Abstract
The FLUXNET2015 dataset provides ecosystem-scale data on CO2, water, and energy exchange between the biosphere and the atmosphere, and other meteorological and biological measurements, from 212 sites around the globe (over 1500 site-years, up to and including year 2014). These sites, independently managed and operated, voluntarily contributed their data to create global datasets. Data were quality controlled and processed using uniform methods, to improve consistency and intercomparability across sites. The dataset is already being used in a number of applications, including ecophysiology studies, remote sensing studies, and development of ecosystem and Earth system models. FLUXNET2015 includes derived-data products, such as gap-filled time series, ecosystem respiration and photosynthetic uptake estimates, estimation of uncertainties, and metadata about the measurements, presented for the first time in this paper. In addition, 206 of these sites are for the first time distributed under a Creative Commons (CC-BY 4.0) license. This paper details this enhanced dataset and the processing methods, now made available as open-source codes, making the dataset more accessible, transparent, and reproducible.
Subject terms: Environmental sciences, Climate sciences, Carbon cycle
| Measurement(s) | net ecosystem exchange • carbon dioxide • water • energy |
| Technology Type(s) | eddy covariance • measurement device |
| Sample Characteristic - Environment | terrestrial biome • atmosphere |
| Sample Characteristic - Location | Earth (planet) |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.12295910
Background & Summary
For over three decades, the eddy covariance technique1 has been used to measure land-atmosphere exchanges of greenhouse gases and energy at sites around the world to study and determine the function and trajectories of both ecosystems and the climate system. The technique allows nondestructive measurement of these fluxes at a high temporal resolution and ecosystem level, making it a unique tool. Based on high frequency (10–20 Hz) measurements of vertical wind velocity and a scalar (CO2, H2O, temperature, etc.), it provides estimates of the net exchange of the scalar over a source footprint area that extends up to hundreds of meters around the point of measurement. Soon after the first consistent measurement sites were operational, regional networks of sites were formed in Europe2 and the US3,4, followed by similar initiatives in other continents5–7. Networks enabled the use of eddy covariance data beyond a single site or ecosystem for cross-site comparisons and regional-to-global studies8–15. These regional networks have evolved into long-term research infrastructures or monitoring activities, such as ICOS, AmeriFlux, NEON, AsiaFlux, ChinaFLUX, and TERN-OzFlux.
FLUXNET was created as a global network of networks16–18, a joint effort among regional networks to harmonize and increase standardization of the data being collected. It made possible the creation of global eddy covariance datasets. The first gap-filled, global FLUXNET dataset, which included derived, partitioned fluxes like photosynthesis and respiration, was the Marconi dataset19 in 2000, with 97 site-years of data, followed by the 2007 LaThuile dataset20 with 965 site-years of data, and finally in 2015 the FLUXNET2015 dataset18,21 (hereafter FLUXNET2015) with 1532 site-years of data. Two main factors limited the numbers of sites and years included in each dataset: data policy and data quality. Willingness to share data under the selected data policy is a major reason why FLUXNET2015 likely only includes between 10–20% of existing sites globally–the total number of existing sites is still unknown. Then there is the evolution of processing pipelines and quality controls, leading to new issues being identified in the data that, if not solved in time, led to leaving out that data. The LaThuile dataset had a more restrictive policy and, in a few cases, previously undiscovered data issues, leading to fewer sites being included in FLUXNET2015.
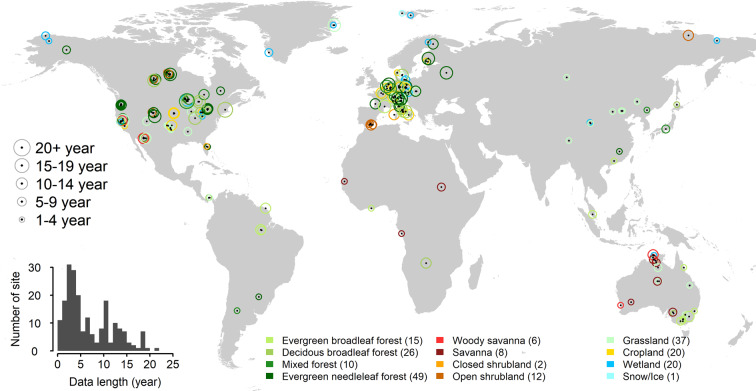
FLUXNET2015 includes for the first time sites with records over two decades long (Fig. 1). The dataset was created through collaborations among many regional networks, with data preparation efforts happening at site, regional network, and global network levels. The global coordination of data preparation activities and data processing was done by a team from the AmeriFlux Management Project (AMP), the European Ecosystem Fluxes Database, and the ICOS Ecosystem Thematic Centre (ICOS-ETC). This team was responsible for the coding efforts, quality checks, and execution of the data processing pipeline. These combined efforts led to a dataset that is standardized with respect to the (1) data products themselves, (2) data distribution formatting, and (3) data quality across sites. The wide application of these datasets in global synthesis and modeling activities highlights their value. At the same time, however, heterogeneity in the data–caused mainly by differences in data collection, flux calculations, and data curation before submission–highlights the need for estimates of data uncertainty and uniform evaluation of data quality.
Fig. 1.
Map of 206 tower sites included in this paper from the 212 sites in the February 2020 release of the FLUXNET2015 dataset. The size of the circle indicates the length of the data record. The color of the circles represents the ecosystem type based on the International Geosphere–Biosphere Programme (IGBP) definition. When overlapping, locations are offset slightly to improve readability. Numbers in parentheses indicate the number of sites in each IGBP group. The inset shows the distribution of data record lengths. See also Supplementary Fig. SM4 for continental scale maps of Australia, Europe, and North America.
The data processing pipeline uses well-established and published methods, with new code implemented for this release as well as code adapted from implementations by the community. The main products in this pipeline are: (1) thorough data quality control checks; (2) calculation of a range of friction velocity thresholds to filter low turbulence periods, allowing an estimate of the uncertainty from this filtering along with the random uncertainty; (3) gap-filling of meteorological and flux measurements, including the use of a downscaled reanalysis data product to fill long gaps in meteorological variables; (4) partitioning of CO2 fluxes into respiration and photosynthesis (gross primary productivity) components using three distinct methods; and (5) calculation of a correction factor for energy fluxes estimating the deviation from energy balance closure for the site. Two features of this pipeline are the ranges of friction velocity thresholds, and the multiple methods for partitioning CO2 fluxes. Both features support a more thorough evaluation of the uncertainty introduced by the processing steps themselves. Our implementation of this pipeline is available as an open-source code package called ONEFlux (Open Network-Enabled Flux processing pipeline)22. The goal of this paper is to describe FLUXNET2015 and additional products, present the details about this processing pipeline, and document the methods used to generate the dataset. Doing so will provide the community of FLUXNET end-users with the technical and practical knowledge necessary to harness the full potential of the FLUXNET data, including data from the FLUXNET2015 release, and data submitted to the network since.
Data Processing Methods
The data contributed by site teams for inclusion in FLUXNET2015 encompassed fluxes, meteorological, environmental, and soil time series at half-hourly or hourly resolutions. Contributed data underwent a uniform data quality control process, with issues addressed in consultation with site teams. Data were then processed using the pipeline (Fig. 2) described in this section. The resulting data products were distributed through the FLUXNET-Fluxdata web portal23, where the usage of the dataset is tracked through a registration of all the requests and details about the user and the data use plan. This information is crucial to better understand the user needs and the impact of the dataset.
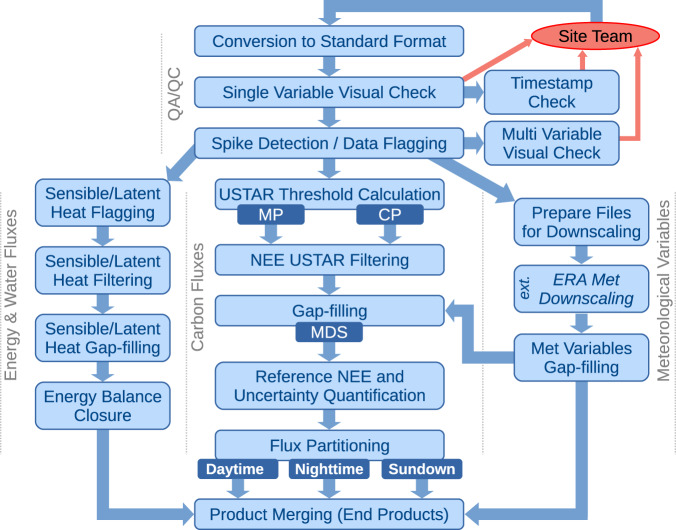
Fig. 2.
The logic of the data processing steps for FLUXNET2015 (details about the different steps and meaning of abbreviations in the text).
Data sources
The first level of processing was completed by the site teams, including the calculation of half-hourly or hourly turbulent fluxes from high-frequency wind and concentration measurements, the averaging of meteorological variables sampled at shorter temporal resolutions, and the site team’s own quality control procedures. The contributed fluxes were required to be submitted separately as turbulent and storage fluxes (components to be added for total flux), and not gap-filled or filtered for low turbulence conditions–see Aubinet et al.24 for details. Control checks were implemented to ensure that this was the case, and to detect additional inconsistencies that site teams were asked to address, for example coherence of the timestamps, consistency across correlated variables, etc25. Starting from these data, we applied the same set of methods for all the remaining processing steps (from filtering to gap-filling and partitioning), increasing uniformity, and allowing quantification of the uncertainty introduced by the processing. The harmonization of data–especially of data quality–was a high priority while creating the dataset, and therefore extensive interaction with the site teams was necessary. Data were contributed through a regional network, and the formats provided by the networks were converted into a standard input for processing. This FLUXNET dataset led to the creation of a new cross-network specification for standard site data and metadata submission formats, now adopted by different regional networks.
Data processing pipeline overview
The data processing pipeline (Fig. 2) is divided into four main processing blocks. The first one is the data quality assurance and quality control (QA/QC) activities our team applied to data from all sites. This part was done with a combination of automated procedures and manual checks for all the variables in the contributed data. The next three blocks were all part of an automated pipeline that was executed separately for each site: the Energy & Water Fluxes processing block encompasses sensible and latent heat variables; the Carbon Fluxes processing block handles the variables for CO2 fluxes like net ecosystem exchange (NEE); and, the Meteorological Variables processing block deals with all the meteorological measurements that are also used in the processing of fluxes and other products. At each processing step, a set of automated pre- and post-conditions are enforced, making sure the inputs and outputs of each step are within the expected behavior. The final step involved merging all the products generated at previous steps, and adding daily through yearly temporal aggregations of most of the variables in the dataset and related quality flags. At this step, automated checks were performed on all the variables to ensure consistency, and the final files with all the contributed and derived data products to be distributed are created. Supplementary Fig. SM2 shows the general steps involved in the processing in the sequence organized in the code, as available in the ONEFlux package22. All the steps have been implemented in the shared code except the Sundown partitioning (see Implementation Approach section for details).
Ensemble of results
The adoption of multiple methods for the same step (e.g., two methods for USTAR threshold calculation, or two or three methods for CO2 flux partitioning, see below in this section for details) is motivated by the existence of different methods in the literature, using different assumptions and potentially having diverging results26,27. On the one hand, this lack of uniformity can represent a problem for synthesis studies. On the other hand, adopting a single method could lead to biases and underestimation of the uncertainty in the methodology. The approach taken here, which simultaneously adopts multiple methods, allows the creation of an ensemble, helping assess uncertainty, and also the suitability of individual methods to a site’s conditions.
Data quality assurance and quality control
Prior to the processing that generated the derived data products (hereafter called post-processing), the data for each site went through QA/QC checks following Pastorello et al.25. All variables included in the dataset underwent checks, and critical variables underwent further scrutiny. These additional checks targeted variables critical to the processing, e.g., flux variables, meteorological variables used in gap-filling, and variables used by uncertainty estimation procedures. The processing did not proceed for sites with pending issues in critical variables.
Critical metadata variables for the post-processing:
The site FluxID in the form CC-SSS (two character country code, three character site identifier within country) – e.g., US-Ha1
The latitude and longitude for the site in the WGS 84 decimal format with at least four decimal points resolution – e.g., 42.5378/−72.1715
Time zone of the site (time series, if time zone changed; timestamps are all local standard time, no daylight savings) – e.g., UTC-5
Height of the gas analyzer – e.g., 30.0 m
Critical data variables for the post-processing, averaged or integrated over 30 or 60 minutes (*required):
*CO2 (µmolCO2 mol−1): Carbon Dioxide (CO2) mole fraction in moist air
*FC (µmolCO2 m−2 s−1): Carbon Dioxide (CO2) turbulent flux (without storage component)
*SC (µmolCO2 m−2 s−1): Carbon Dioxide (CO2) storage flux measured with a vertical profile system, optional if tower shorter than 3 m
*H (W m−2): sensible heat turbulent flux, without storage correction
*LE (W m−2): latent heat turbulent flux, without storage correction
*WS (m s−1): horizontal wind speed
*USTAR (m s−1): friction velocity
*TA (deg C): air temperature
*RH (%): relative humidity (range 0–100%)
*PA (kPa): atmospheric pressure
G (W m−2): ground heat flux, not mandatory, but needed for the energy balance closure calculations
NETRAD (W m−2): net radiation, not mandatory, but needed for the energy balance closure calculations
*SW_IN (W m−2): incoming shortwave radiation
SW_IN_POT (W m−2): potential incoming shortwave radiation (top of atmosphere theoretical maximum radiation), calculated based on the site coordinates22
PPFD_IN (µmolPhotons m−2 s−1): incoming photosynthetic photon flux density
P (mm): precipitation total of each 30 or 60 minute period
LW_IN (W m−2): incoming (down-welling) longwave radiation
SWC (%): soil water content (volumetric), range 0–100%
TS (deg C): soil temperature
File format standardization
To process hundreds of sites, we needed consistent file formats that supported the input data and metadata. This led to multi-network agreements and creation of formats for data and metadata contribution to the regional networks28,29. These formats have now been adopted by networks in Europe and the Americas and by some instrument manufacturers, and are under consideration by other regional networks. In addition, automated extraction and conversion tools for direct format translation were implemented to work with data in older formats.
Data QA/QC steps
Data quality was checked before the processing started. If issues were identified that could not be resolved by the network-level data team, the site team was asked to suggest a course of action or send a new version of the data addressing the quality issue identified. The main data QA/QC steps were: single-variable checks, multi-variable checks, specialized checks, and automatic checks. Single-variable checks look at patterns in the time series of one variable at a time, for long- and short-term trends and other issues. Multi-variable checks look at the relationships among correlated variables (e.g., different radiation variables) to identify periods with disagreements. Specialized checks test for common issues in EC and meteorological data, like timestamp shifts or sensor deterioration. During this phase, a time series of top-of-the-atmosphere potential radiation (SW_IN_POT) is also computed, using latitude/longitude coordinates and time22. These three types of checks are detailed in Pastorello et al.25. The automated checks apply variable-specific despiking routines adapted from Papale et al.30 and apply a set of range controls per variable. This last step creates a series of flags that were discussed with the site managers for corrections and resubmissions and then used to filter the data in subsequent steps.
Meteorological products
The main processing applied to meteorological data was gap-filling by two independent methods: Marginal Distribution Sampling31 (MDS) and ERA-Interim32. Data gap-filled by MDS (applied to all variables that are gap-filled) are identified by the _F_MDS suffix. Data gap-filled using ERA-Interim downscaling (six variables that are available in the reanalysis dataset) have a _F_ERA suffix. The final gap-filled time series for variables combines both of these methods (indicated by an unqualified _F suffix), following a data-quality-based selection approach (see below). For SW_IN, in case of gaps or in case the variable was not measured, we performed a calculation from PPFD_IN when available, calculating the conversion factor from the periods of overlap of the two measurements (and assuming a factor 0.48 J (µmol photon)−1 when the sensors did not run in parallel at the site).
MDS
The MDS method, introduced in Reichstein et al.31, is applied to all variables that may be gap-filled. It works by seeking meteorological conditions physically and temporally similar to the ones for the missing data point(s). The restrictions on the size of the time window and which variables must be available are incrementally relaxed until a suitable set of records is found to fill the gap in the target variable. All values of the target variable satisfying the current set of conditions are averaged to generate the fill value. The method was applied as described in the original implementation31, using SW_IN, TA and VPD as drivers. The basic three scenarios for the time when the target variable is missing are: (i) all three drivers are available; (ii) only SW_IN is available; and (iii) all three drivers are missing. Based on the available co-located variables, a search for similar conditions is started, keeping the searching window as small as possible to avoid changes in other slow-changing drivers (phenology, water availability, etc.). The more variables missing and the larger the time window, the lower the confidence indicated by the _F_MDS_QC flag. The values for this flag are (0–3): _F_MDS_QC = 0 (measured); _F_MDS_QC = 1 (filled with high confidence); _F_MDS_QC = 2 (filled with medium confidence); _F_MDS_QC = 3 (filled with low confidence). For details on the implementation, see the original paper31 and the ONEFlux source code22.
ERA-Interim
This method is based on the ERA-Interim (ERA-I) Reanalysis global atmospheric product33,34 created by the European Centre for Medium‐Range Weather Forecasts (ECMWF)–ERA stands for ECMWF Re-Analysis. Applied to the subset of variables that are also available in the ERA-Interim product, the method involves a spatial and temporal downscaling process using the measured variable at the site. The ERA-I variables that were used are: air temperature at 2 m (t2m, K), incoming shortwave solar radiation at the surface (Sw, W m−2), dew point temperature at 2 m (dt2m, K), wind speed horizontal components at 10 m (u10 and v10, m s−1), total precipitation (Pr, m of water per time step), and incoming longwave solar radiation at the surface (Lw, W m−2). The gap-filling procedure harmonizes units, identifies periods that are long enough to allow a linear relationship to be built, a simple debiasing of the linear relationship, evaluation of the diurnal cycle in the subset of variables, and other evaluations of the results to identify potential missing or incorrect information (e.g., coordinate or temporal mismatches). The linear relationships are built taking into account instantaneous and averaged variables, and then applied to the whole ERA-I record, generating the spatially (coordinate-based) and temporally (diurnal cycle-based) downscaled version of each variable. The method was applied as in the original implementation; additional details can be found in Vuichard and Papale32.
Final gap-filled product
Measured or high quality gap-filled records using MDS (_F_MDS_QC < 2) are used in the final gap-filled products (_F suffixed variables, without _MDS or _ERA). If the variable has a low quality gap-fill flag (2 or 3), the ERA-I product is used instead. The final quality flag (_F_QC) is 0 for measured, 1 for high quality fill using MDS, and 2 for data gap-filled with the ERA-I downscaled product. A gap-filled version of CO2 concentration is also generated (CO2_F_MDS) using the MDS method as described above, including the corresponding quality flag.
