Here, we report the complete-genome assemblies of biofilm isolates 201A and 204H. They possess six and seven plasmids, respectively, with a size ranging from 44 kb to 159 kb. Genomic comparisons place the two strains into one new species belonging to the genus Leisingera as novel representatives of the Roseobacter group.
ABSTRACT
Here, we report the complete-genome assemblies of biofilm isolates 201A and 204H. They possess six and seven plasmids, respectively, with a size ranging from 44 kb to 159 kb. Genomic comparisons place the two strains into one new species belonging to the genus Leisingera as novel representatives of the Roseobacter group.
ANNOUNCEMENT
The novel isolates 201A and 204H were originally isolated from biofilms near tubes of the polychaete Hydroides elegans (Marina del Rey, CA, USA). Samples were cultured in artificial seawater tryptone (ASWT) agar medium (1) and incubated at 28°C for 48 h. The genome sequences of the isolates 201A and 204H place the two strains into one novel species within the genus Leisingera (Fig. 1), belonging to the Roseobacter lineage (Rhodobacteraceae), a widespread Alphaproteobacteria group (2, 3). Other Roseobacter isolates were shown to induce the metamorphosis of H. elegans (4) and the coral Porites astroides (5). These novel isolates recovered from biofilms near tubeworms, along with their relatedness to bacterial species known to induce animal metamorphosis, present interesting future investigation targets (6).
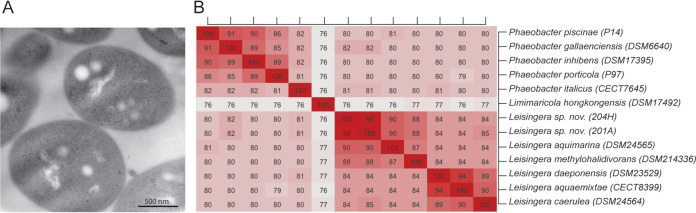
FIG 1.
(A) Transmission electron micrograph of Leisingera sp. nov. strain 201A. (B) Average nucleotide identity (ANI) genome-based distance matrix of Leisingera sp. strains 201A and 204H relative to available Leisingera species genomes and closely related Roseobacter species. All values are given in percentages.
Genomic DNA from isolates 201A and 204H was extracted with a Zymo fungal/bacterial DNA miniprep kit and submitted to McGill University and Genome Quebec Innovation Centre (Montreal, Quebec, Canada) for Pacific Biosciences RS II sequencing (7). The DNA libraries were prepared following the Pacific Biosciences 20-kb template preparation with the SMRTbell template prep kit 1.0 reagents (Pacific Biosciences, Menlo Park, CA, USA). Large-insert DNA (≥20 kb) was sheared using g-TUBEs (Covaris, Inc., Woburn, MA, USA) and size selected using a BluePippin system (Sage Science, Inc., Beverly, MA, USA) following the manufacturer’s recommendations. Libraries were sequenced using the DNA sequencing kit 4.0 v2 and single-molecule real-time (SMRT) cells v3. Reads were filtered and assembled using SMRT Analysis v2.3 (PacBio) with default settings. We obtained a total of 77,942 reads covering a total of 797 Mb for isolate 201A and 70,939 reads totaling 709 Mb for isolate 204H. The mean subread length and N50 value were 8,299 bp and 12,584 bp, respectively. The final assembly performed using the HGAP workflow (8) resulted in a gapless circular chromosome (4,382,007 nucleotides [nt]) and six different closed circular plasmids (159,657 nt, 144,623 nt, 137,742 nt, 128,224 nt, 113,581 nt, and 71,922 nt) for strain 201A and a circular chromosome (4,246,063 nt) and seven associated gapless circular plasmids (151,185 nt, 148,812 nt, 140,681 nt, 124,119 nt, 122,006 nt, 59,982 nt, and 44,085 nt) for strain 204H. For each contig, ends were compared using BLAST (9) to (i) identify overlaps and (ii) confirm the uniqueness of the sequence to that locus. Edges were trimmed manually. The 5.1-Mb genomes had a total GC content of 62% with 72 RNAs for both strains. Annotation was performed with the Rapid Annotations using Subsystems Technology (RAST) server (10). RAST predicted 5,606 and 5,188 coding sequences for isolates 201A and 204H, respectively.
