Abstract
This study aimed to investigate the dissemination and characteristics of blaKPC, blaNDM, blaOXA-48-like, blaIMP, and blaVIM among the carbapenem-resistant Enterobacteriaceae (CRE) strains isolated from adult and children patients. A total of 935 non-duplicate CRE strains were collected from 36 hospitals in 24 provinces or cities across China from 2016 to 2018. Antimicrobial susceptibility testing was performed by broth microdilution method and carbapenemase genes blaKPC, blaNDM, blaOXA-48-like, blaIMP, and blaVIM were screened by PCR and confirmed by DNA sequencing. Overall, carbapenemases were produced in 97.4% (911/935) of CRE strains, including KPC-2 (51.6%, 482/935), NDM (35.7%, 334/935), and OXA-48-like carbapenemases (7.3%, 68/935). Overall, the most prevalent carbapenemase gene was blaKPC-2 among Klebsiella pneumoniae (64.6%, 457/709) and the CRE strains isolated from adult patients (70.3%, 307/437), and blaNDM among Escherichia coli (96.0%, 143/149) and the CRE strains from children (49.0%, 247/498). The blaOXA-232-positive carbapenem-resistant K. pneumoniae (9.3%, 66/709) were all isolated from children. Sixteen strains were positive for blaIMP and 9 strains produced multiple carbapenemases. No strain was positive for blaVIM. Most of the CRE strains (>90%) were resistant to cephalosporins and carbapenems, more than half (>50%) were resistant to aminoglycosides and fluoroquinolones, but the majority (95.8 and 98.4%) were susceptible to polymyxin B and tigecycline. Ceftazidime-avibactam showed excellent in vitro activity against blaKPC-2 and blaOXA-48-like positive strains (100% susceptible). In China, KPC-2, NDM, and OXA-48-like carbapenemases were predominant among CRE clinical isolates. The most prevalent carbapenemase gene was blaKPC-2 among K. pneumoniae isolates from adult patients, and blaNDM among E. coli isolates from children.
Keywords: carbapenem-resistant Enterobacteriaceae, blaKPC-2, blaNDM, blaOXA-48-like, blaIMP
Introduction
Enterobacteriaceae are opportunistic pathogens causing severe hospital-acquired infections (Feil, 2016). The spread of carbapenemase-producing Enterobacteriaceae (CPE) has been a global threat to public health. Carbapenems have conventionally been used for treating infections caused by extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae, and are still considered as last resort antibiotics to date (van Duin and Doi, 2016). According to the data from China Antimicrobial Surveillance Network (CHINET, www.chinets.com), the resistance rate of K. pneumoniae to meropenem and imipenem rapidly increased from 2.9 and 3.0% in 2005 to 26.3 and 25% in 2018, respectively. In Europe, carbapenem-resistant K. pneumoniae are most widespread in the Mediterranean and Balkan countries with a prevalence of 60% in Greece and 40% in Italy, respectively (Perez and Villegas, 2015; Feil, 2016). The production of carbapenemases including KPC, NDM, and OXA-48-like is the most common resistance mechanism among carbapenem-resistant Enterobacteriaceae clinical isolates (Nordmann et al., 2012; Goodman et al., 2016). The blaKPC-positive Enterobacteriaceae were widespread in the United States, Latin America, Italy, Greece, the Middle East, and China (Albiger et al., 2015; Feil, 2016; Villegas et al., 2016; Iovleva and Doi, 2017). The blaNDM-positive Enterobacteriaceae were widespread in India, Pakistan, Bangladesh, Italy, Poland, Denmark, Latin America, and African countries (Yong et al., 2009; Albiger et al., 2015; van Duin and Doi, 2016). The blaOXA−48−like-positive strains remained rare in the US, in contrast to the prevalence in Turkey, Spain, France, Belgium, Romania, Middle East, Africa, Asia, and South America as well (Albiger et al., 2015). These infections are usually associated with very poor prognosis and high mortality, especially in neonates or high-risk immunocompromised patients (Falagas et al., 2014; Feil, 2016). In China, the presence of blaKPC and blaNDM is responsible for phenotypic resistance in most of the CRE strains (Zhang et al., 2017; Wang et al., 2018). Most researches currently focus on the dissemination of carbapenemases among CRE strains isolated from adult patients, while only a few are available to investigate the distribution of carbapenemases among CRE strains isolated from children. To obtain the comprehensive characteristic of carbapenemases among CRE isolated from both adults and children patients in China, we conducted this study to characterize the dissemination and characteristics of carbapenemases (including KPC, NDM OXA-48, IMP, and VIM) among CRE clinical isolates and the susceptibility to antimicrobial agents.
