Abstract
Background
Tumour budding and poorly differentiated clusters (PDC) represent forms of tumour invasion. We hypothesised that T-cell densities (reflecting adaptive anti-tumour immunity) might be inversely associated with tumour budding and PDC in colorectal carcinoma.
Methods
Utilising 915 colon and rectal carcinomas in two U.S.-wide prospective cohort studies, and multiplex immunofluorescence combined with machine learning algorithms, we assessed CD3, CD4, CD8, CD45RO (PTPRC), and FOXP3 co-expression patterns in lymphocytes. Tumour budding and PDC at invasive fronts were quantified by digital pathology and image analysis using the International tumour Budding Consensus Conference criteria. Using covariate data of 4,420 incident colorectal cancer cases, inverse probability weighting (IPW) was integrated with multivariable logistic regression analysis that assessed the association of T-cell subset densities with tumour budding and PDC while adjusting for selection bias due to tissue availability and potential confounders, including microsatellite instability status.
Findings
Tumour budding counts were inversely associated with density of CD3+CD8+ [lowest vs. highest: multivariable odds ratio (OR), 0.50; 95% confidence interval (CI), 0.35–0.70; Ptrend < 0.001] and CD3+CD8+CD45RO+ cells (lowest vs. highest: multivariable OR, 0.44; 95% CI, 0.31–0.63; Ptrend < 0.001) in tumour epithelial region. Tumour budding levels were associated with higher colorectal cancer-specific mortality (multivariable hazard ratio, 2.13; 95% CI, 1.57–2.89; Ptrend < 0.001) in Cox regression analysis. There were no significant associations of PDC with T-cell subsets.
Interpretation
Tumour epithelial naïve and memory cytotoxic T cell densities are inversely associated with tumour budding at invasive fronts, suggesting that cytotoxic anti-tumour immunity suppresses tumour microinvasion.
Keywords: adenocarcinoma; artificial intelligence, clinical outcomes; epithelial mesenchymal transition; host-tumour interaction; molecular pathological epidemiology
Abbreviations: AJCC, American Joint Committee on Cancer; CI, confidence interval; CIMP, CpG island methylator phenotype; CMS, consensus molecular subtype; EMT, epithelial mesenchymal transition; FFPE, formalin-fixed paraffin-embedded; HPFS, Health Professionals Follow-up Study; HR, hazard ratio; IPW, inverse probability weighting; ITBCC, International Tumour Budding Consensus Conference; LINE-1, long-interspersed nucleotide element-1; MSI, microsatellite instability; NHS, Nurses’ Health Study; OR, odds ratio; PCR, polymerase chain reaction; PDCs, poorly differentiated clusters; SD, standard deviation; TMA, tissue microarray; TNM, tumour, node, and metastases
Research in context.
Evidence before this study
The adaptive immune response can restrict tumour growth, while tumour cell invasion can lead to tumour progression. Tumour budding, which is a histological manifestation of initiating invasion and metastasis cascade along the tumour invasive front, is an independent prognostic factor in colorectal cancer. It is formally defined as a single tumour cell or a cluster of fewer than five tumour cells dissociated from the main tumour at the invasive front by the International tumour Budding Consensus Conference (ITBCC) criteria. The adaptive immune response, represented by cytotoxic T cells, plays a crucial role in suppressing tumour invasion and metastasis, and a high density of tumour infiltrating cytotoxic T cells is associated with favourable prognosis in colorectal cancer. However, the interplay between anti-tumour immunity and tumour budding in the colorectal cancer microenvironment has not been adequately elucidated in terms of specific T-cell subsets driving the associations.
Added value of this study
We utilised two U.S. nationwide prospective cohort studies with covariate data of 4420 colorectal cancer cases and a molecular pathological epidemiology database of 915 cases to evaluate relationship between tumour budding and T-cell densities. Using this database, we hound that tumour budding count was inversely associated with density of CD3+CD8+ and CD3+CD8+CD45RO+ cells in tumour intraepithelial region, independent of any tumour molecular features such as microsatellite instability (MSI) status, CpG island methylator phenotype (CIMP) status, long-interspersed nucleotide element-1 (LINE-1) methylation, CTNNB1 (catenin beta 1) and CDH1 (cadherin 1, E-cadherin) expression, and KRAS, BRAF, and PIK3CA mutations. Additionally, our findings provide supporting evidence that high tumour budding grade was associated with higher colorectal cancer-specific mortality independent of tumour intraepithelial cytotoxic T-cell density.
Implication of all the available evidence
Our finding underlines that cytotoxic T cells play a crucial role of anti-tumour immunity in suppressing microinvasion. Our study, based on two large U.S. nationwide prospective cohorts, also represents an extensive validation of the prognostic value of ITBCC evaluation of tumour budding in colorectal cancer, independent of tumour molecular features and immune cell densities.
Alt-text: Unlabelled box
1. Introduction
The tumour invasive front is an important interface of tumour-host interactions and its properties are thought to regulate tumour progression. In particular, tumour budding is a histological manifestation of initiating invasion and metastasis cascade along the tumour invasive front [1]. By International tumour Budding Consensus Conference (ITBCC) criteria, it is formally defined as a single tumour cell or a cluster of fewer than five tumour cells dissociated from the main tumour at the invasive front, whereas clusters of five or more tumour cells without gland formation are defined as poorly differentiated clusters (PDCs) [2,3]. Studies have demonstrated that tumour budding was an independent prognostic factor in colorectal cancer [2].
In recent years, immunotherapy has emerged as a promising therapeutic modality for cancer [4,5]. Accumulating evidence suggests that adaptive immune response, represented by cytotoxic T cells, plays a crucial role in suppressing tumour invasion and metastasis [6], and a high density of tumour infiltrating cytotoxic T cells is associated with favourable prognosis in colorectal cancer [7]. A few studies have shown that anti-tumour immune response might restrict tumour budding and indicated that a combined budding-immune cell score might be a stronger predictor of survival than either parameter alone [6,8,9]. However, the interplay between anti-tumour immunity and tumour budding/PDCs in the colorectal cancer microenvironment has not been adequately elucidated in terms of specific T-cell subsets driving the associations, as well as tumour molecular features such as microsatellite instability (MSI) status, CpG island methylator phenotype (CIMP) status, long-interspersed nucleotide element-1 (LINE-1) methylation, CTNNB1 (catenin beta 1) and CDH1 (cadherin 1, E-cadherin) expression, and KRAS, BRAF, and PIK3CA mutations.
In this study, we utilised two U.S. nationwide prospective cohort studies with covariate data of 4420 colorectal cancer cases and a molecular pathological epidemiology database of 915 cases to evaluate relationship between tumour budding, PDCs, and T-cell densities. We evaluated tumour budding using ITBCC criteria [2] and PDCs using a corresponding approach [3], and we characterised densities and location of specific T-cell subsets with multiplex immunofluorescence assay by simultaneously measuring expression levels of CD3, CD4, CD8, CD45RO (PTPRC), and FOXP3 in immune cells in tumour intraepithelial and stromal regions. Using this database, we tested the hypothesis that specific T-cell subsets in tumour are inversely associated with tumour budding at the invasive front. In addition, we analysed the prognostic role of tumour budding and PDCs in these cohorts, as well as their prognostic interactions with specific T-cell subsets.
2. Materials and methods
2.1. Study population
We collected data on colorectal cancer cases within two prospective cohort studies in the U.S.: the Nurses’ Health Study (NHS; 121,701 women aged 30–55 years, who have been followed since 1976) and the Health Professionals Follow-up Study (HPFS; 51,529 men aged 40–75 years, who have been followed since 1986) [10]. Every two years, questionnaires have been sent to participants to update information on their lifestyle factors and medical history, including incidence of colorectal cancer. For both cohorts, the response rate for each follow-up questionnaire has been more than 90%. The National Death Index was used to ascertain deaths of study participants and to identify unreported lethal cases of colorectal cancer. Medical records were reviewed by study physicians to confirm the diagnosis of colorectal cancer and to record tumour characteristics [e.g., size, location, and the American Joint Committee on Cancer (AJCC) tumour, node, and metastases (TNM) classification] as well as the causes of death for deceased participants.
Formalin-fixed paraffin-embedded (FFPE) tissue blocks were collected from hospitals where participants were diagnosed colorectal cancer and underwent tumour resection. We included 915 patients with available data on T-cell densities, tumour budding, and PDCs in colorectal cancer tissue. On the basis of the colorectal continuum model, both colon and rectal carcinomas were included [11]. Patients were followed until death, or until the end of follow-up (January 1, 2014 for HPFS; May 31, 2014 for NHS), whichever came first. We used covariate data of 4420 incident colorectal cancer cases to adjust for selection bias in the 915 cases. A previous study has shown that there are no substantial demographic or clinical differences between cases with tumour tissue and cases without tumour tissue 12, 13, 14.
