Abstract
Amino groups derived from naturally abundant amino acids or (di)amines can be used as “shuttles” in nature for oxygen transfer to provide intermediates or products comprising N-O functional groups such as N-hydroxy, oxazine, isoxazolidine, nitro, nitrone, oxime, C-, S-, or N-nitroso, and azoxy units. To this end, molecular oxygen is activated by flavin, heme, or metal cofactor-containing enzymes and transferred to initially obtain N-hydroxy compounds, which can be further functionalized. In this review, we focus on flavin-dependent N-hydroxylating enzymes, which play a major role in the production of secondary metabolites, such as siderophores or antimicrobial agents. Flavoprotein monooxygenases of higher organisms (among others, in humans) can interact with nitrogen-bearing secondary metabolites or are relevant with respect to detoxification metabolism and are thus of importance to understand potential medical applications. Many enzymes that catalyze N-hydroxylation reactions have specific substrate scopes and others are rather relaxed. The subsequent conversion towards various N-O or N-N comprising molecules is also described. Overall, flavin-dependent N-hydroxylating enzymes can accept amines, diamines, amino acids, amino sugars, and amino aromatic compounds and thus provide access to versatile families of compounds containing the N-O motif. Natural roles as well as synthetic applications are highlighted.
|
Key points • N-O and N-N comprising natural and (semi)synthetic products are highlighted. • Flavin-based NMOs with respect to mechanism, structure, and phylogeny are reviewed. • Applications in natural product formation and synthetic approaches are provided. |
Graphical abstract.
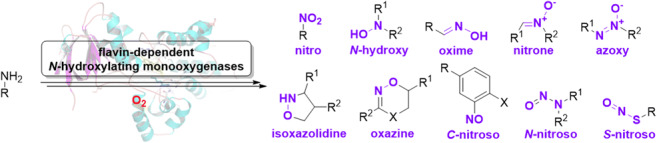
.
Keywords: Biotransformation, N-Hydroxylases, Flavoproteins, Siderophores, Bioactive compounds, Biocatalysis, Monooxygenases, Phylogenetics
Introduction
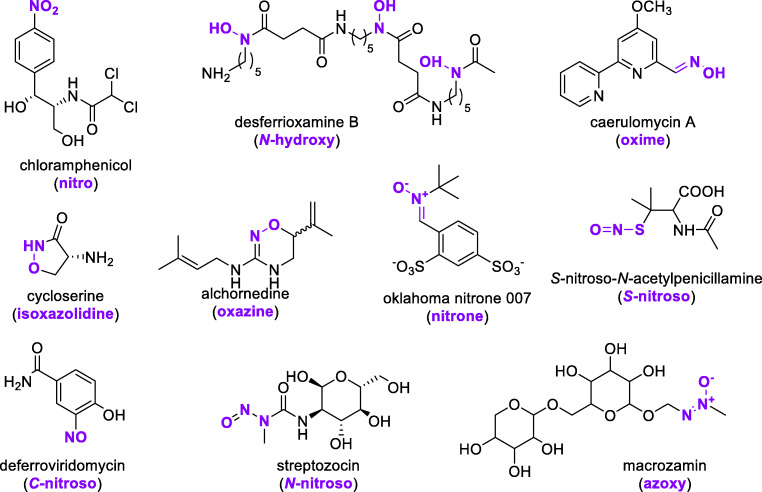
Nature provides access to numerous complex molecules with a variety of activities. The respective function is based on the molecular character of individual compounds and often shaped by their functional groups (Waldman et al. 2017; Davison and Sperry 2017). Among those, many comprise heteroatoms as N, O, S, and P or even a combination thereof. Because heteroatom-containing functional groups not vastly abundant in biological systems usually provide special functions, they have been moving into the focus of recent biotechnological research due to their potential for novel applications (Sulzbach and Kunjapur 2020). Herein, we will focus especially on N-O containing functional groups (mainly N-hydroxy; others are oxime, azoxy, oxazine, nitrone, C-nitroso, N-nitroso, S-nitroso, nitro, isoxazolidine), which are often found in secondary metabolites. Those natural products are interesting for many reasons, as they comprise antibiotics, bioactive agents, flavor and fragrance molecules, food additives, pharmaceutical precursors or products, siderophores and metallophores, enzyme inhibitors, and toxins, among others.
The formation of the various N-O linkages (Fig. 1) in organic molecules is often based on the conversion of soft nucleophilic amines by a form of activated (molecular) oxygen and combined with potential maturation reactions. Sometimes, overoxidation alone leads to a mixture of N-O functional groups in a compound. Nevertheless, a variety of enzymes have evolved to selectively activate oxygen to transfer it onto target compounds, e.g., onto amino groups. Here, especially flavin-dependent monooxygenases, heme-dependent P450 monooxygenases, peroxygenases, and other metal-dependent mono- and dioxygenases can be mentioned. Successive oxidation reactions might change the redox state of respective compounds to yield the final N-O or in other cases N-N functionality containing molecules.
Fig. 1.
N-O functional groups in representative and already used naturally occurring or bio-produced compounds. The formation of N-N bonds can also be initially promoted by intermediate N-O bond formation, and thus, more complex structures can be obtained (vide infra). These structures are only representatives and often many derivatives occur due to natural diversity or synthetic modifications to provide more significant effects in desired fields
In this review, we mainly focus on the properties of flavin-dependent N-hydroxylating monooxygenases (NMOs) and present recent findings about these enzymes since the last reviews six and more years ago (Olucha and Lamb 2011; Robinson and Sobrado 2012; Huijbers et al. 2014).
We first depict the catalytic mechanism and structural properties of NMOs, followed by recent advances and results from in-depth enzymology studies especially in the context of siderophore synthesis, where NMOs catalyze the N-hydroxylation of defined small substrates (i.e., diamino acids and diamines) at the beginning of biosynthetic pathways. Here, structural diversification typically happens in variable downstream conversions. Furthermore, we highlight NMOs that accept more complex substrates, as late diversification stages of complex molecules or involved in the metabolization of bioactive compounds. Lastly, we describe NMOs involved in the formation of N-N and N=N bonds and reveal which common biological pathways are used by microorganisms to produce this great structural diversity. Few NMOs are currently exploited for biotechnological applications, leaving the vast majority of known—but hitherto uncharacterized in terms of enzymatic performance—enzymes as potential research target aiming at the production of complex chemical compounds.
Catalytic mechanism of flavin-dependent NMOs
Flavin-dependent NMOs (EC 1.14.13.x) produce numerous important products, but only a few family members have been thoroughly characterized to date. NMOs belong to the single-component group B flavoprotein monooxygenases (Fraaije and van Berkel 2006; van Berkel et al. 2006; Olucha and Lamb 2011; Huijbers et al. 2014). It needs to be mentioned that some related group B enzymes are also able to perform N-hydroxylations. Those enzymes are typically designated as FMOs, a term which has been introduced for flavin-containing monooxygenases being involved in mammalian detoxification processes (EC 1.14.13.8) (Ziegler 1988; van Berkel and Müller 1991; Ziegler 2002; van Berkel et al. 2006; Huijbers et al. 2014; Mascotti et al. 2015). Therefore, the abbreviation FMO is not synonymous to NMO but represents a specific subgroup of (mostly) eukaryotic group B flavin-containing enzymes that catalyze the monooxygenation of carbon-bound reactive heteroatoms including nitrogen, sulfur, phosphorous, selenium, and iodine (van Berkel et al. 2006). Also, the group of YUCCA and related enzymes from plants form a subgroup of group B flavin-containing monooxygenases and thus are related to type I BVMOs, NMOs, and FMOs (Huijbers et al. 2014).
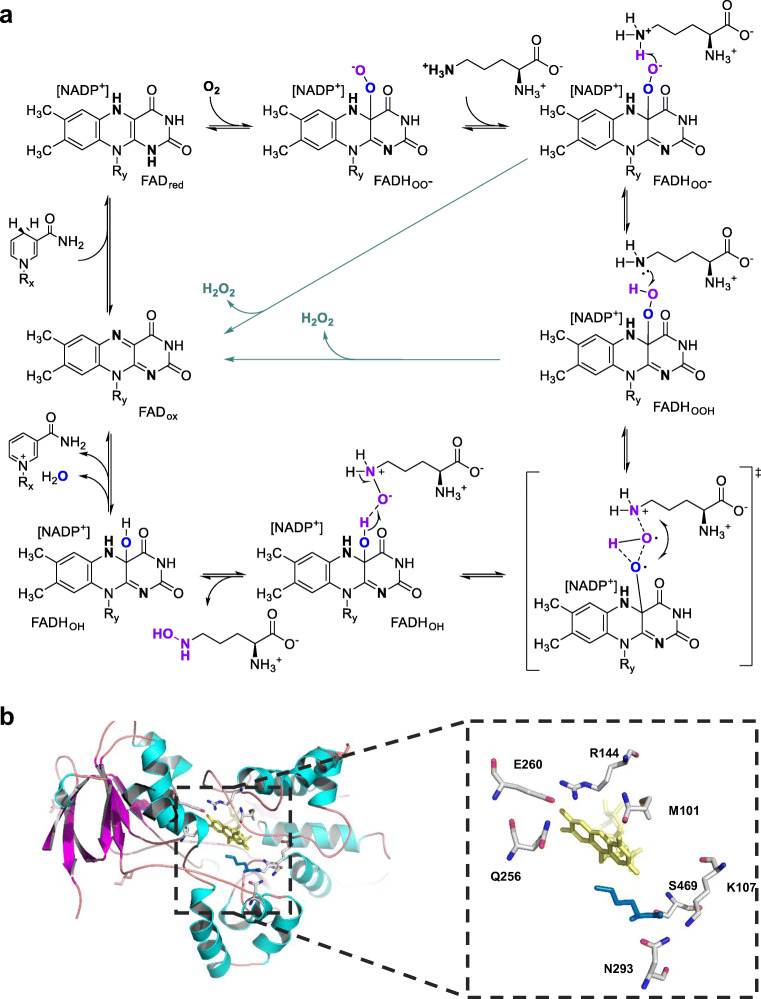
Members of group B flavoprotein monooxygenases contain two Rossmann-type dinucleotide binding domains to harbor the flavin adenine dinucleotide (FAD) and nicotinamide adenine dinucleotide (NAD(P)H) cofactors and keep the pyridine nucleotide bound during the oxidative half-reaction. In general, the mechanism of NMOs is similar to that of group B Baeyer-Villiger monooxygenases (BVMOs) (Ryerson et al. 1982; Sheng et al. 2001; Ballou and Entsch 2013) and can be described as follows (Fig. 2a) (Olucha and Lamb 2011; Bufkin and Sobrado 2017):
Fig. 2.
a Catalytic mechanism of flavin-dependent NMOs as reported for SidA and PvdA. The order of entry of oxygen and ornithine, and the ease of formation of FADHOOH from FADHOO- depends on the enzyme (for details see text). b Three-dimensional model of the crystal structure of SidA from A. fumigatus (PDB code: 4B69) (Franceschini et al. 2012). The SidA structure is colored according to secondary structural elements. The FAD cofactor is shown in yellow and the substrate ornithine in marine blue. A zoom into the active site indicates, next to the flavin and the substrate, several important amino acid residues
In its resting state, the enzyme harbors an oxidized FAD (FADox), as determined for SidA from Aspergillus fumigatus (Alfieri et al. 2008; Chocklett and Sobrado 2010; Romero et al. 2012a; Franceschini et al. 2012; Robinson et al. 2013; Robinson et al. 2014b) and PvdA from Pseudomonas aeruginosa (Meneely et al. 2009). Catalysis is initiated by the binding of NADPH to FADox. Then, FADox is reduced to FADH− (FADred) (Fig. 2a). For both SidA and PvdA, this step is among the slowest in the overall catalysis cycle (Meneely et al. 2009; Mayfield et al. 2010; Romero et al. 2012a). Furthermore, PvdA is highly specific for NADPH (Meneely and Lamb 2007), whereas SidA accepts both coenzymes with a preference for NADPH (Chocklett and Sobrado 2010).
Generating the FADred-NADP+ complex allows binding and activation of molecular oxygen to form the stable, long-lived C4a-hydroperoxflavin (FADHOOH, Fig. 2a) (Massey 1994). The rate of formation of this oxygenation species is moderately enhanced by the binding of ornithine (Frederick et al. 2011). In PvdA, the formation of FADHOOH from C4a-peroxyflavin (FADHOO-) is dependent on substrate binding (Meneely et al. 2009), while in SidA, FADHOOH immediately forms independent of substrate binding (van Berkel et al. 2011; Frederick et al. 2011; Romero et al. 2012b). Based on solvent kinetic isotope effects, density functional theory analysis, and the structural arrangement of the active site, it was argued that in SidA, the 2’OH of the ribose of NADP+ delivers the proton for the conversion of FADHOO- into FADHOOH (Robinson et al. 2014b).
