Abstract
Biofuel production from renewable and sustainable resources is playing an increasingly important role within the fuel industry. Among biofuels, bioethanol has been most widely used as an additive for gasoline. Higher alcohols can be blended at a higher volume compared to ethanol and generate lower greenhouse gas (GHG) emissions without a need to change current fuel infrastructures. Thus, these fuels have the potential to replace fossil fuels in support of more environmentally friendly processes. This review summarizes the efforts to enhance bioalcohol production in engineered Escherichia coli over the last 5 years and analyzes the current challenges for increasing productivities for industrial applications.
Keywords: Escherichia coli, C2–C6 bioalcohol, ethanol, butanol, isobutanol, isopropanol, isopentanol, isopentenol
Introduction
The production of biofuels from renewable resources has gained significant attention due to the rising energy crisis and environmental concerns. Currently, bioethanol is widely used, and Grand View Research, Inc., reported that the global ethanol market size could reach $115.65 billion by 2025, growing at a compound annual growth rate (CAGR) of 6.7%1. In addition, the microbial production of higher alcohols (especially C3–C6) has gained traction over the last decade. The use of higher alcohols such as isopropanol or isobutanol would not require changes to current biofuel refinery or transportation processes as these alcohols can be blended at higher volumes in gasoline compared to ethanol (e.g., 16% for isopropanol/isobutanol versus 10% for ethanol), resulting in lower greenhouse gas (GHG) emissions (Andersen et al., 2010; Slating and Kesan, 2012). However, higher alcohols, except n-butanol, are not commonly produced at high yields in microbes. With the development of molecular biology techniques and metabolic engineering strategies, model systems, such as Escherichia coli (Jojima et al., 2008; Inokuma et al., 2010; Lan and Liao, 2013; Matsubara et al., 2016) and Saccharomyces cerevisiae (Park et al., 2014; Shi et al., 2016), have been modified to synthesize bioalcohols.
Escherichia coli is a well-studied model microorganism which has several advantageous traits for bioalcohol production including fast growth in inexpensive mineral media, the ability to utilize a wide range of substrates from biomass, and detailed genetic information and diverse genetic tools for gene manipulation. However, there are still challenges using engineered E. coli for industrial applications such as the need to improve tolerance to bioalcohols, efficient utilization of low-cost substrates, and productivity toward advanced alcohols (Jojima et al., 2008; Inokuma et al., 2010; Lan and Liao, 2013; Matsubara et al., 2016). Recently, the rapid expansion of genome engineering strategies, synthetic biology techniques, and high-throughput tools have enabled their application to study advanced bioalcohols production and to further investigate the mechanisms of alcohol resistance. This review will summarize recent progress in metabolic engineering of E. coli for C2–C6 alcohol-derived biofuel production (Figure 1), introduce new synthetic biology methods and genome engineering strategies for in-depth studies of alcohol tolerance, and analyze the current challenges for increasing productivity for industrial applications.
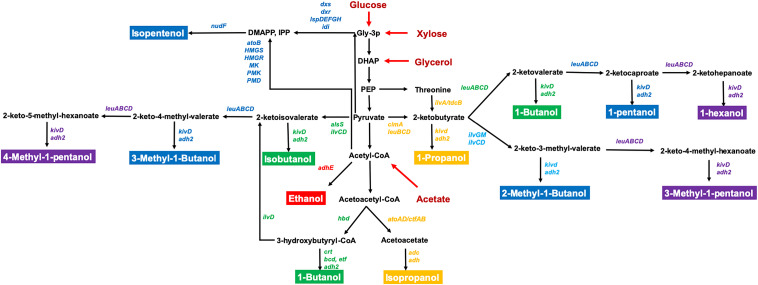
FIGURE 1.
The metabolic pathways for the production of C2–C6 bioalcohol in E. coli. The genes directly related to bioalcohol synthesis are labeled in different colors. C2 (red); C3 (orange); C4 (green); C5 (blue); C6 (purple). The substrates are labeled in red. Relevant reactions are represented by the name of the gene(s) coding for the enzyme(s): adh2/adhE, alcohol dehydrogenase; alsS, acetolactate synthase; atoB, acetyl-CoA acyltransferase; cimA, citramalate synthase; dxr, 1-deoxy-D-xylulose 5-phosphate reductoisomerase; dxs, 1-deoxy-D-xylulose 5-phosphate synthase; HMGR, 3-hydroxy-3-methylglutaryl-CoA reductase; HMGS, 3-hydroxy-3-methylglutaryl-CoA synthase; idi, isopentenylpyrophosphate isomerase; ilvA, threonine deaminase; ilvC, acetohydroxy acid isomeroreductase; ilvD, dihydroxy acid dehydratase; ilvGM, acetohydroxybutanoate synthase; ispD, 4-diphosphocytidyl-2-methylerythritol synthase; ispE, 4-diphosphocytidyl-2-methylerythritol kinase; ispF, 2-methylerythritol 2,4-cyclodiphosphate synthase; ispG, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase; ispH, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase; kivd, ketoisovalerate decarboxylase; leuA, 2-isopropylmalate synthase; leuB, 3-isopropylmalate dehydrogenase; leuCD, 2-isopropylmalate isomerase; MK, mevalonate kinase; nudF, prenyl phosphatase; PMD, phosphomevalonate decarboxylase; PMK, phosphomevalonate kinase; DHAP, dihydroxyacetone-phosphate; DMAPP, dimethylallyl pyrophosphate; Gly-3-P, glyceraldehyde-3-phosphate; IPP, isopentenyl pyrophosphate; PEP, phosphoenolpyruvate.
Ethanol (C2 Bioalcohol) Production in E. Coli
The current chassis for industrial ethanol production is S. cerevisiae due to its ability to produce ethanol from glucose at 95% maximum theoretical yield (Krishnan et al., 1999). The bacterium Zymomonas mobilis has also been proposed for use in industrial ethanol production since it also produces ethanol from glucose at 95% of maximum theoretical yield and has a higher specific ethanol productivity than S. cerevisiae (Zhao et al., 2014). Next generation biofuels and biochemicals aim to use lignocellulosic biomass, which contains both glucose and xylose, as an attractive source of non-food carbohydrates for production (Table 1) (Wang L. et al., 2019). However, neither S. cerevisiae nor Z. mobilis natively utilize xylose. In order to produce bioethanol from cellulosic feedstocks, the cellulose-degrading bacterium Clostridium thermocellum has also been used as a chassis for industrial production because it is able to directly ferment cellulose into ethanol. However, C. thermocellum only generates ethanol from cellulose (and also only natively utilizes glucose) at 75% of the maximum theoretical yield, resulting in lower ethanol yields compared to S. cerevisiae or Z. mobilis (Tian et al., 2016). Furthermore, C. thermocellum has low tolerance to ethanol (Herrero and Gomez, 1980) and has limited genetic tools, making it difficult and time consuming to engineer this strain for increased ethanol production (Tripathi et al., 2010), although CRISPR-Cas-based genome editing systems were recently developed (Walker et al., 2020).