Informed consent was obtained from all study participants, and the study was approved by the institutional review boards of the Brigham and Women's Hospital and Harvard T.H. Chan School of Public Health (Boston, MA), and those of participating registries as required.
2.2. Assessment of tumour morphology and immunohistochemistry
A single pathologist (S.O.), who was unaware of other data, performed a centralised review of haematoxylin and eosin-stained tissue sections from all colorectal carcinoma cases [15]. Tumour differentiation was categorised as well to moderate or poor (> 50% vs. ≤ 50% glandular area, respectively). Four components of lymphocytic reaction to tumours, including tumour-infiltrating lymphocytes, intratumoural periglandular reaction, peritumoural lymphocytic reaction, and Crohn's-like lymphoid reaction were scored as 0, 1+, 2+, 3+, and graded as negative/low (0), intermediate (1+), or high (2+, 3+) [15].
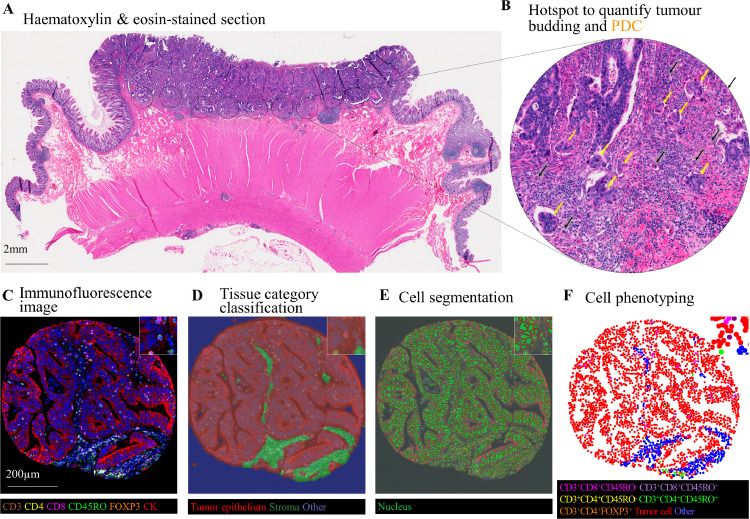
Tumour budding was assessed according to recommendations of the ITBCC [2] and PDCs were assessed according to similar conventional criteria (Fig. 1) [3]. A single hotspot at the invasive front, as well as tumour centre, was identified for counting, and tumour budding was categorised into three grades; low (Bd1, 0–4 buds/0.785 mm2), intermediate (Bd2, 5–9 buds/0.785 mm2), and high (Bd3, ≥10 buds/0.785 mm2). Similarly, PDCs, consisting of five or more tumour cells without gland formation, were categorised into three grades; low (0–4 clusters/0.785 mm2), intermediate (5–9 clusters/0.785 mm2), and high (≥10 clusters/0.785 mm2). All cases were assessed by a single pathologist (J.P.V.) blinded to other data. A consecutively selected subset (n= 140) within our overall case set was independently reviewed by a second pathologist (D.J.P.), and the correlation coefficient by Spearman's correlation test between the two pathologists was 0.79 (continuous, buds/0.785 mm2) for tumour budding and 0.61 (continuous, PDCs/0.785 mm2) for PDC (both P < 0.0001). Weighted-Kappa was 0.66 (3 categories) for tumour budding and 0.33 (3 categories) for PDCs (both P < 0.0001). As previously described [16, 17], immunohistochemistry was performed to evaluate membranous CDH1 expression and nuclear CTNNB1 expression in tumour cells, using anti-CDH1 (clone NCH-38, 1:75 dilution, Dako, Carpinteria, CA, USA) and anti-CTNNB1 antibodies (clone 14, 1:400 dilution; BD Transduction Laboratories, Franklin Lakes, NJ, USA), respectively.
Fig. 1.
Quantification of tumour budding and poorly differentiated clusters (PDCs) using International tumour Budding Consensus Conference (ITBCC) criteria and the densities of specific T-cell subsets using multiplex immunofluorescence. (a)-(b) Evaluation of tumour budding and PDCs using haematoxylin and eosin-stained sections. (a) Whole slide images were scanned at medium power to identify the most intensive areas of budding and PDCs. (b) The number of tumour buds (black arrowheads) and PDCs (yellow arrow head)/20x microscope field (0.785 mm2) were counted. (c)-(f) Evaluation of the densities and location of T cells with multiplex immunofluorescence. Machine learning-based image processing (c), included tissue category classification (d), cell segmentation (e), and cell phenotyping (f) to identify different T-cell subsets in intraepithelial and stromal regions. Abbreviations: ITBCC, International Tumour Budding Consensus Conference; PDC, Poorly differentiated clusters.
2.3. Assessment of T-cell densities
We constructed tissue microarrays (TMAs) consisting of up to four tumour cores [18,19]. We developed a multiplex immunofluorescence assay to simultaneously measure the expression of CD3, CD4, CD8, CD45RO (one isoform of PTPRC gene products), and FOXP3 in immune cells within intraepithelial and stromal regions. The following antibody/fluorophore combinations were used for the staining: anti-CD3 (clone F7.2.38; Dako; Agilent Technologies, Carpenteria, CA)/Opal-520, anti-FOXP3 (clone 206D, Biolegend, San Diego, CA)/Opal-540, anti-CD45RO (clone UCHL1, Dako)/Opal-650, anti-CD8 (clone C8/144B, Dako)/Opal-570, anti-CD4 (clone 4B12, Dako)/Opal-690, anti-KRT (Keratins, pan-keratins; mixture of clones AE1/AE3, Dako and C11, Cell signalling, Denvers, MA)/Opal-620 (Supplementary fig. S1). Digital images of all TMA cores were acquired at 200x magnification using an automated multispectral imaging system (Vectra 3.0, Akoya Biosciences, Hopkinton, MA). Using supervised machine learning algorithms (Inform 2.4.1, Akoya Biosciences), T-cell subset densities were calculated through the process of tissue segmentation (classifying tissue regions into tumour epithelium, stroma, and other), cell segmentation (detecting cells and their nuclear, cytoplasmic, and membranous compartments), and cell phenotyping (classifying cells based on cell phenotypic features including fluorophore intensities and cytomorphology) (Fig. 1). The data were extracted at the single-cell level and used to calculate T-cell subset densities within tumour epithelial and stromal regions. T-cell densities were initially classified into quartile categories (C1-C4). If more than 25% of all cases had zero density (0/mm2) of a specific cell type, these cases were grouped together (C1 category), and the remaining (non-zero) cases were divided into tertiles (C2-C4). For sensitivity analysis, T-cell densities were binarised (high vs. low; with a cut-point at a median value).
2.4. Analyses of tumour molecular characteristics
Genomic DNA was extracted from FFPE colorectal carcinoma tissue blocks. MSI status was determined by polymerase chain reaction (PCR) of 10 microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67, and D18S487); MSI-high was defined as the presence of instability in ≥ 30% of the markers, as previously described [11, 20]. Methylation status of eight CIMP-specific promoters (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1) [21] was determined by MethyLight assay using bisulphite-treated DNA, as previously described [22]. CIMP-high was defined as ≥ 6 methylated promoters of eight promoters, and CIMP-low/negative as 0–5 methylated promoters, as previously described [11, 23]. LINE-1 methylation levels were measured by pyrosequencing using bisulphite-treated DNA, as previously described [24,11]. PCR and pyrosequencing were performed for KRAS (codons 12, 13, 61 and 146), BRAF (codon 600), and PIK3CA (exons 9 and 20) 25, 26, 27.
2.5. Statistical analyses
All statistical analyses were performed using SAS software (version 9.4, SAS Institute, Cary, NC, USA). All P values were two-sided. We used the stringent two-sided α level of 0.005 to decrease false positive findings and improve the reproducibility [28]. Our primary hypothesis testing was the assessment of the association of intraepithelial and stromal T-cell densities (four ordinal categories, C1-C4) with extent of tumour budding and PDCs (tertile categories) in the multivariable ordinal logistic analyses. All the other hypotheses were tested as secondary analyses. We used the Chi-squared test to assess the associations of clinicopathological characteristics with tumour budding, as well as PDC grade. Moreover, we used Spearman's correlation test to assess the relationships of the raw densities of T-cell subsets with the number of tumour buds, as well as PDCs. In the survival analyses, cumulative survival probabilities were estimated with the Kaplan-Meier method the differences between categories were compared using the log-rank test for trend. Survival time was defined as the period from diagnosis of colorectal cancer to death or the end of follow-up, whichever came first. For the analyses of colorectal cancer-specific mortality, deaths due to other causes were censored. Univariable and multivariable Cox proportional hazard regression analyses were conducted to assess the mortality hazard ratios (HRs). To reduce the potential bias due to the availability of tumour tissue samples, we used the inverse probability weighting (IPW) method for logistic regression, Cox regression, and Kaplan-Meier analyses utilising covariate data of 4420 incident colon and rectal carcinoma cases in the two prospective cohort studies [12, 13, 14,29]. Detailed description of the statistical methods is included in Supplementary methods.