Formation of the ternary complex between FADHOOH, NADP+, and substrate allows the selective N-hydroxylation of the substrate (Fig. 2a). In the case of SidA and PvdA, ornithine is oxidized to N5-hydroxyornithine. During this step, hydroxy-FAD (FADHOH) is formed, which decays to FADox and H2O. Finally, after the release of N6-hydroxyornithine and NADP+, FADox is ready for the next cycle.
The stabilization of the reactive C4a-hydroperoxyflavin is crucial for this mechanism in order to prevent unproductive formation of hydrogen peroxide (designated as uncoupling) and wasting the reducing equivalents of NADPH. In the case of SidA and PvdA, which are both highly specific for the N5-hydroxylation of l-ornithine, uncoupling occurs with a range of substrate analogs, including among others d-ornithine, l-arginine, and l-lysine. Structural studies of PvdA indicated that an optimal orientation of the amino group of the substrate is required to prevent uncoupling (Olucha et al. 2011). The bound NADP+ plays a critical role here, because upon FAD reduction, its nicotinamide ring changes position, thereby creating a pocket for the formation and stabilization of the C4a-hydroperoxyflavin (Olucha et al. 2011).
The lysine hydroxylase MbsG from Mycobacterium smegmatis features a somewhat different kinetic mechanism (Robinson et al. 2014b). Here, lysine binds before the pyridine nucleotide, resulting in a substrate-NADPH-FADox ternary complex. Substrate binding slightly decreases the rate of flavin reduction, but hardly influences the rate of the reaction with oxygen. Another difference between SidA and PvdA is that MbsG has a slight preference for NADH (Robinson and Sobrado 2012).
The crystal structure of PvdA (PDB code 3S5W) was the first NMO structure solved (Olucha et al. 2011). This structure of the ternary complex with bound l-ornithine and NADP(H) revealed that the active site is located at the interface of three domains. Next to the FAD- and NADPH-binding domains, the enzyme contains a small helical domain for binding the ornithine substrate. The PvdA structure also revealed that Arg240 and Ser286 were in H-bond distance of the 2′-phosphate group of the adenosine ribose in NADPH, thereby determining the coenzyme specificity.
Crystallographic analysis of the SidA tetramer allowed the three-dimensional structure determination of seven SidA-ligand complexes, including the binary and ternary complexes of FADox and FADred bound to ornithine, lysine, arginine, and NADP+ (Franceschini et al. 2012). The structure of the enzyme-substrate complex as depicted in Fig. 2b is highly similar to the corresponding complex of PvdA (Olucha et al. 2011). For more details about the function of the active site residues highlighted in Fig. 2b, we refer to the original articles, which gave clues about the strict selectivity of both enzymes for l-ornithine, the essential role of NADP+ in stabilizing the C4a-hydroperoxyflavin, and the active site environment where the hydroxylation of the substrate takes place.
Site-directed mutagenesis of SidA was then used to investigate the predicted role of certain critical residues. Changing Ser257, involved in binding the pyrophosphate moiety of NADPH, to Ala revealed that this serine was important for the correct positioning of NADP+, thereby stabilizing the C4a-hydroperoxyflavin (Shirey et al. 2013). Replacement of Arg279, which interacts with the 2′-phosphate of NADPH, by Ala or Glu resulted in a different coenzyme specificity and strong uncoupling of hydroxylation. The R279A variant showed no coenzyme preference, while R279E preferred NADH as coenzyme. The fact that the changes in coenzyme preference were mainly due to changes in coenzyme binding strength corroborated the hypothesis that the positive charge at position 279 is crucial for the tight binding of NADPH (Robinson et al. 2014a).
Four residues involved in ornithine binding were individually changed to Ala (Robinson et al. 2015). The Lys107Ala variant lost its hydroxylation activity, indicating that the ionic interaction between Lys107 and the substrate carboxylate is essential for catalysis. Mutation of Asn293 and Ser469 to Ala strongly weakened the binding of ornithine. A similar effect was observed with the N323A variant. Besides interacting with the substrate, Asn323 also interacts with the nicotinamide ribose of NADPH. The crystal structure of the N323A variant complexed with ornithine and NADP+ revealed a disordered binding mode of the nicotinamide ribose group, while kinetic experiments showed that this mutated variant was much faster than wild-type SidA in reducing the flavin cofactor. The fact that Asn323 facilitates substrate binding at the expense of hindering flavin reduction clearly demonstrates the delicate balance of the enzyme-ligand interaction network in the SidA active site.
NMOs involved in siderophore biosynthesis
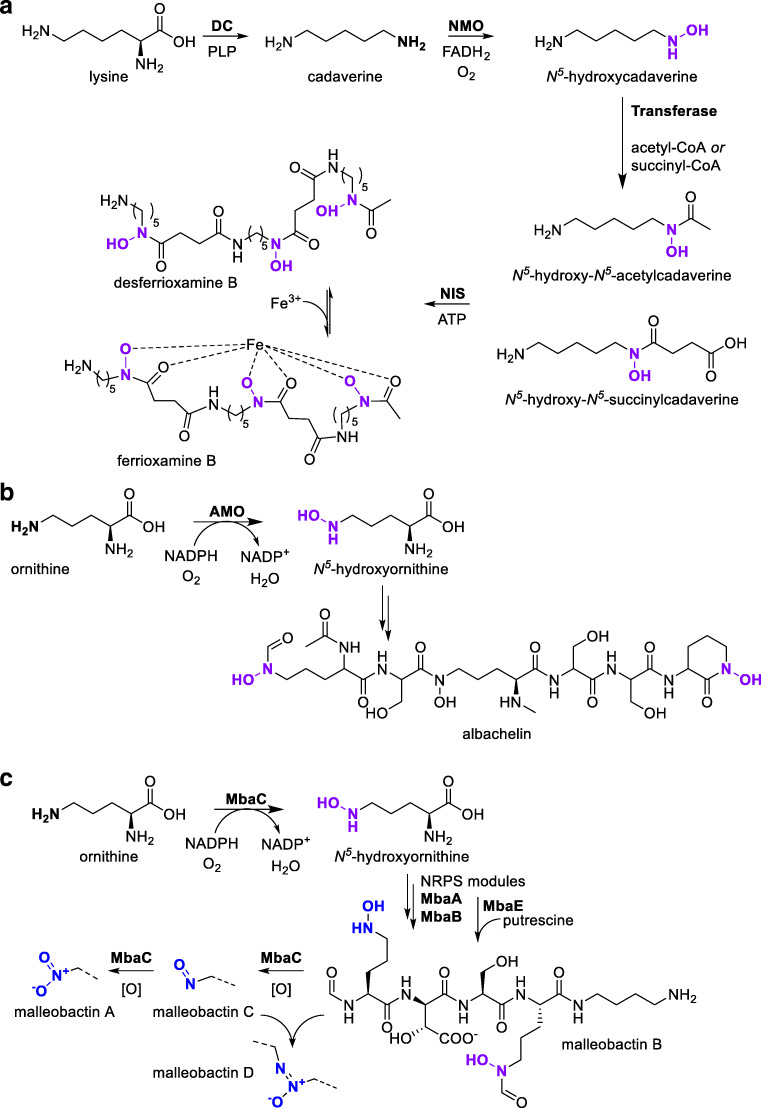
Siderophores (greek: sidero = iron; phore = carrier) are secondary metabolites produced by many organisms (plant, bacteria, and fungi) to sequester iron in a physiological context (Kem and Butler 2015; Carroll and Moore 2018; Hofmann et al. 2020). Often stress, such as iron limitation or ecological pressure, leads to a response of producing and secreting secondary metabolites. Other related natural compounds are specific for zinc, gold, or vanadium or even unspecific with respect to the target metal or metalloid and are thus often called metallophores (Johnston et al. 2013). All these metal-chelating natural products can be synthesized by two different routes. One involves non-ribosomal peptide synthetases (NRPSs), whereas the other one is NRPS-independent and thus designated as NIS-pathway. Many secondary metabolites, including some siderophores, are synthesized through NIS-routes, which can start from simple precursors such as amino acids. The most prominent compound to be mentioned as representative for biosynthesis and application is desferrioxamine B (DFOB) (Fig. 3a). It is produced by various actinobacteria and often the major component of a metabolite cocktail of related desferrioxamines or bisucaberins secreted by the producer strain (Senges et al. 2018; Schwabe et al. 2018; Proença et al. 2019). The production starts from lysine via cadaverine as a substrate of an N-hydroxylating flavoprotein to yield N5-hydroxycadaverine. Subsequently, a substrate-relaxed acyl-transferase comes into play and produces various intermediates, which result in a variety of DFOB- or bisucaberin-like molecules via an ATP-consuming assembly line (Ronan et al. 2018).
Fig. 3.
a Biosynthesis of desferrioxamine B and its iron chelation product ferrioxamine. b First, the pyridoxal phosphate (PLP)–dependent decarboxylase (DC) provides the substrate for the flavin N-hydroxylase (NMO), which activates molecular oxygen via reduced FAD. Then, activated acyl or succinyl residues are transferred to provide the substrate for the NRPS-independent ATP utilizing synthetase (NIS). b Biosynthesis of albachelin. A flavin-dependent N-hydroxylase (AMO) catalyzes the production of N5-hydroxyornithine, multiple subsequent steps follow. c Malleobactins are synthesized by the pathogenic bacterium Burkholderia pseudomallei in which the activity of the NMO MbaC leads to four different siderophore structures
We focus here on routes involving flavoproteins as one of the first biosynthetic steps. Enzymes studied so far are listed in Table 1. Typical precursors to bacterial siderophores are the amino acids ornithine and lysine. These and their decarboxylation products, cadaverine and putrescine, can selectively be hydroxylated by NMOs. Their mechanism of action is similar to the above example for N-hydroxylating ornithine monooxygenases. The resulting N-hydroxy-diamines and amino acids can be further acetylated by corresponding (typically formyl tetrahydrofolate-dependent) acetyltransferases to prevent overoxidation towards nitro functions, or directly used in the assembly line of siderophores, as presented for DFOB in Fig. 3a. Besides formyl tetrahydrofolate–dependent formylation (e.g., in coelichelin or rhodochelin), downstream biosynthetic routes enclose acylation or condensation by NRPS pathways towards various products.
Table 1.
