Abstract
The strains of the Komagataeibacter genus have been shown to be the most efficient bacterial nanocellulose producers. Although exploited for many decades, the studies of these species focused mainly on the optimisation of cellulose synthesis process through modification of culturing conditions in the industrially relevant settings. Molecular physiology of Komagataeibacter was poorly understood and only a few studies explored genetic engineering as a strategy for strain improvement. Only since recently the systemic information of the Komagataeibacter species has been accumulating in the form of omics datasets representing sequenced genomes, transcriptomes, proteomes and metabolomes. Genetic analyses of the mutants generated in the untargeted strain modification studies have drawn attention to other important proteins, beyond those of the core catalytic machinery of the cellulose synthase complex. Recently, modern molecular and synthetic biology tools have been developed which showed the potential for improving targeted strain engineering. Taking the advantage of the gathered knowledge should allow for better understanding of the genotype–phenotype relationship which is necessary for robust modelling of metabolism as well as selection and testing of new molecular engineering targets. In this review, we discuss the current progress in the area of Komagataeibacter systems biology and its impact on the research aimed at scaled-up cellulose synthesis as well as BNC functionalisation.
|
Key points • The accumulated omics datasets advanced the systemic understanding of Komagataeibacter physiology at the molecular level. • Untargeted and targeted strain modification approaches have been applied to improve nanocellulose yield and properties. • The development of modern molecular and synthetic biology tools presents a potential for enhancing targeted strain engineering. • The accumulating omic information should improve modelling of Komagataeibacter’s metabolism as well as selection and testing of new molecular engineering targets. |
Keywords: Komagataeibacter, Bacterial nanocellulose, Systems biology, Strain engineering, Molecular physiology, Composites
Introduction
Bacterial nanocellulose (bionanocellulose, BNC), composed of β-1-4-linked glucose units, is a natural biopolymer produced by a wide variety of bacteria. Dimensions of BNC fibres give this polymer properties of a nanomaterial. Due to its unique features, BNC has found applications in various areas of biotechnology, medicine and industry (reviewed in Jang et al. 2017; García and Prieto 2019; Gorgieva and Trček 2019; Wang et al. 2019). It also constitutes a promising material in food applications, e.g. as rheology modifier, fat replacer, or in the immobilisation of probiotics and enzymes (reviewed in Azeredo et al. 2019). Despite many valuable features and a wide array of applications, products made of BNC are not yet present in our everyday life. This is because BNC production performance is still relatively low since its large-scale industrial fermentation process is more complex and highly capital intensive, especially in comparison with plant cellulose–based processes and pulp and paper industry. Therefore, much of an effort has been devoted to design an economical process for BNC production by optimisation of culture medium composition, cultivation mode and process parameters, bacterial strain, as well as postproduction purification (Krystynowicz et al. 2002; Jozala et al. 2016; Islam et al. 2017). Further improvements in these areas are necessary for increasing BNC production performance and making it more cost-effective.
The most efficient bacterial cellulose producers are acetic acid bacteria (AAB) of the Komagataeibacter genus, which synthesise high amounts of this biopolymer from a wide variety of carbon and nitrogen sources (Islam et al. 2017). K. xylinus is the bacterium in which the molecular mechanism of BNC synthesis was first revealed and studied. The biochemical pathway and the core catalytic machinery responsible for cellulose synthesis have been delineated (Ross et al. 1991; Saxena et al. 1994; Römling 2002; Velasco-Bedrán and López-Isunza 2007; Jacek et al. 2019a). Polymerisation of UDP-glucose into cellulose is catalysed by the bacterial cellulose synthase (BCS) enzyme complex. Two types of cellulose synthase operons are encoded by the Komagataeibacter genomes, i.e. type I and type II (Umeda et al. 1999). The type I cellulose synthase operon (bcsI) comprises four genes, bcsAI, bcsBI, bcsCI and bcsDI. The bcsI operon is flanked by accessory genes (cmcAx, ccpAx and bglAx), which modulate the process of cellulose synthesis (Römling and Galperin 2015; Jedrzejczak-Krzepkowska et al. 2016). The type II operon (bcsII) is probably responsible for the production of acylated cellulose due to the presence of an acyltransferase gene within its structure (Umeda et al. 1999). Expression of the cellulose biosynthesis operon was found to be constitutive, however shown to fluctuate depending on growth phase and environmental conditions (Augimeri and Strap 2015; Hernández-Arriaga et al. 2019; Ryngajłło et al. 2019a). Cellulose synthase enzyme activation occurs at the posttranslational level and is mediated by c-di-GMP (bis-(3′,5′)-cyclic di-guanosine-mono-phosphate), which binds to the PilZ domain of the BcsA subunit and activates it allosterically (Ross et al. 1985; Fujiwara et al. 2013; Römling et al. 2013). The cellular level of c-di-GMP is controlled by diguanylate cyclases (DGCs) and c-di-GMP-specific phosphodiesterases (PDEs) (Tal et al. 1998; Römling 2012).
The development of high-throughput omics techniques created an opportunity to study the Komagataeibacter species as systems and to venture beyond the core catabolic machinery of cellulose synthase. In this review, we discuss the current progress in the systemic understanding of Komagataeibacter physiology at the molecular level. We review the high-throughput studies which provided a global overview of different functional layers of a Komagataeibacter cell. The examples of untargeted and targeted approaches as well as genetic engineering strategies for strain improvement in terms of BNC synthesis intensification and features modification are also further presented.
Systems-level perspective on the physiology of the Komagataeibacter species
Bacteria of the Komagataeibacter genus are considered as the model species for studying cellulose biosynthesis process. However, it cannot be regarded as a model species for molecular analysis since systems biology of Komagataeibacter is a relatively young research field, when compared with other model bacteria species, such as Escherichia coli, Corynebacterium sp., Bacillus sp. or Pseudomonas sp. Over the past decade, the first complete genome sequences became available for this genus and information of other omes started to accumulate. Application of the modern high-throughput omics techniques should enable obtaining of more of the systems-level information for the Komagataeibacter spp. and rapidly advance them to truly ‘model species’ status for studying molecular aspects of BNC synthesis.
The sequenced Komagataeibacter genomes
The first full Komagataeibacter genome representation deposited in the GenBank database was for K. hansenii ATCC 23769 in 2010 (Iyer et al. 2010). Sequencing of the Komagataeibacter genomes accelerated with the appearance of the next-generation sequencing technology, and currently there are 54 genome sequences available (accession date March 2020, NCBI). The majority of these genomes were assembled based on short reads originating from such technologies as Illumina and consists of multiple (usually, several hundreds) of contigs. However, it is difficult to obtain an assembly of a high quality, i.e. low number of contigs, using this approach, since these genomes were shown to contain a large number of insertion sequences (IS), which often contribute to contig breaks during assembling (Coucheron 1993; Ryngajłło et al. 2019b). The low-quality genomes can be used for taxonomical analysis; however, they are of a limited use for the analyses which strongly depend on the integrity of annotations, such as comparative genomics (Ryngajłło et al. 2019b). Fortunately, the Komagataeibacter genome collection is becoming enriched in complete genome sequences such as those generated using PacBio SMRT or a combination of Illumina and Oxford Nanopore MinION sequencing technologies.
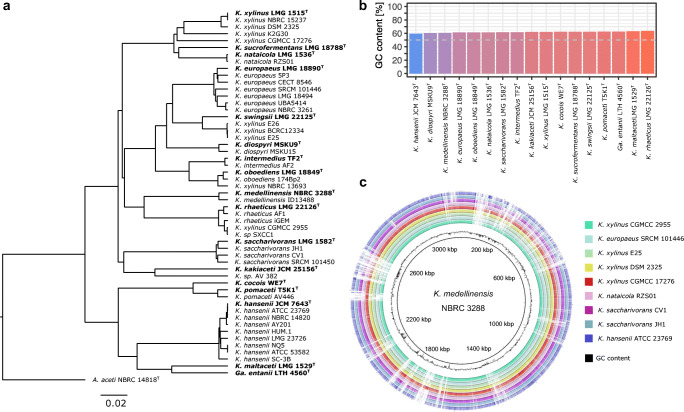
The majority of the type strains of the Komagataeibacter species have been recently sequenced (Škraban et al. 2018) (Table 1). In total, the NCBI genome assembly collection represents 16 Komagataeibacter species: K. xylinus, K. sucrofermentans, K. nataicola, K. europaeus, K. swingsii, K. diospyri, K. intermedius, K. oboediens, K. medellinensis, K. rhaeticus, K. saccharivorans, K. kakiaceti, K. cocois, K. pomaceti, K. hansenii and K. maltaceti (Table 1). In the NCBI database, there is also deposited the genome of a Gluconacetobacter entanii LTH 4560 strain which was not reclassified to the Komagataeibacter genus since it is no longer available at the strain repository. The availability of the type strain genome sequences greatly aids phylogenetic classification of a new cellulose-producing isolate based on the whole-genome alignment measures such as, e.g., Average Nucleotide Identity (ANI) values (Goris et al. 2007; Richter and Rosselló-Móra 2009) or MUM index (MUMi; Deloger et al. 2009; Ryngajłło et al. 2019b). Based on the whole-genome comparison, it can be noticed that some of the sequenced Komagataeibacter strains were misclassified (Fig. 1a). These are mostly the various K. xylinus species, which do not form a clade with the K. xylinus LMG 1515 type strain. This is mainly because, formerly, the diversity of the cellulose producing AAB was underestimated and it was common to call a ‘xylinus’ strain any new isolate. What is characteristic of all the Komagataeibacter genomes is the high % GC content which is the highest for the K. rhaeticus LMG 22126 (63.5 %) and the lowest for the K. hansenii JCM 7643 (59.3%) (Fig. 1b). There are currently 11 genomes with closed chromosomal sequences (Fig. 1c). Of these, eight are complete genome assemblies consisting of one chromosome and several plasmids (Table 2); three genomes have been assembled at the chromosome level only (K. hansenii ATCC 23769, K. rhaeticus iGEM, K. xylinus CGMCC 2955). Based on the complete genome assemblies, the average number of genes encoded by a Komagataeibacter genome is 3571, which include 3327 protein coding genes. Comparative genomics analysis employing 19 complete and draft sequences has predicted the core genome constituting 1578 orthologous gene clusters (present in every Komagataeibacter genome; Ryngajłło et al. 2019b). A similar analysis involving only seven complete Komagataeibacter genomes has found 1719 core genes which make up 21.9% of a non-redundant set of all genes in these genomes (Jang et al. 2019). These results suggest that the Komagataeibacter species have more similar genome contents and share a greater portion of the core genes than other notable groups of bacterial species, such as Mycoplasma or Bifidobacterium (Jang et al. 2019).
Table 1.