3. Results
We included 915 colorectal cancer patients with available data on tumour budding grade and T-cell densities among 4420 incident colorectal cancer cases in the two prospective cohort studies. The tumour budding grade at the invasive front was low, intermediate, and high in 538 (59%), 204 (22%), and 173 (19%) cases, respectively. Table 1 summarises clinical, pathological, and molecular characteristics according to tumour budding at the invasive front. Tumour budding grade was positively associated with depth of tumour invasion (pT stage) (P < 0.001), the number of positive lymph nodes (pN stage) (P < 0.001), AJCC disease stage (P < 0.001), and poor tumour differentiation (P < 0.001) and inversely associated with tumour-infiltrating lymphocytes (P < 0.001), intratumoural periglandular lymphocytic reaction (P= 0.001), and peritumoural lymphocytic reaction (P < 0.001). PDC grade at invasive front was positively associated with depth of tumour invasion (pT stage) (P < 0.001), the number of positive lymph nodes (pN stage) (P < 0.001), AJCC disease stage (P < 0.001), poor tumour differentiation (P < 0.001), and BRAF mutation (P= 0.002) (Supplementary table S1). Tumour budding and PDCs at invasive front were positively correlated with each other [Spearman correlation coefficient (r) = 0.73, P < 0.001]. There was also a strong correlation between tumour budding at the invasive front and tumour centre (r= 0.86, P < 0.001), as well as PDCs at the invasive front and tumour centre (r= 0.81, P < 0.001). Considering the well-established ITBCC criteria for the evaluation of tumour budding and the strong correlations of the number of tumour buds between at tumour invasive front and at tumour centre, the following analyses were conducted with only tumour budding at the invasive front.
Table 1.
Clinical, pathological, and molecular characteristics of colorectal cancer cases according to tumour budding at invasive front .
| Tumour budding at invasive front |
|||||
|---|---|---|---|---|---|
| Total No. | Low | Intermediate | High | ||
| Characteristicsa | (n= 915) | (n= 538) | (n= 204) | (n= 173) | P valueb |
| Sex | 0.40 | ||||
| Female (NHS) | 503 (55%) | 297 (55%) | 105 (51%) | 101 (58%) | |
| Male (HPFS) | 412 (45%) | 241 (45%) | 99 (49%) | 72 (42%) | |
| Mean age ± SD (years) | 69.1 ± 8.8 | 69.4 ± 8.8 | 69.2 ± 8.9 | 68.1 ± 8.6 | 0.23 |
| Year of diagnosis | 0.74 | ||||
| 1995 or before | 290 (32%) | 168 (31%) | 64 (31%) | 58 (34%) | |
| 1996–2000 | 303 (33%) | 174 (32%) | 67 (33%) | 62 (36%) | |
| 2001–2014 | 322 (35%) | 196 (36%) | 73 (36%) | 53 (31%) | |
| Family history of colorectal cancer in a first-degree relative | 0.76 | ||||
| Absent | 715 (79%) | 425 (79%) | 161 (80%) | 129 (77%) | |
| Present | 192 (21%) | 112 (21%) | 41 (20%) | 39 (23%) | |
| Tumour location | 0.12 | ||||
| cecum | 165 (18%) | 106 (20%) | 34 (17%) | 25 (15%) | |
| Ascending to transverse colon | 299 (33%) | 168 (31%) | 64 (31%) | 67 (39%) | |
| Descending to sigmoid colon | 268 (29%) | 149 (28%) | 73 (36%) | 46 (27%) | |
| Rectum | 179 (20%) | 112 (21%) | 33 (16%) | 34 (20%) | |
| pT stage (depth of tumour invasion) | < 0.001 | ||||
| pT1 (submucosa) | 66 (7.8%) | 60 (12%) | 3 (1.6%) | 3 (2.0%) | |
| pT2 (muscularis propria) | 176 (21%) | 125 (25%) | 32 (17%) | 19 (12%) | |
| pT3 (subserosa) | 560 (66%) | 303 (60%) | 141 (76%) | 116 (75%) | |
| pT4 (serosa or other organs) | 46 (5.4%) | 20 (3.9%) | 10 (5.4%) | 16 (10%) | |
| pN stage (number of positive lymph nodes) |
< 0.001 | ||||
| pN0 (0) | 504 (61%) | 354 (71%) | 98 (54%) | 52 (36%) | |
| pN1 (1–3) | 202 (25%) | 97 (20%) | 58 (32%) | 47 (33%) | |
| pN2 ( ≥ 4) | 115 (14%) | 46 (9.3%) | 24 (13%) | 45 (31%) | |
| AJCC disease stage | < 0.001 | ||||
| I | 193 (23%) | 152(30%) | 27 (14%) | 14 (8.8%) | |
| II | 281 (33%) | 187 (37%) | 60 (31%) | 34 (21%) | |
| III | 251 (29%) | 121(24%) | 64 (34%) | 66 (42%) | |
| IV | 126 (15%) | 41 (8.2%) | 40 (21%) | 45 (28%) | |
| Tumour differentiation | < 0.001 | ||||
| Well to moderate | 830 (91%) | 503 (94%) | 186 (92%) | 141 (82%) | |
| Poor | 84 (9.2%) | 34 (6.3%) | 18 (8.8%) | 32 (19%) | |
| MSI status | 0.046 | ||||
| Non-MSI-high | 736 (83%) | 421 (81%) | 165 (83%) | 150 (89%) | |
| MSI-high | 154 (17%) | 102 (20%) | 33 (17%) | 19 (11%) | |
| CIMP status | 0.71 | ||||
| Low/negative | 693 (81%) | 407 (80%) | 151 (83%) | 135 (83%) | |
| High | 159 (19%) | 99 (20%) | 32 (17%) | 28 (17%) | |
| Mean LINE-1 methylation | 62.5 ± 9.6 | 63.0 ± 9.9 | 62.1 ± 9.0 | 61.6 ± 9.5 | 0.20 |
| level ± SD (%) | |||||
| KRAS mutation | 0.41 | ||||
| Wild-type | 524 (59%) | 313 (60%) | 110 (55%) | 101 (59%) | |
| Mutant | 364 (41%) | 205 (40%) | 90 (45%) | 69 (41%) | |
| BRAF mutation | 0.50 | ||||
| Wild-type | 759 (85%) | 451 (86%) | 168 (85%) | 140 (82%) | |
| Mutant | 137 (15%) | 76 (14%) | 30 (15%) | 31 (18%) | |
| PIK3CA mutation | 0.035 | ||||
| Wild-type | 699 (84%) | 407 (83%) | 146 (79%) | 146 (90%) | |
| Mutant | 137 (16%) | 82 (17%) | 38 (21%) | 17 (10%) | |
| Membrane CDH1 (E-cadherin) expression | 0.97 | ||||
| Intact | 272 (47%) | 159 (47%) | 60 (48%) | 53 (46%) | |
| Lost | 306 (53%) | 180 (53%) | 65 (52%) | 61 (54%) | |
| Nuclear CTNNB1 (β-catenin) expression | 0.17 | ||||
| Negative | 473 (54%) | 292 (56%) | 96 (48%) | 85 (53%) | |
| Positive | 407 (46%) | 228 (44%) | 102 (52%) | 77 (48%) | |
| Tumour-infiltrating lymphocytes | < 0.001 | ||||
| Negative/low | 651 (72%) | 352 (66%) | 163 (80%) | 136 (80%) | |
| Intermediate | 151 (17%) | 103 (19%) | 29 (14%) | 19 (11%) | |
| High | 102 (11%) | 76 (14%) | 11 (5.4%) | 15 (8.8%) | |
| Intratumoural periglandular reaction | 0.002 | ||||
| Negative/low | 126 (14%) | 59 (11%) | 33 (16%) | 34 (20%) | |
| Intermediate | 670 (74%) | 393 (74%) | 152 (75%) | 125 (73%) | |
| High | 109 (12%) | 79 (15%) | 18 (8.9%) | 12 (7.0%) | |
| Peritumoural lymphocytic reaction | 0.001 | ||||
| Negative/low | 145 (16%) | 69 (13%) | 37 (18%) | 39 (23%) | |
| Intermediate | 620 (69%) | 362 (68%) | 141 (69%) | 117 (68%) | |
| High | 138 (15%) | 98 (19%) | 25 (12%) | 15 (8.8%) | |
| Crohn's-like lymphoid reaction | 0.28 | ||||
| Negative/low | 577 (74%) | 314 (71%) | 145 (78%) | 118 (79%) | |
| Intermediate | 139 (18%) | 87 (20%) | 30 (16%) | 22 (15%) | |
| High | 60 (7.7%) | 39 (8.9%) | 11 (5.9%) | 10 (6.7%) | |
Percentage indicates the proportion of patients with a specific clinical, pathological, or molecular characteristic among all patients or in the strata of tumour budding at invasive front.