N-Hydroxylating flavoprotein monooxygenases (NMOs) involved in siderophore biosynthesis. Most NMOs utilize ornithine or lysine or their decarboxylation product putrescine or cadaverine as substrates;, few have been reported to use alternative substrates—all use FAD as cofactor and NAD(P)H as coenzyme
| Siderophore | Organism | NMO-substrate | NMO | Accession no. | References |
|---|---|---|---|---|---|
| Ornithine N-hydroxylases | |||||
| Pseudobactin | Pseudomonas sp. B10 | Ornithine | PsbA | Q9F8X0, AAG27518 | Ambrosi et al. (2000) |
| Pyoverdine PAO | Pseudomonas aeruginosa PAO1 | Ornithine | PvdA | Q51548, WP_003114509 | Ge and Seah (2006) |
| Ornibactin | Burkholderia cepacia K56–2 | Ornithine | PvdA Bc | O51940, AAB94515 | Sokol et al. (1999) |
| Cupriachelin | Cupriavidus necator H16 | Ornithine | PvdA Re | Q0K0K9, CAJ96465 | Kurth et al. (2019) |
| Taiwachelin | Cupriavidus taiwanensis | Ornithine | TaiO | YP_002007894, WP_012356056 | Kreutzer and Nett (2012) |
| Malleobactin | Burkholderia pseudomallei | Ornithine | MbaA Bp2 | YP_001066209, WP_004535763 | Alice et al. (2006) |
| Malleobactin A, B, C, D | Burkholderia thailandensis | Ornithine | MbaC | WP_011402367 | Franke et al. (2013) |
| Ferrichrome | Aspergillus nidulans | Ornithine | SidA | Q7Z8P5, AAP56238 | Eisendle et al. (2003) |
| Ferricrocin, ferrichrome | Aspergillus fumigatus | Ornithine | SidA | E9QYP0, AAT84594 | Hissen et al. (2005); Chocklett and Sobrado (2010) |
| Deferriferrichrysin | Aspergillus oryzae | Ornithine | Dff | Q2TZB2, BAC15565 | Yamada et al. (2003) |
| Kutznerides | Kutzneria sp. 744 | Ornithine | KtzI | ABV56589 | Fujimori et al. (2007) |
| Rhodochelin | Rhodococcus jostii RHA1 | Ornithine | Rmo | ABG96502 | Bosello et al. (2012) |
| Fuscachelin | Thermobifida fusca | Ornithine | FscE | PZN64719 | Dimise et al. (2008) |
| Erythrochelin | Saccharopolyspora erythraea NRRL 2338 | Ornithine | EtcB | PFG94420 | Robbel et al. (2011) |
| Thermochelin | Thermocrispum agreste | Ornithine | TheA | WP_028847741 | Heine et al. (2017) |
| Albachelin | Amycolatopsis alba DSM 44262 | Ornithine | AMO | OXM54439 | Bufkin and Sobrado (2017) |
| Coelichelin | Streptomyces ambofaciens ATCC 23877 | Ornithine | CchB | A0ACR3 | Pohlmann and Marahiel (2008) |
| Vicibactin | Rhizobium etli CFN 42 | Ornithine | VbsO | Q2JYJ0 | Heemstra et al. (2009) |
| Mixed substrate N-hydroxylases | |||||
| Aerobactin | Shigella flexneri | Lysine | IucD | Q9XCH1 | Thariath et al. (1993) |
| Aerobactin | Escherichia coli | Lysine | Lys Ec | P11295 | Herrero et al. (1988) |
| Mycobactin | Mycobacterium smegmatis | Lysine | MbsG | CKI16899 | Robinson et al. (2014b) |
| Mycobactins | Mycobacterium tuberculosis, Mycobacterium sp. MCS | Lysine or acyl-lysine | MbtG | Q1B6B3 | Madigan et al. (2012) |
| Nocobactin | Nocardia farcinica IFM 10152 | Lysine | NbtG | BAD55606 | Binda et al. (2015) |
| Bisucaberin | Aliivibrio salmonicida LFI1238 | Cadaverine | BibB | CAQ81032 | Kadi et al. (2008) |
| Desferroxamine | Streptomyces scabiei 87.22 | Cadaverine | DesB | C9Z469 | Barona-Gómez et al. (2004) |
| Desferrioxamine | Erwinia amylovora CFBP1430 | Cadaverine | DfoA | D4I246, CBA23306 | Salomone-Stagni et al. (2018) |
| Desferroxamine | Gordonia rubripertincta CWB2 | Diamines | GorA | AOR50757 | Esuola et al. (2016) |
| Alcaligin | Bordetella bronchiseptica RB50 | Putrescine | AlcA | Q44740 | Kang et al. (1996) |
| Putrabactin | Shewanella oneidensis | Putrescine | PubA | WP_011072933 | Li et al. (2016) |
| Putrabactin | Shewanella putrefaciens 95 | Putrescine | SpPMO (PubA) | QBX90611 | Saroja et al. (2019) |
| Rhizobactin1021 | Sinorhizobium meliloti 1021 | 1,3-Diaminopropane | RhbE | Q9Z3Q8 | Lynch et al. (2001) |
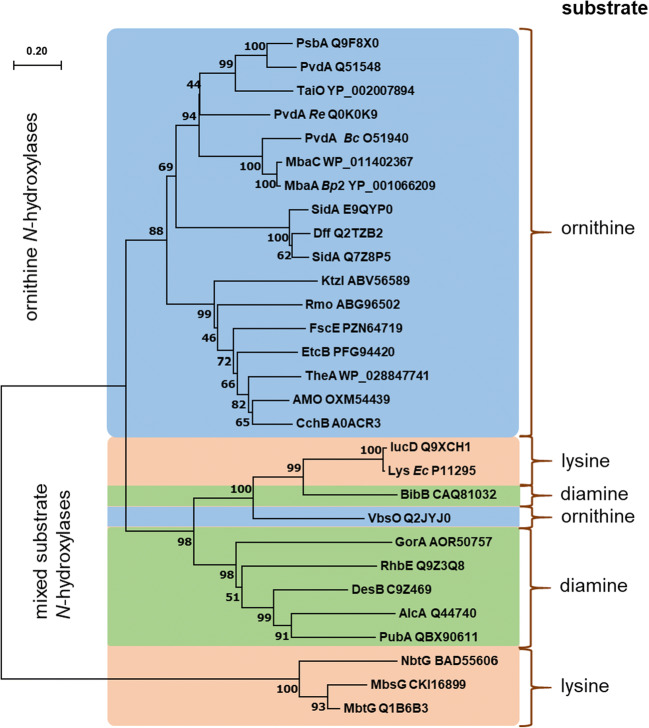
The enzymes mentioned in Table 1 can be classified according to their amino acid sequence and clustered in phylogenetic trees as done earlier (Franke et al. 2013; Esuola et al. 2016) and presented comprehensively in Fig. 4.
Fig. 4.
Minimum evolution distance tree of N-hydroxylating flavoprotein monooxygenases from bacteria and fungi in analogy to earlier studies (Franke et al. 2013; Esuola et al. 2016). Evolutionary distances were computed using the JTT matrix–based method and are given in the units of the number of amino acid substitutions per site (see scale bar). The accession numbers and protein designations are given according to Table 1 and references cited therein. It is worth to mention the most intensively studied representatives: Aspergillus fumigatus SidA (E9QYP0) and Pseudomonas aeruginosa PvdA (Q51548), respectively. The substrate of each NMO is given and respective parts of the tree are color-coded
On the basis of amino acid sequences, one can distinguish lysine from ornithine N-hydroxylases (Fig. 4). Only one enzyme with a close relation to lysine N-hydroxylases accepts ornithine as a substrate: VbsO (Heemstra et al. 2009). This enzyme converts solely l-ornithine, while the d-form and both lysine enantiomers yield no product. Lysine acts as a non-substrate effector of VbsO, leading to the uncoupling of hydroxylation (see also Fig. 2).
Among the lysine N-hydroxylases, two groups exist: one of mycobacterial and nocardia strains and another from Escherichia coli and related bacteria. Both are clearly separated, and the mycobacterial representatives seem most distant to all other N-hydroxylases. Another group of enzymes, diamine accepting N-hydroxylases (Table 1 and Fig. 4), seems closely related to the lysine N-hydroxylating NMOs from E. coli and related strains. Thus, we propose an evolutionary linkage that could be experimentally validated through mutagenesis studies to switch their substrate spectrum from diamine to amino acid or vice versa. Furthermore, among all NMOs, the relation is dependent on the origin of these enzymes. Thus, fungi, mycobacteria, pseudomonads, Burkholderia and E. coli (always with closely related microorganisms) form small subgroups in the phylogenetic tree.
Many enzymes share a similar pattern of activity, and the final product of metabolic pathways they are part of is typically defined by the subsequent enzymatic steps and respective substrates. Functional groups that connect NMO-based routes, often N-O bonds, are crucial for biological function. This is true for all the siderophores produced via such a route since the N-O bond is usually part of a hydroxamate functional unit that allows metal chelation. This hydroxamate unit can be located terminally as an open or ring-like structure or simply in the middle of a siderophore molecule, and several such units can be present in one single molecule.
Recently, the biosynthesis of albachelin was described (Fig. 3b) (Kodani et al. 2015). This siderophore is produced by an actinobacterium named Amycolatopsis alba under iron starvation. The involved NMO is designated AMO and prefers ornithine as substrate and NADPH as an electron donor. NADH does bind and allows conversion but yields only unproductive hydrogen peroxide formation. The same is true for lysine, which is only an effector and increases uncoupling as reported for other ornithine converting NMOs, such as VbsO (vide supra).
The biosynthesis of malleobactin A–D involves the NMO MbaC and leads to products with various N-O functional units such as N-hydroxy, C-nitroso, nitro, and azoxy (Fig. 3c) (Franke et al. 2013; Hedges and Ryan 2020). MbaC produces N5-hydroxy ornithine, which is used to assemble different malleobactins. An interesting example constitutes 2-amino-5-nitropentanoic acid, produced from ornithine as an intermediate to malleobactin A in Burkholderia pseudomallei (Franke et al. 2013). Interestingly in this case, the formation of a terminal nitro group is made possible, even though this oxidation would typically be circumvented in other metabolic pathways by immediate acetylation of the formed N-hydroxy group, thereby also creating a bidentate ligand for metal chelation. It is postulated that the NMO in this case does support the conversion of N-hydroxy group towards the nitro group (Fig. 4b) (Franke et al. 2013).
The diverse nature of siderophores and their structural as well as functional elements provide access towards many applications, as has been extensively reviewed (Saha et al. 2016; Su et al. 2018; Albelda-Berenguer et al. 2019; Řezanka et al. 2019; Hofmann et al. 2020) and will not be discussed in detail here.
NMOs with structurally complex substrates
While flavin-based NMOs related to siderophore biosynthesis typically employ the same “simple” substrates to build up highly complex structures, other NMOs have been disclosed that are able to employ structurally quite diverse substrates, ranging from small nonproteinogenic amino acids to large and complex units. The investigation of these enzymes can reveal valuable insights for future biotechnological applications in hitherto unexploited fields.
NMOs for diverse bacterial substrates
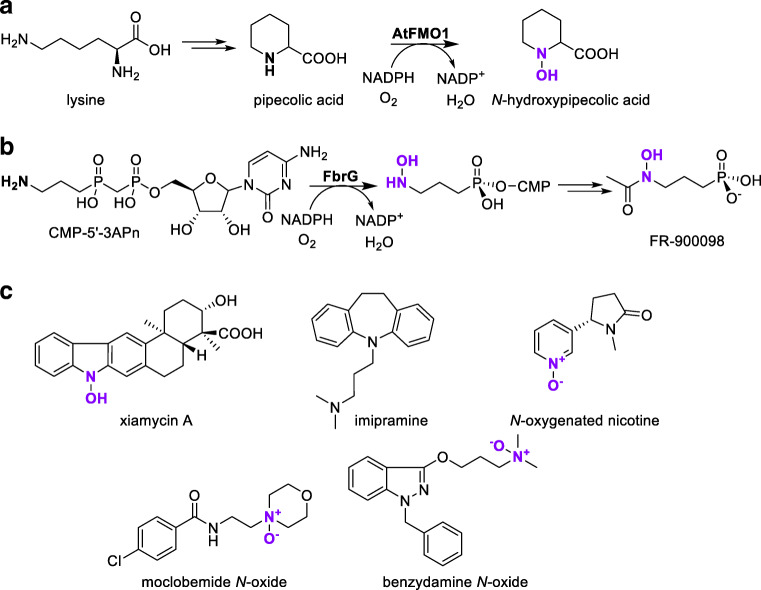
A flavin-dependent NMO named AtFMO1 from the plant Arabidopsis thaliana hydroxylates pipecolic acid to N-hydroxypipecolic acid (Hartmann and Zeier 2018; Hartmann et al. 2018). This reaction is part of a pathogen-inducible catabolic pathway of lysine that has a central function in systemic acquired resistance (Fig. 5a).
Fig. 5.
aAtFMO1-catalyzed hydroxylation of pipecolic acid to N-hydroxypipecolic acid, which provides systemic acquired resistance for plants. b FrbG-catalyzed hydroxylation of the N-acetyl-3-aminopropylphosphonate in the biosynthetic pathway towards the antimalarial agent FR-900098. c Xiamycin A (XMA) is N-hydroxylated by XiaK. hFMO1 catalyzes the N-oxygenation of imipramine, whereas hFMO3 catalyzes the N-oxygenation of nicotine as well as of moclobemide to produce the human drug metabolite moclobemide-N-oxide. N-oxygenation of the anti-inflammatory drug benzydamine by a flavin-containing monooxygenase located in the lungs produces benzydamine-N-oxide
Recently, the groups of Zhao and Nair disclosed the crystal structure and characterization of an FAD-dependent NMO from Streptomyces rubellomurinus, FrbG, that catalyzes the hydroxylation of an aminopropylphosphonate unit within a more complex CMP conjugate (Nguyen et al. 2019). FrbG shares structural similarities with group B FMOs and takes part in the biosynthesis of the antimalarial agent FR-900098, which requires a hydroxamate (Fig. 5b).
The structure obtained from crystallization of FrbG contains an FAD prosthetic group and NADPH coenzyme co-crystallized together, displaying a proper orientation of the nicotinamide ring stacking with the flavin isoalloxazine moiety for optimal hydride transfer. Contrary to group B FMOs (Fig. 2b), the NADPH-binding domain also confers substrate recognition, with the cytidine-5′-monophosphate moiety being crucial for substrate specificity. A conformational reorganization likely occurs after flavin reduction by NADPH (Nguyen et al. 2019), similar to the “moonlighting” effect observed with group B FMOs, i.e., their ability to take over more than one function (Alfieri et al. 2008). The discovery of new types of N-hydroxylases such as FrbG through cloning and sequencing of biosynthetic gene clusters paves the way for metabolite synthesis.
