Abstract
The effectiveness of two different rapid methods involving the 3M™ molecular detection assay Listeria and the 3M™ Petrifilm environmental Listeria Plate were evaluated for the rapid detection of Listeria from naturally contaminated vegetables and chicken-processing environments against the standard culture-based method. A total of 178 samples were examined for the presence of Listeria. A total of 47/178 (26.4%) by standard ISO culture-based method (EN ISO 11290-1), 42/178 (23.6%) by 3M™ MDA Listeria and 40/178 (22.5%) by 3M™ Petrifilm EL Plate showed positive results, respectively. The accuracy, sensitivity, specificity, positive predictive value, and negative predictive value for 3M™ MDA Listeria and 3M™ Petrifilm EL Plate were 97.2, 89.4, 99.3, 97.7, 96.4% and 96.1, 85.1, 100.0, 100.0, 94.9%, respectively. Based on the Cohen’s Kappa value, there was a complete and robust concordance between 3M™ MDA Listeria (0.911) and 3M™ Petrifilm EL Plates (0.894) as compared to the standard culture-based method.
Keywords: 3M™ MDA listeria, 3M™ petrifilm EL plate, Listeria
Introduction
The genus Listeria is a foodborne bacterium with a significant burden on both food industry and public health services. Over the years, there has been an increased concern about Listeria and its role in food safety worldwide. Most cases of Listeria outbreak have been reported from industrial countries which include Spain, Austria, and Germany (De Castro et al., 2012; Fretz et al., 2010). Reports from East Asia and other developing countries are scarce, perhaps due to the under-diagnosed and under-reported cases (Ponniah et al., 2010). In the United States, more than 2000 cases of listeriosis are estimated to occur yearly, with over 250 deaths (Scallan et al., 2011). The economic burden of Listeria infection, including costs of health care, productivity loss, and declined quality of life, is estimated to a total of USD 2.04 billion and the cost per case is expected to be nearly USD 1,282,000 (Byrd-Bredbenner et al., 2013). On top of that, sales of recalled brands declined around 22% after recalls due to Listeria (Thomsen et al., 2006).
The ubiquitous nature of Listeria in a different environment and the ability of Listeria spp. to grow or survive under extreme environmental conditions (pH and temperature) enable this bacteria to proliferate in various types of food products, including vegetables, poultry, cheese, ice cream, raw milk, ready-to-eat foods, meat and fish foodstuffs (Churchill et al., 2006; Pesavento et al., 2010). As this pathogen causes serious illness in humans, many countries have adopted a zero-tolerance policy or stringent requirements in foods (Yue, 2014). These stringent requirements by regulatory agencies have provided an impetus to develop detection methods for Listeria which are rapid, accurate and sensitive. Detection of Listeria is usually used to verify sanitisation programs and as an indicator of possible contamination by pathogenic Listeria monocytogenes (3M™ Microbiology, 2012). Faster and more accurate detection of Listeria can prevent the introduction of contaminated foods into the markets and recalls (Jongvanich, 2016).
The conventional methods for detecting Listeria using the US FDA’s Bacteriological Analytical Manual (BAM) and EN ISO 11290-1 standard methods are relatively time-consuming, requiring 3–5 days, laborious, expensive and may not be sensitive to meet market requirements (Hitchins and Jinneman, 2003; Lee et al., 2015). To overcome these drawbacks of culture-based methods, rapid and molecular-based detection methods; such as polymerase chain reaction (PCR) and enzyme-linked immunosorbent assays (ELISA) have been developed (Oktay and Heperkan, 2006; Feldsine et al., 2008).
Recently, user-friendly rapid detection methods, such as the 3M™ molecular detection assay Listeria (3M™ MDA or commercialized LAMP-based system) and 3M™ Petrifilm™ environmental Listeria (EL) plate methods have been reported to be useful as screening tools for the detection of Listeria in a variety of foods and related processing environment (Nyachuba and Donnelly, 2007; Bird et al., 2015). The 3M™ MDA system is a recent innovation that has been significant in the detection of common foodborne pathogens such as Escherichia coli O157: H7, Listeria, and Salmonella (Loff et al., 2014). 3M™ MDA utilises the combination of loop-mediated isothermal amplification assay and bioluminescence for the real-time qualitative detection of Listeria (Bird et al., 2015). This is the first commercially available assay of its kind that combines two technologies. 3M™ MDA is relatively simple, robust, and fast method that provides presumptive results in just 29 ± 2 h of enrichment using 3M™ Demi Fraser broth base or 3M™ modified Listeria recovery broth. After enrichment, samples are analysed using the 3M™ MDA Listeria, and the expected positive results are displayed in real-time, while negative results are shown after completion of the assay (75 min) on the 3M™ molecular detection system (Bird et al., 2015).
The 3M™ Petrifilm™ EL Plate, a non-enrichment alternative method, overcome some of the shortcomings of culture-based methods and reduces the high cost of the molecular technique. The 3M™ petrifilm EL plate method can be used for the detection of potentially injured Listeria on the food processing environmental surfaces (Nyachuba and Donnelly, 2007).
