Abstract
The dataset presented in this data paper supports “Breaking down insect stoichiometry into chitin-based and internal elemental traits: Patterns and correlates of continent-wide intraspecific variation in the largest European saproxylic beetle” (Orłowski et al. 2020). Here we present the supplementary data and description of methods on the following: (1) mass of elytra and abdomens across 28 local Stag Beetle Lucanus cervus populations in Europe. (2) Population origin and coverage of six major land-cover types, including transport infrastructure, measured in three radii (500 m, 1000 m and 5000 m) around the sampling sites of these populations. (3) The relationship between the mass and concentrations of elements measured in abdomens and elytra in 28 Stag Beetle populations and major land-cover types around the sampling sites.
Keywords: Elemental composition, Trace elements, Internal metal concentrations, Exoskeleton, Chitin-bound metals
Specifications table
| Subject | Ecotoxicology, Ecology, Biological Sciences |
| Specific subject area | Elemental composition, Saproxylic beetles |
| Type of data | Tables and Figures |
| How data was acquired | Through field work and laboratory work |
| Data format | Raw, filtered and analysed |
| Parameters for data collection | Investigation of abdomens (n = 124) and elytra (n = 271) of adult Stag Beetles collected at 28 localities situated in eight European countries from Central Spain to Western Russia. |
| Description of data collection | The measurement of concentrations of 12 chemical elements (Ca, Mg, K, Na, Mn, Fe, Zn, Cu, As, Cd, Pb and Ni), and relating these to insect size and local habitat conditions. |
| Data source location | Institution: Research Station of the Institute of the Agricultural and Forest Environment, Polish Academy of Sciences City/Town/Region: Turew, Wielkopolska Country: Poland |
| Data accessibility Related research article |
All raw data are given along with the article as Appendix 1. G. Orłowski, L. Mróz, M. Kadej, A. Smolis, D. Tarnawski, J. Karg, A. Campanaro, M. Bardiani, D. J. Harvey, M. Méndez, A. Thomaes, A. Vrezec, K. Ziomek, A. L. Rudecki, D. Mader, Breaking down insect stoichiometry into chitin-based and internal elemental traits: Patterns and correlates of continent-wide intraspecific variation in the largest European saproxylic beetle. Environ. Poll. 262, 2020, 114,064. |
Value of the data
-
•
This is the first published set of data on the variability of elemental traits (ETs), i.e. the concentrations of essential and non-essential elements, measured in the elytra and abdomens of Stag Beetles across the species’ distributional range in Europe.
-
•
This dataset fills gaps in the environmental data relating to the accumulation of non-essential (toxic) elements in Stag Beetles.
-
•
The data help to understand the differentiation between the elemental pool of metals accumulated in the exoskeleton and internal organs, including the residual body fat of holometabolous insects.
-
•
The data can be used to determine the extent and impact of the coverage of major land-cover types, including transport infrastructure, on ETs of the exoskeletons and internal organs of beetles.
-
•
The data may be useful for explaining the sources of variation in internal and exoskeletal ETs of beetles and other insects, and provide a basis for further ecotoxicology work.
1. Data description
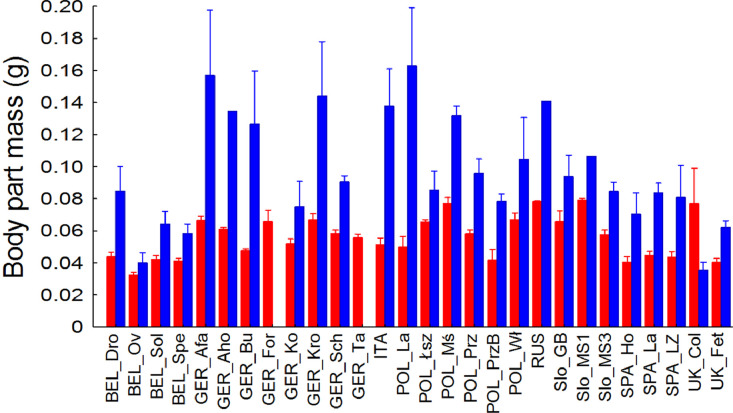
The data presented here (Photo 1; Fig. 1; Table 1, Table 2, Table 3, Table 4; Appendix A) constitute the basis for the article by Orłowski et al. [1]. The dataset provides detailed information on: (1) the masses of elytra and abdomens across 28 local Stag Beetle Lucanus cervus (Photo 1) populations in Europe (Fig. 1); (2) the population origin and coverage of six major land-cover types, including transport infrastructure, measured in three radii (500 m, 1000 m and 5000 m) around the sampling sites of these populations (Tables 1 and 2); (3) the relationship between the mass and concentrations of elements measured in abdomens and elytra in 28 Stag Beetle populations and major land-cover types around the sampling sites (Tables 3 and 4). The raw data with the concentrations of 12 chemical elements measured in elytra and abdomens of Stag Beetles from 28 European populations are presented in Appendix A.
Photo 1.
The European Stag Beetle Lucanus cervus. Top: Sexual dimorphism; left to right – male imago, body size 46 mm, and female imago, body size 37 mm. Bottom: dissection of a teneral imago, body size 36 mm, showing the fat body (white), and part of the alimentary canal. The midgut, 45 mm, has food remnants from the larval stage, the hind gut, ∼24 mm, is clear. The hind gut is relatively short, a characteristic of sap feeders. In the adult, food intake, if any, is restricted to sap runs; but it can be tempted to sugary liquids. Photos© Maria Fremlin.
Fig. 1.
Comparison (average ± SE) of mass of elytra (red columns; n = 271) and abdomens (blue columns; n = 124) across 28 local Stag Beetle Lucanus cervus populations in Europe. Notes: (1) The masses of elytra (F27,243 = 2.94, P < 0.001) and abdomens (F25,98 = 4.18, P < 0.001) both vary significantly among these populations. Similarly, the masses of both these body parts differ significantly per individual population across all these sites (body part × population interaction term: F27,367 = 2.41, P < 0.001). (2) The masses of elytra and abdomens (for each n = 26) were both positively correlated with the longitudes (for georeferences, see Table 1) of the sampling sites (Pearson r = 0.583 and 0.491, P = 0.002 and 0.011; respectively), but not with their latitudes (r = 0.228 and 0.056, P = 0.262 and 0.787; respectively). However, because of the small sample size and no sexing of the beetles, the results of this analysis should be treated with caution: they are presented only for the purposes of this paper, and may not represent the actual variation in body size of L. cervus across Europe.
Table 1.
Population origin and coverage of six major land-cover types, including transport infrastructure, measured in three radii (500 m / 1000 m / 5000 m) around the sampling sites of 28 Stag Beetle Lucanus cervus populations; for sample size, see Table 2. Note: the total areas in consecutive radii are 500 m = 78.5 ha, 1000 m = 314 ha and 5000 m = 7 850 ha. 1Includes artificial non-agricultural areas such as lawns, and sport and leisure areas.
| Country | Site = Village or town name/Conventional description | Population (acronym) | Latitude, longitude | Urban/ built-up areas (ha) | Wood/park/forest (ha) | Agriculturalland/ cultivation (ha) | Grassland/ open habitats with low vegetation (ha)1 | Roads/railways (ha) | Water/ lakes/ river (ha) |
|---|---|---|---|---|---|---|---|---|---|
| Belgium | Speelberg (suburban) | BEL_Spe | 50.761152, 4.530245 | 27.9/29.2/3838.3 | 1.7/50.2/1120.8 | 47.1/197.9/2539.6 | 0.0/23.9/211.4 | 1.8/11.0/67.6 | 0.0/1.9/72.4 |
| Overijse (Putterstr) (suburban) | BEL_Ov | 50.777305, 4.545712 | 27.3/112.8/2809.4 | 19.7/61.1/1777.6 | 26.0/124.5/3056.3 | 0.0/0.0/135.3 | 1.8/4.6/39.7 | 3.7/11.0/31.7 | |
| Solheide (suburban) | BEL_Sol | 50.774054, 4.535564 | 64.9/ 167.7/2924.9 | 3.2/57.8/1832.2 | 1.9/57.9/2862.3 | 4.9/19.3/142.3 | 1.9/6.6/56.0 | 1.8/4.7/32.4 | |
| Drogenberg (suburban) | BEL_Dro | 50.771654, 4.528396 | 40.9/151.1/2945.3 | 25.7/85.8/1998.7 | 0.0/44.2/2622.2 | 7.4/18.0/158.4 | 1.5/7.3/62.9 | 2.9/7.6/62.6 | |
| Germany | Bullay (suburban) | GER_Bu | 50.053653, 7.133485 | 31.8/92.0/2447.8 | 3.3/89.2/2230.5 | 3.4/60.4/1509.5 | 7.9/30.0/690.7 | 4.9/3.6/29.5 | 27.2/38.9/971.5 |
| Alf (Holzplatz) (suburban) | GER_Aho | 50.050175, 7.104856 | 9.6/34.3/1184.8 | 61.8/229.5/5737.3 | 4.9/33.2/829.0 | 0.0/0.0/0.0 | 2.1/16.4/82.0 | 0.0/0.7/16.9 | |
| Alf (Fabrik) (suburban) | GER_Afa | 50.058964, 7.106158 | 1.6/33.6/840.8 | 68.8/195.1/4877.6 | 0.0/61.6/1538.8 | 5.8/22.8/571.0 | 1.3/0.0/0.0 | 1.1/0.9/21.8 | |
| Samtgemeinde Schüttorf (suburban) | GER_Sch | 52.327063, 7.257903 | 3.1/39.1/994.5 | 36.6/91.6/2288.9 | 31.2/120.5/3012.1 | 5.3/43.7/1093.7 | 2.1/11.1/262.5 | 0.1/7.9/198.3 | |
| Koblenz (suburban) | GER_Ko | 50.348776, 7.580741 | 18.2/199.7/4992.7 | 50.4/82.8/2068.8 | 0.0/0.0/0.0 | 1.9/6.4/159.3 | 8.0/25.0/624.4 | 0.0/0.2/4.8 | |
| Tairnbach (suburban) | GER_Ta | 49.255642, 8.750627 | 9.7/38.6/964.2 | 51.2/138.1/3452.2 | 13.5/108.0/2699.9 | 2.7/18.8/469.0 | 1.4/1.9/48.7 | 0.1/8.6/216.0 | |
| Kronau (suburban) | GER_Kro | 49.215231, 8.620142 | 52.0/125.3/3132.6 | 1.6/37.0/926.1 | 16.0/129.5/2714.8 | 8.0/0.0/0.0 | 0.9/5.6/140.2 | 0.0/16.6/414.5 | |
| Forst (suburban) | GER_For | 49.147766, 8.575710 | 33.2/84.2/2120.8 | 25.3/105.7/2641.6 | 5.7/83.3/2082.5 | 6.9/25.4/635.5 | 3.1/9.6/225.9 | 4.3/5.7/143.6 | |
| Italy | Marmirolo (forest) | ITA | 45.200637, 10.745660 | 0.7/1.5/1078.9 | 59.6/214.8/238.8 | 0.0/82.1/6071.8 | 17.8/13.7/90.7 | 0.0/0.0/0.0 | 0.4/1.9/391.8 |
| Poland | Przemków, Beech Forest reserve | POL_PrzB | 51.508000, 15.693300 | 0.0/0.0/175.4 | 78.5/289.9/3747.0 | 0.0/0.0/3927.6 | 0.0/24.1/27.1/ | 0.0/0.0/0.0 | 0.0/0.0/0.0 |
| Przemków (forest) | POL_Prz | 51.516150, 15.861000 | 0.0/0.0/56.0 | 78.5/314.0/6845.0 | 0.0/0.0/949.1 | 0.0/0.0/0.0 | 0.0/0.0/0.0 | 0.0/0.0/0.0 | |
| Mścigniew (forest) | POL_Mś | 51.914931, 16.326594 | 0.0/0.0/321.2 | 77.6/297.7/5015.7 | 0.0/13.9/2092.2 | 0.0/0.0/0.0 | 0.9/2.4/11.9 | 0.0/0.0/409.0 | |
| Włoszakowice (forest) | POL_Wł | 51.897786, 16.336065 | 0.0/0.0/201.0 | 78.5/314.7/5473.4 | 0.0/0.0/2123.0 | 0.0/0.0/0.0 | 0.0/0.0/0.0 | 0.0/0.0/52.5 | |
| Ławszowa Jeziory (forest) | POL_Łsz | 51.397417, 15.455117 | 0.0/0.0/26.1 | 78.5/314.0/880.1 | 0.0/0.0/499.1 | 0.0/0.0/444.7 | 0.0/0.0/0.0 | 0.0/0.0/0.0 | |
| Lasowice Milicz (forest) | POL_La | 51.435584, 17.254254 | 0.0/0.0/79.5 | 77.6/312.0/6836.7 | 0.0/0.0/923.9 | 0.0/0.0/0.0 | 0.9/2.0/9.9 | 0.0/0.0/0.0 | |
| Russia | Kursk (forest) | RUS | 51.146944, 36.430000 | 0.0/0.0/148.5 | 14.3/160.4/852.1 | 57.7/109.0/4876.9 | 6.5/44.7/1972.5 | 0.0/0.0/0.0 | 0.0/0.0/0.0 |
| Slovenia | Gornja Bistrica (forest) | Slo_GB | 46.547811, 16.260903 | 0.7/27.5/686.3 | 69.4/202.1/5069.9 | 3.0/59.2/1479.9 | 0.0/9.2/230.4 | 0.0/1.4/34.1 | 5.4/14.7/367.8 |
| Murska šuma 1 (forest) | Slo_MS1 | 46.493156, 16.52374 | 0.0/0.0/0.0 | 62.0/283.2/7080.4 | 0.0/0.0/0.0 | 8.7/10.6/265.2 | 0.0/0.0/0.0 | 7.8/20.2/504.4 | |
| Murska šuma 3 (forest) | Slo_MS3 | 46.504614, 16.510087 | 0.0/2.9/72.8 | 41.2/159.9/3997.3 | 37.3/146.8/3671.0 | 0.0/0.0/0.0 | 0.0/0.0/0.0 | 0.0/4.4/108.9 | |
| Spain | La Laguna, Asturias (forest) | SPA_La | 43.514092, −5.958645 | 9.3/15.6/389.9 | 32.6/102.5/2562.3 | 33.1/141.1/3527.1 | 2.0/50.6/1265.8 | 1.5/4.2/105.0 | 0.0/0.0/0.0 |
| Lozoya river Spain (forest) | SPA_LZ | 40.900844, −3.457267 | 0.0/0.0/0.0 | 15.3/34.1/65.8 | 0.0/0.0/0.0 | 60.0/271.1/6778.2 | 1.0/3.6/90.1 | 2.3/5.1/981.7 | |
| Horcajo de la Sierra (forest) | SPA_Ho | 41.068931, −3.579413 | 1.9/8.2/205.9 | 8.8/82.7/1011.7 | 24.7/0.0/1054.8 | 41.8/219.6/5490.0 | 1.4/3.5/87.7 | 0.0/0.0/0.0 | |
| United Kingdom | Colchester (urban) | UK_Col | 51.883472, 0.883527 | 58.8/235.3/5881.5 | 3.7/19.0/476.1 | 0.0/0.0/0.0 | 11.4/51.0/1275.1 | 4.6/8.7/217.3 | 0.0/0.0/0.0 |
| Fetcham (urban) | UK_Fet | 51.297159, −0.351009 | 37.5/162.1/4052.4 | 14.5/33.2/860.0 | 9.4/58.0/1451.2 | 5.3/46.9/1172.2 | 7.9/10.2/255.0 | 4.0/3.6/88.8 |
Table 2.
Descriptive statistics (median; range) of concentrations of 12 chemical elements measured in elytra (white rows) and abdomens (blue rows) of Stag Beetles Lucanus cervus from 28 European populations of the species; see Table 1 for population acronyms; the last two rows contain data pooled across all the populations.