To compare categorical data between the tumour budding grade, the chi-square test was performed. To compare continuous variables, an analysis of variance was performed.
Abbreviations: AJCC, American Joint Committee on Cancer; CIMP, CpG island methylator phenotype; HPFS, Health Professionals Follow-up Study; LINE-1, long-interspersed nucleotide element-1; MSI, microsatellite instability; NHS, Nurses’ Health Study; SD, standard deviation.
Table 2 shows the correlation between intraepithelial and stromal T-cell densities and tumour budding at the invasive front. Intraepithelial CD3+, CD3+CD8+, and CD3+CD8+CD45RO+ T-cell densities were inversely correlated with the density of tumour buds at the invasive front (P≤ 0.003); the strongest negative correlation (r = −0.16, P < 0.001) was observed with CD3+CD8+CD45RO+ cells, the most specifically defined T-cell subset of the three, representing cytotoxic memory T-cell phenotype. A similar correlation of T-cell densities with tumour budding at tumour centre was observed. Density of PDCs at the invasive front or tumour centre did not show statistically significant correlation with the densities of any T-cell subset (Supplementary table S2).
Table 2.
Correlation between intraepithelial and stromal T-cell densities and tumour budding at invasive front.
| Tumour buddinga | |
|---|---|
| Intraepithelial region | |
| CD3+ cells | r = −0.10, P= 0.003 |
| CD3+CD4+cells | N.S. |
| CD3+CD8+cells | r = −0.14, P < 0.001 |
| CD3+CD4+FOXP3+ cells | N.S. |
| CD3+CD4+CD45RO+ cells | N.S. |
| CD3+CD4+CD45RO− cells | N.S. |
| CD3+CD8+CD45RO+ cells | r = −0.16, P < 0.001 |
| CD3+CD8+CD45RO− cells | N.S. |
| Stromal region | |
| CD3+ cells | N.S. |
| CD3+CD4+cells | N.S. |
| CD3+CD8+cells | N.S. |
| CD3+CD4+FOXP3+ cells | N.S. |
| CD3+CD4+CD45RO+ cells | N.S. |
| CD3+CD4+CD45RO− cells | N.S. |
| CD3+CD8+CD45RO+ cells | N.S. |
| CD3+CD8+CD45RO− cells | N.S. |
Correlation coefficient and P value were calculated by the Spearman's correlation test between T-cell densities (cells/mm2; as continuous variables) and tumour budding at invasive front (number of tumour buds/0.785 mm2, as continuous variable).
Abbreviation: N.S., not significant.
In our primary hypothesis testing, we used a logistic regression analysis to assess the association of intraepithelial T-cell densities with the extent of tumour budding (Table 3 and Supplementary table S3). Sensitivity analyses of the association of binary categories of intraepithelial T-cell densities with the extent of tumour budding were shown in Supplementary table S4. In multivariable analysis, intraepithelial CD3+CD8+ T-cell density [lowest vs. highest density category: multivariable odds ratio (OR) 0.50, 95% confidence interval (CI), 0.35–0.70; Ptrend < 0.001] and intraepithelial CD3+CD8+CD45RO+ cell density (lowest vs. highest density category: multivariable OR 0.44, 95% CI, 0.31–0.63; Ptrend < 0.001) were inversely associated with the extent of tumour budding. Similar results were obtained by multivariable logistic regression analysis without the IPW (Supplementary table S5). As MSI status and T-cell densities in tumour tissue were strongly correlated (Supplementary table S6), we conducted the analysis to see the association of T-cell densities with tumour budding in strata of MSI status. There was no significant interaction between MSI status and intraepithelial CD3+CD8+ or CD3+CD8+CD45RO+ density in relation to tumour budding (P > 0.24) (Supplementary table S7).
Table 3.
Inverse probability weighting (IPW)-adjusted logistic regression analysis to assess the associations of intraepithelial T-cell densities (predictor) with tumour budding at invasive front (outcome) .
| Univariable OR (95% CI)b,c | Multivariable OR (95% CI)b,c | |
|---|---|---|
| Intraepithelial CD3+ cell density (cells/mm2) | ||
| C1 (lowest) | 1 (referent) | 1 (referent) |
| C2 (second) | 0.75 (0.52–1.09) | 0.74 (0.51–1.08) |
| C3 (third) | 0.62 (0.43–0.89) | 0.61 (0.43–0.88) |
| C4 (highest) | 0.64 (0.44–0.93) | 0.60 (0.41–0.88) |
| Ptrendd | 0.010 | 0.005 |
| Intraepithelial CD3+CD8+ cell density (cells/mm2)a | ||
| C1 (zero) | 1 (referent) | 1 (referent) |
| C2 (low) | 0.92 (0.64–1.32) | 0.92 (0.64–1.32) |
| C3 (intermediate) | 0.57 (0.40–0.82) | 0.57 (0.40–0.81) |
| C4 (high) | 0.52 (0.37–0.73) | 0.50 (0.35–0.70) |
| Ptrendd | < 0.001 | < 0.001 |
| Intraepithelial CD3+CD8+CD45RO+ cell density (cells/mm2)a | ||
| C1 (zero) | 1 (referent) | 1 (referent) |
| C2 (low) | 0.73 (0.51–1.06) | 0.73 (0.50–1.05) |
| C3 (intermediate) | 0.58 (0.41–0.82) | 0.57 (0.40–0.81) |
| C4 (high) | 0.47 (0.33–0.67) | 0.44 (0.31–0.63) |
| Ptrendd | < 0.001 | < 0.001 |
As intraepithelial CD3+CD8+ (377 cases) and CD3+CD8+CD45RO+ (424 cases) cell densities were 0/mm2 (consisting the largest category C1), the remaining cases were divided into tertiles according to density (C2-C4).
IPW was applied to reduce a bias due to the availability of tumour tissue after cancer diagnosis (see “Statistical analysis” subsection for details).
The multivariable ordinal logistic regression model initially included age, sex, year of diagnosis, family history of colorectal cancer, tumour location, microsatellite instability, CpG island methylator phenotype, long-interspersed nucleotide element-1 methylation level, KRAS, BRAF, and PIK3CA mutations, CDH1 expression, and CTNNB1 expression. A backward elimination with a threshold P of 0.05 was used to select variables for the final model.
Ptrend was calculated by the linear trend across the ordinal categories of the intraepithelial T-cell densities (four ordinal predictor variables: C1-C4) in the IPW-adjusted ordinal logistic regression model for the number of tumour buds/0.785 mm2 at invasive front (tertile variables).