Moreover, both 3M™ MDA and 3M™ Petrifilm™ EL plate have been validated as a First Action Official MethodsSM through AOAC International (Benesh et al., 2013; Bird et al., 2015). Studies have been performed on this system with various foods in the USA and Europe (Yue, 2014). However, whether these methods are effective for faster and reliable detection of Listeria in naturally-contaminated leafy vegetables, chicken, and their related processing environments has not been established. This study aimed to evaluate 3M™ MDA Listeria and 3M™ Petrifilm™ EL plate for Listeria detection in naturally contaminated leafy vegetables, chicken carcasses and environmental samples obtained from selected fresh markets against the standard ISO culture-based method (EN ISO 11290-1).
Materials and methods
Sample collection
A total of 178 samples of raw leafy vegetables, chicken and their related processing environments were collected from different fresh markets in Kedah and Penang, Malaysia from November 2015 to April 2016. The vegetables selected were amaranth green (Amaranthus dubius), bean sprout (Vigna radiata), amaranth red (Amaranthus tricolor), coriander (Coriandrum sativum), green leaf lettuce (Lactuca sativa), iceberg lettuce (Lactuca sativa), mint (Mentha arvensis), spring onion (Allium fistulosum), winged bean (Psophocarpus tetragonolobus), laksa leaves (Poligonum minus) Indian pennywort (Centenella asiatica), Japanese parsley (Oenanthe stolonifera), wild parsley (Cosmos caudatus), water spinach (Ipomoea aquatica), Chinese flowering cabbage (Brassica rapa var. parachinensis) and sweet basil (Ocimum basilicum). These vegetables were collected on sterile plastic bags. Both whole chicken carcasses and chicken cuts were sampled using the swab sampling method described by Gill et al. (2005) using 3M™ sponge-sticks to swab the inner-outer surfaces of the samples. For environmental samples (floor, butcher apron, chopping board, transport crate, knife, display table, drum, defeathering machine, drain crevices, and cage) were swabbed correctly based on the manual instructions using the 3M™ sponge-sticks. Surfaces of samples were swabbed concurrently with two swabs as 3M™ MDA Listeria and culture-based methods required pre-enrichment in 3M™ Demi Fraser broth while the 3M™ Petrifilm EL plate required primary enrichment in buffered peptone water (BPW). Sterile Schott Duran® bottles were used for the collection of all the water samples (wash water, scalding tank water, bench water, and drain water). Samples were transported to the laboratory on ice in polystyrene box and processed immediately upon arrival to the laboratory.
Culture-based method
Isolation and identification of Listeria were carried out using the modified EN ISO 11290-1 reference method as described by Becker et al. (2006). Briefly, samples (25 g/25 ml/3M™ sponge-sticks) were enriched with 225 ml of 3M™ Demi Fraser broth (primary enrichment) in a stomacher bag and 3M™ sample collection bag, respectively. The samples were thoroughly homogenised using a stomacher (Seward Stomacher® 400C) followed by incubation at 37 °C for up to 24 h. After enrichment, a loopful of primary enrichment broth was streaked onto chromogenic Listeria agar (Oxoid, UK) and incubated at 37 °C for 48 h. A hundred μl of 3M™ Demi Fraser broth was transferred to 10 ml Fraser broth (Oxoid, UK) as a secondary enrichment broth and incubated at 37 °C for 48 h. Then, a loopful of enriched Fraser broth was streaked onto Listeria selective agar (Oxoid, UK) and PALCAM agar (Oxoid, UK). These selective agars were then incubated for up to 48 h at 37 °C. Suspected colonies were picked and purified by streaking onto tryptic soy agar (Oxoid, UK) supplemented with 0.6% yeast extract (Oxoid, UK) (TSAYE), followed by incubation at 37 °C for 24 h. Well isolated colonies from TSAYE were subjected to biochemical tests, including, catalase, oxidase, urease, Listeria motility test, and triple sugar iron test for the identification confirmation.
Confirmation of Listeria isolates by Polymerase Chain Reaction
Further confirmation of Listeria was done by polymerase chain reaction (PCR). The DNA was prepared by a direct boiling suspension of cell lysates (Thong et al., 2011). The primer pair of Forward 5′GCT GAA GAG ATT GCG AAA GAA G 3′ and Reverse 5′ CAA AGA AAC CTT GGA TTT GCG G3′, was used in the amplification of a 370 bp region of the prs gene (Doumith et al., 2004). The conditions of PCR consisted of an initial denaturation of 95 °C for 4 min and 30 cycles of 95 °C, 1 min, 52 °C, 45 s, 72 °C, 2 min and a final elongation of 72 °C for 8 min. The PCR amplicon was stained with EZ-Vision® One DNA dye (Amresco, USA) on 1.0% agarose gel and was run for 55 min at 100 V, and viewed using Gel Doc XR + System (Bio-Rad, USA). A DNA molecular ladder 100 bp was used as a marker.