| Population | Number of samples | Ca | Mg | K | Na | Mn | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Med | Min | Max | Med | Min | Max | Med | Min | Max | Med | Min | Max | Med | Min | Max | |||||||
| BEL_Dro | 8 | 378.1 | 185.4 | 516.7 | 593.1 | 455.1 | 889.0 | 2068.1 | 1133.2 | 2293.6 | 273.7 | 215.9 | 442.9 | 6.6 | 5.0 | 7.7 | |||||
| BEL_Dro | 3 | 812.6 | 465.6 | 984.7 | 5342.7 | 3268.8 | 5537.5 | 7085.7 | 6123.4 | 9135.3 | 713.7 | 707.0 | 1043.4 | 14.0 | 9.2 | 17.7 | |||||
| BEL_Ov | 4 | 452.8 | 317.3 | 1765.0 | 437.6 | 311.2 | 627.1 | 1765.6 | 784.2 | 2847.4 | 561.0 | 198.4 | 1288.7 | 5.9 | 2.9 | 23.9 | |||||
| BEL_Ov | 2 | 930.8 | 567.2 | 1294.5 | 2121.0 | 1313.9 | 2928.1 | 5839.1 | 1816.1 | 9862.1 | 1065.0 | 353.1 | 1776.9 | 20.3 | 19.3 | 21.3 | |||||
| BEL_Sol | 12 | 510.1 | 290.2 | 2672.5 | 554.5 | 348.7 | 2036.5 | 1919.9 | 661.1 | 2683.9 | 247.7 | 119.8 | 447.2 | 17.3 | 6.4 | 96.2 | |||||
| BEL_Sol | 6 | 1324.2 | 664.9 | 2509.7 | 3220.7 | 809.6 | 10,347.7 | 6390.8 | 945.5 | 7658.4 | 627.4 | 294.4 | 687.9 | 22.6 | 12.1 | 31.0 | |||||
| BEL_Spe | 33 | 476.5 | 257.8 | 3629.5 | 570.1 | 333.8 | 1307.9 | 2466.7 | 496.6 | 4925.9 | 398.0 | 142.6 | 630.0 | 13.6 | 6.2 | 44.6 | |||||
| BEL_Spe | 19 | 669.9 | 445.2 | 2940.5 | 2817.1 | 1514.1 | 9611.3 | 6458.7 | 3247.0 | 9743.8 | 821.1 | 457.9 | 1283.0 | 18.9 | 11.9 | 62.4 | |||||
| GER_Afa | 4 | 302.3 | 200.4 | 481.9 | 692.2 | 641.2 | 777.4 | 3917.8 | 2070.6 | 5828.2 | 753.1 | 646.1 | 795.4 | 34.3 | 26.6 | 48.8 | |||||
| GER_Afa | 2 | 425.7 | 364.2 | 487.2 | 2951.6 | 1920.0 | 3983.3 | 10,678.2 | 10,293.7 | 11,062.7 | 1993.7 | 1744.2 | 2243.2 | 64.7 | 54.9 | 74.5 | |||||
| GER_Aho | 4 | 357.9 | 351.9 | 367.8 | 542.4 | 417.1 | 661.7 | 2754.2 | 2704.0 | 2997.1 | 498.0 | 423.7 | 528.7 | 24.2 | 19.7 | 27.9 | |||||
| GER_Aho | 1 | 483.3 | 483.3 | 483.3 | 4175.6 | 4175.6 | 4175.6 | 6214.7 | 6214.7 | 6214.7 | 986.8 | 986.8 | 986.8 | 35.4 | 35.4 | 35.4 | |||||
| GER_Bu | 6 | 393.9 | 286.6 | 420.9 | 732.8 | 471.2 | 1050.9 | 5182.3 | 4326.2 | 7253.5 | 899.5 | 755.1 | 1022.2 | 21.0 | 16.9 | 24.3 | |||||
| GER_Bu | 3 | 1100.4 | 1006.0 | 1662.5 | 8016.3 | 5888.1 | 10,211.5 | 11,909.8 | 11,175.9 | 15,927.7 | 2520.9 | 1949.1 | 2572.8 | 64.2 | 49.7 | 85.6 | |||||
| GER_For | 6 | 417.3 | 335.6 | 588.8 | 432.5 | 292.9 | 637.0 | 3252.2 | 2595.0 | 3777.8 | 805.8 | 594.9 | 1103.9 | 11.5 | 9.9 | 20.5 | |||||
| GER_Ko | 11 | 359.2 | 295.1 | 482.5 | 704.7 | 392.3 | 926.3 | 2317.8 | 1075.7 | 3836.8 | 750.6 | 486.4 | 1066.3 | 20.6 | 9.5 | 25.2 | |||||
| GER_Ko | 4 | 583.4 | 387.1 | 720.8 | 3207.3 | 2106.6 | 8601.8 | 10,087.6 | 4454.2 | 11,908.2 | 1538.4 | 1009.0 | 1798.1 | 22.2 | 18.3 | 32.5 | |||||
| GER_Kro | 9 | 474.9 | 376.2 | 1573.6 | 693.0 | 368.6 | 1012.9 | 3218.7 | 2032.3 | 4358.5 | 875.6 | 748.2 | 1930.6 | 12.9 | 10.9 | 23.4 | |||||
| GER_Kro | 2 | 704.6 | 676.1 | 733.1 | 2158.9 | 1551.9 | 2765.9 | 8982.1 | 8167.2 | 9797.1 | 2081.2 | 2003.0 | 2159.5 | 16.6 | 15.9 | 17.3 | |||||
| GER_Sch | 6 | 360.1 | 338.0 | 407.7 | 654.9 | 576.5 | 819.9 | 2847.3 | 2539.6 | 3877.3 | 872.0 | 622.8 | 1300.9 | 15.0 | 9.8 | 24.8 | |||||
| GER_Sch | 2 | 675.2 | 639.6 | 710.8 | 5921.5 | 2612.1 | 9230.8 | 10,510.0 | 6717.5 | 14,302.5 | 1778.1 | 1402.0 | 2154.2 | 37.7 | 24.7 | 50.7 | |||||
| GER_Ta | 18 | 378.7 | 265.1 | 1092.2 | 578.2 | 464.8 | 1083.2 | 2559.8 | 808.8 | 5686.5 | 886.9 | 335.2 | 1237.3 | 12.1 | 6.6 | 19.8 | |||||
| ITA | 11 | 598.5 | 311.8 | 3755.3 | 819.3 | 504.7 | 1689.0 | 1294.8 | 977.8 | 3348.4 | 628.1 | 279.0 | 1204.0 | 21.1 | 16.4 | 31.3 | |||||
| ITA | 7 | 837.8 | 521.8 | 4634.5 | 4482.7 | 2495.9 | 6416.5 | 6016.0 | 5425.5 | 14,147.8 | 826.2 | 600.9 | 2612.2 | 17.6 | 10.4 | 29.2 | |||||
| POL_La | 4 | 341.7 | 242.3 | 1765.1 | 664.4 | 582.5 | 1074.0 | 2246.3 | 1570.2 | 4884.9 | 353.0 | 104.2 | 1237.5 | 38.8 | 19.0 | 84.3 | |||||
| POL_La | 3 | 361.0 | 266.3 | 439.2 | 5903.1 | 1507.2 | 13,474.6 | 11,696.2 | 6895.8 | 12,331.8 | 959.4 | 876.2 | 2003.1 | 29.6 | 13.3 | 31.9 | |||||
| POL_Łsz | 12 | 285.0 | 251.6 | 383.3 | 563.1 | 447.8 | 641.6 | 3966.5 | 3260.4 | 6522.9 | 724.8 | 540.0 | 1158.1 | 16.0 | 6.9 | 20.2 | |||||
| POL_Łsz | 6 | 504.2 | 231.3 | 1200.0 | 2119.4 | 974.7 | 4091.6 | 11,715.1 | 6245.5 | 29,765.8 | 1844.1 | 1069.0 | 4018.0 | 63.4 | 23.9 | 109.0 | |||||
| POL_Mś | 6 | 286.7 | 246.7 | 848.1 | 480.8 | 421.9 | 625.1 | 3721.6 | 3252.4 | 4969.1 | 959.7 | 725.7 | 1088.3 | 19.9 | 12.5 | 27.0 | |||||
| POL_Mś | 3 | 469.4 | 456.4 | 662.5 | 1913.4 | 1379.7 | 1970.2 | 12,022.9 | 11,369.6 | 17,210.4 | 1954.6 | ` | 2022.9 | 35.7 | 28.1 | 53.0 | |||||
| POL_Prz | 30 | 335.4 | 219.3 | 2444.6 | 524.1 | 120.2 | 764.9 | 3249.1 | 529.3 | 5485.5 | 822.5 | 82.1 | 1725.2 | 21.7 | 11.1 | 321.0 | |||||
| POL_Prz | 15 | 512.5 | 216.4 | 1300.3 | 2551.8 | 956.5 | 5599.7 | 15,762.7 | 1126.3 | 20,761.3 | 2081.6 | 81.2 | 2677.8 | 78.7 | 27.8 | 623.2 | |||||
| POL_PrzB | 4 | 483.6 | 337.6 | 572.8 | 898.0 | 559.2 | 1152.0 | 3157.5 | 2793.8 | 8913.1 | 626.8 | 564.3 | 694.3 | 30.0 | 28.4 | 35.3 | |||||
| POL_PrzB | 2 | 387.1 | 297.3 | 476.9 | 2315.3 | 1592.8 | 3037.9 | 11,812.4 | 8778.8 | 14,846.0 | 1308.0 | 1071.3 | 1544.7 | 39.6 | 34.9 | 44.2 | |||||
| POL_Wł | 10 | 382.1 | 229.5 | 513.2 | 599.2 | 451.2 | 872.5 | 3445.8 | 2511.6 | 4616.5 | 1314.3 | 701.8 | 1744.7 | 29.3 | 19.4 | 41.5 | |||||
| POL_Wł | 6 | 489.3 | 327.5 | 755.4 | 2472.3 | 780.6 | 9191.9 | 10,039.3 | 3625.7 | 15,077.2 | 1601.5 | 791.3 | 2410.5 | 44.2 | 26.4 | 56.9 | |||||
| RUS | 2 | 290.3 | 262.0 | 318.6 | 544.2 | 505.3 | 583.2 | 2807.9 | 2650.6 | 2965.1 | 1891.9 | 1571.4 | 2212.5 | 8.4 | 7.2 | 9.6 | |||||
| RUS | 1 | 922.7 | 922.7 | 922.7 | 1534.5 | 1534.5 | 1534.5 | 9521.6 | 9521.6 | 9521.6 | 2356.4 | 2356.4 | 2356.4 | 19.1 | 19.1 | 19.1 | |||||
| Slo_GB | 4 | 250.6 | 166.6 | 355.8 | 158.8 | 143.7 | 163.7 | 534.8 | 404.5 | 846.2 | 126.1 | 119.3 | 137.0 | 9.1 | 7.8 | 11.1 | |||||
| Slo_GB | 2 | 502.5 | 437.1 | 567.9 | 521.6 | 391.3 | 651.9 | 903.8 | 789.5 | 1018.2 | 227.4 | 196.8 | 258.0 | 14.5 | 14.3 | 14.7 | |||||
| Slo_MS1 | 2 | 231.7 | 207.7 | 255.7 | 229.7 | 190.2 | 269.3 | 576.5 | 464.0 | 688.9 | 151.6 | 119.9 | 183.3 | 11.4 | 11.3 | 11.5 | |||||
| Slo_MS1 | 1 | 290.0 | 290.0 | 290.0 | 414.4 | 414.4 | 414.4 | 688.0 | 688.0 | 688.0 | 188.4 | 188.4 | 188.4 | 20.4 | 20.4 | 20.4 | |||||
| Slo_MS3 | 10 | 313.1 | 248.1 | 434.1 | 208.6 | 156.5 | 366.8 | 1141.4 | 898.2 | 1608.5 | 211.6 | 165.4 | 351.5 | 12.9 | 9.6 | 21.9 | |||||
| Slo_MS3 | 5 | 579.3 | 519.5 | 623.7 | 577.2 | 424.1 | 823.2 | 2341.1 | 2201.5 | 2880.4 | 439.9 | 358.3 | 452.2 | 18.9 | 15.1 | 27.0 | |||||
| SPA_Ho | 7 | 296.6 | 267.1 | 642.4 | 926.8 | 371.3 | 1007.5 | 2250.6 | 1993.2 | 4658.9 | 969.2 | 371.6 | 1044.7 | 17.1 | 13.8 | 23.3 | |||||
| SPA_Ho | 4 | 506.4 | 384.7 | 606.2 | 2914.3 | 1813.3 | 4954.0 | 8765.3 | 5153.2 | 12,765.2 | 1244.9 | 1010.0 | 1543.9 | 29.6 | 23.0 | 42.6 | |||||
| SPA_La | 17 | 415.3 | 271.5 | 534.4 | 810.0 | 656.8 | 1040.5 | 1559.0 | 854.5 | 2737.9 | 717.8 | 356.6 | 1239.5 | 15.5 | 7.9 | 33.0 | |||||
| SPA_La | 8 | 445.4 | 368.7 | 686.5 | 3092.1 | 1755.2 | 7268.8 | 5793.4 | 4691.8 | 8711.0 | 1111.1 | 834.7 | 2666.1 | 19.1 | 9.0 | 22.5 | |||||
| SPA_LZ | 5 | 278.2 | 222.3 | 359.0 | 1091.9 | 609.0 | 1177.8 | 2511.0 | 1634.7 | 2745.5 | 731.2 | 678.5 | 1660.9 | 20.6 | 18.6 | 23.3 | |||||
| SPA_LZ | 3 | 399.0 | 391.3 | 556.4 | 5496.8 | 1258.7 | 6007.1 | 9316.9 | 5145.2 | 9947.4 | 1084.3 | 1065.1 | 1707.9 | 32.0 | 21.3 | 42.0 | |||||
| UK_Col | 14 | 329.1 | 27.6 | 711.4 | 683.7 | 24.3 | 1080.0 | 2536.4 | 207.2 | 5760.6 | 884.0 | 37.5 | 1410.8 | 15.5 | 0.8 | 22.8 | |||||
| UK_Col | 7 | 1315.0 | 781.5 | 2684.7 | 4165.9 | 720.0 | 16,348.5 | 7025.6 | 2231.3 | 14,496.1 | 902.2 | 556.0 | 2736.4 | 44.1 | 20.3 | 70.9 | |||||
| UK_Fet | 12 | 329.4 | 258.9 | 659.0 | 500.1 | 435.2 | 576.8 | 3755.4 | 2300.4 | 7172.3 | 979.8 | 611.5 | 2027.7 | 13.3 | 9.0 | 21.2 | |||||
| UK_Fet | 7 | 1160.6 | 891.3 | 1680.7 | 3957.3 | 2869.0 | 8756.4 | 15,183.8 | 11,191.2 | 20,611.8 | 2867.6 | 2090.9 | 5717.1 | 30.1 | 23.0 | 51.2 | |||||
| Totals | 271 | 367.5 | 27.6 | 3755.3 | 591.2 | 24.3 | 2036.5 | 2683.9 | 207.2 | 8913.1 | 707.5 | 37.5 | 2212.5 | 17.2 | 0.8 | 321.0 | |||||
| Totals | 124 | 610.3 | 216.4 | 4634.5 | 3016.2 | 391.3 | 16,348.5 | 8349.1 | 688.0 | 29,765.8 | 1206.2 | 81.2 | 5717.1 | 26.9 | 9.0 | 623.2 | |||||
| Population | Fe | Zn | Cu | As | Cd | Pb | Ni | ||||||||||||||
| Med | Min | Max | Med | Min | Max | Med | Min | Max | Med | Min | Max | Med | Min | Max | Med | Min | Max | Med | Min | Max | |
| BEL_Dro | 29.6 | 17.3 | 52.7 | 11.1 | 0.0 | 38.6 | 4.0 | 3.2 | 4.5 | 0.6 | 0.24 | 2.2 | 0.008 | 0.004 | 0.022 | 0.385 | 0.23 | 1.72 | 0.17 | 0.044 | 0.31 |
| BEL_Dro | 156.7 | 153.5 | 323.1 | 170.6 | 156.3 | 210.9 | 18.6 | 12.5 | 27.4 | 2.7 | 2.68 | 16.9 | 0.019 | 0.018 | 0.094 | 0.572 | 0.55 | 1.17 | 0.42 | 0.217 | 0.64 |
| BEL_Ov | 125.6 | 31.9 | 966.7 | 51.5 | 5.9 | 54.5 | 7.7 | 7.4 | 9.5 | 3.8 | 0.94 | 4.3 | 0.035 | 0.024 | 0.082 | 1.152 | 0.80 | 3.58 | 5.00 | 1.999 | 9.20 |
| BEL_Ov | 544.7 | 221.1 | 868.2 | 322.2 | 96.6 | 547.8 | 36.0 | 13.2 | 58.8 | 34.2 | 17.25 | 51.1 | 0.056 | 0.054 | 0.059 | 1.639 | 1.48 | 1.80 | 2.20 | 1.782 | 2.62 |
| BEL_Sol | 135.5 | 7.1 | 2220.8 | 14.1 | 3.0 | 43.5 | 6.7 | 3.5 | 17.1 | 1.4 | 0.39 | 5.7 | 0.020 | 0.006 | 0.171 | 1.114 | 0.43 | 9.80 | 1.08 | 0.358 | 3.49 |
| BEL_Sol | 282.3 | 146.2 | 943.6 | 239.9 | 97.2 | 348.9 | 35.9 | 21.3 | 95.7 | 19.4 | 2.26 | 88.0 | 0.052 | 0.040 | 0.138 | 1.719 | 0.84 | 32.14 | 0.58 | 0.339 | 0.90 |
| BEL_Spe | 32.2 | 11.7 | 1141.9 | 14.5 | 0.0 | 73.4 | 5.3 | 3.8 | 15.6 | 1.4 | 0.21 | 8.7 | 0.026 | 0.008 | 0.094 | 0.714 | 0.23 | 7.86 | 0.37 | 0.075 | 6.29 |
| BEL_Spe | 103.2 | 45.9 | 1243.1 | 218.5 | 110.7 | 526.0 | 28.6 | 10.5 | 156.1 | 22.5 | 1.55 | 186.9 | 0.066 | 0.030 | 0.370 | 0.991 | 0.55 | 3.95 | 0.42 | 0.150 | 7.60 |
| GER_Afa | 317.3 | 69.6 | 1075.1 | 29.9 | 8.8 | 54.3 | 6.7 | 5.4 | 6.9 | 2.2 | 1.52 | 2.9 | 0.015 | 0.010 | 0.019 | 0.607 | 0.34 | 1.45 | 1.21 | 0.957 | 1.82 |
| GER_Afa | 812.4 | 783.3 | 841.5 | 170.9 | 123.9 | 217.9 | 58.9 | 21.7 | 96.1 | 9.6 | 1.09 | 18.0 | 0.026 | 0.021 | 0.031 | 1.183 | 0.93 | 1.43 | 1.41 | 1.405 | 1.41 |
| GER_Aho | 44.0 | 28.9 | 64.2 | 6.1 | 5.5 | 7.0 | 3.9 | 3.7 | 4.8 | 0.8 | 0.61 | 0.9 | 0.012 | 0.006 | 0.014 | 0.299 | 0.14 | 0.43 | 0.33 | 0.172 | 0.46 |
| GER_Aho | 297.1 | 297.1 | 297.1 | 119.0 | 119.0 | 119.0 | 20.2 | 20.2 | 20.2 | 1.3 | 1.27 | 1.3 | 0.017 | 0.017 | 0.017 | 0.223 | 0.22 | 0.22 | 0.31 | 0.307 | 0.31 |
| GER_Bu | 96.7 | 73.4 | 303.1 | 40.2 | 20.8 | 54.0 | 10.5 | 7.2 | 16.8 | 4.5 | 2.28 | 8.5 | 0.013 | 0.013 | 0.018 | 0.718 | 0.44 | 3.79 | 1.03 | 0.572 | 1.48 |
| GER_Bu | 668.1 | 167.0 | 1125.7 | 193.1 | 119.7 | 302.1 | 136.4 | 106.5 | 219.1 | 18.5 | 0.09 | 38.5 | 0.017 | 0.007 | 0.048 | 1.744 | 0.39 | 2.10 | 0.80 | 0.172 | 1.24 |
| GER_For | 28.3 | 14.7 | 72.0 | 30.2 | 13.3 | 53.4 | 3.3 | 2.9 | 3.6 | 0.9 | 0.45 | 1.3 | 0.005 | 0.003 | 0.007 | 0.541 | 0.27 | 0.94 | 0.31 | 0.154 | 1.30 |
| GER_Ko | 26.9 | 13.6 | 102.3 | 8.3 | 0.8 | 46.7 | 4.7 | 3.0 | 8.8 | 1.4 | 0.23 | 11.6 | 0.013 | 0.004 | 0.027 | 0.269 | 0.15 | 1.19 | 0.26 | 0.169 | 0.98 |
| GER_Ko | 123.3 | 76.9 | 215.6 | 247.3 | 91.7 | 407.2 | 70.4 | 53.4 | 125.3 | 21.7 | 7.58 | 59.4 | 0.040 | 0.016 | 0.065 | 0.334 | 0.18 | 1.05 | 0.28 | 0.056 | 0.41 |
| GER_Kro | 35.1 | 23.7 | 121.5 | 8.5 | 4.0 | 17.5 | 4.7 | 3.5 | 5.7 | 0.7 | 0.51 | 2.4 | 0.006 | 0.003 | 0.011 | 0.534 | 0.32 | 1.23 | 0.50 | 0.370 | 0.86 |
| GER_Kro | 122.5 | 67.3 | 177.8 | 153.1 | 145.2 | 161.0 | 49.4 | 43.2 | 55.6 | 14.2 | 1.60 | 26.7 | 0.017 | 0.006 | 0.027 | 0.237 | 0.20 | 0.27 | 0.23 | 0.224 | 0.24 |
| GER_Sch | 24.6 | 13.2 | 59.8 | 8.8 | 0.2 | 22.0 | 5.9 | 3.1 | 7.6 | 4.5 | 0.35 | 5.6 | 0.017 | 0.012 | 0.020 | 0.387 | 0.15 | 0.58 | 0.54 | 0.389 | 1.05 |
| GER_Sch | 270.2 | 221.0 | 319.5 | 247.5 | 223.4 | 271.6 | 114.9 | 51.8 | 178.0 | 97.6 | 57.44 | 137.7 | 0.098 | 0.059 | 0.137 | 0.961 | 0.64 | 1.29 | 0.64 | 0.510 | 0.77 |
| GER_Ta | 30.1 | 11.8 | 83.2 | 5.0 | 0.000 | 32.2 | 3.6 | 2.8 | 5.9 | 0.8 | 0.22 | 3.1 | 0.007 | 0.004 | 0.019 | 0.489 | 0.26 | 1.08 | 0.41 | 0.101 | 1.45 |
| ITA | 24.1 | 13.6 | 289.1 | 13.7 | 6.6 | 60.2 | 4.3 | 2.9 | 4.7 | 1.4 | 0.40 | 2.5 | 0.020 | 0.013 | 0.031 | 0.438 | 0.24 | 32.60 | 0.27 | 0.134 | 0.43 |
| ITA | 140.4 | 68.6 | 469.7 | 148.7 | 88.3 | 283.5 | 36.9 | 25.6 | 134.7 | 14.7 | 3.34 | 39.0 | 0.029 | 0.013 | 0.083 | 0.198 | 0.02 | 1.60 | 0.08 | 0.054 | 0.54 |
| POL_La | 45.7 | 30.0 | 76.2 | 27.9 | 12.2 | 105.3 | 4.0 | 3.8 | 17.1 | 1.2 | 0.84 | 2.5 | 0.016 | 0.007 | 0.064 | 0.784 | 0.42 | 1.41 | 1.20 | 0.933 | 2.00 |
| POL_La | 217.5 | 110.5 | 366.5 | 143.4 | 101.8 | 257.3 | 30.3 | 21.7 | 98.5 | 17.6 | 1.57 | 28.4 | 0.020 | 0.019 | 0.113 | 0.557 | 0.27 | 1.05 | 0.30 | 0.239 | 6.04 |
| POL_Łsz | 25.1 | 16.0 | 45.1 | 14.3 | 7.7 | 44.1 | 4.8 | 3.1 | 7.5 | 1.8 | 0.64 | 7.1 | 0.009 | 0.003 | 0.018 | 0.424 | 0.24 | 0.70 | 0.38 | 0.065 | 1.02 |
| POL_Łsz | 151.0 | 98.0 | 193.2 | 235.4 | 166.5 | 300.9 | 59.1 | 41.8 | 188.7 | 19.3 | 4.35 | 66.9 | 0.038 | 0.024 | 0.083 | 0.668 | 0.44 | 1.26 | 0.38 | 0.274 | 4.92 |
| POL_Mś | 35.1 | 23.6 | 280.3 | 21.7 | 17.0 | 33.6 | 4.6 | 4.3 | 5.6 | 1.1 | 0.25 | 4.2 | 0.036 | 0.021 | 0.098 | 0.867 | 0.68 | 30.77 | 0.38 | 0.264 | 0.64 |
| POL_Mś | 130.4 | 102.5 | 203.3 | 212.5 | 211.9 | 232.8 | 155.4 | 123.6 | 181.2 | 22.8 | 4.61 | 79.6 | 0.045 | 0.021 | 0.052 | 0.622 | 0.59 | 0.70 | 0.20 | 0.152 | 0.22 |
| POL_Prz | 15.8 | 8.9 | 134.0 | 16.7 | 5.4 | 54.1 | 4.6 | 3.4 | 129.6 | 1.2 | 0.38 | 12.7 | 0.020 | 0.008 | 0.118 | 0.353 | 0.14 | 4.23 | 0.17 | 0.020 | 1.81 |
| POL_Prz | 85.4 | 45.8 | 431.0 | 232.8 | 54.6 | 457.8 | 73.5 | 22.9 | 98.5 | 40.2 | 0.00 | 78.0 | 0.046 | 0.025 | 0.184 | 0.689 | 0.24 | 13.02 | 0.11 | 0.031 | 1.74 |
| POL_PrzB | 197.8 | 75.7 | 310.0 | 26.8 | 19.4 | 45.2 | 8.7 | 6.1 | 19.3 | 1.5 | 0.002 | 3.0 | 0.078 | 0.000 | 0.212 | 2.445 | 2.11 | 3.16 | 1.18 | 1.018 | 1.45 |
| POL_PrzB | 397.5 | 382.3 | 412.7 | 259.2 | 212.2 | 306.1 | 82.9 | 61.3 | 104.4 | 32.6 | 29.16 | 36.0 | 0.132 | 0.115 | 0.150 | 2.237 | 2.06 | 2.42 | 1.18 | 0.677 | 1.68 |
| POL_Wł | 26.3 | 18.3 | 50.9 | 6.5 | 4.9 | 10.3 | 4.6 | 3.6 | 7.6 | 1.2 | 0.77 | 2.6 | 0.009 | 0.005 | 0.047 | 0.437 | 0.32 | 0.75 | 0.30 | 0.206 | 0.74 |
| POL_Wł | 124.4 | 105.4 | 216.0 | 217.9 | 72.8 | 273.0 | 59.6 | 31.9 | 211.2 | 19.3 | 11.61 | 144.7 | 0.020 | 0.003 | 0.051 | 0.962 | 0.48 | 1.35 | 0.21 | 0.015 | 3.05 |
| RUS | 24.0 | 19.1 | 28.9 | 12.0 | 12.0 | 12.1 | 4.2 | 4.2 | 4.3 | 0.2 | 0.19 | 0.2 | 0.005 | 0.004 | 0.006 | 0.263 | 0.22 | 0.30 | 0.19 | 0.158 | 0.22 |
| RUS | 124.2 | 124.2 | 124.2 | 180.8 | 180.8 | 180.8 | 52.4 | 52.4 | 52.4 | 2.5 | 2.48 | 2.5 | 0.023 | 0.023 | 0.023 | 0.310 | 0.31 | 0.31 | 0.22 | 0.217 | 0.22 |
