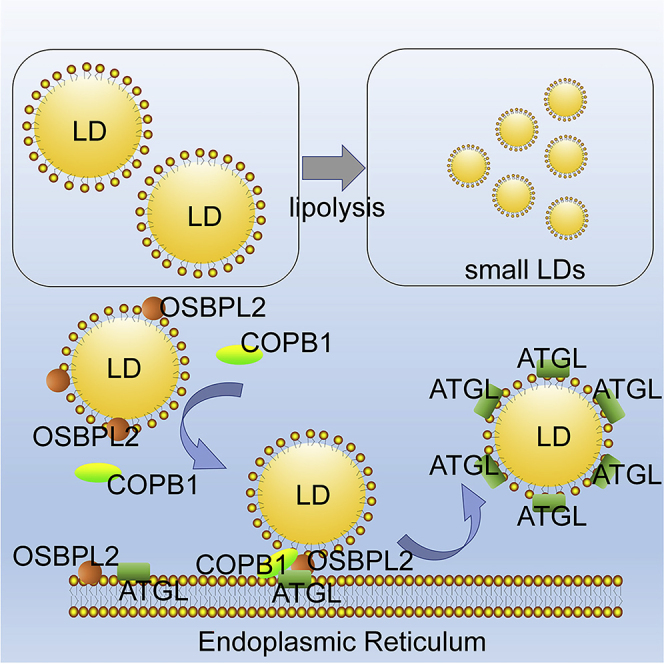
Summary
The accumulation of giant lipid droplets (LDs) increases the risk of metabolic disorders including obesity and insulin resistance. The lipolysis process involves the activation and transfer of lipase, but the molecular mechanism is not completely understood. The translocation of ATGL, a critical lipolysis lipase, from the ER to the LD surface is mediated by an energy catabolism complex. Oxysterol-binding protein-like 2 (OSBPL2/ORP2) is one of the lipid transfer proteins that regulates intracellular cholesterol homeostasis. A recent study has proven that Osbpl2−/− pigs exhibit hypercholesterolemia and obesity phenotypes with an increase in adipocytes. In this study, we identified that OSBPL2 links the endoplasmic reticulum (ER) with LDs, binds to COPB1, and mediates ATGL transport. We provide important insights into the function of OSBPL2, indicating that it is required for the regulation of lipid droplet lipolysis.
Subject Areas: Biological Sciences, Cell Biology, Molecular Biology
Graphical Abstract
Highlights
-
•
LD lipolysis is impaired in OSBPL2/osbpl2b-mutant HepG2 cells and zebrafish
-
•
OSBPL2 interacts with COPB1, a subunit of the COPI complex located on LDs
-
•
Altered COPI complexes on LDs may perturb the trafficking of lipolysis lipase ATGL
Biological Sciences; Cell Biology; Molecular Biology
Introduction
Lipid droplets (LDs) are dynamic organelles into which excess amounts of biochemical energy are deposited in the form of triacylglycerols (TAGs), together with cholesteryl esters (CEs) (Walther and Farese, 2012). A single, large lipid droplet often occupies almost the entire cytoplasmic compartment in white adipose tissues (WATs) for TAG storage, whereas a series of tiny LDs exist in brown adipose tissues (BATs), from which TAGs are mobilized and fatty acids (FAs) are released when energy is needed. The accumulation of enlarged LDs in the liver may lead to obesity, hepatic steatosis, cardiovascular disease, and other metabolic diseases (Gao et al., 2019). Obesity, known as a chronic progressive disease, as declared by the World Obesity Federation and other organizations (Bray et al., 2017), is characterized by an abnormal or excessive accumulation of fat that negatively affects the optimal state of health (Ryan and Kahan, 2018).
The triacylglycerols (TAGs) maintained in LDs are regulated by multiple enzymes, such as glycerol-3-phosphate-O-acyltransferase (GPAT), 1-acylglycerol-3-phosphate-O-acyltransferase (AGPAT), phosphatidic acid phosphatase (PAP, also known as lipin), and diacylglycerol acyltransferase (DGAT) (Onal et al., 2017; Tan et al., 2014). TAGs lipolysis is catalyzed step-by-step with patatin-like phospholipase domain-containing 2 (PNPLA, also known as ATGL), followed by the action of hormone-sensitive lipase (HSL) and monoacyl glycerol lipase (MGL) (Missaglia et al., 2019; Zechner et al., 2012). Multiple proteins that are primarily located in the endoplasmic reticulum (ER) regulate TAG synthesis and the lipolysis of LDs and are transferred through membrane bridges between the ER and the LDs. It has been reported that the establishment of membrane bridges between the ER and LDs is triggered by coat protein complex I (COPI) (Thiam et al., 2013; Wilfling et al., 2013, 2014) and mediates the transport of ATGL to the LD surface (Beller et al., 2008; Ellong et al., 2011; Lee et al., 2004; Soni et al., 2009). The lack of ATGL results in a defect in lipolysis and leads to the accumulation of TAGs in adipose tissue (AT), heart, and muscle (Haemmerle et al., 2006; Schoenborn et al., 2006).
Oxysterol-binding protein (OSBP) and OSBP-related proteins (ORPs) constitute a large family of lipid transfer proteins (LTPs) that bind cholesterol, oxysterols, and anionic phospholipids through a conserved hydrophobic binding domain (Antonny et al., 2018; Jamecna et al., 2019). Additional membrane-targeting motifs allow OSBP/ORPs to simultaneously associate with a diverse set of protein and lipid partners in organelle membranes, thus positioning the lipid-binding domain in close apposition to donor and acceptor membranes to facilitate transfer or signaling functions. OSBPL2, also known as ORP2, is a member of this large family of LTPs and contains an FFAT (diphenylalanine in an acidic tract) motif for ER targeting and a functional OSBP-related domain (ORD) for lipid binding and transfer (Wang et al., 2019b). OSBPL2 is reported to be involved in the metabolism of neutral lipids (Hynynen et al., 2009; Kentala et al., 2018), but the molecular mechanism of how OSBPL2 regulates energy metabolism has not yet been fully elucidated.
Herein, we showed that LDs were enlarged in OSBPL2−/− HepG2 cells with defective lipolysis. Furthermore, the AT in Osbpl2b−/- zebrafish models presented with larger LDs and the resultant obesity phenotypes, which suggested that OSBPL2 mutation altered the ER and LD connections that mediate lipid and protein trafficking. We showed that OSBPL2/ORP2 binds to COPB1 and participated in the transfer of proteins initially localized in the ER. Accordingly, OSBPL2 deficiency caused reduced transfer of ATGL from the ER to LDs, which led to a decrease in lipolysis and caused obesity. Overall, our study provided important insights into the function of OSBPL2, and the results strongly suggest that OSBPL2 could be a promising target for the treatment of obesity in humans.
Results
OSBPL2 Deficiency Is Associated with an Increase in LD Size and the Dynamic Changes in the ER and LDs
LDs are cellular organelles that are the major sites of fat storage in tissues. Each LD is composed of hydrophobic neutral lipid cores containing triacylglycerols (TAGs) and cholesteryl esters, an encapsulating monolayer membrane of phospholipids and a series of surface proteins involved in lipid synthesis, lipolysis, and many other functions. The LD composition and size are different in diverse tissues (Zweytick et al., 2000). TAGs are found in white adipose tissues (WAT) in the form of single giant LDs and several smaller droplets in the liver. The increased size of LDs provides an efficient form of fat storage, in terms of surface-to-volume ratio, to prevent lipotoxicity and hepatic steatosis (Fei et al., 2011).
To study the biological function of OSBPL2 in lipid metabolism, we generated OSBPL2-deficient HepG2 cells by using the CRISPR-Cas9 gene-editing technique through which amino acid sequence is truncated and altered (Figures S1A and S1B). The western blot analysis results showed that the OSBPL2/ORP2 band almost completely disappeared in OSBPL2−/− cells compared with that of the wide-type (WT) cells (Figure S1C).