3M™ molecular detection assay Listeria
3M™ molecular detection assay (MDA) is an alternative method for the rapid detection of Listeria. This experiment was carried out according to the manufacturer’s protocol (www.3M.com/3MMolecularDetectionSystem). Briefly, after 28 ± 2 h of enrichment in Demi Fraser Broth, the samples were mixed thoroughly and 20 μl was transferred into lysis tube and placed in the heating block (100 ± 1 °C) for 15 min and promptly cooled in pre-chilled molecular detection chill block (3M™, USA) (− 20 °C) for 10 min. After cooling, the lysis tubes were inverted three times and left to stand at room temperature for 5 min to allow the settling of resin. Subsequently, 20 μl of the lysates were taken into the reagent tubes that contained a pellet, and it was mixed well by pipetting. The samples were then loaded onto the molecular detection speed loader tray (3M™, USA) and labelled into the molecular detection software (3M™, USA) followed by the sequence in the speed loader tray and then the speed loader tray was transferred to 3M™ molecular detection instrument. Two standard reference strains; Listeria positive control (L. monocytogenes ATCC® 19,112™) and negative control (Salmonella Typhimurium ATCC® 14028™), as well as kit control (reagent and negative) were run simultaneously in the 3M™ MDA Listeria. The detection of Listeria was in real-time, and the negative result was displayed after the 90 min run.
3M™ petrifilm™ environmental Listeria (EL) plate
Alternative 3M™ Petrifilm EL plates were used for the detection of Listeria. Approximately, 15 ml of buffered peptone water (Oxoid, UK) were added to the samples and were homogenised thoroughly, after the samples were left at room temperature (20–25 °C) for 90 min. 3M™ Petrifilm EL Plate was placed on a level surface, and 3 ml of the sample were placed onto the petrifilm by lifting the top film. The top film was gently rolled down by using 3M™ Petrifilm Flat Spreader (3M™, USA), to spread the samples evenly and also to prevent trapping of air bubbles. It was then placed on a flat surface at room temperature (20–25 °C) to allow the cold-soluble gel to form. Upon the formation of a gel, the petrifilms were incubated at 37 °C for 30 h. Red-violate colonies were considered as presumptive Listeria and streaked onto Palcam and Oxford agars and subsequently confirmed by biochemical tests and PCR.
Statistical analysis
The alternative methods were validated against ISO reference method, whereby performance indicator such as sensitivity (SE), accuracy (AC), specificity (SP), negative predictive value (NPV) and positive predictive value (PPV) were analysed as described in previous studies (Eijkelkamp et al., 2009; Abirami et al., 2016). Cohen’s kappa (κ) which explained the statistical concordance between two alternative detection methods was calculated as described by NordVal Validation Protocol for alternative microbiological purposes (Anonymous 2009) and was analysed using SPSS software (Version 22.0, IBM Corp, Armonk, NY). Kappa values were categorised as follows: < 0.01 indicates no concordance, 0.1–0.4 shows weak concordance, 0.41–0.60 for clear concordance, 0.61–0.80 for strong concordance, and 0.81–1.00 for nearly complete agreement (Viera and Garrett, 2005).
Results and discussion
Leafy vegetables, chicken and environmental samples from fresh markets were tested for Listeria by culture-based method, 3M™ MDA Listeria, and 3M™ petrifilm EL plates methods. Results in Table 1 show that 47/178 (26.4%), 42/178 (23.6%) and 40/178 (22.5%) of the samples examined were positive for Listeria by the standard culture-based method, 3M™ MDA and 3M™ petrifilm EL plates, respectively. The results for AC, SE, SP, PPV, and NPV were calculated and are presented in Table 2. The AC, SE, SP, PPV and NPV for 3M™ MDA Listeria and 3M™ petrifilm EL plates were 97.2, 89.4, 99.3, 97.7, 96.4% and 96.1, 85.1, 100.0, 100.0, 94.9%, respectively. The Kappa value for 3M™ MDA Listeria (0.831) and 3M™ petrifilm EL plates (0.797), showed that there was a strong concordance, and nearly complete agreement between and as compared to the standard EN ISO 11290-1 method.
Table 1.