Another bacterial NMO called XiaK, identified from the biosynthetic gene cluster of the indolosesquiterpene xiamycin A (XMA) from a Streptomyces strain, catalyzes the hydroxylation of XMA (Fig. 5c). The groups of Zhang and Liu confirmed XiaK to be a flavin-dependent enzyme functioning as an N-hydroxylase in the biosynthesis of indolosesquiterpene intermediates (Zhang et al. 2017). Recombinant expression and characterization of XiaK showed that it only catalyzed the hydroxylation of XMA and that the N-hydroxylated product decomposed to other compounds in a possible enzyme free radical–mediated mechanism. This highlights the possibility that isolated compounds from microorganisms may not be “true” secondary metabolites but non-enzymatic derivative products of a biosynthesized compound.
FMOs from human/mammalian cells with NMO activity
The human family of FMOs includes five known enzymes, hFMO1–5. As already introduced above, these FMOs belong to the same group of flavoprotein monooxygenases and are able to oxygenate carbon-linked heteroatoms (van Berkel et al. 2006). The five hFMO enzymes exhibit a tissue-specific expression pattern in adults (Perez-Paramo et al. 2019): hFMO1 is expressed in kidneys and hFMO3–5 in the liver, whereas hFMO2 is mainly expressed in the lungs. hFMO5, which was shown to catalyze Baeyer-Villiger oxidation reactions (Fiorentini et al. 2016; Fiorentini et al. 2017), is now commercially available (Gecco Biotech B.V.).
Recently, hFMO3 was reported to exhibit activity towards nicotine (Fig. 5c) (Perez-Paramo et al. 2019). The substrate versatility of this enzyme makes it an attractive catalyst for future applications with complex substrates and towards the synthesis of drug metabolites. Other hFMOs are also involved in nicotine detoxification processes through N-oxygenation in CYP2A6-deficient humans, with hFMO1-3 displaying higher activity and hFMO4-5 lower activity (Perez-Paramo et al. 2019). Furthermore, hFMO1 catalyzes the N-oxygenation of imipramine, among others (Fig. 5c) (Furnes and Schlenk 2004).
A drawback of using hFMOs in biocatalytic applications is their low production levels using recombinant expression in Escherichia coli. Nevertheless, Hanlon et al. managed to produce a sufficient amount of hFMO3 for preparative biotransformation with a large-scale cultivation of recombinant E. coli harboring the gene encoding for hFMO3 (Hanlon et al. 2012). In this way, the authors were able to produce milligram amounts of the drug metabolite moclobemide-N-oxide (Fig. 5c). Such distinctly produced drug metabolites can e.g. be used in the detailed study of metabolites’ mode of action, among others.
Another example is the anti-inflammatory drug benzydamine, metabolized by an FMO located in the lungs, possibly hFMO2 (Fig. 5c) (Störmer et al. 2000). Benzydamine was recently used as a substrate to observe pulmonary FMO activity in rats to assess its metabolic fate (Yilmaz et al. 2019).
NMOs promoting the formation of N-N bonds
N-N bonds are abundant in countless biological compounds (Blair and Sperry 2013; Waldman et al. 2017). Diazo compounds especially have versatile biological functions and are present in numerous biological systems, as has recently been extensively reviewed (Nawrat and Moody 2011; Mix et al. 2016). There are many ways of producing N-N and N=N bonds; NMOs are involved in some of them in different manners. It is especially interesting to see that the biocatalytic pathways of N-N bond containing biomolecules, and thus, the involved enzymes have only been scarcely explored in the past and are gaining increasing attention in recent literature. General pathways have been explored, leading to biomolecules with diverse structures and functions, of which a few are highlighted here.
Amino acid N-hydroxylation and subsequent nitrous acid production
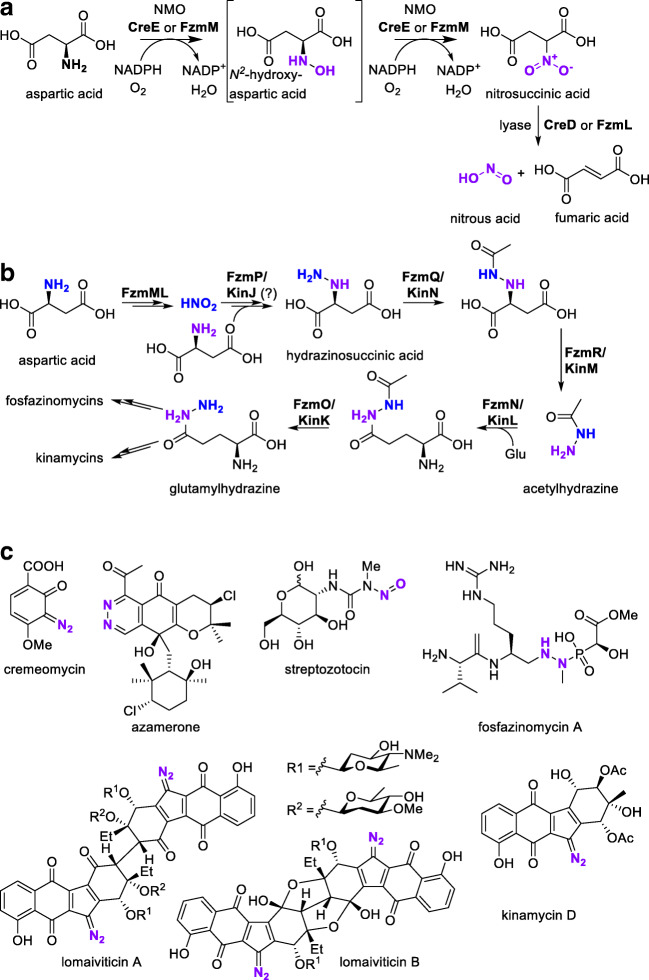
One of the most prominent ways in which NMOs are involved in the generation of N-N bonds is by liberation of nitrous acid starting from the N2-hydroxylation of aspartic acid (Fig. 6a). A stepwise over-oxidation of the amino group to a nitro function by a flavin-based NMO results in the formation of nitrosuccinic acid via the instable N2-hydroxyaspartic acid intermediate (Wang et al. 2018). The oxidation step is followed by a lyase reaction, resulting in cleavage of the nitro group and producing fumaric acid as a second product. The liberated nitrous acid is discussed to react with a primary amine of a second substrate molecule in a non-enzymatic way, thus forming a hydrazine-like intermediate with an N-N bond which can be further modified to produce manifold products. Both the NMO and the lyase are well conserved and widespread in different organisms. It should be mentioned that not only flavin-containing NMOs are capable of liberating small NO compounds; there is a variety of biosynthetic processes that involve “free” NO throughout organisms (Caranto 2019). Several nitrous acid-derived products have recently been reported involving this general biosynthetic pathway, most of them having potential activities as antibiotics or even anti-proliferative agents (Fig. 6c, Table 2, section A).
Fig. 6.
a Release of nitrous acid from aspartic acid, promoted by a double hydroxylation and lyase reaction. b Biosynthesis of fosfazinomycins and kinamycins via the same initial intermediates. c Examples of biomolecules for which biosynthesis involves the NMO-driven generation of nitrous acid
Table 2.
Reported compounds for which the biosynthesis involves either (A) an NMO-induced nitrous acid release or (B) an NMO-catalyzed N-hydroxylation and subsequent condensation reaction to generate (natural) products with complex functional groups
| Compound | Organism | NMO | Additional reaction remarks | References | |
|---|---|---|---|---|---|
| A. NMO-induced nitrous acid release | |||||
| Further relevant enzymes | Coupling partner for HNO2 | ||||
| Cremeomycin | Actinomyces cremeus NRRL3241 | CreE | CreD (lyase) | 3-Amino-2-hydroxy-4-methoxybenzoate | Waldman et al. (2015); Sugai et al. (2016) |
| Fosfazinomycin | Streptomyces sp. NRRL S-149 | FzmM | FzmL | Aspartate | Gao et al. (2014); Huang et al. (2016) |
| Kinamycin | Streptomyces murayamaensis | FzmM | FzmL | Aspartate | Wang et al. (2018) |
| Lomaiviticin | Salinispora sp. | Strop2198 | Kersten et al. (2013) | ||
| Azamerone | Streptomyces sp. CNQ-766 | n.d. | SF2415B1 after C-amination | Amino dihydroquinone intermediate | Winter et al. (2009) |
| Streptozotocin | Streptomyces achromogenes | Unknown | citrulline derivative | Le Goff and Ouazzani (2014) | |
| Triacsins | Streptomyces aureofaciens ATCC31442 | Tri21 (Asp hydroxylase) | Tri16 (lyase) | Hydrazinoacetate derivative | Twigg et al. (2019) |
| Alanosine | Streptomyces alanosinicus DSM 40606 | AlnM | AlnN | (Diaminopropionate bound to AlnI via PCP) | Wang et al. (2020) |
| B. NMO-catalyzedN-hydroxylation and subsequent condensation reactions | |||||
| Substrate | N-OH condensates with | ||||
| Piperazic acid | Actinomycete bacteria | PzbA homologs | Ornithine (ω position) | Itself (α position) | Hu et al. (2019) |
| Kutznerine | Kutzneria sp. 744 | KtzI | Ornithine (ω position) | Itself (α position) | Fujimori et al. (2007) |
| S56-p1 (hydrazinoacetic acid) | Streptomyces sp. SoC090715LN-17 | Spb38 | Lysine | Glycine | Matsuda et al. (2018) |
| Valanymicin | Streptomyces Viridifaciens MG456-hF10 | VlmH | Isobutylamine | Serine | Parry and Li (1997); Tao et al. (2003); Garg et al. (2008); Garg et al. (2009) |
| Pyridazomycin | Streptomyces violaceoniger sp. griseofuscus | Unknown | Ornithine | Oxaloacetate | Bockholt et al. (1994) |
| Triacsins | Streptomyces aureofaciens ATCC31442 | Tri26 | Lysine | Glycine | Twigg et al. (2019) |
| Alanosine | Streptomyces alanosinicus DSM 40606 | AlnG | l-Diaminopropionate bound to AlnI via PCP | NOx | Wang et al. (2020) |
Cremeomycin (Fig. 6c) is one of the best-investigated examples of an antibiotic with a diazo function (Waldman et al. 2015; Sugai et al. 2016). Initially isolated from Actinomyces cremeus (NRRL3241), it adversely affects Gram-positive and Gram-negative bacteria and has antifungal and even antitumor activities (Bergy and Pyke 1967). The diazo-containing moiety of cremeomycin is built up by a late-stage diazotization of an amino function by nitrous acid, itself generated by the typical cascade: aspartic acid is hydroxylated/over-oxidized by the NMO CreE; the formed nitrosuccinic acid liberates nitrous acid by aid of the lyase CreD (Waldman and Balskus 2018).
Fosfazinomycins and kinamycins (Fig. 6b, c) are two antibiotic classes produced by and initially isolated from Streptomyces strains (S. lavendofoliae 630 and S. murayamaensis, respectively) (Ito et al. 1970; Kuroda et al. 1980). Despite their structural diversity, they share the same initial steps of biosynthesis (Fig. 6b) (Wang et al. 2018). Aspartate is hydroxylated by the NMO FzmM to nitrosuccinate in two oxidative stages; liberation of nitrous acid is catalyzed by FzmL, a 3-carboxymuconate cycloisomerase. In multiple subsequent enzymatic steps, the central intermediate glutamylhydrazine is generated via hydrazinosuccinate and acetylhydrazine (Huang et al. 2016). The identified FzmM shows homology to an FAD(NAD)-dependent oxidoreductase from Streptomyces davawensis JCM 4913 (WP_015660731; 56% amino acid identity, 65% similarity) (Gao et al. 2014).
Lomaiviticins (Fig. 6c) are structurally similar to kinamycins, making their biosynthetic origin very likely similar. They are regarded as highly potent antitumor active agents and were initially isolated from Salinispora pacifica (Mix et al. 2016). A potential gene cluster for lovamaiviticin biosynthesis was found in Salinispora tropica CNB-440. The gene strop2198 was assumed to code for a putative FAD-dependent monooxygenase based on homology to Streptomyces albaduncus JagF (CBH32087; 61% amino acid identity 52% similarity) (Kersten et al. 2013).