Energy and water products
The main data products associated with energy and water fluxes are the gap-filled versions of the data and the estimation of a version ensuring the energy balance closure and estimating its uncertainty–for a description of the issue see Stoy et al.35 Turbulent energy fluxes (sensible and latent heat, H and LE, respectively) are gap-filled using the MDS method31 described above. From LE, it is possible to calculate the water flux (evapotranspiration) using the latent heat of vaporization. An energy balance corrected version of LE and H is also created, a data product often needed when data are used in model parameterization and validation for which the closure of the energy balance is prescribed. There is no general agreement on the reasons and approaches to correct the imbalance in the energy budget within EC measurements. In this product, the methodology used to calculate the energy balance corrected fluxes is based on the assumption that the Bowen ratio is correct36. Fluxes are corrected by multiplying the original, gap-filled LE and H data by an energy balance closure correction factor (EBC_CF, in the dataset). The correction factor is calculated starting from the half-hours where all the variables needed to estimate energy balance closure are available (measured NETRAD and G, and measured or good-quality gap-filled H and LE). The correction factor for each single half-hour is calculated as in Eq. (1), but is not applied directly.
| 1 |
First, to avoid transient conditions, the calculated EBC_CF time series is filtered by removing values outside of 1.5 times its own interquartile range. Then, the correction factor used in the calculations is obtained using one of three methods, applied hierarchically (see also diagram in Supplementary Fig. SM3):
EBC_CF Method 1: For each half-hour, a sliding window of ±15 days (31 days total) is used to select half-hours between time periods 22:00–02:30 and 10:00–14:30 (local standard time). These time-of-day restrictions aim at removing sunrise and sunset time periods, when changes in ecosystem heat storage (not measured) are more significant, preventing energy balance closures. For all half-hours meeting these criteria, the corresponding EBC_CFs are selected and used to calculate the corrected values of H and LE for the half-hour processed (center of the sliding window), generating a pool of values for each of these two variables. From each of these two pools, the 25th, 50th (median), and 75th percentiles are extracted for their corresponding variables, generating the values for H_CORR25, H_CORR, H_CORR75 and LE_CORR25, LE_CORR, LE_CORR75. If fewer than five EBC_CF values are present in the sliding window, Method 2 is used for the half-hour. (Note on temporal aggregations: for DD the sliding window size is ±7 days and the EBC_CF are calculated from the daily average values of G, NETRAD, H and LE. For WW, MM, and YY the EBC_CFs are calculated from corresponding average fluxes of the period analysed, but no percentiles are computed. For WW, MM, and YY, Method 1 fails if less than 50% of half-hours within the window have measured values for all four component variables.)
EBC_CF Method 2: For the current half-hour, EBC_CF is calculated as the average of the EBC_CF values used to calculate the H_CORR and LE_CORR with Method 1 within a sliding window of ±5 days and ±1 hour of the time-of-day of the current timestamp. H_CORR and LE_CORR are calculated and the corresponding _CORR25 and _CORR75 percentiles are not generated. If no EBC_CF is available, Method 3 is used for the current half-hour. (Note on temporal aggregations: differing sliding windows are: DD: ±2 weeks, WW: ±2 weeks, MM: ±1 month, and YY: ±1 year.)
EBC_CF Method 3: An approach like Method 2 is applied but using a sliding window of ±5 days for the same half-hour in the previous and next years, with the current EBC_CF being calculated from the average of the EBC_CF values used to calculate the H_CORR and LE_CORR. H_CORR and LE_CORR are calculated and the corresponding _CORR25 and _CORR75 percentiles are not generated. In case this method also cannot be applied due to missing values, the energy balance closure corrected fluxes are not calculated. (Note on temporal aggregations: differing sliding windows are: DD: ±2 weeks, WW: ±2 weeks, MM: ±1 month, and YY: ±2 years.)
H and LE Random Uncertainty
The random uncertainty for H and LE is also estimated at half-hourly resolution, based on the method introduced by Hollinger & Richardson37 and then aggregated at the other temporal resolutions. The random uncertainty (indicated by the suffix _RANDUNC) in the measurements is estimated using one of two methods, applied hierarchically:
H-LE-RANDUNC Method 1 (direct standard deviation method): For a sliding window of ±5 days and ±1 hour of the time-of-day of the current timestamp, the random uncertainty is calculated as the standard deviation of the measured fluxes. The similarity in the meteorological conditions evaluated as in the MDS gap-filling method31 and a minimum of five measured values must be present; otherwise, method 2 is used.
H-LE-RANDUNC Method 2 (median standard deviation method): For the same sliding window of ±5 days and ±1 hour of the time-of-day of the current timestamp, random uncertainty is calculated as the median of the random uncertainty (calculated with H-LE-RANDUNC Method 1) of similar fluxes, i.e., within the range of ±20% and not less than 10 W m–2.
The joint uncertainty for H and LE is computed from the combination of the uncertainty from the energy balance closure correction factor and random uncertainty.
| 2 |
These variables are identified by the _JOINTUNC suffix and are computed for H as in Eq. (2), and similarly for LE. (Note on temporal aggregations: joint uncertainties for H and LE are recomputed at HH and DD resolutions separately, and not generated for WW, MM, and YY resolutions.)
CO2 products
The processing steps applied to CO2 fluxes were: calculation of net ecosystem exchange (NEE) from CO2 turbulent and storage fluxes, applying a spike detection algorithm, filtering for low turbulence conditions using multiple friction velocity (USTAR) thresholds, gap-filling of all NEE time series generated by an ensemble of USTAR thresholds, estimation of random uncertainty, and partitioning of NEE into its ecosystem respiration (RECO) and gross primary production (GPP) components.
Calculation of NEE
CO2 storage fluxes (SC) express the change of CO2 concentration below the measurement level of the eddy covariance system within the half-hour. NEE was calculated as the sum of the CO2 turbulent fluxes (FC) and SC. Both FC and SC are part of the required data contributed by site teams. SC is usually estimated using a profile system38. If SC was not provided or missing, two cases were implemented: for measurement heights lower than 3 m and short canopies, the SC term was considered to be 0; for taller towers/canopies, a discrete estimation based on the top measurement of CO2 concentration was used to compute SC39.
Despiking of NEE
Although the processing of high frequency data into half-hourly fluxes usually includes steps to remove spikes from instantaneous measurements, spikes can also occur in the half-hourly data. The method described in Papale et al.30, based on the median absolute deviation (MAD) with z = 5.5, was applied to filter NEE for residual spikes that were removed.
USTAR threshold estimation and filtering
Filtering for low turbulence conditions is necessary when there is not enough turbulence, causing the ecosystem flux to be transported by advective flows and missed by both the eddy covariance system and the storage profile, resulting in underestimated fluxes. Despite different approaches having been tested to measure and quantify horizontal and vertical advection40, the most often used method to avoid the underestimation of fluxes is removing the data points potentially affected by strong advection1. These points are identified using the friction velocity (USTAR) as an indicator of turbulence strength, defining a threshold value under which NEE measurements are discarded and replaced by gap-filled estimates.
This USTAR threshold is linked to the canopy structure, measurement height, wind regimes, and other factors specific to an individual site. It is estimated using nighttime NEE measurements (only ecosystem respiration), based on the dependency between USTAR and NEE at similar temperatures and periods of the year (main drivers of ecosystem respiration). Under these conditions, NEE is assumed (and expected) to be independent from USTAR, which is not a driver of respiration. However, in most sites below a certain USTAR threshold value, NEE is found to increase with USTAR; this USTAR value is selected as the threshold to define conditions with reduced risk of flux underestimation. Different methods have been proposed to estimate the USTAR thresholds and the related uncertainty as to how the approach works at a specific site1.
CP and MP USTAR threshold methods
Two methods to calculate USTAR thresholds were used: change-point-detection (CP) proposed by Barr et al.41 and a modified version of the moving-point-transition (MP) described originally by Reichstein et al.31 and Papale et al.30. Both methods are similar in terms of data selection, preparation and grouping and aim to estimate the USTAR threshold value. Measurements collected when USTAR is below the threshold are removed. The difference between these methods is in how this threshold value is estimated. For both methods, the nighttime data of a full year are divided in four three-month periods (seasons) and 7 temperature classes (of equal size in terms of number of observations). For each season/temperature group the data are divided into 20 USTAR classes (also with equal number of observations) and the average NEE for each USTAR class is computed. The calculation of the threshold uses each of the methods (see below for details on their differences). For each season, the median value of the 7 temperature classes is calculated and a final threshold is defined by selecting the maximum of the 4 seasonal values.
The CP method uses two linear regressions between NEE and USTAR, the second with an imposed zero slope. The change point is defined as where the two lines cross, i.e., constraining the shape of the NEE-USTAR dependency. The method is extensively used to detect temporal discontinuities in climatic data. Details can be found in Barr et al.41.
For the MP method30,31, the mean NEE value in each of the 20 USTAR classes is compared to the mean NEE measured in the 10 higher USTAR classes. The threshold selected is the USTAR class in which the average nighttime NEE reaches more than 99% of the average NEE at the higher USTAR classes. An improvement of the MP method was implemented here for robustness over noisy data, by adding a second step to the original MP implementation: when a threshold is selected, it was tested to ensure it was also valid for the following USTAR class. In other words, assuming that Eq. (3) holds, where x is one of the 20 USTAR classes and is the average NEE for that USTAR class.
| 3 |
The USTAR value associated to the xth-class was selected as threshold only if Eq. (4) also holds, to confirm that the plateau where NEE is USTAR-independent was reached. If not, the search for the plateau and threshold continued toward higher USTAR values.
| 4 |
Bootstrapping USTAR threshold estimation
For each of the two methods, a bootstrapping technique was used. The full dataset (year of measurement) was re-sampled 100 times with the possibility to select the same data point multiple times (i.e., with replacement), creating 100 versions of the dataset. The threshold values were calculated for each of them, obtaining 100 threshold values per method (CP and MP) and year, for a total of 200 USTAR threshold estimates for each year. This process and next steps are illustrated in Fig. 3. These 200 threshold values represent the uncertainty in the threshold estimation that could also impact the uncertainty of NEE. It is worth noting that there is not always a direct relationship between the threshold and NEE uncertainties. It is possible, for instance, that a small variability in the thresholds has a strong effect on NEE or, conversely, with NEE almost insensitive to the threshold value. This is related to the site characteristics (USTAR variability) and to the level of difficulty in filling the gaps created by the filtering.
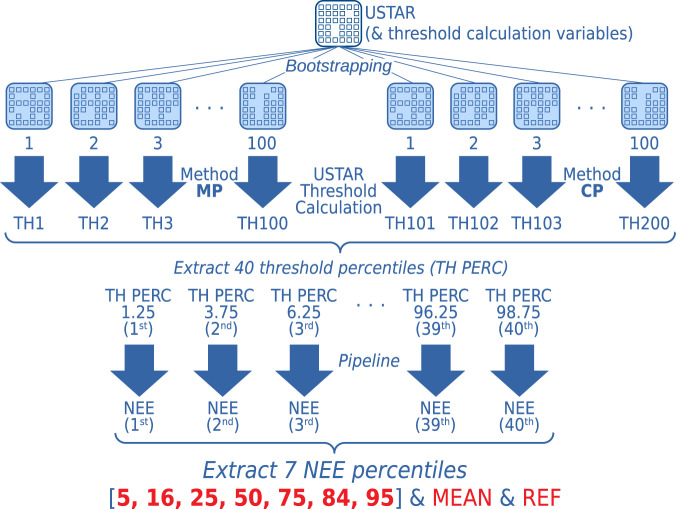
Fig. 3.
To identify and remove data collected under low turbulence conditions, under which advective fluxes could lead to an underestimation of fluxes, filtering based on the USTAR threshold was used. In order to estimate the uncertainty in the USTAR threshold calculation, a bootstrapping approach was implemented, with a selection of values representative of the distribution included in the final data products. From the (up to) 200 thresholds from the combined bootstrapping of the two methods, 40 percentiles are extracted. All the subsequent steps of the pipeline are applied to all 40 versions. For each of the final output products (e.g., NEE, as illustrated here), seven percentiles representative of the distribution are included.
There are cases where not enough data are present to calculate a USTAR threshold (for both the CP and MP methods) or where it is not possible to identify a clear change point (CP method only). This leads to the uncertainty being underestimated (fewer or no USTAR threshold values available). This should be considered as a general indication of difficulties in the application of the USTAR filtering for the specific sites or years. Sites and years where these conditions occurred are reported in the SUCCESS_RUN variable in the AUXNEE product (values 1: threshold found, 0: failed/no threshold found).
Variable and constant variants of the USTAR threshold methods
To calculate the uncertainty in NEE due to the uncertainty in the selected USTAR threshold, all the threshold values obtained with the two methods and the bootstrapping were pooled together, from which 40 representative values were extracted: from the percentile 1.25 of the series to the percentile 98.75, with a step of 2.5, i.e., [1.25:2.5:98.75]. When long time series (multi-years) are processed, it is possible to extract the 40 representative thresholds for each of the years. The threshold is a function of slow-changing dynamics (height of canopy, height of measurement, roughness), but a threshold changing every year could introduce false interannual variability. On the other hand, a constant threshold across all the years would not represent changes in the ecosystem structure and EC system setup. For this reason, two approaches were implemented:
Variable USTAR Threshold (VUT): The thresholds found for each year and the years immediately before and after (if available) have been pooled together, and from their joint population, the final 40 thresholds extracted. With that, the USTAR thresholds vary from year to year; however, they are still influenced by neighboring years. This is identified in FLUXNET2015 variables by the “_VUT” suffix;
Constant USTAR Threshold (CUT): Across years, all the thresholds found have been pooled together and the final 40 thresholds extracted from this dataset. With that, all years were filtered with the same USTAR threshold. This is identified in FLUXNET2015 variables by the “_CUT” suffix.
If the dataset includes up to two years of data, the two methods give the same result, and only the _VUT is generated.
For both the VUT and CUT approaches, 40 NEE datasets have been created, filtering the original NEE time series using 40 different USTAR values estimated as explained above. The values of the thresholds are reported in the AUXNEE product file. These 40 NEE versions have been used as the basis for all the derived variables provided. An example of the variability of the two methods (CP and MP) is shown in Fig. 4, contrasting the distribution of the bootstrapped results for each method, showing comparable values for some years and divergent values for other years (of the same site). This highlights the importance of applying both methods in this ensemble-like way.
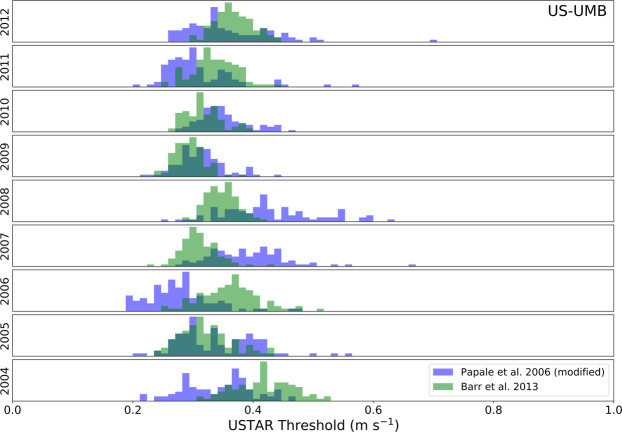
Fig. 4.
Example of the distribution of USTAR thresholds calculated for each year using the MP30 method in blue and CP41 method in green for the US-UMB site (dark green where they overlap). All these thresholds were pulled together to extract the CUT final 40 thresholds, while for the VUT thresholds, each year was pulled with the two immediately before and after (e.g., 2005 + 2006 + 2007 to extract the 40 thresholds to be used to filter 2006). Note that the level of agreement between methods and between subsequent years is variable, justifying the approach that propagates this variability into uncertainty in NEE.
Filtering NEE based on USTAR thresholds
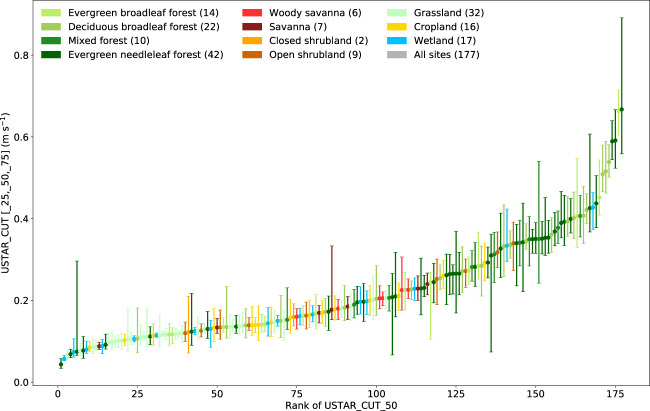
The USTAR thresholds are applied to daytime and nighttime data, removing NEE values collected when USTAR is below the threshold and removing also the first half-hour with high turbulence after a period of low turbulence to avoid false emission pulses due to CO2 accumulated under the canopy and not detected by the storage system (in particular, when a profile is not available at the site). The USTAR filtering is not applied to H and LE, because it has not been proved that when there are CO2 advective fluxes, these also impact energy fluxes, specifically due to the fact that when advection is in general large (nighttime), energy fluxes are small. Figure 5 shows the range of thresholds found (interquartile ranges) across sites in FLUXNET2015. While some sites had low thresholds and low variability in the USTAR thresholds, others show large ranges of values in some more extreme cases (indicating difficulties in estimating the “real” threshold).
Fig. 5.
Ranked USTAR thresholds based on median threshold and error bars showing 25th to 75th percentiles of the 40 thresholds calculated with the Constant USTAR Threshold (CUT) method – only computed for sites with 3 or more years, so only 177 sites out of the 206 are shown. Colors show different ecosystem classes based on the site’s IGBP.
Gap-filling of NEE
Existing gaps from instrument or power failures are further increased after QC and USTAR filtering. The time series with gaps need to be filled, especially before aggregated values can be calculated (from daily to annual). Moffat et al.26 compared different gap-filling methods for CO2 concluding that most of the methods currently available perform sufficiently well with respect to the general uncertainty associated with the measurements. The method implemented here is the Marginal Distribution Sampling (MDS) method already described in the meteorological products.
Selection of reference NEE variables
After filtering NEE using the 40 USTAR thresholds and gap-filling, 40 complete (gap-free) NEE time series were available for each site. For each half hour, it is possible to use the 40 values to estimate the NEE uncertainty resulting from the USTAR threshold estimation (reported as percentiles of the NEE distribution, identified by the “_XX” numeric suffix) and the average value (identified in the dataset by the “_MEAN” suffix). Since the average value has a smoothing effect on the time series, an additional reference value of NEE was selected and identified in FLUXNET2015 variables by the “_REF” suffix, in an attempt to identify which of the 40 NEE realizations was the most representative of the ensemble. The “_REF” NEE was selected among the 40 different NEE instances in this way: (1) the Nash–Sutcliffe model efficiency coefficient42 was calculated between each NEE instance and the remaining 39; (2) the reference NEE has been selected as the one with the highest model efficiency coefficients sum, i.e., the most similar to the other 39. Note that determining the reference NEE is done independently for variables using VUT and CUT USTAR thresholds, as well as for each temporal aggregation. Therefore, the version selected as REF could be different for different temporal resolutions. For instance, NEE_VUT_REF at half-hourly resolution might have been generated using a different USTAR threshold than NEE_VUT_REF at daily resolution. Information on which threshold values were used for each version and temporal aggregation can be found in the auxiliary products for NEE processing (AUXNEE). In addition to the reference NEE, the NEE instance obtained by filtering the data with the median value of the USTAR thresholds distribution is also included. This NEE is identified in FLUXNET2015 variables by the “_USTAR50” suffix (for both CP and MP methods, and both VUT and CUT approaches) and is stable across temporal aggregation resolutions. Individual percentiles of the USTAR thresholds distribution are reported in the AUXNEE file (40 instances for CUT and 40 per year for VUT).