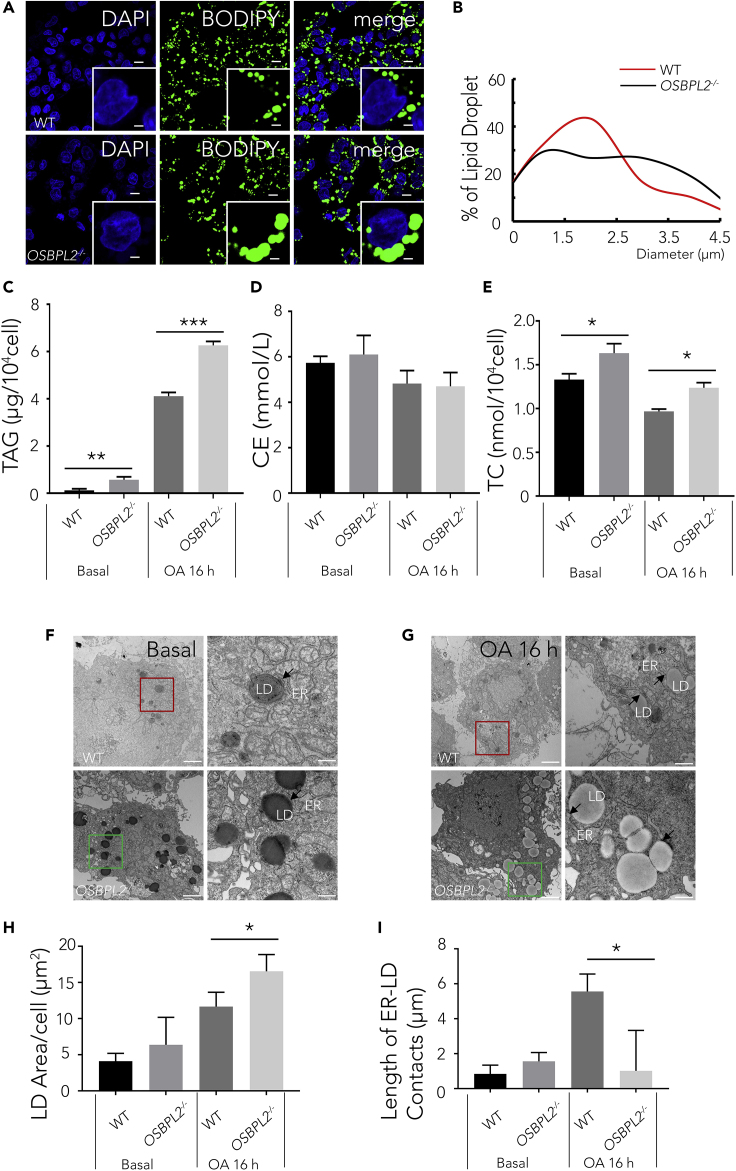
To clarify the biological effect of OSBPL2 deletion, we treated HepG2 cells with oleic acid (OA), a lipogenic stimulus (Kaushik and Cuervo, 2015), for 16 h. Compared with those in the WT cells, more large LDs were formed in OSBPL2−/− cells (Figures 1A and 1B). To obtain images of the LD in the WT cells and OSBPL2−/− cells, confocal microscopy was used, and fluorescence staining of LDs was performed with BODIPY 493/503 to delineate LD sizes and distribution patterns (Figure 1A). Using this approach, we confirmed an increase in number of large LDs and a decrease in number of small LDs in the OSBPL2−/− cells (Figure 1A). As described (Qi et al., 2016), we also measured the LD size with ImageJ software and analyzed the LD distribution in the WT cells and OSBPL2−/− cells. The population of LDs with a diameter of 3–4 μm increased, whereas the population of LDs with a diameter 1–2 μm decreased in the OSBPL2−/− cells (Figure 1B). These results indicated that OSBPL2 deficiency caused LD accumulation with an increase in large LDs and a decrease in small LDs. In addition, the TAG, CE, and TC were measured to find whether OSBPL2/ORP2 regulates TAG or CE synthesis. We found that the TAG level increased in OSBPL2−/− cells compared with that in WT cells treated with OA for 16 h using a TAG biochemical detection kit (Figure 1C), but there were no significant differences in CE levels between WT cells and OSBPL2−/− cells (Figure 1D). The above results indicated that OSBPL2 deletion had little impact on the CE level but TAG in LDs. Consistent with previous data (Wang et al., 2019a; Zhang et al., 2019), we also found that OSBPL2 deficiency increased the total cholesterol (TC) level (Figure 1E). The above indicates that OSBPL2 deficiency may cause an increase of free cholesterol (FC) but not CE.
Figure 1.
OSBPL2deficiency Enlarges Intracellular Lipid Droplets and Alters the Length of the ER–LD Contacts
(A) Confocal imaging of the WT cells and OSBPL2−/− cells using BODIPY 493/503 (green) and DAPI (blue) fluorescence staining. Scale bar, 10 μm (inserts, 2.5 μm).
(B) Distribution of LDs in the HepG2 cells measured using ImageJ software.
(C) The concentration of TAG in the WT cells and OSBPL2−/− cells.
(D) The concentration of CE in the WT cells and OSBPL2−/− cells.
(E) The concentration of TC in the WT cells and OSBPL2−/− cells.
(F and G) TEM images of LDs and the ER in the WT cells and OSBPL2−/− cells. The scale bars in the original and magnified images represent 2 μm and 500 nm, respectively.
(H) Total LD area per cell.
(I) Total length of ER–LD contacts per cell.
HepG2 cells were treated with or without 400 μM OA for 16 h (A). Data were collected from at least 2,000 LDs (B). The arrows indicated the ER–LD contacts; the lengths of ER–LD contacts were measured by ImageJ software within a 30 nm distance (F and G). Data were collected from 16 to 32 cells, with four images from each set analyzed (H and I). All the data represent the mean ± SD; t test significance (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
As is well known, LDs originate in the ER (Ben M'barek et al., 2017; Choudhary et al., 2015); when they become mature, LDs eventually separate from the ER through budding and enter into the cytoplasm (Gross and Silver, 2014; Jacquier et al., 2011; Ohsaki et al., 2014). Recent studies indicate that inter-organelle membrane contact sites (MCSs) are associated with lipid metabolic reactions (Phillips and Voeltz, 2016). Previous studies have also shown that OSBPL2/ORP2 is located on the surface of LDs and at ER–LD contact sites (Hynynen et al., 2009; Kentala et al., 2018). To clarify the role of OSBPL2/ORP2 on the dynamic changes observed in ER and LDs, we used transmission electron microscopy to observe LDs and the ER in OSBPL2−/− cells and WT cells grown with or without added OA loading (Figures 1F and 1G). Generally, the total LD area and the length of the ER-LD contacts were both increased when cells were supplemented with OA for 16 h. Consistent with previous fluorescence staining results (Figures 1A and 1B), we also found that the LD area in the OSBPL2−/− cells was larger than that in the WT cells upon OA treatment (Figure 1H). However, the length of the ER-LD contacts was expanded in the WT cells, whereas no significant change was observed in the OSBPL2−/− cells (Figure 1I), indicating that the absence of OSBPL2/ORP2 inhibited the dynamic changes in the ER-LD contact sites. Our findings are consistent with the results reported in Huh7 cells (Kentala et al., 2018). Increasing evidence suggests that the ER-LD contact sites and membrane extensions mediate the transport of neutral lipids that are synthesized in the ER and the proteins residing the ER that are needed for LD growth, lipogenesis, and lipolysis (Joshi et al., 2017; Kory et al., 2016; Walther et al., 2017; Wilfling et al., 2013). Our results suggested that the altered ER-LD contacts in the OSBPL2−/− cells affected lipolysis and thus caused LD accumulation.
OSBPL2/ORP2 Plays an Important Role in Lipolysis but Not in Lipogenesis
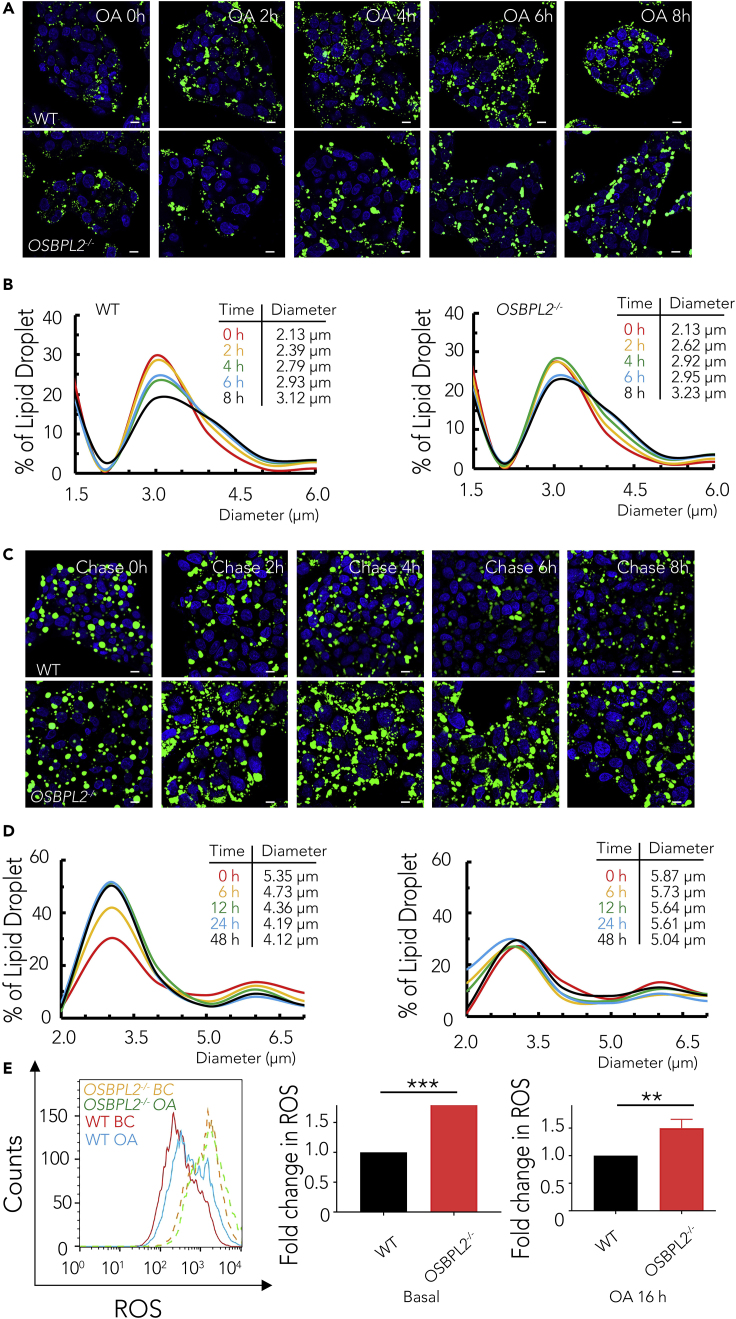
The amount of LDs is known to be determined by the balance between lipogenesis and lipolysis. LD diameters increase within hours during lipogenesis progress, whereas LDs shrink as core lipids are catabolized to liberate FAs during lipolysis (Walther and Farese, 2012). Thus, we next examined whether OSBPL2/ORP2 plays a role in LD formation or the progression of LDs breakdown. We first evaluated dynamic LD formation over an 8-h-period of treatment with OA (Figure 2A). We found no apparent differences in the changes in LD size distribution in the OSBPL2−/− cells, compared with the change in LD distribution in the WT cells (Figure 2B). Next, we analyzed the key enzymes of TAG synthesis by measuring the mRNA expression level, including that of glycerol-3-phosphate O-acyltransferase (GPAT), 1-acylglycerol-3-phosphate O-acyltransferase (AGPAT), phosphatidic acid phosphatase (PAP), and diacylglycerol acyltransferase (DGAT). The qRT-PCR results showed that there were also no significant differences in the mRNA expression levels of these lipogenesis-related genes (Figure S2A and Table S1).