TABLE 1.
Select examples of engineering E. coli for bioalcohol production.
| Alcohol | General strategy | Method | Titer (g/L) | Yield (g/g) | References |
| Ethanol | Improve substrate utilization | (i) Ethanologenic E. coli strains carry the Z. mobilis pdc, adhA, and adhE genes (ii) Deletion of xylR gene to make the LYglc1 strain (iii) Deletion of ptsI, ptsG, galP, and glk; expression of XylR* to make the LYxyl3 strain (iv) Co-culture of LYglc1 and LYxyl3 strains |
46 | 0.45 | Wang L. et al., 2019 |
| Isopropanol | Improve precursor accumulation; decrease the metabolic flux to TCA | Construction of BW25113 (ΔlacI, ΔgltA) with plasmid pTA17 (PLlacO1::thl, atoAB, adc, and adhE), pTA965 (PLlacO1::tetR, PLtetO1::gltA.LAA), and pTA1251 (PLlacO1::poxB, acs, PlacIq::lacI) | 3.8 | Not reported | Soma et al., 2017 |
| Isobutanol | Directed evolution with alcohol-biosensor-based selection | (i) BmoR-based biosensor used in an atmospheric and room temperature plasma (ARTP) mutagenesis library to screen for increased isobutanol production (ii) Fed-batch fermentation with gas-stripping |
56.6 | Not reported | Yu et al., 2019 |
| n-butanol | Inactivation of byproduct pathway; improve substrate utilization; improve cofactor supply; adaptive evolution for improved cell growth; optimize the expression of pathway genes |
(i) Gene knockout of hyc-hyp, fdhF, poxB, pck, fumB, fumAC, tdcD, mdh, focA, ppc, mgsA, yieP, stpA, yqeG, yagM in BW25113 (ii) Integration of the fdh gene into the genome with PydfZ (iii) Adaptive evolution for fast anaerobic cell growth (iv) RBS library for the phaA, hbd, crt, ter, and adhE2 genes |
20 | 0.34 | Dong et al., 2017 |
| 2-methyl-1-butanol and 3-methyl-1-butanol | Inactivation of byproduct pathway; improve substrate utilization; optimize the expression of pathway genes |
(i) Construction of E. coli AY3 (BW25113, ΔglnA, ΔgdhA, ΔlsrA, pYX68 (ilvE-ilvA-sdaB), pYX90 (alsS-ilvC-ilvD-avtA), pYX97 (leuDH-kivd-yqhD) (ii) Construction of E. coli BLF2 [E. coli B Δldh, pLF101 (alsS-ilvC-ilvD), pLF102 (kivd-yqhD)] (iii) Co-culture of AY3 and BLF2 with an inoculation ratio of 1:4 using distillers’ grains with solubles |
2.2 (two alcohols mixture) | Not reported | Liu et al., 2017 |
| Pentanol | Inactivation of byproduct pathway; optimize the expression of pathway genes |
(i) Construction of BW25113 (ΔilvB ΔilvI ΔleuA) transformed with plasmid pAFC52 (cimAΔ2, leuBCD) and pGC22 [leuA (G462D), kivd (V461G), yqhD] (ii) In situ extraction using oleyl alcohol |
4.3 | Not reported | Chen et al., 2017 |
| Isoprenol | Improve precursor accumulation; optimize the expression of pathway genes |
(i) Construction of AK26 (E. coli DH1), transformed with plasmids JBEI-17081 (pA5c-AtoB-HMGS_Sa-HMGR_Sa) and JBEI-17844 (pTrc99a-PMDsc_HKQ-MKmm) (ii) Fed-batch cultures with a solvent overlay | 10.8 | 0.105 | Kang et al., 2019 |
Escherichia coli is additionally investigated as a host for ethanol production because it has a large variety of metabolic engineering tools available for strain modification, it can grow in higher concentrations of ethanol (Zaldivar et al., 2000), and it naturally ferments both glucose and xylose, although the presence of glucose still leads to carbon catabolite repression (CCR) when xylose is present (Liu et al., 2018). Thus, Flores et al. (2019) developed an E. coli co-culture strategy for conversion of glucose-xylose mixtures to ethanol. One strain, LYglc1, was engineered to only utilize glucose by deleting the xylose-specific transcriptional activator, XylR. The other strain, LYxyl3, was engineered to only utilize xylose by mutating XylR to remove CCR and by deleting genes (ΔptsI ΔptsG ΔgalP glk::kanR) required for glucose transport and metabolism. The strains are ethanologenic due to insertion of the pdc, adhA, and adhE genes from Z. mobilis. The LYglc1 and LYxyl3 strains were co-cultured at an optimum ratio of 1:500, enhancing the sugar utilization rate and ethanol productivity by 50 and 28%, respectively, when compared to a monoculture of the parent strain, LY180. Using this system, they achieved productivity of 0.49 g/L/h, with a final ethanol titer of 46 g/L at 90% of maximum theoretical yield. In a different approach, Sun et al. (2018) constructed a “two-phase-two-temperature” strategy using temperature inducible promoters to control the glucose metabolic pathway in E. coli. The final strain, B0013-2021HPA (ΔptsG ΔmanZ Δglk; ptsG expressed under the control of tandem λ pL and pR promoters), utilized all sugars but glucose for cell growth at 34°C, whereas it fermented all sugars to ethanol at 42°C. In addition, Z. mobilis pdc and adhB genes were introduced to increase ethanol production. As a result, this strain produced 127 g ethanol from 260.9 g mixed sugars from corncob hydrolysate with a productivity of 4.06 g/L/h.
In addition to strategies to improve ethanol yield, there have been several studies to understand ethanol-induced stress and improve ethanol tolerance in E. coli (Liu et al., 2016; Cao et al., 2017; Lupino et al., 2018). Cao et al. (2017) systematically analyzed the mechanism for ethanol-induced stress and found that ethanol damages cell wall and membrane integrity, decreases the cross-membrane proton gradient and related ATP synthesis, and changes protein functions by direct binding. Genes that are upregulated in response to ethanol stress include: osmBC and ompCGLR in response to osmotic stress; gadABE and asr in response to acid stress; rpoE, degP, asnB, and opgG in response to envelope stress; groSL, grpE, and metA in response to heat-shock stress; and the OxyR and SoxRS regulons in response to ROS. Therefore, these cellular processes and genes are good targets for engineering strains with increased ethanol tolerance. Expression of heterologous genes can also lead to increased ethanol tolerance. Liu et al. (2016) expressed the yajC gene, which encodes a subunit of a protein translocase complex, from Lactobacillus buchneri in E. coli, and increased tolerance up to 4% ethanol.