Random uncertainty for NEE
In addition to the uncertainty estimates based on multiple thresholds for USTAR filtering, the random uncertainty for NEE is also estimated based on the method used by Hollinger & Richardson37. Variables expressing random uncertainty are identified by the suffix _RANDUNC. One of two methods are used to estimate random uncertainty, applied hierarchically:
NEE-RANDUNC Method 1 (direct standard deviation method): For a sliding window of ±7 days and ±1 hour of the time-of-day of the current timestamp, the random uncertainty is calculated as the standard deviation of the measured fluxes. The similarity in the meteorological conditions evaluated as in the MDS gap-filling method31 and a minimum of five measured values must be present; otherwise, method 2 is used.
NEE-RANDUNC Method 2 (median standard deviation method): For a sliding window of ±5 days and ±1 hour of the time-of-day of the current timestamp, random uncertainty is calculated as the median of the random uncertainty (calculated with NEE-RANDUNC Method 1) of similar fluxes, i.e., within the range of ±20% and not less than 2 µmolCO2 m–2 s–1. (Note on temporal aggregations: differing sliding windows are: WW: ±2 weeks, MM: ±1 month, and YY: ±2 years.)
The joint uncertainty for NEE is computed from the combination of the uncertainty from multiple USTAR thresholds and random uncertainty. These variables identified by the _JOINTUNC suffix and are computed for NEE filtered using the VUT method as in Eq. (5), and similarly for NEE filtered with the CUT method.
| 5 |
The 16th and 84th percentiles are used because they are equivalent to ±1 Standard Deviation in case of a normal distribution. (Note on temporal aggregations: joint uncertainties for NEE are recomputed at all temporal resolutions.)
CO2 flux partitioning in GPP and RECO
Partitioning CO2 fluxes from NEE into estimates of its two main components, Gross Primary Production (GPP) and Ecosystem Respiration (RECO), was done by parameterizations of models using measured data. All sites were partitioned with the nighttime fluxes method31 (_NT suffixes) and the daytime fluxes method43 (_DT suffixes), while a third method, sundown reference respiration44 (_SR suffixes), was applied to all sites meeting the method’s requirements (e.g., high quality storage measurement).
The nighttime method uses nighttime data to parameterize a respiration-temperature model that is then applied to the whole dataset to estimate RECO. GPP is then calculated as the difference between RECO and NEE. The parameterization uses short windows of time (14 days) to account for the dynamic of other important respiration drivers such as water, substrate availability, and phenology (see Reichstein et al.31 for details on the implementation and ONEFlux22 for the code).
The daytime method uses daytime and nighttime data to parameterize a model with one component based on a light-response curve and vapor pressure deficit for GPP, and a second component using a respiration-temperature relationship similar to the nighttime method. In this case, NEE becomes a function of both GPP and RECO, both of which are estimated by the model. Similarly to the nighttime method, the parameterization is done for short windows (8 days) to take into consideration other slower-changing factors (see Lasslop et al.43 for details on the implementation and ONEFlux22 for the code).
For forest sites where a CO2 concentration profile for storage fluxes was available, an additional RECO estimate was calculated using the method from van Gorsel et al.44, with variables identified by the _SR suffix. In this method, the parameterization of a respiration-temperature model is based solely on data acquired just after sundown, aiming at excluding the measurements potentially affected by advection and also assuming that in the first hour of the evening the advective transport is not yet established.
The sundown partitioning method requires that the NEE is not filtered for low turbulence conditions (USTAR), and for this reason it was applied only to the original time series. The nighttime and daytime methods instead require NEE filtered for low turbulence conditions. For this reason they were applied to all the 40 NEE versions resulting from the 40 USTAR thresholds, obtaining 40 versions of GPP and RECO for each of the two partitioning methods, propagating the uncertainty from NEE to GPP and RECO. This has been done for both the CUT and VUT filtering methods.
Similarly to NEE, the 40 GPP and RECO estimates (for each method and for CUT and VUT) have been used to calculate the percentiles of their distribution for each timestep (describing their uncertainty due to the NEE uncertainty). The average value (_MEAN) and the reference value use the same model efficiency approach used for NEE for each temporal aggregation. Similarly, NEE filtered with the median USTAR value (_USTAR50) has been partitioned into GPP and RECO. Information on the threshold values used for all versions of GPP and RECO (_NT and _DT, _VUT and _CUT, HH to YY resolutions) are in the auxiliary files for NEE processing (AUXNEE). Variables for reference GPP and RECO are also identified by a _REF suffix. The two methods for the partitioning (three for the cases in which the sundown method is applied) are not merged in any way, because their difference is informative with the respect to the uncertainty of the methods, as in the case of a model comparison exercise.
Implementation approach
To increase the traceability of changes between versions of datasets and reduce uncertainty stemming from choices made at implementation time, we favored using original code implementations or thoroughly validated re-implementations of original codes. Thus, our code organization strings together loosely coupled components which implement each step, with clear-cut interfaces between steps. This modular approach eases the maintenance and change efforts for any individual step, but adds complexity to evaluating changes for the entire pipeline. Different programming languages (Python, C, MATLAB and IDL, plus PV-WAVE for FLUXNET2015) were used to implement the different steps, all connected using a controller code that makes appropriate calls in the correct order. The ONEFlux22 code collection replaced the PV-WAVE code with a re-implementation in Python, and also collates most of these steps into a cohesive pipeline (see also the Code Availability section). The IDL code, which applies the sundown partitioning method44, is not yet currently implemented in ONEFlux, because some additional testing and development are needed to make it robust and more suitable for general application. Implementation details of individual steps are discussed next, with references to the outputs each step identified by an execution sequential number and the step name–e.g., 01_qc_visual contains the results of the first processing step, the visual check step. Each of these steps correspond to a code module. Supplementary Fig. SM2 shows the steps and their inter-dependencies.
Steps implemented in python
The main controller code for ONEFlux is implemented in Python. Besides being the glue code that executes each step, pre- and post-checks are also executed before and after each step. These checks guarantee that the input data meet the minimum requirements to run the step, that the minimum expected outputs were generated by the execution of the step, and that any errors or exception conditions were handled correctly. Information about execution is recorded in a log for the entire pipeline, along with logs for individual steps. Besides the controller code, two of the three flux partitioning steps were re-implemented in Python (the nighttime and daytime methods, 10_nee_partition_nt and 11_nee_partition_dt), together with other specific steps such as data preparation for the uncertainty estimates (12_ure_input), and the creation and checking of final products (99_fluxnet2015). The original flux partitioning implementation in PV-WAVE was used for the LaThuile2007 and FLUXNET2015 datasets. Also, the tool for the downscaling of the ERA-I meteorological data is implemented in Python and runs on a server connected to the ERA data.
Steps implemented in C
Several steps are implemented in the C programming language, allowing better control over execution performance of these steps. These steps include:
automated QA/QC flagging (02_qc_auto), USTAR threshold estimation using the MP method (04_ustar_mp), the filtering and gap-filling of meteorological data, including the merging with the ERA-I downscaled data (07_meteo_proc), the filtering and gap-filling of CO2 fluxes (08_nee_proc), the filtering, gap-filling, and energy corrections of energy fluxes (09_energy_proc), and the computation of uncertainty products (12_ure). The source codes and the compiled executables are provided for steps implemented in C, as well as build procedures in make/Makefile format.
Steps implemented in MATLAB
The estimation of USTAR thresholds using the CP method (05_ustar_cp) is the only step implemented in MATLAB. It is distributed both as source code and compiled code to be used with the MATLAB Runtime Environment, such that it does not require a license purchase.
Data Records
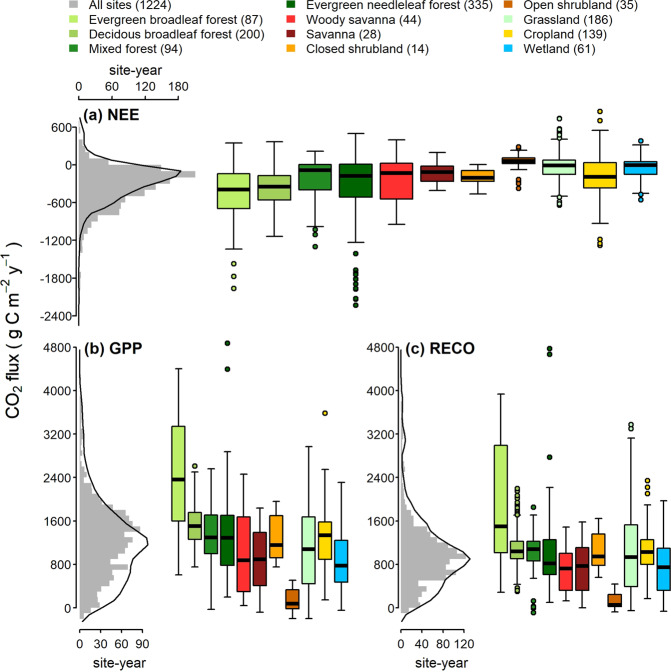
The FLUXNET2015 portion presented in this paper contains 1496 site-years of data from 206 sites45–250, characterizing ecosystem-level carbon and energy fluxes in diverse ecosystems across the globe (Fig. 1, Supplementary Fig. SM1251,252), spanning from the early 1990s to 2014, with 69 sites having decade-long records. The dataset covers the distribution of ecosystem fluxes as reported in the recent meta-analyses253,254 (Fig. 6).
Fig. 6.
Distribution of the yearly (a) net ecosystem exchange (NEE), (b) gross primary production (GPP), and (c) ecosystem respiration (RECO) in FLUXNET2015. Only data with QC flag (NEE_VUT_REF_QC) higher than 0.5 are shown here. The values are reference NEE, GPP, and RECO based on the Variable USTAR Threshold (VUT) and selected reference for model efficiency (REF). GPP and RECO are based on the nighttime partitioning (NT) method. The grey histogram (bin width 100 gC m−2 y−1) shows the flux distribution in 1224 of the available site-years; negative GPP and RECO values are kept to preserve distributions, see Data processing methods section for details. Black lines show the distribution curves based on published data253,254. The boxplots show the flux distribution (i.e., 25th, 50th, and 75th percentiles) for vegetation types defined and color-coded according to IGBP (International Geosphere–Biosphere Programme) definitions. Circles represent data points beyond the 1.5-times interquartile range (25th to 75th percentile) plus the 75th percentile or minus 25th percentile (whisker). Numbers in parentheses indicate the number of site-years used in each IGBP group. The NO-Blv site from the snow/ice IGBP group is not shown in the boxplots.
The dataset is distributed in files separated by sites, by temporal aggregation resolutions (e.g., hourly, weekly), and by data products (e.g., FULLSET with all the variables and SUBSET designed for less experienced users). All data files for a site are available for download as a single ZIP file archive with site-specific DOI. The file-naming conventions details these options for each file (Table 1). Site metadata are also available as a single file containing metadata for all sites, detailed later in this section and Supplementary Table SM8. Note that DOIs are assigned at the site level, one DOI per site for all of that site’s products. A DOI was not assigned to the whole FLUXNET2015 dataset, since this would make citation and assigning credit imprecise and hard to track.
Table 1.
The template of file naming conventions, including the field, field definition, and the possible options.
| File Name Conventions | ||
|---|---|---|
| FLX_[SITE_ID]_FLUXNET2015_[DATA_PRODUCT]_[RESOLUTION]_[FIRST_YEAR]-[LAST_YEAR]_ [SITE_VERSION]-[CODE_VERSION].[EXT] | ||
| Field | Definition | Possible options |
| SITE_ID | FLUXNET site ID in the format CC-SSS (CC is two-letter country code, SSS is three-character site-level identifier) | |
| DATA_PRODUCT | Grouping of variables from release included in file. |
• SUBSET: Core set of variables with quality and uncertainty information needed for general uses of the data • FULLSET: All variables, including all quality and uncertainty information, and key variables from intermediate processing steps • AUXMETEO: Auxiliary variables related to meteorological downscaling • AUXNEE: Auxiliary variables related to NEE, RECO, and GPP processing • ERAI: Full record (1989–2014) of ERA-Interim downscaled meteorological variables for the site |
| RESOLUTION | Temporal resolution of data product |
• HH: Half-Hourly time steps • HR: Hourly time steps • DD: Daily time steps • WW: Weekly time steps • MM: Monthly time steps • YY: Yearly time steps |
|
FIRST_YEAR LAST_YEAR |
First and last years of eddy covariance flux data | |
|
SITE_VERSION CODE_VERSION |
Version string in integer. SITE_VERSION indicates the version of the original dataset for the site used; CODE_VERSION indicates the version of the code of the data processing pipeline used to process the dataset for the site | |
| EXT | File extension |
• csv: Comma-separated values in a text file (ASCII) • zip: Archive file with all temporal resolutions for the same site and data product |
|
Examples of file names and structures: FLX_US-Ha1_FLUXNET2015_FULLSET_HH_1992-2012_1-3.zip - FLX_US-Ha1_FLUXNET2015_FULLSET_HH_1992-2012_1-3.csv - FLX_US-Ha1_FLUXNET2015_FULLSET_DD_1992-2012_1-3.csv - FLX_US-Ha1_FLUXNET2015_FULLSET_WW_1992-2012_1-3.csv - FLX_US-Ha1_FLUXNET2015_FULLSET_MM_1992-2012_1-3.csv - FLX_US-Ha1_FLUXNET2015_FULLSET_YY_1992-2012_1-3.csv - FLX_US-Ha1_FLUXNET2015_ERAI_HH_1992-2012_1-3.csv - FLX_US-Ha1_FLUXNET2015_ERAI_DD_1992-2012_1-3.csv - FLX_US-Ha1_FLUXNET2015_ERAI_WW_1992-2012_1-3.csv - FLX_US-Ha1_FLUXNET2015_ERAI_MM_1992-2012_1-3.csv - FLX_US-Ha1_FLUXNET2015_ERAI_YY_1992-2012_1-3.csv - FLX_US-Ha1_FLUXNET2015_AUXMETEO_1992-2012_1-3.csv - FLX_US-Ha1_FLUXNET2015_AUXNEE_1992-2012_1-3.csv | ||
Examples of file names from a zipped file of a single site are provided.
The FLUXNET2015 dataset provides data at five temporal resolutions. Site teams contribute either half-hourly (HH) or hourly (HR) datasets, depending on the integration/aggregation time decided by the site managers and function of the characteristics of the turbulence. References to half-hourly in this paper also apply to hourly data, unless explicitly stated otherwise. Half-hourly data are the basis of all the processing done for this dataset and are the finest grained temporal resolution provided. Coarser aggregations are generated uniformly from half-hourly data within the data processing pipeline. The other standard temporal aggregations are: daily (DD), weekly (WW), monthly (MM), and yearly (YY).
The complete output from the data processing pipeline includes over 200 variables–among which are measured and derived data, quality flags, uncertainty quantification variables, and results from intermediate data processing steps. The variable names follow the naming conventions of <BASENAME>_<QUALIFIER>, where BASENAME describes the physical quantities (e.g., TA, NEE, Table 2) and QUALIFIER describes the information of processing methods (e.g., VUT, CUT), uncertainties (e.g., RANDUNC), and quality flags (e.g., QC) (see Supplementary Table SM1).
Table 2.
List of the variable basenames, descriptions, available resolutions and units.
| Basename | Description | Units by Resolution | ||||
|---|---|---|---|---|---|---|
| HH/HR | DD | WW | MM | YY | ||
| TA | Air temperature | deg C | ||||
| SW_IN_POT | Shortwave radiation, incoming, potential (top of atmosphere) | W m−2 | ||||
| SW_IN | Shortwave radiation | W m−2 | ||||
| LW_IN | Longwave radiation, incoming | W m−2 | ||||
| VPD | Vapor Pressure saturation Deficit | hPa | ||||
| PA | Atmospheric pressure | kPa | ||||
| P | Precipitation | mm | mm d−1 | mm y−1 | ||
| WS | Wind speed | m s−1 | ||||
| WD | Wind direction | Decimal degrees | n/a | |||
| RH | Relative humidity | % | n/a | |||
| USTAR | Friction velocity | m s−1 | ||||
| NETRAD | Net radiation | W m−2 | ||||
| PPFD_IN | Photosynthetic photon flux density, incoming | µmolPhoton m−2 s−1 | ||||
| PPFD_DIF | Photosynthetic photon flux density, diffuse incoming | µmolPhoton m−2 s−1 | ||||
| PPFD_OUT | Photosynthetic photon flux density, outgoing | µmolPhoton m−2 s−1 | ||||
| SW_DIF | Shortwave radiation, diffuse incoming | W m−2 | ||||
| SW_OUT | Shortwave radiation, outgoing | W m−2 | ||||
| LW_OUT | Longwave radiation, outgoing | W m−2 | ||||
| CO2 | CO2 mole fraction | µmolCO2 mol−1 | ||||
| TS | Soil temperature | deg C | ||||
| SWC | Soil water content | % | ||||
| G | Soil heat flux | W m−2 | ||||
| LE | Latent heat flux | W m−2 | ||||
| H | Sensible heat flux | W m−2 | ||||
| NEE | Net Ecosystem Exchange | µmolCO2 m−2 s−1 | gC m−2 d−1 | gC m−2 y−1 | ||
| RECO | Ecosystem Respiration | µmolCO2 m−2 s−1 | gC m−2 d−1 | gC m−2 y−1 | ||
| GPP | Gross Primary Production | µmolCO2 m−2 s−1 | gC m−2 d−1 | gC m−2 y−1 | ||
Separate units are listed if different units are used in different temporal aggregation resolutions. n/a indicates a variable is not provided at the specified resolution.
To serve the users with an easier-to-use data product, we created two variants with different selections of variables for data distribution: the FULLSET with all the results and variables; and the SUBSET, designed to help non-expert users, with a reduced set of variables that should fit most needs.
- FULLSET: variables generated by the processing such as uncertainty quantification variables, all variants of the data products, all quality information flags, and many variables generated by intermediate processing steps to allow in-depth understanding of individual processing steps and their effect in the final data products. A summary of the main variable basenames is in Table 2, while a full list of variables is provided in Supplementary Table SM1. Key features of the FULLSET version are:
- Meteorological variables filled with multiple gap-filling methods (e.g., MDS, ERA) are provided separately.
- NEE versions filtered with two different methods of extracting the USTAR thresholds (i.e., CUT, VUT) are provided. Multiple percentiles and reference NEE are also provided.
- GPP and RECO partitioned from NEE filtered with VUT and CUT methods, using both daytime and nighttime partitioning methods (i.e., NT, DT). Multiple percentiles and reference GPP and RECO are provided.
- LE and H gap-filled, adjusted and non-adjusted for energy balance closure, are both provided.
- Random, methodological, and joint uncertainties for NEE, GPP, RECO, LE, and H are provided.
- SUBSET: Includes a subset of the data product. The selection of the variables for this data product was done based on the expected usage for most users and to help less experienced users. Although the number of variables used is reduced, they are still accompanied by a set of quality flags and uncertainty quantification variables essential to correctly interpret the data. Key features of the SUBSET version are:
- Only the consolidated gap-filled meteorological variables are provided.
- Only the REF version of NEE filtered with the VUT method is provided. Selected percentiles and reference NEE are also provided.
- GPP and RECO (only REF versions) partitioned from NEE filtered with only the VUT method, using both daytime and nighttime partitioning methods, are provided. Selected percentiles and reference GPP and RECO are also provided.