| Slo_GB | 25.5 | 20.8 | 26.2 | 15.7 | 7.8 | 29.1 | 6.3 | 5.3 | 8.7 | 2.8 | 0.42 | 6.1 | 0.006 | 0.003 | 0.008 | 0.260 | 0.15 | 0.34 | 0.35 | 0.302 | 1.15 |
| Slo_GB | 132.9 | 113.0 | 152.7 | 65.4 | 27.6 | 103.2 | 42.8 | 37.8 | 47.8 | 12.2 | 1.29 | 23.1 | 0.013 | 0.010 | 0.016 | 0.252 | 0.17 | 0.33 | 0.56 | 0.384 | 0.74 |
| Slo_MS1 | 39.4 | 36.3 | 42.5 | 28.0 | 25.8 | 30.2 | 5.2 | 5.1 | 5.3 | 4.7 | 4.43 | 4.9 | 0.009 | 0.009 | 0.009 | 0.210 | 0.20 | 0.22 | 0.66 | 0.526 | 0.80 |
| Slo_MS1 | 817.7 | 817.7 | 817.7 | 55.7 | 55.7 | 55.7 | 40.9 | 40.9 | 40.9 | 12.6 | 12.56 | 12.6 | 0.039 | 0.039 | 0.039 | 0.550 | 0.55 | 0.55 | 0.30 | 0.297 | 0.30 |
| Slo_MS3 | 33.0 | 23.8 | 47.1 | 20.3 | 10.4 | 36.1 | 6.7 | 5.4 | 12.0 | 1.9 | 1.02 | 5.8 | 0.015 | 0.011 | 0.023 | 0.375 | 0.19 | 1.80 | 0.50 | 0.169 | 4.40 |
| Slo_MS3 | 130.4 | 120.4 | 286.4 | 57.0 | 43.9 | 71.5 | 43.2 | 32.3 | 59.1 | 5.9 | 1.99 | 18.9 | 0.080 | 0.057 | 0.105 | 0.311 | 0.22 | 0.34 | 0.42 | 0.158 | 1.13 |
| SPA_Ho | 145.3 | 90.8 | 267.5 | 19.5 | 16.1 | 30.5 | 5.0 | 4.0 | 6.9 | 2.7 | 0.35 | 37.3 | 0.017 | 0.005 | 0.070 | 0.674 | 0.44 | 0.80 | 0.37 | 0.169 | 0.85 |
| SPA_Ho | 471.0 | 131.8 | 935.5 | 228.8 | 159.3 | 315.0 | 66.7 | 46.3 | 75.2 | 15.9 | 0.97 | 70.0 | 0.055 | 0.020 | 0.093 | 0.680 | 0.19 | 0.76 | 0.51 | 0.269 | 0.71 |
| SPA_La | 50.0 | 24.7 | 322.5 | 13.7 | 3.5 | 156.6 | 4.7 | 2.9 | 9.1 | 0.8 | 0.04 | 5.1 | 0.041 | 0.021 | 0.202 | 0.838 | 0.36 | 9.81 | 0.18 | 0.020 | 1.58 |
| SPA_La | 166.1 | 42.7 | 819.7 | 142.3 | 86.6 | 345.4 | 31.2 | 8.6 | 80.1 | 12.4 | 0.52 | 83.5 | 0.090 | 0.059 | 0.586 | 0.641 | 0.36 | 1.21 | 0.19 | 0.107 | 1.11 |
| SPA_LZ | 92.5 | 78.9 | 108.0 | 23.9 | 15.8 | 29.3 | 4.7 | 4.4 | 5.2 | 7.8 | 1.45 | 20.5 | 0.025 | 0.014 | 0.065 | 0.427 | 0.31 | 0.62 | 0.47 | 0.066 | 5.96 |
| SPA_LZ | 223.9 | 161.1 | 372.2 | 156.5 | 124.0 | 195.7 | 54.4 | 48.8 | 76.1 | 119.5 | 26.77 | 254.9 | 0.042 | 0.016 | 0.061 | 0.346 | 0.32 | 0.72 | 0.16 | 0.143 | 0.37 |
| UK_Col | 42.5 | 7.8 | 160.4 | 17.5 | 0.6 | 75.4 | 5.7 | 1.2 | 15.0 | 2.7 | 0.56 | 45.0 | 0.041 | 0.004 | 0.093 | 1.183 | 0.23 | 4.99 | 0.07 | 0.003 | 0.63 |
| UK_Col | 351.5 | 161.8 | 2522.3 | 422.0 | 116.4 | 658.6 | 74.0 | 48.6 | 105.8 | 56.6 | 1.88 | 232.0 | 0.107 | 0.048 | 0.161 | 10.8 | 3.92 | 32.67 | 1.10 | 0.220 | 2.24 |
| UK_Fet | 46.0 | 29.5 | 99.3 | 33.9 | 13.7 | 60.9 | 8.0 | 4.5 | 19.5 | 3.5 | 1.86 | 12.4 | 0.015 | 0.009 | 0.031 | 0.664 | 0.41 | 1.99 | 0.64 | 0.422 | 4.96 |
| UK_Fet | 268.4 | 188.2 | 648.1 | 388.0 | 330.4 | 493.1 | 101.6 | 74.9 | 251.7 | 75.4 | 35.54 | 134.7 | 0.058 | 0.026 | 0.111 | 1.806 | 0.69 | 3.72 | 0.48 | 0.229 | 4.49 |
| All popul | 35.0 | 7.1 | 2220.8 | 15.8 | 0.0 | 156.6 | 4.8 | 1.2 | 129.6 | 1.4 | 0.002 | 45.0 | 0.016 | 0.000 | 0.212 | 0.547 | 0.14 | 32.60 | 0.39 | 0.003 | 9.20 |
| All popul | 161.1 | 42.7 | 2522.3 | 211.4 | 27.6 | 658.6 | 54.5 | 8.6 | 251.7 | 23.0 | 0.002 | 254.9 | 0.050 | 0.003 | 0.586 | 0.766 | 0.02 | 32.67 | 0.38 | 0.015 | 7.60 |
| B) Additional single measurements of the elemental composition of other body parts of two male Stag Beetles from two populations. | |||||||||||||||||||||
| Population acronym | Body part | Ca | Mg | K | Na | Mn | Fe | Zn | Cu | As | Cd | Pb | Ni | ||||||||
| POL_PrzB | head with mandibles | 162.7 | 443.6 | 5017.1 | 715.4 | 20.3 | 156.6 | 87.7 | 3.96 | 1.91 | 0.02 | 0.47 | 0.33 | ||||||||
| POL_PrzB | pronotum with legs | 261.7 | 599.3 | 4850.7 | 948.8 | 22.3 | 218.8 | 108.2 | 4.62 | 1.35 | 0.02 | 0.90 | 0.71 | ||||||||
| POL_Mś | wings | 583.7 | 750.1 | 9635.1 | 1633.3 | 251.1 | 492.3 | 43.3 | 505.8 | 2.85 | 0.27 | 3.01 | 0.93 | ||||||||
| POL_Mś | head with antenna | 206.3 | 600.5 | 6407.8 | 1060.8 | 16.1 | 23.5 | 76.1 | 4.75 | 7.04 | 0.01 | 1.02 | 0.30 | ||||||||
| POL_Mś | pronotum with legs | 263.1 | 634.5 | 5344.5 | 965.3 | 18.1 | 23.1 | 77.1 | 4.70 | 0.76 | 0.01 | 1.76 | 0.96 | ||||||||
| POL_Mś | mandibles | 179.4 | 323.4 | 1318.4 | 529.3 | 22.1 | 5.9 | 6.3 | 9.03 | 1.22 | 0.02 | 0.14 | 0.09 | ||||||||
| POL_Mś | abdomen with legs | 217.9 | 1395.6 | 8428.0 | 1078.9 | 21.2 | 302.2 | 58.0 | 2.52 | 0.44 | 0.01 | 0.07 | 0.10 | ||||||||
Note: from two populations (GER_For, GER_Ta) only the elytra were sampled.
Table 3.
Spearman rank correlation coefficients (rs) testing the relationship between the size of elytra and abdomens (= sample mass) and concentrations of 12 chemical elements measured in various Stag Beetle Lucanus cervus populations (see Fig. 1 and Table 1 for the origin of the populations) with sample sizes n > 10; the statistically significant relationships are in bold; * P ≤ 0.05, ** P < 0.01, *** P < 0.001.
| Sample/population acronym | Number of samples | Ca | Mg | K | Na | Mn | Fe | Zn | Cu | As | Cd | Pb | Ni |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ELYTRA | |||||||||||||
| All populations | 271 | −0.299*** | −0.251*** | 0.109 | 0.195*** | 0.071 | −0.281*** | −0.214*** | −0.350*** | −0.262*** | −0.461*** | −0.478*** | −0.175** |
| BEL_Sol | 12 | 0.091 | 0.112 | 0.322 | 0.196 | 0.189 | −0.014 | −0.259 | −0.336 | 0.126 | −0.427 | −0.503 | −0.706** |
| BEL_Spe | 33 | −0.218 | −0.274 | −0.122 | −0.079 | −0.115 | 0.080 | −0.050 | −0.417* | −0.130 | −0.412* | −0.164 | −0.156 |
| GER_Ko | 11 | 0.382 | 0.173 | 0.209 | 0.164 | −0.282 | −0.336 | −0.364 | −0.555 | −0.709* | −0.636* | −0.436 | −0.227 |
| GER_Ta | 18 | −0.172 | −0.459 | −0.020 | 0.267 | −0.115 | −0.366 | 0.121 | −0.441 | −0.156 | 0.117 | −0.232 | −0.521* |
| ITA | 11 | 0.345 | 0.345 | −0.555 | −0.700* | −0.055 | 0.564 | −0.500 | −0.645* | −0.055 | −0.318 | −0.409 | 0.436 |
| POL_Łsz | 12 | −0.049 | −0.042 | 0.483 | 0.000 | 0.594* | 0.350 | 0.161 | −0.035 | −0.252 | 0.007 | −0.734** | −0.636* |
| POL_Prz | 30 | −0.459* | 0.041 | 0.055 | 0.223 | 0.334 | 0.131 | −0.365* | −0.107 | −0.001 | −0.503** | −0.310 | −0.458* |
| POL_Wł | 10 | −0.912*** | −0.505 | −0.255 | 0.097 | −0.918*** | −0.894*** | −0.243 | −0.322 | −0.723* | −0.365 | −0.705* | −0.274 |
| Slo_MS3 | 10 | 0.024 | 0.644* | 0.413 | 0.298 | 0.973*** | −0.347 | 0.480 | −0.030 | −0.681* | −0.328 | −0.681* | 0.407 |
| SPA_La | 17 | −0.735*** | −0.554* | 0.294 | 0.098 | 0.017 | −0.400 | 0.529* | −0.664** | −0.289 | −0.127 | −0.483* | −0.228 |
| UK_Col | 14 | −0.714** | −0.490 | −0.789*** | −0.244 | −0.516 | −0.811*** | −0.644* | −0.587* | −0.719** | −0.484 | −0.732** | −0.226 |
| UK_Fet | 12 | 0.350 | 0.007 | 0.699* | 0.524 | 0.070 | 0.811** | 0.196 | −0.203 | 0.874*** | 0.105 | −0.168 | 0.469 |
| ABDOMENS | |||||||||||||
| All populations | 124 | −0.312*** | 0.042 | 0.327*** | 0.295*** | −0.004 | −0.123 | −0.355*** | 0.041 | −0.127 | −0.513*** | −0.515*** | −0.457*** |
| BEL_Spe | 19 | −0.282 | −0.212 | 0.226 | 0.116 | 0.142 | 0.516* | −0.075 | −0.253 | 0.026 | 0.023 | 0.181 | −0.325 |
| POL_Prz | 15 | −0.425 | 0.129 | 0.232 | 0.407 | −0.504 | −0.429 | −0.600* | 0.454 | 0.604* | −0.357 | −0.471 | −0.404 |
Additional comment on the data inTable 3.
Given the apparent sexual dimorphism in body size in Stag Beetles, our findings suggest that sex-related differences in ETs for some of these metals can occur in this species. Sex-related differences in the contents of major nutrients and trace elements have been described in dimorphic insects, and it has been hypothesized that this can have certain consequences for their reproduction [15]. To date, given that female Stag Beetles are the smallest individuals [9; see Photo 1], we can speculate that they tend to accumulate relatively more Zn, As, Cd and Ni in both abdomens and elytra, and more Pb and Mg in their abdomens and elytra, respectively. At the same time, higher abdomen K concentrations may be indicative of larger males. These findings are therefore a clear incentive for further detailed assessments under controlled conditions of how ETs in different body parts of beetles and other insects vary with sex and body size heterogeneity – the rule in a population but often overlooked in the stoichiometry/elemental study of insects – and relating this to reproductive indices and landscape heterogeneity.
Table 4.
Pearson correlation coefficients with original P-values testing the relationship between the mass and concentrations of elements measured in abdomens and elytra in 28 Stag Beetle Lucanus cervus populations (data average per population) and six major land-cover types determined within three radii of 500 m, 1000 m and 5000 m (for clarity, each radius is shown against a different background) around the sampling sites (i.e. exact population location; for details, see Table 1). * P ≤ 0.05, ** P < 0.01, *** P < 0.001; the results meeting the FDR-adjusted P-value are indicated in bold.
| Element / body part | Radius | Urban/built-up area | Woods/ parks | Agriculturalland | Grassland/ open natural habitats | Roads/railways | Water |
|---|---|---|---|---|---|---|---|
| Abdomen mass | 500 m | −0.517** | 0.492* | −0.174 | −0.046 | −0.332 | 0.031 |
| 1000 m | −0.382 | 0.492* | −0.056 | −0.306 | −0.294 | 0.047 | |
| 5000 m | −0.461* | 0.361 | 0.159 | −0.378 | −0.439* | 0.193 | |
| Elytra mass | 500 m | −0.379 | 0.398* | −0.174 | −0.131 | −0.277 | −0.122 |
| 1000 m | −0.258 | 0.421* | −0.206 | −0.296 | −0.219 | −0.041 | |
| 5000 m | −0.191 | 0.398* | −0.164 | −0.133 | −0.313 | 0.085 | |
| Abdomen Ca | 500 m | 0.697*** | −0.605*** | 0.037 | 0.170 | 0.429* | 0.252 |
| 1000 m | 0.557** | −0.510** | 0.201 | 0.160 | 0.140 | 0.167 | |
| 5000 m | 0.628*** | −0.596** | 0.265 | 0.172 | 0.244 | 0.087 | |
| Elytra Ca | 500 m | 0.221 | −0.081 | 0.069 | −0.048 | −0.092 | −0.172 |
| 1000 m | 0.123 | −0.112 | 0.355 | −0.254 | −0.007 | −0.005 | |
| 5000 m | 0.217 | −0.280 | 0.493 | −0.440 | −0.024 | 0.059 | |
| Abdomen Mg | 500 m | 0.460* | −0.323 | −0.203 | 0.188 | 0.545** | 0.262 |
| 1000 m | 0.381 | −0.294 | −0.132 | 0.254 | 0.369 | 0.061 | |
| 5000 m | 0.361 | −0.261 | −0.151 | 0.165 | 0.411 | −0.017 | |
| Elytra Mg | 500 m | 0.071 | −0.192 | −0.129 | 0.484* | 0.154 | −0.168 |
| 1000 m | 0.005 | −0.236 | −0.110 | 0.489* | 0.113 | −0.276 | |
| 5000 m | 0.057 | −0.462* | 0.148 | 0.161 | 0.217 | −0.131 | |
| Abdomen K | 500 m | −0.056 | 0.153 | −0.218 | −0.023 | 0.247 | −0.062 |
| 1000 m | −0.071 | 0.172 | −0.284 | 0.023 | −0.020 | −0.300 | |
| 5000 m | −0.004 | 0.002 | −0.033 | −0.024 | −0.041 | −0.190 | |
| Elytra K | 500 m | −0.014 | 0.109 | −0.191 | −0.065 | 0.230 | 0.148 |
| 1000 m | −0.026 | 0.164 | −0.237 | 0.015 | −0.069 | −0.101 | |
| 5000 m | −0.003 | 0.056 | −0.080 | −0.022 | −0.104 | −0.111 | |
| Abdomen Na | 500 m | −0.003 | −0.033 | 0.043 | 0.055 | 0.366 | 0.048 |
| 1000 m | 0.069 | −0.050 | −0.040 | 0.104 | 0.021 | −0.121 | |
| 5000 m | 0.088 | −0.173 | 0.033 | 0.165 | 0.082 | −0.061 | |
| Elytra Na | 500 m | −0.175 | −0.112 | 0.237 | 0.203 | 0.098 | −0.157 |
| 1000 m | −0.128 | −0.052 | −0.028 | 0.219 | −0.078 | −0.241 | |
| 5000 m | −0.062 | −0.282 | 0.152 | 0.219 | 0.008 | −0.158 | |
| Abdomen Mn | 500 m | −0.174 | 0.303 | −0.324 | −0.145 | −0.049 | 0.055 |
| 1000 m | −0.200 | 0.373 | −0.359 | −0.143 | −0.209 | −0.154 | |
| 5000 m | −0.185 | 0.411* | −0.312 | −0.113 | −0.294 | −0.223 | |
| Elytra Mn | 500 m | −0.305 | 0.483* | −0.417* | −0.173 | −0.156 | −0.207 |
| 1000 m | −0.336 | 0.496 | −0.360 | −0.202 | −0.181 | −0.377 | |
| 5000 m | −0.315 | 0.445* | −0.129 | −0.435* | −0.338 | −0.286 | |
| Abdomen Fe | 500 m | 0.290 | −0.259 | −0.176 | 0.264 | 0.247 | 0.429* |
| 1000 m | 0.272 | −0.182 | −0.194 | 0.300 | −0.080 | 0.277 | |
| 5000 m | 0.213 | −0.030 | −0.321 | 0.375 | 0.015 | −0.006 | |
| Elytra Fe | 500 m | 0.254 | −0.181 | −0.049 | 0.059 | 0.040 | 0.103 |
| 1000 m | 0.251 | −0.192 | 0.095 | 0.135 | −0.122 | 0.026 | |
| 5000 m | 0.091 | −0.126 | 0.104 | 0.135 | −0.095 | −0.113 | |
| Abdomen Zn | 500 m | 0.536** | −0.358 | −0.003 | 0.012 | 0.586** | −0.097 |
| 1000 m | 0.445* | −0.359 | −0.097 | 0.222 | 0.305 | −0.277 | |
| 5000 m | 0.580** | −0.408* | −0.070 | 0.252 | 0.403* | −0.353 | |
| Elytra Zn | 500 m | 0.113 | −0.077 | −0.128 | −0.003 | 0.152 | 0.407* |
| 1000 m | −0.011 | 0.016 | −0.085 | 0.083 | −0.327 | 0.153 | |
| 5000 m | −0.006 | 0.060 | −0.083 | 0.119 | −0.184 | −0.045 | |
| Abdomen Cu | 500 m | −0.015 | 0.039 | −0.190 | −0.004 | 0.380 | 0.307 |
| 1000 m | 0.001 | 0.072 | −0.239 | 0.082 | −0.001 | 0.090 | |
| 5000 m | 0.062 | −0.030 | −0.110 | 0.077 | 0.062 | 0.267 | |
| Elytra Cu | 500 m | 0.241 | −0.088 | −0.101 | −0.265 | 0.211 | 0.445* |
| 1000 m | 0.147 | 0.005 | −0.061 | −0.018 | −0.178 | 0.236 | |
| 5000 m | 0.115 | 0.086 | −0.037 | −0.071 | −0.127 | −0.093 | |
| Abdomen As | 500 m | 0.102 | −0.297 | −0.058 | 0.428* | 0.283 | −0.056 |
| 1000 m | 0.070 | −0.371 | −0.137 | 0.531** | 0.177 | −0.054 | |
| 5000 m | 0.159 | −0.375 | −0.270 | 0.424* | 0.430* | 0.221 | |
| Elytra As | 500 m | 0.190 | −0.454* | −0.107 | 0.648*** | 0.398* | 0.303 |
| 1000 m | 0.190 | −0.442* | −0.328 | 0.639*** | 0.108 | 0.190 | |
| 5000 m | 0.242 | −0.336 | −0.559** | 0.636*** | 0.398* | 0.165 | |
| Abdomen Cd | 500 m | 0.178 | −0.195 | 0.291 | −0.139 | 0.071 | −0.283 |
| 1000 m | 0.019 | −0.182 | 0.225 | 0.280 | 0.046 | −0.301 | |
| 5000 m | 0.079 | −0.167 | 0.211 | 0.138 | 0.188 | −0.394* | |
| Elytra Cd | 500 m | 0.077 | −0.010 | −0.070 | −0.080 | −0.053 | −0.199 |
| 1000 m | −0.108 | 0.018 | −0.087 | 0.192 | −0.116 | −0.315 | |
| 5000 m | −0.023 | −0.120 | 0.191 | −0.096 | −0.024 | −0.285 | |
| Abdomen Pb | 500 m | 0.615** | −0.370 | −0.211 | 0.070 | 0.299 | −0.048 |
| 1000 m | 0.437* | −0.263 | −0.170 | 0.157 | 0.066 | −0.115 | |
| 5000 m | 0.508** | −0.196 | −0.133 | 0.140 | 0.171 | −0.294 | |
| Elytra Pb | 500 m | 0.003 | 0.142 | −0.196 | −0.099 | −0.086 | −0.109 |
| 1000 m | −0.136 | 0.177 | −0.032 | −0.128 | −0.169 | −0.198 | |
| 5000 m | −0.020 | −0.078 | 0.346 | −0.334 | −0.166 | 0.167 | |
| Abdomen Ni | 500 m | 0.155 | 0.056 | −0.041 | −0.332 | 0.068 | 0.022 |
| 1000 m | 0.104 | 0.083 | −0.011 | −0.197 | −0.133 | −0.081 | |
| 5000 m | 0.119 | 0.193 | −0.122 | −0.017 | −0.130 | −0.358 | |
| Elytra Ni | 500 m | 0.234 | −0.211 | 0.172 | −0.103 | 0.075 | 0.202 |
| 1000 m | 0.211 | −0.233 | 0.229 | −0.091 | −0.053 | 0.242 | |
| 5000 m | 0.125 | −0.126 | 0.070 | 0.046 | 0.013 | 0.032 |
2. Experimental design, materials and method
2.1. Species and populations
This data applies to adult Stag Beetles collected at 28 localities (hereafter referred to as 'local populations') situated in eight European countries ranging from Central Spain to Western Russia (Fig. 1; for georeferences, see Table 1). Due to the scarcity of their primary larval food source, i.e. a lack of dead wood resources, Stag Beetles, in common with the community of saproxylic invertebrates as a whole, are an endangered species and have, owing to their iconic status, been identified as good indicators of woodland quality (see [2]).