Detection of Listeria in naturally contaminated samples using 3M™ MDA, 3M™ petrifilm environmental Listeria plate and EN ISO 11290-1 methods
| Samples | Number of samples/number of total samples (%) | ||||||
|---|---|---|---|---|---|---|---|
| EN ISO 11290-1 | 3M™ MDA | 3M™ petrifilm EL plate | |||||
| Positive | True positive | False positive | False negative | True positive | False positive | False negative | |
| Chicken samples | |||||||
| Chicken cuts | 1/10 (10.0) | 1/10 (10.0) | 0/10 (0) | 0/10 (0) | 1/10 (10) | 0/10 (0) | 0/10 (0) |
| Whole chicken | 2/9 (22.2) | 2/9 (22.2) | 0/9 (0) | 0/9 (0) | 2/9 (22.2) | 0/9 (0) | 0/9 (0) |
| Environmental samples | |||||||
| Display table | 2/11 (18.2) | 1/11 (9.10) | 0/11 (0) | 1/11 (9.10) | 1/11(9.10) | 0/11 (0) | 1/11 (9.10) |
| Chopping board | 3/9 (33.3) | 2/9 (22.2) | 0/9 (0) | 1/9 (11.1) | 3/9 (33.3) | 0/9 (0) | 0/9 (0) |
| Wash water | 2/9 (22.2) | 2/9 (22.2) | 0/9 (0) | 0/9 (0) | 2/9 (22.2) | 0/9 (0) | 0/9 (0) |
| Drain water | 4/9 (44.4) | 4/9 (44.4) | 0/9 (0) | 0/9 (0) | 3/9 (33.3) | 0/9 (0) | 1/9 (11.1) |
| Butcher apron | 2/8 (25.0) | 2/8 (25.0) | 0/8 (0) | 0/8 (0) | 2/8 (25.0) | 0/8 (0) | 0/8 (0) |
| Knife | 2/8 (25.0) | 2/8 (25.0) | 0/8 (0) | 0/8 (0) | 1/8 (12.5) | 0/8 (0) | 1/8 (12.5) |
| Transport crate | 2/8 (25.0) | 2/8 (25.0) | 0/8 (0) | 0/8 (0) | 2/8 (25.0) | 0/8 (0) | 0/8 (0) |
| Floor | 4/7 (57.1) | 4/7 (71.4) | 1/7 (14.2) | 0/7 (0) | 4/7 (57.1) | 0/7 (0) | 0/7 (0) |
| Scalding tank | 1/7 (14.2) | 1/7 (14.2) | 0/7 (0) | 0/7 (0) | 1/7 (14.2) | 0/7 (0) | 0/7 (0) |
| Defeathering machine | 1/7 (14.2) | 1/7 (14.2) | 0/7 (0) | 0/7 (0) | 1/7 (14.2) | 0/7 (0) | 0/7 (0) |
| Cage | 0/7 (0) | 0/7 (0) | 0/7 (0) | 0/7 (0) | 0/7 (0) | 0/7 (0) | 0/7 (0) |
| Drain crevices | 0/6 (0) | 0/6 (0) | 0/6 (0) | 0/6 (0) | 0/6 (0) | 0/6 (0) | 0/6 (0) |
| Bench wash water | 0/6 (0) | 0/6 (0) | 0/6 (0) | 0/6 (0) | 0/6 (0) | 0/6 (0) | 0/6 (0) |
| Drum | 0/5 (0) | 0/5 (0) | 0/5 (0) | 0/5 (0) | 0/5 (0) | 0/5 (0) | 0/5 (0) |
| Vegetables samples | |||||||
| Amaranth green | 2/4 (50.0) | 2/4 (50.0) | 0/4 (0) | 0/4 (0) | 2/4 (50.0) | 0/4 (0) | 0/4 (0) |
| Winged bean | 2/4 (50.0) | 2/4 (50.0) | 0/4 (0) | 0/4 (0) | 2/4 (50.0) | 0/4 (0) | 0/4 (0) |
| Laksa leaves | 1/4 (25.0) | 0/4 (0) | 0/4 (0) | 1/4 (25.0) | 0/4 (0) | 0/4 (0) | 1/4 (25.0) |
| Wild parsley | 1/4 (25.0) | 1/4 (25.0) | 0/4 (0) | 0/4 (0) | 1/4 (25.0) | 0/4 (0) | 0/4 (0) |
| Amaranth red | 1/3 (33.3) | 1/3 (33.3) | 0/3 (0) | 0/3 (0) | 1/3 (33.3) | 0/3 (0) | 0/3 (0) |
| Coriander | 2/3 (66.7) | 2/3 (66.6) | 0/3 (0) | 0/3 (0) | 2/3 (66.7) | 0/3 (0) | 0/3 (0) |
| Water spinach | 2/3 (66.7) | 2/3 (66.6) | 0/3 (0) | 0/3 (0) | 0/3 (0) | 0/3 (0) | 2/3 (66.7) |
| Iceberg lettuce | 1/3 (33.3) | 0/3 (0) | 0/3 (0) | 1/3 (33.3) | 0/3 (0) | 0/3 (0) | 1/3 (33.3) |
| Spring onion | 2/3 (66.7) | 2/3 (66.6) | 0/3 (0) | 0/3 (0) | 2/3 (66.6) | 0/3 (0) | 0/3 (0) |
| Indian pennywort | 2/3 (66.7) | 2/3 (66.6) | 0/3 (0) | 0/3 (0) | 2/3 (66.6) | 0/3 (0) | 0/3 (0) |
| Green leaf lettuce | 2/3 (66.7) | 2/3 (66.7) | 0/3 (0) | 0/3 (0) | 2/3 (66.6) | 0/3 (0) | 0/3 (0) |
| Chinese f. cabbage | 2/3 (66.7) | 2/3 (66.6) | 0/3 (0) | 0/3 (0) | 2/3 (66.6) | 0/3 (0) | 0/3 (0) |
| Japanese parsley | 1/3 (33.3) | 0/3 (0) | 0/3 (0) | 1/3 (33.3) | 1/3 (33.3) | 0/3 (0) | 0/3 (0) |
| Bean sprout | 0/3 (0) | 0/3 (0) | 0/3 (0) | 0/3 (0) | 0/3 (0) | 0/3 (0) | 0/3 (0) |
| Mint | 0/3 (0) | 0/3 (0) | 0/3 (0) | 0/3 (0) | 0/3 (0) | 0/3 (0) | 0/3 (0) |
| Sweet basil | 0/3 (0) | 0/3 (0) | 0/3 (0) | 0/3 (0) | 0/3 (0) | 0/3 (0) | 0/3 (0) |
| Total | 47/178 (26.4) | 42/178(23.6) | 1/178 (0.6) | 5/178 (2.80) | 40/178 (22.5) | 0/178 (0) | 7/178 (3.93) |
The EN ISO 11290-1 method is the reference method. True positive: Samples are considered true positive when the sample is positive by all the detection methods. False-positive: Samples considered as false positive when the sample is positive either by 3M™ MDA or 3M ™ petrifilm EL plate, but negative by the reference method; False negative: Samples are considered as false negative when the sample is negative by 3M™ MDA or 3M™ petrifilm EL plate but positive by the reference method
NB the value inside the brackets is the percentage
Table 2.