- LE and H gap-filled, adjusted and non-adjusted for energy balance closure, are both provided.
- Random and methodological uncertainties for NEE, GPP, RECO, LE, and H are provided.
The variable proposed in the SUBSET product is NEE_VUT_REF since it maintains the temporal variability (as opposed to the MEAN NEE), it is representative of the ensemble, and the VUT method is sensitive to possible changes of the canopy (density and height) and site setup, which can have an impact on the turbulence and consequently on the USTAR threshold. The RECO and GPP products in SUBSET are calculated from the corresponding NEE variables filtered with the VUT method, generating RECO_NT_VUT_REF and RECO_DT_VUT_REF for RECO, and GPP_NT_VUT_REF and GPP_DT_VUT_REF for GPP. It is important to use both daytime (DT) and nighttime (NT) variables, and consider their difference as uncertainty.
Auxiliary data products provide extra information on specific parameters of the data processing pipeline. The groups of products are:
- AUXMETEO: Auxiliary data product containing information about the downscaling of meteorological variables using the ERA-Interim reanalysis data product (TA, PA, VPD, WS, P, SW_IN, and LW_IN). Variables in these files relate to the linear regression and error/correlation estimates for each data variable used in the downscaling.
- Parameters:
- ERA_SLOPE: the slope of linear regression
- ERA_INTERCEPT: intercept point of linear regression
- ERA_RMSE: root mean square error between site data and downscaled data
- ERA_CORRELATION: correlation coefficient of linear fit
- AUXNEE: Auxiliary data product with variables resulting from the processing of NEE (mainly related to USTAR filtering) and generation of RECO and GPP. Variables in this product include success/failure of execution of USTAR filtering methods, USTAR thresholds applied to different versions of variables, and percentile/threshold pairs with best model efficiency results.
- Variables:
- USTAR_MP_METHOD: Moving Point Test USTAR threshold method run
- USTAR_CP_METHOD: Change Point Detection USTAR threshold method run
- NEE_USTAR50_[UT]: 50th percentile of USTAR thresholds obtained from bootstrapping and sed to generate NEE_USTAR50_[UT] (with UT either CUT or VUT)
- NEE_[UT]_REF: USTAR threshold used to calculate the reference NEE, using model efficiency approach (with UT either CUT or VUT)
- [PROD]_[ALG]_[UT]_REF: USTAR threshold used to filter the NEE that was used to produce the reference product PROD (RECO or GPP), selected using model efficiency approach, using algorithm ALG (NT, DT) (with UT either CUT or VUT)
- Parameters:
- SUCCESS_RUN: 1 if a run of a method (USTAR_MP_METHOD or USTAR_CP_METHOD) was successful, 0 otherwise
- USTAR_PERCENTILE: percentile of USTAR thresholds from bootstrapping at USTAR filtering step
- USTAR_THRESHOLD: USTAR threshold value corresponding to USTAR_PERCENTILE
- [RR]_USTAR_PERCENTILE: percentile of USTAR thresholds from bootstrapping at USTAR filtering step at resolution RR (HH, DD, WW, MM, YY)
- [RR]_USTAR_THRESHOLD: USTAR threshold value corresponding to USTAR_PERCENTILE at resolution RR (HH, DD, WW, MM, YY)
ERAI: Auxiliary data product containing full record (1989–2014) of downscaled meteorological variables using the ERA-Interim reanalysis data product, including TA, PA, VPD, WS, P, SW_IN, and LW_IN.
The FLUXNET2015 metadata are included in a single file (FLX_AA-Flx_BIF_[RESOLUTION]_[YYYYMMDD].xlsx) for all sites for each data product resolution (see Table 1 for resolution options). The metadata follow the Biological, Ancillary, Disturbance, and Metadata (BADM255,256) standards and are provided in the BADM Interchange Format29 (BIF). Table 3 illustrates the type of metadata included with selected metadata variables (See full lists and descriptions of the metadata in Supplementary Tables SM2–SM7). Height and instrument models for the flux variables, as well as soil temperature and moisture depths, are reported in the Variable Information metadata.
Table 3.
Metadata types and selected variables. See Supplementary Tables SM2–SM7 for a full list of metadata with descriptions. Variables collected from or generated for all sites are in bold.
| Metadata Type | Selected Metadata Variables |
|---|---|
|
Site General Info (25 variables) |
SITE_ID: Unique site identifier (CC-sss, where CC is the country code) SITE_NAME: Site name SITE_DESC: Site description LOCATION_LAT: Latitude of site LOCATION_LONG: Longitude of site FLUX_MEASUREMENTS_VARIABLE: Flux variables measured at the site IGBP: Vegetation type based on International Geosphere-Biosphere Programme classification UTC_OFFSET: Offset from UTC of site data TEAM_MEMBER_NAME: Team member name MAT, MAP: Mean annual temperature and precipitation TOWER_TYPE, TOWER_POWER: Type of tower and power type |
|
DOI (12 variables) |
DOI: Digital Object Identifier (DOI) for the flux-met data product DOI_CONTRIBUTOR_NAME: Name of contributor to the development of data (and associated info) DOI_ORGANIZATION: Organization contributing to the data |
|
Reference publications (4 variables) |
REFERENCE_PAPER: Reference for understanding the site REFERENCE_DOI: DOI of the reference REFERENCE_USAGE: Suggested usage of the reference |
|
Canopy Height (2 variables) |
HEIGHTC: Canopy height. In a forest ecosystem, canopy height is the distribution of overstory trees that see light at the top of the canopy. Note: The reported value is representative of the mean of such a distribution. HEIGHTC_DATE: Date of canopy height observation |
|
Variable Information (5 variables) Note: Variable Information groups are only reported for variables with data. |
VAR_INFO_VARNAME: Variable name VAR_INFO_UNIT: Variable unit VAR_INFO_DATE: Start date for reported variable information VAR_INFO_HEIGHT: Height/depth of observation (meters) VAR_INFO_MODEL: Model(s) used to collect observation. |
Technical Validation
Eddy covariance measurements offer a direct method to estimate trace gas or energy exchanges between surface and atmosphere at an ecosystem scale (approximately up to 1 km around the measurement point). This makes eddy covariance difficult to compare with other methods. Nonetheless, eddy covariance data have been extensively used in numerous scientific papers and studies that indirectly validate their reliability and usefulness. Hundreds of articles have been published based on eddy covariance measurements; examples of multi-site studies using FLUXNET2015 data include Jung et al.15, Tramontana et al.257, and Keenan et al.258.
Eddy covariance data were evaluated with respect to other methods such as inventory and chambers by Campioli et al.259, who showed that “EC [eddy covariance] biases are not apparent across sites, suggesting the effectiveness of standard post-processing procedures. Our results increase confidence in EC …”. The approach of Campioli et al.259 requires sites that have several additional (and rare) pieces of information; therefore, it is not generally applicable, particularly not across the sites used in this study. However, the eddy covariance site teams co-authoring this paper have compared and technically validated their measurements with respect to knowledge of their site. Unavoidably, measurement and processing uncertainties exist, and can be large for certain sites and ecosystem conditions. However, in general, flux values provided in this dataset are consistent with expectations, and eddy covariance remains one of the more reliable techniques for assessing land-air exchanges at ecosystem scales.
Usage Notes
Detailed documentation on how to use and interpret FLUXNET2015 is available online at https://fluxnet.fluxdata.org/data/fluxnet2015-dataset/. Here, we present some of the main points to guide the usage of the data.
Risks in the application of standard procedures
When standardized procedures are applied across different sites, the possible differences owing to data treatment are avoided or minimized; this is one of the main goals of FLUXNET2015 and ONEFlux. However, there is also the risk and possibility that the standard methods don’t work properly or as expected at specific sites and under certain conditions. This is particularly true for the CO2 flux partitioning, which as with all models is based on assumptions that could not always be not valid. For this reason, it could be necessary to contact the site PIs that are listed in Supplementary Table SM9.
Using the QC flags
There are quality-control flag variables in the dataset to help users filter and interpret variables, especially for gap-filled and process knowledge-based variables. These flags are described in the variable documentation (Supplementary Table SM1). It is highly recommended that one carefully considers the QC flags when using the data.
Percentile variants for fluxes and reference values
For most flux variables, there are reference values and percentile versions of the variables to help understand some of the uncertainty in the record. For NEE, RECO, and GPP, the percentiles are generated from the bootstrapping of the USTAR threshold estimation step, i.e., they characterize the variability from a range of values obtained as USTAR thresholds. In addition, three different reference values are provided (“_MEAN”, “_USTAR50” and “_REF”) in order to cover different user needs. In general the “_REF” version should be the most representative, particularly if related to the percentiles. It is, however, important to clearly refer to which NEE version is used in order to ensure reproducibility. For the energy balance corrected H and LE variables, the percentiles indicate the variability due to the uncertainty in the correction factor applied. Similarly to NEE, there are gap-filled and energy balance corrected versions of H and LE variables; therefore, it is also important to clearly refer to which version is used. The SUBSET version of the dataset includes a reduced number of variables, selected for non-expert users. We encourage users to carefully evaluate their requirements and options in the dataset, and if needed to contact regional networks, site teams, or even co-authors of this article for help and recommendations. For more detail, see the Methods section above.
Temporally aggregated resolutions
All data products are provided at multiple temporal resolutions where feasible. The finest resolution is either hourly or half-hourly (indicated by the filename tags HR and HH, respectively). These data are then aggregated into daily (DD), 7-day weekly (WW), monthly (MM), and yearly (YY) resolutions, with appropriate aggregations for each variable, such as averaging for TA and summation for P.
Timestamps
Timestamps in the data and metadata files use the format YYYYMMDDHHMM, truncated at the appropriate resolution (e.g., YYYYMMDD for a date or YYYYMM for a month). Two formats of time associated with a record are used: (1) single timestamp, where the meaning of the timestamp is unambiguous, and, (2) a pair of timestamps, indicating the start and end of the period the timestamps represent.
Time zones
To allow more direct site comparability, all time variables are reported in local standard time (i.e., without daylight saving time). The time zone information with respect to UTC time is reported in the site metadata.
Numeric resolution
The floating point numbers are maintained at their original resolution throughout processing steps, using double precision for the majority of cases, and are truncated at up to nine decimal places in the distributed files for numbers between 0.0 and 1.0, and at up to five decimal places for larger numbers.
Column ordering
The order of columns is not always the same in different files (e.g., different sites). User data-processing routines should use the variable label (which is always consistent) and not the order of occurrence of that variable in the file. Timestamps are the only exception and will always be the first variable(s)/column(s) of the data file. This applies to text file data representations (i.e., CSV formatted).
Missing data
Missing data values are indicated with −9999, without decimal points, independent of the cause of the missing value.
Known issues
A list of known issues and limitations relevant to the dataset is maintained online: http://fluxnet.fluxdata.org/data/fluxnet2015-dataset/known-issues/.
Releases of the FLUXNET2015 dataset
The original FLUXNET2015 release was in December 2015, followed by incremental releases in July 2016 and November 2016, and, finally, a release in February 2020 with fixes and additional metadata as described in this paper. More information on the releases can be found in the online change log: http://fluxnet.fluxdata.org/change-log/. A newer release replaces all previous ones, and only the newest release is available for direct download. Access to previous versions can be obtained upon request.
Support to FLUXNET2015 data users
Scientists and staff responsible for the creation of the dataset offer support to data users and can be reached at fluxdata-support@fluxdata.org.
Updates and future versions
There is strong interest and engagement in order to ensure the availability of new data (new sites and new years), keeping the open policy and the high quality data that we tried to reach with this work. We expect that the processing pipeline and QA/QC procedures will continue evolving, in support of new products. However, the amount of both technical and coordination work, along with difficulty securing long-term international funding, hamper creation of new versions of the dataset. There are ongoing discussions among regional networks and FLUXNET on this coordination, but currently there is no plan for a follow-up version of FLUXNET2015.
Supplementary information
Acknowledgements
We thank the many people who helped with generating high quality data through the years at all sites, and the funding sources to the sites and networks, for the data collection, curation, and sharing that make this dataset possible. The eddy covariance data were acquired and shared by the following networks: AmeriFlux, AfriFlux, AsiaFlux, CarboAfrica, CarboEuropeIP, CarboItaly, CarboMont, ChinaFlux, Fluxnet-Canada, GreenGrass, ICOS, KoFlux, LBA, NECC, OzFlux-TERN, TCOS-Siberia, Swiss FluxNet and USCCC. The ERA-Interim reanalysis data were provided by ECMWF and processed by Laboratoire des sciences du climat et de l’environnement (LSCE). The FLUXNET eddy covariance data processing and harmonization was carried out by the European Fluxes Database Cluster and the AmeriFlux Management Project (with support by European Union H2020 projects and U.S. Department of Energy Office of Science, respectively), with contributions from the Carbon Dioxide Information Analysis Center, ICOS Ecosystem Thematic Centre, and OzFlux, ChinaFlux, and AsiaFlux offices.
Author contributions
Dario Papale, Gilberto Pastorello, Margaret Torn, and Deb Agarwal conceived of and organized the FLUXNET2015 dataset. Eleonora Canfora, You-Wei Cheah, Danielle Christianson, Housen Chu, Dario Papale, Gilberto Pastorello, Diego Polidori, Cristina Poindexter, and Carlo Trotta were responsible for quality checking, post-processing, and creating data products. You-Wei Cheah, Danielle Christianson, Abdelrahman Elbashandy, Marty Humphrey, Gilberto Pastorello, Diego Polidori, Alessio Ribeca, Carlo Trotta, and Catharine van Ingen were responsible for code and software preparation and implementation (processing pipeline and data distribution platform). Eleonora Canfora, You-Wei Cheah, Jiquan Chen, Danielle Christianson, Housen Chu, Peter Isaac, Dario Papale and Leiming Zhang managed the flux data and metadata collections. Gilberto Pastorello, Dario Papale, Deb Agarwal, Sebastien Biraud, You-Wei Cheah, Danielle Christianson, Housen Chu, and Margaret Torn conceived of the paper and prepared the first draft that was reviewed and commented on by all the coauthors. All the coauthors collected, processed and contributed the data, also participated in the quality assessment and correction of errors. The link between sites and coauthors is provided in Supplementary Table SM9.
Code availability
The ONEFlux collection of codes used to create data intercomparable with FLUXNET2015 has been packaged to be executed as a complete pipeline and is available in both source-code and executable forms under a 3-clause BSD license on GitHub: https://github.com/AmeriFlux/ONEFlux. The complete environment to run this pipeline requires a GCC compatible C compiler (or capability to run pre-compiled Windows, Linux, and/or Mac executables), a MATLAB Runtime Environment, and a Python interpreter with a few numeric and scientific packages installed. All of these can be obtained at no cost.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Deceased: Ray Leuning.
Contributor Information
Gilberto Pastorello, Email: gzpastorello@lbl.gov.
Dario Papale, Email: darpap@unitus.it.
Supplementary information
is available for this paper at 10.1038/s41597-020-0534-3.
References
- 1.Aubinet, M., Vesala, T. & Papale, D. (Eds.). Eddy Covariance: A Practical Guide to Measurement and Data Analysis (Springer Netherlands, Dordrecht, 2012).
- 2.Aubinet, M. et al. Estimates of the Annual Net Carbon and Water Exchange of Forests: The EUROFLUX Methodology, in Advances in Ecological Research (ed. Fitter, A. H., Raffaelli, D. G.), pp. 113–175 (Elsevier, 1999).
- 3.Law B. AmeriFlux Network aids global synthesis. Eos Trans. AGU. 2007;88:286–286. doi: 10.1029/2007EO280003. [DOI] [Google Scholar]
- 4.Novick KA, et al. The AmeriFlux network: A coalition of the willing. Agr. Forest Meteorol. 2018;249:444–456. doi: 10.1016/j.agrformet.2017.10.009. [DOI] [Google Scholar]
- 5.Yamamoto S, et al. Findings through the AsiaFlux network and a view toward the future. J. Geogr. Sci. 2005;15:142–148. doi: 10.1007/BF02872679. [DOI] [Google Scholar]
- 6.Yu G-R, et al. Overview of ChinaFLUX and evaluation of its eddy covariance measurement. Agr. Forest Meteoro. 2006;137:125–137. doi: 10.1016/j.agrformet.2006.02.011. [DOI] [Google Scholar]
- 7.Beringer J, et al. An introduction to the Australian and New Zealand flux tower network –OzFlux. Biogeosciences. 2016;13:5895–5916. doi: 10.5194/bg-13-5895-2016. [DOI] [Google Scholar]
- 8.Williams, C.A. et al. Climate and vegetation controls on the surface water balance: Synthesis of evapotranspiration measured across a global network of flux towers. Water Resour. Res. 48, W06523 (2012).
- 9.Vargas R, et al. Drought Influences the Accuracy of Simulated Ecosystem Fluxes: A Model-Data Meta-analysis for Mediterranean Oak Woodlands. Ecosystems. 2013;16:749–764. doi: 10.1007/s10021-013-9648-1. [DOI] [Google Scholar]
- 10.Shao J, et al. Biotic and climatic controls on interannual variability in carbon fluxes across terrestrial ecosystems. Agr. Forest Meteorol. 2015;205:11–22. doi: 10.1016/j.agrformet.2015.02.007. [DOI] [Google Scholar]
- 11.Balzarolo M, et al. On the relationship between ecosystem-scale hyperspectral reflectance and CO2 exchange in European mountain grasslands. Biogeosciences. 2015;12:3089–3108. doi: 10.5194/bg-12-3089-2015. [DOI] [Google Scholar]
- 12.Wolf S, et al. Warm spring reduced carbon cycle impact of the 2012 US summer drought. Proc Natl Acad Sci USA. 2016;113:5880–5885. doi: 10.1073/pnas.1519620113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung M, et al. Compensatory water effects link yearly global land CO2 sink changes to temperature. Nature. 2017;541:516–520. doi: 10.1038/nature20780. [DOI] [PubMed] [Google Scholar]
- 14.Besnard S, et al. Quantifying the effect of forest age in annual net forest carbon balance. Environ. Res. Lett. 2018;13:124018. doi: 10.1088/1748-9326/aaeaeb. [DOI] [Google Scholar]
- 15.Jung M, et al. The FLUXCOM ensemble of global land-atmosphere energy fluxes. Sci. Data. 2019;6:74. doi: 10.1038/s41597-019-0076-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baldocchi D, et al. FLUXNET: A New Tool to Study the Temporal and Spatial Variability of Ecosystem-Scale Carbon Dioxide, Water Vapor, and Energy Flux Densities. Bull. Amer. Meteor. Soc. 2001;82:2415–2434. doi: 10.1175/1520-0477(2001)082<2415:FANTTS>2.3.CO;2. [DOI] [Google Scholar]
- 17.Chu H, Baldocchi DD, John R, Wolf S, Reichstein M. Fluxes all of the time? A primer on the temporal representativeness of FLUXNET. J. Geophys. Res. Biogeosci. 2017;122:289–307. doi: 10.1002/2016JG003576. [DOI] [Google Scholar]
- 18.Pastorello, G. et al. A New Data Set to Keep a Sharper Eye on Land-Air Exchanges. Eos98, 2017 Apr 17 (2017).
- 19.Falge, E. et al. FLUXNET Marconi Conference Gap-Filled Flux and Meteorology Data, 1992–2000. ORNL DAAC, 10.3334/ornldaac/811 (2000).