C3–C4 Alcohol Tolerance and Production in E. Coli
C3–C4 alcohols such as propanol, isopropanol, 1-butanol, and isobutanol are higher alcohols which have similar fuel properties (Clomburg and Gonzalez, 2010). Several species of Clostridium have been evaluated for butanol and isopropanol production, but cannot be used for industrial application mainly due to low fermentation yield and titer (Survase et al., 2011; Xue and Cheng, 2019). Alternative organisms such as E. coli have also been engineered toward the goal of industrial production of C3–C4 alcohols (Jojima et al., 2008; Inokuma et al., 2010; Lan and Liao, 2013; Matsubara et al., 2016). E. coli metabolic pathways have shared intermediate metabolites, which reduce central metabolites such as acetyl-CoA and pyruvate into more electron-rich compounds and higher carbon acyl-CoA and 2-keto acids (Saini et al., 2016; Heo et al., 2017; Ohtake et al., 2017; Soma et al., 2017; Nitta et al., 2019).
Precursor accumulation is one of the limiting steps for isopropanol production (Clomburg and Gonzalez, 2010; Soma et al., 2017). The gltA gene is involved in synthesizing isocitrate from oxaloacetate. Soma et al. (2014) developed a metabolic toggle switch (MTS) method by expressing the gltA gene under the PLtetO1 promoter and the TetR repressor under the PLlacO1 promoter; this system allows metabolic flux from the TCA cycle to be redirected toward isopropanol production in an inducible manner. Then, they introduced a plasmid that overexpressed the native poxB and acs genes under the PLlacO1 promoter for conversion of excess pyruvate to acetyl-CoA (Soma et al., 2017). The resulting isopropanol titer was up to 3.8 g/L, a titer 4.4-fold higher than that of the parent strain. Our group applied a CRISPR-based genome engineering strategy to generate ribosome binding site (RBS) libraries (903 mutants in total) for genes in the synthetic pathway for the production of isopropanol (thl, atoDA, adc, and adh), and were able to identify a high isopropanol producer, PA14, that generated 7.1 g/L at 24 h, with a yield of 0.75 mol/mol glucose. In particular, we found that higher expression levels of adc and adh led to increased isopropanol production (Liang et al., 2017).
Other efforts have focused on producing bioalcohols from alternate carbon sources. To enable utilization of acetate as the sole carbon source for isopropanol production, Yang et al. (2020) first constructed the isopropanol pathway by combining genes from Clostridium acetobutylicum (thlA, adc), E. coli (atoDA), and Clostridium beijerinckii (adh). In addition, they replaced the promoter of the native ack-pta genes to improve the acetate kinase and phosphotransacetylase (ACK-PTA) pathway and overexpressed the native nadK gene to increase NADH supply. The highest concentration and yield of isopropanol reached was 1.47 g/L and 0.56 g/g acetate. Other efforts have also focused on improving tolerance to isopropanol. Horinouchi et al. (2017) performed adaptive laboratory evolution (ALE) in E. coli and identified five mutations (relA, marC, proQ, yfgO, and rraA) with enhanced isopropanol tolerance up to 27 g/L. Transcriptome analysis revealed that genes related to amino acid biosynthesis, iron homeostasis, and energy metabolisms are related to isopropanol tolerance. Zhou et al. (2019) used a targeted deletion approach and demonstrated that isopropanol tolerance could be increased by inactivation of the acetoacetyl-CoA transferase genes and atoDA, enabling growth in 500 mM isopropanol.
Improved fermentative production of isobutanol, a non-native alcohol pathway, has been achieved by metabolic engineering approaches in E. coli strains (Blombach and Eikmanns, 2011). Recently, Song et al. (2018) overexpressed the native acs, pckA, and maeB genes to increase acetate uptake, resulting in 26% increased isobutanol titers using acetate as the sole carbon source. Other efforts have focused on increasing isobutanol production from glucose. Liang et al. (2018) introduced the heterologous Entner–Doudoroff (ED) pathway from Z. mobilis to increase glucose transformation to pyruvate for enhanced precursor accumulation. The resulting E. coli strain, ED02, produced 13.67 g/L isobutanol with a productivity of 0.456 g/L/h, a 56.8 and 88.1% improvement over the parent strain, respectively. Ghosh et al. (2019) used the Optimization by Selection and Sequencing (OptSSeq) strategy to regulate expression levels of genes in the isobutanol synthetic pathway. They found that the optimum levels of pathway enzymes (AlsS, IlvC, IlvD, Kivd, and AdhA) were a molar ratio of 2.5:6.7:2:1:5.2, which led to 3 g/h/gDCW of isobutanol production. In addition, Yu et al. (2019) used a BmoR-based biosensor to screen for improved isobutanol producing strains from an atmospheric and room temperature plasma (ARTP) mutagenesis library. The best isolated variant produced twofold more isobutanol than the wild-type, and the titer of isobutanol reached 56.5 g/L, with a productivity of 0.533 g/L/h, during fed-batch fermentation.
Clostridium species have long been employed for n-butanol production through their acetone–butanol–ethanol (ABE) pathway (Xue and Cheng, 2019). However, due to the lack of available genetic tools for Clostridia, these species are currently not robust candidates as industrial chassis (Abdelaal et al., 2019). Therefore, in recent years, many efforts have been applied toward engineering E. coli for n-butanol production (Dong et al., 2016). Over the last 5 years, the utilization of renewable and cheap substrates from agricultural residues and crude glycerol waste streams have been a target for n-butanol production studies in E. coli. Abdelaal et al. (2019) integrated butanol pathway genes (hbd, crt, adhE2, ter, and atoB) into xylose-utilizing host SSK42 (E. coli B PgapAPDH ΔldhA ΔfrdA ΔpfB), and the final strain produced 4.3 g/L butanol using xylose as the sole carbon source. Similarly, Saini et al. (2015, 2017) first integrated butanol pathway genes (phaA, hbd, crt, ter, and adhE2) into a BL21-based host strain with byproduct gene deletions (ΔptsG, ΔpoxB1, ΔldhA, ΔfrdA, and ΔadhE) for glycerol conversion to butanol. In addition, they increased NADH regeneration by overexpression of genes related to NADH-production (aceEF, lpdA, zwf, pgl, and udhA). As a result, the engineered strain produced 6.9 g/L n-butanol from 20 g/L crude glycerol under microaerobic conditions, increasing productivity fivefold compared to the strain without modification of NADH supply.