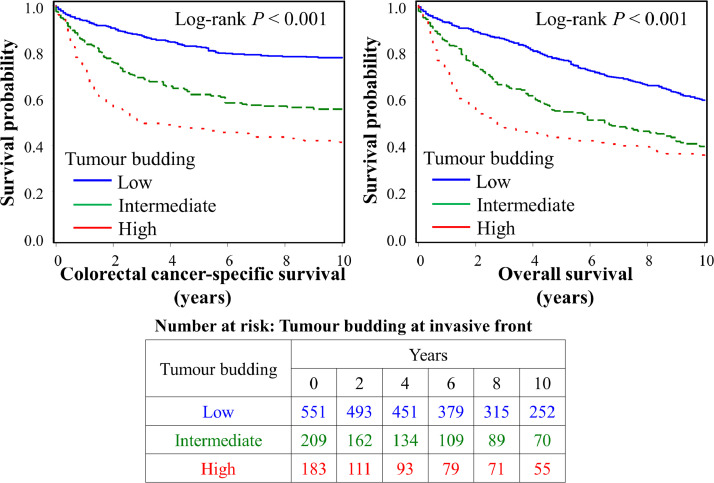
In the survival analyses, using a dataset of 943 cases with available survival data, we examined the prognostic impact of the tumour budding and PDC grade, and their survival interactions with T-cell densities. During the median follow-up time of 12.3 years (interquartile range, 8.7 to 16.3 years), 577 all-causes deaths, including 296 colorectal cancer-specific deaths, were observed. Kaplan-Meier analysis shows that both high tumour budding and PDC grade at the invasive front were associated with higher colorectal cancer specific and overall mortality (Log-rank P < 0.001) (Fig. 2 and Supplementary fig. S2). Multivariable Cox regression models (Table 4 and Supplementary table S8) indicated that tumour budding showed an adverse prognostic association independent of tumour molecular features, patient characteristics, as well as lymphocytic reaction patterns and intraepithelial CD3+CD8+CD45RO+ T-cell density, whereas PDC grade did not remain in the final multivariable Cox regression model (Supplementary table S8). Compared to patients with low tumour budding grade, those with high tumour budding grade were associated with shorter colorectal cancer-specific survival (multivariable HR 2.13 95% CI, 1.57–2.89; Ptrend < 0.001). Considering the strong association of CD3+CD8+CD45RO+ T-cell density with favourable prognosis (Supplementary fig. S3), we examined the prognostic association of tumour budding at invasive front in strata of intraepithelial CD3+CD8+ and CD3+CD8+CD45RO+ T-cell densities. High-grade tumour budding was associated with poor prognosis in patients with both low and high densities of those T-cell subsets (Pinteraction > 0.56) (Supplementary table S9). In a multivariable model without tumour budding as a covariate, high PDC grade was associated with shorter colorectal cancer-specific survival (multivariable HR 1.97; 95% CI, 1.36–2.84; Ptrend < 0.001) (Supplementary table S10). Considering the strong associations of both tumour budding and T-cell densities with patient survival, we conducted the survival analyses of the composite variables of tumour budding and intratumoural CD3+CD8+CD45RO+ T-cell density. Tumours with low-grade tumour budding and high intratumoural CD3+CD8+CD45RO+ T-cell densities were associated with favourable colorectal cancer-specific survival compared to tumours with high-grade tumour budding and low intratumoural CD3+CD8+CD45RO+ T-cell densities (Supplementary fig. S4).
Fig. 2.
Inverse probability weighting (IPW)-adjusted Kaplan-Meier curves of colorectal cancer-specific and overall survival according to tumour budding at invasive front.
The P values were calculated using the weighted log-rank test for trend (two-sided). The table (bottom) shows the number of patients who remained alive and at risk of death at each time point after the diagnosis of colorectal cancer. .
Table 4.
Tumour budding at invasive front and patient survival with inverse probability weighting.
| Colorectal cancer-specific survivala |
Overall survivala |
||||||
|---|---|---|---|---|---|---|---|
| No. ofcases | No. ofevents | UnivariableHR (95% CI) | MultivariableHR (95% CI)b | No. ofevents | UnivariableHR (95% CI) | MultivariableHR (95% CI)b | |
| Tumour budding (n= 943) | |||||||
| Low | 551 | 109 | 1 (referent) | 1 (referent) | 212 | 1 (referent) | 1 (referent) |
| Intermediate | 209 | 77 | 2.33 (1.72–3.15) | 1.75 (1.30–2.37) | 115 | 1.86 (1.47–2.36) | 1.50 (1.15–1.94) |
| High | 183 | 97 | 3.71 (2.78–4.97) | 2.13 (1.57–2.89) | 114 | 2.44 (1.88–3.16) | 1.81 (1.38–2.38) |
| Ptrendc | < 0.001 | < 0.001 | < 0.001 | < 0.001 | |||
IPW was applied to reduce a bias due to the availability of tumour tissue after cancer diagnosis (see “Statistical analysis” subsection for details).
The multivariable Cox regression model initially included age, sex, year of diagnosis, family history of colorectal cancer, tumour location, tumour grade, AJCC disease stage, microsatellite instability, CpG island methylator phenotype, long-interspersed nucleotide element-1 methylation level, KRAS, BRAF, and PIK3CA mutations, CDH1 expression, CTNNB1 expression, tumour-infiltrating lymphocytes, intratumoural periglandular reaction, peritumoural lymphocytic reaction, Crohn's-like lymphoid reaction, intraepithelial CD3+CD8+CD45RO+T-cell densities, and poorly differentiated clusters. A backward elimination with a threshold P of 0.05 was used to select variables for the final models.
Ptrend was calculated by the linear trend across the ordinal categories of tumour budding grade at invasive front (low, intermediate, and high) in the IPW-adjusted Cox regression model.
Abbreviations: CI, confidence interval; HR, hazard ratio; IPW, inverse probability weighting.
4. Discussion
We conducted this study to test the hypothesis that specific subset of T-cell densities in tumour might be inversely associated with tumour budding in colorectal cancer. Utilising a molecular pathologic epidemiology database of 915 colorectal carcinomas among 4420 incident colorectal cancer cases within two U.S. nationwide prospective cohort studies, we found that the densities of intraepithelial CD3+CD8+ and CD3+CD8+CD45RO+ T cells were inversely associated with the extent of tumour budding at invasive front. Our findings suggest that anti-tumour immunity based on cytotoxic T cells may suppress microinvasion. High-grade tumour budding was associated with shorter colorectal cancer-specific survival independent of tumour molecular features, disease stage, PDC grade, lymphocytic reaction, and cytotoxic T cell density, supporting the value of tumour budding as an independent prognostic factor in colorectal cancer.
CD8+ T cells, known as cytotoxic T lymphocytes, play an integral part of the adaptive immune response against pathogens, as well as tumour cells. Peri and intratumoural CD8+ T cells have previously been identified as a major favourable prognostic factor in colorectal cancer [30]. CD8+CD45RO+ memory T cells, which is a subpopulation of cytotoxic T cells, may respond quickly to a previously encountered antigen stimulus [31], and our previous study indicated that intratumoural CD45RO+ cells had prognostic value independent of other (CD3+, CD8+, and FOXP3+) T-cell subsets [18]. Cytotoxic T cells can perform antigen-specific lysis of tumour cells, leading to tumour cell apoptosis, by exocytosis of granules containing PRF1 (perforin 1), GZMA (granzyme A), and GZMB (granzyme B) [32]. Moreover, cytotoxic T-cell surface protein FASLG (Fas cell surface death receptor ligand) may induce cell death by binding FAS (Fas cell surface death receptor) molecules expressed on the tumour cells [33]. Accordingly, high density of T cells in tumour were correlated with low frequency of invasion and metastasis in colorectal cancer [30]. In line with these clinical and experimental studies, our findings suggest that high densities of intraepithelial CD3+CD8+ and CD3+CD8+CD45RO+ cytotoxic T cells are associated with low tumour budding grade, potentially reflecting the suppression of tumour progression by cytotoxic anti-tumour immunity.
Tumour budding is one of the initial steps of cancer invasion and metastasis, as budding cells migrate through the extracellular matrix, invade lymphovascular structures, and form metastatic tumour colonies in lymph nodes and distant organs 34, 35, 36. During this process called epithelial mesenchymal transition (EMT), activation of WNT/CTNNB1 signalling occurs in tumour cells, which lose expression of epithelial markers such as CDH1 expression and instead express genes more commonly associated with mesenchymal cells, such as CDH2 (cadherin 2, N-cadherin) and VIM (vimentin) [2,37,38]. Emerging evidence links EMT with increased stemness, therapeutic resistance, and escape from immune surveillance [37,39]. In non-small cell lung cancer, EMT was associated with increased expression of multiple immunosuppressive cytokines including IL10 (interleukin 10) and TGFB1 (transforming growth factor beta 1) [38]. Tumour cells that underwent EMT had reduced susceptibility to cytotoxic T cell mediated lysis with PRF1 and GZMB proteins [40,41]. A consensus molecular subtype (CMS) classification classifies colorectal cancer into four main groups [42]: the CMS4 subtype is characterised by stromal infiltration, matrix remodelling, and overexpression of EMT-related and TGFB pathway genes [42], as well as low immune activation [38, 39]. Our finding of the inverse association between tumour budding and intraepithelial CD3+CD8+ and CD3+CD8+CD45RO+ cytotoxic T-cell densities suggest that cytotoxic anti-tumour immunity can suppress tumour budding; however, an alternate explanation is that tumour budding cells with the EMT phenotype do not elicit a T-cell response, and thus they can evade detection and are more likely to lead to a poor prognosis.