Stag Beetles are known to have been present for many decades in some of the regions covered by this data article, for example, Belgium [2], Germany [3], the UK [4], Italy [5] and Poland (D. Tarnawski, A. Smolis, M. Kadej – unpubl. results). Furthermore, some of the Stag Beetle populations included in this study have already been extensively examined, from a biometric standpoint (data from several local European populations collated in [4, 9]), pattern of distribution (Belgium: [2]), movement (telemetry) of individuals (Germany: [3]; Italy: [5]), mating behaviour [6] and a pan-European monitoring programme [7].
The beetles used in this data article were already dead and were acquired from relatively small areas (up to c. 100 m in diameter); they included roadkill victims, predatory remains or those dead from unknown causes, most probably natural death after mating (see Table 1). All the Stag Beetles were collected by researchers very familiar with the appearance of this species, so confusion with other species of large beetles can be ruled out. Stag Beetles from most of the populations examined here, formed the basis of a recent study on the genetic diversity of this species in Europe, and many of the individual beetles were from an existing collection [8]. In addition, although there are four other members of genus Lucanus in Europe – L. (Pseudolucanus) barbarossa, L. pontbrianti, L. tetraodon and L. ibericus – the materials used for this article were collected outside the geographical ranges of those species.
For the analysis we used intact abdomens (n = 124) with dry internal organs and gut contents, and both elytra (n = 271); other parts (legs) of the majority of these beetles were also used in the genetic study by Cox et al. [8]. The samples were dried and kept at room temperature until the chemical analysis.
Female Stag Beetles more often move around only on the ground and are less capable of dispersing, and they often succumb to predators [3, 4, 9].
Since in most cases only fragments of the beetles were available, reliable sexing of all of them was impossible; for this reason we did not analyse sex-related differences. However, taking into account the large differences in elytra masses (a function of beetle size) measured in individual populations (up to 2.7-fold between extreme values, as in the Przemków forest, Poland; see Fig. 1), our samples may contain remains of both sexes. Even though Stag Beetles demonstrate a normal size distribution [4], male Stag Beetles in Europe can be up to c. 30% larger than females (see Photo 1), and even among males nearly 3-fold differences in body length can occur [9].
2.2. Land-cover types inhabited by the individual populations
For each sampling site, defined here as the central point of the sampling area, we measured the areas (ha) of several land-cover classes lying within three radii of that point (500 m, 1000 m and 5000 m) using Arc-View software from satellite imagery maps taken from Google Maps and other supporting information sources of local land-use types. These classes were ultimately pooled into six major land-cover types based primarily on the ability of Stag Beetles to utilize them: urban (different types of built-up areas, including industrial buildings); wooded (woods, parks and forests); agricultural (arable land; croplands and different cultivations); grassland (meadows, pastures and other open habitats with low vegetation, such as steppe-like habitats in Spain and Russia); transport infrastructure (tarred roads and highways, and railways); water (lakes, gravel pits and rivers). We assessed the effects of these six major land-cover types on the elytra and abdomen masses (hereafter referred to simply as 'body size') and ETs of Stag Beetles between successive radii. Generally, we did not prioritize any of the land-cover types or radii, because each could be regarded as causing variation in the body size and ETs of Stag Beetles. However, with a knowledge of Stag Beetle biology and ecology, one can make a number of predictions. First of all, deciduous woodland is the key land-cover type (resource) for the persistence of a Stag Beetle population in mainland Europe [2], though not in the UK, where the overwhelming majority of populations live in anthropogenic habitats, which suggests their opportunistic/synanthropic occurrence [4, 9]. Secondly, these beetles are poor dispersers: the home ranges of both sexes are very small, and adults do not move more than 500 m from their reproduction sites [3, 5]. More importantly, we assumed that because Stag Beetles do not need to feed in the adult stage, the quality of the larval habitat (the amount of coverage of wooded area representing the availability of dead wood) may be crucial in explaining their elemental traits and size (this latter point is discussed in [4, 9]). Consequently, Stag Beetles living in poor-quality habitats, i.e. urban areas with small percentages of wooded areas where natural decaying wood is scarce and larvae often feed on various anthropogenic woody substitutes, such as railway sleepers, fence posts and the like in urban gardens, as in the UK, may be smaller in size than those from extensive woodlands (cf. [9]). Thus, we expected that the contribution of two major land-cover types (urban and wooded, representing a negative and a positive influence, respectively), especially in the smallest, 500 m radius, would be stronger than in the other two, larger radii (1000 m and 5000 m). These latter two radii may represent the scale of spatial drift or bioaccumulation into topsoil of metallic pollutants, such as agrochemicals (from crop fields/cultivations or recreational areas) or traffic-related metals (from transport infrastructure and vehicle emissions). Importantly, since railway sleepers are impregnated with creosote, they are sources of both inorganic and organic contaminants that are accumulated in local food webs [10]. Data from the UK suggest, however, that Stag Beetles rarely use railway sleepers [11].
2.3. Chemical analysis
Most of the samples showed no signs of adhering extraneous material; examined under the binocular microscope, the smooth chitin surfaces of the inner and outer layers of the elytra and the abdominal segment surfaces were clean. Nevertheless, the samples were thoroughly cleaned (dusted) with a stream of air from a rubber blower. Any impurities like sand grains, fragments of plant tissue, leaf litter or streaks of dirt on the chitin surfaces were also removed mechanically with a soft plastic bristle brush and blown clean with a stream of air. The samples were then dried at 50 °C to constant weight, and weighed using a digital analytical balance (ONYX-220 FAWAG) with ± 0.0001 g precision.
The entire elytra and abdomens (0.0159 – 0.312 g of dry weight) were digested in 3 ml nitric acid (ultrapure, 65%, Merck) and 0.5 ml perchloric acid (ultrapure, 62%, Merck) in a CEM Mars 5 Corporation microwave oven [12]. The digestion procedure was conducted in three temperature steps (110 °C/10 min, 185 °C/25 min, 200 °C/15 min) at a maximum pressure of 200 psi. After dilution to 10 ml, the digests were analysed for Ca, Fe, K, Mg, Mn, Na and Zn using FAAS and an AVANTA PM GBC Atomic Absorption Spectrophotometer (Ca, K, Na in the emission mode of the spectrophotometer), and As, Cd, Cu, Ni and Pb using ETAAS with a Graphite Furnace and a Perkin-Elmer PinAAcle 900Z Atomic Absorption Spectrophotometer [13]. All the elements were determined against standards (Atomic Absorption Standard Solutions from Sigma Chemical Co.) and blanks containing the same matrix as the samples and were subjected to the same procedure. All the results for the abdomens and elytra were calculated on a dry weight basis. The accuracy of the methods applied for the determination of the elements in the samples was checked by the analysis of Certified Reference Materials. We used DC73348 LGC standards of bush branches and leaves as certified reference materials. The recovery rates were from 94% to 103%.
2.4. Statistical analysis
The statistical analysis involved four major stages of data exploration. Initially, we compared the masses of elytra and abdomens and the metal concentrations (in ppm d.w.) measured in both body parts across the studied populations. As we found substantial variation in both masses and ETs between elytra and abdomens (see Results in [1]), the ETs of each of these two body parts were analysed separately.
We used a general linear model (in Statistica 12; Data Analysis Software System, Version 12.1. Tulsa: USA) to assess whether elytra and abdomen ETs were related to the masses of these body parts (see Results in [1]). Owing to the large differences in ETs amongst populations, we repeated this analysis using the Spearman rank correlation coefficients to test the relationship between the sample mass and elytra, and abdomen metal concentrations within individual populations with sample sizes n > 10 (Table 3).
We used Pearson correlation coefficients to assess the influence of local major land-cover types measured within three radii (500 m, 1000 m and 5000 m) around the sampling sites on the masses and elemental traits of abdomens and elytra (Table 4). Owing to the large number of tests within a particular radius (n = 6), we applied the false discovery rate (FDR) procedure to adjust the original P-values using the classical one-stage method in the statistical software spreadsheet [14]. To meet the assumption of normality (assessed by the Kolmogorov-Smirnov test) required for linear modelling, we log-transformed the following variables before the analysis: elytra and abdomen Pb concentrations; coverage of built-up areas, agricultural land, grassland, transport infrastructure and water measured within 500 m; coverage of built-up areas, grassland and water measured within 1000 m; and grassland, transport infrastructure and water measured within 5000 m.
Declaration of Competing Interest
The authors declared they do not have anything to disclose regarding conflict of interest with respect to this manuscript.
Acknowledgments
We are grateful to Maria Fremlin for her comments and discussion, and for making her unpublished results available to us, which we used in the interpretation of our findings. Our thanks also go to Peter Senn for improving the English, to Marcus Rink for providing us with samples from Germany.
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article. The research was not financed from external grants.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2020.105935.
Contributor Information
Grzegorz Orłowski, Email: orlog@poczta.onet.pl.
Lucyna Mróz, Email: lucyna.mroz@uwr.edu.pl.
Marcin Kadej, Email: marcin.kadej@uwr.edu.pl.
Adrian Smolis, Email: adrian.smolis@uwr.edu.pl.
Dariusz Tarnawski, Email: dariusz.tarnawski@uwr.edu.pl.
Jerzy Karg, Email: turew@poczta.onet.pl.
Deborah J. Harvey, Email: d.harvey@rhul.ac.uk.
Marcos Méndez, Email: marcos.mendez@urjc.es.
Arno Thomaes, Email: arno.thomaes@inbo.be.
Al Vrezec, Email: al.vrezec@nib.si.
Detlef Mader, Email: dr.detlef.mader@web.de.
Appendix A. The raw data – concentrations of 12 chemical elements measured in elytra and abdomens of Stag Beetles Lucanus cervus from 28 European populations of the species; see Table 1 for population acronyms
| Country | Population (acronym) | Body part | Sample mass (g) | Ca ppm | Mg ppm | K ppm | Na ppm | Mn ppm | Fe ppm | Zn ppm | Cu ppm | As ppm | Cd ppm | Pb ppm | Ni ppm |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BEL | BEL_Dro | abdomen | 0.0737 | 812.6 | 3268.8 | 6123.4 | 713.70 | 13.98 | 153.5 | 156.30 | 18.63 | 2.676 | 0.018 | 0.5716 | 0.419 |
| BEL | BEL_Dro | abdomen | 0.1149 | 465.6 | 5342.7 | 9135.3 | 1043.43 | 9.16 | 156.7 | 170.56 | 27.36 | 2.737 | 0.019 | 1.1652 | 0.639 |
| BEL | BEL_Dro | abdomen | 0.0657 | 984.7 | 5537.5 | 7085.7 | 707.00 | 17.73 | 323.1 | 210.85 | 12.47 | 16.932 | 0.094 | 0.5467 | 0.217 |
| BEL | BEL_Dro | elytra | 0.0524 | 291.3 | 889.0 | 1455.3 | 265.27 | 7.69 | 33.8 | 16.16 | 4.06 | 0.677 | 0.007 | 0.2338 | 0.044 |
| BEL | BEL_Dro | elytra | 0.0516 | 185.4 | 584.7 | 1133.2 | 215.89 | 5.64 | 24.6 | 0.00 | 3.17 | 0.316 | 0.005 | 0.3393 | 0.088 |
| BEL | BEL_Dro | elytra | 0.0525 | 468.7 | 597.9 | 2160.2 | 274.86 | 5.12 | 24.0 | 11.06 | 3.42 | 0.330 | 0.004 | 0.3643 | 0.114 |
| BEL | BEL_Dro | elytra | 0.0462 | 373.1 | 455.1 | 2039.5 | 277.27 | 7.03 | 17.3 | 6.02 | 3.77 | 0.242 | 0.007 | 0.3945 | 0.194 |
| BEL | BEL_Dro | elytra | 0.0370 | 264.6 | 613.0 | 2293.6 | 419.19 | 6.36 | 48.6 | 38.59 | 4.52 | 2.212 | 0.022 | 1.7233 | 0.154 |
| BEL | BEL_Dro | elytra | 0.0364 | 426.8 | 514.3 | 1977.1 | 442.86 | 7.69 | 52.7 | 31.58 | 4.54 | 1.645 | 0.012 | 1.2455 | 0.245 |
| BEL | BEL_Dro | elytra | 0.0313 | 516.7 | 591.2 | 2151.9 | 272.52 | 5.01 | 43.8 | 11.21 | 4.29 | 0.673 | 0.011 | 0.6654 | 0.315 |
| BEL | BEL_Dro | elytra | 0.0426 | 383.2 | 594.9 | 2096.8 | 247.42 | 6.84 | 25.4 | 10.79 | 3.98 | 0.612 | 0.009 | 0.3749 | 0.200 |
| BEL | BEL_Ov | elytra | 0.0301 | 330.4 | 514.3 | 2344.8 | 891.03 | 5.21 | 194.0 | 53.47 | 7.97 | 4.305 | 0.046 | 1.3242 | 9.197 |
| BEL | BEL_Ov | elytra | 0.0310 | 575.1 | 627.1 | 2847.4 | 1288.71 | 2.89 | 31.9 | 49.57 | 7.43 | 3.731 | 0.024 | 0.9790 | 4.840 |
| BEL | BEL_Ov | elytra | 0.0310 | 317.3 | 311.2 | 1186.4 | 230.97 | 6.50 | 57.1 | 5.85 | 7.46 | 3.878 | 0.024 | 0.8013 | 1.999 |
| BEL | BEL_Ov | elytra | 0.0375 | 1765.0 | 361.0 | 784.2 | 198.40 | 23.89 | 966.7 | 54.53 | 9.49 | 0.941 | 0.082 | 3.5839 | 5.165 |
| BEL | BEL_Ov | abdomen | 0.0464 | 567.2 | 2928.1 | 9862.1 | 1776.94 | 19.31 | 221.1 | 547.75 | 58.84 | 51.145 | 0.059 | 1.8007 | 1.782 |