Agreement of validation parameters between 3M™ MDA or 3M ™ petrifilm EL plate and EN ISO 11290-1 in the detection of Listeria in vegetable, chicken and related processing environmental samples
| Performance indicator | 3M™ MDA | 3M™ Petrifilm ELP |
|---|---|---|
| SEa (%) | 89.4 (76.9–96.4)f | 85.1 (71.7–93.8)f |
| SPb (%) | 99.3 (96.4–99.9) | 100.00 (97.2–100.00) |
| NPVc (%) | 96.4 (92.2–98.4) | 94.9 (90.4–97.4) |
| PPVd (%) | 97.7 (85.6–99.7) | 100.0 (97.3–100.0) |
| ACe (%) | 97.2 | 96.1 |
| Cohen’s Kappa Index | 0.911 | 0.894 |
Tpos, true positive; Tneg, true negative; Fpos, false positive; Fneg, false negative
aSE, sensitivity [Tpos/(Tpos + Fneg)] × 100%
bSP, specificity [Tneg/(Tneg + Fpos)] × 100%
cNPV, negative predictive value [Tneg/(Tneg + Fneg)] × 100%
dPPV, positive predictive value [Tpos/(Tpos + Fpos)] × 100%
eAC, accuracy [(Tpos + Tneg)/Total Number of Samples]
fValues in brackets indicates 95% confidence interval
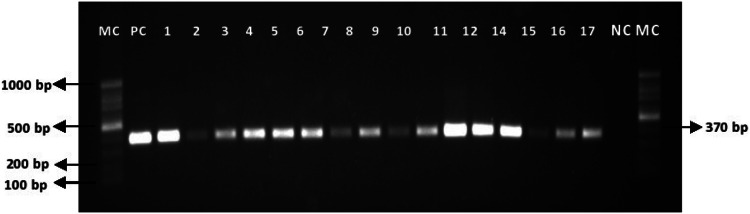
The occurrence of Listeria in foods and their related processing environments has been a major health concern. Thus, there are urgent needs for alternative methods that are culture-independent, simple, sensitive and cost-effective. These assays have the potential to provide rapid and reliable screening results to prevent the dissemination of Listeria along the food chain (Shannon et al., 2007; Straub and Chandler, 2003). The final confirmation of the presumptive Listeria isolated was done by PCR (Fig. 1).
Fig. 1.
A representative gel of PCR amplified products of the 370 bp region in the prs gene for identification of Listeria (genus) from samples study. Lane MC shows the 100 bp DNA marker. Lane PC shows the positive control (L. monocytogenes ATCC® 19112™), Lane 1 to Lane 17 are the representatives of Listeria from the vegetables, chicken and environmental samples. Lane NC as a negative control (Salmonella Typhimurium ATCC® 14028™) of the PCR reaction
In this study, both 3M™ MDA and the 3M™ petrifilm EL plates have the potential for detecting Listeria in naturally contaminated raw leafy vegetables, chicken and their processing environments which have been validated by AOAC. Overall, the 3M™ MDA Listeria showed high specificity (99.3%), accuracy (97.2%) and nearly complete agreement (κ = 0.911) with standard EN ISO 11290-1 method. Vongkamjan et al. (2015) stated that 3M™ MDA provides fast and reliable results for monitoring and detection of Listeria in the food processing plant environment. In their study, they examined 222 environmental samples obtained from fish processing plants, for the presence of Listeria and reported that the sensitivity, specificity, accuracy and the positive predictive value were 87.0%, 97.6%, 95.3%, 89.4%, respectively. According to Jongvanich (2016), 21/60 and 20/60 naturally contaminated chicken meat and environmental samples were positive for Listeria by the reference method and the 3M™ MDA Listeria, respectively. On the other hand, the 3M™ MDA detected Listeria with 93.65% specificity, 91.30% sensitivity, and 92.66% accuracy, respectively.
In another study, Mikš-Krajnik et al. (2016) evaluated the 3M™ MDA for the rapid detection of L. monocytogenes by using a cocktail of three strains at low inocula (100, 101, 102 CFU/100 cm2), inoculated onto polyethene and stainless steel surfaces by comparing against the standard culture-based method. Their finding showed that at 102 CFU/100 cm2, L. monocytogenes were detected in all samples by both methods (3M™ MDA and reference methods). The specificity, sensitivity, and accuracy of both methods were 100%. In addition, 3M™ MDA Listeria was used for the detection of Listeria in effluent water, meat, seafood and for routine hygiene monitoring in industrial plant environments, and observed that this method could be considered as a reliable alternative method (Fortes et al., 2013; Loff et al., 2014; Vongkamjan et al., 2015).