Materials and Methods
Clinical Strains
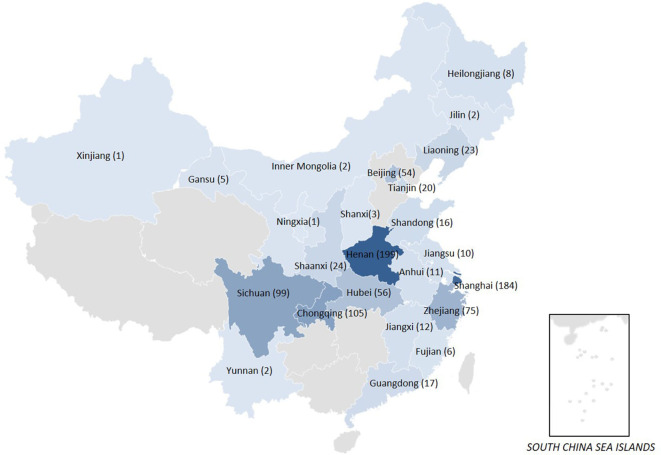
From January 2016 to December 2018, a total of 935 non-duplicate sequential CRE strains were collected from 36 hospitals in 24 provinces or cities across China (Figure 1), including K. pneumoniae (n = 709, 75.8%), E. coli (n = 149, 15.9%), Enterobacter cloacae (n = 36, 3.9%), Citrobacter freundii (n = 14, 1.5%), Serratia marcescens (n = 8, 0.9%), Enterobacter aerogenes (n = 7, 0.7%), Klebsiella oxytoca (n = 7, 0.7%), Morganella morganii (n = 3, 0.3%), Proteus vulgaris (n = 1, 0.1%), Providencia rettgeri (n = 1, 0.1%). In this study, 46.7% (437/935) of CRE strains were collected from adult patients and 53.3% (498/935) from children patients. The Enterobacteriaceae strains resistant to at least one of the carbapenem antibiotics (ertapenem, meropenem, doripenem, or imipenem) or producing a carbapenemase (an enzyme that can make them resistant to carbapenem antibiotics) were defined as CRE by Centers for Disease Control and Prevention of USA (https://www.cdc.gov/hai/organisms/cre/technical-info.html#Definition). These CRE strains were isolated from sputum (27.5%), blood (27.1%), urine (17.0%), secreta (6.9%), bile (5.0%), ascites (3.2%), catheter (2.8%), drainage (2.8%), pus (1.4%) and other aseptic body fluid (6.4%). Species identification was confirmed by MALDI-TOF/MS system (bioMérieux, France). E. coli ATCC 25922, E. coli ATCC 35218, and K. pneumoniae ATCC 700603 were tested as the quality control strains for antimicrobial susceptibility testing.
Figure 1.
The map of CRE clinical strains collected from 24 provinces or cities in China.
Antimicrobial Susceptibility Testing (AST)
AST was performed by the broth microdilution method recommended by the Clinical and Laboratory Standards Institute. Minimum inhibitory concentrations (MICs) of piperacillin, cefoperazone-sulbactam, piperacillin-tazobactam, cefazolin, cefuroxime, ceftazidime, ceftriaxone, ceftazidime-avibactam, cefepime, cefmetazole, aztreonam, ertapenem, imipenem, meropenem, amikacin, gentamicin, ciprofloxacin, levofloxacin, trimethoprim-sulfamethoxazole, polymyxin B, nitrofurantoin, tigecycline were determined. The MIC breakpoints for Enterobacteriaceae (susceptible, ≤2 mg/L; resistant, ≥8 mg/L) issued by the Food and Drug Administration were used as the breakpoints for tigecycline.
Detection of Carbapenemase and mcr-1 Genes
All the CRE strains were tested for the presence of the most common carbapenemase genes (blaKPC, blaNDM, blaOXA-48-like, blaIMP, and blaVIM) by polymerase chain reaction (PCR) with specific primers and conditions as described previously (Poirel et al., 2011; Liu et al., 2016). The colistin resistance gene mcr-1 was also detected by PCR, as previously described (Liu et al., 2016). The positive PCR amplicons were sequenced and compared with the reported sequences from GenBank by Blast (www.ncbi.nlm.nih.gov/blast/).
Statistical Analysis
Descriptive statistics were used to summarize the epidemiologic characteristics of CRE strains. For categorical variables, the percentage of CRE strains in each category was calculated. All analyses were performed using WHONET (version 5.6) and the IBM SPSS Statistics (version 21).
