have approved the manuscript that is enclosed.
Declaration of competing interest
The authors declared that they have no conflicts of interest to this work.
Acknowledgements
This work was supported by grants from the Science and Technology Program of Anhui Province (No.1704A07020066).
References
- 1.Jung K., Saif L.J. Porcine epidemic diarrhea virus infection: etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 2015;204:134–143. doi: 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol. J. 2015;12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan B., Jiao D., Zhao X., Pang F., Xiao Q., Yu Z. Characterization of Chinese porcine epidemic diarrhea virus with novel insertions and deletions in genome. Sci. Rep. 2017;7:44209. doi: 10.1038/srep44209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian Y., Yu Z., Cheng K., Liu Y., Huang J., Xin Y. Molecular characterization and phylogenetic analysis of new variants of the porcine epidemic diarrhea virus in Gansu, China in 2012. Viruses. 2013;5:1991–2004. doi: 10.3390/v5081991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L., Byrum B., Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg. Infect. Dis. 2014;20:917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevenson G.W., Hoang H., Schwartz K.J., Burrough E.R., Sun D., Madson D. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- 8.Schierack P., Nordhoff M., Pollmann M., Weyrauch K.D., Amasheh S., Lodemann U. Characterization of a porcine intestinal epithelial cell line for in vitro studies of microbial pathogenesis in swine. Histochem. Cell Biol. 2006;125:293–305. doi: 10.1007/s00418-005-0067-z. [DOI] [PubMed] [Google Scholar]
- 9.Curry S.M., Schwartz K.J., Yoon K.J., Gabler N.K., Burrough E.R. Effects of porcine epidemic diarrhea virus infection on nursery pig intestinal function and barrier integrity. Vet. Microbiol. 2017;211:58–66. doi: 10.1016/j.vetmic.2017.09.021. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y., Lee C. Porcine epidemic diarrhea virus induces caspase-independent apoptosis through activation of mitochondrial apoptosis-inducing factor. Virology. 2014;460–461:180–193. doi: 10.1016/j.virol.2014.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee C., Kim Y., Jeon J.H. JNK and p38 mitogen-activated protein kinase pathways contribute to porcine epidemic diarrhea virus infection. Virus Res. 2016;222:1–12. doi: 10.1016/j.virusres.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X., Xu Y., Zhang Q., Yang F., Yin Z., Wang L. Porcine epidemic diarrhea virus infections induce apoptosis in Vero cells via a reactive oxygen species (ROS)/p53, but not p38 MAPK and SAPK/JNK signalling pathways. Vet. Microbiol. 2019;232:1–12. doi: 10.1016/j.vetmic.2019.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun P., Wu H., Huang J., Xu Y., Yang F., Zhang Q. Porcine epidemic diarrhea virus through p53-dependent pathway causes cell cycle arrest in the G0/G1 phase. Virus Res. 2018;253:1–11. doi: 10.1016/j.virusres.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu X.G., Zhang H.L., Zhang Q., Dong J., Huang Y., Tong D.W. Porcine epidemic diarrhea virus M protein blocks cell cycle progression at S-phase and its subcellular localization in the porcine intestinal epithelial cells. Acta Virolog. 2015;59:265–275. doi: 10.4149/av_2015_03_265. [DOI] [PubMed] [Google Scholar]
- 15.Zakrzewski S.S., Richter J.F., Krug S.M., Jebautzke B., Lee I.F., Rieger J. Improved cell line IPEC-J2, characterized as a model for porcine jejunal epithelium. PLoS One. 2013;8 doi: 10.1371/journal.pone.0079643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altawaty T., Liu L., Zhang H., Tao C., Hou S., Li K. Lack of LTβR increases susceptibility of IPEC-J2 cells to porcine epidemic diarrhea virus. Cells. 2018;7 doi: 10.3390/cells7110222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin H., Li B., Chen L., Ma Z., He K., Fan H. Differential protein analysis of IPEC-J2 cells infected with porcine epidemic diarrhea virus pandemic and classical strains elucidates the pathogenesis of infection. J. Proteome Res. 2017;16:2113–2120. doi: 10.1021/acs.jproteome.6b00957. [DOI] [PubMed] [Google Scholar]