According to Abirami et al. (2016), false-negative and false-positive results can cause slight changes in the sensitivity and specificity of 3M™ MDA. They reported a false positive sample was detected by 3M™ MDA, and their findings agreed with those published previously by other researchers (Loff et al., 2014; Vongkamjan et al., 2015). The 3M™ MDA employs numerous set of primers, usually ranging between 4 and 6 at a higher concentration compared to concentrations used for conventional PCR based methods, which may likely develop a non-specific amplification induced by the formation of dimers, giving false positive outcomes (Wang et al., 2015). Another reason might be due to the possibility of amplification of DNA from dead or lethally injured cells, which cannot be detected by the culture-based standard method (Lim et al., 2015). For example, a false positive sample from the floor sample was detected by 3M™ MDA Listeria, because dead cells of Listeria might have been amplified, giving false-positive results.
Similarly, false-negative results obtained in this study, as in the case of the display table, wash water and laksa leaves could be due to the single enrichment step being used in the 3M™ MDA Listeria. According to Vongkamjan et al. (2015), while both ISO reference and 3M™ MDA Listeria methods require enrichment step, for the 3M™ MDA Listeria method, the enrichment step is only for 26–30 h while for the culture-based method is 48 h in selective media which might favour the growth or resuscitation of stressed Listeria to sufficient levels to facilitate detection. Another explanation for false-negative results can also be attributed to the insufficient quantities of lysate transferred to the tubes and the possible occurrence of cross-contamination during the transfer of resin from one tube to another (Loff et al., 2014). The speed in obtaining a real-time presumptive result and the substantial agreement with the reference method, suggest that 3M™ MDA is fast and a reliable alternative method for detecting Listeria in several food matrices (Yue, 2014).
In the current study, both 3M™ petrifilm EL plates and 3M™ MDA Listeria exhibited high specificity and accuracy for the detection of Listeria. The ready-to-use plating system (3M™ Petrifilm EL Plates) showed higher sensitivity and specificity compared to previous studies, as zero false-positive results were observed in this finding. Groves and Donnelly (2005) evaluated the efficacy of 3M™ petrifilm EL plate for the detection of Listeria from environmental samples, compared to a culture-based method and stated that the specificity and accuracy of both methods were similar. In another study, Horter and Lubrant (2004) evaluated both 3M™ petrifilm EL plates and EN ISO 11290-1 reference method for enumeration of Listeria on surfaces. Using many bacterial strains consisting of Listeria (n = 51) and non-Listeria (n = 37), three surface types, and nine sampling devices, concluded that 3M™ Petrifilm EL Plates method was cost-effective and rapid with high sensitivity (98%) and 100% specificity. Also, in the presence of background flora, both methods were effective in the quantitative recovery of Listeria from environmental surfaces. This finding and those from previous studies (Horter and Lubrant, 2004; Groves and Donnelly, 2005) showed that the 3M™ petrifilm EL plate, without an enrichment step, may provide an alternative for the detection and recovery of sublethally injured Listeria from the foods and environmental sources. The 3M™ petrifilm EL plate allows for relatively fast testing, unlike the culture-based method which requires a minimum duration of 4–5 days to complete (Norton et al., 2000) while 3M™ Petrifilm EL Plate method takes 29 ± 2 h to provide results (Horter and Lubrant, 2004; 3M™ Microbiology, 2012). Thus, over 80 h can be saved when using the 3M™ petrifilm EL plate method.
As compared to 3M™ MDA Listeria, the petrifilm EL plates are cost-effective, require little equipment and know-how. The commercialised LAMP-based system uses bioluminescence and isothermal amplification of nucleic acid sequences for sensitive, accurate and faster detection of Listeria and is thus appropriate for screening a large number of sample at the same time. In comparison to standard culture-based methods, the commercial LAMP-based system (3M™MDA Listeria) provides presumptive positive results within a shorter period (29 ± 2 h).
This study showed that both 3M™ methods are faster, specific, accurate, and comparable to the culture-based reference method in terms of accuracy, sensitivity, and specificity. Both the commercial LAMP-based system and ready-to-use plating system are easy to handle and provide possible positive results, rapidly. Overall, the rapid detection of Listeria in varieties of foods and their related processing environments is considered significant to develop effective control measures to reduce the occurrence and distribution of Listeria to retail establishments. Alternatively, microbiological methods may help the industry and diagnostic laboratories to find new ways of getting reliable results more efficiently in a shorter period.
Acknowledgements
The first author is thankful to USM-IPS for providing a USM Global fellowship. This work was supported by the Universiti Sains Malaysia, with a University Research Grants (1001/PTEKIND/811289).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mustapha Goni Abatcha, Email: mustygoni@gmail.com.
Pei Ling Tan, Email: pltan0310@gmail.com.
Li- Oon Chuah, Email: nateliechuah@gmail.com.
Gulam Rusul, Email: gulam@usm.my.
Mohd Esah Effarizah, Email: effarizah@usm.my.