- 20.FLUXNET-Fluxdata Portal. LaThuile Dataset, https://fluxnet.fluxdata.org/data/la-thuile-dataset/ (2007).
- 21.FLUXNET-Fluxdata Portal. FLUXNET2015 Dataset, https://fluxnet.fluxdata.org/data/fluxnet2015-dataset/ (2020).
- 22.Pastorello, G. et al. ONEFlux: Open Network-Enabled Flux processing pipeline. AmeriFlux Management Project, European Ecosystem Fluxes Database, ICOS Ecosystem Thematic Center, https://github.com/AmeriFlux/ONEFlux/ (2019).
- 23.FLUXNET-Fluxdata Portal. Fluxdata - The Data Portal serving the FLUXNET community, https://fluxnet.fluxdata.org/ (2020).
- 24.Aubinet, M. et al. Nighttime Flux Correction, in Eddy Covariance (eds. Aubinet, M., Vesala, T. & Papale, D.), pp. 133–157, (Springer Netherlands, Dordrecht, 2012).
- 25.Pastorello, G. et al. Observational Data Patterns for Time Series Data Quality Assessment, in Proc. IEEE 10th International Conference on e-Science, Sao Paulo, Brazil, pp. 271–278 (IEEE, 2014).
- 26.Moffat AM, et al. Comprehensive comparison of gap-filling techniques for eddy covariance net carbon fluxes. Agr. Forest Meteorol. 2007;147:209–232. doi: 10.1016/j.agrformet.2007.08.011. [DOI] [Google Scholar]
- 27.Desai AR, et al. Cross-site evaluation of eddy covariance GPP and RE decomposition techniques. Agr. Forest Meteorol. 2008;148:821–838. doi: 10.1016/j.agrformet.2007.11.012. [DOI] [Google Scholar]
- 28.AmeriFlux. Data Variables and Format, https://ameriflux.lbl.gov/data/aboutdata/data-variables/ (2020).
- 29.AmeriFlux. BADM Data Templates, https://ameriflux.lbl.gov/data/badm-data-templates/ (2020).
- 30.Papale D, et al. Towards a standardized processing of Net Ecosystem Exchange measured with eddy covariance technique: algorithms and uncertainty estimation. Biogeosciences. 2006;3:571–583. doi: 10.5194/bg-3-571-2006. [DOI] [Google Scholar]
- 31.Reichstein M, et al. On the separation of net ecosystem exchange into assimilation and ecosystem respiration: review and improved algorithm. Global Change Biol. 2005;11:1424–1439. doi: 10.1111/j.1365-2486.2005.001002.x. [DOI] [Google Scholar]
- 32.Vuichard N, Papale D. Filling the gaps in meteorological continuous data measured at FLUXNET sites with ERA-Interim reanalysis. Earth Syst. Sci. Data. 2015;7:157–171. doi: 10.5194/essd-7-157-2015. [DOI] [Google Scholar]
- 33.Dee DP, et al. The ERA-Interim reanalysis: configuration and performance of the data assimilation system. Q.J.R. Meteorol. Soc. 2011;137:553–597. doi: 10.1002/qj.828. [DOI] [Google Scholar]
- 34.European Centre for Medium-Range Weather Forecasts. ERA-Interim is a global atmospheric reanalysis dataset, https://www.ecmwf.int/en/forecasts/datasets/reanalysis-datasets/era-interim (2020).
- 35.Stoy PC, et al. A data-driven analysis of energy balance closure across FLUXNET research sites: The role of landscape scale heterogeneity. Agr. Forest Meteorol. 2013;171–172:137–152. doi: 10.1016/j.agrformet.2012.11.004. [DOI] [Google Scholar]
- 36.Foken T. The Energy Balance Closure Problem: An Overview. Ecol. Appl. 2008;18:1351–1367. doi: 10.1890/06-0922.1. [DOI] [PubMed] [Google Scholar]
- 37.Hollinger DY, Richardson AD. Uncertainty in eddy covariance measurements and its application to physiological models. Tree Physiol. 2005;25:873–885. doi: 10.1093/treephys/25.7.873. [DOI] [PubMed] [Google Scholar]
- 38.Finnigan J. The storage term in eddy flux calculations. Agr. Forest Meteorol. 2006;136:108–113. doi: 10.1016/j.agrformet.2004.12.010. [DOI] [Google Scholar]
- 39.Nicolini G, et al. Impact of CO2 storage flux sampling uncertainty on net ecosystem exchange measured by eddy covariance. Agr. Forest Meteorol. 2018;248:228–239. doi: 10.1016/j.agrformet.2017.09.025. [DOI] [Google Scholar]
- 40.Aubinet M, et al. Direct advection measurements do not help to solve the night-time CO2 closure problem: Evidence from three different forests. Agr. Forest Meteorol. 2010;150:655–664. doi: 10.1016/j.agrformet.2010.01.016. [DOI] [Google Scholar]
- 41.Barr AG, et al. Use of change-point detection for friction–velocity threshold evaluation in eddy-covariance studies. Agr. Forest Meteorol. 2013;171–172:31–45. doi: 10.1016/j.agrformet.2012.11.023. [DOI] [Google Scholar]
- 42.Nash JE, Sutcliffe JV. River flow forecasting through conceptual models part I — A discussion of principles. J. Hydrol. 1970;10:282–290. doi: 10.1016/0022-1694(70)90255-6. [DOI] [Google Scholar]
- 43.Lasslop G, et al. Separation of net ecosystem exchange into assimilation and respiration using a light response curve approach: critical issues and global evaluation. Glob. Change Biol. 2010;16:187–208. doi: 10.1111/j.1365-2486.2009.02041.x. [DOI] [Google Scholar]
- 44.van Gorsel E, et al. Estimating nocturnal ecosystem respiration from the vertical turbulent flux and change in storage of CO2. Agr. Forest Meteorol. 2009;149:1919–1930. doi: 10.1016/j.agrformet.2009.06.020. [DOI] [Google Scholar]
- 45.Garcia A, 2016. FLUXNET2015 AR-SLu San Luis. FLUXNET; Instituto Nacional de Tecnología Agropecuaria (INTA) [DOI]
- 46.Posse G, Lewczuk N, Richter K, Cristiano P. 2016. FLUXNET2015 AR-Vir Virasoro. FLUXNET; Instituto Nacional de Tecnología Agropecuaria. [DOI]
- 47.Wohlfahrt G, Hammerle A, Hörtnagl L. 2016. FLUXNET2015 AT-Neu Neustift. FLUXNET; University of Innsbruck. [DOI]
- 48.Beringer J, Hutley L. 2016. FLUXNET2015 AU-Ade Adelaide River. FLUXNET; Monash University; Charles Darwin University. [DOI]
- 49.Cleverly J, Eamus D. 2016. FLUXNET2015 AU-ASM Alice Springs. FLUXNET; University of Technology Sydney. [DOI]
- 50.Meyer W, 2016. FLUXNET2015 AU-Cpr Calperum. FLUXNET; University of Adelaide. [DOI]
- 51.Pendall E, Griebel A. 2016. FLUXNET2015 AU-Cum Cumberland Plains. FLUXNET; Western Sydney University. [DOI]
- 52.Beringer J, Hutley L. 2016. FLUXNET2015 AU-DaP Daly River Savanna. FLUXNET; Monash University; Charles Darwin University. [DOI]
- 53.Beringer J, Hutley P. 2016. FLUXNET2015 AU-DaS Daly River Cleared. FLUXNET; University of Western Australia; Charles Darwin University; Monash University. [DOI]
- 54.Beringer J, Hutley L. 2016. FLUXNET2015 AU-Dry Dry River. FLUXNET; Monash University; University of Western Australia; Charles Darwin University. [DOI]
- 55.Schroder I, Zegelin S, Palu T, Feitz A. 2016. FLUXNET2015 AU-Emr Emerald. FLUXNET; CSIRO; Geoscience Australia. [DOI]
- 56.Beringer J, Hutley L. 2016. FLUXNET2015 AU-Fog Fogg Dam. FLUXNET; Monash University; Charles Darwin University. [DOI]
- 57.Macfarlane C, 2016. FLUXNET2015 AU-Gin Gingin. FLUXNET; Edith Cowan University (Centre for Ecosystem Management) [DOI]
- 58.Macfarlane C, Prober S, Wiehl G. 2016. FLUXNET2015 AU-GWW Great Western Woodlands, Western Australia, Australia. FLUXNET; CSIRO. [DOI]
- 59.Beringer J, Hutley L. 2016. FLUXNET2015 AU-How Howard Springs. FLUXNET; Charles Darwin University; University of Western Australia; Monash University. [DOI]
- 60.Ewenz C, Stevens R, Grigson G. 2016. FLUXNET2015 AU-Lox Loxton. FLUXNET; South Australian Research and Development Institute (SARDI) [DOI]
- 61.Beringer J, Hutley L. 2016. FLUXNET2015 AU-RDF Red Dirt Melon Farm, Northern Territory. FLUXNET; Monash University; Charles Darwin University. [DOI]
- 62.Beringer J, 2016. FLUXNET2015 AU-Rig Riggs Creek. FLUXNET; Monash University. [DOI]
- 63.Liddell M. 2016. FLUXNET2015 AU-Rob Robson Creek, Queensland, Australia. FLUXNET; James Cook University. [DOI]
- 64.Beringer J, Hutley L. 2016. FLUXNET2015 AU-Stp Sturt Plains. FLUXNET; University of Western Australia; Charles Darwin University; Monash University. [DOI]
- 65.Cleverly J, Eamus D. 2016. FLUXNET2015 AU-TTE Ti Tree East. FLUXNET; University of Technology Sydney. [DOI]
- 66.Woodgate W, Van Gorsel E, Leuning R. 2016. FLUXNET2015 AU-Tum Tumbarumba. FLUXNET; CSIRO. [DOI]
- 67.Beringer, J., Hutley, L., McGuire, D. & U, P. FLUXNET2015 AU-Wac Wallaby Creek. FLUXNET; Monash University; University of California Davis; Charles Darwin University; University of Alaska Fairbanks; University of Melbourne, 10.18140/flx/1440127 (2016).
- 68.Beringer J, 2016. FLUXNET2015 AU-Whr Whroo. FLUXNET; Monash University. [DOI]
- 69.Arndt S, Hinko-Najera N, Griebel A. 2016. FLUXNET2015 AU-Wom Wombat. FLUXNET; University of Melbourne, School of Ecosystem and Forest Sciences. [DOI]
- 70.Beringer J, Walker J. 2016. FLUXNET2015 AU-Ync Jaxa. FLUXNET; University of Western Australia; Monash University. [DOI]
- 71.Neirynck J, 2016. FLUXNET2015 BE-Bra Brasschaat. FLUXNET; University of Antwerp. [DOI]
- 72.De Ligne A, 2016. FLUXNET2015 BE-Lon Lonzee. FLUXNET; University of Liege - Gembloux Agro-Bio Tech. [DOI]
- 73.De Ligne, A. et al. FLUXNET2015 BE-Vie Vielsalm. FLUXNET; University of Liege - Gembloux Agro-Bio Tech; University catholic of Louvain-la-Neuve, 10.18140/flx/1440130 (2016).
- 74.Saleska S. 2016. FLUXNET2015 BR-Sa1 Santarem-Km67-Primary Forest. FLUXNET; University of Arizona. [DOI]
- 75.Goulden M. 2016. FLUXNET2015 BR-Sa3 Santarem-Km83-Logged Forest. FLUXNET; University of California - Irvine. [DOI]
- 76.McCaughey H. 2016. FLUXNET2015 CA-Gro Ontario - Groundhog River, Boreal Mixedwood Forest. FLUXNET; Queen’s University. [DOI]
- 77.Amiro B. 2016. FLUXNET2015 CA-Man Manitoba - Northern Old Black Spruce (former BOREAS Northern Study Area) FLUXNET; University of Manitoba. [DOI]
- 78.Goulden M. 2016. FLUXNET2015 CA-NS1 UCI-1850 burn site. FLUXNET; University of California - Irvine. [DOI]
- 79.Goulden M. 2016. FLUXNET2015 CA-NS2 UCI-1930 burn site. FLUXNET; University of California - Irvine. [DOI]
- 80.Goulden M. 2016. FLUXNET2015 CA-NS3 UCI-1964 burn site. FLUXNET; University of California - Irvine. [DOI]
- 81.Goulden M. 2016. FLUXNET2015 CA-NS4 UCI-1964 burn site wet. FLUXNET; University of California - Irvine. [DOI]
- 82.Goulden M. 2016. FLUXNET2015 CA-NS5 UCI-1981 burn site. FLUXNET; University of California - Irvine. [DOI]
- 83.Goulden M. 2016. FLUXNET2015 CA-NS6 UCI-1989 burn site. FLUXNET; University of California - Irvine. [DOI]
- 84.Goulden M. 2016. FLUXNET2015 CA-NS7 UCI-1998 burn site. FLUXNET; University of California - Irvine. [DOI]
- 85.Black T. 2016. FLUXNET2015 CA-Oas Saskatchewan - Western Boreal, Mature Aspen. FLUXNET; The University of British Columbia. [DOI]
- 86.Black T. 2016. FLUXNET2015 CA-Obs Saskatchewan - Western Boreal, Mature Black Spruce. FLUXNET; The University of British Columbia. [DOI]
- 87.Margolis H. 2016. FLUXNET2015 CA-Qfo Quebec - Eastern Boreal, Mature Black Spruce. FLUXNET; Université Laval. [DOI]
- 88.Amiro B. 2016. FLUXNET2015 CA-SF1 Saskatchewan - Western Boreal, forest burned in 1977. FLUXNET; University of Manitoba. [DOI]
- 89.Amiro B. 2016. FLUXNET2015 CA-SF2 Saskatchewan - Western Boreal, forest burned in 1989. FLUXNET; University of Manitoba. [DOI]
- 90.Amiro B. 2016. FLUXNET2015 CA-SF3 Saskatchewan - Western Boreal, forest burned in 1998. FLUXNET; University of Manitoba; Canadian Forest Service. [DOI]
- 91.Arain M. 2016. FLUXNET2015 CA-TP1 Ontario - Turkey Point 2002 Plantation White Pine. FLUXNET; McMaster University. [DOI]
- 92.Arain M. 2016. FLUXNET2015 CA-TP2 Ontario - Turkey Point 1989 Plantation White Pine. FLUXNET; McMaster University. [DOI]
- 93.Arain M. 2016. FLUXNET2015 CA-TP3 Ontario - Turkey Point 1974 Plantation White Pine. FLUXNET; McMaster University. [DOI]
- 94.Arain M. 2016. FLUXNET2015 CA-TP4 Ontario - Turkey Point 1939 Plantation White Pine. FLUXNET; McMaster University. [DOI]
- 95.Arain M. 2016. FLUXNET2015 CA-TPD Ontario - Turkey Point Mature Deciduous. FLUXNET; McMaster University. [DOI]
- 96.Nouvellon Y. 2016. FLUXNET2015 CG-Tch Tchizalamou. FLUXNET; Centre de coopération internationale en recherche agronomique pour le développement. [DOI]
- 97.Hörtnagl L, 2016. FLUXNET2015 CH-Cha Chamau. FLUXNET; ETH Zurich. [DOI]
- 98.Hörtnagl L, 2016. FLUXNET2015 CH-Dav Davos. FLUXNET; ETH Zurich. [DOI]
- 99.Hörtnagl L, 2016. FLUXNET2015 CH-Fru Früebüel. FLUXNET; ETH Zurich. [DOI]
- 100.Hörtnagl L, 2016. FLUXNET2015 CH-Lae Laegern. FLUXNET; ETH Zurich. [DOI]
- 101.Ammann C. 2016. FLUXNET2015 CH-Oe1 Oensingen grassland. FLUXNET; Agroscope Zuerich. [DOI]
- 102.Hörtnagl L, 2016. FLUXNET2015 CH-Oe2 Oensingen crop. FLUXNET; ETH Zurich. [DOI]
- 103.Zhang J, Han S. 2016. FLUXNET2015 CN-Cha Changbaishan. FLUXNET; IAE Chinese Academy of Sciences. [DOI]
- 104.Dong G. 2016. FLUXNET2015 CN-Cng Changling. FLUXNET; Shanxi University. [DOI]
- 105.Shi P, Zhang X, He Y. 2016. FLUXNET2015 CN-Dan Dangxiong. FLUXNET; IGSNRR Chinese Academy of Sciences. [DOI]
- 106.Zhou G, Yan J. 2016. FLUXNET2015 CN-Din Dinghushan. FLUXNET; SCIB Chinese Academy of Sciences. [DOI]
- 107.Chen S. 2016. FLUXNET2015 CN-Du2 Duolun_grassland (D01) FLUXNET; Institute of Botany, Chinese Academy of Sciences. [DOI]
- 108.Shao C. 2016. FLUXNET2015 CN-Du3 Duolun Degraded Meadow. FLUXNET. [DOI]
- 109.Li Y. 2016. FLUXNET2015 CN-Ha2 Haibei Shrubland. FLUXNET; NWIPB Chinese Academy of Sciences. [DOI]
- 110.Tang Y, Kato T, Du M. 2016. FLUXNET2015 CN-HaM Haibei Alpine Tibet site. FLUXNET; National Institute for Environmental Studies. [DOI]
- 111.Wang H, Fu X. 2016. FLUXNET2015 CN-Qia Qianyanzhou. FLUXNET; IGSNRR Chinese Academy of Sciences. [DOI]
- 112.Shao C. 2016. FLUXNET2015 CN-Sw2 Siziwang Grazed (SZWG) FLUXNET. [DOI]
- 113.Sigut L, 2016. FLUXNET2015 CZ-BK1 Bily Kriz forest. FLUXNET; Global Change Research Institute CAS. [DOI]
- 114.Sigut L, 2016. FLUXNET2015 CZ-BK2 Bily Kriz grassland. FLUXNET; Global Change Research Institute CAS. [DOI]
- 115.Dušek J, Janouš D, Pavelka M. 2016. FLUXNET2015 CZ-wet Trebon (CZECHWET) FLUXNET; Global Change Research Institute CAS. [DOI]
- 116.Bernhofer C, 2016. FLUXNET2015 DE-Akm Anklam. FLUXNET; TU Dresden. [DOI]
- 117.Brümmer C, Lucas-Moffat A, Herbst M, Kolle O. 2016. FLUXNET2015 DE-Geb Gebesee. FLUXNET; Thünen Institute of Climate-Smart Agriculture, Braunschweig. [DOI]
- 118.Bernhofer C, 2016. FLUXNET2015 DE-Gri Grillenburg. FLUXNET; TU Dresden. [DOI]
- 119.Knohl A, 2016. FLUXNET2015 DE-Hai Hainich. FLUXNET; University of Goettingen, Bioclimatology. [DOI]
- 120.Bernhofer C, 2016. FLUXNET2015 DE-Kli Klingenberg. FLUXNET; TU Dresden. [DOI]
- 121.Lindauer M, 2016. FLUXNET2015 DE-Lkb Lackenberg. FLUXNET; Karlsruhe Institute of Technology, IMK-IFU. [DOI]
- 122.Knohl A, 2016. FLUXNET2015 DE-Lnf Leinefelde. FLUXNET; University of Goettingen, Bioclimatology. [DOI]
- 123.Bernhofer C, 2016. FLUXNET2015 DE-Obe Oberbärenburg. FLUXNET; TU Dresden. [DOI]
- 124.Schmidt M, Graf A. 2016. FLUXNET2015 DE-RuR Rollesbroich. FLUXNET; IBG-3 Agrosphäre, Research Centre Jülich GmbH. [DOI]
- 125.Schmidt M, Graf A. 2016. FLUXNET2015 DE-RuS Selhausen Juelich. FLUXNET; IBG-3 Agrosphäre, Research Centre Jülich GmbH. [DOI]
- 126.Schneider K, Schmidt M. 2016. FLUXNET2015 DE-Seh Selhausen. FLUXNET; University of Cologne. [DOI]
- 127.Klatt J, Schmid H, Mauder M, Steinbrecher R. 2016. FLUXNET2015 DE-SfN Schechenfilz Nord. FLUXNET; Karlsruhe Institute of Technology, IMK-IFU. [DOI]
- 128.Bernhofer C, 2016. FLUXNET2015 DE-Spw Spreewald. FLUXNET; TU Dresden. [DOI]
- 129.Bernhofer C, 2016. FLUXNET2015 DE-Tha Tharandt. FLUXNET; TU Dresden. [DOI]
- 130.Sachs T, Wille C, Larmanou E, Franz D. 2016. FLUXNET2015 DE-Zrk Zarnekow. FLUXNET; GFZ German Research Centre for Geosciences. [DOI]
- 131.Pilegaard K, Ibrom A. 2016. FLUXNET2015 DK-Eng Enghave. FLUXNET; Technical University of Denmark (DTU) [DOI]
- 132.Olesen J. 2016. FLUXNET2015 DK-Fou Foulum. FLUXNET; Danish Institute of Agricultural Sciences. [DOI]
- 133.Ibrom A, Pilegaard K. 2016. FLUXNET2015 DK-Sor Soroe. FLUXNET; Technical University of Denmark (DTU) [DOI]
- 134.Poveda F, 2016. FLUXNET2015 ES-Amo Amoladeras. FLUXNET; Estación Experimental de Zona Áridas (EEZA, CSIC) [DOI]
- 135.Reverter B, Perez-Cañete E, Kowalski A. 2016. FLUXNET2015 ES-LgS Laguna Seca. FLUXNET; Universidad de Granada. [DOI]
- 136.Cañete E, 2016. FLUXNET2015 ES-LJu Llano de los Juanes. FLUXNET; University of Granada. [DOI]
- 137.Reverter B, Perez-Cañete E, Kowalski A. 2016. FLUXNET2015 ES-Ln2 Lanjaron-Salvage logging. FLUXNET; Universidad de Granada. [DOI]
- 138.Mammarella I, 2016. FLUXNET2015 FI-Hyy Hyytiala. FLUXNET; University of Helsinki. [DOI]
- 139.Lohila A, 2016. FLUXNET2015 FI-Jok Jokioinen. FLUXNET; Finnish Meteorological Institute. [DOI]
- 140.Lohila A, 2016. FLUXNET2015 FI-Let Lettosuo. FLUXNET; Finnish Meteorological Institute. [DOI]
- 141.Aurela M, 2016. FLUXNET2015 FI-Lom Lompolojankka. FLUXNET; Finnish Meteorological Institute. [DOI]
- 142.Aurela M, 2016. FLUXNET2015 FI-Sod Sodankyla. FLUXNET; Finnish Meteorological Institute. [DOI]
- 143.Berveiller D, 2016. FLUXNET2015 FR-Fon Fontainebleau-Barbeau. FLUXNET; CNRS. [DOI]
- 144.Buysse P, 2016. FLUXNET2015 FR-Gri Grignon. FLUXNET; French National Institute for Agricultural Research. [DOI]
- 145.Berbigier P, Loustau D. 2016. FLUXNET2015 FR-LBr Le Bray. FLUXNET; INRA - UMR ISPA. [DOI]
- 146.Ourcival J. 2016. FLUXNET2015 FR-Pue Puechabon. FLUXNET; CNRS. [DOI]
- 147.Bonal D, Burban B. 2016. FLUXNET2015 GF-Guy Guyaflux (French Guiana) FLUXNET; INRA. [DOI]
- 148.Valentini, R. et al. FLUXNET2015 GH-Ank Ankasa. FLUXNET; Euro Mediterranean Center for Climate Change - Viterbo; University of Tuscia - Vietrbo, 10.18140/flx/1440229 (2016).
