A coding-complete genome sequence of a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) isolate was revealed. The sample for the virus was isolated from a female patient from Dhaka, Bangladesh, suffering from coronavirus disease-2019 (COVID-19).
ABSTRACT
A coding-complete genome sequence of a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) isolate was revealed. The sample for the virus was isolated from a female patient from Dhaka, Bangladesh, suffering from coronavirus disease-2019 (COVID-19).
ANNOUNCEMENT
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a member of the Coronaviridae family and Betacoronavirus genus, is the causative agent of pandemic coronavirus disease-2019 (COVID-19). In Bangladesh, the rate of positive cases and the death toll from COVID-19 are increasing at an alarming rate (https://corona.gov.bd/). To understand the genomic characteristics of SARS-CoV-2 in Bangladesh, several isolates have been sequenced and deposited in GISAID (https://www.gisaid.org/). However, those isolates have been sequenced using a next-generation sequencing platform, except for the one we are reporting. In this study, we sequenced the viral genome by Sanger sequencing technology, which is a gold standard method and is necessary for thorough genomic analysis (1).
The isolate (SARS-CoV-2/human/BGD/NIB_01/2020) was collected from an oropharyngeal specimen on 11 May 2020. The patient was a 28-year-old saleswoman who tested positive (via reverse transcriptase PCR [RT-PCR]) for COVID-19 with symptoms of cough, mild fever, and throat congestion. (all applicable international, national, and/or institutional guidelines for the care and use of animals were followed; ethical approval number NIBREC2020-01). The viral RNA was extracted directly from the patient’s specimen using the PureLink viral RNA/DNA minikit (Invitrogen). The viral RNA was then converted into cDNA using a SuperScript VILO cDNA synthesis kit (Invitrogen).
To cover the whole genome of the virus, 48 pairs of primers were designed by following two conditions: (i) their sequence is conserved among all the available SARS-CoV-2 isolates, and (ii) the terminal of the amplicons will overlap the adjacent amplicon (Table 1). These primers underwent PCR and generated 96 amplicons, which were visualized using 1.5% agarose gel electrophoresis. The PCR products were then purified using the PureLink PCR purification kit (Thermo Fisher Scientific, USA). These purified amplicons were finally sequenced with 2× coverage using the Sanger dideoxy method by “ABI 3500” with a BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems, USA).
TABLE 1.
Information about primers, amplicon size, and overlapping length
| Amplicon | Primer | Sequence | Product size (bp) | Overlapping length (bp) |
|---|---|---|---|---|
| 1 | Forward | AGGTTTATACCTTCCCAGG | 765 | 131 |
| 1 | Reverse | CACCACTGCTATGTTTAGTG | ||
| 2 | Forward | CGCAAGGTTCTTCTTCGTA | 797 | |
| 2 | Reverse | AGACTATGCTCAGGTCCTAC | 100 | |
| 3 | Forward | AAGAAGGTGCCACTACTTG | 794 | |
| 3 | Reverse | GTTAGTTAGCCACTGCGAA | 114 | |
| 4 | Forward | ACTGAGACTCATTGATGCTA | 788 | |
| 4 | Reverse | TCACACTCTTGTAACCTTGC | 130 | |
| 5 | Forward | AGAAAAGTACTGTGCCCTTG | 783 | |
| 5 | Reverse | ACCACTGTTGGTTTTACCTT | 131 | |
| 6 | Forward | GATGGAACTTACACCAGTTG | 781 | |
| 6 | Reverse | GTCACTAACAAGAGTGGCAG | 110 | |
| 7 | Forward | GAAGAAGTTACAACAACTCTGG | 718 | |
| 7 | Reverse | AAACTGTAGCTGGCACTTTG | 136 | |
| 8 | Forward | TGTAGCGTCACTTATCAACA | 746 | |
| 8 | Reverse | CAGTGGCAAGATAACAGTTG | 158 | |
| 9 | Forward | CTCTACGTGTTGAGGCTTTT | 720 | |
| 9 | Reverse | CATCCGTAATAGGACCTTTGT | 130 | |
| 10 | Forward | TTGTGCTAGTGAGTACACTG | 760 | |
| 10 | Reverse | AATGTCTCCTACAACTTCGG | 150 | |
| 11 | Forward | TGATGTACTGAAGTCAGAGG | 737 | |
| 11 | Reverse | AATAGCCTTCTCTGTAACCAG | 90 | |
| 12 | Forward | TTCTTTAATCTACTCAACCGC | 706 | |