Figure 2.
OSBPL2 Deficiency Impairs Efficient Lipolysis and Increases ROS Levels
(A) Confocal imaging with BODIPY 493/503 (green); DAPI (blue) fluorescence staining of the WT cells and OSBPL2−/− cells during lipogenesis. Scale bar, 10 μm.
(B) The dynamic changes of LDs on lipogenesis in the HepG2 cells.
(C) Confocal imaging with BODIPY 493/503 (green); DAPI (blue) fluorescence staining of the WT cells and OSBPL2−/− cells during lipolysis. Scale bar, 10 μm.
(D) The dynamic changes of LDs on lipolysis in the HepG2 cells.
(E) Mitochondrial ROS levels in the WT and OSBPL2−/− HepG2 cells.
HepG2 cells were treated with OA at 400 μM for the indicated time points in (A) and (C). LDs were measured using ImageJ software. Data were collected from at least 2,000 LDs in each treatment, and the dynamic changes in LD size were analyzed by Gaussian distribution curves using GraphPad Prism software with data from (B) and (D). All the data represent the mean ± SD; t test significance (∗∗p < 0.01, ∗∗∗p < 0.001).
Next, we focused on the progress of LD breakdown to determine whether OSBPL2 deficiency impacted the effect of lipolysis. The cells were cultured in OA-containing medium for 24 h, followed by a chase of lipids for 48 h in fatty-acid-free medium (OPTI-MEM, 5% fatty-acid-free BSA) to induce TAG breakdown. The size of most LDs in the WT cells decreased over time, whereas only a small fraction of supersized LDs were changed in the OSBPL2−/− cells (Figure 2C). The peak value of the curves representing the size of most of the LDs in fatty-acid-free medium was changed at the time of TAG breakdown (Hynynen et al., 2009), suggesting that the mobilization of TAGs was slowed in the absence of OSBPL2 (Figure 2D). To confirm the effect of OSBPL2/ORP2 on lipolysis, we measured the expression of lipolysis-related genes, such as carnitine palmitoyltransferase 1 (CPT1), carnitine palmitoyltransferase 2 (CPT2), acetyl-CoA carboxylase (ACC), PNPLA (also known as ATGL), hormone-sensitive lipase (HSL), and COPI coat complex subunit beta 1 (COPB1), with qRT-PCR. These genes play important roles in TAG mobilization, energy metabolism, and lipase trafficking to regulate lipolysis. As shown in Figure S2B and Table S1, OSBPL2 deficiency led to the upregulated expression of ACC, GPD2, CPT1, and CPT2. In addition, considering that excessive TAGs might induce intracellular reactive oxygen species (ROS) production (de Mello et al., 2018; Newsholme et al., 2016), we detected ROS levels by flow cytometry. As expected, the ROS levels of OSBPL2−/− cells were ∼80% higher than those of WT cells under basic condition and ∼50% higher than those of WT cells treated with OA (Figure 2E).
Taken together, these data suggest that intracellular OSBPL2 deletion impacts the progress of lipolysis rather than LD formation or morphological changes in LDs.
OSBPL2/ORP2 Is Required for the Transport of ATGL from the ER to LDs
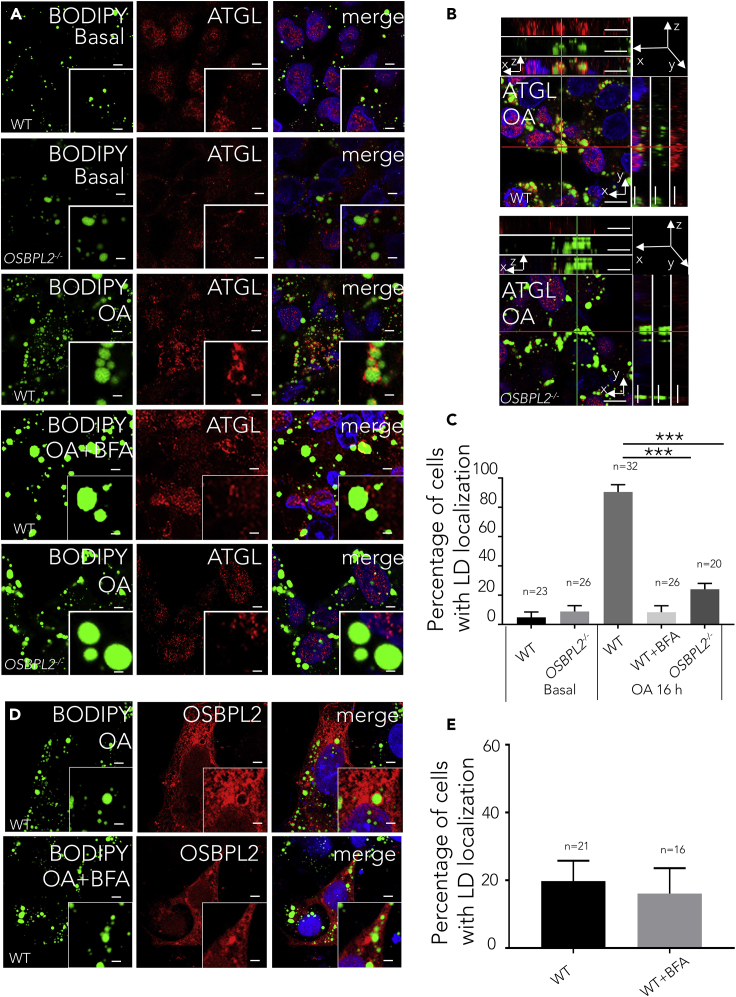
To address how lipolysis is regulated by OSBPL2/ORP2, we focused on the rate-limiting lipases of lipolysis more closely. In the lipolysis process, the LD volume decreased as core lipids were catabolized sequentially by ATGL, HSL, and monoacylglycerol lipase (Walther and Farese, 2012). Of the three lipases, ATGL is the key lipase for the first rate-limiting step (Walther and Farese, 2012; Zechner et al., 2017). Consistent with previous reports (Sugihara et al., 2019), our findings verified that ATGL attached to the LDs in the HepG2 cells treated with OA (Figure 3A). It was observed that there were no significant differences in ATGL expression levels between the WT cells and OSBPL2−/− cells (Figure S2B), so we measured ATGL to assess whether similar results would be obtained at protein expression level. We found no significant differences in the protein expression level of ATGL in OSBPL2−/− cells compared with the level in the WT cells (Figure S2C).
Figure 3.
OSBPL2 Deficiency Leads to a Handicap of ATGL Transport from the ER to LDs.
(A) Confocal imaging of the WT or OSBPL2−/− HepG2 cells using BODIPY 493/503 (green), DAPI (blue), and anti-ATGL (red) immunostaining. Scale bar, 5 μm (inserts, 2.5 μm).
(B) A reconstructed 3D image of the WT and OSBPL2−/− HepG2 cells treated with OA using BODIPY 493/503 (green), DAPI (blue), and anti-ATGL (red) immunostaining. Scale bar, 10 μm.
(C) Percentage of cells that showed the localization of endogenous ATGL on LDs.
(D) Confocal images showing the localizations of RFP-OSBPL2 in the WT cells treated with or without BFA using BODIPY 493/503 (green) and DAPI (blue). Scale bar, 5 μm (inserts, 2.5 μm).
(E) Percentage of cells expressing RFP-tagged OSBPL2 that showed LD localization.
HepG2 cells were treated with 400 μM OA for 16 h (A and B). The main portion is indicated in the xy image. The two rectangular panels (top and right) are xz and yz images, respectively (B). HepG2 cells were transfected with RFP-tagged OSBPL2 for 24 h, then treated with 400 μM OA and 10 ng/mL BFA (D). All the data represent the mean ± SD; t test significance (∗∗∗p < 0.001).
We next focused on the location of endogenous ATGL to determine whether OSBPL2 deletion altered the distribution of ATGL on the LDs. Previous studies in Drosophila S2 cells and HeLa cells demonstrated that ATGL is transferred from the ER membrane to LD surface by the COPI machinery (Soni et al., 2009; Wilfling et al., 2014). Using z stack of confocal scanning after cells received OA supplement, we observed an abundant of ATGL wrapped around LDs in WT cells, whereas few ATGL proteins were visibly attached to the LDs in the OSBPL2−/− cells. This result in the OSBPL2−/− cells was similar to that in the WT cells treated with brefeldin A (BFA, an inhibitor of COPI machinery), which caused a disruption of ATGL trafficking to LDs (Figures 3A–3C). We blocked the COPI machinery with BFA to verify the attachment of OSBPL2/ORP2 to the LDs. Using the method as described (Du et al., 2020), the WT cells were transfected with RFP-tagged OSBPL2/ORP2 and the localization of OSBPL2/ORP2 to the LDs was quantified. The results indicated that the degree of co-localization of OSBPL2/ORP2 with LDs was unaffected, suggesting that the localization of OSBPL2/ORP2 on the LDs is independent of the COPI machinery (Figures 3D and 3E).