Results
In vitro Antimicrobial Susceptibility
Most of the CRE strains (>90%) were resistant to cephalosporins, piperacillin, cefoperazone-sulbactam, piperacillin-tazobactam, aztreonam, and carbapenems. Overall, 61.4, 50.1, and 45.2% of the strains were susceptible to ceftazidime-avibactam, amikacin, and trimethoprim-sulfamethoxazole, respectively, followed by gentamicin (31.8%), levofloxacin (22.9%), ciprofloxacin (19%), and nitrofurantoin (18.8%). Polymyxin B and tigecycline showed excellent antibacterial activity against CRE strains (95.8 and 98.4% susceptible, respectively) (Table 1). Ceftazidime-avibactam had potent activity against both KPC-2-producing and OXA-48-like producing Enterobacteriaceae (100% susceptible) and inhibited all of blaKPC-2-positive and blaOXA-48-like-positive strains at 8 mg/L. However, all NDM-producing Enterobacteriaceae were resistant to ceftazidime-avibactam (MIC90 > 32 mg/L). The MICs of ceftazidime-avibactam were higher than 32 mg/L against IMP- and multi-carbapenemase producing Enterobacteriaceae (KPC and NDM co-producers, NDM and OXA-48 co-producer). Most of the blaNDM-positive strains were susceptible to amikacin (86.2% susceptible) (Table 1).
Table 1.
Antimicrobial susceptibility testing results of clinical CRE strains (MICs, mg/L).
| Antimicrobial agent | All CRE (n = 935) | KPC-producers (n = 482) | NDM-producers (n = 334) | OXA-48-like producers (n = 68) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC | MIC50 | MIC90 | %R | %S | MIC50 | MIC90 | %R | %S | MIC50 | MIC90 | %R | %S | MIC50 | MIC90 | %R | %S | |
| range | |||||||||||||||||
| Piperacillin | 4->256 | >256 | >256 | 98.9 | 0.9 | >256 | >256 | 99.4 | 0.2 | >256 | >256 | 99.7 | 0.3 | >256 | >256 | 100 | 0 |
| Cefoperazone-sulbactam | 1->128 | >128 | >128 | 98.3 | 1.2 | >128 | >128 | 98.1 | 1.2 | >128 | >128 | 99.4 | 0 | >128 | >128 | 100 | 0 |
| Piperacillin-tazobactam | 2->256 | >256 | >256 | 97.2 | 1.5 | >256 | >256 | 98.8 | 0.6 | >256 | >256 | 99.4 | 0 | >256 | >256 | 100 | 0 |
| Cefazolin | 32->32 | >32 | >32 | 100 | 0 | >32 | >32 | 100 | 0 | >32 | >32 | 100 | 0 | >32 | >32 | 100 | 0 |
| Cefuroxime | 2->64 | >64 | >64 | 99.9 | 0.1 | >64 | >64 | 100 | 0 | >64 | >64 | 100 | 0 | >64 | >64 | 100 | 0 |
| Ceftazidime | 0.5->32 | >32 | >32 | 98.6 | 0.7 | >32 | >32 | 98.1 | 0.8 | >32 | >32 | 99.7 | 0 | >32 | >32 | 100 | 0 |
| Ceftriaxone | 0.12-64 | >32 | >32 | 99.4 | 0.6 | >32 | >32 | 99.4 | 0.6 | >32 | >32 | 99.7 | 0.3 | >32 | >32 | 100 | 0 |
| Ceftazidime-avibactam | 0.25->32 | 2 | >32 | 38.6 | 61.4 | 2 | 4 | 0 | 100 | >32 | >32 | 100 | 0 | 0.5 | 4 | 0 | 100 |
| Cefepime | 0.25->32 | >32 | >32 | 98.1 | 0.9 | >32 | >32 | 97.9 | 1 | >32 | >32 | 99.4 | 0 | >32 | >32 | 100 | 0 |
| Cefmetazole | 1->64 | >64 | >64 | 92.7 | 4.5 | >64 | >64 | 92.3 | 5.6 | >64 | >64 | 97.6 | 1.2 | 64 | >64 | 73.5 | 13.2 |
| Aztreonam | 0.25->128 | >128 | >128 | 93.2 | 4.2 | >128 | >128 | 99 | 0.8 | >128 | >128 | 85.3 | 7.8 | >128 | >128 | 100 | 0 |
| Ertapenem | 0.25->32 | >32 | >32 | 98.9 | 1 | >32 | >32 | 99 | 1 | >32 | >32 | 99.7 | 0.3 | >32 | >32 | 100 | 0 |
| Imipenem | 0.12->16 | >16 | >16 | 96.1 | 2.1 | >16 | >16 | 99.2 | 0.6 | 16 | >16 | 99.4 | 0.3 | >16 | >16 | 73.5 | 17.6 |
| Meropenem | 0.12->16 | >16 | >16 | 97 | 1.9 | >16 | >16 | 98.1 | 1.5 | >16 | >16 | 99.7 | 0.3 | >16 | >16 | 85.3 | 4.4 |
| Amikacin | 1->128 | 16 | >128 | 49.6 | 50.1 | >128 | >128 | 69.7 | 29.9 | 1 | >128 | 13.8 | 86.2 | >128 | >128 | 100 | 0 |
