Peptidoglycan is a conserved extracellular macromolecule that protects bacterial cells from turgor pressure. Peptidoglycan of Gram-positive bacteria serves as a scaffold for the attachment of polymers that provide defined bacterial interactions with their environment. One such polymer, B. anthracis SCWP, is pyruvylated at its distal end to serve as a receptor for secreted proteins bearing the S-layer homology domain. Repeat units of SCWP carry three galactoses in B. anthracis. Glycosylation is a recurring theme in nature and often represents a means to mask or alter conserved molecular signatures from intruders such as bacteriophages. Several glycosyltransferase families have been described based on bioinformatics prediction, but few have been studied. Here, we describe the glycosyltransferases that mediate the galactosylation of B. anthracis SCWP.
KEYWORDS: Bacillus anthracis, Gal substitution, envelope biogenesis, glycosyltransferase, secondary cell wall polymer, undecaprenol
ABSTRACT
Bacillus anthracis, the causative agent of anthrax disease, elaborates a secondary cell wall polysaccharide (SCWP) that is required for the retention of surface layer (S-layer) and S-layer homology (SLH) domain proteins. Genetic disruption of the SCWP biosynthetic pathway impairs growth and cell division. B. anthracis SCWP is comprised of trisaccharide repeats composed of one ManNAc and two GlcNAc residues with O-3–α-Gal and O-4–β-Gal substitutions. UDP-Gal, synthesized by GalE1, is the substrate of galactosyltransferases that modify the SCWP repeat. Here, we show that the gtsE gene, which encodes a predicted glycosyltransferase with a GT-A fold, is required for O-4–β-Gal modification of trisaccharide repeats. We identify a DXD motif critical for GtsE activity. Three distinct genes, gtsA, gtsB, and gtsC, are required for O-3–α-Gal modification of trisaccharide repeats. Based on the similarity with other three-component glycosyltransferase systems, we propose that GtsA transfers Gal from cytosolic UDP-Gal to undecaprenyl phosphate (C55-P), GtsB flips the C55-P-Gal intermediate to the trans side of the membrane, and GtsC transfers Gal onto trisaccharide repeats. The deletion of galE1 does not affect growth in vitro, suggesting that galactosyl modifications are dispensable for the function of SCWP. The deletion of gtsA, gtsB, or gtsC leads to a loss of viability, yet gtsA and gtsC can be deleted in strains lacking galE1 or gtsE. We propose that the loss of viability is caused by the accumulation of undecaprenol-bound precursors and present an updated model for SCWP assembly in B. anthracis to account for the galactosylation of repeat units.
IMPORTANCE Peptidoglycan is a conserved extracellular macromolecule that protects bacterial cells from turgor pressure. Peptidoglycan of Gram-positive bacteria serves as a scaffold for the attachment of polymers that provide defined bacterial interactions with their environment. One such polymer, B. anthracis SCWP, is pyruvylated at its distal end to serve as a receptor for secreted proteins bearing the S-layer homology domain. Repeat units of SCWP carry three galactoses in B. anthracis. Glycosylation is a recurring theme in nature and often represents a means to mask or alter conserved molecular signatures from intruders such as bacteriophages. Several glycosyltransferase families have been described based on bioinformatics prediction, but few have been studied. Here, we describe the glycosyltransferases that mediate the galactosylation of B. anthracis SCWP.