- 149.Hansen B. 2016. FLUXNET2015 GL-NuF Nuuk Fen. FLUXNET; University of Copenhagen; University of Aarhus; Asiaq - Greenland Survey. [DOI]
- 150.Lund M, Jackowicz-Korczyński M, Abermann J. 2016. FLUXNET2015 GL-ZaF Zackenberg Fen. FLUXNET; Aarhus University. [DOI]
- 151.Lund M, Jackowicz-Korczyński M, Abermann J. 2016. FLUXNET2015 GL-ZaH Zackenberg Heath. FLUXNET; Aarhus University. [DOI]
- 152.Magliulo V, 2016. FLUXNET2015 IT-BCi Borgo Cioffi. FLUXNET; CNR ISAFOM. [DOI]
- 153.Sabbatini S, Arriga N, Papale D. 2016. FLUXNET2015 IT-CA1 Castel d’Asso1. FLUXNET; University of Tuscia - Vietrbo. [DOI]
- 154.Sabbatini S, Arriga N, Gioli B, Papale D. 2016. FLUXNET2015 IT-CA2 Castel d’Asso2. FLUXNET; CNR IBIMET; University of Tuscia - Vietrbo. [DOI]
- 155.Sabbatini S, Arriga N, Matteucci G, Papale D. 2016. FLUXNET2015 IT-CA3 Castel d’Asso 3. FLUXNET; University of Tuscia - Vietrbo; CNR IBAF. [DOI]
- 156.Matteucci G. 2016. FLUXNET2015 IT-Col Collelongo. FLUXNET; Istituto di Ecologia e Idrologia Forestale CNR. [DOI]
- 157.Fares S, Savi F, Conte A. 2016. FLUXNET2015 IT-Cp2 Castelporziano 2. FLUXNET; Council for Agricultural Research and Economics. [DOI]
- 158.Valentini R, 2016. FLUXNET2015 IT-Cpz Castelporziano. FLUXNET; University of Tuscia - Vietrbo. [DOI]
- 159.Gruening C, Goded I, Cescatti A, Pokorska O. 2016. FLUXNET2015 IT-Isp Ispra ABC-IS. FLUXNET; European Commission - Joint Research Centre. [DOI]
- 160.Cescatti A, Marcolla B, Zorer R, Gianelle D. 2016. FLUXNET2015 IT-La2 Lavarone2. FLUXNET; Centro di Ecologia Alpina. [DOI]
- 161.Gianelle D, Zampedri R, Cavagna M, Sottocornola M. 2016. FLUXNET2015 IT-Lav Lavarone. FLUXNET; Edmund Mach Foundation. [DOI]
- 162.Gianelle D, Cavagna M, Zampedri R, Marcolla B. 2016. FLUXNET2015 IT-MBo Monte Bondone. FLUXNET; Edmund Mach Foundation. [DOI]
- 163.Spano D, 2016. FLUXNET2015 IT-Noe Arca di Noe - Le Prigionette. FLUXNET; University of Sassari; CNR-Ibimet Sassari. [DOI]
- 164.Manca G, Goded I. 2016. FLUXNET2015 IT-PT1 Parco Ticino forest. FLUXNET; European Commission - DG Joint Research Centre. [DOI]
- 165.Montagnani L, Minerbi S. 2016. FLUXNET2015 IT-Ren Renon. FLUXNET; Autonomous Province of Bolzano, Forest Services. [DOI]
- 166.Valentini R, 2016. FLUXNET2015 IT-Ro1 Roccarespampani 1. FLUXNET; University of Tuscia - Vietrbo. [DOI]
- 167.Papale D, 2016. FLUXNET2015 IT-Ro2 Roccarespampani 2. FLUXNET; University of Tuscia - Vietrbo. [DOI]
- 168.Gruening C, Goded I, Cescatti A, Pokorska O. 2016. FLUXNET2015 IT-SR2 San Rossore 2. FLUXNET; European Commission - Joint Research Centre. [DOI]
- 169.Gruening C, 2016. FLUXNET2015 IT-SRo San Rossore. FLUXNET; European Commission - Joint Research Centre. [DOI]
- 170.Cremonese E, Galvagno M, Di Cella U, Migliavacca M. 2016. FLUXNET2015 IT-Tor Torgnon. FLUXNET; Environmental Protection Agency of Aosta Valley. [DOI]
- 171.Kotani A. 2016. FLUXNET2015 JP-MBF Moshiri Birch Forest Site. FLUXNET; Nagoya University. [DOI]
- 172.Kotani A. 2016. FLUXNET2015 JP-SMF Seto Mixed Forest Site. FLUXNET; Nagoya University. [DOI]
- 173.Kosugi Y, Takanashi S. 2016. FLUXNET2015 MY-PSO Pasoh Forest Reserve (PSO) FLUXNET; FRIM(Forest Research Institute of Malaysia); Kyoto University. [DOI]
- 174.Dolman H, 2016. FLUXNET2015 NL-Hor Horstermeer. FLUXNET; Vrije Universiteit Amsterdam. [DOI]
- 175.Moors E, Elbers J. 2016. FLUXNET2015 NL-Loo Loobos. FLUXNET; ALTERRA/Wageningen Environmental Research. [DOI]
- 176.Wolf S, Eugster W, Buchmann N. 2016. FLUXNET2015 PA-SPn Sardinilla Plantation. FLUXNET; ETH Zurich. [DOI]
- 177.Wolf S, Eugster W, Buchmann N. 2016. FLUXNET2015 PA-SPs Sardinilla-Pasture. FLUXNET; ETH Zurich. [DOI]
- 178.Merbold L, Rebmann C, Corradi C. 2016. FLUXNET2015 RU-Che Cherski. FLUXNET; Max-Planck Institute for Biogeochemistry. [DOI]
- 179.Dolman H, 2016. FLUXNET2015 RU-Cok Chokurdakh. FLUXNET; Vrije Universiteit Amsterdam. [DOI]
- 180.Varlagin A, Kurbatova J, Vygodskaya N. 2016. FLUXNET2015 RU-Fyo Fyodorovskoye. FLUXNET; A.N. Severtsov Institute of Ecology and Evolution. [DOI]
- 181.Belelli L, Papale D, Valentini R. 2016. FLUXNET2015 RU-Ha1 Hakasia steppe. FLUXNET; University of Tuscia - Vietrbo. [DOI]
- 182.Ardö J, El Tahir B, ElKhidir H. 2016. FLUXNET2015 SD-Dem Demokeya. FLUXNET; LUND UNIVERSITY. [DOI]
- 183.Christensen T. 2016. FLUXNET2015 SJ-Adv Adventdalen. FLUXNET; NATEKO; Lund University. [DOI]
- 184.Boike, J. et al. FLUXNET2015 SJ-Blv Bayelva, Spitsbergen. FLUXNET; University of Oslo, Department of Geosciences, 0316 OSLO, Norway; Universität Bayreuth, Department of Earth Sciences, 95440 Bayreuth, Germany; Alfred Wegener Institute, Helmholtz Centre for Polar and Marine Research, Periglacial Research Unit, 14473 Potsdam, Germany, 10.18140/flx/1440242 (2016).
- 185.Tagesson T, Ardö J, Fensholt R. 2016. FLUXNET2015 SN-Dhr Dahra. FLUXNET; Lund University. [DOI]
- 186.Billesbach D, Bradford J, Torn M. 2016. FLUXNET2015 US-AR1 ARM USDA UNL OSU Woodward Switchgrass 1. FLUXNET; Lawerence Berkeley National Lab; U.S. Department of Agriculture; University of Nebraska. [DOI]
- 187.Billesbach D, Bradford J, Torn M. 2016. FLUXNET2015 US-AR2 ARM USDA UNL OSU Woodward Switchgrass 2. FLUXNET; Lawrence Berkeley National Lab; U.S. Department of Agriculture; University of Nebraska. [DOI]
- 188.Torn M. 2016. FLUXNET2015 US-ARb ARM Southern Great Plains burn site- Lamont. FLUXNET; Lawrence Berkeley National Laboratory. [DOI]
- 189.Torn M. 2016. FLUXNET2015 US-ARc ARM Southern Great Plains control site- Lamont. FLUXNET; Lawrence Berkeley National Laboratory. [DOI]
- 190.Biraud S, Fischer M, Chan S, Torn M. 2016. FLUXNET2015 US-ARM ARM Southern Great Plains site- Lamont. FLUXNET; Lawrence Berkeley National Laboratory. [DOI]
- 191.Zona D, Oechel W. 2016. FLUXNET2015 US-Atq Atqasuk. FLUXNET; San Diego State University. [DOI]
- 192.Goldstein A. 2016. FLUXNET2015 US-Blo Blodgett Forest. FLUXNET; University of California, Berkeley. [DOI]
- 193.Bowling D. 2016. FLUXNET2015 US-Cop Corral Pocket. FLUXNET; University of Utah. [DOI]
- 194.Chen J, Chu H. 2016. FLUXNET2015 US-CRT Curtice Walter-Berger cropland. FLUXNET; University of Toledo/Michigan State University. [DOI]
- 195.Massman B. 2016. FLUXNET2015 US-GBT GLEES Brooklyn Tower. FLUXNET; USDA Forest Service. [DOI]
- 196.Massman B. 2016. FLUXNET2015 US-GLE GLEES. FLUXNET; USDA Forest Service. [DOI]
- 197.Meyers T. 2016. FLUXNET2015 US-Goo Goodwin Creek. FLUXNET; NOAA/ARL. [DOI]
- 198.Munger J. 2016. FLUXNET2015 US-Ha1 Harvard Forest EMS Tower (HFR1) FLUXNET; Harvard University. [DOI]
- 199.Matamala R. 2016. FLUXNET2015 US-IB2 Fermi National Accelerator Laboratory- Batavia (Prairie site) FLUXNET; Argonne National Laboratory. [DOI]
- 200.Zona D, Oechel W. 2016. FLUXNET2015 US-Ivo Ivotuk. FLUXNET; San Diego State University. [DOI]
- 201.Drake B, Hinkle R. 2016. FLUXNET2015 US-KS1 Kennedy Space Center (slash pine) FLUXNET; Smithsonian Environmental Research Center; University of Central Florida. [DOI]
- 202.Drake B, Hinkle R. 2016. FLUXNET2015 US-KS2 Kennedy Space Center (scrub oak) FLUXNET; Smithsonian Environmental Research Center; University of Central Florida. [DOI]
- 203.Fares S. 2016. FLUXNET2015 US-Lin Lindcove Orange Orchard. FLUXNET; Entecra. [DOI]
- 204.Desai A. 2016. FLUXNET2015 US-Los Lost Creek. FLUXNET; University of Wisconsin. [DOI]
- 205.Meyers T. 2016. FLUXNET2015 US-LWW Little Washita Watershed. FLUXNET; NOAA/ARL. [DOI]
- 206.Law B. 2016. FLUXNET2015 US-Me1 Metolius - Eyerly burn. FLUXNET; Oregon State University. [DOI]
- 207.Law B. 2016. FLUXNET2015 US-Me2 Metolius mature ponderosa pine. FLUXNET; Oregon State University. [DOI]
- 208.Law B. 2016. FLUXNET2015 US-Me3 Metolius-second young aged pine. FLUXNET; Oregon State University. [DOI]
- 209.Law B. 2016. FLUXNET2015 US-Me4 Metolius-old aged ponderosa pine. FLUXNET; Oregon State University. [DOI]
- 210.Law B. 2016. FLUXNET2015 US-Me5 Metolius-first young aged pine. FLUXNET; Oregon State University. [DOI]
- 211.Law B. 2016. FLUXNET2015 US-Me6 Metolius Young Pine Burn. FLUXNET; Oregon State University. [DOI]
- 212.Novick K, Phillips R. 2016. FLUXNET2015 US-MMS Morgan Monroe State Forest. FLUXNET; Indiana University. [DOI]
- 213.Sturtevant C, 2016. FLUXNET2015 US-Myb Mayberry Wetland. FLUXNET; University of California, Berkeley. [DOI]
- 214.Suyker A. 2016. FLUXNET2015 US-Ne1 Mead - irrigated continuous maize site. FLUXNET; University of Nebraska - Lincoln. [DOI]
- 215.Suyker A. 2016. FLUXNET2015 US-Ne2 Mead - irrigated maize-soybean rotation site. FLUXNET; University of Nebraska - Lincoln. [DOI]
- 216.Suyker A. 2016. FLUXNET2015 US-Ne3 Mead - rainfed maize-soybean rotation site. FLUXNET; University of Nebraska - Lincoln. [DOI]
- 217.Blanken P, 2016. FLUXNET2015 US-NR1 Niwot Ridge Forest (LTER NWT1) FLUXNET; University of Colorado. [DOI]
- 218.Chen J, Chu H, Noormets A. 2016. FLUXNET2015 US-Oho Oak Openings. FLUXNET; University of Toledo/Michigan State University. [DOI]
- 219.Bohrer G. 2016. FLUXNET2015 US-ORv Olentangy River Wetland Research Park. FLUXNET; The Ohio State University. [DOI]
- 220.Desai A. 2016. FLUXNET2015 US-PFa Park Falls/WLEF. FLUXNET; University of Wisconsin. [DOI]
- 221.Kobayashi H, Suzuki R. 2016. FLUXNET2015 US-Prr Poker Flat Research Range Black Spruce Forest. FLUXNET; Japan Agency for Marine-Earth Science and Technology. [DOI]
- 222.Kurc S. 2016. FLUXNET2015 US-SRC Santa Rita Creosote. FLUXNET; University of Arizona. [DOI]
- 223.Scott R. 2016. FLUXNET2015 US-SRG Santa Rita Grassland. FLUXNET; United States Department of Agriculture. [DOI]
- 224.Scott R. 2016. FLUXNET2015 US-SRM Santa Rita Mesquite. FLUXNET; United States Department of Agriculture. [DOI]
- 225.Ewers B, Pendall E. 2016. FLUXNET2015 US-Sta Saratoga. FLUXNET; University of Wyoming. [DOI]
- 226.Desai A. 2016. FLUXNET2015 US-Syv Sylvania Wilderness Area. FLUXNET; University of Wisconsin. [DOI]
- 227.Baldocchi D, Ma S. 2016. FLUXNET2015 US-Ton Tonzi Ranch. FLUXNET; University of California, Berkeley. [DOI]
- 228.Szutu D, Baldocchi D, Eichelmann E, Knox S. 2016. FLUXNET2015 US-Tw1 Twitchell Wetland West Pond. FLUXNET; University of California, Berkeley. [DOI]
- 229.Baldocchi D. 2016. FLUXNET2015 US-Tw2 Twitchell Corn. FLUXNET; University of California, Berkeley. [DOI]
- 230.Szutu D, Baldocchi D. 2016. FLUXNET2015 US-Tw3 Twitchell Alfalfa. FLUXNET; University of California, Berkeley. [DOI]
- 231.Sanchez C, 2016. FLUXNET2015 US-Tw4 Twitchell East End Wetland. FLUXNET; University of California, Berkeley. [DOI]
- 232.Baldocchi D. 2016. FLUXNET2015 US-Twt Twitchell Island. FLUXNET; University of California, Berkeley. [DOI]
- 233.Gough C, Bohrer G, Curtis P. 2016. FLUXNET2015 US-UMB Univ. of Mich. Biological Station. FLUXNET; Ohio State University; Virginia Commonwealth University. [DOI]
- 234.Gough C, Bohrer G, Curtis P. 2016. FLUXNET2015 US-UMd UMBS Disturbance. FLUXNET; Ohio State University; Virginia Commonwealth University. [DOI]
- 235.Baldocchi D, Ma S, Xu L. 2016. FLUXNET2015 US-Var Vaira Ranch- Ione. FLUXNET; University of California, Berkeley. [DOI]
- 236.Desai A. 2016. FLUXNET2015 US-WCr Willow Creek. FLUXNET; University of Wisconsin. [DOI]
- 237.Scott R. 2016. FLUXNET2015 US-Whs Walnut Gulch Lucky Hills Shrub. FLUXNET; United States Department of Agriculture. [DOI]
- 238.Chen J. 2016. FLUXNET2015 US-Wi0 Young red pine (YRP) FLUXNET; Michigan State University. [DOI]
- 239.Chen J. 2016. FLUXNET2015 US-Wi1 Intermediate hardwood (IHW) FLUXNET; Michigan State University. [DOI]
- 240.Chen J. 2016. FLUXNET2015 US-Wi2 Intermediate red pine (IRP) FLUXNET; Michigan State University. [DOI]
- 241.Chen J. 2016. FLUXNET2015 US-Wi3 Mature hardwood (MHW) FLUXNET; Michigan State University. [DOI]
- 242.Chen J. 2016. FLUXNET2015 US-Wi4 Mature red pine (MRP) FLUXNET; Michigan State University. [DOI]
- 243.Chen J. 2016. FLUXNET2015 US-Wi5 Mixed young jack pine (MYJP) FLUXNET; Michigan State University. [DOI]
- 244.Chen J. 2016. FLUXNET2015 US-Wi6 Pine barrens #1 (PB1) FLUXNET; Michigan State University. [DOI]
- 245.Chen J. 2016. FLUXNET2015 US-Wi7 Red pine clearcut (RPCC) FLUXNET; Michigan State University. [DOI]
- 246.Chen J. 2016. FLUXNET2015 US-Wi8 Young hardwood clearcut (YHW) FLUXNET; Michigan State University. [DOI]
- 247.Chen J. 2016. FLUXNET2015 US-Wi9 Young Jack pine (YJP) FLUXNET; Michigan State University. [DOI]
- 248.Scott R. 2016. FLUXNET2015 US-Wkg Walnut Gulch Kendall Grasslands. FLUXNET; United States Department of Agriculture. [DOI]
- 249.Chen J, Chu H. 2016. FLUXNET2015 US-WPT Winous Point North Marsh. FLUXNET; University of Toledo/Michigan State University. [DOI]
- 250.Kutsch W, Merbold L, Kolle O. 2016. FLUXNET2015 ZM-Mon Mongu. FLUXNET; Max-Planck Institute for Biogeochemistry. [DOI]
- 251.Harris I, Jones PD, Osborn TJ, Lister DH. Updated high-resolution grids of monthly climatic observations - the CRU TS3.10 Dataset. Int. J. Climatol. 2014;34:623–642. doi: 10.1002/joc.3711. [DOI] [Google Scholar]
- 252.Channan, S., Collins, K. & Emanuel, W. Global mosaics of the standard MODIS land cover type data. University of Maryland and the Pacific Northwest National Laboratory, College Park, Maryland, USA (2014).