Azamerone (Fig. 6c) is another compound with an uncommon N-N bond. The potential topoisomerase inhibitor was first isolated from actinomycetes MAR4 (Cho et al. 2006). The biosynthesis of azamerone was initially believed to occur homologous to pyridazomycin (cf. below), but isotope labeling studies indicated that the origin of the diazo function in confirmed precursors to the compound stems from nitric acid, therefore involving the N2-hydroxylation of aspartic acid by an NMO (Winter et al. 2009).
In the case of streptozotocin (Fig. 6C), an anticancer antibiotic known since the 1950s, it is also believed that the nitrosamine functional group originates from a central nitrous acid as intermediate. However, the biocatalytic pathway has not been elucidated to date (Le Goff and Ouazzani 2014).
N-Hydroxylation and condensation with amino or hydroxy functions
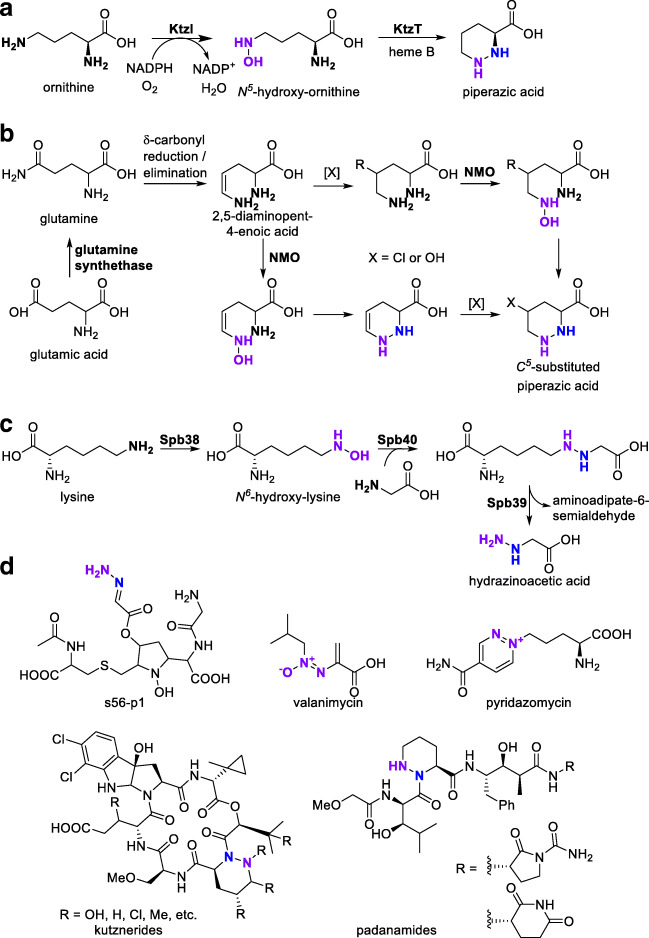
The second typical way to generate N-N bonds in biosynthesis with the aid of flavin-NMOs is the condensation of an NMO-generated hydroxylamine with carboxy functions. Subsequent rearrangement steps can result in new N-N bond containing molecules (Table 2, section B).
Piperazic acid is one of the most common N-N containing building blocks of natural products, being found in numerous natural compounds with activities ranging from antibiotics to immunosuppressants. All piperazic acid-containing molecules known to date come from Actinomycete bacteria isolated from diverse environments (Morgan et al. 2019), but the structural and functional variety is remarkable: kutznerides, padanamides, himastatins, monamycines, polyoxypeptin and sanglifehrins, to name a few, contain piperazic acid (Fig. 7a). A recent review demonstrates the versatile incorporation of piperazic acid into natural products (Morgan et al. 2019).
Fig. 7.
a Biosynthesis of piperazic acid. b Biosynthesis of substituted piperazic acids via two proposed pathways. c Biosynthesis of hydrazinoacetic acid as a building block for more complex biomolecules. d Diverse N-N bond containing biomolecules: s56-p1 is derived from hydrazinoacetic acid, kuznerides, and padanamides contain piperazic acid building blocks. Valanimycin and pyridazomycin are further recently investigated examples
The synthesis of piperazic acid starts with the N5-hydroxylation of ornithine by a flavin-based NMO (Fig. 7a) (Neumann et al. 2012). N5-hydroxyornithine itself does not cyclize spontaneously (Le Goff and Ouazzani 2014). Instead, a heme-dependent protein, typically termed piperazate synthase, fuses the two nitrogen atoms in a condensation reaction, creating piperazic acid (Du et al. 2017). The enzymatic pair is highly conserved in the gene clusters responsible for the synthesis of different piperazic acid–containing molecules (Morgan et al. 2019).
Depending on the final product, the enzyme nomenclature varies. The best-described case, with a detailed functional and structural investigation of the flavin-based NMO, is probably the production of kutznerine (Fig. 7d) by Kutzneria sp. 744, where the two enzymes are designated as N-hydroxylase KtzI and piperazate synthase KtzT (Neumann et al. 2012; Setser et al. 2014; Du et al. 2017). A recent bioinformatic study revealed, next to numerous stand-alone enzyme pairs, the existence of 11 chimeric two-enzyme pairs with unique position-specific amino acid utilization patterns compared with the stand-alone homologs (Hu et al. 2019).
Furthermore, derivatives of piperazic acid, such as 5-hydroxy-, 5-chloro-, and dehydro-piperazic acid can be produced and incorporated into more complex structures such as piperazimycins. There are two theories about how these derivatives are produced from glutamic acid (Fig. 7b) (Miller et al. 2007): the central intermediate 2,5-diaminopent-4-enoic acid is hydroxylated at the N5 position and, subsequently, addition of a heteroatom to the double bond occurs. Alternatively, N-hydroxylation takes place after the addition of the heteroatom to the double bond (Oelke et al. 2011; Handy and Sello 2017). Alternatively, downstream modification of piperazic acid has been discussed after it has been synthesized from ornithine in its cyclic form (Jiang et al. 2011).
Hydrazinoacetic acid is another building block which has been reported in different biosynthetic pathways. It is proposed to be an intermediate in the biosynthesis of a complex natural compound s56-p1, isolated from Streptomyces lividans through heterologous expression (Fig. 7c). It is built from lysine, which is transformed into N6-hydroxylysine by the flavin-dependent hydroxylase Spb38, followed by condensation with glycine through Spb40. The intermediate hydrazine adduct is then oxidized at the C6-N bond by Spb39, which was identified as flavin-dependent d-amino acid oxidase homolog. This reaction results in liberation of hydrazinoacetic acid, which is expected to be directly incorporated into the target compound s56-p1 (Matsuda et al. 2018). The gene cluster was identified and annotated based on homology annotation, with the closest homology for all three genes originating from Catenulispora acidiphila DSM 44928 (Sbp38: 85% amino acid identity, 90% similarity to the respective lysine monooxygenase) (Matsuda et al. 2017).
For valanomycin (Fig. 7d), a versatile azoxy compound from Streptomyces viridifaciens MG456-hF10, biosynthesis involves the decarboxylation of valine and N-hydroxylation of the intermediate isobutylamine. Based on functional and homology annotation, the VlmH/R enzyme pair was denoted as FAD/NADPH-dependent isobutylamine hydroxylase and FAD reductase, respectively. Here, vlmH encodes for an enzyme of 378 amino acids homologous to an isobutylamine N-hydroxylase from Streptomyces avermitilis (BAB69230, 51% amino acid identity and 67% similarity) (Garg et al. 2002). In subsequent steps to generate the -N+(O−)=N- unit, different mechanisms have initially been proposed for a reaction of isobutylhydroxylamine with serine (Tao et al. 2003; Garg et al. 2008; Garg et al. 2009); the condensation of the N-OH function with the serine-carboxy function and successive rearrangement has been widely accepted in the literature (Le Goff and Ouazzani 2014).
The biosynthesis of pyridazomycin (Fig. 7d) is not well understood, but it is believed that pyridazomycin originates from ornithine, oxaloacetate, and glycine, making the intermediate formation of N5-hydroxyornithine which condensates with oxalacetate a very probable hypothesis (Bockholt et al. 1994; Wermuth 2011).
Compounds with more than one NMO involved in biosynthesis
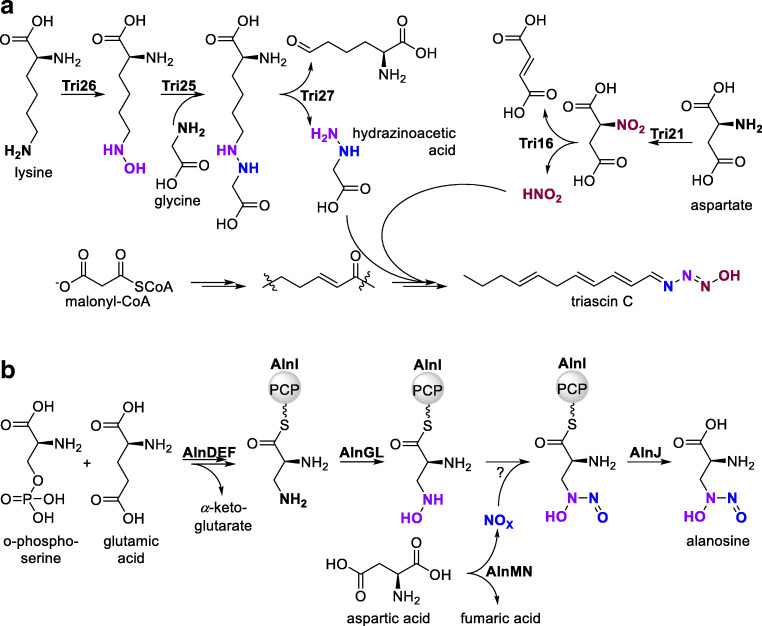
Triacsins are another class of compounds with highly interesting properties and peculiar biosynthetic origin first isolated from Streptomyces aureofaciens ATCC 31442 (Twigg et al. 2019). The compounds bear an N-hydroxytriazene unit terminally bound to a poly-unsaturated C11 alkyl chain (Fig. 8a). The compounds therefore structurally mimic fatty acids and bear multifaceted biological functions, ranging from acetyl-CoA-synthetase inhibition to antimalarial and antiviral activities. For the biosynthesis of the N-hydroxytriazene functional group, two independent N-hydroxylation steps are required (Fig. 8a). Using [15N] isotope labeling and homology annotation, the enzyme Tri21 was recently identified as a flavin-dependent NMO, putatively catalyzing aspartate overoxidation finally leading to nitrous acid liberation (Twigg et al. 2019). The second N-hydroxylation step was ascribed to Tri26, a putative lysine monooxygenase. Subsequent condensation of the N6-hydroxy function with glycine produces first an internal hydrazine unit and next hydrazinoacetic acid. The fatty acid backbone and the two nitrogen components are then assembled in multiple, not yet fully understood steps. Both NMOs involved in this pathway were annotated as flavin-dependent N-hydroxylases, with Tri21 showing homology to an FAD/NAD(P) binding protein from Salinispora pacifica (WP_018723641; 67% amino acid identity, 75% similarity) and Tri26 being homologous to N-hydroxylase MbtG in Streptomyces ipomoeae (WP_009311506, 81% amino acid identity, 89% similarity) (Krithika et al. 2006).
Fig. 8.
a The biosynthesis of triacsins involves two N-hydroxylation steps catalyzed by two different NMOs. b Two N-hydroxylase-catalyzed reaction steps are also involved in the biosynthesis of alanosine
Another important intermediate in the biosynthesis of more complex structures is the non-proteinogenic amino acid alanosine, itself discussed to have antibiotic, antiviral and anticancer activities (Fig. 8b) (Ng et al. 2019). Its biosynthesis was elucidated from the alanosine pathway in Streptomyces alanosinicus sp. (Ng et al. 2019; Wang et al. 2020). A gene pair alnM/N was identified with homology to the above-described creD/E and fzmM/N pairs, with AlnM being annotated as aspartate-converting amine hydroxylase, eventually promoting the liberation of NO2−. The gene alnM was found as 76% identity homolog to a similar gene from Streptomyces kanamyceticus (WP_055544225). Interestingly, a second N-hydroxylation step is discussed within the biosynthesis pathway involving enzymes denoted as AlnG and AlnL. The former was described as a putative flavin-dependent acyl-CoA dehydrogenase, which operates in the oxidative direction to activate oxygen for N4-hydroxylation of a diaminopropane unit (Wang et al. 2020). AlnG was found as 89% identity homolog to a gene from Streptomyces hirstus (WP_055594947). Overall, 14 gene clusters with relation to the l-alanosine gene cluster were identified in Streptomyces and Saccharothrix sp., all containing genes similar to alnG but none with genes for the alnM/N pair (Wang et al. 2020).