Characteristics of Komagataeibacter type strains and their genome sequences
| Type strain | RefSeq assembly accession | Total genome size (Mb) | GC (%) | Assembly level | Isolation source | Cellulose synthesis? | Number of sequenced strains | References |
|---|---|---|---|---|---|---|---|---|
|
K. xylinus LMG 1515 |
GCF_003207795.1 | 3.66 | 62.22 | Contig | Mountain ash berry | + | 5 | (Gosselé et al. 1983; Yamada et al. 2012) |
|
K. sucrofermentans LMG 18788 |
GCF_003207865.1 | 3.36 | 62.33 | Contig | Black cherry, Japan | + | 1 | (Toyosaki et al. 1995; Yamada et al. 2012) |
|
K. nataicola LMG 1536 |
GCF_003207795.1 | 3.67 | 61.48 | Contig | Nata de coco, Philippines | + | 2 | (Lisdiyanti et al. 2006; Yamada et al. 2012) |
|
K. europaeus LMG 18890 |
GCF_000285295.1 | 4.23 | 61.26 | Scaffold | A submerged culture vinegar generator, Germany | − | 7 | (Sievers et al. 1992; Yamada et al. 2012) |
|
K. swingsii LMG 22125 |
GCF_003207895.1 | 3.73 | 62.40 | Contig | Apple juice, Italy | + | 1 | (Dellaglio et al. 2005; Yamada et al. 2012) |
|
K. diospyri MSKU9 |
GCF_006538165.1 | 3.76 | 60.43 | Contig | Persimmon, Thailand | + | 2 | (Naloka et al. 2019) |
|
K. intermedius TF2 |
GCF_000964425.1 | 3.88 | 61.60 | Scaffold | Kombucha beverage, Switzerland | + | 2 | (Boesch et al. 1998; Yamada et al. 2012) |
|
K. oboediens LMG 18849 |
GCF_003207815.1 | 3.78 | 61.35 | Contig | Submerged red wine vinegar, Germany | − | 2 | (Sokollek et al. 1998; Yamada et al. 2012) |
|
K. medellinensis NBRC 3288 |
GCF_000182745.2 | 3.51 | 60.58 | Complete genome | Vinegar, Japan | − | 2 | (Castro et al. 2013) |
|
K. rhaeticus LMG 22126 |
GCF_003207855.1 | 3.47 | 63.49 | Contig | Apple juice, Italy | + | 3 | (Dellaglio et al. 2005; Yamada et al. 2012) |
|
K. saccharivorans LMG 1582 |
GCF_003207825.1 | 3.35 | 61.59 | Contig | Beet juice, Germany | − | 4 | (Lisdiyanti et al. 2006; Yamada et al. 2012) |
|
K. kakiaceti JCM 25156 |
GCF_000613305.1 | 3.13 | 62.14 | Contig | Kaki vinegar, Japan | + | 1 | (Iino et al. 2012) |
|
K. cocois WE7 |
GCF_003311635.1 | 3.41 | 62.27 | Scaffold | Contaminated coconut milk, China | + | 1 | (Liu et al. 2018a) |
|
K. pomaceti T5K1 |
GCF_003207955.1 | 3.45 | 62.53 | Contig | Apple cider vinegar, Slovenia | + | 2 | (Škraban et al. 2018) |
|
K. hansenii JCM 7643 |
GCF_000964405.1 | 3.71 | 59.28 | Scaffold | Vinegar, Israel | − | 9 | (Gosselé et al. 1983; Yamada et al. 2012) |
|
K. maltaceti LMG 1529 |
GCF_003206475.1 | 3.64 | 63.18 | Contig | Malt vinegar brewery acetifier | − | 1 | (Slapšak et al. 2013) |
|
Ga. entanii LTH 4560 |
GCF_003206495.1 | 3.60 | 62.57 | Contig | High-acid spirit industrial vinegar, Germany | − | 1 | (Schüller et al. 2000; Yamada et al. 2012) |
+, cellulose synthesis
Fig. 1.
Sequence similarity of the Komagataeibacter genomes. a Phylogenetic tree based on ANI analysis performed using PYANI (0.2.9) python program employing BLAST+ program (Camacho et al. 2009; Pritchard et al. 2016). The UPGM tree based on ANI − 1 values was calculated using phangorn R package (Schliep 2011). The Acetobacter aceti 14818 was used as an outgroup. The tree was drawn in FigTree program (v.1.4.4). The scale bar represents the sequence divergence. b GC content of the type Komagataeibacter genomes. c Alignment of the closed chromosomal sequences of the Komagataeibacter genomes using BRIG (Alikhan et al. 2011)
Table 2.
General properties of the complete Komagataeibacter genomes deposited at the NCBI database
| Strain | RefSeq assembly accession | Number of chromosome sequences | Number of plasmid sequences | Chromosome size (Mb) | Plasmid sizes (kbp) | Protein coding genes | Pseudogenes | rRNA | tRNA | Other RNA |
|---|---|---|---|---|---|---|---|---|---|---|
|
K. xylinus E25 |
GCF_000550765.1 | 1 | 5 | 3.45 | 336.14; 87.18; 26.30; 5.53; 2.22 | 3408 | 224 | 15 | 57 | 4 |
|
K. xylinus CGMCC 17276 |
GCF_009834365.1 | 1 | 3 | 3.53 | 233.86; 190.75; 31.01 | 3599 | 170 | 15 | 57 | 4 |
|
K. xylinus DSM 2325 |
GCF_004006375.1 | 1 | 2 | 3.35 | 368.64; 5.31 | 3330 | 182 | 13 | 56 | 4 |
|
K. nataicola RZS01 |
GCF_002009295.1 | 1 | 6 | 3.49 | 102.28; 39.91; 38.68; 37.91; 30.55; 25.77 | 3351 | 168 | 15 | 59 | 4 |
|
K. europaeus SRCM 101446 |
GCF_002173515.1 | 1 | 3 | 3.45 | 309.05; 37.20; 2.90 | 3294 | 138 | 15 | 58 | 4 |
|
K. medellinensis NBRC 3288 |
GCF_000182745.2 | 1 | 7 | 3.14 | 255.87; 76.07; 28.57; 4.78; 4.62; 4.26; 2.22 | 3108 | 195 | 15 | 57 | 4 |
|
K. saccharivorans CV1 |
GCF_003546645.1 | 1 | 6 | 3.19 | 217.54; 159.31; 127.84; 42.58; 26.65; 1.87 | 3277 | 144 | 15 | 57 | 4 |
|
K. saccharivorans JH1 |
GCF_004348195.1 | 1 | 14 | 3.10 | 221.73; 183.53; 92.69; 59.56; 41.06; 5.32; 4.90; 3.05; 2.81; 2.29; 2.00; 1.46; 1.09; 0.53 | 3251 | 123 | 15 | 57 | 4 |
Various phylogenetic analyses based on the Komagataeibacter genomes have highlighted a strong separation of the K. hansenii strains from the other species (Škraban et al. 2018; Jang et al. 2019; Ryngajłło et al. 2019b). Several of the predicted functional features of the Komagataeibacter genomes followed this phylogenetic separation. It has been shown that the structure of the first cellulose synthase operon (bcsI) differs between these two groups, whereas in the K. hansenii genomes it has been shown that the bcsA and bcsB genes are fused (Saxena et al. 1994; Matsutani et al. 2015; Ryngajłło et al. 2019b). Moreover, the presence of the acetan biosynthesis gene cluster, which consists of 17 genes, was predicted in the majority of the Komagataeibacter genomes (Ryngajłło et al. 2019b). However, in the K. hansenii spp., the acetan cluster was absent, which suggested that they do not produce this exopolysaccharide (EPS) or produce one of a different composition. Moreover, it has been shown that the number of genes encoding enzymes metabolising c-di-GMP, the allosteric activator of cellulose synthase subunit A (bcsA), is much lower in the K. hansenii spp., which suggested that the c-di-GMP-based regulatory network is much less expanded in these species. What is more, it has been discovered that the presence of the CRISPR-Cas gene cluster is one of the distinctive features of the K. hansenii spp.
Further comparative genomics studies should benefit from the clarified taxonomy of the Komagataeibacter species. Recruitment of the genomic information and revisiting of the large number of physiological reports published in the past may highlight more common as well as distinctive functional features which characterise these species and explain the difference in BNC yield and structure.
Global gene expression studies
While a genome sequence only gives information on the genetic content of a cell, studies involving investigation of gene expression patterns provide an insight and deepen the understanding of gene functions. The employment of RNA sequencing (RNA-seq) is particularly useful for gaining a global overview of dynamic changes at the transcriptional level. The first global transcriptome of a Komagataeibacter sp. has been published and involves the study of effect of ethanol supplementation in the culture of K. xylinus E25 (Kubiak et al. 2014; Ryngajłło et al. 2019a). This study has expanded the understanding of the positive effect of ethanol on BNC synthesis by revealing the gene expression changes triggering this effect, such as up-regulation of UDP-glucose synthesis pathway and down-regulation of glycolysis and acetan biosynthesis. Moreover, metabolic pathways responsible for ethanol metabolism were also delineated. Several of the previously not described genomic regions, such as acetoin metabolism and iron uptake, were also found to be important during the growth in ethanol-containing medium.
Recent availability of complete Komagataeibacter genomes should encourage more of RNA-seq-based transcriptomic studies which should give a better insight into regulatory pathways which are not well understood in the Komagataeibacter species. For example, up to 150 DNA-binding proteins are predicted in the K. hansenii ATCC 23769 genome (based on the predictions from MIST3 and P2RP database). One of the Crp/Fnr transcription factors was previously shown to be important for cellulose synthesis since it positively regulated the expression of the bglAx gene of the bcsI operon (Deng et al. 2013). However, apart from the Crp/Fnr, the gene targets of other transcription factors remain unknown. What is more, we do not know if the bglAx and the bcsI genes are the only components of the Crp/Fnr regulon. A chromatin Immunoprecipitation Sequencing (ChIP-Seq) study investigating Crp/Fnr DNA binding profile could clarify if its impact on global gene expression expands beyond the known gene targets.
Other RNA-seq approaches present a great potential for the analysis of the structure and dynamics of Komagataeibacter transcriptomes. For instance, sequencing of primary transcriptomes which involves enrichment of native transcripts bearing a 5′-triphosphate group, in another AAB Gluconobacter oxydans 621H, has shown to be useful for correction of start codons, prediction of operons as well as for identification of promoter motifs, 5′-untranslated regions (UTRs), ribosome binding sites (RBS) and cis-regulatory elements such as riboswitches (Kranz et al. 2018). Currently, the annotation of the Komagataeibacter genomes is being done mostly through automated prediction which is based on sequence homology and which could miss or incorrectly annotate genes and other functional genome elements.
Proteomic studies
The measurement of cellular mRNA levels gives only partial information about gene expression since it also depends on post-transcriptional regulatory effects which influence mRNA lifetime. Identification and quantification of cellular protein content complements this picture and further improves understanding of the phenotype. A study involving K. xylinus E25 investigated and compared the protein profiles of wild-type (Cel+) and cellulose non-producing cells (cellulose-negative or Cel−, discussed later) using two-dimensional (2D) gel electrophoresis, peptide mass fingerprinting using MALDI-TOF mass spectrometry (MS) as well as by partial peptide sequencing with ion trap electrospray MS (Krystynowicz et al. 2005). This study revealed that the disappearance of cellulose-producing ability was related to the lack of two enzymes necessary for UDP-glucose synthesis: phosphoglucomutase and glucose-1-phosphate uridylyltransferase. Another study collated the protein profiles between a K. xylinus K3 strain isolated from Kombucha and its spontaneous mutant of lower cellulose productivity using 2D gel electrophoresis and MALDI-MS (Nguyen et al. 2010). Three out of five of the up-regulated protein spots in the mutant strain were selected and identified: chaperonin GroEL, short-chain dehydrogenase/reductase (SDR) and deoxythymidine diphosphate (dTDP)-4-dehydrorhamnose 3,5-epimerase. Specifically, the last protein was considered likely to be responsible for the reduced cellulose synthesis by the mutant strain due to its involvement in acetan biosynthesis and competition for the same carbon substrates as cellulose synthesis process.
A metaproteomics approach was undertaken to study the bacteria involved in the production of high-acid spirit vinegar using culture-independent method (Andrés-Barrao et al. 2016). Through 16S RNA and housekeeping genes (dnaK, groEL, rpoB) sequencing, it was found that the bacterial population of the culture was homogenous and belonged to unknown Komagataeibacter species which clustered closed to K. hansenii and Ga. entanii in a phylogenetic tree. The study analysed differentially expressed proteins between low- and high-acidity cultures using 2D-differential in-gel electrophoresis (2D-DIGE) and MS. Out of 2000 proteins detected, 32 most abundant and significantly up-regulated proteins from the high-acid condition were selected and identified. The majority of these proteins were involved in stress response, the TCA cycle and other metabolic processes.
The presented proteomic studies employ gel-based technologies, which are known to be more laborious and of limited scope and resolution. Application of more high-throughput approach by, e.g., coupling of liquid chromatography to MS (LC–MS), should allow for more efficient and broader applications.