Maintaining cofactor balance and resolving free CoA imbalance are important for CoA-dependent n-butanol production (Nitta et al., 2019). To this end, various groups have overexpressed formate dehydrogenase (Fdh) for NADH regeneration under endogenous fermentation regulatory elements (FREs) control (Wen and Shen, 2016), knocked out the pgi gene for increased NADH by activation of the pentose phosphate pathway (PPP) (Saini et al., 2016), decreased carbon flux from acetyl-CoA to the TCA cycle, and improved NADH and CoA supply by downregulation of citrate synthase (Saini et al., 2016; Heo et al., 2017), knocked out genes from the glyoxylate shunt for increased CoA accumulation (Nitta et al., 2019), and optimized AdhE2 activity for CoA recycling and supplemented with cysteine for increased CoA supply (Ohtake et al., 2017). All of these approaches resulted in increased n-butanol production. In addition, Dong et al. (2017) developed a completely chromosomally engineered E. coli strain capable of producing butanol efficiently (Table 1). They first integrated the butanol pathway genes into a BW25113-based host strain with deleted byproduct genes. They then modified the expression of fdh to increase NADH regeneration and improve anaerobic cell growth by adaptive evolution. The final strain, which also had an optimized butanol pathway (from RBS libraries for pathway genes), produced 20 g/L n-butanol at 83% of theoretical yield, the highest titer achieved compared to the above studies. Low n-butanol tolerance is also limiting for the economic viability of n-butanol production. A number of studies have identified strategies that improve tolerance: (1) mutation of genes related to cis-regulatory elements (yqjA, yabI, and rob) or the efflux pump subunit, acrB (Jeong et al., 2017; He et al., 2019); (2) disruption of the transmembrane protein, TqsA (He et al., 2019); (3) disruption of succinylglutamate desuccinylase (AstE) (Guo et al., 2019); (4) overexpression of the chaperone protein SecB or its mutation, SecBT10A (Xu et al., 2019); and (5) overexpression of the membrane-targeted tilapia metallothionein, OmpC-TMT (Chin et al., 2017).
C5–C6 Alcohol Production and Tolerance in E. Coli
C5 alcohols such as 1-pentanol, 2-methyl-1-butanol, and 3-methyl-1-butanol (isopentanol) and C6 alcohols such as 1-hexanol, 3-methyl-1-pentanol, and 4-methyl-1-pentanol represent a useful class of chemicals with potential application as biofuels (Wang et al., 2017). In E. coli, iterative keto-acid elongation resulted in C5, C6, and even longer chain (C7–C8) alcohols (Wang et al., 2017). Recently, Eiben et al. (2020) constructed a new isopentanol production pathway in E. coli XX03 (BW25113 ΔadhE ΔldhA ΔfrdBC) by introducing the isovaleryl-CoA pathway (LiuC, AibAB, and AibC) from Myxococcus xanthus and a butyryl-CoA reductase (AdhE2) from C. acetobutylicum, and the resulting strain produced 80.5 mg/L isopentanol after 36 h under microaerobic conditions. In addition, Chen et al. (2017) constructed a 1-pentanol production pathway by controlling the keto acid elongation cycle [BW25113 ΔilvB ΔilvI ΔleuA expressing cimAΔ2, leuBCD, leuA (G462D), kivd (V461G), yqhD] and identifying a new mutation in the ketoisovalerate decarboxylase, KivD V461D, that preferentially tuned the KivD from Lactococcus lactis specificity toward 1-pentanol synthesis. The titer of 1-pentanol reached 4.3 g/L and comprised 90% of the total alcohol content. However, the titer, yield, and productivities for C5-C6 bioalcohol production in E. coli are still too low for industrial applications. Recently, Chen and coworkers employed an adaptive evolution method to increase the tolerance of E. coli to isopropanol, isobutanol, and isopentanol, identifying that upregulated RpoS can increase general alcohol resistance (Wang et al., 2020), a strategy that may also be relevant for tolerance to other C5–C6 alcohols.
The biosynthesis of isopentenols, including isoprenol and prenol, provides an additional route to the production of C5 alcohols (George et al., 2015). Unlike other C5–C6 alcohols, isopentenols are synthesized from the isoprenoid pathway precursor metabolites isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP) (Figure 1). To improve the availability of IPP/DMAPP, Tian et al. (2019) constructed a CRISPRi-mediated multiplex repression system to knockdown genes asnA, gldA, and prpE which are involved in asparagine production, glycerol utilization, and propionyl-CoA synthesis, respectively, resulting in 18-24% higher isopentenol. Kang et al. (2016) developed “IPP-bypass” mevalonate pathways using mevalonate diphosphate decarboxylase (PMD) and phosphatase (AphA) for isopentenol production. They then further improved PMD activity through high-throughput enzyme screening (Kang et al., 2017) and optimized the origin and expression level of genes in this IPP-bypass pathway (Kang et al., 2019). Their final engineered strain had an isoprenol titer of 10.8 g/L in a fed-batch fermentation (Kang et al., 2019). Other efforts have focused on producing isoprenol from cellulosic feedstocks. Wang et al. (2018) initially utilized switchgrass hydrolysate derived from ionic liquid pretreatment for isoprenol production, but the remaining ionic liquids in the hydrolysate required multiple washes to decrease its toxicity to E. coli. Thus, they used an adaptive evolution strategy to improve tolerance to ionic liquids, and the adapted E. coli strain produced 1.06 g/L which is 6.6-fold more isoprenol compared to the parent strain in the presence of ionic liquids (Wang et al., 2019b). In addition, they found that NaCl enhanced the tolerance of E. coli to ionic liquids. MG1655 reached an OD600 that was ∼twofold higher with 200 mM NaCl than without NaCl (Wang et al., 2019a).