Based on earlier discussion, we confirmed that the expression of ATGL was not altered both at the mRNA and protein levels, and we also found an abnormal localization of ATGL on the LDs in the OSBPL2−/− cells, suggesting that OSBPL2 deficiency interfered with the distribution of ATGL, but not its abundance, on the LDs. The results implied that OSBPL2/ORP2 is required in the transport of ATGL and is possibly an underlying protein involved in the COPI machinery that mediates ATGL during lipolysis.
OSBPL2/ORP2 Binds to COPB1 and Downregulates COPB1 Expression Levels
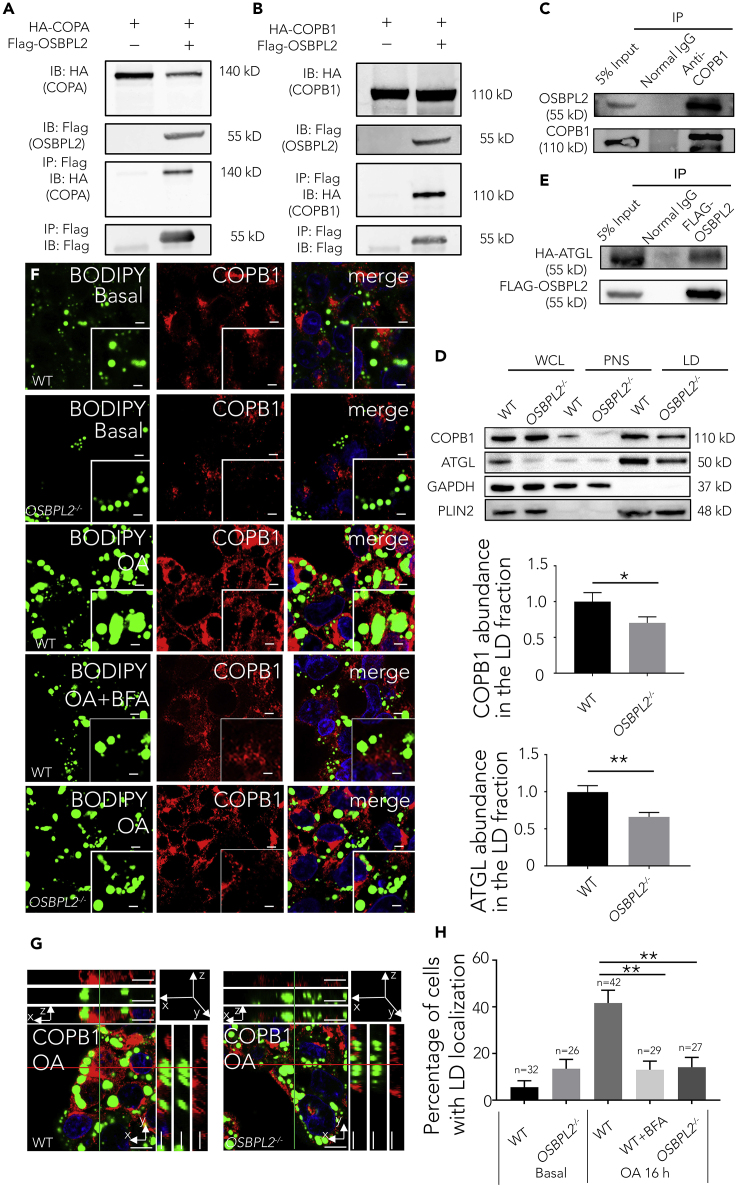
Given that OSBPL2 deficiency hindered ATGL transport, we sought to determine its intrinsic regulatory mechanisms. We had previously identified 47 proteins by proteomic analysis of the OSBPL2/ORP2 interactome and found COPA and COPB1, both COPI coat complex subunits, among the binding partners of OSBPL2/ORP2. FLAG-tagged OSBPL2/ORP2 was previously constructed and expressed in HEK293Ta cells, and then, the protein complex was pulled down with anti-FLAG magnetic beads and analyzed by mass spectrometry. The proteomic analysis revealed an interaction between OSBPL2/ORP2 and candidate proteins, which prompted us to confirm that the two proteins are associated with each other at the molecular level. Co-immunoprecipitation (Co-IP) experiments showed a specific interaction of FLAG-tagged OSBPL2/ORP2 with transfected COPA in HEK293Ta cells (Figure 4A), and we also found a specific interaction of FLAG-tagged OSBPL2/ORP2 with transfected COPB1 in HEK293Ta cells (Figure 4B). Because COPB1, but not COPA, is associated with lipid droplet accumulation (Kociucka et al., 2016; Wilfling et al., 2014), another laboratory suggested that COPB1 might play an important role in confining COPI orientation relative to the membrane (Yu et al., 2012), and our qPCR results also indicated that knocking out OSBPL2 caused changes in COPB1. Similarly, the physical interaction of endogenous OSBPL2/ORP2 with endogenous COPB1 was validated in HepG2 cells (Figure 4C). More interestingly, we found that OSBPL2 deficiency led to an increase in COPB1 expression, OSBPL2 overexpression significantly decreased the expression level of COPB1, and OSBPL2 transfection in OSBPL2−/− cells also reduced COPB1 expression (Figures S3A–S3C). Moreover, a similar trend was observed among OA-treated cells (Figures S3D–S3G).
Figure 4.
OSBPL2 Binds to COPB1, and OSBPL2 Deficiency Leads to Hindered COPB1 Localization to the LDs
(A) Co-IP assay verified the interaction of FLAG-tagged OSBPL2 with HA-tagged COPA in the HEK293Ta cells.
(B) Co-IP assay verified the interaction of FLAG-tagged OSBPL2 with HA-tagged COPB1 in the HEK293Ta cells.
(C) Co-IP assay verified the interaction of endogenous OSBPL2 with endogenous COPB1 in HepG2 cells.
(D) Western blot of whole-cell lysate (WCL), post nuclear supernatant (PNS), and LD proteins in the WT and OSBPL2−/− cells.
(E) Co-IP assays verified the interaction of FLAG-tagged OSBPL2 with HA-tagged ATGL in the HEK293Ta cells.
(F) Confocal imaging of the WT cells or OSBPL2−/− cells using BODIPY 493/503 (green), DAPI (blue), and anti-COPB1 (red) immunostaining. Scale bar, 5 μm (inserts, 2.5 μm).
(G) A reconstructed 3D image of the WT cells and OSBPL2−/− cells treated with OA using BODIPY 493/503 (green), DAPI (blue), and anti-COPB1 (red) immunostaining. Scale bar, 10 μm.
(H) Percentage of cells that show the localization of endogenous COPB1 on LDs.
HepG2 cells were treated with 400 μM OA for 16 h and treated with or without 10 ng/mL BFA (F). The main portion is shown in the xy image. The two rectangular panels (top and right) are the xz and yz images, respectively (G). All the data represent the mean ± SD; t test significance (∗p < 0.05, ∗∗p < 0.01).
Although we have demonstrated that OSBPL2/ORP2 binds to COPB1 and that OSBPL2 deletion upregulates COPB1 expression levels, how OSBPL2/ORP2 regulates the expression of COPB1 and how OSBPL2/ORP2 regulates ATGL transport remain unknown. We suggest that an underlying molecular mechanism combines OSBPL2/ORP2 with COPB1 and ATGL to facilitate ATGL localization on LDs.
OSBPL2/ORP2 Is Required for COPB1 Localization to LD Surfaces and ATGL Transport from the ER and Attachment to LDs
It has been reported that the transport of GPAT4, which originates in the ER, to LDs requires COPB1 (Wilfling et al., 2014). Similar to GPAT4, ATGL transport from the ER to LDs also depends on the COPI machinery; therefore, we compared the localization of COPB1 in the OSBPL2−/− cells with that in the WT cells. The cells were treated with OA for 16 h, and the lysate was then divided into whole-cell lysate (WCL), post nuclear (PNS), and LD fractions as described (Liu et al., 2016). Similar to the finding with ATGL, fewer COPB1 molecules were targeted to LDs in the OSBPL2−/− cells, as determined by western blotting (Figure 4D). In addition, Co-IP was performed, and the results showed that HA-tagged ATGL was detected in the FLAG-tagged OSBPL2/ORP2 complexes but was absent in the negative control (Figure 4E), supporting the supposition that ATGL specifically interacts with OSBPL2/ORP2. As shown by fluorescence microscopy, we found that COPB1 wrapped around the LDs in the WT cells but not in the WT cells treated with BFA or in the OSBPL2−/− cells (Figures 4F–4H) supplemented with OA, implying that OSBPL2/ORP2 is required for COPB1 localization on the LDs. The data suggested that OSBPL2/ORP2 binds to COPB1 and that the complex is involved in ATGL transport. Finally, we knocked down COPB1 using siRNA to confirm that the disrupted expression of COPB1 leads to blocked ATGL transport. As expected, confocal scanning imaging showed that the extent of ATGL localization to the LDs was reduced when COPB1 was knocked down (Figures S3H–S3J).