| BEL | BEL_Ov | abdomen | 0.0337 | 1294.5 | 1313.9 | 1816.1 | 353.12 | 21.27 | 868.2 | 96.58 | 13.15 | 17.248 | 0.054 | 1.4783 | 2.621 |
| BEL | BEL_Sol | elytra | 0.0415 | 867.0 | 348.7 | 856.1 | 119.76 | 60.72 | 1043.1 | 37.90 | 6.90 | 1.464 | 0.123 | 4.9231 | 1.680 |
| BEL | BEL_Sol | elytra | 0.0389 | 1175.8 | 771.8 | 998.9 | 143.70 | 96.16 | 2220.8 | 43.55 | 5.98 | 1.796 | 0.171 | 9.7953 | 2.490 |
| BEL | BEL_Sol | elytra | 0.0306 | 509.7 | 397.6 | 950.8 | 221.90 | 13.54 | 154.9 | 15.82 | 12.04 | 0.988 | 0.038 | 2.1829 | 3.493 |
| BEL | BEL_Sol | elytra | 0.0324 | 510.6 | 555.6 | 948.1 | 217.59 | 12.10 | 113.0 | 12.32 | 10.37 | 1.242 | 0.028 | 1.1185 | 3.076 |
| BEL | BEL_Sol | elytra | 0.0405 | 332.4 | 544.0 | 2324.0 | 282.47 | 17.70 | 132.1 | 25.10 | 17.06 | 0.807 | 0.028 | 1.4729 | 1.166 |
| BEL | BEL_Sol | elytra | 0.0403 | 343.7 | 586.0 | 2273.9 | 277.42 | 12.78 | 48.6 | 18.32 | 6.49 | 0.679 | 0.017 | 0.5585 | 2.160 |
| BEL | BEL_Sol | elytra | 0.0598 | 290.2 | 447.9 | 2683.9 | 318.06 | 6.37 | 25.1 | 8.09 | 4.83 | 0.711 | 0.016 | 0.4341 | 0.991 |
| BEL | BEL_Sol | abdomen | 0.0693 | 680.9 | 2580.8 | 7658.4 | 650.94 | 22.46 | 327.0 | 348.92 | 95.65 | 43.313 | 0.138 | 32.1371 | 0.904 |
| BEL | BEL_Sol | abdomen | 0.0700 | 2509.7 | 3323.3 | 6349.6 | 554.29 | 22.72 | 943.6 | 307.15 | 30.24 | 15.232 | 0.046 | 1.5826 | 0.651 |
| BEL | BEL_Sol | abdomen | 0.0756 | 664.9 | 3118.1 | 5873.4 | 642.59 | 12.15 | 151.9 | 161.17 | 21.34 | 5.803 | 0.046 | 0.8448 | 0.499 |
| BEL | BEL_Sol | abdomen | 0.0780 | 1628.2 | 8412.9 | 7426.5 | 687.95 | 22.40 | 354.5 | 210.41 | 44.67 | 87.951 | 0.058 | 1.8548 | 0.459 |
| BEL | BEL_Sol | abdomen | 0.0675 | 1020.1 | 10,347.7 | 6432.0 | 612.15 | 23.89 | 237.6 | 269.30 | 22.71 | 23.488 | 0.040 | 11.4131 | 0.339 |
| BEL | BEL_Sol | abdomen | 0.0249 | 2158.9 | 809.6 | 945.5 | 294.38 | 31.04 | 146.2 | 97.19 | 41.53 | 2.257 | 0.090 | 1.3601 | 0.902 |
| BEL | BEL_Sol | elytra | 0.0455 | 1410.1 | 1099.8 | 1772.9 | 225.93 | 19.94 | 138.9 | 10.90 | 7.57 | 3.975 | 0.014 | 0.8394 | 0.551 |
| BEL | BEL_Sol | elytra | 0.0484 | 2672.5 | 2036.5 | 2432.4 | 269.42 | 22.91 | 199.8 | 33.50 | 9.04 | 5.745 | 0.023 | 0.9722 | 0.417 |
| BEL | BEL_Sol | elytra | 0.0408 | 381.2 | 545.3 | 2098.2 | 431.86 | 16.47 | 24.5 | 6.23 | 4.48 | 1.948 | 0.008 | 1.1094 | 0.358 |
| BEL | BEL_Sol | elytra | 0.0409 | 293.1 | 568.6 | 2067.0 | 447.19 | 16.98 | 7.1 | 2.96 | 3.54 | 1.799 | 0.006 | 0.9755 | 0.462 |
| BEL | BEL_Sol | elytra | 0.0480 | 547.9 | 553.5 | 661.1 | 170.42 | 19.83 | 251.5 | 9.58 | 6.21 | 0.389 | 0.018 | 1.3017 | 0.674 |
| BEL | BEL_Spe | elytra | 0.0386 | 395.7 | 548.4 | 3815.5 | 496.89 | 17.70 | 20.7 | 55.17 | 9.13 | 0.734 | 0.067 | 0.8843 | 0.316 |
| BEL | BEL_Spe | elytra | 0.0396 | 368.9 | 494.5 | 3278.2 | 421.72 | 14.99 | 22.7 | 27.50 | 6.86 | 0.496 | 0.026 | 0.8624 | 4.548 |
| BEL | BEL_Spe | elytra | 0.0240 | 465.8 | 654.0 | 3879.4 | 630.00 | 37.80 | 70.4 | 16.64 | 8.65 | 2.493 | 0.038 | 7.8629 | 0.797 |
| BEL | BEL_Spe | elytra | 0.0267 | 683.6 | 1307.9 | 4925.9 | 570.41 | 33.56 | 44.2 | 35.80 | 15.56 | 3.423 | 0.094 | 0.4399 | 0.330 |
| BEL | BEL_Spe | elytra | 0.0509 | 257.8 | 333.8 | 2805.9 | 319.45 | 13.64 | 18.1 | 7.13 | 4.45 | 1.772 | 0.032 | 0.5059 | 0.360 |
| BEL | BEL_Spe | elytra | 0.0508 | 394.5 | 732.8 | 2466.7 | 292.91 | 13.23 | 31.1 | 10.72 | 8.10 | 1.660 | 0.017 | 0.2685 | 0.218 |
| BEL | BEL_Spe | elytra | 0.0336 | 495.3 | 540.0 | 3048.0 | 402.38 | 8.33 | 17.0 | 12.60 | 8.60 | 1.025 | 0.030 | 0.4836 | 0.372 |
| BEL | BEL_Spe | elytra | 0.0341 | 584.1 | 568.0 | 3113.1 | 410.56 | 13.14 | 58.4 | 13.84 | 10.74 | 1.433 | 0.031 | 6.6728 | 0.330 |
| BEL | BEL_Spe | elytra | 0.0492 | 429.1 | 638.0 | 2628.8 | 391.87 | 10.47 | 29.3 | 12.05 | 7.32 | 0.295 | 0.022 | 0.5599 | 0.198 |
| BEL | BEL_Spe | elytra | 0.0486 | 2299.8 | 616.3 | 747.9 | 142.59 | 21.66 | 357.6 | 14.69 | 8.66 | 0.582 | 0.075 | 1.6379 | 0.761 |
| BEL | BEL_Spe | elytra | 0.0384 | 3629.5 | 1273.1 | 1274.9 | 179.43 | 44.63 | 1141.9 | 73.42 | 9.24 | 0.209 | 0.087 | 4.0011 | 1.217 |
| BEL | BEL_Spe | elytra | 0.0427 | 1496.5 | 664.4 | 922.5 | 184.31 | 27.28 | 44.5 | 13.32 | 4.70 | 0.371 | 0.039 | 0.4578 | 0.315 |
| BEL | BEL_Spe | elytra | 0.0479 | 323.4 | 517.1 | 2548.2 | 243.42 | 15.67 | 19.2 | 11.37 | 3.96 | 2.400 | 0.021 | 0.2677 | 0.075 |
| BEL | BEL_Spe | elytra | 0.0332 | 383.6 | 585.5 | 2854.1 | 475.30 | 14.17 | 20.5 | 17.13 | 4.88 | 0.895 | 0.026 | 0.4426 | 0.148 |
| BEL | BEL_Spe | abdomen | 0.0762 | 696.6 | 2275.3 | 5916.5 | 794.09 | 22.19 | 99.7 | 184.98 | 11.52 | 1.549 | 0.043 | 1.2270 | 0.407 |
| BEL | BEL_Spe | abdomen | 0.1017 | 575.2 | 9611.3 | 9674.1 | 1011.60 | 18.94 | 136.8 | 184.64 | 23.48 | 35.748 | 0.094 | 1.1794 | 0.193 |
| BEL | BEL_Spe | abdomen | 0.0470 | 782.6 | 2037.4 | 7633.2 | 911.28 | 16.68 | 86.4 | 287.98 | 20.76 | 51.817 | 0.112 | 0.6688 | 0.312 |
| BEL | BEL_Spe | abdomen | 0.0607 | 525.2 | 2817.1 | 6395.1 | 544.48 | 28.42 | 79.2 | 128.77 | 10.50 | 3.951 | 0.158 | 1.8839 | 1.053 |
| BEL | BEL_Spe | abdomen | 0.0384 | 620.2 | 1681.9 | 6458.7 | 848.44 | 17.21 | 65.9 | 205.76 | 21.66 | 11.473 | 0.112 | 0.9909 | 1.873 |
| BEL | BEL_Spe | abdomen | 0.0441 | 614.5 | 4759.2 | 9743.8 | 1282.99 | 31.24 | 118.8 | 525.97 | 68.69 | 186.893 | 0.081 | 1.1138 | 0.287 |
| BEL | BEL_Spe | abdomen | 0.0410 | 734.9 | 2339.1 | 6447.7 | 766.34 | 17.76 | 45.9 | 305.75 | 156.07 | 17.457 | 0.370 | 0.7766 | 0.274 |
| BEL | BEL_Spe | abdomen | 0.0521 | 669.9 | 5287.4 | 6584.6 | 969.87 | 16.55 | 158.3 | 250.13 | 43.95 | 149.820 | 0.080 | 1.2084 | 1.072 |
| BEL | BEL_Spe | abdomen | 0.0263 | 685.8 | 2652.8 | 3247.0 | 460.08 | 19.59 | 92.4 | 149.06 | 84.76 | 60.236 | 0.055 | 1.1171 | 1.555 |
| BEL | BEL_Spe | abdomen | 0.0597 | 676.4 | 2782.3 | 7376.9 | 966.33 | 20.26 | 103.2 | 301.77 | 28.56 | 22.531 | 0.067 | 0.8063 | 0.289 |
| BEL | BEL_Spe | abdomen | 0.0585 | 511.5 | 3121.2 | 4160.4 | 457.95 | 26.23 | 202.7 | 126.17 | 17.02 | 1.936 | 0.044 | 0.9852 | 0.421 |
| BEL | BEL_Spe | abdomen | 0.0377 | 789.2 | 4031.6 | 4451.4 | 752.79 | 29.71 | 53.3 | 184.87 | 24.05 | 7.568 | 0.048 | 1.2062 | 7.601 |
| BEL | BEL_Spe | elytra | 0.0442 | 415.6 | 581.5 | 3005.6 | 425.34 | 13.18 | 37.8 | 19.16 | 5.99 | 1.498 | 0.010 | 0.7090 | 0.542 |
| BEL | BEL_Spe | elytra | 0.0419 | 447.9 | 546.4 | 2773.4 | 436.52 | 13.63 | 13.4 | 7.80 | 4.94 | 1.014 | 0.011 | 0.8850 | 0.283 |
| BEL | BEL_Spe | elytra | 0.0329 | 583.8 | 763.8 | 1704.2 | 373.25 | 21.79 | 16.7 | 9.19 | 5.41 | 1.003 | 0.013 | 1.0919 | 6.290 |
| BEL | BEL_Spe | elytra | 0.0351 | 647.9 | 1046.2 | 1839.3 | 375.21 | 20.74 | 32.2 | 22.06 | 5.68 | 1.514 | 0.016 | 0.5765 | 0.257 |
| BEL | BEL_Spe | elytra | 0.0509 | 1220.6 | 441.3 | 658.7 | 304.91 | 20.46 | 60.5 | 25.91 | 4.08 | 3.577 | 0.026 | 0.7442 | 0.996 |
| BEL | BEL_Spe | elytra | 0.0276 | 437.0 | 560.9 | 1349.5 | 255.07 | 11.77 | 43.8 | 14.47 | 5.53 | 8.742 | 0.035 | 2.0585 | 3.149 |
| BEL | BEL_Spe | elytra | 0.0294 | 1690.7 | 391.8 | 496.6 | 242.52 | 18.67 | 53.7 | 0.00 | 4.99 | 0.657 | 0.046 | 0.8961 | 0.548 |
| NET | BEL_Spe | abdomen | 0.0626 | 590.5 | 1802.3 | 5082.0 | 731.31 | 21.65 | 192.3 | 262.42 | 37.01 | 62.953 | 0.066 | 0.7313 | 0.811 |
| NET | BEL_Spe | abdomen | 0.1361 | 2940.5 | 1514.1 | 4457.3 | 764.88 | 62.38 | 1243.1 | 110.74 | 15.64 | 31.322 | 0.069 | 3.9484 | 2.180 |
| NET | BEL_Spe | elytra | 0.0485 | 855.3 | 570.1 | 2365.7 | 428.45 | 22.63 | 434.0 | 22.95 | 5.72 | 3.460 | 0.029 | 1.6355 | 3.369 |
| NET | BEL_Spe | elytra | 0.0437 | 618.1 | 378.9 | 2224.9 | 428.38 | 19.73 | 186.5 | 18.27 | 5.30 | 2.955 | 0.026 | 1.1375 | 2.723 |
| NET | BEL_Spe | elytra | 0.0463 | 451.4 | 489.8 | 2352.1 | 414.04 | 13.06 | 59.4 | 17.51 | 5.63 | 4.517 | 0.012 | 0.6136 | 1.118 |
| NET | BEL_Spe | elytra | 0.0461 | 353.8 | 465.4 | 2248.6 | 411.93 | 10.93 | 54.9 | 16.27 | 4.58 | 4.214 | 0.009 | 0.7141 | 1.288 |
| NET | BEL_Spe | elytra | 0.0459 | 409.6 | 409.4 | 1780.6 | 361.22 | 11.71 | 134.2 | 12.13 | 4.57 | 2.526 | 0.016 | 0.9207 | 0.565 |
| NET | BEL_Spe | elytra | 0.0558 | 476.5 | 574.2 | 2291.3 | 362.01 | 21.08 | 260.8 | 16.91 | 3.87 | 0.684 | 0.020 | 0.9142 | 0.648 |
| BEL | BEL_Spe | abdomen | 0.0695 | 445.2 | 2053.3 | 9181.9 | 1227.05 | 16.28 | 90.8 | 326.44 | 52.18 | 25.553 | 0.049 | 0.8906 | 0.150 |
| BEL | BEL_Spe | abdomen | 0.0365 | 637.2 | 4998.6 | 7371.5 | 1048.77 | 13.81 | 77.5 | 218.46 | 10.83 | 10.364 | 0.030 | 0.7027 | 0.595 |
| BEL | BEL_Spe | abdomen | 0.0456 | 796.0 | 3314.2 | 4095.9 | 775.88 | 17.19 | 160.5 | 225.81 | 45.79 | 16.071 | 0.049 | 1.9196 | 0.615 |
| BEL | BEL_Spe | abdomen | 0.0478 | 752.8 | 3850.0 | 6913.4 | 821.13 | 11.95 | 161.1 | 166.31 | 29.09 | 9.129 | 0.031 | 0.7528 | 0.288 |
| BEL | BEL_Spe | abdomen | 0.0673 | 552.2 | 3179.6 | 7953.2 | 1185.29 | 12.32 | 113.1 | 353.76 | 56.18 | 32.645 | 0.052 | 0.5510 | 0.211 |
| BEL | BEL_Spe | elytra | 0.0511 | 314.4 | 674.9 | 2894.9 | 599.61 | 10.08 | 11.7 | 8.76 | 3.78 | 0.515 | 0.008 | 0.7306 | 0.106 |
| BEL | BEL_Spe | elytra | 0.0499 | 966.2 | 600.2 | 2753.7 | 593.79 | 11.22 | 13.2 | 2.91 | 3.89 | 0.387 | 0.010 | 0.2294 | 0.104 |
| BEL | BEL_Spe | elytra | 0.0466 | 550.0 | 491.3 | 2470.3 | 425.11 | 11.30 | 26.4 | 14.54 | 4.26 | 0.822 | 0.010 | 0.2659 | 0.276 |
| BEL | BEL_Spe | elytra | 0.0447 | 283.5 | 528.3 | 1976.2 | 397.99 | 9.27 | 26.0 | 10.56 | 3.84 | 0.650 | 0.011 | 0.2868 | 0.088 |
| BEL | BEL_Spe | elytra | 0.0308 | 540.0 | 731.7 | 2485.2 | 366.23 | 6.18 | 16.9 | 1.18 | 4.04 | 1.816 | 0.011 | 0.3816 | 0.468 |
| BEL | BEL_Spe | elytra | 0.0313 | 508.9 | 736.1 | 2359.9 | 353.67 | 6.44 | 12.8 | 6.57 | 4.47 | 1.404 | 0.042 | 0.2292 | 0.076 |
| GER | GER_Afa | abdomen | 0.1158 | 487.2 | 1920.0 | 11,062.7 | 1744.21 | 54.94 | 841.5 | 217.88 | 96.13 | 18.044 | 0.031 | 0.9314 | 1.405 |
| GER | GER_Afa | abdomen | 0.1978 | 364.2 | 3983.3 | 10,293.7 | 2243.17 | 74.46 | 783.3 | 123.92 | 21.69 | 1.092 | 0.021 | 1.4337 | 1.412 |
| GER | GER_Afa | elytra | 0.0590 | 200.4 | 700.5 | 5297.6 | 728.47 | 48.79 | 1075.1 | 54.35 | 6.92 | 2.862 | 0.019 | 1.4545 | 1.815 |
| GER | GER_Afa | elytra | 0.0667 | 227.6 | 641.2 | 5828.2 | 777.66 | 39.63 | 502.7 | 48.62 | 6.45 | 2.743 | 0.016 | 0.8211 | 1.139 |
| GER | GER_Afa | elytra | 0.0702 | 481.9 | 777.4 | 2070.6 | 795.44 | 29.04 | 131.9 | 11.20 | 6.90 | 1.714 | 0.014 | 0.3931 | 0.957 |
| GER | GER_Afa | elytra | 0.0698 | 377.0 | 683.9 | 2538.0 | 646.13 | 26.64 | 69.6 | 8.84 | 5.36 | 1.517 | 0.010 | 0.3429 | 1.273 |
| GER | GER_Aho | abdomen | 0.1346 | 483.3 | 4175.6 | 6214.7 | 986.78 | 35.36 | 297.1 | 119.00 | 20.24 | 1.272 | 0.017 | 0.2228 | 0.307 |
| GER | GER_Aho | elytra | 0.0608 | 353.8 | 491.4 | 2997.1 | 523.03 | 26.71 | 38.0 | 6.97 | 3.71 | 0.732 | 0.012 | 0.4335 | 0.461 |
| GER | GER_Aho | elytra | 0.0599 | 351.9 | 417.1 | 2743.0 | 528.71 | 27.86 | 28.9 | 5.45 | 3.81 | 0.605 | 0.006 | 0.1430 | 0.172 |
| GER | GER_Aho | elytra | 0.0642 | 362.0 | 661.7 | 2765.4 | 472.90 | 21.63 | 64.2 | 6.41 | 4.78 | 0.913 | 0.013 | 0.3549 | 0.428 |
| GER | GER_Aho | elytra | 0.0591 | 367.8 | 593.3 | 2704.0 | 423.69 | 19.71 | 50.1 | 5.73 | 4.09 | 0.943 | 0.014 | 0.2435 | 0.222 |
| GER | GER_Bu | abdomen | 0.1899 | 1006.0 | 8016.3 | 11,175.9 | 1949.13 | 49.66 | 668.1 | 119.67 | 136.44 | 0.093 | 0.017 | 1.7435 | 0.802 |
| GER | GER_Bu | abdomen | 0.0781 | 1100.4 | 5888.1 | 15,927.7 | 2520.87 | 85.61 | 1125.7 | 302.07 | 106.47 | 38.489 | 0.048 | 2.1010 | 1.241 |
| GER | GER_Bu | abdomen | 0.1117 | 1662.5 | 10,211.5 | 11,909.8 | 2572.85 | 64.17 | 167.0 | 193.11 | 219.07 | 18.498 | 0.007 | 0.3867 | 0.172 |
| GER | GER_Bu | elytra | 0.0507 | 411.1 | 1050.9 | 4539.5 | 804.34 | 21.43 | 73.4 | 20.76 | 10.48 | 2.367 | 0.013 | 0.7119 | 0.637 |
| GER | GER_Bu | elytra | 0.0513 | 409.5 | 962.8 | 4326.2 | 772.32 | 22.92 | 116.2 | 20.99 | 10.43 | 2.284 | 0.018 | 0.4553 | 1.485 |
| GER | GER_Bu | elytra | 0.0452 | 420.9 | 807.6 | 4430.4 | 755.09 | 24.28 | 303.1 | 34.00 | 12.13 | 3.835 | 0.013 | 0.7611 | 1.023 |
| GER | GER_Bu | elytra | 0.0476 | 378.3 | 658.0 | 5825.1 | 994.75 | 16.94 | 128.4 | 51.09 | 16.80 | 5.146 | 0.014 | 0.4411 | 1.036 |
| GER | GER_Bu | elytra | 0.0466 | 286.6 | 471.2 | 7145.5 | 1019.96 | 17.55 | 75.8 | 54.01 | 7.19 | 8.488 | 0.018 | 0.7240 | 1.033 |
| GER | GER_Bu | elytra | 0.0451 | 322.7 | 478.9 | 7253.5 | 1022.17 | 20.61 | 77.2 | 46.41 | 8.03 | 8.080 | 0.013 | 3.7934 | 0.572 |
| GER | GER_For | elytra | 0.0971 | 335.6 | 637.0 | 3189.6 | 1103.91 | 20.53 | 24.8 | 22.56 | 3.11 | 0.451 | 0.003 | 0.4678 | 0.325 |
| GER | GER_For | elytra | 0.0681 | 440.0 | 292.9 | 2848.7 | 594.86 | 12.33 | 65.1 | 37.85 | 2.91 | 0.939 | 0.007 | 0.6136 | 0.298 |
| GER | GER_For | elytra | 0.0634 | 394.6 | 353.2 | 3777.8 | 827.13 | 13.07 | 31.9 | 53.44 | 3.20 | 1.288 | 0.004 | 0.7238 | 1.300 |
| GER | GER_For | elytra | 0.0679 | 588.8 | 409.3 | 3594.4 | 827.98 | 9.90 | 72.0 | 46.15 | 3.36 | 0.979 | 0.006 | 0.9388 | 0.458 |
| GER | GER_For | elytra | 0.0496 | 440.4 | 480.5 | 2595.0 | 768.55 | 10.61 | 18.3 | 17.08 | 3.58 | 0.578 | 0.005 | 0.4191 | 0.154 |
| GER | GER_For | elytra | 0.0482 | 356.3 | 455.6 | 3314.7 | 784.44 | 10.46 | 14.7 | 13.30 | 3.57 | 0.825 | 0.004 | 0.2690 | 0.158 |
| GER | GER_Ko | abdomen | 0.1227 | 496.5 | 2106.6 | 8984.9 | 1489.89 | 18.35 | 129.3 | 181.77 | 53.42 | 13.056 | 0.016 | 0.2129 | 0.150 |
| GER | GER_Ko | abdomen | 0.0635 | 387.1 | 2994.5 | 4454.2 | 1008.98 | 25.57 | 215.6 | 91.66 | 59.51 | 59.365 | 0.065 | 0.4553 | 0.414 |
| GER | GER_Ko | abdomen | 0.0585 | 670.3 | 8601.8 | 11,190.2 | 1798.12 | 32.55 | 76.9 | 312.75 | 125.35 | 7.577 | 0.039 | 0.1806 | 0.056 |
| GER | GER_Ko | abdomen | 0.0552 | 720.8 | 3420.0 | 11,908.2 | 1586.96 | 18.87 | 117.2 | 407.20 | 81.25 | 30.327 | 0.040 | 1.0512 | 0.401 |