- 18.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klopfenstein D.V., Zhang L., Pedersen B.S., Ramírez F., Warwick Vesztrocy A., Naldi A. GOATOOLS: a Python library for Gene Ontology analyses. Sci. Rep. 2018;8:10872. doi: 10.1038/s41598-018-28948-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin C.W., Lin K.H., Hsieh T.H., Shiu S.Y., Li J.Y. Severe acute respiratory syndrome coronavirus 3C-like protease-induced apoptosis. FEMS Immunol. Med. Microbiol. 2006;46:375–380. doi: 10.1111/j.1574-695X.2006.00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chulu J.L., Huang W.R., Wang L., Shih W.L., Liu H.J. Avian reovirus nonstructural protein p17-induced G(2)/M cell cycle arrest and host cellular protein translation shutoff involve activation of p53-dependent pathways. J. Virol. 2010;84:7683–7694. doi: 10.1128/JVI.02604-09. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Lee H.M., Lee B.J., Tae J.H., Kweon C.H., Lee Y.S., Park J.H. Detection of porcine epidemic diarrhea virus by immunohistochemistry with recombinant antibody produced in phages. J. Vet. Med. Sci. 2000;62:333–337. doi: 10.1292/jvms.62.333. [DOI] [PubMed] [Google Scholar]
- 23.Lee Y.J., Lee C. Porcine deltacoronavirus induces caspase-dependent apoptosis through activation of the cytochrome c-mediated intrinsic mitochondrial pathway. Virus Res. 2018;253:112–123. doi: 10.1016/j.virusres.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Junwei G., Baoxian L., Lijie T., Yijing L. Cloning and sequence analysis of the N gene of porcine epidemic diarrhea virus LJB/03. Virus Gene. 2006;33:215–219. doi: 10.1007/s11262-005-0059-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu X., Zhang H., Zhang Q., Huang Y., Dong J., Liang Y. Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet. Microbiol. 2013;164:212–221. doi: 10.1016/j.vetmic.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shuai K., Liu B. Regulation of JAK-STAT signalling in the immune system. Nat. Rev. Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 27.Xue S., Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee A.S.-Y., Kerr R.B.K., Whelan S.P.J. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc. Natl. Acad. Sci. U. S. A. 2013;110:324–329. doi: 10.1073/pnas.1216454109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guadet P., Livestone M.S., Lewis S.E., Thomas P.D. Phylogenetic-based propagation of functional annotations within the gene ontology consortium. Briefings Bioinf. 2011;12:449–462. doi: 10.1093/bib/bbr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.George A., Panda S., Kudmulwar D., Chhatbar S.P., Nayak S.C., Krishnan H.H. Hepatitis C virus NS5A binds to the mRNA cap-binding eukaryotic translation initiation 4F (eIF4F) complex and up-regulates host translation initiation machinery through eIF4E-binding protein 1 inactivation. J. Biol. Chem. 2012;287:5042–5058. doi: 10.1074/jbc.M111.308916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ain A., Itahana K., O’Keefe K., Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol. 2004;24:7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu X., Zhang H., Zhang Q., Huang Y., Dong J., Liang Y. Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet. Microbiol. 2013;164:212–221. doi: 10.1016/j.vetmic.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun M., Ma J., Yu Z., Pan Z., Lu C., Yao H. Identification of two mutation sites in spike and envelope proteins mediating optimal cellular infection of porcine epidemic diarrhea virus from different pathways. Vet. Res. 2017;48:44. doi: 10.1186/s13567-017-0449-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarz D.S., Blower M.D. The endoplasmic reticulum: structure, function and response to cellular signaling. Cell. Mol. Life Sci. 2016;73:79–94. doi: 10.1007/s00018-015-2052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borzeniewski K. Regulation of oxidative phosphorylation through parallel activation. Biophys. Chem. 2007;129:93–110. doi: 10.1016/j.bpc.