Together, the results showed that OSBPL2/ORP2 located to the LDs independent of COPI machinery, but it was bound to both COPB1 and COPA, which are the subunits of COPI that mediate ATGL trafficking for lipolysis. OSBPL2 deletion reduced the location of COPB1 on the LDs and caused a hindrance to ATGL transport that was similar to that observed after COPB1 knockdown by siRNA. Furthermore, ATGL was suggested to interact with OSBPL2/ORP2. Thus, we conclude that OSBPL2/ORP2 is located on LDs and binds COPB1, which is in a complex that possibly transfers ATGL from the ER to LDs for lipolysis.
Aggravated Obesity Phenotypes and Abnormal Behavior in Osbpl2b-Deficient Zebrafish
The results from our previous study showed that OSBPL2−/− Bama mini pigs exhibited obvious obesity and hypercholesterolemia phenotypes (Yao et al., 2019), and the established adult osbpl2b−/− zebrafish have also shown abnormal lipid-metabolism-related characteristics. The osbpl2b−/− zebrafish in this study were used to test whether altered osbpl2b disturbs energy homeostasis in vivo. There are two genes (osbpl2a and osbpl2b) in zebrafish that are homologous with human OSBPL2, and osbpl2b was proven to be the orthologous gene of human OSBPL2 (OMIM: 606731) in our previous study, sharing identity of 71.2% with human OSBPL2 amino acid sequences (Liu et al., 2016). The osbpl2b−/− zebrafish were previously generated using the CRISPR/Cas9 gene-editing technique, through which a 5-nucleotide (GAGCT) deletion was generated in exon 6 and resulted in a truncated protein. This mutation in osbpl2b caused hearing impairment and lipid-metabolism-related characteristics (results are not shown). Adult WT zebrafish and osbpl2b−/− zebrafish were allocated to groups and fed with the same basic chow (BC), for the WT-BC group and MT-BC group, or the same high-fat diet (HFD), for the WT-HFD group and MT-HFD group. Deletion of osbpl2b seemed to increase somatic growth and body weight, which were measured every month (Figures 5A and 5B). As described (Minchin et al., 2015), we next stained zebrafish with BODIPY 493/503, anesthetized the fish, and observed the fat distribution in the fish bodies using a fluorescence stereo microscope. Although there were no significant differences in the total adipose tissue (AT) area among the four groups as shown Figure 5C, we found that the livers of the MT group were larger and contained more fat attached on the surface, as observed when the fish were dissected (Figure 5D). Next, to determine the TAGs content accurately, we divided the adipocytes of the zebrafish body into three regions: miscellaneous AT (misc. AT), subcutaneous adipose tissue (SAT), and visceral adipose tissue (VAT). We then measured the LD size in adipocytes of different regions to determine whether the osbpl2b mutation altered fat distribution in zebrafish. Fluorescence staining with BODIPY 493/503 showed that the HFD treatment did not cause significant differences in miscellaneous AT in the WT-HFD group and MT-HFD group but caused a substantial increase in the LD area in the miscellaneous AT in the MT-BC group compared with that in the WT-BC group (Figures 5E and 5F). Furthermore, we found that SAT was increased in the MT group compared with the SAT level in the WT group fed either a BC or HFD (Figures 5E and 5G). Similar to SAT, VAT increased in the MT-HFD group; however, VAT in the MT-BC group did not expand obviously (Figures 5E and 5H). More interestingly, we observed abnormal behavior in the osbpl2b−/− zebrafish, especially the MT-HFD group, which tended to live near the top of the water, whereas most of the WT zebrafish fed a BC or HFD lived in the middle of the fish tank (Figures S4A, S4B, and Video S1); this observation led us to consider possible lipid toxicity in the liver (Botha et al., 2019; Wang et al., 2015). Further data showed that osbpl2b deficiency led to an increase in food intake (Figure S4C).
Figure 5.
osbpl2b Deficiency Alters Somatic Growth and Exacerbates Fat Accumulation in Adult Zebrafish
(A) Body weight curves.
(B) Mean length of the fish bodies. The data represent the mean ± SD; t test significance (∗p < 0.05).
(C) Fluorescence imaging of total AT using BODIPY 493/503 staining. Scale bar: 2,000 μm.
(D) A representative view images of the liver.
(E) Fluorescence imaging of Misc. AT, VAT, and SAT using BODIPY 493/503 staining. Scale bar, 50 μm.
(F–H) Distribution of LDs in Misc. AT, SAT, and VAT measured using ImageJ software.
The WT-HFD group (left) and MT-HFD group (right) were maintained in buffered reverse osmosis water.
The results suggest that osbpl2b mutation could cause lipotoxicity in the liver and an obesity-like phenotype.
Reduced ER–LD Contact Area Lengths and Decreased Localization of COPB1 on LDs in the osbpl2b-Deficient Zebrafish
The results of our in vitro experiments suggested that OSBPL2/ORP2 is located on LDs and COPB1 is bound in a complex that possibly transfers ATGL from the ER to LDs. We next focused on whether this molecular mechanism could mediate lipolysis in zebrafish. Fluorescence staining of fat distribution showed an increase in VAT and SAT in the osbpl2b−/− zebrafish, and using Masson dye staining, we verified the morphological changes induced by osbpl2b deletion. Similar to the results shown in Figures 5E–5H, we found that SAT was increased in the osbpl2b−/- zebrafish (Figures 6A–6C). H&E staining of liver tissue revealed exacerbated fat accumulation and edema with inflammation in the MT-HFD group (Figure 6D). As we suggested that OSBPL2 depletion hinders the ATGL localization to the LD surface, we sought to determine whether a similar effect could be observed in the osbpl2b−/− zebrafish. We observed LDs and the ER by transmission electron microscopy to evaluate the role of osbpl2b in the dynamic changes in the ER and LD contact in zebrafish. Similar to the results found in vitro (Figures 1F–1I), the length of the ER–LD contacts in liver tissue increased only for the WT-HFD group and did not change significantly in the MT-HFD group (Figures 6E and 6F). Furthermore, by detecting the extracted LD-associated proteins in the liver by western blotting, we detected a decrease in Copb1 targeted to the LDs in the MT-HFD group compared with that in the WT-HFD group (Figures 6G and 6H), suggesting that the mutation of osbpl2b in zebrafish led to a decrease of Copb1 on the LDs.
Figure 6.
osbpl2b Deficiency Leads to Steatohepatitis and Reduces the Length of the ER–LD Contacts in the Liver
(A) Masson's trichrome-stain zebrafish indicated the anatomical localization of SAT and liver. Muscle (m), swim bladder (sb), VAT, SAT, and liver are indicated with arrows. Scale bar: 1,000 μm.
(B) Image of SAT using Masson's trichrome staining. Scale bar, 50 μm.
(C) Distribution of LDs in SAT measured using ImageJ software.
(D) H&E-stained histological sections indicating the morphology of the liver. Scale bar, 50 μm.
(E) TEM imaging of the LDs and the ER in the livers. The scale bars in the original and magnified images represent 2 μm and 500 nm, respectively.
(F) Total length of ER–LD contacts per cell.
(G) Western blot of LD proteins in the WT-HFD and MT-HFD zebrafish.
(H) Copb1 abundance in the LD fraction, which represented the quantification of images shown in G.
The arrows indicated the ER–LD contacts, the lengths of ER–LD contacts were measured by ImageJ software within a 30 nm distance (E). Data were collected from 16 to 32 cells, with four images from each set analyzed (F). All the data represent the mean ± SD; t test significance (∗p < 0.05).
These data obtained from zebrafish experiments are similar to the in vitro results and suggest a possible molecular mechanism: OSBPL2/ORP2 binds COPB1 in a complex that transfers ATGL from the ER to LDs for lipolysis, which explains why OSBPL2 deficiency causes fat accumulation and energy metabolic disorder.
Discussion
Our laboratory and others had previously identified that OSBPL2 was associated with autosomal dominant nonsyndromic hearing loss (Thoenes et al., 2015; Wu et al., 2019; Xing et al., 2015). To confirm the consistent effect of OSBPL2/ORP2 in hearing loss by both genotype and phenotype, we successfully simulated human hearing loss phenotypes in both Bama mini pig and zebrafish models. Especially in the OSBPL2-disrupted pigs, we reproduced not only a deafness phenotype similar to that of humans but also a hypercholesterolemia phenotype (Yao et al., 2019). In subsequent studies of OSBPL2/ORP2 functions, we found that OSBPL2 was downregulated by 25-hydroxycholesterol (Wang et al., 2019c). In OC1 cells, the deletion of OSBPL2 led to increased cholesterol biosynthesis and upregulated ROS production (Wang et al., 2019a). In HeLa cells, OSBPL2 deficiency was also proven to upregulate intracellular cholesterol and cholesteryl ester (Zhang et al., 2019). Recently, it was suggested that the deletion of OSBPL2 in HuH7 cells reduce glucose absorption and glycogen synthesis and inhibited glycolysis (Kentala et al., 2018). In addition, others (Hynynen et al., 2009; Olkkonen et al., 2019) have suggested that OSBPL2/ORP2 impacts cellular TAG and carbohydrate metabolism. These results indicate that OSBPL2/ORP2 has versatile functionality.