The novel isolates 201A and 204H had less than a 95% average amino acid identity/average nucleotide identity (AAI/ANI) and less than a 70% genome-to-genome distance (GGDH) from their closest neighbor, Leisingera methylohalidivorans (11, 12) (Fig. 1). The cutoffs used for the delimitation of bacterial species are more than 95% AAI and more than 70% GGDH (13). We conclude that the two isolates belong to a novel species of the genus Leisingera.
Data availability.
The genome sequencing and assembly projects for Leisingera sp. nov. strains 201A and 204H have been deposited in DDBJ/EMBL/GenBank under accession number PRJNA515513. The raw sequencing data have been deposited under SRA accession numbers SRS6579670 and SRS6579671, respectively.
ACKNOWLEDGMENTS
We gratefully acknowledge the assistance of Ingrid Niesman, director of the SDSU EM facility, for helping with the microbial transmission electron microscopy (TEM). We thank Pacific Biosciences for donating reagents for portions of this work conducted at the 2015 and 2016 Cold Spring Harbor Laboratory Course “Advanced Sequencing Technologies and Applications.” We also thank the sequencing group at the McGill University and Genome Quebec Innovation Centre.
This material is based upon work supported by the National Science Foundation under grant number 1942251 and supported by funds provided by the Office of Naval Research (N00014-14-1-0340 and N00014-16-1-2135 to N.J.S.), Alfred P. Sloan Foundation, Sloan Research Fellowship (to N.J.S.), and San Diego State University (to N.J.S.).
REFERENCES
- 1.Shikuma NJ, Pilhofer M, Weiss GL, Hadfield MG, Jensen GJ, Newman DK. 2014. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science 343:529–533. doi: 10.1126/science.1246794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo H, Moran MA. 2014. Evolutionary ecology of the marine Roseobacter clade. Microbiol Mol Biol Rev 78:573–587. doi: 10.1128/MMBR.00020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner-Döbler I, Biebl H. 2006. Environmental biology of the marine Roseobacter lineage. Annu Rev Microbiol 60:255–280. doi: 10.1146/annurev.micro.60.080805.142115. [DOI] [PubMed] [Google Scholar]
- 4.Lau SCK, Tsoi MMY, Li X, Plakhotnikova I, Wu M, Wong P-K, Qian P-Y. 2004. Loktanella hongkongensis sp. nov., a novel member of the α-Proteobacteria originating from marine biofilms in Hong Kong waters. Int J Syst Evol Microbiol 54:2281–2284. doi: 10.1099/ijs.0.63294-0. [DOI] [PubMed] [Google Scholar]
- 5.Sharp KH, Sneed JM, Ritchie KB, McDaniel L, Paul VJ. 2015. Induction of larval settlement in the reef coral Porites astreoides by a cultivated marine Roseobacter strain. Biol Bull 228:98–107. doi: 10.1086/BBLv228n2p98. [DOI] [PubMed] [Google Scholar]
- 6.Cavalcanti GS, Alker AT, Delherbe N, Malter KE, Shikuma NJ. The influence of bacteria on animal metamorphosis. Annu Rev Microbiol, in press. [DOI] [PubMed] [Google Scholar]
- 7.Rhoads A, Au KF. 2015. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics 13:278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 9.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 10.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:206–214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez-R LM, Konstantinidis KT. 2014. Bypassing cultivation to identify bacterial species. Microbe 9:111–118. doi: 10.1128/microbe.9.111.1. [DOI] [Google Scholar]
- 12.Rodriguez-R L, Konstantinidis K. 2016. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr doi: 10.7287/peerj.preprints.1900v1. [DOI] [Google Scholar]
- 13.Thompson CC, Chimetto L, Edwards R. a, Swings J, Stackebrandt E, Thompson FL. 2013. Microbial genomic taxonomy. BMC Genomics 14:913. doi: 10.1186/1471-2164-14-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequencing and assembly projects for Leisingera sp. nov. strains 201A and 204H have been deposited in DDBJ/EMBL/GenBank under accession number PRJNA515513. The raw sequencing data have been deposited under SRA accession numbers SRS6579670 and SRS6579671, respectively.