INTRODUCTION
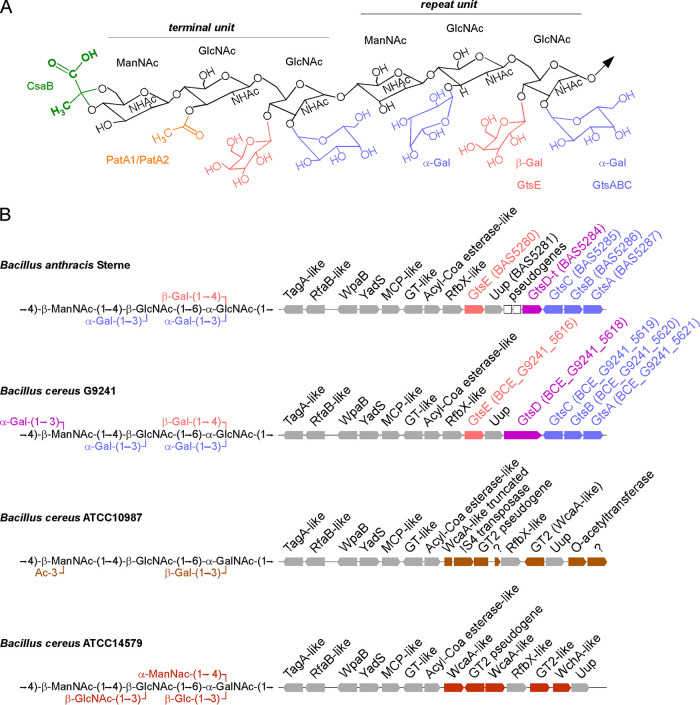
The secondary cell wall polysaccharide (SCWP) of Bacillus anthracis consists of 6 to 12 repeats of a trisaccharide unit described as [→4)-β-ManNAc-(1→4)-β-GlcNAc-(1→6)-α-GlcNAc-(1→] (Fig. 1A) (1). Ketal pyruvyl modification at O-4 and O-6 of the terminal ManNAc and acetylation at O-3 of the penultimate GlcNAc serve to retain the surface layer (S-layer) proteins (SLPs) Sap and EA1 and another 22 Bacillus surface layer (BSL) proteins bearing three S-layer homology (SLH) domains (Fig. 1A) (2–6). Sap and EA1 are endowed with self-oligomerizing domains and assemble into distinct S-layers depending on the growth phase of the bacterial culture (7–9). In the absence of csaB (cell surface-anchoring B), the gene that encodes the pyruvyl transferase of SCWP, all SLPs and BSLs are released into the extracellular milieu, and bacilli remain tethered in long chains following cell division (2–4). This phenotype was attributed to the loss of two secreted hydrolases, BslO and BslS, that cleave septal peptidoglycan between select daughter cells to form chains of four bacilli (10, 11). The SCWP also serves as a ligand for murein hydrolases of bacteriophages such as PlyL and PlyG (12, 13). However, these hydrolases lack an SLH domain. PlyG of γ-phage binds to the SCWPs from B. anthracis and Bacillus cereus G9241 but not those from B. cereus ATCC 10987 and ATCC 14579 (12, 14). Using synthetic oligosaccharides with various galactosyl modifications, Mo and colleagues reported that high-affinity binding of PlyL and PlyG is modulated by Gal modification of GlcNAc residues (15). In B. anthracis, repeating units are modified with α-Gal at O-3 and β-Gal at O-4 of α-GlcNAc and with α-Gal at O-3 of β-GlcNAc (Fig. 1) (1). In the related isolate B. cereus G9241 and other B. cereus isolates causing anthrax-like disease, the trisaccharide repeat carries an additional α-Gal substitution at O-3 of ManNAc (Fig. 1B) (16). B. anthracis CDC684 elaborates an SCWP devoid of all galactosyl modifications but retains ketal pyruvyl and acetyl modifications at the distal end of the SCWP (5); this strain assembles an S-layer but is avirulent (17). We recently showed that the UDP-glucose 4-epimerase, GalE1, is necessary for the conversion of UDP-glucose to UDP-galactose (UDP-Gal) in B. anthracis (18). The SCWPs of strains lacking galE1 were devoid of galactosyl modifications, supported the partial binding of γ-phage murein hydrolases, and retained all SLPs and BSLs in the envelope (18). Bacilli lacking galE1 also displayed reduced capsulation with poly-γ–d-glutamic acid, resulting in decreased virulence in a mouse model of anthrax (18). Here, we identify the enzymes responsible for α-Gal and β-Gal modifications by comparing predicted gene clusters of SCWP between bacilli with structurally related trisaccharide repeats. We use a genetic approach to confirm our prediction.