- 253.Baldocchi D, Chu H, Reichstein M. Inter-annual variability of net and gross ecosystem carbon fluxes: A review. Agr. Forest Meteorol. 2018;249:520–533. doi: 10.1016/j.agrformet.2017.05.015. [DOI] [Google Scholar]
- 254.Baldocchi DD. How eddy covariance flux measurements have contributed to our understanding of Global Change Biology. Glob. Change Biol. 2019;26:242–260. doi: 10.1111/gcb.14807. [DOI] [PubMed] [Google Scholar]
- 255.Law, B. E. et al. Terrestrial Carbon Observations: Protocols for Vegetation Sampling and Data Submission. Report No.55 (Global Terrestrial Observing System, FAO, Rome, 2008).
- 256.Papale, D., Canfora, E. & Polidori, D. ICOS Ecosystem Instructions for Use the ICOS BADM (Version 20171013), ICOS Ecosystem Thematic Centre, 10.18160/6m8s-fy7m (2017).
- 257.Tramontana G, et al. Predicting carbon dioxide and energy fluxes across global FLUXNET sites with regression algorithms. Biogeosciences. 2016;13:4291–4313. doi: 10.5194/bg-13-4291-2016. [DOI] [Google Scholar]
- 258.Keenan TF, et al. Widespread inhibition of daytime ecosystem respiration. Nat. Ecol. Evol. 2019;3:407–415. doi: 10.1038/s41559-019-0809-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Campioli M, et al. Evaluating the convergence between eddy-covariance and biometric methods for assessing carbon budgets of forests. Nat. Commun. 2016;7:13717. doi: 10.1038/ncomms13717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Garcia A, 2016. FLUXNET2015 AR-SLu San Luis. FLUXNET; Instituto Nacional de Tecnología Agropecuaria (INTA) [DOI]
- Posse G, Lewczuk N, Richter K, Cristiano P. 2016. FLUXNET2015 AR-Vir Virasoro. FLUXNET; Instituto Nacional de Tecnología Agropecuaria. [DOI]
- Wohlfahrt G, Hammerle A, Hörtnagl L. 2016. FLUXNET2015 AT-Neu Neustift. FLUXNET; University of Innsbruck. [DOI]
- Beringer J, Hutley L. 2016. FLUXNET2015 AU-Ade Adelaide River. FLUXNET; Monash University; Charles Darwin University. [DOI]
- Cleverly J, Eamus D. 2016. FLUXNET2015 AU-ASM Alice Springs. FLUXNET; University of Technology Sydney. [DOI]
- Meyer W, 2016. FLUXNET2015 AU-Cpr Calperum. FLUXNET; University of Adelaide. [DOI]
- Pendall E, Griebel A. 2016. FLUXNET2015 AU-Cum Cumberland Plains. FLUXNET; Western Sydney University. [DOI]
- Beringer J, Hutley L. 2016. FLUXNET2015 AU-DaP Daly River Savanna. FLUXNET; Monash University; Charles Darwin University. [DOI]
- Beringer J, Hutley P. 2016. FLUXNET2015 AU-DaS Daly River Cleared. FLUXNET; University of Western Australia; Charles Darwin University; Monash University. [DOI]
- Beringer J, Hutley L. 2016. FLUXNET2015 AU-Dry Dry River. FLUXNET; Monash University; University of Western Australia; Charles Darwin University. [DOI]
- Schroder I, Zegelin S, Palu T, Feitz A. 2016. FLUXNET2015 AU-Emr Emerald. FLUXNET; CSIRO; Geoscience Australia. [DOI]
- Beringer J, Hutley L. 2016. FLUXNET2015 AU-Fog Fogg Dam. FLUXNET; Monash University; Charles Darwin University. [DOI]
- Macfarlane C, 2016. FLUXNET2015 AU-Gin Gingin. FLUXNET; Edith Cowan University (Centre for Ecosystem Management) [DOI]
- Macfarlane C, Prober S, Wiehl G. 2016. FLUXNET2015 AU-GWW Great Western Woodlands, Western Australia, Australia. FLUXNET; CSIRO. [DOI]
- Beringer J, Hutley L. 2016. FLUXNET2015 AU-How Howard Springs. FLUXNET; Charles Darwin University; University of Western Australia; Monash University. [DOI]
- Ewenz C, Stevens R, Grigson G. 2016. FLUXNET2015 AU-Lox Loxton. FLUXNET; South Australian Research and Development Institute (SARDI) [DOI]
- Beringer J, Hutley L. 2016. FLUXNET2015 AU-RDF Red Dirt Melon Farm, Northern Territory. FLUXNET; Monash University; Charles Darwin University. [DOI]
- Beringer J, 2016. FLUXNET2015 AU-Rig Riggs Creek. FLUXNET; Monash University. [DOI]
- Liddell M. 2016. FLUXNET2015 AU-Rob Robson Creek, Queensland, Australia. FLUXNET; James Cook University. [DOI]
- Beringer J, Hutley L. 2016. FLUXNET2015 AU-Stp Sturt Plains. FLUXNET; University of Western Australia; Charles Darwin University; Monash University. [DOI]
- Cleverly J, Eamus D. 2016. FLUXNET2015 AU-TTE Ti Tree East. FLUXNET; University of Technology Sydney. [DOI]
- Woodgate W, Van Gorsel E, Leuning R. 2016. FLUXNET2015 AU-Tum Tumbarumba. FLUXNET; CSIRO. [DOI]
- Beringer J, 2016. FLUXNET2015 AU-Whr Whroo. FLUXNET; Monash University. [DOI]
- Arndt S, Hinko-Najera N, Griebel A. 2016. FLUXNET2015 AU-Wom Wombat. FLUXNET; University of Melbourne, School of Ecosystem and Forest Sciences. [DOI]
- Beringer J, Walker J. 2016. FLUXNET2015 AU-Ync Jaxa. FLUXNET; University of Western Australia; Monash University. [DOI]
- Neirynck J, 2016. FLUXNET2015 BE-Bra Brasschaat. FLUXNET; University of Antwerp. [DOI]
- De Ligne A, 2016. FLUXNET2015 BE-Lon Lonzee. FLUXNET; University of Liege - Gembloux Agro-Bio Tech. [DOI]
- Saleska S. 2016. FLUXNET2015 BR-Sa1 Santarem-Km67-Primary Forest. FLUXNET; University of Arizona. [DOI]
- Goulden M. 2016. FLUXNET2015 BR-Sa3 Santarem-Km83-Logged Forest. FLUXNET; University of California - Irvine. [DOI]
- McCaughey H. 2016. FLUXNET2015 CA-Gro Ontario - Groundhog River, Boreal Mixedwood Forest. FLUXNET; Queen’s University. [DOI]
- Amiro B. 2016. FLUXNET2015 CA-Man Manitoba - Northern Old Black Spruce (former BOREAS Northern Study Area) FLUXNET; University of Manitoba. [DOI]
- Goulden M. 2016. FLUXNET2015 CA-NS1 UCI-1850 burn site. FLUXNET; University of California - Irvine. [DOI]
- Goulden M. 2016. FLUXNET2015 CA-NS2 UCI-1930 burn site. FLUXNET; University of California - Irvine. [DOI]
- Goulden M. 2016. FLUXNET2015 CA-NS3 UCI-1964 burn site. FLUXNET; University of California - Irvine. [DOI]
- Goulden M. 2016. FLUXNET2015 CA-NS4 UCI-1964 burn site wet. FLUXNET; University of California - Irvine. [DOI]
- Goulden M. 2016. FLUXNET2015 CA-NS5 UCI-1981 burn site. FLUXNET; University of California - Irvine. [DOI]
- Goulden M. 2016. FLUXNET2015 CA-NS6 UCI-1989 burn site. FLUXNET; University of California - Irvine. [DOI]
- Goulden M. 2016. FLUXNET2015 CA-NS7 UCI-1998 burn site. FLUXNET; University of California - Irvine. [DOI]
- Black T. 2016. FLUXNET2015 CA-Oas Saskatchewan - Western Boreal, Mature Aspen. FLUXNET; The University of British Columbia. [DOI]
- Black T. 2016. FLUXNET2015 CA-Obs Saskatchewan - Western Boreal, Mature Black Spruce. FLUXNET; The University of British Columbia. [DOI]
- Margolis H. 2016. FLUXNET2015 CA-Qfo Quebec - Eastern Boreal, Mature Black Spruce. FLUXNET; Université Laval. [DOI]
- Amiro B. 2016. FLUXNET2015 CA-SF1 Saskatchewan - Western Boreal, forest burned in 1977. FLUXNET; University of Manitoba. [DOI]
- Amiro B. 2016. FLUXNET2015 CA-SF2 Saskatchewan - Western Boreal, forest burned in 1989. FLUXNET; University of Manitoba. [DOI]
- Amiro B. 2016. FLUXNET2015 CA-SF3 Saskatchewan - Western Boreal, forest burned in 1998. FLUXNET; University of Manitoba; Canadian Forest Service. [DOI]
- Arain M. 2016. FLUXNET2015 CA-TP1 Ontario - Turkey Point 2002 Plantation White Pine. FLUXNET; McMaster University. [DOI]
- Arain M. 2016. FLUXNET2015 CA-TP2 Ontario - Turkey Point 1989 Plantation White Pine. FLUXNET; McMaster University. [DOI]
- Arain M. 2016. FLUXNET2015 CA-TP3 Ontario - Turkey Point 1974 Plantation White Pine. FLUXNET; McMaster University. [DOI]
- Arain M. 2016. FLUXNET2015 CA-TP4 Ontario - Turkey Point 1939 Plantation White Pine. FLUXNET; McMaster University. [DOI]
- Arain M. 2016. FLUXNET2015 CA-TPD Ontario - Turkey Point Mature Deciduous. FLUXNET; McMaster University. [DOI]
- Nouvellon Y. 2016. FLUXNET2015 CG-Tch Tchizalamou. FLUXNET; Centre de coopération internationale en recherche agronomique pour le développement. [DOI]
- Hörtnagl L, 2016. FLUXNET2015 CH-Cha Chamau. FLUXNET; ETH Zurich. [DOI]
- Hörtnagl L, 2016. FLUXNET2015 CH-Dav Davos. FLUXNET; ETH Zurich. [DOI]
- Hörtnagl L, 2016. FLUXNET2015 CH-Fru Früebüel. FLUXNET; ETH Zurich. [DOI]
- Hörtnagl L, 2016. FLUXNET2015 CH-Lae Laegern. FLUXNET; ETH Zurich. [DOI]
- Ammann C. 2016. FLUXNET2015 CH-Oe1 Oensingen grassland. FLUXNET; Agroscope Zuerich. [DOI]
- Hörtnagl L, 2016. FLUXNET2015 CH-Oe2 Oensingen crop. FLUXNET; ETH Zurich. [DOI]
- Zhang J, Han S. 2016. FLUXNET2015 CN-Cha Changbaishan. FLUXNET; IAE Chinese Academy of Sciences. [DOI]
- Dong G. 2016. FLUXNET2015 CN-Cng Changling. FLUXNET; Shanxi University. [DOI]
- Shi P, Zhang X, He Y. 2016. FLUXNET2015 CN-Dan Dangxiong. FLUXNET; IGSNRR Chinese Academy of Sciences. [DOI]
- Zhou G, Yan J. 2016. FLUXNET2015 CN-Din Dinghushan. FLUXNET; SCIB Chinese Academy of Sciences. [DOI]
- Chen S. 2016. FLUXNET2015 CN-Du2 Duolun_grassland (D01) FLUXNET; Institute of Botany, Chinese Academy of Sciences. [DOI]
- Shao C. 2016. FLUXNET2015 CN-Du3 Duolun Degraded Meadow. FLUXNET. [DOI]
- Li Y. 2016. FLUXNET2015 CN-Ha2 Haibei Shrubland. FLUXNET; NWIPB Chinese Academy of Sciences. [DOI]
- Tang Y, Kato T, Du M. 2016. FLUXNET2015 CN-HaM Haibei Alpine Tibet site. FLUXNET; National Institute for Environmental Studies. [DOI]
- Wang H, Fu X. 2016. FLUXNET2015 CN-Qia Qianyanzhou. FLUXNET; IGSNRR Chinese Academy of Sciences. [DOI]
- Shao C. 2016. FLUXNET2015 CN-Sw2 Siziwang Grazed (SZWG) FLUXNET. [DOI]
- Sigut L, 2016. FLUXNET2015 CZ-BK1 Bily Kriz forest. FLUXNET; Global Change Research Institute CAS. [DOI]
- Sigut L, 2016. FLUXNET2015 CZ-BK2 Bily Kriz grassland. FLUXNET; Global Change Research Institute CAS. [DOI]
- Dušek J, Janouš D, Pavelka M. 2016. FLUXNET2015 CZ-wet Trebon (CZECHWET) FLUXNET; Global Change Research Institute CAS. [DOI]
- Bernhofer C, 2016. FLUXNET2015 DE-Akm Anklam. FLUXNET; TU Dresden. [DOI]
- Brümmer C, Lucas-Moffat A, Herbst M, Kolle O. 2016. FLUXNET2015 DE-Geb Gebesee. FLUXNET; Thünen Institute of Climate-Smart Agriculture, Braunschweig. [DOI]
- Bernhofer C, 2016. FLUXNET2015 DE-Gri Grillenburg. FLUXNET; TU Dresden. [DOI]
- Knohl A, 2016. FLUXNET2015 DE-Hai Hainich. FLUXNET; University of Goettingen, Bioclimatology. [DOI]
- Bernhofer C, 2016. FLUXNET2015 DE-Kli Klingenberg. FLUXNET; TU Dresden. [DOI]
- Lindauer M, 2016. FLUXNET2015 DE-Lkb Lackenberg. FLUXNET; Karlsruhe Institute of Technology, IMK-IFU. [DOI]
- Knohl A, 2016. FLUXNET2015 DE-Lnf Leinefelde. FLUXNET; University of Goettingen, Bioclimatology. [DOI]
- Bernhofer C, 2016. FLUXNET2015 DE-Obe Oberbärenburg. FLUXNET; TU Dresden. [DOI]
- Schmidt M, Graf A. 2016. FLUXNET2015 DE-RuR Rollesbroich. FLUXNET; IBG-3 Agrosphäre, Research Centre Jülich GmbH. [DOI]
- Schmidt M, Graf A. 2016. FLUXNET2015 DE-RuS Selhausen Juelich. FLUXNET; IBG-3 Agrosphäre, Research Centre Jülich GmbH. [DOI]
- Schneider K, Schmidt M. 2016. FLUXNET2015 DE-Seh Selhausen. FLUXNET; University of Cologne. [DOI]
- Klatt J, Schmid H, Mauder M, Steinbrecher R. 2016. FLUXNET2015 DE-SfN Schechenfilz Nord. FLUXNET; Karlsruhe Institute of Technology, IMK-IFU. [DOI]
- Bernhofer C, 2016. FLUXNET2015 DE-Spw Spreewald. FLUXNET; TU Dresden. [DOI]
- Bernhofer C, 2016. FLUXNET2015 DE-Tha Tharandt. FLUXNET; TU Dresden. [DOI]
- Sachs T, Wille C, Larmanou E, Franz D. 2016. FLUXNET2015 DE-Zrk Zarnekow. FLUXNET; GFZ German Research Centre for Geosciences. [DOI]
- Pilegaard K, Ibrom A. 2016. FLUXNET2015 DK-Eng Enghave. FLUXNET; Technical University of Denmark (DTU) [DOI]
- Olesen J. 2016. FLUXNET2015 DK-Fou Foulum. FLUXNET; Danish Institute of Agricultural Sciences. [DOI]
- Ibrom A, Pilegaard K. 2016. FLUXNET2015 DK-Sor Soroe. FLUXNET; Technical University of Denmark (DTU) [DOI]
- Poveda F, 2016. FLUXNET2015 ES-Amo Amoladeras. FLUXNET; Estación Experimental de Zona Áridas (EEZA, CSIC) [DOI]
- Reverter B, Perez-Cañete E, Kowalski A. 2016. FLUXNET2015 ES-LgS Laguna Seca. FLUXNET; Universidad de Granada. [DOI]
- Cañete E, 2016. FLUXNET2015 ES-LJu Llano de los Juanes. FLUXNET; University of Granada. [DOI]
- Reverter B, Perez-Cañete E, Kowalski A. 2016. FLUXNET2015 ES-Ln2 Lanjaron-Salvage logging. FLUXNET; Universidad de Granada. [DOI]
- Mammarella I, 2016. FLUXNET2015 FI-Hyy Hyytiala. FLUXNET; University of Helsinki. [DOI]
- Lohila A, 2016. FLUXNET2015 FI-Jok Jokioinen. FLUXNET; Finnish Meteorological Institute. [DOI]
- Lohila A, 2016. FLUXNET2015 FI-Let Lettosuo. FLUXNET; Finnish Meteorological Institute. [DOI]
- Aurela M, 2016. FLUXNET2015 FI-Lom Lompolojankka. FLUXNET; Finnish Meteorological Institute. [DOI]
- Aurela M, 2016. FLUXNET2015 FI-Sod Sodankyla. FLUXNET; Finnish Meteorological Institute. [DOI]
- Berveiller D, 2016. FLUXNET2015 FR-Fon Fontainebleau-Barbeau. FLUXNET; CNRS. [DOI]
- Buysse P, 2016. FLUXNET2015 FR-Gri Grignon. FLUXNET; French National Institute for Agricultural Research. [DOI]
- Berbigier P, Loustau D. 2016. FLUXNET2015 FR-LBr Le Bray. FLUXNET; INRA - UMR ISPA. [DOI]
- Ourcival J. 2016. FLUXNET2015 FR-Pue Puechabon. FLUXNET; CNRS. [DOI]