Metabolomic profiling
Metabolomics is commonly employed to give a global overview of changes in the metabolic status of an organism in response to environmental fluctuations, nutrition availability or genetic disturbances. While extracellular metabolites, such as gluconic or acetic acid, have been often measured in the cultures of the Komagataeibacter species, a recent study in K. xylinus CGMCC 2955 (Gluconacetobacter in the original work) investigated the differences between static and agitated cultures by measuring their internal metabolomic profiles using gas chromatography coupled with mass spectrometry (GC–MS) (Liu et al. 2015). In total, 79 intracellular metabolites were detected and quantified in both culture types. The results of multivariate data analysis showed clear separation of the samples from the tested cultures, what indicated that the cells in the static culture were metabolically distinct from those in the agitated one. It was shown that metabolites of the central carbon metabolism and amino acids biosynthesis pathways displayed significant changes in the samples from different culturing methods and sampling times. The metabolites responsible for distinguishing the samples were mainly trehalose, phosphate and gluconic acid. The agitated culture promoted cell growth and viability; however, the static culture was characterised by the highest conversion rate of glucose to BNC. Moreover, the initial accumulation of trehalose and amino acids (proline, glutamic acid, alanine, l-valine and l-threonine) in the agitated culture at the highest rotational speed was interpreted to fulfil a synergistic protective role during adaptation to the environmental stress caused by the culture condition, such as the hydrodynamic stress. At the molecular level, it has been shown that trehalose plays an important role in protein stabilisation by influencing the water environment surrounding a protein and promoting maintenance of their native state (Singer and Lindquist 1998; Ruhal et al. 2013; Olsson et al. 2016). In case of proline, its role in stress protection against various environmental factors is well documented and includes osmo- and thermo-protection, maintenance of protein stability and oxidative stress resistance (Christgen and Becker 2019). It has been shown in E. coli that proline oxidative metabolism generates hydrogen peroxide and thus increases oxidative stress tolerance via a preadaptive effect which involves induction of pathway for H2O2 removal and maintenance of redox homeostasis (Zhang et al. 2015). Due to the higher availability of oxygen in the agitated cultures of K. xylinus CGMCC 2955, degradation of glucose to gluconic acid proceeded very fast. The following rapid consumption of the gluconic acid in the cytosol may have resulted in generation of the oxidative stress in these cultures due to the increased electron flux through the respiratory chain.
It has been observed that the sufficient and uniform aeration of the agitated culture favours selection of cellulose non-producing cells, in comparison with the static culture (Park et al. 2003; Krystynowicz et al. 2005). The conclusion which can be drawn from the studies employing agitated cultures is that BNC production is a transient feature of the Komagataeibacter species and is maintained as long as it confers a survival advantage. Since these are obligate aerobic bacteria, formation of a cellulose pellicle in a liquid culture is advantageous since it enables the retention of cells close to the air–liquid interface where the concentration of oxygen is high and there is access to the nutrients from the medium.
More studies investigating exo- and intra-metabolic profiles under various conditions, relevant for cellulose biosynthesis, will definitely bring a valuable insight into the systemic understanding of a Komagataeibacter cell. Moreover, in the case of some of the metabolites, targeted approaches for isolation and quantification are needed. For the Komagataeibacter species, it is particularly important to study the profile of synthesised exopolysaccharides. Production of such EPS as acetan not only competes with the cellulose synthesis pathway for carbon precursors but was also shown to interfere with cellulose network structure, i.e. fibril assembly and crystallisation (Ishida et al. 2002; Fang and Catchmark 2014). The monosaccharide composition and yield of EPS synthesised has been found to vary depending on strain and carbon source (Fang and Catchmark 2015). Further research involving the impact of EPS synthesis by the Komagataeibacter species should contribute to engineering of BNC membranes of a custom structure and properties.
Genome-scale metabolism modelling
Genome-scale metabolic models (GEMs), which employ both the genomic and biochemical information to describe gene–protein–reaction associations, have been shown to be particularly useful for systems-level metabolic studies (Gu et al. 2019). The recent availability of complete genome sequences of the Komagataeibacter strains stimulated reconstruction of GEMs to better understand their metabolism and to predict metabolic capabilities of the BNC producers. A GEM has been reconstructed for K. nataicola RZS01 and comprised 771 genes, 2014 reactions and 2035 metabolites (Zhang et al. 2017). Through constraint-based analysis, this model was used to characterise and evaluate cellular metabolism and to predict genes and reactions necessary for growth. The analysis revealed that glycerol is the most optimal carbon source for the highest BNC production. Moreover, the minimisation of metabolic adjustment (MOMA) algorithm was used to predict eight overexpression targets for increasing BNC production; however, these candidates were not experimentally tested. In the case of K. hansenii ATCC 23769, only the core metabolic model was reconstructed which included 74 reactions and 68 metabolites (de Souza et al. 2018). The metabolic flux distribution of this model was calculated using flux balance analysis (FBA) approach and in silico simulations were performed to predict the growing abilities on different substrates and the minimal medium capable of supporting BNC production. A more expanded GEM was constructed for Komagataeibacter xylinus DSM 2325 (Jang et al. 2019). This GEM was based on genome sequence and experimental growth data and contained information on 686 metabolic genes, 1810 reactions and 1712 metabolites. It was also used to predict gene overexpression targets for increased BNC production. Heterologous overexpression of two of these targets, glucose-6-phosphate isomerase (pgi) and phosphogluconate dehydrogenase (gnd) brought enhanced BNC production, which was 2-fold for the strain overexpressing the E. coli pgi gene. In another study of this team, exploiting this GEM and through simulated FBA, it was found that the inclusion of a reaction catalysed by phosphofructokinase (pfkA), which is absent in the Komagataeibacter spp., improves cellulose production rate and specific growth rate (discussed later; Gwon et al. 2019).
The functionality of GEMs greatly depends on the annotation status of a genome. Advancement in functional genomics of the Komagataeibacter species should improve GEMs and allow for generation of more precise models and the subsequent selection of a higher number of gene targets for further engineering.
Untargeted and targeted approaches for improving BNC synthesis efficiency or properties
Two main approaches, untargeted and targeted, for modification of a cellulose producer have been employed. The untargeted methods usually involve induction of changes in culturing conditions or mutagenisation and require generation and testing of a large number of clones. Despite being laborious and time consuming, these approaches have successfully been employed to obtain producers of improved BNC yield. At the same time, the studies which were coupled with genetic analysis of the mutant strains enhanced the understanding of Komagataeibacter molecular physiology. On the other hand, rational strain design through targeted approaches is usually more efficient and may yield cellulose of specifically desired features. However, this approach would greatly benefit from the understanding of a whole system at a molecular level because, at present, cellular response to introduced changes is hardly predictable. We will discuss the studies employing both of these approaches for obtaining mutants producing BNC of improved yield and properties.
Untargeted approaches
The early untargeted studies explored the natural phenotypic variability of some of the Komagataeibacter strains which often leads to spontaneous disappearance of cellulose-synthesising ability through the formation of cellulose non-producing cells (Table 3). It has been suggested that these phenotypic changes are induced due to the stress factors, e.g. increased oxygen concentration, high concentration of ethanol in the medium or an increase in temperature during cultivation (Gatenholm et al. 2013; Taweecheep et al. 2019a). So far, the reasons for phenotypic variability at the molecular level are not well understood. Coucheron and co-workers pointed to IS1031 insertion elements as the cause of K. xylinus production instability, although insertions of these elements were detected only in some of the cellulose non-producing mutants (Coucheron 1991). In other studies, genetic analyses of DNA of Cel− found mutations in genes encoding proteins involved in cellulose biosynthesis pathway as the possible reasons for disappearance of BNC production ability (Table 3). The cellulose negative phenotype may spontaneously appear as well as be reversed. It has often been reported that after several passages of Cel− forms in a static culture, it is possible to restore BNC biosynthesis ability (Matsutani et al. 2015; Taweecheep et al. 2019b). Genomic DNA analyses of such cells (called ‘revertants’) have identified point mutations (insertions or deletions) in the bcsB and bcsC genes which caused the frameshifts restoring the BcsB or BcsC activity. It is interesting that the point mutations in bcsCI not only resulted in the restoration of the BNC biosynthesis capability but also caused up to 6.5-fold increase in BNC yield (Taweecheep et al. 2019b). Moreover, the same study has shown that a single mutation in the bcsCI gene may not only improve BNC biosynthesis but also lead to changes in its structure and properties. The authors received two converters (R37-4 and R37-9) synthesising BNC of 2-fold higher fibre density and improved mechanical properties, in comparison to the parental strain.
Table 3.
Effect of mutation on BNC biosynthesis and/or BNC properties
| Strain | Disrupted gene or a fragment | Mutation | Mutation conditions or type of mutagenesis | Phenotypic effects | References | |||
|---|---|---|---|---|---|---|---|---|
| Culture conditions or environment effect on the formation of cellulose-non-producing (Cel−) forms | ||||||||
| K. xylinus E25 | Cel− | Fragment downstream of ORF encoding phosphoglucomutase | A single nucleotide (T) deletion. Inhibited expression of phosphoglucomutase and glucose-1-phosphate uridylyltransferase | Repeated 3 times passages under agitated culture conditions (SH medium, 48 h, 30 °C, 90 rpm) of the parental strain | No BNC production | (Krystynowicz et al. 2005) | ||
| K. europaeus | LMG 18494 | bcsCI (cellulose synthase subunit CI) | Large deletion (1900-bp deletion) | Isolation from vinegar | No BNC production | (Andrés-Barrao et al. 2011) | ||
| LMG 18890 | bcsCI | 5-bp deletion | ||||||
| Komagataeibacter medellinensis | NBRC 3288 (parental strain) | bcsBI (cellulose synthase subunit BI) | 17-bp deletion | Frameshift. Gene disruption by stop codon introduction | Isolation from vinegar | No BNC production | (Ogino et al. 2011) | |
| R1 | Revertants | a1-bp (C) deletion at the 277486th position of the gene. Frameshift and BcsBI restoration | Repeated static culture of the parental strain | Restoration of BNC production | (Matsutani et al. 2015) | |||
| R2 | a1-bp (C) deletion at the 277491th position of the gene, frameshift and BcsBI restoration | |||||||
| K. oboediens |
MSKU 3 (wild type) |
bcsCI (cellulose synthase subunit CI) | – | Isolated from rotten fruit samples in Thailand; thermotolerant strain (growth up to 39 °C) | BNC yield of 0.33 g/L. BNC fibril diameter size: 70.52 nm; density: 0.43 ± 0.05 g cm−3. Mechanical properties of BNC; tensile strength: 73.94 ± 16.94 MPa, Young’s modulus: 5.83 ± 0.69 | (Taweecheep et al. 2019a, b) | ||
|
E3 (parental strain for revertants) |
bA single nucleotide (T) insertion at the 2135th position of bcsCI | Frameshift. Gene disruption by stop codon | Repeated static cultivation of MSKU 3 strain for 63 passages (YPGD1A medium containing increasing concentration of ethanol) | No BNC production. High acetic acid production ability | ||||
| R30-3 | Revertants | 2-bp (GC) insertion at the 2145th position. Frameshift and two amino acid alterations (N713E and Q714P) and one amino acid addition (A715) | Repeated static cultivation of E3 strain (YPGD1A3E medium) | Increased BNC yield (2.15 g/L). BNC fibril diameter size: 65.9 nm; density 0.42 ± 0.02 g cm−3. Reduced mechanical properties of BNC (ca. 2-fold ↓); tensile strength: 43.56 ± 10.98 MPa, Young’s modulus: 3.60 ± 0.40 | ||||