| Gentamicin | 1->128 | 128 | >128 | 67.9 | 31.8 | >128 | >128 | 83.8 | 16 | 1 | 128 | 40.4 | 59.3 | >128 | >128 | 100 | 0 |
| Ciprofloxacin | 0.06->8 | >8 | >8 | 78.4 | 19 | >8 | >8 | 95.6 | 3.7 | 8 | >8 | 53.6 | 41.3 | >8 | >8 | 100 | 0 |
| Levofloxacin | 0.06->16 | >16 | >16 | 76.3 | 22.9 | >16 | >16 | 94.6 | 4.8 | 4 | >16 | 49.4 | 49.7 | >16 | >16 | 100 | 0 |
| Trimethoprim- Sulfamethoxazole | 0.25->32 | 32 | >32 | 54.8 | 45.2 | 1 | >32 | 47.9 | 52.1 | >32 | >32 | 54.5 | 45.5 | >32 | >32 | 100 | 0 |
| Polymyxin B | 0.125->16 | 0.25 | 1 | 4 | 95.8 | 0.25 | 1 | 4.4 | 95.4 | 0.25 | 1 | 3.6 | 96.1 | 0.5 | 0.5 | 1.5 | 98.5 |
| Nitrofurantoin | 4->128 | >128 | >128 | 64.1 | 18.8 | >128 | >128 | 92.9 | 4.6 | 64 | >128 | 22.8 | 41.9 | 128 | >128 | 64.7 | 8.8 |
| Tigecycline | 0.12–8 | 0.5 | 2 | 0.3 | 98.4 | 0.5 | 2 | 0.4 | 97.7 | 0.5 | 1 | 0.3 | 99.1 | 1 | 2 | 0 | 100 |
CRE, carbapenem-resistant Enterobacteriaceae; MIC50/90, 50%/90% minimum inhibitory concentration; %R, % of isolates resistant; %S, % of isolates susceptible.
Prevalence of blaKPC, blaNDM, blaOXA-48, blaIMP, and blaVIM Carbapenemase and mcr-1 Genes
Carbapenemase gene was positive in 97.4% (911/935) of the CRE strains, including blaKPC-2 in 51.6% (482/935), blaNDM in 35.7% (334/935), blaOXA-48-like in 7.3% (68/935), blaIMP in 1.7% (16/935), blaKPC and blaNDM in 1.0% (9/935), blaNDM-24 and blaOXA-48 in 0.1% (1/935), blaNDM-1 and blaIMP-4 in 0.1% (1/935) of the strains (Table 2). KPC-2 was the most prevalent carbapenemase among K. pneumoniae (64.5%, 457/709) and S. marcescens (100%, 8/8) strains. NDM-5 was the predominant type carbapenemase among E. coli (74.5%, 111/149), E. cloacae (66.7%, 24/36) and C. freundii (64.3%, 9/14). Among all OXA-48-like producing K. pneumoniae, PCR and DNA sequencing results showed the presence of blaOXA-232 (97.1%, 66/68) and blaOXA-48 (2.9%, 2/68) (Table 2).
Table 2.
Prevalence of different carbapenemase genes among 935 CRE strains.
| Species | Strains tested, N | blaKPC-2, n (%) | blaNDM, n (%) | blaOXA-48-like, n (%) | blaIMP, n (%) | Two genes, n (%) | Any gene, n (%) |
|---|---|---|---|---|---|---|---|
| K. pneumoniae | 709 | 457 (64.5) | blaNDM-1, 64 (9.0) | blaOXA-48, 2 (0.3) | blaIMP-4, 6 (0.8) | blaKPC-2+blaNDM-1, 6 (0.8) | 693 (97.7) |
| blaNDM-5, 85 (12.0) | blaOXA-232, 66 (9.3) | blaIMP-69, 3 (0.4) | blaKPC-2+blaNDM-5, 1 (0.1) | ||||
| blaNDM-3, 1 (0.1) | blaNDM-1+blaIMP-4, 1 (0.1) | ||||||
| blaNDM-24+blaOXA-48, 1 (0.1) | |||||||
| E. coli | 149 | 4 (2.7) | blaNDM-1, 31 (20.8) | 147 (98.7) | |||
| blaNDM-5, 111 (74.5) | |||||||
| blaNDM-3, 1 (0.7) | |||||||
| E. cloacae | 36 | 3 (8.3) | blaNDM-1, 24 (66.7) | blaIMP-4, 4 (11.1) | blaKPC-2+blaNDM-1, 1 (2.8) | 36 (100) | |
| blaNDM-5, 3 (8.3) | blaIMP-6, 1 (2.8) | ||||||
| C. freundii | 14 | 3 (21.4) | blaNDM-1, 9 (64.3) | 12 (85.7) | |||
| S. marcescens | 8 | 8 (100) | 8 (100) | ||||
| E. aerogenes | 7 | 1 (14.3) | blaNDM-1, 1 (14.3) | 3 (42.9) | |||
| blaNDM-5, 1 (14.3) | |||||||
| K. oxytoca | 7 | 3 (42.9) | blaNDM-1, 2 (28.6) | blaIMP-4, 1 (14.3) | blaKPC-2+blaNDM-1, 1 (14.3) | 7 (100) | |
| M. morganii | 3 | 2 (66.7) | blaNDM-1, 1 (33.3) | 3 (100) | |||
| P. vulgaris | 1 | 1 (100) | 1 (100) | ||||
| P. rettgeri | 1 | blaIMP-4, 1 (100) | 1 (100) | ||||
| Total | 935 | 482 (51.6) | 334 (35.7) | 68 (7.3) | 16 (1.7) | 11 (1.2) | 911 (97.4) |
CRE, carbapenem-resistant Enterobacteriaceae.