PDCs and tumour buds form a continuum of histological features with an arbitrarily set cut-point of 5 tumour cells to differentiate the two [2, 3]. Therefore, PDCs and tumour budding share many tumour molecular features, as well as clinical and pathological features including mortality of colorectal cancer patients [3]. However, we found that PDC grade, but not budding grade, was significantly associated with BRAF mutation, while tumour budding, but not PDC grade, had a tendency towards an inverse association with MSI-high phenotype. This is in line with some previous studies that tumour budding was inversely associated with MSI-high status [43,44], and BRAF mutations were more frequent among tumours with high PDC grade [45]. Emerging studies show that PDCs encompass heterogeneous morphologies such as micropapillary and medullary-like components, mucinous differentiation, and solid growth [43,46, 47, 48]. Poor differentiation has frequently been associated with MSI-high status and high densities of tumour-infiltrating lymphocytes and T cells, contributing to the favourable prognosis [18, 49]. In our study, while tumour budding was inversely associated with the densities of intraepithelial CD3+CD8+ and CD3+CD8+CD45RO+ cytotoxic T cells, no significant associations were observed between T-cell densities and PDCs. Additionally, PDC grade showed less prognostic power than tumour budding. We suggest that, although the two phenomena are correlated, the morphological diversity of PDCs compared to tumour budding might account for these divergent results.
The TNM classification system remains the gold standard for stratification of colorectal cancer patients into prognostic subgroups, while tumour budding is thought to be an independent poor prognostic factor [50]. Recently, tumour budding has been included as an additional prognostic factor to the eighth edition of the AJCC Cancer Staging Manual. Similar to tumour budding, PDCs are associated with high tumour grade, high lymph node and distant metastasis, and higher patient mortality in colorectal cancer [3]. Our study is one of the largest so far to simultaneously evaluate the prognostic value of tumour budding, using the ITBCC criteria, and PDCs in colorectal cancer. Notably, our findings show that tumour budding grade had stronger prognostic power than PDC grade, which was also independent of tumour molecular features, lymphocytic reaction scores and cytotoxic T-cell density. Taken together, our findings support the value of tumour budding as an independent prognostic factor in colorectal cancer. Although intratumour heterogeneity should be considered, counting tumour budding in the colorectal cancer biopsy specimens may also be useful for the preoperative prediction of patient mortality, considering the previous promising reports 51, 52, 53, as well as high correlation we observed between intratumoural budding and budding at the advancing edge.
There is increasing evidence supporting the prognostic significance of tumour infiltrating immune cells in colorectal cancer. The prognostic value of the Immunoscore, based on counting the densities of CD3+ and CD8+ cells in the invasive margin and tumour centre, has been validated in a recent multicentre study [30, 54]. It is of note that the previous studies using the Immunoscore [54, 55] did not evaluate the value of examining tumour edges and tumour centre, compared to examining a comparable number of separate areas in the tumour centre. Our results indicate that microscopic tumour invasive edges (including tumour budding) are present in tumour centre areas to a similar degree to gross tumour invasive edges. Therefore, our study can underscore the value of examining immune cells in tumour centre areas containing microscopic tumour invasive edges that are ubiquitous across tumour areas (regardless of gross tumour edges or tumour centre areas). A few recent studies have shown the combined measurements of tumour-infiltrating lymphocytes and tumour budding may have higher prognostic power than either measurement alone [56, 57]. Our results are in accordance with these reports, as lymphocytic infiltrates and tumour budding had prognostic value independent of each other. Taken together, these results support the significance of simultaneous measurements of tumour budding and T-cell infiltration as prognostic biomarkers, potentially providing more comprehensive estimate of the tumour-host interactions than either parameter alone.
Our study has several limitations. First, we assessed T-cell densities using TMAs, which only allow for the examination of a small tumour region. Due to intratumour heterogeneity, use of TMAs also led to potential misclassification of T-cell densities; because this heterogeneity is expected to have a nearly random distribution, it would tend to drive our results towards the null hypothesis. Additionally, the TMAs were constructed from two to up to four tumour tissue cores including tumour centre and tumour margin from each tumour [18]. In a separate, ongoing study, we have determined that examination of at least two TMA cores can provide reasonably accurate immune cell density measurements when compared to more extensive sampling (unpublished data). These suggest that T-cell densities based on the TMAs can represent the overall T-cell densities of each tumour, supporting our finding that inverse association of cytotoxic T cells and tumour budding in the tumour microenvironment. Second, despite using the ITBCC criteria, the evaluation of tumour budding and PDCs was still subjective. The data used in this study was based on visual evaluation by one pathologist. we evaluated the interobserver agreement between two pathologists in a subset of 140 cases. The agreement between two pathologists was relatively high for tumour budding (Spearman's correlation coefficient 0.79; weighted kappa 0.66) but lower for PDCs (Spearman's correlation coefficient 0.61; weighted kappa 0.33), which may favour the reproducibility of tumour budding evaluation over PDC evaluation. Nonetheless, the agreement we observed for PDCs is still in line with a multicentre study for PDC grading, where the median weighted Kappa value amongst 12 institutions was 0.52 (range, 0.23 to 0.77) [46]. Finally, the data on cancer recurrence were not available in this study. However, given that median time to colorectal cancer recurrence is approximately 10 to 20 months [58], colorectal cancer-specific survival is expected to be an accurate surrogate of clinical outcome in a population-based study with long-term follow-up. Lastly, data on cancer treatment were limited. However, high-grade tumour budding was associated with poor survival even considering that the patient with high-grade tumour budding was potentially treated aggressively.
The strength of the study included the availability of data pertaining to many potential confounding factors in this rich molecular pathological epidemiology [59], 60, [61], 62 dataset, which were adjusted for in the logistic regression model and Cox regression models. In addition to clarifying the relationships between tumour budding and cytotoxic T-cell response, our study represents one of the most extensive validations of the prognostic significance of ITBCC grading of tumour budding, as well as an investigation of the molecular correlates of tumour budding and PDCs. The study population was recruited from a large number of hospitals throughout the U.S., which facilitates the generalisability of our results. Moreover, multiplex immunofluorescence enabled simultaneous examination of multiple T-cell markers and identification of specific T-cell subsets not possible with single marker approaches. Indeed, our analysis indicated that a specific type of T cell, CD3+CD8+CD45RO+ cytotoxic memory T cell, defined by a combination of three markers, had the highest inverse association with tumour budding. Importantly, single marker CD45RO immunohistochemistry is not adequate for accurate identification of this population, as CD45RO may also be expressed by other immune cell types including macrophages and B cells [63].
In conclusion, we have shown an inverse association of intraepithelial CD3+CD8+ and CD3+CD8+CD45RO+ cytotoxic T cells with the extent of tumour budding, suggesting a role for cytotoxic anti-tumour immunity in suppressing microinvasion. Our study, based on two large U.S. nationwide prospective cohorts, also represents an extensive validation of the prognostic value of ITBCC evaluation of tumour budding in colorectal cancer, independent of tumour molecular features and immune cell densities.
Acknowledgments
Acknowledgments
We would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
Funding sources
This work was supported by U.S. National Institutes of Health (NIH) grants (P01 CA87969 to M.J. Stampfer; UM1 CA186107 to M.J. Stampfer; P01 CA55075 to W.C. Willett; UM1 CA167552 to W.C. Willett; U01 CA167552 to W.C. Willett and L.A. Mucci; P50 CA127003 to C.S.F.; R01 CA118553 to C.S.F.; R01 CA169141 to C.S.F.; R01 CA137178 to A.T.C.; K24 DK098311 to A.T.C.; R35 CA197735 to S.O.; R01 CA151993 to S.O.; R01 CA248857 to S.O.; K07 CA188126 to X.Z., and R01 CA225655 to J.K.L.); by Nodal Award (2016–20) from the Dana-Farber Harvard Cancer Centre (to S.O.); by Stand Up to Cancer Colorectal Cancer Dream Team Translational Research Grant (SU2C-AACR-DT22–17 to C.S.F. and M.G.), administered by the American Association for Cancer Research, a scientific partner of SU2C; and by grants from the Project P Fund, The Friends of the Dana-Farber Cancer Institute, Bennett Family Fund, and the Entertainment Industry Foundation through National Colorectal Cancer Research Alliance and SU2C. K.F. was supported by fellowship grants from the Uehara Memorial Foundation and Grant of The Clinical Research Promotion Foundation (2018). K.A. and T.U. were supported by a grant from Overseas Research Fellowship (201860083 to K.A.; 201960541 to T.U.) from Japan Society for the Promotion of Science. K.H. was supported by fellowship grants from the Uehara Memorial Foundation and the Mitsukoshi Health and Welfare Foundation. J.A.M. research is supported by the Douglas gray Woodruff Chair fund, the Guo Shu Shi Fund, Anonymous Family Fund for Innovations in Colorectal Cancer, P fund, and the George Stone Family Foundation. M.G. is supported by an ASCO Conquer Cancer Foundation Career Development Award. A.T.C. is a Stuart and Suzanne Steele MGH Research Scholar. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure of interests
J.A.M. has received institutional research funding from Boston Biomedical, has served as an advisor/consultant to Ignyta and COTA Healthcare, and served on a grant review panel for the National Comprehensive Cancer Network funded by Taiho Pharmaceutical. M.G. receives research funding from Bristol-Myers Squibb and Merck. This study was not funded by any of these commercial entities. A.T.C. previously served as a consultant for Bayer Healthcare and Pfizer Inc.. This study was not funded by Bayer Healthcare or Pfizer Inc.. C.S.F. previously served as a consultant for Agios, Bain Capital, Bayer, Celgene, Dicerna, Five Prime Therapeutics, Gilead Sciences, Eli Lilly, Entrinsic Health, Genentech, KEW, Merck, Merrimack Pharmaceuticals, Pfizer Inc, Sanofi, Taiho, and Unum Therapeutics; C.S.F. also serves as a Director for CytomX Therapeutics and owns unexercised stock options for CytomX and Entrinsic Health. No other conflicts of interest exist. The other authors declare that they have no conflicts of interest.