References
- 3M Microbiology. 3M™ Petrifilm™ Environmental Listeria Plates interpretation guide.Availablefrom:https://multimedia.3m.com/mws/media/278947O/3m-petrifilm-environmental-listeria-plate-interpretation-guide.pdf. Accessed June 13, 2016 (2012)
- Abirami N, Nidaullah H, Chuah LO, Shamila-Syuhada AK, Chandraprasad S, Huda N, Hasmaizal H, Rusul G. Evaluation of commercial loop-mediated isothermal amplification-based kit and ready-to-use plating system for detection of Salmonella in naturally contaminated poultry and their processing environment. Food Control. 2016;70:74–78. doi: 10.1016/j.foodcont.2016.05.035. [DOI] [Google Scholar]
- Anonymous. Protocol for the validation of alternative microbiological methods. NV-DOC. In: D-2004–01–01 (2009)
- Becker B, Schuler S, Lohneis M, Sabrowski A, Curtis GD, Holzapfel WH. Comparison of two chromogenic media for the detection of Listeria monocytogenes with the plating media recommended by EN/DIN 11290–1. Int. J. Food Microbiol. 2006;109:127–131. doi: 10.1016/j.ijfoodmicro.2006.01.030. [DOI] [PubMed] [Google Scholar]
- Benesh DL, Crowley ES, Bird PM. 3M™ Petrifilm™ Environmental Listeria Plate. J. AOAC Int. 2013;96:225–228. doi: 10.5740/jaoacint.GovVal02. [DOI] [PubMed] [Google Scholar]
- Bird P, Flannery J, Crowley E, Agin J, Goins D, Monteroso L, Benesh D. Evaluation of 3M™ Molecular Detection Assay (MDA) Listeria for the Detection of Listeria species in Selected Foods and Environmental Surfaces: Collaborative Study, First Action 2014.06. J. AOAC Int. 2015;98:993–1002. doi: 10.5740/jaoacint.15-026. [DOI] [PubMed] [Google Scholar]
- Byrd-Bredbenner C, Berning J, Martin-Biggers J, Quick V. Food safety in home kitchens: a synthesis of the literature. Int. J. Environ. Res. Public Health. 2013;10:4060–4085. doi: 10.3390/ijerph10094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill RL, Lee H, Hall JC. Detection of Listeria monocytogenes and the toxin listeriolysin O in food. J. Microbiol. Methods. 2006;64:141–170. doi: 10.1016/j.mimet.2005.10.007. [DOI] [PubMed] [Google Scholar]
- De Castro V, Escudero J, Rodriguez J, Muniozguren N, Uribarri J, Saez D, Vazquez J. Listeriosis outbreak caused by Latin-style fresh cheese, Bizkaia, Spain, August 2012. Euro. Surveill. 2012;17:42. [PubMed] [Google Scholar]
- Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 2004;42:3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijkelkamp J, Aarts H, Van der Fels-Klerx H. Suitability of rapid detection methods for Salmonella in poultry slaughterhouses. Food Anal. Methods. 2009;2(1):1–13. doi: 10.1007/s12161-008-9040-5. [DOI] [Google Scholar]
- Feldsine PT, Kerr DE, Shen GS, Lienau AH. Comparative validation study to demonstrate the equivalence of a minor modification to AOAC Method 997.03 [Visual Immunoprecipitate (VIP) for Listeria] to the reference culture methods. J. AOAC. Int. 2008;91:365–369. doi: 10.1093/jaoac/91.2.365. [DOI] [PubMed] [Google Scholar]
- Fortes ED, David J, Koeritzer B, Wiedmann M. Validation of the 3M molecular detection system for the detection of Listeria in meat, seafood, dairy, and retail environments. J. Food Prot. 2013;76:874–878. doi: 10.4315/0362-028X.JFP-12-552. [DOI] [PubMed] [Google Scholar]
- Fretz R, Sagel U, Ruppitsch W, Pietzka A, Stöger A, Huhulescu S, Heuberger S, Pichler J, Much P, Pfaff G. Listeriosis outbreak caused by acid curd cheese ‘Quargel’, Austria and Germany 2009. Euro. Surveill. 2010;15:19477. [PubMed] [Google Scholar]
- Gill C, Badoni M, Moza L, Barbut S, Griffiths M. Microbiological sampling of poultry carcass portions by excision, rinsing, or swabbing. J. Food Prot. 2005;68:2718–2720. doi: 10.4315/0362-028X-68.12.2718. [DOI] [PubMed] [Google Scholar]
- Groves E, Donnelly CW. Comparison of 3M petrifilm TM environmental Listeria plate vs standard methods in detecting Listeria from environmental surfaces. IAFP Annual Meeting. Abstract nr P5–20 (2005)
- Hitchins AD, Jinneman K. Detection and enumeration of Listeria monocytogenes in foods. Bacteriological analytical manual online: US Food and Drug Administration; 2003. [Google Scholar]
- Horter B, Lubrant H. 3M petrifilm environmental Listeria plate for rapid enumeration of Listeria from environmental surfaces. In IAFP Annual Meeting (pp. P138) (2004)
- Jongvanich S. Detection of Listeria species in naturally and artificially contaminated chicken meat and environment samples: A comparison of the reference method to 3M™ molecular detection assay 2 Listeria. In IAFP 2016 Annual Meeting (2016)