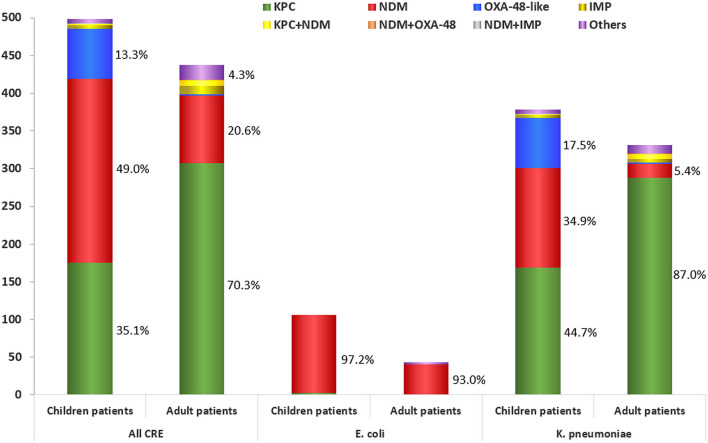
Of the CRE strains isolated from adult patients, 70.3% (307/437) were KPC-2-producers; 20.6% (90/437) were NDM-producers (including 12.1% of NDM-1-producers, 8.2% of NDM-5-producers, and 0.2% of NDM-3-producer); and 0.5% (2/437) were OXA-48-producers (Table 3, Figure 2) (P < 0.01). However, of the CRE strains isolated from children, 49.0% (244/498) were NDM-producers (including 32.9% of NDM-5-producers, 15.9% of NDM-1-producers and 0.2% of NDM-3-producer); 35.1% (175/498) were KPC-2-producers and 13.3% (66/498) were OXA-232-producers (Table 3, Figure 2) (P < 0.01). The blaOXA-232-positive K. pneumoniae were only isolated from children patients while blaOXA-48-positive K. pneumoniae were isolated from adults. One polymyxin B resistant E. coli was positive for mcr-1 with co-producing blaNDM-5.
Table 3.
Distribution of different carbapenemase genes in 935 CRE strains isolated from adults and children patients.
| Carbapenemase genes | All CRE, n (%) | E. coli, n (%) | K. pneumoniae, n (%) | |||
|---|---|---|---|---|---|---|
| From children | From adults | From children | From adults | From children | From adults | |
| blaKPC-2 | 175 (35.1) | 307 (70.3) | 3 (2.8) | 1 (2.3) | 169 (44.7) | 288 (87.0) |
| blaNDM-1 | 79 (15.9) | 53 (12.1) | 21 (19.8) | 10 (23.3) | 50 (13.2) | 14 (4.2) |
| blaNDM-5 | 164 (32.9) | 36 (8.2) | 82 (77.4) | 29 (67.4) | 81 (21.4) | 4 (1.2) |
| blaNDM-3 | 1 (0.2) | 1 (0.2) | 1 (2.3) | 1 (0.3) | ||
| blaOXA-48 | 2 (0.5) | 2 (0.6) | ||||
| blaOXA-232 | 66 (13.3) | 66 (17.5) | ||||
| blaIMP-4 | 3 (0.6) | 9 (2.1) | 2 (0.5) | 4 (1.2) | ||
| blaIMP-6 | 1 (0.2) | |||||
| blaIMP-69 | 2 (0.4) | 1 (0.2) | 2 (0.5) | 1 (0.3) | ||
| blaKPC-2+blaNDM-1 | 2 (0.4) | 6 (1.4) | 1 (0.3) | 5 (1.5) | ||
| blaKPC-2+blaNDM-5 | 1 (0.2) | 1 (0.3) | ||||
| blaNDM-1+ blaIMP-4 | 1 (0.2) | 1 (0.3) | ||||
| blaNDM-24+ blaOXA-48 | 1 (0.2) | 1 (0.3) | ||||
| Others | 5 (1.0) | 19 (4.3) | 2 (4.7) | 5 (1.3) | 11 (3.3) | |
| Total | 498 | 437 | 106 | 43 | 378 | 331 |
CRE, carbapenem-resistant Enterobacteriaceae.
Figure 2.
Carbapenemase distribution among the carbapenem-resistant Enterobacteriaceae strains isolated from adult and children patients.