| 12 | Reverse | CTGTAGTGACAAGTCTCTCG | 118 | |
| 13 | Forward | ATGCTAATGGAGGTAAAGGC | 701 | |
| 13 | Reverse | ACAACTATCGCCAGTAACTTC | 115 | |
| 14 | Forward | CTTTTATTTCAGCAGCTCGG | 714 | |
| 14 | Reverse | GTGCGTAATATCGTGCCA | 133 | |
| 15 | Forward | GCTGATTTTGACACATGGTT | 812 | |
| 15 | Reverse | GGTAAGAATGAGTAAACTGGTG | 196 | |
| 16 | Forward | CCTATTGGTGCTTTGGACATA | 727 | |
| 16 | Reverse | AACCCTCAACTTTACCAGATG | 146 | |
| 17 | Forward | CTTGTTGTCATCTCGCAAAG | 767 | |
| 17 | Reverse | TCGATTGAGAAACCACCTGT | 112 | |
| 18 | Forward | TTGTTGACAGGCAAACAGC | 770 | |
| 18 | Reverse | ACCATCATCATACACAGTTCT | 121 | |
| 19 | Forward | TGACATGGTTGGATATGGTTG | 794 | |
| 19 | Reverse | GTTTATGTCTACAGCACCCT | 172 | |
| 20 | Forward | AATTGTGGGCTCAATGTGT | 787 | |
| 20 | Reverse | GCAACAGGACTAAGCTCATTA | 155 | |
| 21 | Forward | GGAAATCCAACAGGTTGTAGA | 795 | |
| 21 | Reverse | ACAGGGTCATTAGCACAAGT | 90 | |
| 22 | Forward | GTTGCCACATAGATCATCCAA | 790 | |
| 22 | Reverse | AACAATACCAGCATTTCGC | 233 | |
| 23 | Forward | GCAGACCTCGTCTATGCTTT | 813 | |
| 23 | Reverse | GCACGTAGTGCGTTTATCT | 147 | |
| 24 | Forward | CCACTTCAGAGAGCTAGGTG | 782 | |
| 24 | Reverse | GTGAGGGTTTTCTACATCACT | 114 | |
| 25 | Forward | ATTGAAATCAATAGCCGCCA | 775 | |
| 25 | Reverse | ATCTGGGTAAGGAAGGTACA | 117 | |
| 26 | Forward | GTCTGAAGCAAAATGTTGGA | 805 | |
| 26 | Reverse | GAGTCTTTCAGTACAGGTGTT | 142 | |
| 27 | Forward | TGTGTGCTAATGGACAAGTT | 784 | |
| 27 | Reverse | TCAAAACACTCTACACGAGC | 132 | |
| 28 | Forward | CTTCTGCTCGCATAGTGTAT | 769 | |
| 28 | Reverse | CAAGAGTGAGCTGTTTCAGT | 191 | |
| 29 | Forward | AATAGGCGTGGTAAGAGAAT | 790 | |
| 29 | Reverse | GTACATAAGTGGTATGAGGTGT | 139 | |
| 30 | Forward | AGCTAGGTTTTTCTACAGGTG | 756 | |
| 30 | Reverse | CTTTGTCACTACAAGGCTGT | 152 | |
| 31 | Forward | GTAGAAAGGTTCAACACATGG | 733 | |
| 31 | Reverse | ATAGAAACTGGTACTTCACCC | 144 | |
| 32 | Forward | GCTTTAGCTTGTGGGTTTAC | 808 | |
| 32 | Reverse | CCACCTAACTGACTATGACT | 139 | |
| 33 | Forward | CAAGAATTTAAACCCAGGAG | 758 | |
| 33 | Reverse | GCATCAGAGACAAAGTCATT | 155 | |
| 34 | Forward | CACATTAACATTAGCTGTACCC | 781 | |
| 34 | Reverse | TGACTAGAGACTAGTGGCA | 182 | |
| 35 | Forward | AAGGGGTACTGCTGTTATGT | 775 | |
| 35 | Reverse | TTAATAGGCGTGTGCTTAGA | 116 | |
| 36 | Forward | TCAGCCTTTTCTTATGGACC | 794 | |
| 36 | Reverse | TCCAAGCTATAACGCAGC | 104 | |
| 37 | Forward | TTAGAGGTGATGAAGTCAGA | 760 | |
| 37 | Reverse | TGTTCAGCCCCTATTAAACA | 149 | |
| 38 | Forward | TAACCAGGTTGCTGTTCTTT | 797 | |
| 38 | Reverse | CAATCATTTCATCTGTGAGCA | 191 | |
| 39 | Forward | CAGATCCATCAAAACCAAGC | 771 | |
| 39 | Reverse | GCAAGAAGACTACACCATGA | 137 | |
| 40 | Forward | TCAGAGCTTCTGCTAATCTTG | 759 | |
| 40 | Reverse | GTAATTTGACTCCTTTGAGC | 137 | |
| 41 | Forward | TTGCCATAGTAATGGTGACA | 798 | |
| 41 | Reverse | AGCTGGTAATAGTCTGAAGTG | 120 | |
| 42 | Forward | GCACAACAAGTCCTATTTCT | 784 | |
| 42 | Reverse | CCATAACAGCCAGAGGAAAA | 170 | |
| 43 | Forward | GCAGATTCCAACGGTACT | 707 | |
| 43 | Reverse | TAGTAACCTGAAAGTCAACG | 117 | |
| 44 | Forward | GCTACAGGATTGGCAACTAT | 785 | |
| 44 | Reverse | TTTCATGTTCGTTTAGGCGT | 174 | |
| 45 | Forward | CACTTTGCTTCACACTCAAA | 791 | |
| 45 | Reverse | TCTGGACTGCTATTGGTGTT | 180 | |
| 46 | Forward | CAGATTCAACTGGCAGTAAC | 793 | |
| 46 | Reverse | TTTCCTTGGGTTTGTTCTGG | 187 | |
| 47 | Forward | CTGCTTGACAGATTGAACCA | 698 | |
| 47 | Reverse | CTTGTGCTATGTAGTTACGAGA | 242 | |
| 48 | Forward | ATGAAACTCAAGCCTTACCG | 518 | |
| 48 | Reverse | CCTTTCGTGCAGGTCAATA |
The raw reads were assembled using DNA Sequence Assembler version 4 (2013) (Heracle BioSoft) and verified with SeqMan Pro version 14.1 (DNAStar, Madison, WI). After assembly, 48 contigs with 94 overlapping regions were obtained. These overlapping regions were visualized using CLC Genomics Workbench version 20.0.4 and merged with EMBOSS: merger (2).