Conclusion and Perspectives
Converting renewable biomass into biofuel using engineered microbial cell factories provides a promising alternative to fossil fuels. To date, synthetic pathways have been constructed for the production of bioalcohols ranging from C2 up to C10 (Jang et al., 2012; Kim et al., 2015). The titer, yield, and productivity for C3–C6 bioalcohol production in engineered E. coli have improved in recent years; however, the industrial application for the production of these bioalcohols still has some way to go compared to ethanol production. Despite these limitations, some companies (e.g., Butamax and Gevo) have begun to approach commercialization of isobutanol bioproduction. However, additional efforts are still required to overcome technological limitations. New strategies for engineering current microbes still need to be identified or developed for: (1) utilization of non-food-based carbon sources, (2) tolerance to inhibitors derived from biomass or the fermentation process, and (3) tolerance to high concentrations of substrate and biofuel products (Bilal et al., 2018; Shanmugam et al., 2020). While it might be possible to engineer a single microbe for industrial use, microbial consortia could be another solution to improve biofuel production. Cultures containing multiple microbes could improve tolerance to different inhibitors and utilize various components from complex carbon sources, further improving the economics of biofuel production (Bernstein and Carlson, 2012; Shong et al., 2012). In addition, feedstock cost is almost half the cost of the fermentation process (Bhatia et al., 2015), so developing strains that can utilize cheaper feedstocks would have a large economic benefit for industrial biofuel production. Moreover, development of new fermentation processes that increase productivity at the industrial level, such as low-cost and efficient in situ product removal (ISPR), could be a new requirement for material and process engineering.
Advances in technologies for DNA synthesis and sequencing have simplified the process of reprogramming metabolism for optimal production of desired chemicals. CRISPR-based technologies have increased the accuracy and speed of gene editing and regulation at genome-scale (Ronda et al., 2015; Garst et al., 2017; Si et al., 2017; Liu R. et al., 2019; Liu Y. et al., 2019). In addition, advanced systems biology tools including genomics, transcriptomics, proteomics, metabolomics, and fluxomics will help facilitate the characterization, analysis, and design of new metabolic pathways for bioalcohol production (Adamczyk and Reed, 2017; McCloskey et al., 2018). Using advanced technologies to gain a deeper understanding of tolerance mechanisms and to refine metabolism for increased titer, yield, and productivity of alcohols from various feedstocks will enable economically viable production of bioalcohols from microbial hosts such as E. coli, with findings from E. coli providing valuable insight into improving these systems in other non-model microbial chassis.
Author Contributions
All authors contributed to the analysis of the literature, compiling the related data, and writing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This project was sponsored by the U.S. Department of Energy (Grant DE-SC0018368) and the Center for Bioenergy Innovation a U.S. Department of Energy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science.
References
- Abdelaal A. S., Jawed K., Yazdani S. S. (2019). CRISPR/Cas9-mediated engineering of Escherichia coli for n-butanol production from xylose in defined medium. J. Ind. Microbiol. Biotechnol. 46 965–975. 10.1007/s10295-019-02180-8 [DOI] [PubMed] [Google Scholar]
- Adamczyk P. A., Reed J. L. (2017). Escherichia coli as a model organism for systems metabolic engineering. Curr. Opin. Syst. Biol. 6 80–88. 10.1016/j.coisb.2017.11.001 [DOI] [Google Scholar]
- Andersen V. F., Anderson J. E., Wallington T. J., Mueller S. A., Nielsen O. J. (2010). Vapor pressures of alcohol-gasoline blends. Energ. Fuels 24 3647–3654. 10.1021/ef100254w [DOI] [Google Scholar]
- Bernstein H. C., Carlson R. P. (2012). Microbial consortia engineering for cellular factories: in vitro to in silico systems. Comput. Struct. Biotechnol. J. 3:e201210017. 10.5936/csbj.201210017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S. K., Shim Y.-H., Jeon J.-M., Brigham C. J., Kim Y.-H., Kim H.-J., et al. (2015). Starch based polyhydroxybutyrate production in engineered Escherichia coli. Bioprocess Biosyst. placecountry-regionEng. 38 1479–1484. 10.1007/s00449-015-1390-y [DOI] [PubMed] [Google Scholar]
- Bilal M., Iqbal H. M. N., Hu H., Wang W., Zhang X. (2018). Metabolic engineering and enzyme-mediated processing: a biotechnological venture towards biofuel production – A review. Renew. Sust. Energ. Rev. 82 436–447. 10.1016/j.rser.2017.09.070 [DOI] [Google Scholar]
- Blombach B., Eikmanns B. J. (2011). Current knowledge on isobutanol production with Escherichia coli, Bacillus subtilis and Corynebacterium glutamicum. Bioeng. Bugs 2 346–350. 10.4161/bbug.2.6.17845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Wei D., Yang Y., Shang Y., Li G., Zhou Y., et al. (2017). Systems-level understanding of ethanol-induced stresses and adaptation in E. coli. Sci. Rep. 7:44150. 10.1038/srep44150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. S., Siao S. W., Shen C. R. (2017). Saturated mutagenesis of ketoisovalerate decarboxylase V461 enabled specific synthesis of 1-pentanol via the ketoacid elongation cycle. Sci. Rep. 7:11284. 10.1038/s41598-017-11624-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin W.-C., Lin K.-H., Liu C.-C., Tsuge K., Huang C.-C. (2017). Improved n-butanol production via co-expression of membrane-targeted tilapia metallothionein and the clostridial metabolic pathway in Escherichia coli. BMC Biotechnol. 17:36. 10.1186/s12896-017-0356-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clomburg J. M., Gonzalez R. (2010). Biofuel production in Escherichia coli: the role of metabolic engineering and synthetic biology. Appl. Microbiol. Biotechnol. 86 419–434. 10.1007/s00253-010-2446-1 [DOI] [PubMed] [Google Scholar]