Our results have shown that OSBPL2/ORP2 is involved in the establishment of the ER-LD membrane contacts. One type of membrane contact sites (MCS) called an ER–LD contacts links adjacent membranes of the ER and apposed LDs without allowing them to fuse to each other. It has been reported that multiple proteins associated with ER–LD contacts are also involved in LD formation (Jackson, 2019). Rab18 and NAG-RINT1-ZW10 (NRZ) located at ER–LD points of a contact are involved in LD biogenesis in adipocytes (Xu et al., 2018). With results similar to those from a previous study (Kentala et al., 2018), we found that OSBPL2 deficiency impacts the dynamic change in the ER and LDs upon fatty acid addition. However, in contrast with the results obtained from cells treated with OA for 3 h, we further investigated whether the total LD area per cell was increased in the OSBPL2−/− cells when cells were treated with OA for 16 h. The inconsistent results may be due to different treatment times and the effect of lipolysis. Our observation was consistent with a report (Hynynen et al., 2009) showing that knocking down OSBPL2 impacts lipolysis. The production of ROS is associated with energy metabolism. The activation or inhibition of the enzymes implicated in ROS production can affect cell structure and function (Quijano et al., 2016). Some hormonal and metabolic adaptations upregulate mitochondrial fatty acid oxidation (mtFAO) to restrain hepatic fat accumulation; however, they may also result in ROS overproduction rather than a reduction in fat accumulation in the liver (Begriche et al., 2013). Furthermore, deficiency in the coactivator of ATGL, comparative gene identification-58 (CGI-58) inhibits the lipolysis effect of ATGL and activates the ROS-inflammasome pathway (Miao et al., 2014). Thus, the enhanced ROS level with LD enlargement appearing from our data can be viewed from another perspective to suggest that OSBPL2 deletion possibly causes abnormal energy metabolism and leads to obesity.
Our data have identified that OSBPL2/ORP2 binds COPB1 in a complex that mediates protein transport. The vesicle coat proteins, such as coat protein I (COPI) and coat protein II (COPII), play an important role in regulating vesicle formation and mediate specific transport routes. COPI vesicles are considered to be mediators of transport back from the Golgi apparatus to the ER, whereas COPII vesicles transport cargo from the ER to the Golgi apparatus. The formation of the COPI complex associated with the ER and LDs requires the localized activation of a small GTPase at the LDs membrane. Guanine-nucleotide exchange factors catalyze and activate small GTPase through the exchange of GDP for GTP (Bethune and Wieland, 2018). The clathrin-coated vesicles (CCVs) containing AP-2 (an adaptor protein complex) are recruited to the plasma membrane (PM) by phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] and cargo proteins (Robinson, 2015). We found that COPB1, a subunit of the COPI complex, is predicted to contain the AP-2 domain, as indicated by the CD-search online software, suggesting that COPB1 is possibly recruited to the PM by PI(4,5)P2, and a recent study has proven that OSBPL2 delivers cholesterol to the PM in exchange for PI(4,5)P2 (Wang et al., 2019b). Oxysterol-binding protein (OSBP) is reported to have the ability to both tether organelles and transport lipids between them, with an FFAT motif that interacts with the ER protein VAP-A and transfer sterol through the lipid transfer domain (ORD) (Mesmin et al., 2013). Here, our evidence supports the position that OSBPL2/ORP2 binds to COPB1 and that OSBPL2/ORP2 localization to LDs is independent of COPI machinery. These findings suggested that OSBPL2/ORP2 linked the ER to the limiting membrane of LDs, which was followed by the recruitment of COPB1.
We have also identified OSBPL2/ORP2 as a lipid transfer protein involved in the transport of ATGL and lipolysis. Given that proteins targeting LDs have been classified into two paths (Kory et al., 2016), class I proteins access LDs through ER-LD contact sites, and class II proteins bind to LDs directly from the cytosol. ATGL is one of the class I proteins considered to be the first embedded in the ER membrane through its hydrophobic hairpin motif, followed by its transfer to LDs. Previous studies have demonstrated that COPI mediates the membrane-trafficking pathway that contributes to the localization of ATGL to LDs via ER-LD membrane bridges (Ellong et al., 2011; Soni et al., 2009). A recent study has proven that Arf1/COPI machinery controls ER-LD connections to mediate TAG storage and catabolism (Wilfling et al., 2014). Similar to those from previous studies, our data demonstrate that OSBPL2 deletion reduces the amount of COPB1 located on LDs and inhibits the establishment of the ER and LD membrane bridge, which in turn results in hindered ATGL transfer for TAG catabolism. In previous reports, nonbilayer lipids, such as cholesterol, can increase membrane tension, which was adverse to the egress of neutral lipids from the bilayer and LD budding (Gao et al., 2019). In our study, the absence of OSBPL2/ORP2 causing cholesterol upregulation can be viewed from another perspective to support the effect on lipolysis.
As the regional distribution and morphology of AT are known to be predictors of metabolic disease (Ahima and Lazar, 2013; Arner and Arner, 2013), our observations in vivo show exacerbated fat accumulation and edema with inflammation in the osbpl2b-deficient liver and an increase in SAT and an expansion of VAT. Generally, high-fat diet-induced fat accumulation increases VAT and is followed by fat redistribution into subcutaneous tissue and other organs. Here, the expansion of VAT suggests that osbpl2b deficiency aggravates the fat accumulation induced by a high-fat diet. SAT expansion was reported to predict impaired glucose metabolism, hyperinsulinemia, and insulin resistance (Ktotkiewski et al., 1975; Lundgren et al., 2007; Stern et al., 1972), and other studies have suggested that increased SAT may store potentially toxic lipids in obese individuals (Unger and Scherer, 2010). Given that zebrafish are known to be predictors by changes in behavior or ventilatory patterns due to their sensitivity to toxic substances (Oh et al., 2018; Wang et al., 2013), the abnormal behavior associated with osbpl2b deficiency that drove some zebrafish to live in the top portion of the water tank was possibly due to an increase in oxygen consumption and lipid toxicity in the liver. We had previously reported that osbpl2b is highly expressed in the central nervous system (CNS) of zebrafish. The markedly increased feeding and the altered swimming behavior of the osbpl2b−/- zebrafish most likely reflected functional impacts of the mutation on the CNS, the regions controlling appetite and satiety, and possibly the swimming behavior.
It has been reported that knocking down mysterin, which specifically eliminates ATGL from LDs, leads to a decrease in LDs in HeLa cells and a decrease in LDs in the head region of zebrafish (Sugihara et al., 2019). In our study, we found that LDs are enlarged in OSBPL2−/− cells, whereas VAT and SAT increase in the osbpl2b−/- zebrafish. We have also found that COPB1 attachment to LDs is reduced both in the OSBPL2−/− cells and in the osbpl2b−/− zebrafish to reduce ATGL localization to the LDs. The observation made via TEM in a previous study indicated that DFCP1 regulates the size of LDs and the formation of ER-LD contacts (Li et al., 2019). In our study, we demonstrated that ER-LD contacts are altered when OSBPL2−/− cells are treated with OA or when osbpl2b−/− zebrafish are fed a high-fat diet. Consistent with this in vitro data, we obtained a series of similar results in vivo, suggesting that OSBPL2 deficiency leads to an enhanced LD phenotype and impairment to ATGL localization on LDs at the molecular level.
Overall, this work presents the underlying molecular mechanism by which (1) OSBPL2/ORP2 binds to COPB1 in complex and maintains COPB1 located on LDs; (2) the OSBPL2-COPB1 complex mediates ATGL transfer from the ER to LDs; (3) ATGL wraps around the LDs during lipolysis; and (4) loss of OSBPL2/ORP2 enlarges LDs and changes lipid metabolism. Although this work has considerably improved our understanding of the function of OSBPL2/ORP2, OSBPL2/ORP2 regulates lipolysis and fat accumulation in human and may be the basis of therapeutic approaches that target OSBPL2 biology in the treatment of metabolic disease.
Limitations of the Study
Although OSBPL2/ORP2 has been proven to have multiple functions, including cholesterol transport, energy metabolism, and auditory function, the underlying molecular mechanism of OSBPL2/ORP2 is not fully understood. We acknowledge that the present study has several limitations, including the determination of the structure of the OSBPL2-COPB1 complex linking the ER and LDs. In addition, the abnormal zebrafish behaviors, such that the osbpl2b mutation causes an increase in food intake, are not understood. Future studies will focus on developing tools to determine the structure of the complex and to clarify the underlying reason for the altered zebrafish behavior.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Xin Cao (caoxin@njmu.edu.cn).
Materials Availability
Materials and protocols used in this study are available from the authors upon request.
Data and Code Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This research was supported by the grants from the National Natural Science Foundation of China (81771000, 31571302), the Key Project of Science and Technology Innovation of Nanjing Medical University (2017NJMUCX001), and the Project of the Laboratory Platform Construction of Department of Finance of Jiangsu Province to XC.
Author Contributions
T.W. and Q.W. contributed equally to the execution of most experiments and the writing of the manuscript. L.L. and X.T. performed immunofluorescence staining, LDs measurements, and the zebrafish husbandry. J.Y. and Y.L performed histochemistry and made the figures. Y.Q., Z.C., and G.X. contributed to the data analysis and chart making. X.C. was responsible for the overall conception and design of the study and for drafting and revising the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: July 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101252.