Discussion
Previous studies have proved that the presence of carbapenemase genes, including blaKPC-2 and blaNDM, was the major mechanism of carbapenem resistance among CRE strains in China, which were the most prevalent in K. pneumoniae and E. coli, respectively (Zhang et al., 2017; Wang et al., 2018). However, the researches on CRE strains isolated from children patients are limited in China. This study provided a comprehensive and updated carbapenemase profile of 935 CRE strains isolated from both adult and children patients. We found that blaKPC-2 (51.6%) and blaNDM (35.7%) were the most common carbapenemase genes among CRE strains, while the emergence of blaOXA-232, blaIMP, and other multi-carbapenemase genes have been increasing in recent years. KPC-2 was the most frequently detected carbapenemase gene in K. pneumoniae, while NDM was the most prevalent one in E. coli. This pattern in China is significantly different from that in Europe. In Europe, the prevalence of OXA-48-like producing Enterobacteriaceae was 38% (333/927), next to KPC- (42%, 393/927), but higher than NDM-producing Enterobacteriaceae (12%, 113/927) (Grundmann et al., 2017). The distribution of carbapenemase-producers also varied with bacterial species. In K. pneumoniae, KPC-producers were the most prevalent, followed by OXA-48-like (37%, 310/850) and NDM-producers (11%, 93/850). In E. coli, OXA-48-like producers were the most prevalent (56%, 43/77), followed by NDM- (26%, 20/77) and KPC-producers (18%, 14/77). K. pneumoniae and E. coli were the two main species in China with a ratio of 5:1 (4:1 in children, 8:1 in adults) in this study, which differed from the prevalence trends (ratio of 11:1) in EuSCAPE (Grundmann et al., 2017).
Notably, KPC-2-producers were widespread in adult patients, followed by NDM-producers, while NDM-producers were prevalent in children patients, followed by KPC-2- and OXA-48-like producers. These findings described the different patterns of carbapenemases among CRE strains from adults and children. In contrast to the previous finding that NDM-1 was the most common carbapenemase among children patients, we have found that NDM-5-producers (32.9%) were most frequently detected CRE strains from children (Tian et al., 2018; Yin et al., 2018; Zhang et al., 2018). The outbreak of NDM-5-producing ST48 K. pneumoniae was first reported in Shanghai (Tian et al., 2018). We speculated that outbreak of NDM-5-producers accounted for the spread of NDM-5 among children patients in this study (Tian et al., 2018; Li et al., 2020). Further study is needed to track the type of plasmids harboring these carbapenemase genes.
Unlike the previous report that few OXA-48-like producing Enterobacteriaceae (0.1%, 2/1801) were detected in China from 2012 to 2016 (Wang et al., 2018), we found 7.3% (68/935) OXA-48-like producing K. pneumoniae between 2016 and 2018. Since the first OXA-232-producing K. pneumoniae isolated from neonate in 2017, the outbreaks of OXA-232-producing Enterobacteriaceae have been successively reported in children patients (Yin et al., 2017; Tian et al., 2018). Subsequently, 10 strains of OXA-232-producing K. pneumoniae were isolated from elderly patients in the intensive care unit in 2019 and the blaOXA-232 was located in a 6.1-kb ColKP3-type non-conjugative plasmid, which was highly similar to the pkNICU5 first reported (similarity about 99%) in 2017 (Yin et al., 2017; Shu et al., 2019). We speculated that the presence of blaOXA-232 on a mobile element and its spread among different strains were responsible for the recent dissemination of OXA-232-producing Enterobacteriaceae, which would make it possible to become the “third epidemic” carbapenemase after KPC-2 and NDM in China (Yin et al., 2017; Tian et al., 2018).
All of the CRE strains were highly resistant to cephalosporins, carbapenems, aminoglycosides, and fluoroquinolones but susceptible to polymyxin B and tigecycline. Ceftazidime-avibactam, launched last year in China, showed excellent in vitro antibacterial activity against both KPC-2- and OXA-48-like producers, but not active against metallo-β-lactamases producers. Most (86.2%) of NDM-producers were susceptible to amikacin. In addition, we found a blaNDM-5 and mcr-1 co-harboring E. coli resistant to polymyxin B. These findings limited the utility of ceftazidime-avibactam and polymyxin B and prompted the development of novel or combinational therapies to combat CRE strains. For example, aztreonam plus meropenem-vaborbactam and aztreonam plus ceftazidime-avibactam showed good antibacterial activity against NDM- and non-OXA-48-like producing Enterobacteriaceae (Biagi et al., 2019). The combination of colistin and amikacin showed consistently bactericidal against NDM-5-bearing mcr-1-positive E. coli, which might be an alternative therapeutic option for the treatment of lethal infections (Zhou et al., 2017).