FIG 1.
Repeat unit structures and scwp2 gene clusters of members of the B. cereus sensu lato group. (A) Structure of terminal and repeat units of B. anthracis SCWP. Acetylation and pyruvylation of the terminal unit are depicted in orange and green, along with the enzymes CsaB and PatA1/PatA2, which are responsible for these substitutions. α-Gal and β-Gal modifications are depicted in blue and red. Ac, acetyl; MCP, methyl-accepting chemotaxis protein. (B) Comparison of scwp2 gene clusters (right) with a description of repeat unit structures (left). Gray blocks represent conserved genes between B. anthracis, B. cereus G9241, B. cereus ATCC 14579, and B. cereus ATCC 10987. Colored blocks indicate genes encoding predicted galactosyltransferases. Functions of genes are provided as described in the PubMed Data Bank and as described previously (20). GT, glycosyltransferase.
RESULTS
Comparative analysis of the sps and scwp2 gene clusters identifies putative glycosyltransferases for α-Gal and β-Gal modifications of SCWP.
B. cereus isolates ATCC 10987 and ATCC 14579 elaborate SCWP with trisaccharide repeats structurally related to those of B. anthracis Sterne and B. cereus G9241 but with distinct branched residues (16, 19) (Fig. 1B). We reasoned that a genomic comparison between these strains could be used to deduce the glycosyltransferases responsible for α-Gal and β-Gal substitutions in B. anthracis Sterne and B. cereus G9241. Four gene clusters have been proposed to contribute to SCWP biosynthesis in B. anthracis: scwp1, scwp2, scwp3, and sps (surface polysaccharide synthesis) (20). Several predicted glycosyltransferases were identified in the sps and scwp2 clusters (20). Four predicted glycosyltransferases (bas5121, bas5124, bas5126, and bas5127 products) are encoded in the sps cluster, along with the conserved GneZ enzyme that converts UDP-GlcNAc to UDP-ManNAc for incorporation into the SCWP backbone and the conserved GalE1 enzyme that provides UDP-Gal for glycosylation (18, 21, 22). However, the bas5121, bas5124, bas5126, and bas5127 genes were ruled out as they were not found in the genome of B. cereus G9241. Furthermore, strains carrying a transposon insertion in the bas5121 gene or a deletion of the genes bas5124 through bas5127 retained the same galactosylation pattern of SCWP as that of wild-type B. anthracis (data not shown).
B. anthracis and B. cereus G9241 share almost identical scwp2 gene clusters, while only some of the genes are conserved in the ATCC 10987 and ATCC 14579 isolates (Fig. 1B). Only the conserved gene wpaB has been examined thus far in this cluster and has been proposed to support the assembly of trisaccharide repeats in B. anthracis (11). Genes located further downstream of wpaB are much more variable and include four predicted glycosyltransferases, bas5280, bas5285, bas5286, and bas5287 (Fig. 1B). This region also encompasses a predicted Uup protein (bas5281 in B. anthracis). In Escherichia coli, Uup has been implicated in the precise excision of transposon elements (23, 24). All four predicted glycosyltransferase genes are conserved between B. anthracis and B. cereus G9241 (Fig. 1B). In addition, the bas5284 gene encodes a 227-amino-acid product that matches the C-terminal end of the 12-transmembrane predicted glycosyltransferase BCE_G9241_5618 (Fig. 1B). Together, either the genes for these five glycosyltransferases are absent in B. cereus ATCC 10987 and ATCC 14579 or their products share very little identity, as should be expected for isolates that display SCWPs with distinct branched patterns. In B. cereus ATCC 10987, an IS4 transposase interrupts a gene with homology to bas5280 (Fig. 1B). This region is also rearranged in B. anthracis with two pseudogenes and a partial glycosyltransferase (bas5284) (Fig. 1B). Thus, the predicted Uup of scwp2 clusters may directly contribute to the shuffling of glycosyltransferase-encoding genes to expand carbohydrate diversity in the envelope of bacilli. The studies described here confirm our predictions. Henceforth, the genes are referred as gtsA (bas5287), gtsB (bas5286), gtsC (bas5285), and gtsE (bas5280). Functional and topology predictions using InterPro and TMHMM analyses suggest that GtsE is a cytosolic glycosyltransferase (Table 1). GtsA, GtsB, and GtsC are membrane proteins that share similarity with three-component glycosylation systems (see below). GtsA and GtsC are predicted to function as glycosyltransferases, while GtsB is predicted to function as a flippase (Table 1). The bas5284 gene in B. anthracis and the BCE_G9241_5618 gene in B. cereus G9241 are referred to as GtsD1 and GtsD, respectively, to reflect the possibility that strain Sterne encodes a truncated glycosyltransferase. Of note, the galE1, gtsA, gtsB, gtsC, and gtsD genes are all accounted for in strain CDC684, and thus, the genetic basis for the lack of galactosylation in this isolate remains unknown.