Supplemental Information
References
- Ahima R.S., Lazar M.A. Physiology. The health risk of obesity--better metrics imperative. Science. 2013;341:856–858. doi: 10.1126/science.1241244. [DOI] [PubMed] [Google Scholar]
- Antonny B., Bigay J., Mesmin B. The oxysterol-binding protein cycle: burning off PI(4)P to transport cholesterol. Annu. Rev. Biochem. 2018;87:809–837. doi: 10.1146/annurev-biochem-061516-044924. [DOI] [PubMed] [Google Scholar]
- Arner E., Arner P. Health and obesity: not just skin deep. Science. 2013;342:558–559. doi: 10.1126/science.342.6158.558-b. [DOI] [PubMed] [Google Scholar]
- Begriche K., Massart J., Robin M.A., Bonnet F., Fromenty B. Mitochondrial adaptations and dysfunctions in nonalcoholic fatty liver disease. Hepatology. 2013;58:1497–1507. doi: 10.1002/hep.26226. [DOI] [PubMed] [Google Scholar]
- Beller M., Sztalryd C., Southall N., Bell M., Jackle H., Auld D.S., Oliver B. COPI complex is a regulator of lipid homeostasis. Plos Biol. 2008;6:e292. doi: 10.1371/journal.pbio.0060292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben M'barek K., Ajjaji D., Chorlay A., Vanni S., Foret L., Thiam A.R. ER membrane phospholipids and surface tension control cellular lipid droplet formation. Dev. Cell. 2017;41:591–604 e597. doi: 10.1016/j.devcel.2017.05.012. [DOI] [PubMed] [Google Scholar]
- Bethune J., Wieland F.T. Assembly of COPI and COPII vesicular coat proteins on membranes. Annu. Rev. Biophys. 2018;47:63–83. doi: 10.1146/annurev-biophys-070317-033259. [DOI] [PubMed] [Google Scholar]
- Botha T.L., Brand S.J., Ikenaka Y., Nakayama S.M.M., Ishizuka M., Wepener V. How toxic is a non-toxic nanomaterial: behaviour as an indicator of effect in Danio rerio exposed to nanogold. Aquat. Toxicol. 2019;215:105287. doi: 10.1016/j.aquatox.2019.105287. [DOI] [PubMed] [Google Scholar]
- Bray G.A., Kim K.K., Wilding J.P.H., World Obesity F. Obesity: a chronic relapsing progressive disease process. A position statement of the World Obesity Federation. Obes. Rev. 2017;18:715–723. doi: 10.1111/obr.12551. [DOI] [PubMed] [Google Scholar]
- Choudhary V., Ojha N., Golden A., Prinz W.A. A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J. Cel. Biol. 2015;211:261–271. doi: 10.1083/jcb.201505067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mello A.H., Costa A.B., Engel J.D.G., Rezin G.T. Mitochondrial dysfunction in obesity. Life Sci. 2018;192:26–32. doi: 10.1016/j.lfs.2017.11.019. [DOI] [PubMed] [Google Scholar]
- Du X., Zhou L., Aw Y.C., Mak H.Y., Xu Y., Rae J., Wang W., Zadoorian A., Hancock S.E., Osborne B. ORP5 localizes to ER-lipid droplet contacts and regulates the level of PI(4)P on lipid droplets. J. Cell Biol. 2020;219:e201905162. doi: 10.1083/jcb.201905162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellong E.N., Soni K.G., Bui Q.T., Sougrat R., Golinelli-Cohen M.P., Jackson C.L. Interaction between the triglyceride lipase ATGL and the Arf1 activator GBF1. PLoS One. 2011;6:e21889. doi: 10.1371/journal.pone.0021889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W., Du X., Yang H. Seipin, adipogenesis and lipid droplets. Trends Endocrinol. Metab. 2011;22:204–210. doi: 10.1016/j.tem.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Gao M., Huang X., Song B.L., Yang H. The biogenesis of lipid droplets: lipids take center stage. Prog. Lipid Res. 2019;75:100989. doi: 10.1016/j.plipres.2019.100989. [DOI] [PubMed] [Google Scholar]
- Gross D.A., Silver D.L. Cytosolic lipid droplets: from mechanisms of fat storage to disease. Crit. Rev. Biochem. Mol. Biol. 2014;49:304–326. doi: 10.3109/10409238.2014.931337. [DOI] [PubMed] [Google Scholar]
- Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 2006;312:734–737. doi: 10.1126/science.1123965. [DOI] [PubMed] [Google Scholar]
- Hynynen R., Suchanek M., Spandl J., Back N., Thiele C., Olkkonen V.M. OSBP-related protein 2 is a sterol receptor on lipid droplets that regulates the metabolism of neutral lipids. J. Lipid Res. 2009;50:1305–1315. doi: 10.1194/jlr.M800661-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C.L. Lipid droplet biogenesis. Curr. Opin. Cell Biol. 2019;59:88–96. doi: 10.1016/j.ceb.2019.03.018. [DOI] [PubMed] [Google Scholar]
- Jacquier N., Choudhary V., Mari M., Toulmay A., Reggiori F., Schneiter R. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 2011;124:2424–2437. doi: 10.1242/jcs.076836. [DOI] [PubMed] [Google Scholar]
- Jamecna D., Polidori J., Mesmin B., Dezi M., Levy D., Bigay J., Antonny B. An intrinsically disordered region in OSBP acts as an entropic barrier to control protein dynamics and orientation at membrane contact sites. Dev. Cell. 2019;49:220–234 e228. doi: 10.1016/j.devcel.2019.02.021. [DOI] [PubMed] [Google Scholar]
- Joshi A.S., Zhang H., Prinz W.A. Organelle biogenesis in the endoplasmic reticulum. Nat. Cell Biol. 2017;19:876–882. doi: 10.1038/ncb3579. [DOI] [PubMed] [Google Scholar]
- Kaushik S., Cuervo A.M. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat. Cell Biol. 2015;17:759–770. doi: 10.1038/ncb3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentala H., Koponen A., Vihinen H., Pirhonen J., Liebisch G., Pataj Z., Kivela A., Li S., Karhinen L., Jaaskelainen E. OSBP-related protein-2 (ORP2): a novel Akt effector that controls cellular energy metabolism. Cell Mol. Life Sci. 2018;75:4041–4057. doi: 10.1007/s00018-018-2850-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kociucka B., Flisikowska T., Mroz D., Szczerbal I. Expression of genes involved in lipid droplet formation (BSCL2, SNAP23 and COPA) during porcine in vitro adipogenesis. J. Appl. Genet. 2016;57:505–510. doi: 10.1007/s13353-016-0350-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kory N., Farese R.V., Jr., Walther T.C. Targeting fat: mechanisms of protein localization to lipid droplets. Trends Cell Biol. 2016;26:535–546. doi: 10.1016/j.tcb.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ktotkiewski M., Sjostrom L., Bjorntorp P., Smith U. Regional adipose tissue cellularity in relation to metabolism in young and middle-aged women. Metabolism. 1975;24:703–710. doi: 10.1016/0026-0495(75)90038-4. [DOI] [PubMed] [Google Scholar]
- Lee M.C., Miller E.A., Goldberg J., Orci L., Schekman R. Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- Li D., Zhao Y.G., Li D., Zhao H., Huang J., Miao G., Feng D., Liu P., Li D., Zhang H. The ER-localized protein DFCP1 modulates ER-lipid droplet contact formation. Cell Rep. 2019;27:343–358 e345. doi: 10.1016/j.celrep.2019.03.025. [DOI] [PubMed] [Google Scholar]
- Liu C., Yao J., Wei Q., Xing G., Cao X. Spatial and temporal expression patterns of Osbpl2a and Osbpl2b during zebrafish embryonic development. Int. J. Pediatr. Otorhinolaryngol. 2016;84:174–179. doi: 10.1016/j.ijporl.2016.02.030. [DOI] [PubMed] [Google Scholar]
- Lundgren M., Svensson M., Lindmark S., Renstrom F., Ruge T., Eriksson J.W. Fat cell enlargement is an independent marker of insulin resistance and 'hyperleptinaemia. Diabetologia. 2007;50:625–633. doi: 10.1007/s00125-006-0572-1. [DOI] [PubMed] [Google Scholar]
- Mesmin B., Bigay J., Moser von Filseck J., Lacas-Gervais S., Drin G., Antonny B. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 2013;155:830–843. doi: 10.1016/j.cell.2013.09.056. [DOI] [PubMed] [Google Scholar]
- Miao H., Ou J., Ma Y., Guo F., Yang Z., Wiggins M., Liu C., Song W., Han X., Wang M. Macrophage CGI-58 deficiency activates ROS-inflammasome pathway to promote insulin resistance in mice. Cell Rep. 2014;7:223–235. doi: 10.1016/j.celrep.2014.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchin J.E., Dahlman I., Harvey C.J., Mejhert N., Singh M.K., Epstein J.A., Arner P., Torres-Vazquez J., Rawls J.F. Plexin D1 determines body fat distribution by regulating the type V collagen microenvironment in visceral adipose tissue. Proc. Natl. Acad. Sci. U S A. 2015;112:4363–4368. doi: 10.1073/pnas.1416412112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missaglia S., Coleman R.A., Mordente A., Tavian D. Neutral lipid storage diseases as cellular model to study lipid droplet function. Cell. 2019;8:187. doi: 10.3390/cells8020187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsholme P., Cruzat V.F., Keane K.N., Carlessi R., de Bittencourt P.I., Jr. Molecular mechanisms of ROS production and oxidative stress in diabetes. Biochem. J. 2016;473:4527–4550. doi: 10.1042/BCJ20160503C. [DOI] [PubMed] [Google Scholar]
- Oh H., Kim C.Y., Ryu B., Kim U., Kim J., Lee J.M., Lee B.H., Moon J., Jung C.R., Park J.H. Respiratory toxicity of polyhexamethylene guanidine phosphate exposure in zebrafish. Zebrafish. 2018;15:460–472. doi: 10.1089/zeb.2018.1571. [DOI] [PubMed] [Google Scholar]