Use of standardised official symbols
We use HUGO (Human Genome Organisation)-approved official symbols for genes and gene products, including BRAF, CACNA1G, CD3, CD4, CD8, CDH1, CDH2, CDKN2A, CRABP1, CTNNB1, FAS, FASLG, FOXP3, GZMA, GZMB, IGF2, IL10, KRAS, MLH1, NEUROG1, PIK3CA, PRF1, PTPRC, RUNX3, SOCS1, TGFB1, VIM, and WNT; all of which are described at www.genenames.org. The official gene symbols are italicised, to differentiate from non-italicised gene product names and non-official names.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102860.
Appendix. Supplementary materials
References
- 1.Zlobec I., Lugli A. Tumour budding in colorectal cancer: molecular rationale for clinical translation. Nat Rev Cancer. 2018;18(4):203–204. doi: 10.1038/nrc.2018.1. [DOI] [PubMed] [Google Scholar]
- 2.Lugli A., Kirsch R., Ajioka Y., Bosman F., Cathomas G., Dawson H. Recommendations for reporting tumor budding in colorectal cancer based on the International Tumor Budding Consensus Conference (ITBCC) 2016. Mod Pathol. 2017;30(9):1299–1311. doi: 10.1038/modpathol.2017.46. [DOI] [PubMed] [Google Scholar]
- 3.Ueno H., Kajiwara Y., Shimazaki H., Shinto E., Hashiguchi Y., Nakanishi K. New criteria for histologic grading of colorectal cancer. Am J Surg Pathol. 2012;36(2):193–201. doi: 10.1097/PAS.0b013e318235edee. [DOI] [PubMed] [Google Scholar]
- 4.Binnewies M., Roberts E.W., Kersten K., Chan V., Fearon D.F., Merad M. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24(5):541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yarchoan M., Johnson B.A., 3rd, Lutz E.R., Laheru D.A., Jaffee E.M. Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer. 2017;17(4):209–222. doi: 10.1038/nrc.2016.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lugli A., Karamitopoulou E., Panayiotides I., Karakitsos P., Rallis G., Peros G. CD8+ lymphocytes/ tumour-budding index: an independent prognostic factor representing a 'pro-/anti-tumour' approach to tumour host interaction in colorectal cancer. Br J Cancer. 2009;101(8):1382–1392. doi: 10.1038/sj.bjc.6605318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allen W.L., Dunne P.D., McDade S., Scanlon E., Loughrey M., Coleman H. Transcriptional subtyping and CD8 immunohistochemistry identifies poor prognosis stage II/III colorectal cancer patients who benefit from adjuvant chemotherapy. JCO Precis Oncol. 2018;2018:1–15. doi: 10.1200/PO.17.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawson H., Christe L., Eichmann M., Reinhard S., Zlobec I., Blank A. Tumour budding/T cell infiltrates in colorectal cancer: proposal of a novel combined score. Histopathology. 2020;76(4):572–580. doi: 10.1111/his.14006. [DOI] [PubMed] [Google Scholar]
- 9.van Wyk H.C., Roseweir A., Alexander P., Park J.H., Horgan P.G., McMillan D.C. The relationship between tumor budding, tumor microenvironment, and survival in patients with primary operable colorectal cancer. Ann Surg Oncol. 2019;26(13):4397–4404. doi: 10.1245/s10434-019-07931-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishihara R., Wu K., Lochhead P., Morikawa T., Liao X., Qian Z.R. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med. 2013;369(12):1095–1105. doi: 10.1056/NEJMoa1301969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamauchi M., Morikawa T., Kuchiba A., Imamura Y., Qian Z.R., Nishihara R. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61(6):847–854. doi: 10.1136/gutjnl-2011-300865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L., Nevo D., Nishihara R., Cao Y., Song M., Twombly T.S. Utility of inverse probability weighting in molecular pathological epidemiology. Eur J Epidemiol. 2018;33(4):381–392. doi: 10.1007/s10654-017-0346-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamada T., Cao Y., Qian Z.R., Masugi Y., Nowak J.A., Yang J. Aspirin Use and Colorectal Cancer Survival According to Tumor CD274 (Programmed Cell Death 1 Ligand 1) Expression Status. J Clin Oncol. 2017;35(16):1836–1844. doi: 10.1200/JCO.2016.70.7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seaman S.R., White I.R. Review of inverse probability weighting for dealing with missing data. Stat Methods Med Res. 2013;22(3):278–295. doi: 10.1177/0962280210395740. [DOI] [PubMed] [Google Scholar]
- 15.Haruki K., Kosumi K., Li P., Arima K., Vayrynen J.P., Lau M.C. An integrated analysis of lymphocytic reaction, tumour molecular characteristics and patient survival in colorectal cancer. Br J Cancer. 2020 doi: 10.1038/s41416-020-0780-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S.A., Inamura K., Yamauchi M., Nishihara R., Mima K., Sukawa Y. Loss of CDH1 (E-cadherin) expression is associated with infiltrative tumour growth and lymph node metastasis. Br J Cancer. 2016;114(2):199–206. doi: 10.1038/bjc.2015.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morikawa T., Kuchiba A., Yamauchi M., Meyerhardt J.A., Shima K., Nosho K. Association of CTNNB1 (beta-catenin) alterations, body mass index, and physical activity with survival in patients with colorectal cancer. JAMA. 2011;305(16):1685–1694. doi: 10.1001/jama.2011.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nosho K., Baba Y., Tanaka N., Shima K., Hayashi M., Meyerhardt J.A. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. J Pathol. 2010;222(4):350–366. doi: 10.1002/path.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan A.T., Ogino S., Fuchs C.S. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356(21):2131–2142. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 20.Ogino S., Nosho K., Kirkner G.J., Kawasaki T., Meyerhardt J.A., Loda M. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58(1):90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogino S., Kawasaki T., Kirkner G.J., Kraft P., Loda M., Fuchs C.S. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9(3):305–314. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogino S., Kawasaki T., Brahmandam M., Cantor M., Kirkner G.J., Spiegelman D. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8(2):209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inamura K., Yamauchi M., Nishihara R., Lochhead P., Qian Z.R., Kuchiba A. Tumor LINE-1 methylation level and microsatellite instability in relation to colorectal cancer prognosis. J Natl Cancer Inst. 2014;106(9) doi: 10.1093/jnci/dju195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irahara N., Nosho K., Baba Y., Shima K., Lindeman N.I., Hazra A. Precision of pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa, and peripheral blood cells. J Mol Diagn. 2010;12(2):177–183. doi: 10.2353/jmoldx.2010.090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogino S., Kawasaki T., Brahmandam M., Yan L., Cantor M., Namgyal C. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7(3):413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao X., Lochhead P., Nishihara R., Morikawa T., Kuchiba A., Yamauchi M. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367(17):1596–1606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imamura Y., Lochhead P., Yamauchi M., Kuchiba A., Qian Z.R., Liao X. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer. 2014;13:135. doi: 10.1186/1476-4598-13-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamin D.J., Berger J.O., Johannesson M., Nosek B.A., Wagenmakers E.J., Berk R. Redefine statistical significance. Nat Hum Behav. 2018;2(1):6–10. doi: 10.1038/s41562-017-0189-z. [DOI] [PubMed] [Google Scholar]
- 29.Xie J., Liu C. Adjusted Kaplan-Meier estimator and log-rank test with inverse probability of treatment weighting for survival data. Stat Med. 2005;24(20):3089–3110. doi: 10.1002/sim.2174. [DOI] [PubMed] [Google Scholar]