- Dong H., Zhao C., Zhang T., Lin Z., Li Y., Zhang Y. (2016). Engineering Escherichia coli cell factories for n-butanol production. Adv. Biochem. Eng. Biotechnol. 155 141–163. 10.1007/10_2015_306 [DOI] [PubMed] [Google Scholar]
- Dong H., Zhao C., Zhang T., Zhu H., Lin Z., Tao W., et al. (2017). A systematically chromosomally engineered Escherichia coli efficiently produces butanol. Metab. Eng. 44 284–292. 10.1016/j.ymben.2017.10.014 [DOI] [PubMed] [Google Scholar]
- Eiben C. B., Tian T., Thompson M. G., Mendez-Perez D., Kaplan N., Goyal G., et al. (2020). Adenosine triphosphate and carbon efficient route to second generation biofuel isopentanol. ACS Synth. Biol. 9 468–474. 10.1021/acssynbio.9b00402 [DOI] [PubMed] [Google Scholar]
- Flores A. D., Ayla E. Z., Nielsen D. R., Wang X. (2019). Engineering a synthetic, catabolically orthogonal coculture system for enhanced conversion of lignocellulose-derived sugars to ethanol. ACS Synth. Biol. 8 1089–1099. 10.1021/acssynbio.9b00007 [DOI] [PubMed] [Google Scholar]
- Garst A. D., Bassalo M. C., Pines G., Lynch S. A., Halweg-Edwards A. L., Liu R., et al. (2017). Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat. Biotechnol. 35 48–55. 10.1038/nbt.3718 [DOI] [PubMed] [Google Scholar]
- George K. W., Thompson M. G., Kang A., Baidoo E., Wang G., Chan L. J. G., et al. (2015). Metabolic engineering for the high-yield production of isoprenoid-based C5 alcohols in E. coli. Sci. Rep. 5:11128. 10.1038/srep11128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh I. N., Martien J., Hebert A. S., Zhang Y., Coon J. J., Amador-Noguez D., et al. (2019). OptSSeq explores enzyme expression and function landscapes to maximize isobutanol production rate. Metab. Eng. 52 324–340. 10.1016/j.ymben.2018.12.008 [DOI] [PubMed] [Google Scholar]
- Guo Y., Lu B., Tang H., Bi D., Zhang Z., Lin L., et al. (2019). Tolerance against butanol stress by disrupting succinylglutamate desuccinylase in Escherichia coli. RSC Adv. 9 11683–11695. 10.1039/C8RA09711A [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Xue T., Ma Y., Zhang J., Wang Z., Hong J., et al. (2019). Identification of functional butanol-tolerant genes from Escherichia coli mutants derived from error-prone PCR-based whole-genome shuffling. Biotechnol. Biofuels 12:73. 10.1186/s13068-019-1405-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo M.-J., Jung H.-M., Um J., Lee S.-W., Oh M.-K. (2017). Controlling citrate synthase expression by CRISPR/Cas9 genome editing for n-butanol production in Escherichia coli. ACS Synth. Biol. 6 182–189. 10.1021/acssynbio.6b00134 [DOI] [PubMed] [Google Scholar]
- Herrero A. A., Gomez R. F. (1980). Development of ethanol tolerance in Clostridium thermocellum: effect of growth temperature. Appl. Environ. Microbiol. 40 571–577. 10.1128/aem.40.3.571-577.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi T., Sakai A., Kotani H., Tanabe K., Furusawa C. (2017). Improvement of isopropanol tolerance of Escherichia coli using adaptive laboratory evolution and omics technologies. J. Biotechnol. 255 47–56. 10.1016/j.jbiotec.2017.06.408 [DOI] [PubMed] [Google Scholar]
- Inokuma K., Liao J. C., Okamoto M., Hanai T. (2010). Improvement of isopropanol production by metabolically engineered Escherichia coli using gas stripping. J. Biosci. Bioeng. 110 696–701. 10.1016/j.jbiosc.2010.07.010 [DOI] [PubMed] [Google Scholar]
- Jang Y.-S., Kim B., Shin J. H., Choi Y. J., Choi S., Song C. W., et al. (2012). Bio-based production of C2–C6 platform chemicals. Biotechnol. Bioeng. 109 2437–2459. 10.1002/bit.24599 [DOI] [PubMed] [Google Scholar]
- Jeong H., Lee S.-W., Kim S. H., Kim E.-Y., Kim S., Yoon S. H. (2017). Global functional analysis of butanol-sensitive Escherichia coli and its evolved butanol-tolerant strain. J. Microbiol. Biotechnol. 27 1171–1179. 10.4014/jmb.1702.02021 [DOI] [PubMed] [Google Scholar]
- Jojima T., Inui M., Yukawa H. (2008). Production of isopropanol by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 77 1219–1224. 10.1007/s00253-007-1246-8 [DOI] [PubMed] [Google Scholar]
- Kang A., George K. W., Wang G., Baidoo E., Keasling J. D., Lee T. S. (2016). Isopentenyl diphosphate (IPP)-bypass mevalonate pathways for isopentenol production. Metab. Eng. 34 25–35. 10.1016/j.ymben.2015.12.002 [DOI] [PubMed] [Google Scholar]
- Kang A., Meadows C. W., Canu N., Keasling J. D., Lee T. S. (2017). High-throughput enzyme screening platform for the IPP-bypass mevalonate pathway for isopentenol production. Metab. Eng. 41 125–134. 10.1016/j.ymben.2017.03.010 [DOI] [PubMed] [Google Scholar]
- Kang A., Mendez-Perez D., Goh E.-B., Baidoo E. E. K., Benites V. T., Beller H. R., et al. (2019). Optimization of the IPP-bypass mevalonate pathway and fed-batch fermentation for the production of isoprenol in Escherichia coli. Metab. Eng. 56 85–96. 10.1016/j.ymben.2019.09.003 [DOI] [PubMed] [Google Scholar]
- Kim S., Clomburg J. M., Gonzalez R. (2015). Synthesis of medium-chain length (C6–C10) fuels and chemicals via β-oxidation reversal in Escherichia coli. J. Ind. Microbiol. Biotechnol. 42 465–475. 10.1007/s10295-015-1589-6 [DOI] [PubMed] [Google Scholar]
- Krishnan M. S., Ho N. W., Tsao G. T. (1999). Fermentation kinetics of ethanol production from glucose and xylose by recombinant Saccharomyces 1400(pLNH33). Appl. Biochem. Biotechnol. 77-79 373–388. 10.1385/abab:78:1-3:373 [DOI] [PubMed] [Google Scholar]
- Lan E. I., Liao J. C. (2013). Microbial synthesis of n-butanol, isobutanol, and other higher alcohols from diverse resources. Bioresour. Technol. 135 339–349. 10.1016/j.biortech.2012.09.104 [DOI] [PubMed] [Google Scholar]