| GER | GER_Ko | elytra | 0.0499 | 412.5 | 686.8 | 3245.8 | 1066.33 | 16.83 | 18.0 | 6.79 | 3.15 | 0.681 | 0.007 | 0.2205 | 0.169 |
| GER | GER_Ko | elytra | 0.0802 | 334.2 | 392.3 | 2861.3 | 486.41 | 16.76 | 13.6 | 3.32 | 3.02 | 0.230 | 0.004 | 0.1496 | 0.567 |
| GER | GER_Ko | elytra | 0.0434 | 319.8 | 673.5 | 2317.8 | 532.03 | 24.26 | 36.6 | 22.86 | 5.35 | 1.660 | 0.014 | 0.2892 | 0.981 |
| GER | GER_Ko | elytra | 0.0435 | 298.8 | 579.3 | 2580.9 | 619.08 | 22.66 | 26.9 | 46.73 | 5.24 | 3.637 | 0.018 | 1.1859 | 0.294 |
| GER | GER_Ko | elytra | 0.0468 | 301.3 | 832.3 | 1336.7 | 662.61 | 20.58 | 94.0 | 8.27 | 8.77 | 11.198 | 0.026 | 0.3795 | 0.251 |
| GER | GER_Ko | elytra | 0.0480 | 295.1 | 820.5 | 1443.5 | 683.33 | 25.20 | 102.3 | 9.83 | 6.30 | 11.561 | 0.027 | 0.4705 | 0.897 |
| GER | GER_Ko | elytra | 0.0544 | 359.2 | 767.6 | 3170.4 | 750.55 | 22.85 | 36.2 | 20.02 | 4.67 | 1.081 | 0.008 | 0.2983 | 0.321 |
| GER | GER_Ko | elytra | 0.0518 | 405.9 | 704.7 | 3836.8 | 780.12 | 22.92 | 40.9 | 17.29 | 5.33 | 1.289 | 0.011 | 0.2686 | 0.177 |
| GER | GER_Ko | elytra | 0.0539 | 386.5 | 733.4 | 1075.7 | 782.37 | 12.05 | 19.9 | 1.57 | 3.53 | 1.343 | 0.010 | 0.2690 | 0.206 |
| GER | GER_Ko | elytra | 0.0534 | 482.5 | 926.3 | 1341.1 | 1026.78 | 13.63 | 21.0 | 2.04 | 4.38 | 1.394 | 0.017 | 0.2301 | 0.216 |
| GER | GER_Ko | elytra | 0.0449 | 394.7 | 699.2 | 1145.4 | 796.44 | 9.48 | 15.6 | 0.81 | 4.25 | 3.265 | 0.013 | 0.2236 | 0.261 |
| GER | GER_Kro | abdomen | 0.1100 | 676.1 | 1551.9 | 8167.2 | 2003.00 | 17.31 | 67.3 | 160.97 | 43.23 | 26.720 | 0.027 | 0.2730 | 0.224 |
| GER | GER_Kro | abdomen | 0.1780 | 733.1 | 2765.9 | 9797.1 | 2159.49 | 15.92 | 177.8 | 145.16 | 55.59 | 1.599 | 0.006 | 0.2016 | 0.239 |
| GER | GER_Kro | elytra | 0.0748 | 376.2 | 693.0 | 4358.5 | 1930.61 | 14.37 | 31.3 | 17.47 | 5.72 | 0.664 | 0.003 | 0.4126 | 0.527 |
| GER | GER_Kro | elytra | 0.0760 | 405.1 | 643.3 | 4188.4 | 1770.39 | 12.53 | 23.7 | 16.56 | 4.71 | 0.695 | 0.004 | 0.7622 | 0.454 |
| GER | GER_Kro | elytra | 0.0730 | 405.4 | 396.5 | 3762.3 | 763.15 | 10.89 | 25.3 | 8.62 | 3.65 | 0.838 | 0.004 | 0.3213 | 0.388 |
| GER | GER_Kro | elytra | 0.0752 | 384.9 | 368.6 | 3915.9 | 757.45 | 12.66 | 25.5 | 8.69 | 4.12 | 0.508 | 0.006 | 0.3864 | 0.666 |
| GER | GER_Kro | elytra | 0.0708 | 1573.6 | 1012.9 | 2264.9 | 748.16 | 20.25 | 121.5 | 8.55 | 4.74 | 0.571 | 0.006 | 0.5488 | 0.664 |
| GER | GER_Kro | elytra | 0.0730 | 1274.4 | 887.7 | 2032.3 | 757.95 | 18.72 | 81.5 | 6.13 | 4.72 | 0.583 | 0.006 | 0.4728 | 0.861 |
| GER | GER_Kro | elytra | 0.0617 | 600.1 | 734.0 | 2777.4 | 1197.08 | 12.89 | 66.6 | 6.08 | 3.53 | 1.570 | 0.005 | 0.5338 | 0.499 |
| GER | GER_Kro | elytra | 0.0521 | 474.9 | 592.9 | 2410.9 | 875.62 | 12.68 | 35.1 | 6.27 | 4.33 | 2.424 | 0.011 | 1.2306 | 0.389 |
| GER | GER_Kro | elytra | 0.0449 | 1266.4 | 954.1 | 3218.7 | 1284.19 | 23.45 | 72.6 | 4.04 | 4.72 | 2.288 | 0.006 | 0.7116 | 0.370 |
| GER | GER_Sch | abdomen | 0.0942 | 639.6 | 9230.8 | 14,302.5 | 2154.25 | 24.73 | 221.0 | 271.59 | 177.98 | 137.706 | 0.137 | 1.2852 | 0.510 |
| GER | GER_Sch | abdomen | 0.0868 | 710.8 | 2612.1 | 6717.5 | 1401.96 | 50.71 | 319.5 | 223.39 | 51.78 | 57.442 | 0.059 | 0.6377 | 0.767 |
| GER | GER_Sch | elytra | 0.0656 | 398.1 | 666.2 | 2873.9 | 1300.91 | 15.02 | 16.3 | 2.03 | 3.60 | 0.413 | 0.013 | 0.2708 | 0.524 |
| GER | GER_Sch | elytra | 0.0637 | 365.7 | 576.5 | 2539.6 | 1118.21 | 14.95 | 13.2 | 0.19 | 3.09 | 0.348 | 0.012 | 0.1452 | 0.411 |
| GER | GER_Sch | elytra | 0.0519 | 407.7 | 819.9 | 3074.9 | 944.70 | 24.82 | 19.7 | 8.63 | 5.50 | 5.376 | 0.017 | 0.5831 | 0.751 |
| GER | GER_Sch | elytra | 0.0547 | 338.0 | 643.7 | 2654.7 | 799.27 | 21.50 | 29.6 | 9.07 | 6.85 | 5.207 | 0.020 | 0.3374 | 1.045 |
| GER | GER_Sch | elytra | 0.0555 | 347.0 | 699.2 | 3877.3 | 717.30 | 11.10 | 59.8 | 22.02 | 7.61 | 5.618 | 0.017 | 0.4357 | 0.558 |
| GER | GER_Sch | elytra | 0.0584 | 354.6 | 588.1 | 2820.8 | 622.77 | 9.78 | 31.5 | 13.26 | 6.23 | 3.887 | 0.019 | 0.5667 | 0.389 |
| GER | GER_Ta | elytra | 0.0458 | 380.8 | 974.7 | 5686.5 | 1101.31 | 18.59 | 17.7 | 32.23 | 5.67 | 2.484 | 0.008 | 0.5104 | 1.450 |
| GER | GER_Ta | elytra | 0.0604 | 265.1 | 528.1 | 4237.1 | 821.69 | 19.84 | 11.8 | 17.03 | 3.83 | 1.486 | 0.008 | 0.3178 | 0.101 |
| GER | GER_Ta | elytra | 0.0568 | 360.6 | 595.8 | 3316.4 | 1237.32 | 14.79 | 13.2 | 2.13 | 3.63 | 2.833 | 0.005 | 0.4975 | 0.400 |
| GER | GER_Ta | elytra | 0.0586 | 333.9 | 554.1 | 3829.0 | 1166.89 | 13.57 | 14.5 | 7.85 | 3.63 | 3.090 | 0.005 | 0.3499 | 0.216 |
| GER | GER_Ta | elytra | 0.0796 | 356.2 | 500.2 | 1843.4 | 719.85 | 14.49 | 12.8 | 3.95 | 3.06 | 0.337 | 0.007 | 0.2561 | 0.173 |
| GER | GER_Ta | elytra | 0.0620 | 366.2 | 516.8 | 1683.5 | 782.26 | 10.66 | 17.7 | 1.17 | 2.75 | 0.219 | 0.007 | 0.3337 | 0.289 |
| GER | GER_Ta | elytra | 0.0443 | 487.0 | 464.8 | 808.8 | 335.21 | 6.57 | 30.9 | 0.27 | 3.23 | 0.400 | 0.012 | 0.4410 | 0.469 |
| GER | GER_Ta | elytra | 0.0503 | 959.6 | 765.8 | 1912.0 | 349.50 | 14.47 | 72.6 | 7.22 | 3.61 | 1.254 | 0.019 | 1.0507 | 1.121 |
| GER | GER_Ta | elytra | 0.0561 | 646.7 | 1083.2 | 3724.8 | 479.14 | 10.78 | 65.2 | 21.57 | 5.87 | 0.631 | 0.010 | 0.5960 | 0.288 |
| GER | GER_Ta | elytra | 0.0504 | 534.6 | 627.1 | 3241.2 | 654.17 | 12.22 | 36.9 | 5.04 | 3.32 | 0.626 | 0.008 | 0.4811 | 0.175 |
| GER | GER_Ta | elytra | 0.0521 | 431.0 | 667.5 | 2950.1 | 643.95 | 11.61 | 13.4 | 7.20 | 3.80 | 0.452 | 0.004 | 0.5356 | 0.399 |
| GER | GER_Ta | elytra | 0.0685 | 903.3 | 496.1 | 2210.4 | 1133.43 | 11.94 | 56.2 | 5.65 | 3.26 | 0.691 | 0.011 | 0.5254 | 0.473 |
| GER | GER_Ta | elytra | 0.0677 | 1092.2 | 486.0 | 1877.6 | 1012.85 | 11.58 | 77.4 | 5.00 | 2.94 | 0.658 | 0.016 | 0.7479 | 0.531 |
| GER | GER_Ta | elytra | 0.0518 | 364.7 | 540.7 | 2532.7 | 897.10 | 11.89 | 29.3 | 0.00 | 3.36 | 2.388 | 0.005 | 0.2739 | 0.430 |
| GER | GER_Ta | elytra | 0.0526 | 376.6 | 594.1 | 2586.9 | 1111.03 | 10.22 | 39.7 | 4.83 | 3.79 | 1.810 | 0.005 | 0.5531 | 1.246 |
| GER | GER_Ta | elytra | 0.0543 | 335.0 | 612.6 | 2688.1 | 913.81 | 8.46 | 27.6 | 5.13 | 3.29 | 1.719 | 0.004 | 0.2949 | 0.333 |
| GER | GER_Ta | elytra | 0.0484 | 360.2 | 562.3 | 2120.8 | 876.65 | 18.05 | 76.0 | 3.00 | 3.67 | 0.747 | 0.005 | 1.0770 | 0.910 |
| GER | GER_Ta | elytra | 0.0435 | 399.1 | 600.8 | 2099.1 | 946.67 | 18.54 | 83.2 | 2.50 | 4.09 | 0.766 | 0.007 | 0.4807 | 0.556 |
| ITA | ITA | elytra | 0.0670 | 640.7 | 869.4 | 978.1 | 456.27 | 25.58 | 37.0 | 6.86 | 2.88 | 1.976 | 0.014 | 0.5857 | 0.347 |
| ITA | ITA | elytra | 0.0674 | 527.4 | 819.3 | 1005.3 | 345.40 | 23.43 | 32.2 | 6.64 | 3.76 | 1.855 | 0.013 | 1.4670 | 0.269 |
| ITA | ITA | elytra | 0.0371 | 636.8 | 1007.2 | 1273.4 | 743.40 | 25.66 | 37.5 | 40.77 | 4.31 | 2.538 | 0.028 | 1.8737 | 0.185 |
| ITA | ITA | elytra | 0.0374 | 598.5 | 875.9 | 1294.8 | 628.07 | 23.66 | 20.3 | 60.18 | 4.71 | 1.357 | 0.027 | 32.5962 | 0.192 |
| ITA | ITA | elytra | 0.0411 | 625.2 | 653.4 | 2762.3 | 744.28 | 17.17 | 16.1 | 13.54 | 4.73 | 1.257 | 0.029 | 0.3165 | 0.271 |
| ITA | ITA | elytra | 0.0398 | 358.6 | 504.7 | 3141.6 | 632.16 | 18.01 | 13.6 | 9.42 | 4.34 | 1.066 | 0.020 | 0.5610 | 0.427 |
| ITA | ITA | elytra | 0.0424 | 311.8 | 613.0 | 3348.4 | 1204.01 | 18.23 | 19.3 | 44.80 | 4.35 | 1.404 | 0.024 | 0.3476 | 0.134 |
| ITA | ITA | elytra | 0.0416 | 313.3 | 595.4 | 3088.8 | 1041.83 | 19.92 | 18.8 | 25.60 | 4.55 | 1.261 | 0.016 | 0.3068 | 0.262 |
| ITA | ITA | elytra | 0.0715 | 2670.4 | 1644.4 | 977.8 | 340.84 | 16.45 | 193.0 | 12.69 | 3.48 | 0.405 | 0.031 | 0.3693 | 0.351 |
| ITA | ITA | elytra | 0.0691 | 3755.3 | 1689.0 | 981.9 | 279.02 | 31.28 | 289.1 | 19.96 | 4.07 | 1.409 | 0.016 | 0.2380 | 0.293 |
| ITA | ITA | elytra | 0.0494 | 357.4 | 632.6 | 2993.2 | 584.41 | 21.09 | 24.1 | 13.72 | 3.25 | 2.061 | 0.019 | 0.4378 | 0.224 |
| ITA | ITA | abdomen | 0.1473 | 4634.5 | 6416.5 | 5487.1 | 686.63 | 25.62 | 469.7 | 147.03 | 25.57 | 8.580 | 0.030 | 0.9919 | 0.536 |
| ITA | ITA | abdomen | 0.1361 | 560.6 | 5324.1 | 6016.0 | 826.16 | 11.44 | 82.8 | 148.66 | 29.63 | 14.716 | 0.019 | 0.1306 | 0.200 |
| ITA | ITA | abdomen | 0.1602 | 851.2 | 4482.7 | 5425.5 | 644.44 | 10.42 | 192.0 | 109.59 | 28.51 | 3.343 | 0.013 | 0.1288 | 0.054 |
| ITA | ITA | abdomen | 0.0851 | 521.8 | 2495.9 | 14,147.8 | 2268.98 | 17.64 | 140.4 | 254.30 | 122.20 | 33.032 | 0.029 | 0.1975 | 0.078 |
| ITA | ITA | abdomen | 0.0866 | 778.5 | 3137.7 | 12,337.1 | 1534.30 | 29.23 | 112.1 | 257.18 | 134.68 | 39.022 | 0.083 | 0.2300 | 0.421 |
| ITA | ITA | abdomen | 0.0904 | 1263.0 | 2760.5 | 13,792.1 | 2612.17 | 27.13 | 68.6 | 283.48 | 61.54 | 25.354 | 0.043 | 1.6034 | 0.082 |
| ITA | ITA | abdomen | 0.2585 | 837.8 | 5081.2 | 5768.2 | 600.85 | 12.35 | 157.0 | 88.33 | 36.92 | 11.197 | 0.013 | 0.0181 | 0.083 |
| POL | POL_La | abdomen | 0.1020 | 439.2 | 13,474.6 | 12,331.8 | 876.18 | 29.65 | 217.5 | 257.28 | 98.48 | 28.391 | 0.113 | 1.0509 | 6.041 |
| POL | POL_La | abdomen | 0.2280 | 266.3 | 5903.1 | 6895.8 | 959.39 | 13.26 | 110.5 | 101.80 | 21.72 | 1.573 | 0.020 | 0.2677 | 0.239 |
| POL | POL_La | abdomen | 0.1586 | 361.0 | 1507.2 | 11,696.2 | 2003.09 | 31.92 | 366.5 | 143.37 | 30.31 | 17.620 | 0.019 | 0.5575 | 0.298 |
| POL | POL_La | elytra | 0.0691 | 316.0 | 582.5 | 2376.9 | 1237.48 | 18.96 | 30.0 | 25.22 | 3.93 | 0.971 | 0.010 | 0.8483 | 0.933 |
| POL | POL_La | elytra | 0.0480 | 242.3 | 1074.0 | 2115.8 | 298.13 | 43.87 | 41.5 | 30.50 | 3.98 | 2.514 | 0.023 | 0.4189 | 1.383 |
| POL | POL_La | elytra | 0.0408 | 1765.1 | 725.3 | 4884.9 | 104.17 | 84.27 | 76.2 | 105.28 | 17.06 | 1.332 | 0.064 | 1.4069 | 2.000 |
| POL | POL_La | elytra | 0.0408 | 367.5 | 603.5 | 1570.2 | 407.84 | 33.76 | 50.0 | 12.16 | 3.79 | 0.843 | 0.007 | 0.7202 | 1.012 |
| POL | POL_Łsz | abdomen | 0.0643 | 589.0 | 4091.6 | 13,106.3 | 2541.52 | 61.31 | 193.2 | 274.19 | 55.88 | 4.350 | 0.067 | 1.2594 | 0.443 |
| POL | POL_Łsz | abdomen | 0.0516 | 419.5 | 1229.3 | 10,323.9 | 1718.41 | 65.55 | 177.7 | 280.93 | 41.77 | 13.574 | 0.029 | 1.0360 | 4.916 |
| POL | POL_Łsz | abdomen | 0.0899 | 351.2 | 974.7 | 6245.5 | 1068.97 | 23.92 | 98.0 | 195.72 | 62.41 | 12.668 | 0.032 | 0.4881 | 0.318 |
| POL | POL_Łsz | abdomen | 0.1149 | 756.9 | 2081.0 | 17,722.4 | 1969.71 | 84.12 | 170.4 | 196.57 | 109.65 | 66.853 | 0.044 | 0.6132 | 0.288 |
| POL | POL_Łsz | abdomen | 0.0679 | 1200.0 | 3644.5 | 29,765.8 | 4017.97 | 109.03 | 117.5 | 300.90 | 188.70 | 24.931 | 0.083 | 0.7238 | 2.342 |
| POL | POL_Łsz | abdomen | 0.1238 | 231.3 | 2157.7 | 9628.4 | 1321.00 | 41.71 | 131.7 | 166.47 | 51.01 | 38.805 | 0.024 | 0.4445 | 0.274 |
| POL | POL_Łsz | elytra | 0.0719 | 265.6 | 551.8 | 6520.4 | 1058.28 | 19.00 | 38.2 | 38.03 | 5.44 | 2.917 | 0.015 | 0.4172 | 0.346 |
| POL | POL_Łsz | elytra | 0.0733 | 253.2 | 503.9 | 6060.0 | 1016.78 | 20.17 | 38.2 | 34.17 | 5.64 | 2.974 | 0.011 | 0.4108 | 0.216 |
| POL | POL_Łsz | elytra | 0.0652 | 315.8 | 476.0 | 3497.0 | 706.75 | 13.57 | 17.5 | 10.76 | 3.71 | 0.640 | 0.011 | 0.4569 | 0.612 |
| POL | POL_Łsz | elytra | 0.0656 | 303.2 | 447.8 | 3665.6 | 654.73 | 14.17 | 19.1 | 13.10 | 3.11 | 0.772 | 0.008 | 0.3479 | 0.305 |
| POL | POL_Łsz | elytra | 0.0650 | 255.3 | 602.6 | 3309.6 | 540.00 | 20.16 | 18.6 | 15.45 | 6.36 | 1.364 | 0.008 | 0.3059 | 0.252 |
| POL | POL_Łsz | elytra | 0.0643 | 260.5 | 574.4 | 3459.3 | 557.54 | 17.42 | 18.2 | 18.44 | 4.34 | 1.597 | 0.010 | 0.4306 | 0.375 |
| POL | POL_Łsz | elytra | 0.0629 | 266.7 | 452.1 | 5967.4 | 1084.74 | 16.03 | 41.2 | 44.05 | 7.50 | 7.106 | 0.018 | 0.7016 | 0.798 |
| POL | POL_Łsz | elytra | 0.0608 | 251.6 | 464.2 | 6522.9 | 1158.06 | 16.03 | 45.1 | 43.98 | 7.41 | 7.019 | 0.011 | 0.4895 | 0.503 |
| POL | POL_Łsz | elytra | 0.0582 | 334.2 | 587.6 | 3413.5 | 667.87 | 8.27 | 16.0 | 10.60 | 4.74 | 2.257 | 0.008 | 0.5473 | 1.020 |
| POL | POL_Łsz | elytra | 0.0585 | 313.1 | 594.5 | 3260.4 | 745.64 | 6.89 | 17.8 | 7.65 | 3.76 | 1.946 | 0.007 | 0.5552 | 0.387 |
| POL | POL_Łsz | elytra | 0.0691 | 321.6 | 576.2 | 4267.4 | 664.69 | 14.91 | 31.1 | 8.23 | 3.95 | 0.771 | 0.005 | 0.2691 | 0.747 |
| POL | POL_Łsz | elytra | 0.0716 | 383.3 | 641.6 | 4464.1 | 742.88 | 16.89 | 39.1 | 11.83 | 4.81 | 0.794 | 0.003 | 0.2426 | 0.065 |
| POL | POL_Mś | elytra | 0.0880 | 246.7 | 490.9 | 4107.7 | 725.68 | 26.98 | 44.9 | 17.05 | 4.26 | 2.120 | 0.021 | 0.6794 | 0.430 |
| POL | POL_Mś | elytra | 0.0880 | 848.1 | 625.1 | 4969.1 | 975.00 | 26.35 | 23.6 | 22.28 | 5.61 | 4.180 | 0.028 | 4.7553 | 0.264 |
| POL | POL_Mś | abdomen | 0.1350 | 469.4 | 1379.7 | 12,022.9 | 2022.89 | 28.12 | 130.4 | 212.46 | 123.64 | 79.567 | 0.045 | 0.6985 | 0.203 |
| POL | POL_Mś | elytra | 0.0710 | 705.3 | 454.3 | 4088.0 | 944.37 | 12.46 | 31.4 | 32.38 | 4.80 | 0.252 | 0.098 | 0.7073 | 0.336 |
| POL | POL_Mś | elytra | 0.0710 | 254.0 | 421.9 | 3252.4 | 771.83 | 14.20 | 33.9 | 33.57 | 4.71 | 0.293 | 0.058 | 30.7702 | 0.507 |
| POL | POL_Mś | abdomen | 0.1400 | 456.4 | 1970.2 | 17,210.4 | 1954.57 | 52.96 | 102.5 | 211.89 | 155.35 | 4.608 | 0.052 | 0.6218 | 0.152 |
| POL | POL_Mś | elytra | 0.0760 | 283.6 | 524.8 | 3355.2 | 1088.29 | 21.22 | 36.2 | 17.04 | 4.53 | 0.745 | 0.023 | 0.9354 | 0.318 |
| POL | POL_Mś | elytra | 0.0690 | 289.7 | 470.6 | 3297.3 | 1073.62 | 18.67 | 280.3 | 21.22 | 4.52 | 1.524 | 0.044 | 0.7985 | 0.640 |
| POL | POL_Mś | abdomen | 0.1200 | 662.5 | 1913.4 | 11,369.6 | 1899.75 | 35.75 | 203.3 | 232.83 | 181.24 | 22.776 | 0.021 | 0.5928 | 0.219 |
| POL | POL_Prz | elytra | 0.0640 | 2444.6 | 599.6 | 4218.1 | 1148.44 | 25.03 | 48.8 | 15.50 | 129.60 | 0.556 | 0.025 | 0.5299 | 0.195 |
| POL | POL_Prz | elytra | 0.0640 | 2177.7 | 598.5 | 3614.2 | 1089.06 | 21.35 | 22.8 | 31.38 | 5.15 | 0.848 | 0.030 | 0.6272 | 0.153 |