TABLE 1.
Bioinformatic analysis of key glycosyltransferases identified in this studya
| Gene | Protein name | Length (amino acids) | Classification or predicted family | Predicted DXD-like motif(s) | Predicted activity | No. of TM segments | Predicted localization of GT domain |
|---|---|---|---|---|---|---|---|
| bas5287 | GtsA | 323 | Dolichol-phosphate-mannose synthetaseb | 94DAD | GT | 2 | Cytoplasmic |
| bas5286 | GtsB | 127 | GtrA family; PF04138 | No | Flippase | 4 | NA |
| bas5285 | GtsC | 479 | GT family C fold; PF13231 | Unknown | GT | 12 | Extracytoplasmic |
| bas5280 | GtsE | 273 | GT family A fold; PF00535 | 86DDD, 162DED | GT | 0 | Cytoplasmic |
Functional and topology predictions were obtained using InterPro and TMHMM analyses. TM, transmembrane; GT, glycosyltransferase; NA, not applicable.
This family of glycosyltransferases has been shown to transfer the sugar moiety from carriers such as UDP‐glucose, UDP‐N‐acetylgalactosamine, GDP‐mannose, or TDP‐rhamnose onto substrates that include cellulose, dolichol phosphate, and teichoic acid.
Deletion of gtsA, gtsB, or gtsC but not of gtsD1 or gtsE leads to loss of viability in B. anthracis.
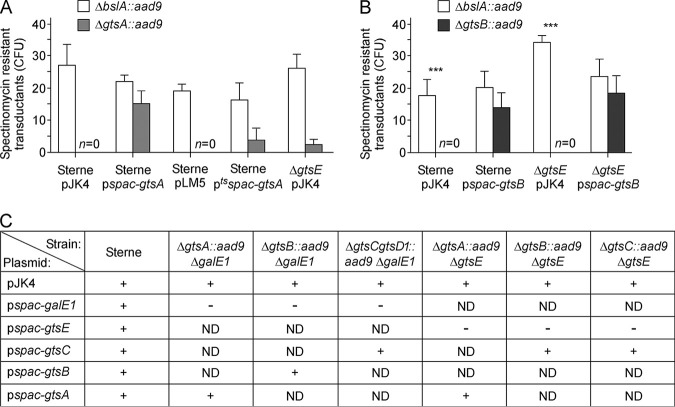
A canonical allelic-replacement approach was first used to generate gene deletions. One-kilobase-pair DNA sequences upstream and downstream of the gene targeted for deletion were ligated and cloned into vector pLM4 that carries a temperature-sensitive replicon. This approach proved successful for the construction of gtsD1 and gtsE mutants. Attempts to inactivate gtsA, gtsB, gtsC, or gtsC-gtsD1 were unsuccessful. To circumvent this problem, a merodiploid gtsA strain was constructed by transforming the wild-type Sterne strain with pgtsA, a plasmid encoding gtsA under the control of the constitutive hprK promoter of plasmid pWWW412. Next, plasmid pLM4-gtsA::aad9, designed for the replacement of gtsA with aad9 (encoding spectinomycin [Spc] resistance), was transformed into Sterne carrying a control vector (pWWW412) or pgtsA. When grown at a nonpermissive temperature, only bacilli carrying pgtsA could be selected on spectinomycin plates, suggesting that recombination of the gtsA::aad9 allele occurred only in the presence of a second copy of gtsA. To further evaluate the gtsA requirement for viability, the new recombinants were subjected to bacteriophage CP-51 lysis for transduction experiments. As a control, a CP-51 phage lysate was also derived from B. anthracis bslA::aad9, a nonessential gene required for virulence (10). Four Sterne recipient strains were generated, carrying vector pJK4 or pLM5 without or with an insertion of gtsA, yielding plasmid pspac-gtsA or ptsspac-gtsA, respectively (Fig. 2A). pLM5 carries a thermosensitive replicon but is otherwise identical to pJK4 with a kanamycin (Kan) resistance gene and