- Liang L., Liu R., Garst A. D., Lee T., Nogué V. S. I., Beckham G. T., et al. (2017). CRISPR EnAbled Trackable genome Engineering for isopropanol production in Escherichia coli. Metab. Eng. 41 1–10. 10.1016/j.ymben.2017.02.009 [DOI] [PubMed] [Google Scholar]
- Liang S., Chen H., Liu J., Wen J. (2018). Rational design of a synthetic Entner–Doudoroff pathway for enhancing glucose transformation to isobutanol in Escherichia coli. J. Ind. Microbiol. Biotechnol. 45 187–199. 10.1007/s10295-018-2017-5 [DOI] [PubMed] [Google Scholar]
- Liu F., Wu W., Tran-Gyamfi M. B., Jaryenneh J. D., Zhuang X., Davis R. W. (2017). Bioconversion of distillers’ grains hydrolysates to advanced biofuels by an Escherichia coli co-culture. Microb. Cell Fact. 16:192. 10.1186/s12934-017-0804-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Liang L., Choudhury A., Garst A. D., Eckert C. A., Oh E. J., et al. (2019). Multiplex navigation of global regulatory networks (MINR) in yeast for improved ethanol tolerance and production. Metab. Eng. 51 50–58. 10.1016/j.ymben.2018.07.007 [DOI] [PubMed] [Google Scholar]
- Liu R., Liang L., Garst A. D., Choudhury A., Nogué V. S. I., Beckham G. T., et al. (2018). Directed combinatorial mutagenesis of Escherichia coli for complex phenotype engineering. Metab. Eng. 47 10–20. 10.1016/j.ymben.2018.02.007 [DOI] [PubMed] [Google Scholar]
- Liu S., Skory C., Qureshi N., Hughes S. (2016). The yajC gene from Lactobacillus buchneri and Escherichia coli and its role in ethanol tolerance. J. Ind. Microbiol. Biotechnol. 43 441–450. 10.1007/s10295-015-1730-6 [DOI] [PubMed] [Google Scholar]
- Liu Y., Wan X., Wang B. (2019). Engineered CRISPRa enables programmable eukaryote-like gene activation in bacteria. Nat. Commun. 10:3693. 10.1038/s41467-019-11479-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupino K. M., Romano K. A., Simons M. J., Gregg J. T., Panepinto L., Cruz G. M., et al. (2018). A recurrent silent mutation implicates fecA in ethanol tolerance by Escherichia coli. BMC Microbiol. 18:36. 10.1186/s12866-018-1180-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara M., Urano N., Yamada S., Narutaki A., Fujii M., Kataoka M. (2016). Fermentative production of 1-propanol from d-glucose, l-rhamnose and glycerol using recombinant Escherichia coli. J. Biosci. Bioeng. 122 421–426. 10.1016/j.jbiosc.2016.03.011 [DOI] [PubMed] [Google Scholar]
- McCloskey D., Xu S., Sandberg T. E., Brunk E., Hefner Y., Szubin R., et al. (2018). Evolution of gene knockout strains of E. coli reveal regulatory architectures governed by metabolism. Nat. Commun. 9:3796. 10.1038/s41467-018-06219-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta K., Laviña W. A., Pontrelli S., Liao J. C., Putri S. P., Fukusaki E. (2019). Metabolome analysis revealed the knockout of glyoxylate shunt as an effective strategy for improvement of 1-butanol production in transgenic Escherichia coli. J. Biosci. Bioeng. 127 301–308. 10.1016/j.jbiosc.2018.08.013 [DOI] [PubMed] [Google Scholar]
- Ohtake T., Pontrelli S., Laviña W. A., Liao J. C., Putri S. P., Fukusaki E. (2017). Metabolomics-driven approach to solving a CoA imbalance for improved 1-butanol production in Escherichia coli. Metab. Eng. 41 135–143. 10.1016/j.ymben.2017.04.003 [DOI] [PubMed] [Google Scholar]
- Park S.-H., Kim S., Hahn J.-S. (2014). Metabolic engineering of Saccharomyces cerevisiae for the production of isobutanol and 3-methyl-1-butanol. Appl. Microbiol. Biotechnol. 98, 9139–9147. 10.1007/s00253-014-6081-0 [DOI] [PubMed] [Google Scholar]
- Ronda C., Maury J., Jakoèiunas T., Jacobsen S. A. B., Germann S. M., Harrison S. J., et al. (2015). CrEdit: CRISPR mediated multi-loci gene integration in Saccharomyces cerevisiae. Microb. Cell Fact. 14:97. 10.1186/s12934-015-0288-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini M., Hong Chen M., Chiang C.-J., Chao Y.-P. (2015). Potential production platform of n-butanol in Escherichia coli. Metab. Eng. 27 76–82. 10.1016/j.ymben.2014.11.001 [DOI] [PubMed] [Google Scholar]
- Saini M., Li S.-Y., Wang Z. W., Chiang C.-J., Chao Y.-P. (2016). Systematic engineering of the central metabolism in Escherichia coli for effective production of n-butanol. Biotechnol. Biofuels 9:69. 10.1186/s13068-016-0467-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini M., Wang Z. W., Chiang C.-J., Chao Y.-P. (2017). Metabolic engineering of Escherichia coli for production of n-butanol from crude glycerol. Biotechnol. Biofuels 10:173. 10.1186/s13068-017-0857-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam S., Ngo H.-H., Wu Y.-R. (2020). Advanced CRISPR/Cas-based genome editing tools for microbial biofuels production: a review. Renew. Energ. 149 1107–1119. 10.1016/j.renene.2019.10.107 [DOI] [Google Scholar]
- Shi S., Si T., Liu Z., Zhang H., Ang E., Zhao H. (2016). Metabolic engineering of a synergistic pathway for n-butanol production in Saccharomyces cerevisiae. Sci. Rep. 6:25675. 10.1038/srep25675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shong J., Jimenez Diaz M. R., Collins C. H. (2012). Towards synthetic microbial consortia for bioprocessing. Curr. Opin. Biotechnol. 23 798–802. 10.1016/j.copbio.2012.02.001 [DOI] [PubMed] [Google Scholar]
- Si T., Chao R., Min Y., Wu Y., Ren W., Zhao H. (2017). Automated multiplex genome-scale engineering in yeast. Nat. Commun. 8:15187. 10.1038/ncomms15187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slating T. A., Kesan J. P. (2012). A legal analysis of the effects of the Renewable Fuel Standard (RFS2) and Clean Air Act on the commercialization of biobutanol as a transportation fuel in the United States. Glob. Change Biol. Bioenergy 4 107–118. 10.1111/j.1757-1707.2011.01146.x [DOI] [Google Scholar]
- Soma Y., Tsuruno K., Wada M., Yokota A., Hanai T. (2014). Metabolic flux redirection from a central metabolic pathway toward a synthetic pathway using a metabolic toggle switch. Metab. Eng. 23 175–184. 10.1016/j.ymben.2014.02.008 [DOI] [PubMed] [Google Scholar]