| R30-12 | 1-bp (C) deletion at the 2149th position. Frameshift and four amino acid substitution (N713E, Q714P, R715A and G716W) | Repeated static cultivation of E3 strain (YPGD medium) | Increased BNC yield (0.53 g/L). BNC fibril diameter size: 70.14 nm; density: 0.54 ± 0.01 g cm−3. Mechanical properties of BNC; tensile strength: 73.94 ± MPa; Young’s modulus: 5.14 ± 0.58 | |||||
| R37-4 | 1-bp (T) deletion at the 2132nd position. Frameshift and a single amino acid substitution (L711R) | Repeated static cultivation of E3 strain (YPGD medium) | Increased BNC yield (1.12 g/L). BNC fibrils diameter size: 59.14 nm; density: 0.72 ± 0.05 g cm−3. Increased mechanical properties of BNC (ca. 2-fold ↑); tensile strength: 158.72 ± 28.29 MPa; Young’s modulus: 8.75 ± 1.54 | |||||
| R37-9 | 1-bp (A) deletion at the 2139th position. Frameshift and a single amino acid substitution (N713D) | Increased BNC yield (0.7 g/L). BNC fibril diameter size: 34.58 nm; increased density (2-fold ↑): 0.85 ± 0.07 g cm−3. Increased mechanical properties of BNC (2-fold ↑); tensile strength: 159.47 ± 29.76 MPa; Young’s modulus: 9.83 ± 0.69 | ||||||
| Transposon mutagenesis | ||||||||
| K. hansenii ATCC 23769 | 5 | acsA (cellulose synthase subunit A) | Insertion site 1893 bp, gene disruption | Tn 5 transposon insertion mutagenesis | No or reduced BNC production | (Deng et al. 2013) | ||
| 10 | acsC (cellulose synthase subunit C) | Insertion site 1984 bp, gene disruption | ||||||
| I-13 | ccpAx (cellulose complementing protein Acetobacter xylinum) | Insertion site 40 bp, gene disruption | ||||||
| I-7 | dgc1 (diguanylate cyclase) | Insertion site 656 bp, gene disruption | ||||||
| V-31 | dgc1 (diguanylate cyclase) | Insertion site 242 bp, gene disruption | ||||||
| II-23 | crp− fnr (transcriptional regulator, Crp/Fnr family protein) | Insertion site 187 bp, gene disruption | ||||||
| K. hansenii ATCC 23769 | I-23 | AlaR (alanine racemase) | Insertion site 628 bp, gene disruption | Tn 5 transposon insertion mutagenesis | Reduced BNC production and crystallinity | (Deng et al. 2015) | ||
| #52 | LDC (lysine decarboxylase) | Insertion site 347 bp, gene disruption | ||||||
| Mutagenisation by chemical and/or physical agents | ||||||||
| K. xylinus ATCC 53582 | plr 15 | bcsA (cellulose synthase subunit AI) | Substitution of nucleotide G to A at position 1345 bp of the gene; A449T substitution in the BcsA (missense mutation) | EMS (ethyl methane sulfonate) mutagenesis and screening on a medium containing10 μM pellicin | Increased BNC synthesis rate; reduced crystallinity; resistance to pellicin | (Salgado et al. 2019) | ||
| K. hansenii | HDM 1–3 (wild type) | – | – | Isolated from rotten Actinida chinesis Planch | BNC yield of 1.43 g/L | (Li et al. 2016) | ||
| Br-12 | Three specific fragments were identified: TonB-dependent transport (TBDT, 779 bp, LCL 80534); exopolysaccharides output protein (PePr, 162 bp, LCL77813); unknown protein (380 bp, LCL57743) | AFLP-based analysis between mutant lines and the wild type | DES (diethyl sulphate) mutagenesis | Decreased BNC yield (0.56 g/L). Increased gluconic acid production (67.53%) | ||||
| Br-3 | nd | DES (diethyl sulphate) mutagenesis and screened and selection of mutant in a medium containing NaBr–NaBrO3 | Increased BNC yield (2.45 g/L). Decreased gluconic acid production (10.23%) | |||||
| Co-5 | Two specific fragments were identified: acsD and galE (UDP-galactose-4-epimerase) | Mutagenesis of the Br-3 mutant by 60Co-γ radiation and screened and selection of mutant in medium containing NaBr–NaBrO3 | Increased BNC yield (3.38 g/L). Decreased gluconic acid production (54.79%) | |||||
YPGD medium (0.5% of glucose, 1% of yeast extract, 1% of polypeptone and 2% of glycerol) containing 1% acetic acid and ethanol (YPGD1A3E)
aAll the mutations were found to occur in a small C-rich region (CCCGGCCC) in bcsBI
bAll the mutations were found to occur in a small region (TGCTGAACCAGCGTGGC) in bcsCI, which seems to be a sensitive region for spontaneous mutation
Mutagenisation with the use of transposons was another approach used to identify genes influencing BNC biosynthesis (Table 3). This type of mutagenisation was applied by Deng et al. and showed that the yield and crystallinity of BNC are influenced by the alaR and LDC genes encoding alanine racemase and lysine decarboxylase, respectively (Deng et al. 2015). AlaR and LDC proteins are required for maintaining of peptidoglycan integrity. Mutants with interrupted alaR and LDC genes synthesised less BNC, which was less crystalline, unevenly distributed, with some regions appearing to contain non-cellulose polysaccharides. Ion chromatography–based analysis showed an increase in the number of monosaccharides (galactose and mannose) associated with non-cellulose polysaccharides in comparison to the parental strain. It would be interesting to further investigate the influence of overexpression of these genes on BNC synthesis and properties.
Another strategy to improve cellulose biosynthesis can be the use of mutagenisation with chemical and/or physical agents (Table 3). Culture acidification due to organic acid production is a known factor limiting growth and BNC biosynthesis by the Komagataeibacter species (De Wulf et al. 1996). Based on these observations, Li and co-workers aimed to select a high-yield BNC producer by screening for low acid yielding mutants using a combined mutagenesis (treatment with diethyl sulphate coupled with 60Co-γ irradiation and proton suicide on medium containing NaBr–NaBrO3) (Li et al. 2016). Two mutants were obtained during this study (Br-3 and a Co-5) which synthesised more BNC (1.5 and 2 times more, respectively) and less gluconic acid (10% and 55% less, respectively) in comparison to the wild-type strain. Moreover, the study generated another mutant, Br-12, which synthesised 61% less BNC and 68% more gluconic acid. By employing amplified fragment length polymorphism (AFLP) analysis, the authors explored the genetic differences between the mutants and the wild type strain (see Table 3). The same authors investigated further the physiological basis of acid stress resistance in K. hansenii HDM1-3 and showed that it is associated with fatty acids variation in the cell membrane (Li et al. 2019). The team observed changes in the expression of genes of fatty acid dehydrogenase (des) and cyclopropane synthase (cfa) which encode the key enzymes for the synthesis of unsaturated fatty acids (namely, oleic acid (C18: lw9c) and cyclopropane fatty acid (C19-cyc)). The results of this study encourage further exploration and the manipulation of fatty acids biosynthetic pathway and propose a novel strategy to improve the efficiency of BNC production by the strains of the Komagataeibacter genus.
Molecular and synthetic biology tools for targeted strain manipulation
Until recently, targeted genetic modifications of Komagataeibacter spp. were uncommon and limited to the use of two expression vector backbones, namely pSA (Tonouchi et al. 1994; Nakai et al. 1999) and, more frequently, pBBR122 (Setyawati et al. 2007, 2009; Yadav et al. 2010; Florea et al. 2016; Liu et al. 2018b). Induction of gene disruption through a homologous recombination was commonly performed with the use of non-replicating plasmid disruption cassette carriers, such as pUC19, pT7Blue, pET14 or pHSG399 (Table 4). At present, one can expect intensification of genetic engineering of different Komagataeibacter strains due to the progress in the application of three new vector backbones for endogenous and heterologous gene expression, namely pTI99, pTSa and pSEVA331Bb (Fang et al. 2015; Florea et al. 2016; Jacek et al. 2018, 2019b; Gwon et al. 2019). Moreover, the performance of various promoters, ribosome binding sites (RBS) and terminators has been characterised in the first two studies applying synthetic biology tools in Komagataeibacter hosts (Florea et al. 2016; Teh et al. 2019). These studies together give a comprehensive guide for the construction of vectors of a high potential to be effective in the numerous species from this genus. These recent achievements include the use of quorum sensing (QS) synthetic biology elements known to be functional in E. coli (Florea et al. 2016; Walker et al. 2018). The first example of employment of these elements was the application of inducible lux promoter and luxR gene for modification of K. rhaeticus strain (Florea et al. 2016). This study showed that the capability of cellulose production was lowered and eventually switched off with increasing concentration of N-acyl homoserine lactone (AHL) in the media. Another study reported the construction of Sender and Receiver recombinant K. rhaeticus strains (Walker et al. 2018). The first strain produced AHL in response to an environmental signal, while the second strain induced recombinant protein expression (red fluorescent protein, RFP) in response to AHL in a concentration-dependent manner. The most recent synthetic biology study was published by Teh et al. and consists of a comprehensive review of performance of several promoters, RBS and terminator sequences tested in the pSEVA plasmid backbone in three Komagataeibacter strains (K. hansenii ATCC 53582, K. rhaeticus iGEM and K. xylinus ATCC 700178). Importantly, the first use of CRISPR interference (CRISPRi), targeting chromosome-encoded acs operon in K. hansenii ATCC 53582, was described in this study (Teh et al. 2019). Summarising, this work paves the way for studies aiming at obtaining of self-immobilised Komagataeibacter hosts expressing biological circuits of desired function and which may be applied as, e.g., biosensors in intelligent packaging material.
Table 4.
Genetic engineering tools used in various Komagataeibacter strains
| Induction of gene expression | ||||
|---|---|---|---|---|
| Vector backbone | Promoters | Derived constructs | Komagataeibacterstrain | References |
| pSA19 | lac |
pSA-SD pSA-SD-S11E |
K. sucrofermentans BPR 2001 | (Nakai et al. 1999) |
| pBBR122 |
Bla lac |
pBla-Vhb-122 pLacDAAO-122 pBla-Vhb-122 pBBR-Glc-NAc |
K. xylinus BCRC 12334 K. xylinus BCRC 12334 K. xylinus CGMCC 2955 K. xylinus 10245 |
(Setyawati et al. 2007) (Setyawati et al. 2009) (Liu et al. 2018b) (Yadav et al. 2010) |
| pTI99 | trc |
pTI99-cdrS pTI99-motAB −motA; −motB |
K. hansenii ATCC 23769 K. hansenii ATCC 23769 K. hansenii ATCC 53582 |
(Fang et al. 2015) (Jacek et al. 2018) (Jacek et al. 2018) |
| pTSa |
tac gapA |
pTSaEX1 −pfk −pgi; −pgk; −fba; −tpi; −gap; −gpm; −pck; −mae; pTSaEX2 |
K xylinus DSM 2325 | (Gwon et al. 2019) |
| pIN01 | tac | pIN01-crp | K xylinus DSM 2325 | (Gwon et al. 2019) |
| pSEVA331Bb |
J23104 lux tet Bad J23100 J23101 J23102 J23104 J23105 J23108 J23109 J23110 J23115 J23119 J23119-A27T |
J-sRNA-331Bb pLux01 pLux02 pTet01 pTet02 pReceiver pSender pSEVA331Bb-mRFP pSEVA331Bb-NAG5-AGM1-UAP1 |
K. rhaeticus iGEM K. rhaeticus iGEM K. rhaeticus iGEM K. hansenii ATCC 53582 K. xylinus ATCC 700178 |
(Florea et al. 2016) a (Walker et al. 2018) (Teh et al. 2019) |
| pLBT | Promoter-less miniTn10 | pLBT::lacZ:kan | K. xylinus ITDI | (Battad-Bernardo et al. 2004) |
| Knock-out or inhibition of gene expression | ||||
| Vector backbone | Method/selection marker | Obtained constructs | Komagataeibacterstrain | References |
| pUC18 |
HR/ampR HR/kanR |
pUCA-EP | K. sucrofermentans BPR 2001 | (Ishida et al. 2002) |
| acsD deletion | K. hansenii ATCC 23769 | (Hu et al. 2010) | ||
| pUC19 | HR/ampR | T-GDH-Amp | K. xylinus BCRC 12334 | (Kuo et al. 2015) |
| pT7Blue | HR/cat-1 | pT7-Blue-PGDHC-I | K. sucrofermentans BPR 2001 | (Shigematsu et al. 2005) |
| pET14B | HR/kanR | motA motB and motAB disruption | K. hansenii ATCC 53582 | (Jacek et al. 2019b) |
| pHSG399 |
HR kanR HR ampR |
Dgc1 cmcAx pSA-ORF2/k (cpcAx deletion) |
K. sucrofermentans BPR 2001 |
(Bae et al. 2004) (Nakai et al. 2013) (Nakai et al. 2002) |
| pSEVA331Bb | sRNA | J-sRNA-331Bb (co-expression of Hfq and sRNA targeting UGPase mRNA) | K. rhaeticus iGEM | (Florea et al. 2016) |
| ter |
ECK120033736 ECK120010818 ECK120010799 ECK120051401 BBa_B0010 L3S2P21 L3S3P21 L3S2P24 L3S2P44 L3S1P47 |
K. rhaeticus iGEM K. hansenii ATCC 53582 K. xylinus ATCC 700178 |
(Teh et al. 2019) | |
| Protein degradation tags |
LAA AAV LVA DAS DAS + 2 |
K. rhaeticus iGEM K. hansenii ATCC 53582 K. xylinus ATCC 700178 |
||
| CRISPRi | pSEVA331Bb-Cas9-3xFLAG-sgRNA | K. hansenii ATCC 53582 | ||
HR homologous recombination, CRISPRi deactivated Caspase9-based system = CRISPR interference, sRNA short RNA mediated, ter translation terminator, kanR kanamycin resistance gene, cat-1 chloramphenicol acetyltransferase gene, ampR ampicillin resistance gene
aOther vector backbones reported here as replicating in K. rhaeticus iGEM but not studied further: pBla-Vhb-122; pSEVA321Bb, pBAV1K-T5-sfGFP
All the vectors used for gene overexpression in the Komagataeibacter strains together with vectors used in gene disruption and deletion approaches are summarised in Table 4. In most cases, the functionality of the constructs was verified in a limited number of strains and their compatibility with other Komagataeibacter species has yet to be tested.