Conclusions
In conclusion, KPC-2, NDM, and OXA-48-like enzymes were the most prevalent carbapenemases among CRE clinical isolates in China. The most prevalent carbapenemase gene was blaKPC-2 among K. pneumoniae isolated from adult patients, and blaNDM among E. coli isolates from both children and adult patients. The blaOXA-232 was only detected among K. pneumoniae isolates from children.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The study protocol was approved by the Institutional Review Board of Huashan Hospital, Fudan University (Number: 2018-408).
Author Contributions
FH and RZ designed the study. RH, QS, SW, and MP performed the experimental work. RH and DY collected the data. FH analyzed the data. All authors read and approved the final manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer YY declared a shared affiliation with one of the authors RZ to the handling editor at time of review.
Acknowledgments
The authors gratefully acknowledge the contributions of the laboratory personnel for assistance in isolates collection tested in the present study. Name list is as follows: Fu Wang, Demei Zhu, and Yan Guo from Huashan Hospital, Fudan University; Mei Kang and Chao He from West China Hospital, Sichuan University; Wen'en Liu and Yanming Li from Xiangya Hospital, Central South University; Yan Jin and Yueling Wang from Shandong Provincial Hospital; Lei Zhu and Jinhua Meng from Children's Hospital of Shanxi; Yunsong Yu and Jie Lin from Sir Run Run Shaw Hospital, Zhejiang University School of Medicine; Bin Shan and Yan Du from the First Affiliated Hospital of Kunming Medical University; Dawen Gou and Jinying Zhao from the First Affiliated Hospital of Harbin Medical University; Yunjian Hu and Xiaoman Ai from Beijing Hospital; Yuxing Ni, Jingyong Sun, and Lianyan Xie from Ruijin Hospital, Shanghai Jiaotong University School of Medicine; Gang Li and Wei Jia from General Hospital of Ningxia Medical University; Shuping Zhou and Jiangwei Ke from Jiangxi Provincial Children's Hospital; Lianhua Wei and Xin Wang from Gansu Provincial Hospital; Yi Li and Shanmei Wang from Henan Provincial People's Hospital; Yuanhong Xu and Yin Huang from the First Affiliated Hospital of Anhui Medical University; Zhongju Chen and Ziyong Sun from Tongji Hospital, Tongji Medical College, Huazhong University of Science & Technology; Chuanqing Wang and Leiyan He from Children's Hospital of Fudan University; Yingchun Xu, Xiangjiang Zhang, and Shuyin Yu from Peking Union Medical College Hospital; Chao Zhuo and Danhong Su from the First Affiliated Hospital of Guangzhou Medical University; Zhaoxia Zhang and Pin Ji from the First Affiliated Hospital of Xinjiang Medical University; Yunzhuo Chu and Sufei Tian from the First Affiliated Hospital of China Medical University; Ruizhong Wang and Hua Fang from Pudong New Area People's Hospital; Kaizhen Weng and Yirong Zhang from Jinjiang Municipal Hospital; Xuesong Xu and Chao Yan from China-Japan Union Hospital, Jilin University; Hong Zhang and Chun Wang from Children's Hospital of Shanghai; Sufang Guo and Yanyan Wang from the First Affiliated Hospital of Inner Mongolia Medical University; Zhidong Hu and Jin Li from Tianjin Medical University General Hospital; Bixia Yu from Zhejiang Ningbo Zhenhai Longsai Hospital; Pin Gong and Miao Song from People's Hospital of Zigui, Hubei Province; Xiangning Huang and Hua Yu from Sichuan Provincial People's Hospital; Lixia Zhang and Juan Ma from Shaanxi Provincial People's Hospital; Jinsong Wu and Yuemei Lu from Shenzhen People's Hospital; Ruyi Guo and Ye Zhu from Quanzhou First Hospital; Junwen Yang from Zhengzhou Children's Hospital; Chengbin Xie from Sichuan Provincial Hospital for Women and Children; Lijun Tian from Xuzhou Children's Hospital; Ping Ma from the Affiliated Hospital of Xuzhou Medical University.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (grant nos. 81871690, 81902101, and 81861138051), the National Mega-project for Innovative Drugs (2019ZX09721001-006-004) and China Antimicrobial Surveillance Network (WI207259).