- Ohsaki Y., Suzuki M., Fujimoto T. Open questions in lipid droplet biology. Chem. Biol. 2014;21:86–96. doi: 10.1016/j.chembiol.2013.08.009. [DOI] [PubMed] [Google Scholar]
- Olkkonen V.M., Koponen A., Arora A. OSBP-related protein 2 (ORP2): Unraveling its functions in cellular lipid/carbohydrate metabolism, signaling and F-actin regulation. J. Steroid Biochem. Mol. Biol. 2019;192:105298. doi: 10.1016/j.jsbmb.2019.01.016. [DOI] [PubMed] [Google Scholar]
- Onal G., Kutlu O., Gozuacik D., Dokmeci Emre S. Lipid droplets in health and disease. Lipids Health Dis. 2017;16:128. doi: 10.1186/s12944-017-0521-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M.J., Voeltz G.K. Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 2016;17:69–82. doi: 10.1038/nrm.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Kapterian T.S., Du X., Ma Q., Fei W., Zhang Y., Huang X., Dawes I.W., Yang H. CDP-diacylglycerol synthases regulate the growth of lipid droplets and adipocyte development. J. Lipid Res. 2016;57:767–780. doi: 10.1194/jlr.M060574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quijano C., Trujillo M., Castro L., Trostchansky A. Interplay between oxidant species and energy metabolism. Redox Biol. 2016;8:28–42. doi: 10.1016/j.redox.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.S. Forty years of clathrin-coated vesicles. Traffic. 2015;16:1210–1238. doi: 10.1111/tra.12335. [DOI] [PubMed] [Google Scholar]
- Ryan D.H., Kahan S. Guideline recommendations for obesity management. Med. Clin. North Am. 2018;102:49–63. doi: 10.1016/j.mcna.2017.08.006. [DOI] [PubMed] [Google Scholar]
- Schoenborn V., Heid I.M., Vollmert C., Lingenhel A., Adams T.D., Hopkins P.N., Illig T., Zimmermann R., Zechner R., Hunt S.C. The ATGL gene is associated with free fatty acids, triglycerides, and type 2 diabetes. Diabetes. 2006;55:1270–1275. doi: 10.2337/db05-1498. [DOI] [PubMed] [Google Scholar]
- Soni K.G., Mardones G.A., Sougrat R., Smirnova E., Jackson C.L., Bonifacino J.S. Coatomer-dependent protein delivery to lipid droplets. J. Cell Sci. 2009;122:1834–1841. doi: 10.1242/jcs.045849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern J.S., Batchelor B.R., Hollander N., Cohn C.K., Hirsch J. Adipose-cell size and immunoreactive insulin levels in obese and normal-weight adults. Lancet. 1972;2:948–951. doi: 10.1016/s0140-6736(72)92474-9. [DOI] [PubMed] [Google Scholar]
- Sugihara M., Morito D., Ainuki S., Hirano Y., Ogino K., Kitamura A., Hirata H., Nagata K. The AAA+ ATPase/ubiquitin ligase mysterin stabilizes cytoplasmic lipid droplets. J. Cell Biol. 2019;218:949–960. doi: 10.1083/jcb.201712120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J.S., Seow C.J., Goh V.J., Silver D.L. Recent advances in understanding proteins involved in lipid droplet formation, growth and fusion. J. Genet. Genomics. 2014;41:251–259. doi: 10.1016/j.jgg.2014.03.003. [DOI] [PubMed] [Google Scholar]
- Thiam A.R., Antonny B., Wang J., Delacotte J., Wilfling F., Walther T.C., Beck R., Rothman J.E., Pincet F. COPI buds 60-nm lipid droplets from reconstituted water-phospholipid-triacylglyceride interfaces, suggesting a tension clamp function. Proc. Natl. Acad. Sci. U S A. 2013;110:13244–13249. doi: 10.1073/pnas.1307685110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoenes M., Zimmermann U., Ebermann I., Ptok M., Lewis M.A., Thiele H., Morlot S., Hess M.M., Gal A., Eisenberger T. OSBPL2 encodes a protein of inner and outer hair cell stereocilia and is mutated in autosomal dominant hearing loss (DFNA67) Orphanet J. Rare Dis. 2015;10:15. doi: 10.1186/s13023-015-0238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger R.H., Scherer P.E. Gluttony, sloth and the metabolic syndrome: a roadmap to lipotoxicity. Trends Endocrinol. Metab. 2010;21:345–352. doi: 10.1016/j.tem.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.C., Chung J., Farese R.V., Jr. Lipid droplet biogenesis. Annu. Rev. Cell Dev. Biol. 2017;33:491–510. doi: 10.1146/annurev-cellbio-100616-060608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.C., Farese R.V., Jr. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Liang Y., Li S., Chang J. Acute toxicity, respiratory reaction, and sensitivity of three cyprinid fish species caused by exposure to four heavy metals. PLoS One. 2013;8:e65282. doi: 10.1371/journal.pone.0065282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Lin C., Yao J., Shi H., Zhang C., Wei Q., Lu Y., Chen Z., Xing G., Cao X. Deletion of OSBPL2 in auditory cells increases cholesterol biosynthesis and drives reactive oxygen species production by inhibiting AMPK activity. Cell Death Dis. 2019;10:627. doi: 10.1038/s41419-019-1858-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ma Q., Qi Y., Dong J., Du X., Rae J., Wang J., Wu W.F., Brown A.J., Parton R.G. ORP2 delivers cholesterol to the plasma membrane in exchange for phosphatidylinositol 4, 5-bisphosphate (PI(4,5)P2) Mol. Cell. 2019;73:458–473 e457. doi: 10.1016/j.molcel.2018.11.014. [DOI] [PubMed] [Google Scholar]
- Wang J., Wang X., Xiong C., Liu J., Hu B., Zheng L. Chronic bisphenol A exposure alters behaviors of zebrafish (Danio rerio) Environ. Pollut. 2015;206:275–281. doi: 10.1016/j.envpol.2015.07.015. [DOI] [PubMed] [Google Scholar]
- Wang Q., Lin C., Zhang C., Wang H., Lu Y., Yao J., Wei Q., Xing G., Cao X. 25-hydroxycholesterol down-regulates oxysterol binding protein like 2 (OSBPL2) via the p53/SREBF2/NFYA signaling pathway. J. Steroid Biochem. Mol. Biol. 2019;187:17–26. doi: 10.1016/j.jsbmb.2018.10.018. [DOI] [PubMed] [Google Scholar]
- Wilfling F., Thiam A.R., Olarte M.J., Wang J., Beck R., Gould T.J., Allgeyer E.S., Pincet F., Bewersdorf J., Farese R.V., Jr. Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. Elife. 2014;3:e01607. doi: 10.7554/eLife.01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F., Wang H., Haas J.T., Krahmer N., Gould T.J., Uchida A., Cheng J.X., Graham M., Christiano R., Frohlich F. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell. 2013;24:384–399. doi: 10.1016/j.devcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Husile H., Yang L., Cao Y., Li X., Huo W., Bai H., Liu Y., Wu Q. A novel pathogenic variant in OSBPL2 linked to hereditary late-onset deafness in a Mongolian family. BMC Med. Genet. 2019;20:43. doi: 10.1186/s12881-019-0781-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing G., Yao J., Wu B., Liu T., Wei Q., Liu C., Lu Y., Chen Z., Zheng H., Yang X. Identification of OSBPL2 as a novel candidate gene for progressive nonsyndromic hearing loss by whole-exome sequencing. Genet. Med. 2015;17:210–218. doi: 10.1038/gim.2014.90. [DOI] [PubMed] [Google Scholar]
- Xu D., Li Y., Wu L., Li Y., Zhao D., Yu J., Huang T., Ferguson C., Parton R.G., Yang H. Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions. J. Cell Biol. 2018;217:975–995. doi: 10.1083/jcb.201704184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Zeng H., Zhang M., Wei Q., Wang Y., Yang H., Lu Y., Li R., Xiong Q., Zhang L. OSBPL2-disrupted pigs recapitulate dual features of human hearing loss and hypercholesterolaemia. J. Genet. Genomics. 2019;46:379–387. doi: 10.1016/j.jgg.2019.06.006. [DOI] [PubMed] [Google Scholar]
- Yu X., Breitman M., Goldberg J. A structure-based mechanism for Arf1-dependent recruitment of coatomer to membranes. Cell. 2012;148:530–542. doi: 10.1016/j.cell.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner R., Madeo F., Kratky D. Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat. Rev. Mol. Cell Biol. 2017;18:671–684. doi: 10.1038/nrm.2017.76. [DOI] [PubMed] [Google Scholar]
- Zechner R., Zimmermann R., Eichmann T.O., Kohlwein S.D., Haemmerle G., Lass A., Madeo F. FAT SIGNALS--lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15:279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Zhang H., Zhang M., Lin C., Wang H., Yao J., Wei Q., Lu Y., Chen Z., Xing G. OSBPL2 deficiency upregulate SQLE expression increasing intracellular cholesterol and cholesteryl ester by AMPK/SP1 and SREBF2 signalling pathway. Exp. Cell Res. 2019;383:111512. doi: 10.1016/j.yexcr.2019.111512. [DOI] [PubMed] [Google Scholar]
- Zweytick D., Athenstaedt K., Daum G. Intracellular lipid particles of eukaryotic cells. Biochim. Biophys. Acta. 2000;1469:101–120. doi: 10.1016/s0005-2736(00)00294-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The WT-HFD group (left) and MT-HFD group (right) were maintained in buffered reverse osmosis water.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.