Improvement in cellulose productivity
Genetic modification of the Komagataeibacter strains has been used to generate a few industrially relevant mutant strains with improved cellulose productivity. These improvements have been achieved either through metabolic pathway modulation (Shigematsu et al. 2005; Kuo et al. 2015; Gwon et al. 2019) or heterologous gene expression (Setyawati et al. 2007; Liu et al. 2018b; Jang et al. 2019).
From a cellulose productivity point of view, a desirable direction of metabolic shift is limitation of production the major side-product from glucose which is gluconic acid. This goal was reached, e.g. via deactivation of a gene encoding membrane-bound glucose dehydrogenase (GDH; Shigematsu et al. 2005; Kuo et al. 2015). Active oxidation of glucose into gluconic acid has two undesired effects in Komagataeibacter cultures: pH lowering and quick loss of glucose from medium. These obstacles were overcome in two GDH-deficient mutants: the GD-1 in the K. xylinus BPR2001 strain (Shigematsu et al. 2005) and, more recently, the GDH-KO in the K. xylinus BCRC 12334 strain (Kuo et al. 2015). Both these strains were obtained by means of homologous recombination (Table 4). The GDH-deficient mutants produced BNC at ca. 2-fold higher yield than the parental strains. What is more, the GD-1 strain was shown to be able to effectively produce BNC on enzymatically saccharified potato pulp (BC yield reached 5.0 g/L without and 7 g/L after ethanol supplementation; Shigematsu et al. 2005).
Another example of a genetically improved cellulose-producing strain was expressing Vitreoscilla haemoglobin (VHb) which assured higher ATP synthesis under hypoxic conditions (static culture or lowered O2 tension in a closed bioreactor) (Setyawati et al. 2007; Liu et al. 2018b). In both studies, the observed increase of cellulose production yield by the VHb+ strains was in the range of 1.5- to 1.7-fold.
A more recent study involving K. xylinus DSM 2325 strain has shown that the reduction of gluconic acid production can be achieved through manipulation of regulatory proteins, besides the direct catabolic enzymes (Gwon et al. 2019). This study generated a strain with reduced gluconic acid synthesis yield (39.2% in the mutant and 64.8% of glucose in the parental strain) after introduction of an additional copy of the crp (Crp/Fnr) gene, expressed from a plasmid, under the tac promoter control. Apart from crp overexpression, the mutant strain (named S.Koma-pfkA/crp) used by Gwon et al. harboured the pfkA gene fused in chromosome (Gwon et al. 2019). Selection of this gene was based on the analysis of the aforementioned GEM for this strain. The authors speculate that this change caused redirection of metabolic flux to the Embden–Meyerhof–Parnas (EMP) pathway, which was predicted to be inactive in their parental strain, enabling higher ATP production and, as a result, an increased BNC production yield (4.3 g/L in mutant; 3.5 g/L in parental strain). Constitutive expression of several other genes from EMP pathway (namely pgi, pgk, fba, tpi, gap, gpm, pck and mae) was also tested but did not influence the final BNC yield. The finally optimised strain expressing pfkA and crp genes (S.Koma-pfkA/crp) gave 4.5 g/L cellulose production rate and was used for batch fermentation in a 30-L bioreactor. After optimisation of fermentation process, the authors obtained 3.3 times higher cellulose production yield by a mutant strain related to the parental one.
When discussing genetic modification achievements in the field of BNC production, one should not overlook the early constructions of strains capable of carbon assimilation from sucrose (Tonouchi et al. 1998a, 1998b; Nakai et al. 1999) or lactose (Battad-Bernardo et al. 2004), both being more economical carbon sources than glucose. Particularly an efficient sucrose-assimilating mutant strain, which showed 2–3-fold increase in cellulose productivity, when compared to the parental strain, was obtained by expression of mung bean sucrose synthase on the pSA plasmid (Table 4) (Nakai et al. 1999). The lactose-assimilating strain (ITz3) harboured transposon-mediated chromosome integration of E. coli lacZ gene (Battad-Bernardo et al. 2004). The ITz3 mutant strain differed from K. xylinus ITDI parental strain by its ability to produce cellulose on media containing lactose and on whey, which is the most common by-product of the dairy industry. The ITz3 strain constitutively expressed enzymatically active beta-galactosidase and achieved BNC synthesis yield from 0.73 g/L on whey up to 1.30 g/L on the PYSL4 medium, even though the parental strain was unable to produce cellulosic pellicle in any of these media.
Modification of BNC structure
Having regard to the diversity and still expanding scopes of BNC applications, it is still essential to take full advantage of its exceptional structure and properties. The development of novel BNC-based materials with unique properties has been the subject of an extensive research over the last two decades. Numerous modification methods have been explored to inquire BNCs with novel or improved functionalities (Hu et al. 2014). Biosynthetic (in situ) or physical/chemical (ex situ) modifications of cellulose membranes constitute the vast majority of all the explored functionalising methods. Genetic engineering tools, however, offer the possibility of introducing non-invasive changes to cellulose, without the risk of unintentionally influencing its native properties.
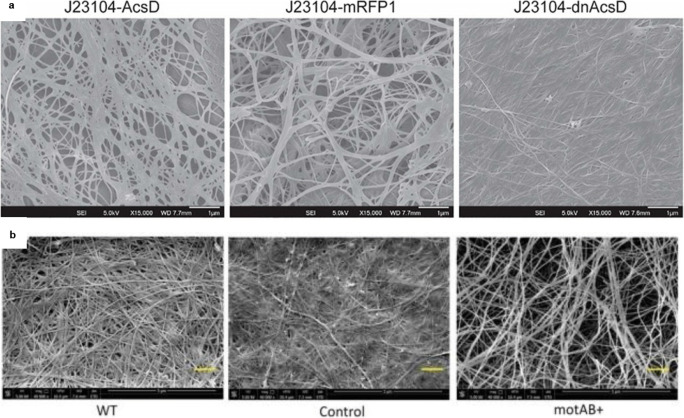
Cellulose, as a highly porous, nanofibrillated material, has been the subject of numerous structural modifications. Up to date, only a very few of them employ recombinant strains, what result from still ongoing research on the role of particular genes involved in metabolism of cellulose synthesis. The main target genes are the constituents of bacterial cellulose synthase (BCS) operon. Teh and co-workers revealed that the dominant negative AcsD (dnAcsD) expression causes the production of a dense cellulose matrix (Fig. 2a; Teh et al. 2019). At the same time, dnAcsD-overexpressing cells secreted thinner fibres than those produced by the wild-type and other bacterial strains tested in the study. It was speculated that the presence of dnAcsD might disrupt the crystallisation of thicker subelementary fibrils after glucan chain extrusion. Such results suggest it might be possible to tailor the structure of BNC by tuning the ratio of wild-type AcsD to dnAcsD.
Fig. 2.
Genetically induced structural changes in BNC network. a A dense cellulose matrix of thin fibres produced by the dominant negative AcsD (dnAcsD) K. hansenii ATCC 53582 mutant (AcsD—cellulose synthase subunit D; strain expressing wild-type AcsD (J23104-AcsD) or mRFP1 (J23104-mRFP1)). Reprinted with permission from ACS Synth. Biol. (2019, 8, 4, 708–723). Copyright (2019) American Chemical Society. b A loose BNC network produced by K. hansenii ATCC 23769 mutants overexpressing the motility-related genes motA and motB (WT—wild-type strain, Control—strain transformed with an empty vector; motAB+—mutant with motA and motB overexpression). Reprinted from Jacek et al. (2018)
Going beyond the operon coding for cellulose synthase, since more and more genomes of cellulose-producing strains are published and analysed, the scientists have been investigating the roles of genes responsible for general cellular behaviour. One example of such approach is the study on the motA and motB genes which build up a proton pump and which are probably involved in cell motility (Jacek et al. 2018, 2019b, 2019c). Jacek et al. subjected K. hansenii ATCC 53582 to disruption and complementation of one or both of the mentioned genes, whereas in K. hansenii ATCC 23769 induced motA and motB genes overexpression. The latter mutation caused bacterial cell elongation (or formation of filaments) and intensification of colony-spreading ability. Microscopic analysis of the BNC membrane has shown a significant loosening of intra-membrane structure and fibre thickening was observed (Fig. 2b; Jacek et al. 2018).The opposite effect was observed in the motA− and motB− disruption mutants which were synthesising more dense and compact cellulose (Jacek et al. 2019b). Yet, both the examples indicate that a change in motility-related genes indirectly influences the nanostructure of cellulose, making it more porous and relaxed when the recombinant cells are elongated, and their movement is intensified. Moreover, the mutant-derived BNC was proved to preserve biocompatibility and chemical properties of BNC of the parental, non-modified strains which still makes it highly valuable in medical applications.
Synthesis of BNC nanocomposites
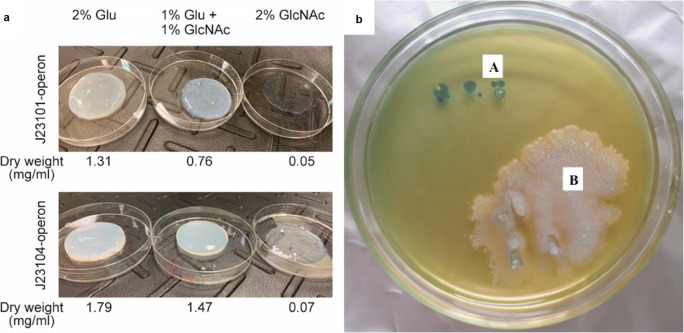
Last but not least, genetic manipulation has been shown to be an attractive approach for producing BNC nanocomposites. Yadav et al. were the first to engineer K. xylinus to rationally redesign the flow of cellular metabolites so as to incorporate N-acetylglucosamine (GlcNAc) sugar residues into glucan chains during the biosynthesis (Yadav et al. 2010). The authors took the advantage of the fact that K. xylinus (formerly Ga. xylinus) can utilise both UDP-glucose and UDP-GlcNAc as substrates. In order to produce a composite of bacterial cellulose and chitin, a recombinant strain of K. xylinus 10245 was developed. An operon composed of three genes from Candida albicans (AGM1, NAG5 and UAP1) responsible for UDP-GlcNAc synthesis was expressed under the control of bla promoter in K. xylinus (Table 4). The modified strain was able to produce activated cytoplasmic UDP-GlcNAc monomers accessible to cellulose synthase to join both glucose and GlcNAc, and synthesise a chimeric polymer. The generated material contained over 18-fold more GlcNAc, as compared to the control BNC; it was more facile to enzyme degradation and displayed improved degradability in vivo. The same concept of de novo synthesis of chitin–cellulose composite was adopted by Teh et al. in order to demonstrate the utility of a genetic toolkit for synthetic biology applications in Acetobacteraceae family (Teh et al. 2019). The three genes responsible for UDP-GlcNAc synthesis were expressed in the wild-type K. xylinus ATCC 700178 under either a weak (J23101) or a strong promoter (J23104) revealing that more cellulose is formed in the latter case. Nevertheless, both systems successfully generated a composite material which synthesis level and quality strongly depends on the content of the both substrates (glucose and GlcNAc) in the medium (Fig. 3a).