Perspectives
In this review, we focused on the remarkable ability of flavin-dependent enzymes to hydroxylate nitrogen-containing compounds, leading to a myriad of possible functional groups which play a major role in the bioactivity of secondary metabolites. The detailed understanding of ornithine-converting NMOs and their mode of action has set the basis for focused research efforts to include this enzyme class in biocatalytic processes. These efforts have especially stimulated processes employing NMOs from the siderophore biosynthetic pathways.
Here, we highlighted the versatility in substrate scope some recently divulged NMOs can have and showcased their broad occurrence in different biosynthetic surroundings. It is especially noteworthy that many of these NMOs have just been discovered from genomics and/or proteomics studies and that they have not been characterized in an enzymological or biocatalytic context. We thus see great potential in these recent developments: From substrate scope to applications, flavin-dependent NMOs are key enzymes that we expect to play an even more prominent role in biotechnology in the near future.
Author contributions
CM and DT conceived and planned the overall reviews’ purpose and structure. CM, TH, and AGB performed primary literature research. CEP and WJHvB added mechanistic details. All authors contributed to the manuscript text and approved the final manuscript.
Funding information
Open Access funding provided by Projekt DEAL. Funding was received as follows: DT, AGB, and CM were supported by the Federal Ministry for Innovation, Science and Research of North Rhine–Westphalia (PtJ-TRI/1411ng006—ChemBioCat). DT and TH were supported by a Junior Research Grant from the German Federal Ministry of Education and Research (BakSolEx 033R147).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Albelda-Berenguer M, Monachon M, Joseph E (2019) Chapter five - siderophores: from natural roles to potential applications. In: Gadd GM, Sariaslani S (eds) Advances in Applied Microbiology. Academic Press, pp 193–225 [DOI] [PubMed]
- Alfieri A, Malito E, Orru R, Fraaije MW, Mattevi A. Revealing the moonlighting role of NADP in the structure of a flavin-containing monooxygenase. Proc Natl Acad Sci. 2008;105:6572–6577. doi: 10.1073/pnas.0800859105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alice AF, López CS, Lowe CA, Ledesma MA, Crosa JH. Genetic and transcriptional analysis of the siderophore malleobactin biosynthesis and transport genes in the human pathogen Burkholderia pseudomallei K96243. J Bacteriol. 2006;188:1551–1566. doi: 10.1128/JB.188.4.1551-1566.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosi C, Leoni L, Putignani L, Orsi N, Visca P. Pseudobactin biogenesis in the plant growth-promoting Rhizobacterium Pseudomonas strain B10: identification and functional analysis of the l-ornithine N5-oxygenase (psbA) gene. J Bacteriol. 2000;182:6233–6238. doi: 10.1128/JB.182.21.6233-6238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou DP, Entsch B (2013) The reaction mechanisms of groups A and B flavoprotein monooxygenases. In: Complex Flavoproteins. Dehydrogenases and Physical Methods. De Gruyter, Berlin/Boston, pp 1–28
- Barona-Gómez F, Wong U, Giannakopulos AE, Derrick PJ, Challis GL. Identification of a cluster of genes that directs Desferrioxamine biosynthesis in Streptomyces coelicolor M145. J Am Chem Soc. 2004;126:16282–16283. doi: 10.1021/ja045774k. [DOI] [PubMed] [Google Scholar]
- Bergy ME, Pyke TR (1967) Cremeomycin and process for making. US3350269A
- Binda C, Robinson RM, del Campo JSM, Keul ND, Rodriguez PJ, Robinson HH, Mattevi A, Sobrado P. An unprecedented NADPH domain conformation in lysine monooxygenase NbtG provides insights into uncoupling of oxygen consumption from substrate hydroxylation. J Biol Chem. 2015;290:12676–12688. doi: 10.1074/jbc.M114.629485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair LM, Sperry J. Natural products containing a nitrogen–nitrogen bond. J Nat Prod. 2013;76:794–812. doi: 10.1021/np400124n. [DOI] [PubMed] [Google Scholar]
- Bockholt H, Beale JM, Rohr J (1994) Biosynthetic investigations on pyridazomycin. Angew Chem Int Ed 33:1648–1651. 10.1002/anie.199416481
- Bosello M, Mielcarek A, Giessen TW, Marahiel MA. An enzymatic pathway for the biosynthesis of the formylhydroxyornithine required for Rhodochelin iron coordination. Biochemistry. 2012;51:3059–3066. doi: 10.1021/bi201837f. [DOI] [PubMed] [Google Scholar]
- Bufkin K, Sobrado P. Characterization of the ornithine hydroxylation step in albachelin biosynthesis. Molecules. 2017;22:1652. doi: 10.3390/molecules22101652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caranto JD. The emergence of nitric oxide in the biosynthesis of bacterial natural products. Curr Opin Chem Biol. 2019;49:130–138. doi: 10.1016/j.cbpa.2018.11.007. [DOI] [PubMed] [Google Scholar]
- Carroll CS, Moore MM. Ironing out siderophore biosynthesis: a review of non-ribosomal peptide synthetase (NRPS)-independent siderophore synthetases. Crit Rev Biochem Mol Biol. 2018;53:356–381. doi: 10.1080/10409238.2018.1476449. [DOI] [PubMed] [Google Scholar]
- Cho JY, Kwon HC, Williams PG, Jensen PR, Fenical W (2006) Azamerone, a terpenoid phthalazinone from a marine-derived bacterium related to the genus Streptomyces (Actinomycetales). Org Lett 8:2471–2474. 10.1021/ol060630r [DOI] [PMC free article] [PubMed]
- Chocklett SW, Sobrado P. Aspergillus fumigatus SidA is a highly specific ornithine hydroxylase with bound flavin cofactor. Biochemistry. 2010;49:6777–6783. doi: 10.1021/bi100291n. [DOI] [PubMed] [Google Scholar]
- Davison EK, Sperry J. Natural products with heteroatom-rich ring systems. J Nat Prod. 2017;80:3060–3079. doi: 10.1021/acs.jnatprod.7b00575. [DOI] [PubMed] [Google Scholar]
- Dimise EJ, Widboom PF, Bruner SD. Structure elucidation and biosynthesis of fuscachelins, peptide siderophores from the moderate thermophile Thermobifida fusca. Proc Natl Acad Sci. 2008;105:15311–15316. doi: 10.1073/pnas.0805451105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y-L, He H-Y, Higgins MA, Ryan KS. A heme-dependent enzyme forms the nitrogen–nitrogen bond in piperazate. Nat Chem Biol. 2017;13:836–838. doi: 10.1038/nchembio.2411. [DOI] [PubMed] [Google Scholar]
- Eisendle M, Oberegger H, Zadra I, Haas H. The siderophore system is essential for viability of Aspergillus nidulans: functional analysis of two genes encoding l-ornithine N5-monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC) Mol Microbiol. 2003;49:359–375. doi: 10.1046/j.1365-2958.2003.03586.x. [DOI] [PubMed] [Google Scholar]
- Esuola CO, Babalola OO, Heine T, Schwabe R, Schlömann M, Tischler D (2016) Identification and characterization of a FAD-dependent putrescine N-hydroxylase (GorA) from Gordonia rubripertincta CWB2. J Mol Catal B Enzym 134:378–389. 10.1016/j.molcatb.2016.08.003
- Fiorentini F, Geier M, Binda C, Winkler M, Faber K, Hall M, Mattevi A. Biocatalytic characterization of human FMO5: unearthing Baeyer–Villiger reactions in humans. ACS Chem Biol. 2016;11:1039–1048. doi: 10.1021/acschembio.5b01016. [DOI] [PubMed] [Google Scholar]
- Fiorentini F, Romero E, Fraaije MW, Faber K, Hall M, Mattevi A. Baeyer–Villiger monooxygenase FMO5 as entry point in drug metabolism. ACS Chem Biol. 2017;12:2379–2387. doi: 10.1021/acschembio.7b00470. [DOI] [PubMed] [Google Scholar]
- Fraaije MW, van Berkel WJH. Biocatalysis in the Pharmaceutical and Biotechnology Industries. 2006. Flavin-containing oxidative biocatalysts; pp. 181–202. [Google Scholar]
- Franceschini S, Fedkenheuer M, Vogelaar NJ, Robinson HH, Sobrado P, Mattevi A. Structural insight into the mechanism of oxygen activation and substrate selectivity of flavin-dependent N-hydroxylating monooxygenases. Biochemistry. 2012;51:7043–7045. doi: 10.1021/bi301072w. [DOI] [PubMed] [Google Scholar]
- Franke J, Ishida K, Ishida-Ito M, Hertweck C. Nitro versus hydroxamate in siderophores of pathogenic bacteria: effect of missing hydroxylamine protection in malleobactin biosynthesis. Angew Chem Int Ed. 2013;52:8271–8275. doi: 10.1002/anie.201303196. [DOI] [PubMed] [Google Scholar]
- Frederick RE, Mayfield JA, DuBois JL. Regulated O2 activation in flavin-dependent monooxygenases. J Am Chem Soc. 2011;133:12338–12341. doi: 10.1021/ja203397s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori DG, Hrvatin S, Neumann CS, Strieker M, Marahiel MA, Walsh CT. Cloning and characterization of the biosynthetic gene cluster for kutznerides. Proc Natl Acad Sci. 2007;104:16498–16503. doi: 10.1073/pnas.0708242104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnes B, Schlenk D. Evaluation of xenobiotic N- and S-oxidation by variant flavin-containing monooxygenase 1 (FMO1) enzymes. Toxicol Sci. 2004;78:196–203. doi: 10.1093/toxsci/kfh079. [DOI] [PubMed] [Google Scholar]
- Gao J, Ju K-S, Yu X, Velásquez JE, Mukherjee S, Lee J, Zhao C, Evans BS, Doroghazi JR, Metcalf WW, van der Donk WA. Use of a phosphonate methyltransferase in the identification of the fosfazinomycin biosynthetic gene cluster. Angew Chem Int Ed. 2014;53:1334–1337. doi: 10.1002/anie.201308363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg RP, Ma Y, Hoyt JC, Parry RJ. Molecular characterization and analysis of the biosynthetic gene cluster for the azoxy antibiotic valanimycin. Mol Microbiol. 2002;46:505–517. doi: 10.1046/j.1365-2958.2002.03169.x. [DOI] [PubMed] [Google Scholar]
- Garg RP, Qian XL, Alemany LB, Moran S, Parry RJ. Investigations of valanimycin biosynthesis: elucidation of the role of seryl-tRNA. Proc Natl Acad Sci. 2008;105:6543–6547. doi: 10.1073/pnas.0708957105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg RP, Alemany LB, Moran S, Parry RJ. Identification, characterization, and bioconversion of a new intermediate in valanimycin biosynthesis. J Am Chem Soc. 2009;131:9608–9609. doi: 10.1021/ja901243p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Seah SYK (2006) Heterologous expression, purification, and characterization of an l-ornithine N5-hydroxylase involved in pyoverdine siderophore biosynthesis in Pseudomonas aeruginosa. J Bacteriol 188:7205–7210. 10.1128/JB.00949-06 [DOI] [PMC free article] [PubMed]
- Handy EL, Sello JK. Structure and synthesis of conformationally constrained molecules containing piperazic acid. In: Lubell WD, editor. Peptidomimetics I. Cham: Springer International Publishing; 2017. pp. 97–124. [Google Scholar]
- Hanlon SP, Camattari A, Abad S, Glieder A, Kittelmann M, Lütz S, Wirz B, Winkler M. Expression of recombinant human flavin monooxygenase and moclobemide-N-oxide synthesis on multi-mg scale. Chem Commun. 2012;48:6001–6003. doi: 10.1039/C2CC17878H. [DOI] [PubMed] [Google Scholar]
- Hartmann M, Zeier J. l-lysine metabolism to N-hydroxypipecolic acid: an integral immune-activating pathway in plants. Plant J. 2018;96:5–21. doi: 10.1111/tpj.14037. [DOI] [PubMed] [Google Scholar]