- Bonal D, Burban B. 2016. FLUXNET2015 GF-Guy Guyaflux (French Guiana) FLUXNET; INRA. [DOI]
- Hansen B. 2016. FLUXNET2015 GL-NuF Nuuk Fen. FLUXNET; University of Copenhagen; University of Aarhus; Asiaq - Greenland Survey. [DOI]
- Lund M, Jackowicz-Korczyński M, Abermann J. 2016. FLUXNET2015 GL-ZaF Zackenberg Fen. FLUXNET; Aarhus University. [DOI]
- Lund M, Jackowicz-Korczyński M, Abermann J. 2016. FLUXNET2015 GL-ZaH Zackenberg Heath. FLUXNET; Aarhus University. [DOI]
- Magliulo V, 2016. FLUXNET2015 IT-BCi Borgo Cioffi. FLUXNET; CNR ISAFOM. [DOI]
- Sabbatini S, Arriga N, Papale D. 2016. FLUXNET2015 IT-CA1 Castel d’Asso1. FLUXNET; University of Tuscia - Vietrbo. [DOI]
- Sabbatini S, Arriga N, Gioli B, Papale D. 2016. FLUXNET2015 IT-CA2 Castel d’Asso2. FLUXNET; CNR IBIMET; University of Tuscia - Vietrbo. [DOI]
- Sabbatini S, Arriga N, Matteucci G, Papale D. 2016. FLUXNET2015 IT-CA3 Castel d’Asso 3. FLUXNET; University of Tuscia - Vietrbo; CNR IBAF. [DOI]
- Matteucci G. 2016. FLUXNET2015 IT-Col Collelongo. FLUXNET; Istituto di Ecologia e Idrologia Forestale CNR. [DOI]
- Fares S, Savi F, Conte A. 2016. FLUXNET2015 IT-Cp2 Castelporziano 2. FLUXNET; Council for Agricultural Research and Economics. [DOI]
- Valentini R, 2016. FLUXNET2015 IT-Cpz Castelporziano. FLUXNET; University of Tuscia - Vietrbo. [DOI]
- Gruening C, Goded I, Cescatti A, Pokorska O. 2016. FLUXNET2015 IT-Isp Ispra ABC-IS. FLUXNET; European Commission - Joint Research Centre. [DOI]
- Cescatti A, Marcolla B, Zorer R, Gianelle D. 2016. FLUXNET2015 IT-La2 Lavarone2. FLUXNET; Centro di Ecologia Alpina. [DOI]
- Gianelle D, Zampedri R, Cavagna M, Sottocornola M. 2016. FLUXNET2015 IT-Lav Lavarone. FLUXNET; Edmund Mach Foundation. [DOI]
- Gianelle D, Cavagna M, Zampedri R, Marcolla B. 2016. FLUXNET2015 IT-MBo Monte Bondone. FLUXNET; Edmund Mach Foundation. [DOI]
- Spano D, 2016. FLUXNET2015 IT-Noe Arca di Noe - Le Prigionette. FLUXNET; University of Sassari; CNR-Ibimet Sassari. [DOI]
- Manca G, Goded I. 2016. FLUXNET2015 IT-PT1 Parco Ticino forest. FLUXNET; European Commission - DG Joint Research Centre. [DOI]
- Montagnani L, Minerbi S. 2016. FLUXNET2015 IT-Ren Renon. FLUXNET; Autonomous Province of Bolzano, Forest Services. [DOI]
- Valentini R, 2016. FLUXNET2015 IT-Ro1 Roccarespampani 1. FLUXNET; University of Tuscia - Vietrbo. [DOI]
- Papale D, 2016. FLUXNET2015 IT-Ro2 Roccarespampani 2. FLUXNET; University of Tuscia - Vietrbo. [DOI]
- Gruening C, Goded I, Cescatti A, Pokorska O. 2016. FLUXNET2015 IT-SR2 San Rossore 2. FLUXNET; European Commission - Joint Research Centre. [DOI]
- Gruening C, 2016. FLUXNET2015 IT-SRo San Rossore. FLUXNET; European Commission - Joint Research Centre. [DOI]
- Cremonese E, Galvagno M, Di Cella U, Migliavacca M. 2016. FLUXNET2015 IT-Tor Torgnon. FLUXNET; Environmental Protection Agency of Aosta Valley. [DOI]
- Kotani A. 2016. FLUXNET2015 JP-MBF Moshiri Birch Forest Site. FLUXNET; Nagoya University. [DOI]
- Kotani A. 2016. FLUXNET2015 JP-SMF Seto Mixed Forest Site. FLUXNET; Nagoya University. [DOI]
- Kosugi Y, Takanashi S. 2016. FLUXNET2015 MY-PSO Pasoh Forest Reserve (PSO) FLUXNET; FRIM(Forest Research Institute of Malaysia); Kyoto University. [DOI]
- Dolman H, 2016. FLUXNET2015 NL-Hor Horstermeer. FLUXNET; Vrije Universiteit Amsterdam. [DOI]
- Moors E, Elbers J. 2016. FLUXNET2015 NL-Loo Loobos. FLUXNET; ALTERRA/Wageningen Environmental Research. [DOI]
- Wolf S, Eugster W, Buchmann N. 2016. FLUXNET2015 PA-SPn Sardinilla Plantation. FLUXNET; ETH Zurich. [DOI]
- Wolf S, Eugster W, Buchmann N. 2016. FLUXNET2015 PA-SPs Sardinilla-Pasture. FLUXNET; ETH Zurich. [DOI]
- Merbold L, Rebmann C, Corradi C. 2016. FLUXNET2015 RU-Che Cherski. FLUXNET; Max-Planck Institute for Biogeochemistry. [DOI]
- Dolman H, 2016. FLUXNET2015 RU-Cok Chokurdakh. FLUXNET; Vrije Universiteit Amsterdam. [DOI]
- Varlagin A, Kurbatova J, Vygodskaya N. 2016. FLUXNET2015 RU-Fyo Fyodorovskoye. FLUXNET; A.N. Severtsov Institute of Ecology and Evolution. [DOI]
- Belelli L, Papale D, Valentini R. 2016. FLUXNET2015 RU-Ha1 Hakasia steppe. FLUXNET; University of Tuscia - Vietrbo. [DOI]
- Ardö J, El Tahir B, ElKhidir H. 2016. FLUXNET2015 SD-Dem Demokeya. FLUXNET; LUND UNIVERSITY. [DOI]
- Christensen T. 2016. FLUXNET2015 SJ-Adv Adventdalen. FLUXNET; NATEKO; Lund University. [DOI]
- Tagesson T, Ardö J, Fensholt R. 2016. FLUXNET2015 SN-Dhr Dahra. FLUXNET; Lund University. [DOI]
- Billesbach D, Bradford J, Torn M. 2016. FLUXNET2015 US-AR1 ARM USDA UNL OSU Woodward Switchgrass 1. FLUXNET; Lawerence Berkeley National Lab; U.S. Department of Agriculture; University of Nebraska. [DOI]
- Billesbach D, Bradford J, Torn M. 2016. FLUXNET2015 US-AR2 ARM USDA UNL OSU Woodward Switchgrass 2. FLUXNET; Lawrence Berkeley National Lab; U.S. Department of Agriculture; University of Nebraska. [DOI]
- Torn M. 2016. FLUXNET2015 US-ARb ARM Southern Great Plains burn site- Lamont. FLUXNET; Lawrence Berkeley National Laboratory. [DOI]
- Torn M. 2016. FLUXNET2015 US-ARc ARM Southern Great Plains control site- Lamont. FLUXNET; Lawrence Berkeley National Laboratory. [DOI]
- Biraud S, Fischer M, Chan S, Torn M. 2016. FLUXNET2015 US-ARM ARM Southern Great Plains site- Lamont. FLUXNET; Lawrence Berkeley National Laboratory. [DOI]
- Zona D, Oechel W. 2016. FLUXNET2015 US-Atq Atqasuk. FLUXNET; San Diego State University. [DOI]
- Goldstein A. 2016. FLUXNET2015 US-Blo Blodgett Forest. FLUXNET; University of California, Berkeley. [DOI]
- Bowling D. 2016. FLUXNET2015 US-Cop Corral Pocket. FLUXNET; University of Utah. [DOI]
- Chen J, Chu H. 2016. FLUXNET2015 US-CRT Curtice Walter-Berger cropland. FLUXNET; University of Toledo/Michigan State University. [DOI]
- Massman B. 2016. FLUXNET2015 US-GBT GLEES Brooklyn Tower. FLUXNET; USDA Forest Service. [DOI]
- Massman B. 2016. FLUXNET2015 US-GLE GLEES. FLUXNET; USDA Forest Service. [DOI]
- Meyers T. 2016. FLUXNET2015 US-Goo Goodwin Creek. FLUXNET; NOAA/ARL. [DOI]
- Munger J. 2016. FLUXNET2015 US-Ha1 Harvard Forest EMS Tower (HFR1) FLUXNET; Harvard University. [DOI]
- Matamala R. 2016. FLUXNET2015 US-IB2 Fermi National Accelerator Laboratory- Batavia (Prairie site) FLUXNET; Argonne National Laboratory. [DOI]
- Zona D, Oechel W. 2016. FLUXNET2015 US-Ivo Ivotuk. FLUXNET; San Diego State University. [DOI]
- Drake B, Hinkle R. 2016. FLUXNET2015 US-KS1 Kennedy Space Center (slash pine) FLUXNET; Smithsonian Environmental Research Center; University of Central Florida. [DOI]
- Drake B, Hinkle R. 2016. FLUXNET2015 US-KS2 Kennedy Space Center (scrub oak) FLUXNET; Smithsonian Environmental Research Center; University of Central Florida. [DOI]
- Fares S. 2016. FLUXNET2015 US-Lin Lindcove Orange Orchard. FLUXNET; Entecra. [DOI]
- Desai A. 2016. FLUXNET2015 US-Los Lost Creek. FLUXNET; University of Wisconsin. [DOI]
- Meyers T. 2016. FLUXNET2015 US-LWW Little Washita Watershed. FLUXNET; NOAA/ARL. [DOI]
- Law B. 2016. FLUXNET2015 US-Me1 Metolius - Eyerly burn. FLUXNET; Oregon State University. [DOI]
- Law B. 2016. FLUXNET2015 US-Me2 Metolius mature ponderosa pine. FLUXNET; Oregon State University. [DOI]
- Law B. 2016. FLUXNET2015 US-Me3 Metolius-second young aged pine. FLUXNET; Oregon State University. [DOI]
- Law B. 2016. FLUXNET2015 US-Me4 Metolius-old aged ponderosa pine. FLUXNET; Oregon State University. [DOI]
- Law B. 2016. FLUXNET2015 US-Me5 Metolius-first young aged pine. FLUXNET; Oregon State University. [DOI]
- Law B. 2016. FLUXNET2015 US-Me6 Metolius Young Pine Burn. FLUXNET; Oregon State University. [DOI]
- Novick K, Phillips R. 2016. FLUXNET2015 US-MMS Morgan Monroe State Forest. FLUXNET; Indiana University. [DOI]
- Sturtevant C, 2016. FLUXNET2015 US-Myb Mayberry Wetland. FLUXNET; University of California, Berkeley. [DOI]
- Suyker A. 2016. FLUXNET2015 US-Ne1 Mead - irrigated continuous maize site. FLUXNET; University of Nebraska - Lincoln. [DOI]
- Suyker A. 2016. FLUXNET2015 US-Ne2 Mead - irrigated maize-soybean rotation site. FLUXNET; University of Nebraska - Lincoln. [DOI]
- Suyker A. 2016. FLUXNET2015 US-Ne3 Mead - rainfed maize-soybean rotation site. FLUXNET; University of Nebraska - Lincoln. [DOI]
- Blanken P, 2016. FLUXNET2015 US-NR1 Niwot Ridge Forest (LTER NWT1) FLUXNET; University of Colorado. [DOI]
- Chen J, Chu H, Noormets A. 2016. FLUXNET2015 US-Oho Oak Openings. FLUXNET; University of Toledo/Michigan State University. [DOI]
- Bohrer G. 2016. FLUXNET2015 US-ORv Olentangy River Wetland Research Park. FLUXNET; The Ohio State University. [DOI]
- Desai A. 2016. FLUXNET2015 US-PFa Park Falls/WLEF. FLUXNET; University of Wisconsin. [DOI]
- Kobayashi H, Suzuki R. 2016. FLUXNET2015 US-Prr Poker Flat Research Range Black Spruce Forest. FLUXNET; Japan Agency for Marine-Earth Science and Technology. [DOI]
- Kurc S. 2016. FLUXNET2015 US-SRC Santa Rita Creosote. FLUXNET; University of Arizona. [DOI]
- Scott R. 2016. FLUXNET2015 US-SRG Santa Rita Grassland. FLUXNET; United States Department of Agriculture. [DOI]
- Scott R. 2016. FLUXNET2015 US-SRM Santa Rita Mesquite. FLUXNET; United States Department of Agriculture. [DOI]
- Ewers B, Pendall E. 2016. FLUXNET2015 US-Sta Saratoga. FLUXNET; University of Wyoming. [DOI]
- Desai A. 2016. FLUXNET2015 US-Syv Sylvania Wilderness Area. FLUXNET; University of Wisconsin. [DOI]
- Baldocchi D, Ma S. 2016. FLUXNET2015 US-Ton Tonzi Ranch. FLUXNET; University of California, Berkeley. [DOI]
- Szutu D, Baldocchi D, Eichelmann E, Knox S. 2016. FLUXNET2015 US-Tw1 Twitchell Wetland West Pond. FLUXNET; University of California, Berkeley. [DOI]
- Baldocchi D. 2016. FLUXNET2015 US-Tw2 Twitchell Corn. FLUXNET; University of California, Berkeley. [DOI]
- Szutu D, Baldocchi D. 2016. FLUXNET2015 US-Tw3 Twitchell Alfalfa. FLUXNET; University of California, Berkeley. [DOI]
- Sanchez C, 2016. FLUXNET2015 US-Tw4 Twitchell East End Wetland. FLUXNET; University of California, Berkeley. [DOI]
- Baldocchi D. 2016. FLUXNET2015 US-Twt Twitchell Island. FLUXNET; University of California, Berkeley. [DOI]
- Gough C, Bohrer G, Curtis P. 2016. FLUXNET2015 US-UMB Univ. of Mich. Biological Station. FLUXNET; Ohio State University; Virginia Commonwealth University. [DOI]
- Gough C, Bohrer G, Curtis P. 2016. FLUXNET2015 US-UMd UMBS Disturbance. FLUXNET; Ohio State University; Virginia Commonwealth University. [DOI]
- Baldocchi D, Ma S, Xu L. 2016. FLUXNET2015 US-Var Vaira Ranch- Ione. FLUXNET; University of California, Berkeley. [DOI]
- Desai A. 2016. FLUXNET2015 US-WCr Willow Creek. FLUXNET; University of Wisconsin. [DOI]
- Scott R. 2016. FLUXNET2015 US-Whs Walnut Gulch Lucky Hills Shrub. FLUXNET; United States Department of Agriculture. [DOI]
- Chen J. 2016. FLUXNET2015 US-Wi0 Young red pine (YRP) FLUXNET; Michigan State University. [DOI]
- Chen J. 2016. FLUXNET2015 US-Wi1 Intermediate hardwood (IHW) FLUXNET; Michigan State University. [DOI]
- Chen J. 2016. FLUXNET2015 US-Wi2 Intermediate red pine (IRP) FLUXNET; Michigan State University. [DOI]
- Chen J. 2016. FLUXNET2015 US-Wi3 Mature hardwood (MHW) FLUXNET; Michigan State University. [DOI]
- Chen J. 2016. FLUXNET2015 US-Wi4 Mature red pine (MRP) FLUXNET; Michigan State University. [DOI]
- Chen J. 2016. FLUXNET2015 US-Wi5 Mixed young jack pine (MYJP) FLUXNET; Michigan State University. [DOI]
- Chen J. 2016. FLUXNET2015 US-Wi6 Pine barrens #1 (PB1) FLUXNET; Michigan State University. [DOI]
- Chen J. 2016. FLUXNET2015 US-Wi7 Red pine clearcut (RPCC) FLUXNET; Michigan State University. [DOI]
- Chen J. 2016. FLUXNET2015 US-Wi8 Young hardwood clearcut (YHW) FLUXNET; Michigan State University. [DOI]
- Chen J. 2016. FLUXNET2015 US-Wi9 Young Jack pine (YJP) FLUXNET; Michigan State University. [DOI]
- Scott R. 2016. FLUXNET2015 US-Wkg Walnut Gulch Kendall Grasslands. FLUXNET; United States Department of Agriculture. [DOI]
- Chen J, Chu H. 2016. FLUXNET2015 US-WPT Winous Point North Marsh. FLUXNET; University of Toledo/Michigan State University. [DOI]
- Kutsch W, Merbold L, Kolle O. 2016. FLUXNET2015 ZM-Mon Mongu. FLUXNET; Max-Planck Institute for Biogeochemistry. [DOI]
Supplementary Materials
Data Availability Statement
The ONEFlux collection of codes used to create data intercomparable with FLUXNET2015 has been packaged to be executed as a complete pipeline and is available in both source-code and executable forms under a 3-clause BSD license on GitHub: https://github.com/AmeriFlux/ONEFlux. The complete environment to run this pipeline requires a GCC compatible C compiler (or capability to run pre-compiled Windows, Linux, and/or Mac executables), a MATLAB Runtime Environment, and a Python interpreter with a few numeric and scientific packages installed. All of these can be obtained at no cost.