- Soma Y., Yamaji T., Matsuda F., Hanai T. (2017). Synthetic metabolic bypass for a metabolic toggle switch enhances acetyl-CoA supply for isopropanol production by Escherichia coli. J. Biosci. Bioeng. 123 625–633. 10.1016/j.jbiosc.2016.12.009 [DOI] [PubMed] [Google Scholar]
- Song H.-S., Seo H.-M., Jeon J.-M., Moon Y.-M., Hong J. W., Hong Y. G., et al. (2018). Enhanced isobutanol production from acetate by combinatorial overexpression of acetyl-CoA synthetase and anaplerotic enzymes in engineered Escherichia coli. Biotechnol. Bioeng. 115 1971–1978. 10.1002/bit.26710 [DOI] [PubMed] [Google Scholar]
- Sun J., Tian K., Wang J., Dong Z., Liu X., Permaul K., et al. (2018). Improved ethanol productivity from lignocellulosic hydrolysates by Escherichia coli with regulated glucose utilization. Microb. Cell Fact. 17:66. 10.1186/s12934-018-0915-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Survase S. A., Jurgens G., van Heiningen A., Granström T. (2011). Continuous production of isopropanol and butanol using Clostridium beijerinckii DSM 6423. Appl. Microbiol. Biotechnol. 91 1305–1313. 10.1007/s00253-011-3322-3 [DOI] [PubMed] [Google Scholar]
- Tian L., Papanek B., Olson D. G., Rydzak T., Holwerda E. K., Zheng T., et al. (2016). Simultaneous achievement of high ethanol yield and titer in Clostridium thermocellum. Biotechnol. Biofuels 9:116. 10.1186/s13068-016-0528-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T., Kang J. W., Kang A., Lee T. S. (2019). Redirecting metabolic flux via combinatorial multiplex CRISPRi-mediated repression for isopentenol production in Escherichia coli. ACS Synth. Biol. 8 391–402. 10.1021/acssynbio.8b00429 [DOI] [PubMed] [Google Scholar]
- Tripathi S. A., Olson D. G., Argyros D. A., Miller B. B., Barrett T. F., Murphy D. M., et al. (2010). Development of pyrF-based genetic system for targeted gene deletion in Clostridium thermocellum and creation of a pta mutant. Appl. Environ. Microbiol. 76 6591–6599. 10.1128/AEM.01484-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. E., Lanahan A. A., Zheng T., Toruno C., Lynd L. R., Cameron J. C., et al. (2020). Development of both type I-B and type II CRISPR/Cas genome editing systems in the cellulolytic bacterium Clostridium thermocellum. Metab. Eng. Commun. 10:e00116. 10.1016/j.mec.2019.e00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Guo Y., Xu Z., Tu R., Wang Q. (2020). Genomic, transcriptomic, and metabolic characterizations of Escherichia coli adapted to branched-chain higher alcohol tolerance. Appl. Microbiol. Biotechnol. 104 4171–4184. 10.1007/s00253-020-10507-0 [DOI] [PubMed] [Google Scholar]
- Wang C., Pfleger B. F., Kim S. W. (2017). Reassessing Escherichia coli as a cell factory for biofuel production. Curr. Opin. Biotechnol. 45 92–103. 10.1016/j.copbio.2017.02.010 [DOI] [PubMed] [Google Scholar]
- Wang L., York S. W., Ingram L. O., Shanmugam K. T. (2019). Simultaneous fermentation of biomass-derived sugars to ethanol by a co-culture of an engineered Escherichia coli and Saccharomyces cerevisiae. Bioresour. Technol. 273 269–276. 10.1016/j.biortech.2018.11.016 [DOI] [PubMed] [Google Scholar]
- Wang S., Cheng G., Dong J., Tian T., Lee T. S., Mukhopadhyay A., et al. (2019a). NaCl enhances Escherichia coli growth and isoprenol production in the presence of imidazolium-based ionic liquids. Bioresour. Technol. Rep. 6 1–5. 10.1016/j.biteb.2019.01.021 [DOI] [Google Scholar]
- Wang S., Cheng G., Dong J., Tian T., Lee T. S., Mukhopadhyay A., et al. (2019b). Tolerance characterization and isoprenol production of adapted Escherichia coli in the presence of ionic liquids. ACS Sustainable Chem. Eng. 7 1457–1463. 10.1021/acssuschemeng.8b05144 [DOI] [Google Scholar]
- Wang S., Zhao W., Lee T. S., Singer S. W., Simmons B. A., Singh S., et al. (2018). Dimethyl sulfoxide assisted ionic liquid pretreatment of switchgrass for isoprenol production. ACS Sustainable Chem. Eng. 6 4354–4361. 10.1021/acssuschemeng.7b04908 [DOI] [Google Scholar]
- Wen R. C., Shen C. R. (2016). Self-regulated 1-butanol production in Escherichia coli based on the endogenous fermentative control. Biotechnol. Biofuels 9:267. 10.1186/s13068-016-0680-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Wu A., Xiao L., Han R., Ni Y. (2019). Enhancing butanol tolerance of Escherichia coli reveals hydrophobic interaction of multi-tasking chaperone SecB. Biotechnol. Biofuels 12:164. 10.1186/s13068-019-1507-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C., Cheng C. (2019). “Chapter two - butanol production by Clostridium,” in Advances in Bioenergy, eds Li Y., Ge X. (Amsterdam: Elsevier; ), 35–77. 10.1016/bs.aibe.2018.12.001 [DOI] [Google Scholar]
- Yang H., Zhang C., Lai N., Huang B., Fei P., Ding D., et al. (2020). Efficient isopropanol biosynthesis by engineered Escherichia coli using biologically produced acetate from syngas fermentation. Bioresour. Technol. 296:122337. 10.1016/j.biortech.2019.122337 [DOI] [PubMed] [Google Scholar]
- Yu H., Wang N., Huo W., Zhang Y., Zhang W., Yang Y., et al. (2019). Establishment of BmoR-based biosensor to screen isobutanol overproducer. Microb. Cell Fact. 18:30. 10.1186/s12934-019-1084-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaldivar J., Martinez A., Ingram L. O. (2000). Effect of alcohol compounds found in hemicellulose hydrolysate on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol. Bioeng. 68 524–530. [DOI] [PubMed] [Google Scholar]
- Zhao N., Bai Y., Liu C.-G., Zhao X.-Q., Xu J.-F., Bai F.-W. (2014). Flocculating Zymomonas mobilis is a promising host to be engineered for fuel ethanol production from lignocellulosic biomass. Biotechnol. J. 9 362–371. 10.1002/biot.201300367 [DOI] [PubMed] [Google Scholar]
- Zhou J., Lu X., Tian B., Wang C., Shi H., Luo C., et al. (2019). Knockout of acetoacetate degradation pathway gene atoDA enhances the toxicity tolerance of Escherichia coli to isopropanol and acetone. 3 Biotech 9:343. 10.1007/s13205-019-1867-5 [DOI] [PMC free article] [PubMed] [Google Scholar]