Fig. 3.
Examples of genetically engineered BNC bionanocomposites. a Images showing the cellulose–chitin copolymer produced under two different promotors, using different concentrations of glucose (Glu) or GlcNAc. Reprinted with permission from ACS Synth. Biol. (2019, 8, 4, 708–723). Copyright (2019) American Chemical Society. b Curdlan–cellulose composite produced by genetically engineered K. hansenii AY201 visualised by aniline blue staining (a—colonies of the mutant strain expressing curdlan synthase; b—colonies of the control strain). Reprinted from Biomacromolecules (2015, 16, 10, 3154–3160)
In 2015, the group of Professor Kondo developed in vivo a curdlan–cellulose bionanocomposite by applying a genetically modified K. hansenii AY201 (Ga. xylinus in the original work; Fang et al. 2015). Curdlan is an exopolysaccharide known in biomedical applications because of its nonionic gelation properties and low toxicity. Genetically engineered strain was obtained by a direct modification of cellulose-synthesising metabolic pathway of bacteria to secrete a mixture of cellulose and curdlan (β-1,3-glucan). Similarly, as during cellulose biosynthesis, curdlan synthesising microorganisms use UDP-glucose as a precursor for its production. To introduce a curdlan biosynthesis system into K. xylinus, the authors expressed curdlan synthase (crdS) gene from the most efficient producer of this biopolymer, Agrobacterium sp. ATCC 31749. The other accessory genes of curdlan excretion were not included in order to provide the exclusive extracellular secretion of curdlan–cellulose nanocomposite directly from a bacterial cell. The resulting strain was shown to simultaneously secrete curdlan and crystalline cellulose nanofibres (see Fig. 3b), providing a composite material of slightly changed surface morphology (curdlan to some extent covered the pores of cellulose) but still with preserved crystallinity. It is believed that the constantly growing state of knowledge of function of Komagataeibacter genes will enable further research on efficient engineering of these bacteria for biomanufacturing of novel cellulose-based materials and numerous commercially valuable products.
Conclusions and future perspectives
This review aimed to discuss the current progress in the systemic understanding of Komagataeibacter physiology. Gaining of this perspective depends on the availability of high-quality omics datasets which improve genome functional annotation. This information is crucial for building of robust genome-scale metabolic models and selection of new targets for genetic engineering. Targeted and untargeted approaches for strain improvement have shown that it is important to investigate genes indirectly related to metabolic pathway of BNC synthesis as well as transcriptional regulators. Moreover, low immunogenicity and good biocompatibility of BNC products is maintained when using recombinant bacteria instead of chemical actions on this material which is especially important in biomedical field. Preserving its native physico-chemical properties, cellulose produced by modified microorganisms may display altered 3D nanostructure, porosity or density. On the other hand, the importance and potential of bacterial nanocellulose may be greatly increased when applying genetically engineered strains in combination with chemical and physical modifications of BNC. The intentional interconnections between specific metabolic pathways in the Komagataeibacter might result in the design of novel, chimeric biopolymers. It seems inevitable that still growing knowledge in the field of genome-dependent metabolic mechanisms influencing the process of BNC biosynthesis will be constantly widening the scope of its possible targeted modifications, as well as its final implementation.
Acknowledgements
We would like to thank Teresa Pankiewicz and Jolanta Płoszyńska for their contribution to the BNC research.
Author contributions
M.R. and S.B. conceived and designed the review. M.R., M.J.-K., K.K., K.L. and S.B. wrote the manuscript. All authors read and approved the manuscript.
Funding information
M.R. acknowledges partial support through MINIATURA2 grant 2018/02/X/NZ1/00998 from Narodowe Centrum Nauki (NCN; National Science Centre, Poland).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Disclaimer
None.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés-Barrao C, Falquet L, Calderon-Copete SP, Descombes P, Pérez RO, Barja F. Genome sequences of the high-acetic acid-resistant bacteria Gluconacetobacter europaeus LMG 18890 T and G. europaeus LMG 18494 (reference strains), G. europaeus 5P3, and Gluconacetobacter oboediens 174Bp2 (isolated from vinegar) J Bacteriol. 2011;193:2670–2671. doi: 10.1128/JB.00229-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés-Barrao C, Saad MM, Cabello Ferrete E, Bravo D, Chappuis ML, Ortega Pérez R, Junier P, Perret X, Barja F. Metaproteomics and ultrastructure characterization of Komagataeibacter spp. involved in high-acid spirit vinegar production. Food Microbiol. 2016;55:112–122. doi: 10.1016/j.fm.2015.10.012. [DOI] [PubMed] [Google Scholar]
- Augimeri RV, Strap JL. The phytohormone ethylene enhances cellulose production, regulates CRP/FNRKx transcription and causes differential gene expression within the bacterial cellulose synthesis operon of Komagataeibacter (Gluconacetobacter) xylinus ATCC 53582. Front Microbiol. 2015;6:1459. doi: 10.3389/fmicb.2015.01459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azeredo HMC, Barud H, Farinas CS, Vasconcellos VM, Claro AM (2019) Bacterial cellulose as a raw material for food and food packaging applications. Front Sustain Food Syst 3. 10.3389/fsufs.2019.00007
- Bae SO, Sugano Y, Ohi K, Shoda M. Features of bacterial cellulose synthesis in a mutant generated by disruption of the diguanylate cyclase 1 gene of Acetobacter xylinum BPR 2001. Appl Microbiol Biotechnol. 2004;65:315–322. doi: 10.1007/s00253-004-1593-7. [DOI] [PubMed] [Google Scholar]
- Battad-Bernardo E, McCrindle SL, Couperwhite I, Neilan BA. Insertion of an E. coli lacZ gene in Acetobacter xylinus for the production of cellulose in whey. FEMS Microbiol Lett. 2004;231:253–260. doi: 10.1016/S0378-1097(04)00007-2. [DOI] [PubMed] [Google Scholar]
- Boesch C, Trček J, Sievers M, Teuber M. Acetobacter intermedius, sp. nov. Syst Appl Microbiol. 1998;21:220–229. doi: 10.1016/S0723-2020(98)80026-X. [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro C, Cleenwerck I, Trček J, Zuluaga R, de Vos P, Caro G, Aguirre R, Putaux JL, Gañá P. Gluconacetobacter medellinensis sp. nov., cellulose- and non-cellulose-producing acetic acid bacteria isolated from vinegar. Int J Syst Evol Microbiol. 2013;63:1119–1125. doi: 10.1099/ijs.0.043414-0. [DOI] [PubMed] [Google Scholar]
- Christgen SL, Becker DF. Role of proline in pathogen and host interactions. Antioxid Redox Signal. 2019;30:683–709. doi: 10.1089/ars.2017.7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coucheron DH. An Acetobacter xylinum insertion sequence element associated with inactivation of cellulose production. J Bacteriol. 1991;173:5723–5731. doi: 10.1128/JB.173.18.5723-5731.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coucheron DH. A family of IS1031 elements in the genome of Acetobacter xylinum: nucleotide sequences and strain distribution. Mol Microbiol. 1993;9:211–218. doi: 10.1111/j.1365-2958.1993.tb01682.x. [DOI] [PubMed] [Google Scholar]
- de Souza SS, de Vasconcellos CJ, Porto LM (2018) Modeling the core metabolism of Komagataeibacter hansenii ATCC 23769 to evaluate nanocellulose biosynthesis. Braz J Chem Eng 35(3). 10.1590/0104-6632.20180353s20170327
- De Wulf P, Joris K, Vandamme EJ (1996) Improved cellulose formation by an Acetobacter xylinum mutant limited in (keto)gluconate synthesis. J Chem Technol Biotechnol 67:376–380. 10.1002/(SICI)1097-4660(199612)67:4<376::AID-JCTB569>3.0.CO;2-J
- Dellaglio F, Cleenwerck I, Felis GE, Engelbeen K, Janssens D, Marzotto M. Description of Gluconacetobacter swingsii sp. nov. and Gluconacetobacter rhaeticus sp. nov., isolated from Italian apple fruit. Int J Syst Evol Microbiol. 2005;55:2365–2370. doi: 10.1099/ijs.0.63301-0. [DOI] [PubMed] [Google Scholar]
- Deloger M, El Karoui M, Petit M-A. A genomic distance based on MUM indicates discontinuity between most bacterial species and genera. J Bacteriol. 2009;191:91–99. doi: 10.1128/JB.01202-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Nagachar N, Xiao C, Tien M, Kao TH. Identification and characterization of non-cellulose-producing mutants of Gluconacetobacter hansenii generated by Tn5 transposon mutagenesis. J Bacteriol. 2013;195:5072–5083. doi: 10.1128/JB.00767-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Nagachar N, Fang L, Luan X, Catchmark JM, Tien M, Kao TH. Isolation and characterization of two cellulose morphology mutants of Gluconacetobacter hansenii ATCC 23769 producing cellulose with lower crystallinity. PLoS One. 2015;10:e0119504. doi: 10.1371/journal.pone.0119504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Catchmark JM. Characterization of water-soluble exopolysaccharides from Gluconacetobacter xylinus and their impacts on bacterial cellulose crystallization and ribbon assembly. Cellulose. 2014;21:3965–3978. doi: 10.1007/s10570-014-0443-8. [DOI] [Google Scholar]
- Fang L, Catchmark JM. Characterization of cellulose and other exopolysaccharides produced from Gluconacetobacter strains. Carbohydr Polym. 2015;115:663–669. doi: 10.1016/j.carbpol.2014.09.028. [DOI] [PubMed] [Google Scholar]
- Fang J, Kawano S, Tajima K, Kondo T. In vivo curdlan/cellulose bionanocomposite synthesis by genetically modified Gluconacetobacter xylinus. Biomacromolecules. 2015;16:3154–3160. doi: 10.1021/acs.biomac.5b01075. [DOI] [PubMed] [Google Scholar]
- Florea M, Hagemann H, Santosa G, Abbott J, Micklem CN, Spencer-Milnes X, De Arroyo GL, Paschou D, Lazenbatt C, Kong D, Chughtai H, Jensen K, Freemont PS, Kitney R, Reeve B, Ellis T. Engineering control of bacterial cellulose production using a genetic toolkit and a new cellulose producing strain. Proc Natl Acad Sci U S A. 2016;113:E3431–E3440. doi: 10.1073/pnas.1522985113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Komoda K, Sakurai N, Tajima K, Tanaka I, Yao M. The c-di-GMP recognition mechanism of the PilZ domain of bacterial cellulose synthase subunit A. Biochem Biophys Res Commun. 2013;431:802–807. doi: 10.1016/j.bbrc.2012.12.103. [DOI] [PubMed] [Google Scholar]