- Hartmann M, Zeier T, Bernsdorff F, Reichel-Deland V, Kim D, Hohmann M, Scholten N, Schuck S, Bräutigam A, Hölzel T, Ganter C, Zeier J. Flavin monooxygenase-generated N-hydroxypipecolic acid is a critical element of plant systemic immunity. Cell. 2018;173:456–469.e16. doi: 10.1016/j.cell.2018.02.049. [DOI] [PubMed] [Google Scholar]
- Hedges JB, Ryan KS (2020) Biosynthetic pathways to nonproteinogenic α-amino acids. Chem Rev 120:3161–3209. 10.1021/acs.chemrev.9b00408 [DOI] [PubMed]
- Heemstra JR, Walsh CT, Sattely ES. Enzymatic tailoring of ornithine in the biosynthesis of the rhizobium cyclic trihydroxamate siderophore vicibactin. J Am Chem Soc. 2009;131:15317–15329. doi: 10.1021/ja9056008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine T, Mehnert M, Schwabe R, Tischler D (2017) Thermochelin, a hydroxamate siderophore from Thermocrispum agreste DSM 44070. Solid State Phenom 262:501–504. 10.4028/www.scientific.net/SSP.262.501
- Herrero M, de Lorenzo V, Neilands JB. Nucleotide sequence of the iucD gene of the pColV-K30 aerobactin operon and topology of its product studied with phoA and lacZ gene fusions. J Bacteriol. 1988;170:56–64. doi: 10.1128/jb.170.1.56-64.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hissen AHT, Wan ANC, Warwas ML, Pinto LJ, Moore MM. The Aspergillus fumigatus siderophore biosynthetic gene sidA, encoding l-ornithine N5-oxygenase, is required for virulence. Infect Immun. 2005;73:5493–5503. doi: 10.1128/IAI.73.9.5493-5503.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M, Retamal-Morales G, Tischler D (2020) Metal binding ability of microbial metallophores and potential applications. Nat Prod Rep published online. 10.1039/C9NP00058E [DOI] [PubMed]
- Hu Y, Qi Y, Stumpf SD, D’Alessandro JM, Blodgett JAV. Bioinformatic and functional evaluation of actinobacterial piperazate metabolism. ACS Chem Biol. 2019;14:696–703. doi: 10.1021/acschembio.8b01086. [DOI] [PubMed] [Google Scholar]
- Huang Z, Abraham Wang K-K, van der Donk WA. New insights into the biosynthesis of fosfazinomycin. Chem Sci. 2016;7:5219–5223. doi: 10.1039/C6SC01389A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbers MME, Montersino S, Westphal AH, Tischler D, van Berkel WJH. Flavin dependent monooxygenases. Arch Biochem Biophys. 2014;544:2–17. doi: 10.1016/j.abb.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Ito S, Matsuya T, Omura S, Otani M, Nakagawa A, Takeshima H, Iwai Y, Ohtani M, Hata T. A new antibiotic, kinamycin. J Antibiot (Tokyo) 1970;23:315–317. doi: 10.7164/antibiotics.23.315. [DOI] [PubMed] [Google Scholar]
- Jiang W, Heemstra JR, Forseth RR, Neumann CS, Manaviazar S, Schroeder FC, Hale KJ, Walsh CT. Biosynthetic chlorination of the piperazate residue in kutzneride biosynthesis by KthP. Biochemistry. 2011;50:6063–6072. doi: 10.1021/bi200656k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CW, Wyatt MA, Li X, Ibrahim A, Shuster J, Southam G, Magarvey NA. Gold biomineralization by a metallophore from a gold-associated microbe. Nat Chem Biol. 2013;9:241–243. doi: 10.1038/nchembio.1179. [DOI] [PubMed] [Google Scholar]
- Kadi N, Song L, Challis GL (2008) Bisucaberin biosynthesis: an adenylating domain of the BibC multi-enzyme catalyzes cyclodimerization of N-hydroxy-N-succinylcadaverine. Chem Commun:5119–5121. 10.1039/B813029A [DOI] [PubMed]
- Kang HY, Brickman TJ, Beaumont FC, Armstrong SK (1996) Identification and characterization of iron-regulated Bordetella pertussis alcaligin siderophore biosynthesis genes. J Bacteriol 178:4877–4884. 10.1128/jb.178.16.4877-4884.1996 [DOI] [PMC free article] [PubMed]
- Kem MP, Butler A. Acyl peptidic siderophores: structures, biosyntheses and post-assembly modifications. BioMetals. 2015;28:445–459. doi: 10.1007/s10534-015-9827-y. [DOI] [PubMed] [Google Scholar]
- Kersten RD, Lane AL, Nett M, Richter TKS, Duggan BM, Dorrestein PC, Moore BS (2013) Bioactivity-guided genome mining reveals the lomaiviticin biosynthetic gene cluster in Salinispora tropica. ChemBioChem 14:955–962. 10.1002/cbic.201300147 [DOI] [PMC free article] [PubMed]
- Kodani S, Komaki H, Suzuki M, Hemmi H, Ohnishi-Kameyama M. Isolation and structure determination of new siderophore albachelin from Amycolatopsis alba. BioMetals. 2015;28:381–389. doi: 10.1007/s10534-015-9842-z. [DOI] [PubMed] [Google Scholar]
- Kreutzer MF, Nett M (2012) Genomics-driven discovery of taiwachelin, a lipopeptide siderophore from Cupriavidus taiwanensis. Org Biomol Chem 10:9338–9343. 10.1039/C2OB26296G [DOI] [PubMed]
- Krithika R, Marathe U, Saxena P, Ansari MZ, Mohanty D, Gokhale RS. A genetic locus required for iron acquisition in Mycobacterium tuberculosis. Proc Natl Acad Sci. 2006;103:2069–2074. doi: 10.1073/pnas.0507924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda Y, Okuhara M, Goto T, Okamoto M, Terano H, Kohsaka M, Aoki H, Imanaka H. Studies on new phosphonic acid antibiotics. J Antibiot (Tokyo) 1980;33:29–35. doi: 10.7164/antibiotics.33.29. [DOI] [PubMed] [Google Scholar]
- Kurth C, Wasmuth I, Wichard T, Pohnert G, Nett M. Algae induce siderophore biosynthesis in the freshwater bacterium Cupriavidus necator H16. BioMetals. 2019;32:77–88. doi: 10.1007/s10534-018-0159-6. [DOI] [PubMed] [Google Scholar]
- Le Goff G, Ouazzani J. Natural hydrazine-containing compounds: biosynthesis, isolation, biological activities and synthesis. Bioorg Med Chem. 2014;22:6529–6544. doi: 10.1016/j.bmc.2014.10.011. [DOI] [PubMed] [Google Scholar]
- Li B, Lowe-Power T, Kurihara S, Gonzales S, Naidoo J, MacMillan JB, Allen C, Michael AJ (2016) Functional identification of putrescine C- and N-hydroxylases. ACS Chem Biol 11:2782–2789. 10.1021/acschembio.6b00629 [DOI] [PubMed]
- Lynch D, O’Brien J, Welch T, Clarke P, Cuív PO, Crosa JH, O’Connell M. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J Bacteriol. 2001;183:2576–2585. doi: 10.1128/JB.183.8.2576-2585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan CA, Cheng T-Y, Layre E, Young DC, McConnell MJ, Debono CA, Murry JP, Wei J-R, Barry CE, Rodriguez GM, Matsunaga I, Rubin EJ, Moody DB. Lipidomic discovery of deoxysiderophores reveals a revised mycobactin biosynthesis pathway in Mycobacterium tuberculosis. Proc Natl Acad Sci. 2012;109:1257–1262. doi: 10.1073/pnas.1109958109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascotti ML, Lapadula WJ, Ayub MJ (2015) The origin and evolution of Baeyer-Villiger monooxygenases (BVMOs): an ancestral family of flavin monooxygenases. PLoS One 10:e0132689. 10.1371/journal.pone.0132689 [DOI] [PMC free article] [PubMed]
- Massey V. Activation of molecular oxygen by flavins and flavoproteins. J Biol Chem. 1994;269:22459–22462. [PubMed] [Google Scholar]
- Matsuda K, Hasebe F, Shiwa Y, Kanesaki Y, Tomita T, Yoshikawa H, Shin-ya K, Kuzuyama T, Nishiyama M. Genome mining of amino group carrier protein-mediated machinery: discovery and biosynthetic characterization of a natural product with unique hydrazone unit. ACS Chem Biol. 2017;12:124–131. doi: 10.1021/acschembio.6b00818. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Tomita T, Shin-ya K, Wakimoto T, Kuzuyama T, Nishiyama M. Discovery of unprecedented hydrazine-forming machinery in bacteria. J Am Chem Soc. 2018;140:9083–9086. doi: 10.1021/jacs.8b05354. [DOI] [PubMed] [Google Scholar]
- Mayfield JA, Frederick RE, Streit BR, Wencewicz TA, Ballou DP, DuBois JL (2010) Comprehensive spectroscopic, steady state, and transient kinetic studies of a representative siderophore-associated flavin monooxygenase. J Biol Chem 285:30375–30388. 10.1074/jbc.M110.157578 [DOI] [PMC free article] [PubMed]
- Meneely KM, Lamb AL. Biochemical characterization of a flavin adenine dinculeotide-dependent monooxygenase, ornithine hydroxylase from Pseudomonas aeruginosa, suggests a novel reaction mechanism. Biochemistry. 2007;46:11930–11937. doi: 10.1021/bi700932q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneely KM, Barr EW, Bollinger JM, Lamb AL. Kinetic mechanism of ornithine hydroxylase (PvdA) from Pseudomonas aeruginosa: substrate triggering of O2 addition but not flavin reduction. Biochemistry. 2009;48:4371–4376. doi: 10.1021/bi900442z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ED, Kauffman CA, Jensen PR, Fenical W. Piperazimycins: cytotoxic hexadepsipeptides from a marine-derived bacterium of the genus Streptomyces. J Org Chem. 2007;72:323–330. doi: 10.1021/jo061064g. [DOI] [PubMed] [Google Scholar]
- Mix KA, Aronoff MR, Raines RT. Diazo compounds: versatile tools for chemical biology. ACS Chem Biol. 2016;11:3233–3244. doi: 10.1021/acschembio.6b00810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan KD, Andersen RJ, Ryan KS. Piperazic acid-containing natural products: structures and biosynthesis. Nat Prod Rep. 2019;36:1628–1653. doi: 10.1039/C8NP00076J. [DOI] [PubMed] [Google Scholar]
- Nawrat CC, Moody CJ. Natural products containing a diazo group. Nat Prod Rep. 2011;28:1426–1444. doi: 10.1039/C1NP00031D. [DOI] [PubMed] [Google Scholar]
- Neumann CS, Jiang W, Heemstra JR Jr, Gontang EA, Kolter R, Walsh CT (2012) Biosynthesis of piperazic acid via N5-hydroxy-ornithine in Kutzneria spp. 744. ChemBioChem 13:972–976 [DOI] [PMC free article] [PubMed]
- Ng TL, McCallum ME, Zheng CR, Wang JX, Wu KJY, Balskus EP (2020) The l-alanosine gene cluster encodes a pathway for diazeniumdiolate biosynthesis. ChemBioChem 21:1155–1160. 10.1002/cbic.201900565 [DOI] [PMC free article] [PubMed]
- Nguyen K, DeSieno MA, Bae B, Johannes TW, Cobb RE, Zhao H, Nair SK. Characterization of the flavin monooxygenase involved in biosynthesis of the antimalarial FR-900098. Org Biomol Chem. 2019;17:1506–1518. doi: 10.1039/C8OB02840K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelke AJ, France DJ, Hofmann T, Wuitschik G, Ley SV. Piperazic acid -containing natural products: isolation, biological relevance and total synthesis. Nat Prod Rep. 2011;28:1445–1471. doi: 10.1039/C1NP00041A. [DOI] [PubMed] [Google Scholar]
- Olucha J, Lamb AL. Mechanistic and structural studies of the N-hydroxylating flavoprotein monooxygenases. Bioorg Chem. 2011;39:171–177. doi: 10.1016/j.bioorg.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olucha J, Meneely KM, Chilton AS, Lamb AL. Two structures of an N-hydroxylating flavoprotein monooxygenase ornithine hydroxylase from Pseudomonas aerugionosa. J Biol Chem. 2011;286:31789–31798. doi: 10.1074/jbc.M111.265876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry RJ, Li W. Purification and characterization of isobutylamine N-hydroxylase from the valanimycin producer Streptomyces viridifaciens MG456-hF10. Arch Biochem Biophys. 1997;339:47–54. doi: 10.1006/abbi.1996.9857. [DOI] [PubMed] [Google Scholar]
- Perez-Paramo YX, Chen G, Ashmore JH, Watson CJW, Nasrin S, Adams-Haduch J, Wang R, Gao Y-T, Koh W-P, Yuan J-M, Lazarus P. Nicotine-N’-oxidation by flavin monooxygenase enzymes. Cancer Epidemiol Prev Biomark. 2019;28:311–320. doi: 10.1158/1055-9965.EPI-18-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlmann V, Marahiel MA. δ-Amino group hydroxylation of l-ornithine during coelichelin biosynthesis. Org Biomol Chem. 2008;6:1843–1848. doi: 10.1039/B801016A. [DOI] [PubMed] [Google Scholar]