The assembled viral genome consists of a single-stranded positive (+) RNA that is 29,724 nucleotides long. The NCBI BLASTN program (3) showed that the genome was mostly similar to SARS-CoV-2/human/BGD/CHRF_0001/2020 (GenBank accession number MT476385.1). From NCBI, the FASTA sequences of 7 mostly similar genomes from Bangladesh, India, Sri Lanka, and the United States were taken along with the reference genome. Another 16 genomes of SARS-CoV-2 that were isolated in Bangladesh were collected from GISAID (https://www.gisaid.org/). The genomes were aligned with MAFFT version 7 using default parameters (4). The phylogenetic tree was constructed using FastTree version 2.1.10 (5) through the Galaxy platform (6). Here, the tree was built by nucleotide alignment using the generalized time-reversable model (GTR) plus the CAT nucleotide evolution model (GTR+CAT). The tree was visualized using iTOL (7), where the tree structure was rerooted on the position of reference isolate SARS-CoV-2 Wuhan-Hu-1.
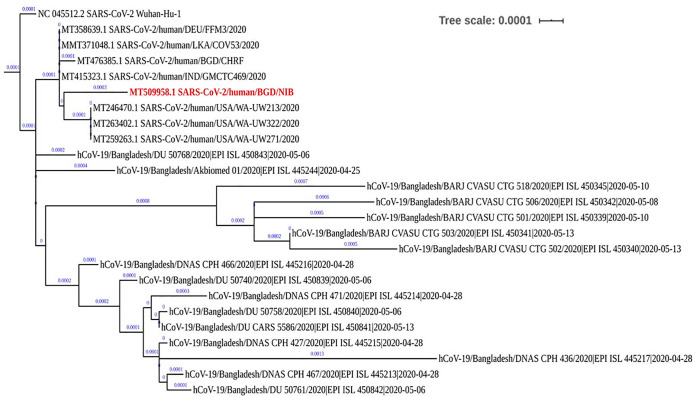
The genome has 8 nucleotide differences from the closest isolate. Interestingly, except for isolate SARS-CoV-2/human/BGD/CHRF0001/2020, the other strains of SARS-CoV-2 from Bangladesh showed separate clades and distant genetic relations. The tree also demonstrated that our viral genome and three isolates from the United States share an ancestor (Fig. 1).
FIG 1.
Phylogenetic analysis of the SARS-CoV-2/human/BGD/NIB_01/2020 isolate. Nucleotide alignment and the GTR+CAT nucleotide evolution model was applied to construct the tree. The tree was visualized using i-TOL. Here, the x axis represents the tree scale. A scale bar with a 0.0001 value is given on the top. The genome (labeled in red) shares a common ancestor with some isolates from the United States.
Data availability.
The complete nucleotide sequence of this SARS-CoV-2 isolate (SARS-CoV-2/human/BGD/NIB_01/2020) has been deposited in GenBank under the accession number MT509958.
ACKNOWLEDGMENTS
We are grateful to the Ministry of Science and Technology for its extensive support during this research work.
This study was funded by the National Institute of Biotechnology, Ministry of Science and Technology, government of the People’s Republic of Bangladesh.
REFERENCES
- 1.Besser J, Carleton HA, Gerner-Smidt P, Lindsey RL, Trees E. 2018. Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clin Microbiol Infect 24:335–341. doi: 10.1016/j.cmi.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Z, Schwartz S, Wagner L, Miller W. 2000. A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 4.Kazutaka K, John R, Kazunori DY. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2: approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Grüning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D. 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46:W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Letunic I, Bork P. 2019. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete nucleotide sequence of this SARS-CoV-2 isolate (SARS-CoV-2/human/BGD/NIB_01/2020) has been deposited in GenBank under the accession number MT509958.