- García C, Prieto MA. Bacterial cellulose as a potential bioleather substitute for the footwear industry. Microb Biotechnol. 2019;12:582–585. doi: 10.1111/1751-7915.13306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenholm P, Höglund K, Johannesson S, Puchades M, Brackmann C, Enejder A, Olsson L (2013) Effect of cultivation conditions on the structure and morphological properties of BNC biomaterials with a focus on vascular grafts. In: Gama M, Paul Gatenholm DK (eds) Bacterial nanocellulose: a sophisticated multifunctional material. CRC Press Taylor & Francis Group, pp 19–42
- Gorgieva S, Trček J. Bacterial cellulose: production, modification and perspectives in biomedical applications. Nanomaterials. 2019;9:E1352. doi: 10.3390/nano9101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- Gosselé F, Swings J, Kersters K, Pauwels P, De Ley J. Numerical analysis of phenotypic features and protein gel electrophoregrams of a wide variety of Acetobacter strains. Proposal for the improvement of the taxonomy of the genus Acetobacter Beijerinck 1898, 215. Syst Appl Microbiol. 1983;4:338–368. doi: 10.1016/S0723-2020(83)80020-4. [DOI] [PubMed] [Google Scholar]
- Gu C, Kim GB, Kim WJ, Kim HU, Lee SY. Current status and applications of genome-scale metabolic models. Genome Biol. 2019;20:121. doi: 10.1186/s13059-019-1730-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwon H, Park K, Chung SC, Kim RH, Kang JK, Ji SM, Kim NJ, Lee S, Ku JH, Do EC, Park S, Kim M, Shim WY, Rhee HS, Kim JY, Kim J, Kim TY, Yamaguchi Y, Iwamuro R, Saito S, Kim G, Jung IS, Park H, Lee C, Lee S, Jeon WS, Jang WD, Kim HU, Lee SY, Im D, Doo SG, Lee SY, Lee HC, Park JH. A safe and sustainable bacterial cellulose nanofiber separator for lithium rechargeable batteries. Proc Natl Acad Sci U S A. 2019;116:19288–19293. doi: 10.1073/pnas.1905527116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Arriaga AM, del Cerro C, Urbina L, Eceiza A, Corcuera MA, Retegi A, Auxiliadora Prieto M. Genome sequence and characterization of the bcs clusters for the production of nanocellulose from the low pH resistant strain Komagataeibacter medellinensis ID13488. Microb Biotechnol. 2019;12:620–632. doi: 10.1111/1751-7915.13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S-Q, Gao Y-G, Tajima K, Sunagawa N, Zhou Y, Kawano S, Fujiwara T, Yoda T, Shimura D, Satoh Y, others (2010) Structure of bacterial cellulose synthase subunit D octamer with four inner passageways. Proc Natl Acad Sci 107:17957–17961 [DOI] [PMC free article] [PubMed]
- Hu W, Chen S, Yang J, Li Z, Wang H. Functionalized bacterial cellulose derivatives and nanocomposites. Carbohydr Polym. 2014;101:1043–1060. doi: 10.1016/j.carbpol.2013.09.102. [DOI] [PubMed] [Google Scholar]
- Iino T, Suzuki R, Tanaka N, Kosako Y, Ohkuma M, Komagata K, Uchimura T. Gluconacetobacter kakiaceti sp. nov., an acetic acid bacterium isolated from a traditional Japanese fruit vinegar. Int J Syst Evol Microbiol. 2012;62:1465–1469. doi: 10.1099/ijs.0.031773-0. [DOI] [PubMed] [Google Scholar]
- Ishida T, Sugano Y, Nakai T, Shoda M. Effects of acetan on production of bacterial cellulose by Acetobacter xylinum. Biosci Biotechnol Biochem. 2002;66:1677–1681. doi: 10.1271/bbb.66.1677. [DOI] [PubMed] [Google Scholar]
- Islam MU, Ullah MW, Khan S, Shah N, Park JK. Strategies for cost-effective and enhanced production of bacterial cellulose. Int J Biol Macromol. 2017;102:1166–1173. doi: 10.1016/j.ijbiomac.2017.04.110. [DOI] [PubMed] [Google Scholar]
- Iyer PR, Geib SM, Catchmark J, Kao T, Tien M. Genome sequence of a cellulose-producing bacterium, Gluconacetobacter hansenii ATCC 23769. J Bacteriol. 2010;192:4256–4257. doi: 10.1128/JB.00588-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacek P, Szustak M, Kubiak K, Gendaszewska-Darmach E, Ludwicka K, Bielecki S. Scaffolds for chondrogenic cells cultivation prepared from bacterial cellulose with relaxed fibers structure induced genetically. Nanomaterials. 2018;8:E1066. doi: 10.3390/NANO8121066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacek P, Dourado F, Gama M, Bielecki S. Molecular aspects of bacterial nanocellulose biosynthesis. Microb Biotechnol. 2019;12:633–649. doi: 10.1111/1751-7915.13386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacek P, Kubiak K, Ryngajłło M, Rytczak P, Paluch P, Bielecki S. Modification of bacterial nanocellulose properties through mutation of motility related genes in Komagataeibacter hansenii ATCC 53582. New Biotechnol. 2019;52:60–68. doi: 10.1016/j.nbt.2019.05.004. [DOI] [PubMed] [Google Scholar]
- Jacek P, Ryngajłło M, Bielecki S. Structural changes of bacterial nanocellulose pellicles induced by genetic modification of Komagataeibacter hansenii ATCC 23769. Appl Microbiol Biotechnol. 2019;103:5339–5353. doi: 10.1007/s00253-019-09846-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang WD, Hwang JH, Kim HU, Ryu JY, Lee SY. Bacterial cellulose as an example product for sustainable production and consumption. Microb Biotechnol. 2017;10:1181–1185. doi: 10.1111/1751-7915.12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang WD, Kim TY, Kim HU, Shim WY, Ryu JY, Park JH, Lee SY. Genomic and metabolic analysis of Komagataeibacter xylinus DSM 2325 producing bacterial cellulose nanofiber. Biotechnol Bioeng. 2019;116:3372–3381. doi: 10.1002/bit.27150. [DOI] [PubMed] [Google Scholar]
- Jedrzejczak-Krzepkowska M, Kubiak K, Ludwicka K, Bielecki S (2016) Chapter 2—bacterial nanocellulose synthesis, recent findings. In: Gama M, Dourado F, Bielecki S (eds) Bacterial nanocellulose: from biotechnology to bio-economy. Elsevier, pp 19–46
- Jozala AF, de Lencastre-Novaes LC, Lopes AM, de Carvalho S-EV, Mazzola PG, Pessoa-Jr A, Grotto D, Gerenutti M, Chaud MV. Bacterial nanocellulose production and application: a 10-year overview. Appl Microbiol Biotechnol. 2016;100:2063–2072. doi: 10.1007/s00253-015-7243-4. [DOI] [PubMed] [Google Scholar]
- Kranz A, Busche T, Vogel A, Usadel B, Kalinowski J, Bott M, Polen T. RNAseq analysis of α-proteobacterium Gluconobacter oxydans 621H. BMC Genomics. 2018;19:24. doi: 10.1186/s12864-017-4415-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystynowicz A, Czaja W, Wiktorowska-Jezierska A, Gonçalves-Miśkiewicz M, Turkiewicz M, Bielecki S. Factors affecting the yield and properties of bacterial cellulose. J Ind Microbiol Biotechnol. 2002;29:189–195. doi: 10.1038/sj.jim.7000303. [DOI] [PubMed] [Google Scholar]
- Krystynowicz A, Koziolkiewicz M, Wiktorowska-Jezierska A, Bielecki S, Klemenska E, Masny A, Plucienniczak A. Molecular basis of cellulose biosynthesis disappearance in submerged culture of Acetobacter xylinum. ACTA Biochim Pol Ed. 2005;52:691–698. doi: 10.18388/abp.2005_3432. [DOI] [PubMed] [Google Scholar]
- Kubiak K, Kurzawa M, Jędrzejczak-Krzepkowska M, Ludwicka K, Krawczyk M, Migdalski A, Kacprzak MM, Loska D, Krystynowicz A, Bielecki S. Complete genome sequence of Gluconacetobacter xylinus E25 strain—valuable and effective producer of bacterial nanocellulose. J Biotechnol. 2014;176:18–19. doi: 10.1016/j.jbiotec.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Kuo CH, Teng HY, Lee CK. Knock-out of glucose dehydrogenase gene in Gluconacetobacter xylinus for bacterial cellulose production enhancement. Biotechnol Bioprocess Eng. 2015;20:18–25. doi: 10.1007/s12257-014-0316-x. [DOI] [Google Scholar]
- Li Y, Tian J, Tian H, Chen X, Ping W, Tian C, Lei H. Mutation-based selection and analysis of Komagataeibacter hansenii HDM1-3 for improvement in bacterial cellulose production. J Appl Microbiol. 2016;121:1323–1334. doi: 10.1111/jam.13244. [DOI] [PubMed] [Google Scholar]
- Li Y, Yan P, Lei Q, Li B, Sun Y, Li S, Lei H, Xie N. Metabolic adaptability shifts of cell membrane fatty acids of Komagataeibacter hansenii HDM1-3 improve acid stress resistance and survival in acidic environments. J Ind Microbiol Biotechnol. 2019;46:1491–1503. doi: 10.1007/s10295-019-02225-y. [DOI] [PubMed] [Google Scholar]
- Lisdiyanti P, Navarro RR, Uchimura T, Komagata K. Reclassification of Gluconacetobacter hansenii strains and proposals of Gluconacetobacter saccharivorans sp. nov. and Gluconacetobacter nataicola sp. nov. Int J Syst Evol Microbiol. 2006;56:2101–2111. doi: 10.1099/ijs.0.63252-0. [DOI] [PubMed] [Google Scholar]
- Liu M, Zhong C, Wu XY, Wei YQ, Bo T, Han PP, Jia SR. Metabolomic profiling coupled with metabolic network reveals differences in Gluconacetobacter xylinus from static and agitated cultures. Biochem Eng J. 2015;101:85–98. doi: 10.1016/j.bej.2015.05.002. [DOI] [Google Scholar]
- Liu LX, Liu SX, Wang YM, Bi JC, Chen HM, Deng J, Zhang C, Hu QS, Li CF. Komagataeibacter cocois sp. nov., a novel cellulose-producing strain isolated from coconut milk. Int J Syst Evol Microbiol. 2018;68:3125–3131. doi: 10.1099/ijsem.0.002947. [DOI] [PubMed] [Google Scholar]
- Liu M, Li S, Xie Y, Jia S, Hou Y, Zou Y, Zhong C. Enhanced bacterial cellulose production by Gluconacetobacter xylinus via expression of Vitreoscilla hemoglobin and oxygen tension regulation. Appl Microbiol Biotechnol. 2018;102:1155–1165. doi: 10.1007/s00253-017-8680-z. [DOI] [PubMed] [Google Scholar]
- Matsutani M, Ito K, Azuma Y, Ogino H, Shirai M, Yakushi T, Matsushita K. Adaptive mutation related to cellulose producibility in Komagataeibacter medellinensis (Gluconacetobacter xylinus) NBRC 3288. Appl Microbiol Biotechnol. 2015;99:7229–7240. doi: 10.1007/s00253-015-6598-x. [DOI] [PubMed] [Google Scholar]
- Nakai T, Tonouchi N, Konishi T, Kojima Y, Tsuchida T, Yoshinaga F, Sakai F, Hayashi T. Enhancement of cellulose production by expression of sucrose synthase in Acetobacter xylinum. Proc Natl Acad Sci U S A. 1999;96:14–18. doi: 10.1073/pnas.96.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai T, Nishiyama Y, Kuga S, Sugano Y, Shoda M. ORF2 gene involves in the construction of high-order structure of bacterial cellulose. Biochem Biophys Res Commun. 2002;295:458–462. doi: 10.1016/S0006-291X(02)00696-4. [DOI] [PubMed] [Google Scholar]
- Nakai T, Sugano Y, Shoda M, Sakakibara H, Oiwa K, Tuzi S, Imai T, Sugiyama J, Takeuchi M, Yamauchi D, Mineyukia Y. Formation of highly twisted ribbons in a carboxymethylcellulase gene-disrupted strain of a cellulose-producing bacterium. J Bacteriol. 2013;195:958–964. doi: 10.1128/JB.01473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naloka K, Yukphan P, Matsutani M, Matsushita K, Theeragool G. Komagataeibacter diospyri sp. nov., a novel species of thermotolerant bacterial nanocellulose-producing bacterium. Int J Syst Evol Microbiol. 2019;70:251–258. doi: 10.1099/ijsem.0.003745. [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Flanagan B, Mikkelsen D, Ramirez S, Rivas L, Gidley MJ, Dykes GA. Spontaneous mutation results in lower cellulose production by a Gluconacetobacter xylinus strain from Kombucha. Carbohydr Polym. 2010;80:337–343. doi: 10.1016/j.carbpol.2009.11.019. [DOI] [Google Scholar]
