We announce the genome sequences of two strains of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) isolated in Israel, one imported by a traveler who returned from Japan and the second strain collected from a patient infected by a traveler returning from Italy. The sequences obtained are valuable as early manifestations for future follow-up of the local spread of the virus in Israel.
ABSTRACT
We announce the genome sequences of two strains of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) isolated in Israel, one imported by a traveler who returned from Japan and the second strain collected from a patient infected by a traveler returning from Italy. The sequences obtained are valuable as early manifestations for future follow-up of the local spread of the virus in Israel.
ANNOUNCEMENT
Since its first occurrence in China, in December 2019 (1, 2), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), belonging to the Coronaviridae family and the Betacoronavirus genus, has been extensively spreading to the rest of the world. The first cases were imported and identified in Israel in late February 2020 with the arrival of passengers from the Diamond Princess cruise ship. Since then, over 20,000 cases (as of June 2020) have been reported in Israel.
In this report, we describe the sequencing of two strains isolated from two subjects, one traveler who returned to Israel from Japan and a patient who was infected by a traveler returning from Italy. These subjects are among the first confirmed coronavirus disease 2019 (COVID-19) cases in Israel. This study was reviewed and approved by the Institutional Review Board at the Chaim Sheba Medical Center. The two patients showed mild symptoms and recovered within a few weeks. The patients were initially identified as positive for COVID-19 by reverse transcriptase quantitative PCR (RT-qPCR). The samples were collected directly from nasopharyngeal swabs. Tubes containing the swabs were vortexed for 2 min, and samples from the transfer buffer were overlaid on Vero E6 cell monolayers (ATCC CRL-1586, 37°C, 5% CO2). After 4 days, RNA was purified from 140 μl of the cell culture supernatant using a QIAamp viral RNA minikit (Qiagen) according to the manufacturer’s protocol. A SMARTer stranded total RNA-Seq kit v2-pico input mammalian (TaKaRa Bio) was used for library construction prior to sequencing. Whole-genome paired-end (PE) sequencing was conducted on a MiSeq instrument using the 300-PE v2 kit (Illumina) with a read length of 150 nucleotides. This produced 1,522,094 and 7,149,870 reads for the Japanese and Italian strains (here, denoted ISR_JP0320 and ISR_IT0320), respectively. FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc) with default settings was used for quality control of the data. Trimming and removal of low-quality reads was performed using Trim Galore! v0.6.3 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with default settings. Reads originating from the Vero E6 host cells were filtered out using Bowtie 2 (3) with default parameters, resulting in 20,218 and 226,488 reads for the ISR_JP0320 and ISR_IT0320 strains, respectively. Genome assembly was performed using SPAdes v3.13.0 (4) with default parameters and produced a single contig for each strain, consisting of 29,851 and 29,870 nucleotides for the ISR_JP0320 and ISR_IT0320 strains, respectively. The overall G+C contents of the sequences were 38.02% and 38.05% for the ISR_JP0320 and ISR_IT0320 strains, respectively. The average coverages were 150× and 932× for the ISR_JP0320 and ISR_IT0320 strains, respectively. Mapping of the resulting reads was conducted using Bowtie 2 (3) against the reference Wuhan strain (GenBank accession number NC_045512) and revealed the existence of 3 mutations for the ISR_JP0320 strain (T5137→C, G11083→T, and G15438→A) and 8 mutations for the ISR_IT0320 strain (C241→T, C313→T, C3037→T, C14408→T, A23403→G, G28881→A, G28882→A, and G28883→C). The Italian strain harbors a documented replacement of D614→G in the spike protein, a replacement which is typical of European strains (https://nextstrain.org/ncov), compared with the strain imported from Japan.
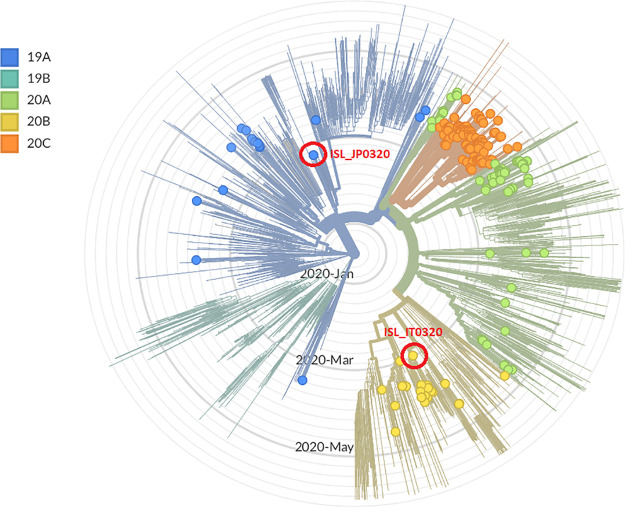
The radial presentation of a phylogenetic tree generated by Nextstrain (5) is presented in Fig. 1 for all the Asian strains available to date in the Nextstrain data set. The phylogeny is rooted relative to early samples from Wuhan, China. The tree displays the two sequenced strains in the context of over 200 sequences of strains collected in Israel in the past 2 months, colored by the different clades designated for SARS-CoV-2 (6). The ISR_JP0320 strain was identified as belonging to clade 19A (Asia), while the ISR_IT0320 strain was identified as belonging to the 20B clade (Europe). These strain sequences represent an early manifestation of the virus in Israel and are therefore important as starting points for studying the epidemiology and evolution of the virus and its circulation in Israel.
FIG 1.
Phylogenetic tree displaying sequenced SARS-CoV-2 strains, according to the designated clades of the virus. The tree was generated by the Nextstrain site using the bioinformatics toolkit Augur, which implements FastTree, which infers approximately maximum-likelihood phylogenetic trees from alignments of nucleotide or protein sequences (5, 7). All strains are colored by their assigned clade, according to the nomenclature established recently in reference 6. Background, strains of the Asia data set available in Nextstrain; dots, all strains collected in Israel; circled in red, the two strains documented in the current study, ISR_JP0320 and ISR_IT0320.
Data availability.
These sequences have been deposited at the GISAID EpiCoV coronavirus SARS-CoV-2 platform database under identifiers EPI_ISL_419211 and EPI_ISL_419210 and in the NCBI GenBank database under the accession numbers MT276597 and MT276598. The raw reads have been submitted to the NCBI Sequence Read Archive under the accession numbers PRJNA635017 and PRJNA626526.
REFERENCES
- 1.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bankevich A, Nurk S, Antipov D, Gurevich A, Dvorkin M, Kulikov AS, Lesin V, Nikolenko S, Pham S, Prjibelski A, Pyshkin A, Sirotkin A, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA. 2018. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rambaut A, Holmes AC, Hill V, O’Toole A, McCrone JT, Ruis C, Du Plessis L, Pybus OG. 2020. A dynamic nomenclature proposal for SARS-CoV-2 to assist genomic epidemiology. bioRxiv https://www.biorxiv.org/content/10.1101/2020.04.17.046086v1. [DOI] [PMC free article] [PubMed]
- 7.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2: approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
These sequences have been deposited at the GISAID EpiCoV coronavirus SARS-CoV-2 platform database under identifiers EPI_ISL_419211 and EPI_ISL_419210 and in the NCBI GenBank database under the accession numbers MT276597 and MT276598. The raw reads have been submitted to the NCBI Sequence Read Archive under the accession numbers PRJNA635017 and PRJNA626526.