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Z., Wang J.C.-Y., Gonzalez-Gutierrez G., Venkatakrishnan B., Asor R., Khaykelson D. Structural differences between the woodchuck hepatitis virus core protein in the dimer and capsid States are consistent with entropic and conformational regulation of assembly. J. Virol. 2019;93 doi: 10.1128/JVI.00141-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li G., Qin Y. Mitochondrial translation factor EF4 regulates oxidative phosphorylation complexes and the production of ROS. Free Radic. Res. 2018;52:1250–1255. doi: 10.1080/10715762.2018.1479063. [DOI] [PubMed] [Google Scholar]
- 38.Halper J., Kjaer M. Basic components of connective tissues and extracellular matrix: elastin, fibrillin, fibulins, fibrinogen, fibronectin, laminin, tenascins and thrombospondins. Adv. Exp. Med. Biol. 2014;802:31–47. doi: 10.1007/978-94-007-7893-1_3. [DOI] [PubMed] [Google Scholar]
- 39.Ahmadi N., Mohamed S., Rahman H.S., Rosli R. Epicatechin and scopoletin-rich Morinda citrifolia leaf ameliorated leukemia via anti-inflammatory, anti-angiogenesis, and apoptosis pathways in vitro and in vivo. J. Food Biochem. 2019;43 doi: 10.1111/jfbc.12868. [DOI] [PubMed] [Google Scholar]
- 40.Casey S.C., Baylot V., Felsher D.W. The MYC oncogene is a global regulator of the immune response. Blood. 2018;131:2007–2015. doi: 10.1182/blood-2017-11-742577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haddadi N., Lin Y., Travis G., Simpson A.M., Nassif N.T., McGowan E.M. PTEN/PTENP1: ‘regulating the regulator of RTK-dependent PI3K/Akt signalling’, new targets for cancer therapy. Mol. Cancer. 2018;17:37. doi: 10.1186/s12943-018-0803-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.H L., B L., L C., Z M., K H., H F. Differential protein analysis of IPEC-J2 cells infected with porcine epidemic diarrhea virus pandemic and classical strains elucidates the pathogenesis of infection. J. Proteome Res. 2017;16:2113–2120. doi: 10.1021/acs.jproteome.6b00957. [DOI] [PubMed] [Google Scholar]
- 43.Guo X., Hu H., Chen F., Li Z., Ye S., Cheng S. iTRAQ-based comparative proteomic analysis of Vero cells infected with virulent and CV777 vaccine strain-like strains of porcine epidemic diarrhea virus. J. Proteomics. 2016;130:65–75. doi: 10.1016/j.jprot.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buchkovich N.J., Yu Y., Zampieri C.A., Alwine J.C. The TORrid affairs of viruses: effects of mammalian DNA viruses on the PI3K-Akt-mTOR signalling pathway. Nat. Rev. Microbiol. 2008;6:266–275. doi: 10.1038/nrmicro1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y.L., Xing X., Cai L.B., Zhu L., Yang X.M., Wang Y.H. Integrin α9 suppresses hepatocellular carcinoma metastasis by rho GTPase signaling. J. Immunol. Res. 2018;2018:4602570. doi: 10.1155/2018/4602570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin C.M., Chen H.H., Lin C.A., Wu H.C., Sheu J.J.-C., Chen H.-J. Apigenin-induced lysosomal degradation of β-catenin in Wnt/β-catenin signaling. Sci. Rep. 2017;7:372. doi: 10.1038/s41598-017-00409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding L.C., Huang X.Y., Zheng F.F., Xie J., She L., Feng Y. FZD2 inhibits the cell growth and migration of salivary adenoid cystic carcinomas. Oncol. Rep. 2016;35:1006–1012. doi: 10.3892/or.2015.3811. [DOI] [PubMed] [Google Scholar]
- 48.Ou H., Chen Z., Xiang L., Fang Y., Xu Y., Liu Q. Frizzled 2-induced epithelial-mesenchymal transition correlates with vasculogenic mimicry, stemness, and Hippo signaling in hepatocellular carcinoma. Cancer Sci. 2019;110:1169–1182. doi: 10.1111/cas.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H., Liu Q., Su W., Wang J., Sun Y., Zhang J. Genome-wide analysis of differentially expressed genes and the modulation of PEDV infection in Vero E6 cells. Microb. Pathog. 2018;117:247–254. doi: 10.1016/j.micpath.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]