| POL | POL_Prz | abdomen | 0.1380 | 434.0 | 3247.3 | 16,084.5 | 1783.12 | 45.53 | 81.6 | 167.24 | 73.48 | 28.065 | 0.050 | 0.6886 | 0.107 |
| POL | POL_Prz | elytra | 0.0570 | 334.3 | 606.3 | 3932.8 | 1269.47 | 22.60 | 22.1 | 13.80 | 5.37 | 0.878 | 0.048 | 0.7095 | 0.410 |
| POL | POL_Prz | elytra | 0.0577 | 297.3 | 462.9 | 3809.4 | 1233.80 | 24.65 | 34.0 | 23.91 | 5.13 | 0.459 | 0.118 | 1.3035 | 0.181 |
| POL | POL_Prz | abdomen | 0.0917 | 512.5 | 2551.8 | 18,606.7 | 2081.57 | 49.95 | 108.4 | 232.80 | 85.48 | 22.753 | 0.052 | 0.8150 | 0.074 |
| POL | POL_Prz | elytra | 0.0734 | 292.9 | 594.4 | 2321.9 | 497.41 | 20.90 | 17.2 | 18.63 | 3.46 | 0.439 | 0.048 | 1.7877 | 0.036 |
| POL | POL_Prz | elytra | 0.0766 | 286.9 | 518.9 | 1975.3 | 434.60 | 20.47 | 13.6 | 6.32 | 4.67 | 0.382 | 0.020 | 0.3259 | 0.020 |
| POL | POL_Prz | abdomen | 0.0658 | 216.4 | 3194.0 | 1692.6 | 144.53 | 137.53 | 170.5 | 54.62 | 22.89 | 0.000 | 0.043 | 2.4831 | 0.234 |
| POL | POL_Prz | elytra | 0.0641 | 324.9 | 764.9 | 3420.4 | 1414.98 | 28.48 | 16.5 | 14.91 | 5.33 | 1.541 | 0.012 | 0.2381 | 0.127 |
| POL | POL_Prz | elytra | 0.0628 | 361.6 | 671.8 | 5485.5 | 1725.16 | 19.62 | 15.4 | 14.26 | 6.41 | 12.694 | 0.021 | 0.2597 | 0.071 |
| POL | POL_Prz | elytra | 0.0339 | 468.1 | 598.9 | 3048.7 | 743.36 | 23.13 | 13.3 | 32.12 | 4.50 | 1.375 | 0.029 | 0.6217 | 0.291 |
| POL | POL_Prz | elytra | 0.0336 | 338.6 | 600.0 | 3229.5 | 742.56 | 24.33 | 12.8 | 19.09 | 4.54 | 1.538 | 0.020 | 0.4132 | 0.262 |
| POL | POL_Prz | elytra | 0.0544 | 367.4 | 571.8 | 4941.4 | 1513.42 | 16.06 | 23.0 | 13.35 | 4.49 | 1.247 | 0.015 | 0.2983 | 0.177 |
| POL | POL_Prz | elytra | 0.0554 | 275.2 | 423.7 | 4391.4 | 1031.05 | 19.21 | 12.3 | 7.64 | 3.54 | 0.848 | 0.012 | 0.3318 | 0.115 |
| POL | POL_Prz | elytra | 0.0638 | 287.2 | 764.0 | 3941.2 | 1347.18 | 21.94 | 15.7 | 9.86 | 3.37 | 1.225 | 0.014 | 0.2456 | 0.077 |
| POL | POL_Prz | elytra | 0.0659 | 336.5 | 565.9 | 4911.4 | 1496.97 | 19.54 | 14.7 | 9.92 | 5.86 | 7.981 | 0.015 | 0.3428 | 0.123 |
| POL | POL_Prz | elytra | 0.0601 | 241.8 | 620.6 | 3362.7 | 707.49 | 17.89 | 13.6 | 17.92 | 5.14 | 4.001 | 0.015 | 0.2656 | 0.078 |
| POL | POL_Prz | elytra | 0.0586 | 219.3 | 421.4 | 3336.7 | 600.68 | 17.58 | 14.3 | 15.49 | 3.61 | 4.294 | 0.020 | 0.2793 | 0.086 |
| POL | POL_Prz | elytra | 0.0684 | 278.4 | 371.6 | 2983.5 | 1037.57 | 25.22 | 11.1 | 7.08 | 4.27 | 1.496 | 0.008 | 0.1517 | 0.023 |
| POL | POL_Prz | elytra | 0.0674 | 314.9 | 336.5 | 3116.9 | 1012.61 | 24.59 | 8.9 | 5.39 | 3.48 | 1.587 | 0.008 | 0.1424 | 0.063 |
| POL | POL_Prz | elytra | 0.0460 | 381.9 | 491.5 | 3116.9 | 851.74 | 23.37 | 13.9 | 11.57 | 4.52 | 2.397 | 0.036 | 0.3795 | 0.213 |
| POL | POL_Prz | elytra | 0.0496 | 498.1 | 512.4 | 3263.1 | 919.96 | 21.45 | 28.0 | 21.96 | 4.86 | 2.784 | 0.023 | 0.5546 | 0.224 |
| POL | POL_Prz | elytra | 0.0590 | 285.4 | 410.0 | 3518.2 | 793.22 | 20.12 | 20.5 | 20.30 | 4.87 | 1.163 | 0.017 | 0.3138 | 0.137 |
| POL | POL_Prz | elytra | 0.0597 | 443.5 | 413.7 | 3385.9 | 783.58 | 23.64 | 15.9 | 16.82 | 4.16 | 1.096 | 0.018 | 0.2139 | 0.140 |
| POL | POL_Prz | elytra | 0.0464 | 351.1 | 518.3 | 2459.4 | 654.09 | 11.10 | 11.2 | 13.56 | 4.28 | 0.771 | 0.049 | 0.2240 | 0.370 |
| POL | POL_Prz | elytra | 0.0462 | 296.7 | 462.9 | 2429.8 | 666.45 | 13.58 | 13.4 | 49.24 | 6.77 | 0.608 | 0.040 | 0.9260 | 0.052 |
| POL | POL_Prz | elytra | 0.0374 | 380.5 | 529.4 | 3143.3 | 750.00 | 11.98 | 23.5 | 16.50 | 3.63 | 1.484 | 0.018 | 0.3127 | 0.275 |
| POL | POL_Prz | elytra | 0.0367 | 1281.8 | 549.3 | 3235.1 | 781.20 | 11.90 | 12.5 | 36.27 | 4.76 | 1.233 | 0.026 | 1.9037 | 0.183 |
| POL | POL_Prz | abdomen | 0.1318 | 568.7 | 4245.2 | 14,296.6 | 2677.85 | 78.69 | 78.6 | 214.99 | 98.51 | 50.441 | 0.034 | 0.4306 | 0.169 |
| POL | POL_Prz | abdomen | 0.0822 | 754.2 | 5599.7 | 20,070.5 | 2137.35 | 27.80 | 54.0 | 320.87 | 54.58 | 28.896 | 0.035 | 0.5190 | 0.062 |
| POL | POL_Prz | abdomen | 0.1246 | 635.0 | 2060.0 | 14,449.5 | 2218.46 | 96.72 | 51.4 | 222.01 | 75.74 | 40.225 | 0.077 | 2.5747 | 0.225 |
| POL | POL_Prz | abdomen | 0.1465 | 340.8 | 1849.9 | 16,010.3 | 1960.20 | 33.56 | 85.4 | 194.95 | 78.28 | 67.542 | 0.025 | 0.3153 | 0.037 |
| POL | POL_Prz | abdomen | 0.0950 | 445.6 | 2361.6 | 15,762.7 | 2139.58 | 122.49 | 82.7 | 263.85 | 69.39 | 71.503 | 0.050 | 0.6480 | 0.073 |
| POL | POL_Prz | abdomen | 0.1010 | 503.4 | 4772.7 | 16,602.0 | 2092.48 | 187.07 | 193.0 | 250.73 | 58.76 | 66.145 | 0.047 | 0.5702 | 0.102 |
| POL | POL_Prz | abdomen | 0.0934 | 431.8 | 2077.5 | 13,743.8 | 1563.60 | 126.27 | 119.7 | 219.62 | 32.10 | 48.667 | 0.046 | 0.7247 | 1.154 |
| POL | POL_Prz | abdomen | 0.1217 | 434.0 | 1451.2 | 9602.1 | 1940.84 | 27.88 | 93.1 | 198.29 | 84.42 | 78.045 | 0.033 | 3.4367 | 0.164 |
| POL | POL_Prz | abdomen | 0.1020 | 513.5 | 2653.4 | 19,667.5 | 2275.78 | 71.70 | 70.8 | 251.02 | 88.80 | 27.575 | 0.046 | 0.2402 | 0.031 |
| POL | POL_Prz | abdomen | 0.0786 | 592.2 | 3123.7 | 20,761.3 | 2375.95 | 45.60 | 45.8 | 319.37 | 26.89 | 42.454 | 0.031 | 0.4588 | 0.086 |
| POL | POL_Prz | abdomen | 0.0271 | 1279.3 | 956.5 | 1126.3 | 120.30 | 623.23 | 431.0 | 303.62 | 59.15 | 2.911 | 0.184 | 13.0152 | 1.736 |
| POL | POL_Prz | abdomen | 0.0409 | 1300.3 | 1638.9 | 1168.7 | 81.17 | 501.95 | 314.7 | 457.76 | 84.43 | 7.211 | 0.135 | 9.9166 | 1.486 |
| POL | POL_Prz | elytra | 0.0873 | 273.4 | 396.7 | 2745.2 | 641.47 | 37.85 | 39.6 | 28.97 | 4.56 | 1.392 | 0.011 | 0.4356 | 0.227 |
| POL | POL_Prz | elytra | 0.0914 | 288.0 | 593.2 | 2829.8 | 1110.39 | 36.27 | 23.9 | 18.80 | 3.72 | 1.319 | 0.010 | 0.3638 | 1.252 |
| POL | POL_Prz | elytra | 0.0514 | 487.1 | 149.9 | 558.9 | 82.10 | 320.96 | 134.0 | 54.14 | 9.14 | 0.439 | 0.086 | 4.2312 | 0.684 |
| POL | POL_Prz | elytra | 0.0515 | 422.9 | 120.2 | 529.3 | 96.31 | 270.32 | 61.4 | 36.65 | 7.29 | 0.535 | 0.082 | 2.3340 | 1.811 |
| POL | POL_PrzB | elytra | 0.0530 | 337.6 | 1152.0 | 8913.1 | 593.58 | 35.29 | 310.0 | 45.20 | 19.35 | 0.000 | 0.000 | 2.5428 | 1.018 |
| POL | POL_PrzB | elytra | 0.0540 | 572.8 | 1124.0 | 2793.8 | 660.00 | 30.49 | 75.7 | 31.82 | 10.88 | 2.967 | 0.212 | 2.3462 | 1.118 |
| POL | POL_PrzB | abdomen | 0.0830 | 297.3 | 3037.9 | 8778.8 | 1071.33 | 34.95 | 382.3 | 212.24 | 104.43 | 36.046 | 0.150 | 2.4167 | 0.677 |
| POL | POL_PrzB | elytra | 0.0300 | 428.1 | 672.0 | 3053.7 | 564.33 | 28.37 | 226.7 | 19.36 | 6.59 | 1.458 | 0.108 | 2.1102 | 1.237 |
| POL | POL_PrzB | elytra | 0.0300 | 539.1 | 559.2 | 3261.3 | 694.33 | 29.49 | 169.0 | 21.78 | 6.11 | 1.487 | 0.048 | 3.1622 | 1.445 |
| POL | POL_PrzB | abdomen | 0.0740 | 476.9 | 1592.8 | 14,846.0 | 1544.73 | 44.19 | 412.7 | 306.14 | 61.33 | 29.163 | 0.115 | 2.0573 | 1.677 |
| POL | POL_Wł | abdomen | 0.1276 | 493.4 | 2311.2 | 15,077.2 | 2135.89 | 26.42 | 216.0 | 208.86 | 132.64 | 17.939 | 0.023 | 1.1149 | 0.223 |
| POL | POL_Wł | abdomen | 0.1155 | 604.5 | 4769.5 | 9841.4 | 2410.48 | 53.04 | 120.8 | 226.85 | 69.49 | 27.415 | 0.020 | 0.6603 | 0.163 |
| POL | POL_Wł | abdomen | 0.2116 | 755.4 | 9191.9 | 10,237.3 | 1633.27 | 39.70 | 111.1 | 123.28 | 49.67 | 13.757 | 0.003 | 0.4840 | 0.015 |
| POL | POL_Wł | abdomen | 0.0899 | 327.5 | 2633.3 | 14,898.4 | 1569.74 | 27.16 | 128.7 | 272.97 | 211.23 | 144.664 | 0.019 | 0.9300 | 0.188 |
| POL | POL_Wł | abdomen | 0.0392 | 356.8 | 780.6 | 3625.7 | 791.33 | 56.86 | 105.4 | 72.85 | 31.94 | 11.605 | 0.019 | 1.3542 | 3.047 |
| POL | POL_Wł | abdomen | 0.0446 | 485.3 | 2192.3 | 5002.4 | 1008.30 | 48.72 | 128.0 | 228.16 | 38.90 | 20.581 | 0.051 | 0.9931 | 0.768 |
| POL | POL_Wł | elytra | 0.0597 | 382.8 | 723.6 | 4511.0 | 1744.72 | 31.52 | 33.8 | 6.49 | 6.01 | 2.575 | 0.014 | 0.7549 | 0.533 |
| POL | POL_Wł | elytra | 0.0571 | 389.2 | 645.6 | 4010.6 | 1418.74 | 33.15 | 21.7 | 4.87 | 4.89 | 2.483 | 0.009 | 0.6777 | 0.465 |
| POL | POL_Wł | elytra | 0.0758 | 271.9 | 451.2 | 3829.9 | 1266.89 | 19.36 | 19.9 | 6.39 | 3.97 | 1.113 | 0.006 | 0.3241 | 0.312 |
| POL | POL_Wł | elytra | 0.0758 | 309.4 | 552.8 | 3920.4 | 1267.68 | 19.80 | 21.5 | 10.06 | 4.31 | 1.189 | 0.008 | 0.4438 | 0.264 |
| POL | POL_Wł | elytra | 0.0819 | 229.5 | 533.6 | 2820.3 | 1360.93 | 20.38 | 20.9 | 5.17 | 3.99 | 0.766 | 0.006 | 0.3633 | 0.737 |
| POL | POL_Wł | elytra | 0.0849 | 239.8 | 491.9 | 2721.6 | 1368.08 | 19.79 | 18.3 | 7.70 | 3.79 | 0.812 | 0.014 | 0.4090 | 0.227 |
| POL | POL_Wł | elytra | 0.0686 | 413.0 | 741.0 | 3061.8 | 1056.12 | 28.57 | 30.9 | 6.53 | 7.59 | 1.249 | 0.005 | 0.3796 | 0.206 |
| POL | POL_Wł | elytra | 0.0701 | 381.4 | 669.7 | 2532.3 | 953.50 | 30.04 | 32.7 | 5.18 | 6.52 | 1.223 | 0.010 | 0.4297 | 0.241 |
| POL | POL_Wł | elytra | 0.0510 | 410.5 | 872.5 | 4616.5 | 1363.73 | 34.48 | 37.1 | 9.49 | 3.64 | 1.186 | 0.011 | 0.5091 | 0.285 |
| POL | POL_Wł | elytra | 0.0456 | 513.2 | 530.5 | 2511.6 | 701.75 | 41.51 | 50.9 | 10.35 | 6.27 | 1.790 | 0.047 | 0.6265 | 0.582 |
| RUS | RUS | abdomen | 0.1410 | 922.7 | 1534.5 | 9521.6 | 2356.38 | 19.14 | 124.2 | 180.81 | 52.42 | 2.482 | 0.023 | 0.3100 | 0.217 |
| RUS | RUS | elytra | 0.0779 | 318.6 | 583.2 | 2965.1 | 2212.45 | 9.63 | 19.1 | 12.12 | 4.29 | 0.189 | 0.004 | 0.3014 | 0.221 |
| RUS | RUS | elytra | 0.0788 | 262.0 | 505.3 | 2650.6 | 1571.45 | 7.25 | 28.9 | 11.98 | 4.20 | 0.193 | 0.006 | 0.2247 | 0.158 |
| SLO | Slo_GB | abdomen | 0.1071 | 567.9 | 391.3 | 1018.2 | 196.83 | 14.75 | 113.0 | 27.57 | 47.75 | 1.295 | 0.016 | 0.1729 | 0.737 |
| SLO | Slo_GB | elytra | 0.0776 | 355.8 | 163.3 | 846.2 | 130.54 | 11.11 | 25.6 | 9.04 | 8.66 | 0.417 | 0.008 | 0.1936 | 0.366 |
| SLO | Slo_GB | elytra | 0.0774 | 289.3 | 163.7 | 649.3 | 119.25 | 9.98 | 20.8 | 7.82 | 6.03 | 0.435 | 0.005 | 0.1523 | 1.146 |
| SLO | Slo_GB | abdomen | 0.0804 | 437.1 | 651.9 | 789.5 | 257.96 | 14.35 | 152.7 | 103.24 | 37.85 | 23.144 | 0.010 | 0.3318 | 0.384 |
| SLO | Slo_GB | elytra | 0.0532 | 166.6 | 154.3 | 420.3 | 121.62 | 7.79 | 25.4 | 29.11 | 5.27 | 6.078 | 0.003 | 0.3255 | 0.302 |
| SLO | Slo_GB | elytra | 0.0546 | 212.0 | 143.7 | 404.5 | 137.00 | 8.21 | 26.2 | 22.38 | 6.57 | 5.140 | 0.007 | 0.3360 | 0.326 |
| SLO | Slo_MS1 | abdomen | 0.1065 | 290.0 | 414.4 | 688.0 | 188.36 | 20.40 | 817.7 | 55.67 | 40.87 | 12.564 | 0.039 | 0.5504 | 0.297 |
| SLO | Slo_MS1 | elytra | 0.0802 | 255.7 | 269.3 | 688.9 | 183.29 | 11.31 | 42.5 | 30.17 | 5.10 | 4.919 | 0.009 | 0.2023 | 0.799 |
| SLO | Slo_MS1 | elytra | 0.0780 | 207.7 | 190.2 | 464.0 | 119.87 | 11.49 | 36.3 | 25.75 | 5.30 | 4.429 | 0.009 | 0.2186 | 0.526 |
| SLO | Slo_MS3 | abdomen | 0.0828 | 623.7 | 662.6 | 2201.5 | 409.90 | 18.94 | 126.7 | 56.99 | 53.52 | 6.032 | 0.078 | 0.2698 | 0.349 |
| SLO | Slo_MS3 | elytra | 0.0556 | 434.1 | 310.8 | 1245.3 | 240.29 | 13.29 | 34.7 | 36.13 | 5.78 | 2.593 | 0.023 | 0.2741 | 0.515 |
| SLO | Slo_MS3 | elytra | 0.0544 | 284.6 | 251.5 | 1164.3 | 216.73 | 12.56 | 25.2 | 34.48 | 5.42 | 2.421 | 0.013 | 0.1871 | 0.245 |
| SLO | Slo_MS3 | abdomen | 0.1012 | 621.9 | 823.2 | 2745.8 | 445.95 | 27.00 | 286.4 | 71.50 | 39.71 | 1.992 | 0.057 | 0.2154 | 1.126 |
| SLO | Slo_MS3 | elytra | 0.0689 | 372.1 | 366.8 | 1608.5 | 351.52 | 21.94 | 23.9 | 34.07 | 7.88 | 1.139 | 0.019 | 0.2426 | 4.396 |
| SLO | Slo_MS3 | elytra | 0.0702 | 330.8 | 331.3 | 1331.3 | 226.64 | 21.54 | 23.8 | 27.23 | 7.10 | 1.021 | 0.013 | 0.2545 | 0.484 |
| SLO | Slo_MS3 | abdomen | 0.0928 | 554.5 | 577.2 | 2341.1 | 439.87 | 15.09 | 170.4 | 71.06 | 32.32 | 18.900 | 0.080 | 0.3108 | 0.421 |
| SLO | Slo_MS3 | elytra | 0.0494 | 248.1 | 161.8 | 898.2 | 177.33 | 11.34 | 32.6 | 20.57 | 7.45 | 5.770 | 0.015 | 0.4200 | 0.434 |
| SLO | Slo_MS3 | elytra | 0.0529 | 295.3 | 156.5 | 922.3 | 183.55 | 11.01 | 35.5 | 17.16 | 8.03 | 4.837 | 0.020 | 1.7953 | 1.053 |
| SLO | Slo_MS3 | abdomen | 0.0676 | 579.3 | 503.8 | 2880.4 | 452.22 | 15.91 | 120.4 | 45.64 | 59.13 | 5.855 | 0.105 | 0.3433 | 0.158 |
| SLO | Slo_MS3 | elytra | 0.0455 | 331.3 | 185.1 | 1118.6 | 197.36 | 9.60 | 26.6 | 10.37 | 5.63 | 1.614 | 0.015 | 0.5365 | 0.169 |
| SLO | Slo_MS3 | elytra | 0.0480 | 336.6 | 229.5 | 1329.0 | 230.00 | 10.97 | 38.5 | 18.15 | 12.03 | 2.090 | 0.017 | 0.7822 | 0.579 |
| SLO | Slo_MS3 | abdomen | 0.0786 | 519.5 | 424.1 | 2257.0 | 358.27 | 21.94 | 130.4 | 43.87 | 43.21 | 5.107 | 0.088 | 0.3276 | 0.553 |
| SLO | Slo_MS3 | elytra | 0.0656 | 253.3 | 185.5 | 1036.4 | 206.40 | 17.93 | 33.4 | 17.89 | 5.70 | 1.367 | 0.014 | 0.3528 | 0.472 |
| SLO | Slo_MS3 | elytra | 0.0656 | 255.5 | 187.7 | 938.3 | 165.40 | 20.83 | 47.1 | 20.11 | 6.40 | 1.465 | 0.011 | 0.3969 | 0.906 |
| SPA | SPA_Ho | abdomen | 0.0805 | 534.9 | 1871.1 | 8327.1 | 1543.85 | 33.53 | 935.5 | 284.98 | 75.17 | 0.974 | 0.020 | 0.6115 | 0.711 |
| SPA | SPA_Ho | abdomen | 0.0439 | 477.9 | 3957.5 | 5153.2 | 1010.02 | 25.77 | 182.7 | 159.31 | 46.27 | 69.977 | 0.051 | 0.7557 | 0.269 |
| SPA | SPA_Ho | abdomen | 0.1032 | 606.2 | 1813.3 | 12,765.2 | 1241.86 | 23.01 | 131.8 | 172.59 | 73.85 | 29.170 | 0.059 | 0.1918 | 0.401 |
| SPA | SPA_Ho | abdomen | 0.0536 | 384.7 | 4954.0 | 9203.6 | 1247.95 | 42.63 | 759.3 | 314.99 | 59.64 | 2.654 | 0.093 | 0.7476 | 0.617 |
| SPA | SPA_Ho | elytra | 0.0510 | 412.6 | 371.3 | 2055.3 | 371.57 | 22.40 | 267.5 | 25.62 | 4.05 | 0.348 | 0.017 | 0.6301 | 0.553 |
| SPA | SPA_Ho | elytra | 0.0282 | 642.4 | 926.8 | 2250.6 | 923.40 | 17.08 | 152.1 | 30.46 | 6.12 | 1.530 | 0.034 | 0.8010 | 0.619 |
| SPA | SPA_Ho | elytra | 0.0283 | 276.7 | 1007.5 | 4658.9 | 1040.99 | 23.35 | 175.3 | 29.50 | 5.03 | 1.905 | 0.017 | 0.7359 | 0.326 |
| SPA | SPA_Ho | elytra | 0.0413 | 283.9 | 530.0 | 3389.1 | 937.77 | 13.83 | 145.3 | 17.58 | 5.51 | 2.652 | 0.005 | 0.6743 | 0.846 |
| SPA | SPA_Ho | elytra | 0.0413 | 354.8 | 599.7 | 3163.4 | 969.25 | 14.92 | 90.8 | 19.04 | 4.68 | 3.101 | 0.008 | 0.4848 | 0.169 |