- Ogino H, Azuma Y, Hosoyama A, Nakazawa H, Matsutani M, Hasegawa A, Otsuyama K, Matsushita K, Fujita N, Shirai M. Complete genome sequence of NBRC 3288, a unique cellulose-nonproducing strain of Gluconacetobacter xylinus isolated from vinegar. J Bacteriol. 2011;193:6997–6998. doi: 10.1128/JB.06158-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson C, Jansson H, Swenson J. The role of trehalose for the stabilization of proteins. J Phys Chem B. 2016;120:4723–4731. doi: 10.1021/acs.jpcb.6b02517. [DOI] [PubMed] [Google Scholar]
- Park JK, Jung JY, Park YH. Cellulose production by Gluconacetobacter hansenii in a medium containing ethanol. Biotechnol Lett. 2003;25:2055–2059. doi: 10.1023/B:BILE.0000007065.63682.18. [DOI] [PubMed] [Google Scholar]
- Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods. 2016;8:12–24. doi: 10.1039/C5AY02550H. [DOI] [Google Scholar]
- Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U. Molecular biology of cellulose production in bacteria. Res Microbiol. 2002;153:205–212. doi: 10.1016/S0923-2508(02)01316-5. [DOI] [PubMed] [Google Scholar]
- Römling U. Cyclic di-GMP, an established secondary messenger still speeding up. Environ Microbiol. 2012;14:1817–1829. doi: 10.1111/j.1462-2920.2011.02617.x. [DOI] [PubMed] [Google Scholar]
- Römling U, Galperin MY. Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol. 2015;23:545–557. doi: 10.1016/j.tim.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P, Aloni Y, Weinhouse C, Michaeli D, Weinberger-Ohana P, Meyer R, Benziman M. An unusual guanyl oligonucleotide regulates cellulose synthesis in Acetobacter xylinum. FEBS Lett. 1985;186:191–196. doi: 10.1016/0014-5793(85)80706-7. [DOI] [PubMed] [Google Scholar]
- Ross P, Mayer R, Benziman M. Cellulose biosynthesis and function in bacteria. Microbiol Rev. 1991;55:35–58. doi: 10.1128/MMBR.55.1.35-58.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhal R, Kataria R, Choudhury B. Trends in bacterial trehalose metabolism and significant nodes of metabolic pathway in the direction of trehalose accumulation. Microb Biotechnol. 2013;6:493–502. doi: 10.1111/1751-7915.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryngajłło M, Jacek P, Cielecka I, Kalinowska H, Bielecki S. Effect of ethanol supplementation on the transcriptional landscape of bionanocellulose producer Komagataeibacter xylinus E25. Appl Microbiol Biotechnol. 2019;103:6673–6688. doi: 10.1007/s00253-019-09904-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryngajłło M, Kubiak K, Jędrzejczak-Krzepkowska M, Jacek P, Bielecki S. Comparative genomics of the Komagataeibacter strains—efficient bionanocellulose producers. Microbiologyopen. 2019;8:e00731. doi: 10.1002/mbo3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado L, Blank S, Esfahani RAM, Strap JL, Bonetta D. Missense mutations in a transmembrane domain of the Komagataeibacter xylinus BcsA lead to changes in cellulose synthesis. BMC Microbiol. 2019;19:216. doi: 10.1186/s12866-019-1577-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena IM, Kudlicka K, Okuda K, Brown RM. Characterization of genes in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum: implications for cellulose crystallization. J Bacteriol. 1994;176:5735–5752. doi: 10.1128/JB.176.18.5735-5752.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller G, Hertel C, Hammes WP. Gluconacetobacter entanii sp. nov., isolated from submerged high-acid industrial vinegar fermentations. Int J Syst Evol Microbiol. 2000;6:2013–2020. doi: 10.1099/00207713-50-6-2013. [DOI] [PubMed] [Google Scholar]
- Setyawati MI, Chien LJ, Lee CK. Expressing Vitreoscilla hemoglobin in statically cultured Acetobacter xylinum with reduced O2 tension maximizes bacterial cellulose pellicle production. J Biotechnol. 2007;132:38–43. doi: 10.1016/j.jbiotec.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Setyawati MI, Chien LJ, Lee CK. Self-immobilized recombinant Acetobacter xylinum for biotransformation. Biochem Eng J. 2009;43:78–84. doi: 10.1016/j.bej.2008.09.002. [DOI] [Google Scholar]
- Shigematsu T, Takamine K, Kitazato M, Morita T, Naritomi T, Morimura S, Kida K. Cellulose production from glucose using a glucose dehydrogenase gene (gdh)-deficient mutant of Gluconacetobacter xylinus and its use for bioconversion of sweet potato pulp. J Biosci Bioeng. 2005;99:415–422. doi: 10.1263/jbb.99.415. [DOI] [PubMed] [Google Scholar]
- Sievers M, Sellmer S, Teuber M. Acetobacter europaeus sp. nov., a main component of industrial vinegar fermenters in Central Europe. Syst Appl Microbiol. 1992;15:386–392. doi: 10.1016/S0723-2020(11)80212-2. [DOI] [Google Scholar]
- Singer MA, Lindquist S. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol Cell. 1998;1:639–648. doi: 10.1016/S1097-2765(00)80064-7. [DOI] [PubMed] [Google Scholar]
- Škraban J, Cleenwerck I, Vandamme P, Fanedl L, Trček J. Genome sequences and description of novel exopolysaccharides producing species Komagataeibacter pomaceti sp. nov. and reclassification of Komagataeibacter kombuchae (Dutta and Gachhui 2007) Yamada et al., 2013 as a later heterotypic synonym of Komagataeib. Syst Appl Microbiol. 2018;41:581–592. doi: 10.1016/j.syapm.2018.08.006. [DOI] [PubMed] [Google Scholar]
- Slapšak N, Cleenwerck I, De Vos P, Trček J. Gluconacetobacter maltaceti sp. nov., a novel vinegar producing acetic acid bacterium. Syst Appl Microbiol. 2013;36:17–21. doi: 10.1016/j.syapm.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Sokollek SJ, Hertel C, Hammes WP. Description of Acetobacter oboediens sp. nov. and Acetobacter pomorum sp. nov., two new species isolated from industrial vinegar fermentations. Int J Syst Bacteriol. 1998;48:935–940. doi: 10.1099/00207713-48-3-935. [DOI] [PubMed] [Google Scholar]
- Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, Cohen A, Sapir S, Ohana P, Benziman M. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol. 1998;180:4416–4425. doi: 10.1128/JB.180.17.4416-4425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taweecheep P, Naloka K, Matsutani M, Yakushi T, Matsushita K, Theeragool G. In vitro thermal and ethanol adaptations to improve vinegar fermentation at high temperature of Komagataeibacter oboediens MSKU 3. Appl Biochem Biotechnol. 2019;189:144–159. doi: 10.1007/s12010-019-03003-3. [DOI] [PubMed] [Google Scholar]
- Taweecheep P, Naloka K, Matsutani M, Yakushi T, Matsushita K, Theeragool G. Superfine bacterial nanocellulose produced by reverse mutations in the bcsC gene during adaptive breeding of Komagataeibacter oboediens. Carbohydr Polym. 2019;226:115243. doi: 10.1016/j.carbpol.2019.115243. [DOI] [PubMed] [Google Scholar]
- Teh MY, Ooi KH, Danny Teo SX, Bin Mansoor ME, Shaun Lim WZ, Tan MH. An expanded synthetic biology toolkit for gene expression control in Acetobacteraceae. ACS Synth Biol. 2019;8:708–723. doi: 10.1021/acssynbio.8b00168. [DOI] [PubMed] [Google Scholar]
- Tonouchi N, Tsuchida T, Yoshlnaga F, Horinouchi S, Beppu T, Beppu T. A host–vector system for a cellulose-producing Acetobacter strain. Biosci Biotechnol Biochem. 1994;58:1899–1901. doi: 10.1271/bbb.58.1899. [DOI] [PubMed] [Google Scholar]
- Tonouchi N, Horinouchi S, Tsuchida T, Yoshinaga F. Increased cellulose production from sucrose by Acetobacter after introducing the sucrose phosphorylase gene. Biosci Biotechnol Biochem. 1998;62:1778–1780. doi: 10.1271/bbb.62.1778. [DOI] [PubMed] [Google Scholar]
- Tonouchi N, Yanase H, Kojima Y, Tsuchida T, Yoshinaga F, Horinouchi S. Increased cellulose production from sucrose with reduced levan accumulation by an Acetobacter strain harboring a recombinant plasmid. Biosci Biotechnol Biochem. 1998;62:833–836. doi: 10.1271/bbb.62.833. [DOI] [PubMed] [Google Scholar]
- Toyosaki H, Kojima Y, Tsuchida T, Hoshino KI, Yamada W, Yoshinaga F. The characterization of an acetic acid bacterium useful for producing bacterial cellulose in agitation cultures: the proposal of Acetobacter xylinum subsp. sucrofermentans subsp. nov. J Gen Appl Microbiol. 1995;41:307–314. doi: 10.2323/jgam.41.307. [DOI] [Google Scholar]
- Umeda Y, Hirano A, Ishibashi M, Akiyama H, Onizuka T, Ikeuchi M, Inoue Y. Cloning of cellulose synthase genes from Acetobacter xylinum JCM 7664: implication of a novel set of cellulose synthase genes. DNA Res. 1999;6:109–115. doi: 10.1093/dnares/6.2.109. [DOI] [PubMed] [Google Scholar]
- Velasco-Bedrán H, López-Isunza F. The unified metabolism of Gluconacetobacter entanii in continuous and batch processes. Process Biochem. 2007;42:1180–1190. doi: 10.1016/j.procbio.2007.05.017. [DOI] [Google Scholar]
- Walker KT, Goosens VJ, Das A, Graham AE, Ellis T. Engineered cell-to-cell signalling within growing bacterial cellulose pellicles. Microb Biotechnol. 2018;12:611–619. doi: 10.1111/1751-7915.13340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tavakoli J, Tang Y. Bacterial cellulose production, properties and applications with different culture methods—a review. Carbohydr Polym. 2019;219:63–76. doi: 10.1016/j.carbpol.2019.05.008. [DOI] [PubMed] [Google Scholar]
- Yadav V, Paniliatis BJ, Shi H, Lee K, Cebe P, Kaplan DL. Novel in vivo-degradable cellulose–chitin copolymer from metabolically engineered Gluconacetobacter xylinus. Appl Environ Microbiol. 2010;76:6257–6265. doi: 10.1128/AEM.00698-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Yukphan P, Lan Vu HT, Muramatsu Y, Ochaikul D, Tanasupawat S, Nakagawa Y. Description of Komagataeibacter gen. nov., with proposals of new combinations (Acetobacteraceae) J Gen Appl Microbiol. 2012;58:397–404. doi: 10.2323/jgam.58.397. [DOI] [PubMed] [Google Scholar]
- Zhang L, Alfano JR, Becker DF. Proline metabolism increases katG expression and oxidative stress resistance in Escherichia coli. J Bacteriol. 2015;197:431–440. doi: 10.1128/JB.02282-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ye C, Xu N, Chen C, Chen X, Yuan F, Xu Y, Yang J, Sun D. Reconstruction of a genome-scale metabolic network of Komagataeibacter nataicola RZS01 for cellulose production. Sci Rep. 2017;7:7911. doi: 10.1038/s41598-017-06918-1. [DOI] [PMC free article] [PubMed] [Google Scholar]