- 30.Mlecnik B., Bindea G., Angell H.K., Maby P., Angelova M., Tougeron D. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity. 2016;44(3):698–711. doi: 10.1016/j.immuni.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 31.Sathaliyawala T., Kubota M., Yudanin N., Turner D., Camp P., Thome J.J. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity. 2013;38(1):187–197. doi: 10.1016/j.immuni.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Budhu S., Loike J.D., Pandolfi A., Han S., Catalano G., Constantinescu A. CD8+ T cell concentration determines their efficiency in killing cognate antigen-expressing syngeneic mammalian cells in vitro and in mouse tissues. J Exp Med. 2010;207(1):223–235. doi: 10.1084/jem.20091279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu J., Powis de Tenbossche C.G., Cane S., Colau D., van Baren N., Lurquin C. Resistance to cancer immunotherapy mediated by apoptosis of tumor-infiltrating lymphocytes. Nat Commun. 2017;8(1):1404. doi: 10.1038/s41467-017-00784-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lugli A., Karamitopoulou E., Zlobec I. Tumour budding: a promising parameter in colorectal cancer. Br J Cancer. 2012;106(11):1713–1717. doi: 10.1038/bjc.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez D.M., Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7(344) doi: 10.1126/scisignal.2005189. re8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slik K., Blom S., Turkki R., Valimaki K., Kurki S., Mustonen H. Combined epithelial marker analysis of tumour budding in stage II colorectal cancer. J Pathol Clin Res. 2019;5(1):63–78. doi: 10.1002/cjp2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terry S., Savagner P., Ortiz-Cuaran S., Mahjoubi L., Saintigny P., Thiery J.P. New insights into the role of EMT in tumor immune escape. Mol Oncol. 2017;11(7):824–846. doi: 10.1002/1878-0261.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chae Y.K., Chang S., Ko T., Anker J., Agte S., Iams W. Epithelial-mesenchymal transition (EMT) signature is inversely associated with T-cell infiltration in non-small cell lung cancer (NSCLC) Sci Rep. 2018;8(1):2918. doi: 10.1038/s41598-018-21061-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dongre A., Weinberg R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20(2):69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 40.Akalay I., Janji B., Hasmim M., Noman M.Z., Andre F., De Cremoux P. Epithelial-to-mesenchymal transition and autophagy induction in breast carcinoma promote escape from T-cell-mediated lysis. Cancer Res. 2013;73(8):2418–2427. doi: 10.1158/0008-5472.CAN-12-2432. [DOI] [PubMed] [Google Scholar]
- 41.Hamilton D.H., Huang B., Fernando R.I., Tsang K.Y., Palena C. WEE1 inhibition alleviates resistance to immune attack of tumor cells undergoing epithelial-mesenchymal transition. Cancer Res. 2014;74(9):2510–2519. doi: 10.1158/0008-5472.CAN-13-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guinney J., Dienstmann R., Wang X., de Reynies A., Schlicker A., Soneson C. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J.W., Shin M.K., Kim B.C. Clinicopathologic impacts of poorly differentiated cluster-based grading system in colorectal carcinoma. J Korean Med Sci. 2015;30(1):16–23. doi: 10.3346/jkms.2015.30.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zlobec I., Bihl M.P., Foerster A., Rufle A., Lugli A. The impact of CpG island methylator phenotype and microsatellite instability on tumour budding in colorectal cancer. Histopathology. 2012;61(5):777–787. doi: 10.1111/j.1365-2559.2012.04273.x. [DOI] [PubMed] [Google Scholar]
- 45.Barresi V., Bonetti L.R., Bettelli S. KRAS, NRAS, BRAF mutations and high counts of poorly differentiated clusters of neoplastic cells in colorectal cancer: observational analysis of 175 cases. Pathology. 2015;47(6):551–556. doi: 10.1097/PAT.0000000000000300. [DOI] [PubMed] [Google Scholar]
- 46.Ueno H., Hase K., Hashiguchi Y., Shimazaki H., Tanaka M., Miyake O. Site-specific tumor grading system in colorectal cancer: multicenter pathologic review of the value of quantifying poorly differentiated clusters. Am J Surg Pathol. 2014;38(2):197–204. doi: 10.1097/PAS.0000000000000113. [DOI] [PubMed] [Google Scholar]
- 47.Barresi V., Branca G., Vitarelli E., Tuccari G. Micropapillary pattern and poorly differentiated clusters represent the same biological phenomenon in colorectal cancer: a proposal for a change in terminology. Am J Clin Pathol. 2014;142(3):375–383. doi: 10.1309/AJCPFEA7KA0SBBNA. [DOI] [PubMed] [Google Scholar]
- 48.Gonzalez R.S., Huh W.J., Cates J.M., Washington K., Beauchamp R.D., Coffey R.J. Micropapillary colorectal carcinoma: clinical, pathological and molecular properties, including evidence of epithelial-mesenchymal transition. Histopathology. 2017;70(2):223–231. doi: 10.1111/his.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogino S., Nosho K., Irahara N., Meyerhardt J.A., Baba Y., Shima K. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res. 2009;15(20):6412–6420. doi: 10.1158/1078-0432.CCR-09-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rogers A.C., Winter D.C., Heeney A., Gibbons D., Lugli A., Puppa G. Systematic review and meta-analysis of the impact of tumour budding in colorectal cancer. Br J Cancer. 2016;115(7):831–840. doi: 10.1038/bjc.2016.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Almangush A., Youssef O., Pirinen M., Sundstrom J., Leivo I., Makitie A.A. Does evaluation of tumour budding in diagnostic biopsies have a clinical relevance? A systematic review. Histopathology. 2019;74(4):536–544. doi: 10.1111/his.13793. [DOI] [PubMed] [Google Scholar]
- 52.Zlobec I., Hädrich M., Dawson H., Koelzer V.H., Borner M., Mallaev M. Intratumoural budding (ITB) in preoperative biopsies predicts the presence of lymph node and distant metastases in colon and rectal cancer patients. Br J Cancer. 2014;110(4):1008–1013. doi: 10.1038/bjc.2013.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rogers A.C., Gibbons D., Hanly A.M., Hyland J.M., O'Connell P.R., Winter D.C. Prognostic significance of tumor budding in rectal cancer biopsies before neoadjuvant therapy. Mod Pathol. 2014;27(1):156–162. doi: 10.1038/modpathol.2013.124. [DOI] [PubMed] [Google Scholar]
- 54.Pages F., Mlecnik B., Marliot F., Bindea G., Ou F.S., Bifulco C. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391(10135):2128–2139. doi: 10.1016/S0140-6736(18)30789-X. [DOI] [PubMed] [Google Scholar]
- 55.Marliot F., Chen X., Kirilovsky A., Sbarrato T., El Sissy C., Batista L. Analytical validation of the Immunoscore and its associated prognostic value in patients with colon cancer. J Immunother Cancer. 2020;8(1) doi: 10.1136/jitc-2019-000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lang-Schwarz C., Melcher B., Haumaier F., Lang-Schwarz K., Rupprecht T., Vieth M. Budding and tumor-infiltrating lymphocytes - combination of both parameters predicts survival in colorectal cancer and leads to new prognostic subgroups. Hum Pathol. 2018;79:160–167. doi: 10.1016/j.humpath.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 57.Lee H., Sha D., Foster N.R., Shi Q., Alberts S.R., Smyrk T.C. Analysis of tumor microenvironmental features to refine prognosis by T, N risk group in patients with stage III colon cancer (NCCTG N0147) (Alliance) Ann Oncol. 2020;31(4):487–494. doi: 10.1016/j.annonc.2020.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meyerhardt J.A., Mayer R.J. Systemic therapy for colorectal cancer. N Engl J Med. 2005;352(5):476–487. doi: 10.1056/NEJMra040958. [DOI] [PubMed] [Google Scholar]
- 59.Ogino S., Chan A.T., Fuchs C.S., Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60(3):397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ogino S., Nowak J.A., Hamada T., Milner D.A., Jr., Nishihara R. Insights into Pathogenic Interactions Among Environment, Host, and Tumor at the Crossroads of Molecular Pathology and Epidemiology. Annu Rev Pathol. 2019;14:83–103. doi: 10.1146/annurev-pathmechdis-012418-012818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hughes L.A.E., Simons C., van den Brandt P.A., van Engeland M., Weijenberg M.P. Lifestyle, Diet, and Colorectal Cancer Risk According to (Epi)genetic Instability: current Evidence and Future Directions of Molecular Pathological Epidemiology. Curr Colorectal Cancer Rep. 2017;13(6):455–469. doi: 10.1007/s11888-017-0395-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang S.T., Cui W.Q., Pan D., Jiang M., Chang B., Sang L.X. Tea polyphenols and their chemopreventive and therapeutic effects on colorectal cancer. World J Gastroenterol. 2020;26(6):562–597. doi: 10.3748/wjg.v26.i6.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jackson S.M., Harp N., Patel D., Zhang J., Willson S., Kim Y.J. CD45RO enriches for activated, highly mutated human germinal center B cells. Blood. 2007;110(12):3917–3925. doi: 10.1182/blood-2007-05-087767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.