- Proença DN, Heine T, Senges CHR, Bandow JE, Morais PV, Tischler D (2019) Bacterial metabolites produced under iron limitation kill pinewood nematode and attract Caenorhabditis elegans. Front Microbiol 10:2166. 10.3389/fmicb.2019.02166 [DOI] [PMC free article] [PubMed]
- Řezanka T, Palyzová A, Faltýsková H, Sigler K (2019) Chapter 5 - Siderophores: amazing metabolites of microorganisms. In: Atta-ur-Rahman (ed) Studies in Natural Products Chemistry. Elsevier, pp 157–188
- Robbel L, Helmetag V, Knappe TA, Marahiel MA. Consecutive enzymatic modification of ornithine generates the hydroxamate moieties of the siderophore erythrochelin. Biochemistry. 2011;50:6073–6080. doi: 10.1021/bi200699x. [DOI] [PubMed] [Google Scholar]
- Robinson RM, Sobrado P. Flavin-dependent monooxygenases in siderophore biosynthesis. In: Hille R, Miller S, Palfey B, editors. Complex Flavoproteins. Berlin: Dehydrogenases and Physical Methods. De Gruyter; 2012. pp. 29–50. [Google Scholar]
- Robinson R, Badieyan S, Sobrado P. C4a-hydroperoxyflavin formation in N-hydroxylating flavin monooxygenases is mediated by the 2′-OH of the nicotinamide ribose of NADP+ Biochemistry. 2013;52:9089–9091. doi: 10.1021/bi4014903. [DOI] [PubMed] [Google Scholar]
- Robinson R, Franceschini S, Fedkenheuer M, Rodriguez PJ, Ellerbrock J, Romero E, Echandi MP, Martin del Campo JS, Sobrado P. Arg279 is the key regulator of coenzyme selectivity in the flavin-dependent ornithine monooxygenase SidA. Biochim Biophys Acta BBA - Proteins Proteomics. 2014;1844:778–784. doi: 10.1016/j.bbapap.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Robinson RM, Rodriguez PJ, Sobrado P. Mechanistic studies on the flavin-dependent N6-lysine monooxygenase MbsG reveal an unusual control for catalysis. Arch Biochem Biophys. 2014;550–551:58–66. doi: 10.1016/j.abb.2014.04.006. [DOI] [PubMed] [Google Scholar]
- Robinson R, Qureshi IA, Klancher CA, Rodriguez PJ, Tanner JJ, Sobrado P. Contribution to catalysis of ornithine binding residues in ornithine N5-monooxygenase. Arch Biochem Biophys. 2015;585:25–31. doi: 10.1016/j.abb.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero E, Fedkenheuer M, Chocklett SW, Qi J, Oppenheimer M, Sobrado P. Dual role of NADP(H) in the reaction of a flavin dependent N-hydroxylating monooxygenase. Biochim Biophys Acta BBA Proteins Proteomics. 2012;1824:850–857. doi: 10.1016/j.bbapap.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Romero E, Robinson R, Sobrado P (2012b) Monitoring the reductive and oxidative half-reactions of a flavin-dependent monooxygenase using stopped-flow spectrophotometry. J Vis Exp:e3803. 10.3791/3803 [DOI] [PMC free article] [PubMed]
- Ronan JL, Kadi N, McMahon SA, Naismith JH, Alkhalaf LM, Challis GL. Desferrioxamine biosynthesis: diverse hydroxamate assembly by substrate-tolerant acyl transferase DesC. Philos Trans R Soc B Biol Sci. 2018;373:20170068. doi: 10.1098/rstb.2017.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryerson CC, Ballou DP, Walsh C. Mechanistic studies on cyclohexanone oxygenase. Biochemistry. 1982;21:2644–2655. doi: 10.1021/bi00540a011. [DOI] [PubMed] [Google Scholar]
- Saha M, Sarkar S, Sarkar B, Sharma BK, Bhattacharjee S, Tribedi P. Microbial siderophores and their potential applications: a review. Environ Sci Pollut Res. 2016;23:3984–3999. doi: 10.1007/s11356-015-4294-0. [DOI] [PubMed] [Google Scholar]
- Salomone-Stagni M, Bartho JD, Polsinelli I, Bellini D, Walsh MA, Demitri N, Benini S. A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora. J Struct Biol. 2018;202:236–249. doi: 10.1016/j.jsb.2018.02.002. [DOI] [PubMed] [Google Scholar]
- Saroja NR, Mohan AHS, Srividya D, Supreetha K (2019) Chaperone-assisted expression and purification of putrescine monooxygenase from Shewanella putrefaciens-95. Protein Expr Purif 157:9–16. 10.1016/j.pep.2019.01.006 [DOI] [PubMed]
- Schwabe R, Anke MK, Szymańska K, Wiche O, Tischler D. Analysis of desferrioxamine-like siderophores and their capability to selectively bind metals and metalloids: development of a robust analytical RP-HPLC method. Res Microbiol. 2018;169:598–607. doi: 10.1016/j.resmic.2018.08.002. [DOI] [PubMed] [Google Scholar]
- Senges CHR, Al-Dilaimi A, Marchbank DH, Wibberg D, Winkler A, Haltli B, Nowrousian M, Kalinowski J, Kerr RG, Bandow JE. The secreted metabolome of Streptomyces chartreusis and implications for bacterial chemistry. Proc Natl Acad Sci. 2018;115:2490–2495. doi: 10.1073/pnas.1715713115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setser JW, Heemstra JR Jr, Walsh CT, Drennan CL (2014) Crystallographic evidence of drastic conformational changes in the active site of a flavin-dependent N-hydroxylase. Biochemistry 53:6063–6077 [DOI] [PMC free article] [PubMed]
- Sheng D, Ballou DP, Massey V. Mechanistic studies of cyclohexanone monooxygenase: chemical properties of intermediates involved in catalysis. Biochemistry. 2001;40:11156–11167. doi: 10.1021/bi011153h. [DOI] [PubMed] [Google Scholar]
- Shirey C, Badieyan S, Sobrado P. Role of Ser-257 in the sliding mechanism of NADP(H) in the reaction catalyzed by the Aspergillus fumigatus flavin-dependent ornithine N5-monooxygenase SidA. J Biol Chem. 2013;288:32440–32448. doi: 10.1074/jbc.M113.487181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol PA, Darling P, Woods DE, Mahenthiralingam E, Kooi C. Role of ornibactin biosynthesis in the virulence of Burkholderia cepacia: characterization of pvdA, the gene encoding l-ornithine N5-oxygenase. Infect Immun. 1999;67:4443. doi: 10.1128/IAI.67.9.4443-4455.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Störmer E, Roots I, Brockmöller J. Benzydamine N-oxidation as an index reaction reflecting FMO activity in human liver microsomes and impact of FMO3 polymorphisms on enzyme activity. Br J Clin Pharmacol. 2000;50:553–561. doi: 10.1046/j.1365-2125.2000.00296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q, Xu G, Guan T, Que Y, Lu H. Mass spectrometry-derived systems biology technologies delineate the system’s biochemical applications of siderophores. Mass Spectrom Rev. 2018;37:188–201. doi: 10.1002/mas.21513. [DOI] [PubMed] [Google Scholar]
- Sugai Y, Katsuyama Y, Ohnishi Y. A nitrous acid biosynthetic pathway for diazo group formation in bacteria. Nat Chem Biol. 2016;12:73–75. doi: 10.1038/nchembio.1991. [DOI] [PubMed] [Google Scholar]
- Sulzbach M, Kunjapur AM (2020) The pathway less traveled: engineering biosynthesis of nonstandard functional groups. Trends Biotechnol 38:532–545. 10.1016/j.tibtech.2019.12.014 [DOI] [PubMed]
- Tao T, Alemany LB, Parry RJ. Valanimycin biosynthesis: investigations of the mechanism of isobutylhydroxylamine incorporation. Org Lett. 2003;5:1213–1215. doi: 10.1021/ol0340989. [DOI] [PubMed] [Google Scholar]
- Thariath A, Socha D, Valvano MA, Viswanatha T (1993) Construction and biochemical characterization of recombinant cytoplasmic forms of the IucD protein (lysine:N6-hydroxylase) encoded by the pColV-K30 aerobactin gene cluster. J Bacteriol 175:589–596. 10.1128/jb.175.3.589-596.1993 [DOI] [PMC free article] [PubMed]
- Twigg FF, Cai W, Huang W, Liu J, Sato M, Perez TJ, Geng J, Dror MJ, Montanez I, Tong TL, Lee H, Zhang W (2019) Identifying the biosynthetic gene cluster for triacsins with an N-hydroxytriazene moiety. ChemBioChem 20:1145–1149. 10.1002/cbic.201800762 [DOI] [PMC free article] [PubMed]
- van Berkel WJH, Müller F. Flavin-dependent monooxygenases with special reference to p-hydroxybenzoate hydroxylase. In: Müller F, editor. Chemistry and biochemistry of flavoenzymes. 1. Boca Raton: CRC Press; 1991. pp. 1–30. [Google Scholar]
- van Berkel WJH, Kamerbeek NM, Fraaije MW. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J Biotechnol. 2006;124:670–689. doi: 10.1016/j.jbiotec.2006.03.044. [DOI] [PubMed] [Google Scholar]
- van Berkel WJH, Montersino S, Tischler D, Kaschabek S, Schlömann M, Gassner GT (2011) Flavins on the move: flavoprotein hydroxylases and epoxidases. In: Flavins and Flavoproteins 2011: proceedings 17th international symposium on Flavins and Flavoproteins 2011, Berkeley, pp 277–288
- Waldman AJ, Balskus EP. Discovery of a diazo-forming enzyme in cremeomycin biosynthesis. J Org Chem. 2018;83:7539–7546. doi: 10.1021/acs.joc.8b00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman AJ, Pechersky Y, Wang P, Wang JX, Balskus EP. The cremeomycin biosynthetic gene cluster encodes a pathway for diazo formation. ChemBioChem. 2015;16:2172–2175. doi: 10.1002/cbic.201500407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman AJ, Ng TL, Wang P, Balskus EP. Heteroatom–heteroatom bond formation in natural product biosynthesis. Chem Rev. 2017;117:5784–5863. doi: 10.1021/acs.chemrev.6b00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K-KA, Ng TL, Wang P, Huang Z, Balskus EP, van der Donk WA. Glutamic acid is a carrier for hydrazine during the biosyntheses of fosfazinomycin and kinamycin. Nat Commun. 2018;9:1–11. doi: 10.1038/s41467-018-06083-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Niikura H, He H-Y, Daniel-Ivad P, Ryan KS. Biosynthesis of the N–N-bond-containing compound l-alanosine. Angew Chem Int Ed. 2020;59:3881–3885. doi: 10.1002/anie.201913458. [DOI] [PubMed] [Google Scholar]
- Wermuth CG. Are pyridazines privileged structures? MedChemComm. 2011;2:935–941. doi: 10.1039/C1MD00074H. [DOI] [Google Scholar]
- Winter JM, Jansma AL, Handel TM, Moore BS. Formation of the pyridazine natural product azamerone by biosynthetic rearrangement of an aryl diazoketone. Angew Chem Int Ed. 2009;48:767–770. doi: 10.1002/anie.200805140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada O, Na Nan S, Akao T, Tominaga M, Watanabe H, Satoh T, Enei H, Akita O (2003) dffA gene from Aspergillus oryzae encodes l-ornithine N5-oxygenase and is indispensable for deferriferrichrysin biosynthesis. J Biosci Bioeng 95:82–88. 10.1016/S1389-1723(03)80153-6 [DOI] [PubMed]
- Yilmaz Y, Williams G, Manevski N, Walles M, Krähenbühl S, Camenisch G. Functional assessment of rat pulmonary flavin-containing monooxygenase activity. Xenobiotica. 2019;49:503–512. doi: 10.1080/00498254.2018.1469804. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Li H, Yu L, Sun Y, Zhu Y, Zhu H, Zhang L, Li S-M, Shen Y, Tian C, Li A, Liu H, Zhang C. Characterization of the flavoenzyme XiaK as an N-hydroxylase and implications in indolosesquiterpene diversification. Chem Sci. 2017;8:5067–5077. doi: 10.1039/C7SC01182B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler DM. Flavin-containing monooxygenases: catalytic mechanism and substrate specificities. Drug Metab Rev. 1988;19:1–32. doi: 10.3109/03602538809049617. [DOI] [PubMed] [Google Scholar]
- Ziegler DM. An overview of the mechanism, substrate specificities, and structure of FMOs. Drug Metab Rev. 2002;34:503–511. doi: 10.1081/DMR-120005650. [DOI] [PubMed] [Google Scholar]