- Lee KM, Runyon M, Herrman TJ, Phillips R, Hsieh J. Review of Salmonella detection and identification methods: Aspects of rapid emergency response and food safety. Food Control. 2015;47:264–276. doi: 10.1016/j.foodcont.2014.07.011. [DOI] [Google Scholar]
- Lim HSY, Zheng Q, Miks-Krajnik M, Turner M, Yuk HG. Evaluation of commercial kit based in loop-mediated isothermal amplification for rapid detection of low levels of uninjured and injured Salmonella on duck meat, bean sprouts, and fishballs in Singapore. J. Food Prot. 2015;78:1203–1207. doi: 10.4315/0362-028X.JFP-14-535. [DOI] [PubMed] [Google Scholar]
- Loff M, Mare L, de Kwaadsteniet M, Khan W. 3M™ molecular detection system versus MALDI-TOF mass spectrometry and molecular techniques for the identification of Escherichia coli 0157: H7, Salmonella spp. & Listeria spp. J. Microbiol. Methods. 2014;101:33–43. doi: 10.1016/j.mimet.2014.03.015. [DOI] [PubMed] [Google Scholar]
- Mikš-Krajnik M, Lim HSY, Zheng Q, Turner M, Yuk HG. Loop-mediated isothermal amplification (LAMP) coupled with bioluminescence for the detection of Listeria monocytogenes at low levels on food contact surfaces. Food Control. 2016;60:237–240. doi: 10.1016/j.foodcont.2015.07.035. [DOI] [Google Scholar]
- Norton DM, McCamey M, Boor KJ, Wiedmann M. Application of the BAX for screening/genus Listeria polymerase chain reaction system for monitoring Listeria species in cold-smoked fish and in the smoked fish processing environment. J. Food Prot. 2000;63:343–346. doi: 10.4315/0362-028X-63.3.343. [DOI] [PubMed] [Google Scholar]
- Nyachuba D, Donnelly C. Comparison of 3M™ Petrifilm™ environmental Listeria plates against standard enrichment methods for the detection of Listeria monocytogenes of epidemiological significance from environmental surfaces. J. Food Sci. 2007;72:M346–M354. doi: 10.1111/j.1750-3841.2007.00554.x. [DOI] [PubMed] [Google Scholar]
- Oktay HI, Heperkan D. Evaluation of ISO method and VIDAS automated system for identifying Listeria and Salmonella in selected foods. J. Rapid Methods Autom. Microbiol. 2006;14:133–145. doi: 10.1111/j.1745-4581.2006.00033.x. [DOI] [Google Scholar]
- Pesavento G, Ducci B, Nieri D, Comodo N, Nostro AL. Prevalence and antibiotic susceptibility of Listeria spp. isolated from raw meat and retail foods. Food Control. 2010;21:708–713. doi: 10.1016/j.foodcont.2009.10.012. [DOI] [Google Scholar]
- Ponniah J, Robin T, Paie MS, Radu S, Ghazali FM, Kqueen CY, Nishibuchi M, Nakaguchi Y, Malakar PK. Listeria monocytogenes in raw salad vegetables sold at retail level in Malaysia. Food Control. 2010;21:774–778. doi: 10.1016/j.foodcont.2009.09.008. [DOI] [Google Scholar]
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17: ( 2011) [DOI] [PMC free article] [PubMed]
- Shannon K, Lee DY, Trevors J, Beaudette L. Application of real-time quantitative PCR for the detection of selected bacterial pathogens during municipal wastewater treatment. Sci. Total Environ. 2007;382:121–129. doi: 10.1016/j.scitotenv.2007.02.039. [DOI] [PubMed] [Google Scholar]
- Straub TM, Chandler DP. Towards a unified system for detecting waterborne pathogens. J. Microbiol. Methods. 2003;53:185–197. doi: 10.1016/S0167-7012(03)00023-X. [DOI] [PubMed] [Google Scholar]
- Thomsen MR, Shiptsova R, Hamm SJ. Sales Responses to recalls for Listeria monocytogenes: Evidence from branded ready-to-eat meats. Appl. Econ. Perspect. Policy. 2006;28:482–493. [Google Scholar]
- Thong K, Lai M, Teh C, Chua K. Simultaneous detection of methicillin-resistant Staphylococcus aureus, Acinetobacter baumannii, Escherichia coli, Klebsiella pneumoniae and Pseudomonas aeruginosa by multiplex PCR. Trop. Biomed. 2011;28:21–31. [PubMed] [Google Scholar]
- Viera AJ, Garrett JM. Understanding interobserver agreement: the kappa statistic. Fam. Med. 2005;37:360–363. [PubMed] [Google Scholar]
- Vongkamjan K, Fuangpaiboon J, Jirachotrapee S, Turner MP. Occurrence and diversity of Listeria spp. in seafood processing plant environments. Food Control. 2015;50:265–272. doi: 10.1016/j.foodcont.2014.09.001. [DOI] [Google Scholar]
- Wang DG, Brewster JD, Paul M, Tomasula PM. Two methods for increased specificity and sensitivity in loop-mediated isothermal amplification. Molecules. 2015;20:6048–6059. doi: 10.3390/molecules20046048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue HLS. Evaluation of Loop Mediated Isothermal DNA Aplification (LAMP) Detection of Salmonella spp. in Foods and Listeria monocytogenes on Environmental Surfaces. Master’s Thesis National University Singapore. https://scholarbank.nus.edu.sg/handle/10635/118603. Accessed on 17/9/2018 (2014)