| SPA | SPA_Ho | elytra | 0.0473 | 296.6 | 978.8 | 1993.2 | 997.67 | 16.81 | 113.5 | 16.12 | 4.51 | 4.800 | 0.015 | 0.4441 | 0.172 |
| SPA | SPA_Ho | elytra | 0.0459 | 267.1 | 992.9 | 2077.2 | 1044.66 | 17.57 | 123.5 | 19.51 | 6.95 | 37.268 | 0.070 | 0.6755 | 0.370 |
| SPA | SPA_La | abdomen | 0.0825 | 534.5 | 3069.4 | 5413.5 | 834.67 | 21.72 | 158.3 | 117.04 | 19.44 | 2.351 | 0.586 | 0.3570 | 0.192 |
| SPA | SPA_La | abdomen | 0.0693 | 686.5 | 3114.8 | 5664.1 | 1238.67 | 21.33 | 380.4 | 139.68 | 46.02 | 32.164 | 0.087 | 0.6168 | 0.112 |
| SPA | SPA_La | abdomen | 0.0771 | 513.6 | 2924.8 | 8711.0 | 2666.15 | 22.52 | 173.8 | 345.39 | 80.06 | 16.812 | 0.123 | 1.2133 | 0.107 |
| SPA | SPA_La | abdomen | 0.0619 | 368.7 | 1755.2 | 4691.8 | 878.51 | 19.36 | 231.0 | 86.60 | 28.75 | 31.481 | 0.136 | 0.8473 | 0.132 |
| SPA | SPA_La | abdomen | 0.1038 | 483.5 | 5246.7 | 6035.4 | 1288.54 | 18.88 | 95.8 | 144.98 | 21.05 | 0.519 | 0.059 | 0.5672 | 1.111 |
| SPA | SPA_La | abdomen | 0.1100 | 402.9 | 1898.8 | 7263.1 | 1397.09 | 8.96 | 142.3 | 214.83 | 33.56 | 83.527 | 0.078 | 0.4659 | 0.194 |
| SPA | SPA_La | abdomen | 0.0931 | 384.0 | 7268.8 | 5922.6 | 983.57 | 10.95 | 42.7 | 189.39 | 8.63 | 1.231 | 0.094 | 0.6644 | 0.277 |
| SPA | SPA_La | abdomen | 0.0725 | 407.4 | 3247.4 | 5081.8 | 933.24 | 16.84 | 819.7 | 126.17 | 37.95 | 7.964 | 0.067 | 1.1002 | 0.381 |
| SPA | SPA_La | elytra | 0.0572 | 273.9 | 697.3 | 2175.0 | 1135.31 | 11.94 | 39.9 | 31.52 | 3.82 | 0.822 | 0.050 | 0.6677 | 0.183 |
| SPA | SPA_La | elytra | 0.0592 | 318.5 | 656.8 | 2516.8 | 1168.75 | 11.16 | 52.5 | 35.36 | 2.96 | 0.771 | 0.029 | 9.8121 | 0.188 |
| SPA | SPA_La | elytra | 0.0567 | 442.5 | 758.1 | 1370.8 | 717.81 | 32.99 | 34.6 | 156.64 | 2.88 | 0.087 | 0.202 | 0.3599 | 0.081 |
| SPA | SPA_La | elytra | 0.0546 | 382.7 | 710.8 | 1539.3 | 715.20 | 22.77 | 24.7 | 7.31 | 3.93 | 0.042 | 0.029 | 0.5387 | 0.318 |
| SPA | SPA_La | elytra | 0.0326 | 469.9 | 777.4 | 2130.2 | 820.55 | 15.46 | 50.0 | 7.79 | 7.00 | 1.120 | 0.055 | 2.4871 | 0.209 |
| SPA | SPA_La | elytra | 0.0358 | 453.8 | 915.1 | 1334.9 | 505.87 | 22.21 | 182.1 | 107.14 | 9.06 | 1.760 | 0.120 | 2.2592 | 0.133 |
| SPA | SPA_La | elytra | 0.0467 | 451.9 | 985.2 | 1359.6 | 718.63 | 13.67 | 29.1 | 27.21 | 4.50 | 0.208 | 0.056 | 1.1245 | 1.583 |
| SPA | SPA_La | elytra | 0.0498 | 415.3 | 964.3 | 1559.0 | 719.08 | 13.94 | 33.3 | 10.69 | 5.10 | 0.191 | 0.041 | 0.7343 | 0.059 |
| SPA | SPA_La | elytra | 0.0413 | 329.3 | 707.8 | 2737.9 | 1212.35 | 7.86 | 32.9 | 10.25 | 5.83 | 4.404 | 0.040 | 0.8421 | 0.484 |
| SPA | SPA_La | elytra | 0.0408 | 367.7 | 661.8 | 2553.5 | 1239.46 | 9.33 | 27.0 | 9.79 | 4.65 | 4.627 | 0.034 | 0.8224 | 0.081 |
| SPA | SPA_La | elytra | 0.0308 | 509.9 | 1016.9 | 1147.2 | 486.36 | 16.36 | 68.8 | 3.54 | 4.40 | 0.249 | 0.035 | 0.9699 | 0.020 |
| SPA | SPA_La | elytra | 0.0353 | 430.9 | 979.0 | 2098.4 | 810.20 | 10.47 | 55.8 | 13.71 | 8.37 | 1.657 | 0.021 | 0.5915 | 1.549 |
| SPA | SPA_La | elytra | 0.0641 | 316.8 | 795.3 | 1668.3 | 356.63 | 19.92 | 41.3 | 14.16 | 3.90 | 0.380 | 0.024 | 0.4708 | 0.102 |
| SPA | SPA_La | elytra | 0.0415 | 384.3 | 959.4 | 1827.8 | 419.28 | 22.67 | 99.8 | 7.29 | 5.57 | 1.616 | 0.028 | 0.8376 | 0.165 |
| SPA | SPA_La | elytra | 0.0355 | 534.4 | 1040.5 | 1381.3 | 520.56 | 26.19 | 322.5 | 19.09 | 5.88 | 2.379 | 0.082 | 3.0525 | 0.259 |
| SPA | SPA_La | elytra | 0.0480 | 271.5 | 810.0 | 1198.2 | 435.83 | 14.93 | 175.4 | 16.89 | 4.90 | 5.108 | 0.055 | 0.8011 | 0.161 |
| SPA | SPA_La | elytra | 0.0286 | 463.8 | 971.7 | 854.5 | 391.61 | 16.84 | 82.2 | 4.23 | 4.73 | 0.256 | 0.044 | 1.1800 | 1.154 |
| SPA | SPA_LZ | abdomen | 0.0413 | 399.0 | 1258.7 | 5145.2 | 1084.26 | 42.03 | 372.2 | 156.45 | 54.43 | 254.859 | 0.061 | 0.7238 | 0.371 |
| SPA | SPA_LZ | abdomen | 0.1067 | 556.4 | 5496.8 | 9316.9 | 1065.14 | 21.31 | 161.1 | 123.97 | 48.80 | 119.494 | 0.042 | 0.3462 | 0.143 |
| SPA | SPA_LZ | abdomen | 0.0941 | 391.3 | 6007.1 | 9947.4 | 1707.86 | 32.02 | 223.9 | 195.66 | 76.14 | 26.769 | 0.016 | 0.3153 | 0.157 |
| SPA | SPA_LZ | elytra | 0.0558 | 335.3 | 609.0 | 2511.0 | 1660.93 | 23.28 | 78.9 | 23.85 | 5.19 | 20.515 | 0.014 | 0.3123 | 0.066 |
| SPA | SPA_LZ | elytra | 0.0361 | 252.5 | 1114.9 | 2745.5 | 794.74 | 18.61 | 88.9 | 15.75 | 4.62 | 7.919 | 0.051 | 0.6185 | 0.466 |
| SPA | SPA_LZ | elytra | 0.0362 | 222.3 | 1091.9 | 2647.4 | 678.45 | 20.11 | 92.5 | 16.38 | 4.85 | 7.831 | 0.065 | 0.3825 | 0.679 |
| SPA | SPA_LZ | elytra | 0.0464 | 359.0 | 1177.8 | 1853.3 | 706.90 | 21.72 | 108.0 | 26.60 | 4.71 | 1.450 | 0.025 | 0.4267 | 0.189 |
| SPA | SPA_LZ | elytra | 0.0430 | 278.2 | 1031.4 | 1634.7 | 731.16 | 20.58 | 99.1 | 29.27 | 4.41 | 1.477 | 0.025 | 0.6081 | 5.960 |
| UK | UK_Clo | abdomen | 0.0466 | 1100.5 | 1792.3 | 2883.2 | 574.46 | 33.65 | 161.8 | 286.41 | 63.38 | 1.876 | 0.129 | 3.9180 | 0.861 |
| UK | UK_Clo | elytra | 0.0493 | 395.1 | 639.7 | 2885.8 | 1410.75 | 17.95 | 73.2 | 41.72 | 6.58 | 0.981 | 0.089 | 2.3256 | 0.613 |
| UK | UK_Clo | elytra | 0.0490 | 335.9 | 642.1 | 2891.8 | 1328.98 | 18.06 | 53.1 | 41.98 | 8.99 | 0.762 | 0.093 | 2.0166 | 0.477 |
| UK | UK_Clo | abdomen | 0.0486 | 781.5 | 4165.9 | 14,496.1 | 2736.42 | 20.28 | 827.2 | 422.01 | 89.04 | 231.982 | 0.079 | 32.6688 | 0.220 |
| UK | UK_Clo | elytra | 0.0516 | 343.3 | 740.9 | 1985.0 | 1131.78 | 17.58 | 26.2 | 11.49 | 4.24 | 0.561 | 0.075 | 0.6464 | 0.003 |
| UK | UK_Clo | elytra | 0.0540 | 302.8 | 773.3 | 2274.8 | 1204.26 | 14.52 | 32.2 | 12.77 | 4.36 | 0.782 | 0.020 | 0.3733 | 0.052 |
| UK | UK_Clo | abdomen | 0.0350 | 1315.0 | 5519.3 | 7025.6 | 986.57 | 53.12 | 1666.6 | 495.40 | 89.83 | 8.863 | 0.069 | 17.8577 | 1.100 |
| UK | UK_Clo | abdomen | 0.0480 | 1897.1 | 16,348.5 | 8371.2 | 702.08 | 70.93 | 2522.3 | 483.37 | 105.85 | 9.517 | 0.129 | 20.5830 | 2.242 |
| UK | UK_Clo | abdomen | 0.0272 | 999.6 | 3703.2 | 8524.1 | 1559.93 | 44.06 | 351.5 | 193.07 | 48.65 | 159.601 | 0.048 | 6.2771 | 0.424 |
| UK | UK_Clo | abdomen | 0.0275 | 2004.5 | 4911.7 | 5612.2 | 902.18 | 48.47 | 199.3 | 658.58 | 73.97 | 56.606 | 0.107 | 4.2580 | 2.040 |
| UK | UK_Clo | abdomen | 0.0159 | 2684.7 | 720.0 | 2231.3 | 555.97 | 26.77 | 280.5 | 116.43 | 70.56 | 57.387 | 0.161 | 10.7706 | 1.252 |
| UK | UK_Clo | elytra | 0.1680 | 70.8 | 70.3 | 207.2 | 50.89 | 1.93 | 7.8 | 0.65 | 1.33 | 1.317 | 0.007 | 0.2569 | 0.051 |
| UK | UK_Clo | elytra | 0.1690 | 155.2 | 24.3 | 208.5 | 37.46 | 2.58 | 12.1 | 3.58 | 1.23 | 0.865 | 0.021 | 0.3143 | 0.050 |
| UK | UK_Clo | elytra | 0.0256 | 460.2 | 677.8 | 5760.6 | 886.72 | 17.06 | 75.0 | 12.76 | 4.89 | 18.799 | 0.053 | 1.3076 | 0.049 |
| UK | UK_Clo | elytra | 0.0250 | 378.0 | 676.8 | 4586.4 | 919.20 | 16.58 | 160.4 | 22.26 | 11.58 | 25.553 | 0.053 | 2.3187 | 0.010 |
| UK | UK_Clo | elytra | 0.0313 | 322.3 | 825.8 | 5203.7 | 1146.33 | 7.51 | 139.6 | 61.85 | 9.54 | 45.001 | 0.040 | 3.6947 | 0.088 |
| UK | UK_Clo | elytra | 0.0344 | 711.4 | 1023.5 | 3569.0 | 600.00 | 18.88 | 141.6 | 58.39 | 13.75 | 2.467 | 0.041 | 4.9854 | 0.584 |
| UK | UK_Clo | elytra | 0.0412 | 238.8 | 1080.0 | 1781.5 | 881.31 | 9.51 | 35.0 | 7.34 | 4.18 | 3.032 | 0.041 | 0.8180 | 0.578 |
| UK | UK_Clo | elytra | 0.0407 | 248.6 | 889.8 | 1626.8 | 673.96 | 9.36 | 24.3 | 24.38 | 3.99 | 3.181 | 0.021 | 1.2505 | 0.568 |
| UK | UK_Clo | elytra | 0.0260 | 542.8 | 689.5 | 2798.0 | 480.77 | 22.83 | 50.0 | 75.39 | 15.03 | 8.993 | 0.049 | 1.1148 | 0.627 |
| UK | UK_Clo | elytra | 0.3120 | 27.6 | 60.0 | 382.0 | 95.99 | 0.75 | 10.5 | 3.68 | 7.73 | 2.857 | 0.004 | 0.2265 | 0.023 |
| UK | UK_Fet | abdomen | 0.0538 | 1680.7 | 3248.0 | 20,611.8 | 5717.10 | 51.21 | 648.1 | 390.42 | 167.02 | 80.656 | 0.058 | 1.7777 | 0.479 |
| UK | UK_Fet | abdomen | 0.0718 | 1043.8 | 2869.0 | 15,644.2 | 2090.95 | 26.99 | 188.2 | 340.33 | 146.18 | 52.155 | 0.111 | 0.6883 | 0.229 |
| UK | UK_Fet | abdomen | 0.0536 | 1250.3 | 3957.3 | 13,328.4 | 2685.45 | 43.67 | 449.1 | 480.14 | 101.64 | 67.574 | 0.042 | 3.7169 | 0.492 |
| UK | UK_Fet | abdomen | 0.0685 | 977.4 | 3220.6 | 17,242.0 | 2867.59 | 30.08 | 266.6 | 387.99 | 251.70 | 134.733 | 0.061 | 1.8056 | 0.428 |
| UK | UK_Fet | abdomen | 0.0790 | 891.3 | 5602.3 | 11,191.2 | 3061.27 | 22.97 | 268.4 | 330.40 | 77.44 | 75.361 | 0.038 | 2.0241 | 0.252 |
| UK | UK_Fet | abdomen | 0.0581 | 1160.6 | 4374.5 | 15,183.8 | 3221.17 | 24.48 | 300.2 | 371.35 | 74.91 | 35.543 | 0.089 | 1.6312 | 4.489 |
| UK | UK_Fet | abdomen | 0.0495 | 1219.5 | 8756.4 | 11,694.2 | 2335.76 | 37.79 | 214.1 | 493.05 | 81.18 | 93.847 | 0.026 | 2.3648 | 0.873 |
| UK | UK_Fet | elytra | 0.0548 | 373.6 | 576.8 | 7172.3 | 2027.74 | 16.55 | 69.3 | 60.94 | 9.55 | 12.386 | 0.014 | 0.5418 | 0.468 |
| UK | UK_Fet | elytra | 0.0550 | 322.0 | 501.4 | 6215.2 | 1736.73 | 16.90 | 67.1 | 49.72 | 8.61 | 10.614 | 0.014 | 0.4104 | 1.901 |
| UK | UK_Fet | elytra | 0.0306 | 339.1 | 498.8 | 2384.6 | 937.58 | 21.23 | 30.1 | 37.57 | 7.02 | 1.863 | 0.016 | 0.6892 | 0.526 |
| UK | UK_Fet | elytra | 0.0302 | 299.2 | 503.0 | 2300.4 | 972.19 | 19.28 | 42.4 | 46.08 | 19.46 | 1.943 | 0.016 | 0.7337 | 0.422 |
| UK | UK_Fet | elytra | 0.0372 | 410.6 | 571.0 | 4141.1 | 1037.10 | 9.03 | 53.2 | 13.66 | 5.70 | 2.646 | 0.013 | 0.7170 | 0.569 |
| UK | UK_Fet | elytra | 0.0364 | 258.9 | 488.6 | 3640.4 | 937.64 | 9.85 | 36.8 | 14.29 | 11.84 | 2.339 | 0.013 | 0.5611 | 0.423 |
| UK | UK_Fet | elytra | 0.0409 | 325.0 | 447.1 | 3870.4 | 611.49 | 10.95 | 43.5 | 31.36 | 8.82 | 2.812 | 0.031 | 0.9526 | 0.679 |
| UK | UK_Fet | elytra | 0.0402 | 371.4 | 456.7 | 4109.9 | 704.98 | 9.75 | 40.3 | 26.19 | 4.53 | 3.340 | 0.023 | 0.4697 | 4.962 |
| UK | UK_Fet | elytra | 0.0407 | 261.2 | 435.2 | 4766.6 | 1022.60 | 11.56 | 56.5 | 42.81 | 4.71 | 5.217 | 0.009 | 0.7917 | 0.715 |
| UK | UK_Fet | elytra | 0.0308 | 275.3 | 556.4 | 3031.8 | 940.26 | 12.00 | 29.5 | 36.14 | 9.49 | 3.757 | 0.014 | 0.5724 | 0.688 |
| UK | UK_Fet | elytra | 0.0438 | 659.0 | 549.0 | 3571.1 | 987.44 | 20.71 | 99.3 | 18.23 | 7.33 | 6.316 | 0.016 | 1.9871 | 2.677 |
| UK | UK_Fet | elytra | 0.0431 | 333.7 | 487.8 | 3442.7 | 1008.35 | 14.55 | 48.5 | 31.72 | 5.40 | 4.793 | 0.019 | 0.6385 | 0.595 |
Appendix B. Supplementary materials
References
- 1.Orłowski G., Mróz L., Kadej M., Smolis A., Tarnawski D., Karg J., Campanaro A., Bardiani M., Harvey D.J., Méndez M., Thomaes A., Vrezec A., Ziomek K., Rudecki A.L., Mader D. Breaking down insect stoichiometry into chitin-based and internal elemental traits: patterns and correlates of continent-wide intraspecific variation in the largest European saproxylic beetle. Environ. Poll. 2020;262 doi: 10.1016/j.envpol.2020.114064. [DOI] [PubMed] [Google Scholar]
- 2.Thomaes A., Kervyn T., Maes D. Applying species distribution modelling for the conservation of the threatened saproxylic Stag Beetle (Lucanus cervus) Biol. Conserv. 2008;141:1400–1410. [Google Scholar]
- 3.Rink M., Sinsch U. Radio-telemetric monitoring of dispersing stag beetles: implications for conservation. J. Zool. 2007;272:235–243. [Google Scholar]
- 4.Harvey D.J., Gange A.C. Size variation and mating success in the Stag Beetle, Lucanus cervus. Physiol. Entomol. 2006;31:218–226. [Google Scholar]
- 5.Tini M., Bardiani M., Chiari S., Campanaro A., Maurizi E., Toni I., Mason F., Audisio P.A., Carpaneto G.M. Use of space and dispersal ability of a flagship saproxylic insect: a telemetric study of the Stag Beetle (Lucanus cervus) in a relict lowland forest. Insect Conserv. Divers. 2018;11:116–129. [Google Scholar]
- 6.Harvey D.J., Hawes C.J., Gange A.C., Finch P., Chesmore D., Farr I. Development of non-invasive monitoring methods for larvae and adults of the Stag Beetle, Lucanus cervus. Insect Conserv. Divers. 2011;4:4–14. [Google Scholar]
- 7.Campanaro A., Zapponi L., Hardersen S., Mendez M., Al Fulaij N., Audisio P., Bardiani M., Carpaneto G.M., Corezzola S., Della Rocca F., Harvey D., Hawes C., Kadej M., Karg J., Rink M., Smolis A., Sprecher E., Thomaes A., Toni I., Vrezec A., Zauli A., Zilioli M., Chiari S. A European monitoring protocol for the stag beetle, a saproxylic flagship species. Insect Conserv. Divers. 2016;9:574–584. [Google Scholar]
- 8.Cox K., McKeown N., Antonini G., Harvey D., Solano E., Van Breusegem A. Phylogeographic structure and ecological niche modelling reveal signals of isolation and postglacial colonisation in the European Stag Beetle. PLoS ONE. 2019;14(4) doi: 10.1371/journal.pone.0215860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harvey D.J., Gange A.C., Hawes C.J., Rink M. Bionomics and distribution of the stag beetle, Lucanus cervus (L.) across Europe. Insect Conserv. Divers. 2011;4:23–38. [Google Scholar]
- 10.Thierfelder T., Sandström E. The creosote content of used railway crossties as compared with European stipulations for hazardous waste. Sci. Total Environ. 2008;402:106–112. doi: 10.1016/j.scitotenv.2008.04.035. [DOI] [PubMed] [Google Scholar]
- 11.Fremlin M. Results of the “Stag Beetle ‘larval incidents’ in private gardens” survey. Essex Nat. 2013;30:94–106. [Google Scholar]
- 12.Matusiewicz H. Wet digestion methods. In: Mester Z., Sturgeon R., editors. Sample Preparation For Trace Element analysis, Comprehensive Analytical Chemistry. Elsevier; Amsterdam: 2003. p. 41. [Google Scholar]
- 13.Lajunen L.H.J., Perämäki P. Royal Society of Chemistry; Cambridge UK: 2004. Spectrochemical Analysis by Atomic Absorption and Emission. [Google Scholar]
- 14.Pike N. Using false discovery rates for multiple comparisons in ecology and evolution. Methods Ecol. Evol. 2011;2:278–282. [Google Scholar]
- 15.Sowa G., Skalski T. Effects of chronic metal exposure on the morphology of beetle's species representing different ecological niches. Bull. Environ. Contam. Toxicol. 2019;102:191–197. doi: 10.1007/s00128-018-02532-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.