References
- Albiger B., Glasner C., Struelens M. J., Grundmann H., Monnet D. L. (2015). Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries, May 2015. Euro. Surveill. 20. 10.2807/1560-7917.ES.2015.20.45.30062 [DOI] [PubMed] [Google Scholar]
- Biagi M., Wu T., Lee M., Patel S., Butler D., Wenzler E. (2019). Searching for the optimal treatment for metallo- and serine-beta-lactamase producing Enterobacteriaceae: Aztreonam in combination with ceftazidime-avibactam or meropenem-vaborbactam. Antimicrob. Agents Chemother. 63:e01426–19. 10.1128/AAC.01426-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falagas M. E., Tansarli G. S., Karageorgopoulos D. E., Vardakas K. Z. (2014). Deaths attributable to carbapenem-resistant Enterobacteriaceae infections. Emerg. Infect. Dis. 20, 1170–1175. 10.3201/eid2007.121004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil E. J. (2016). Enterobacteriaceae: joining the dots with pan-European epidemiology. Lancet Infect Dis. 17, 118–119. 10.1016/S1473-3099(16)30333-4 [DOI] [PubMed] [Google Scholar]
- Goodman K. E., Simner P. J., Tamma P. D., Milstone A. M. (2016). Infection control implications of heterogeneous resistance mechanisms in carbapenem-resistant Enterobacteriaceae (CRE). Expert. Rev. Anti. Infect. Ther. 14, 95–108. 10.1586/14787210.2016.1106940 [DOI] [PubMed] [Google Scholar]
- Grundmann H., Glasner C., Albiger B., Aanensen D. M., Tomlinson C. T., Andrasevic A. T., et al. (2017). Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect. Dis. 17, 153–163. 10.1016/S1473-3099(16)30257-2 [DOI] [PubMed] [Google Scholar]
- Iovleva A., Doi Y. (2017). Carbapenem-resistant Enterobacteriaceae. Clin. Lab. Med. 37, 303–315. 10.1016/j.cll.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yu T., Tao X. Y., Hu Y. M., Wang H. C., Liu J. L., et al. (2020). Emergence of an NDM-5-producing Escherichia coli sequence type 410 clone in infants in a children's hospital in China. Infect. Drug Resist. 13, 703–710. 10.2147/IDR.S244874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Nordmann P., Dortet L., Poirel L. (2012). Carbapenem resistance in Enterobacteriaceae: here is the storm!. Trends Mol. Med. 18, 263–272. 10.1016/j.molmed.2012.03.003 [DOI] [PubMed] [Google Scholar]
- Perez F., Villegas M. V. (2015). The role of surveillance systems in confronting the global crisis of antibiotic-resistant bacteria. Curr. Opin. Infect. Dis. 28, 375–383. 10.1097/QCO.0000000000000182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Walsh T. R., Cuvillier V., Nordmann P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Micr. Infec. Dis. 70, 119–123. 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Shu L., Dong N., Lu J., Zheng Z., Hu J., Zeng W., et al. (2019). Emergence of OXA-232 carbapenemase-producing Klebsiella pneumoniae that carries a pLVPK-like virulence plasmid among elderly patients in China. Antimicrob. Agents Chemother. 63:e02246-18. 10.1128/AAC.02246-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Pan F., Wang C., Sun Y., Zhang H. (2018). Resistance phenotype and clinical molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae among pediatric patients in Shanghai. Infect Drug Resist. 11, 1935–1943. 10.2147/IDR.S175584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duin D., Doi Y. (2016). The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 8, 460–469. 10.1080/21505594.2016.1222343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas M. V., Pallares C. J., Escandón-Vargas K., Hernández-Gómez C., Correa A., Álvarez C., et al. (2016). Characterization and clinical impact of bloodstream infection caused by carbapenemase-producing Enterobacteriaceae in seven latin American Countries. PLoS ONE 11:e0154092. 10.1371/journal.pone.0154092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wang X., Wang H., Ouyang P., Jin C., Wang R., et al. (2018). Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae data from a longitudinal large-scale CRE study in China (2012–2016). Clin. Infect. Dis. 67, S196–S205. 10.1093/cid/ciy660 [DOI] [PubMed] [Google Scholar]
- Yin D., Dong D., Li K., Zhang L., Liang J., Yang Y., et al. (2017). Clonal dissemination of OXA-232 carbapenemase-producing Klebsiella pneumoniae in neonates. Antimicrob. Agents Chemother. 61:e00385–17. 10.1128/AAC.00385-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin D., Zhang L., Wang A., He L., Cao Y., Hu F., et al. (2018). Clinical and molecular epidemiologic characteristics of carbapenem-resistant Klebsiella pneumoniae infection/colonization among neonates in China. J. Hosp. Infect. 100, 21–28. 10.1016/j.jhin.2018.05.005 [DOI] [PubMed] [Google Scholar]
- Yong D., Toleman M. A., Giske C. G., Cho H. S., Sundman K., Lee K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Liu L., Zhou H., Chan E. W., Li J., Fang Y., et al. (2017). Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMed. 19, 98–106. 10.1016/j.ebiom.2017.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Chen D., Xu G., Huang W., Wang X. (2018). Molecular epidemiology and drug resistant mechanism in carbapenem-resistant Klebsiella pneumoniae isolated from pediatric patients in Shanghai, China. PLoS ONE 13:e0194000. 10.1371/journal.pone.0194000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Tao M., Feng Y., Yang R., Liao X., Liu Y., et al. (2017). Increased activity of colistin in combination with amikacin against Escherichia coli co-producing NDM-5 and MCR-1. J. Antimicrob. Chemoth. 72, 1723–1730. 10.1093/jac/dkx038 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.