an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible spac promoter. Transductants were enumerated as CFU per agar plate using a growth medium supplemented with spectinomycin, kanamycin, and IPTG. The bslA::aad9 allele could be transduced in all four recipient strains (Fig. 2A). In contrast, the gtsA::aad9 allele could be transduced only in merodiploid pspac-gtsA or ptsspac-gtsA strains (Fig. 2A), suggesting that B. anthracis is not viable in the absence of gtsA. A similar approach revealed that gtsB is also required for growth (Fig. 2B).
FIG 2.
B. anthracis gtsA and gtsB are indispensable for growth. (A and B) CP-51 phage lysates were prepared from B. anthracis ΔbslA::aad9 (control), ΔgtsA::aad9 (A), or ΔgtsB::aad9 (B) for crossing into the wild type harboring empty vector pJK4 or pLM5 and merodiploid recipient strains or mutant strains, as indicated. Transductants were selected by plating and incubated for 30 h at 30°C. The average numbers of colonies and associated standard errors from 3 independent experimental determinations are shown. n=0 indicates that no colonies were obtained. (C) Summary of results of complementation studies in various strains. Following the transformation of strains with the indicated plasmids, bacteria were plated onto selective medium. + and − indicate the formation and absence of colonies on a plate, respectively. ND, not determined.
Deletion of gtsA, gtsB, or gtsC is tolerated in some backgrounds.
We wondered whether the loss of viability associated with the deletion of gtsA, gtsB, or gtsC may be caused by the accumulation of reaction intermediates. In the absence of galE1, the SCWP of B. anthracis is no longer modified with Gal residues, as the UDP-Gal substrate is missing (18). Using the temperature-sensitive replication vector pLM4, gtsA, gtsB, and the gtsC-gtsD1 combination were successfully replaced with aad9 in a strain deleted for galE1 (ΔgalE1). Attempts to transform these strains with the galE1-complementing plasmid were unsuccessful (Fig. 2C), further supporting a model whereby the loss of gtsA, gtsB, or gtsC is permissible only in the absence of the UDP-Gal substrate. The gtsA::aad9 allele could also be recombined in a strain lacking gtsE with low efficiency (ΔgtsE/pJK4) (Fig. 2A). The ΔgtsA::aad9 ΔgtsE double mutant could be transformed with a plasmid bearing gtsA, but not gtsE, placed under the control of the spac promoter (Fig. 2C). Double ΔgtsC::aad9 ΔgtsE and triple ΔgtsC-gtsD1::aad9 ΔgtsE mutant strains were also obtained and similarly could be transformed with a plasmid bearing gtsC but not gtsE (Fig. 2C). However, attempts to replace wild-type gtsB with a mutant allele (ΔgtsB::aad9) in the ΔgtsE background remained unsuccessful (Fig. 2B). We surmise that the loss of GtsA, GtsB, or GtsC disrupts SCWP assembly and leads to the toxic accumulation of biosynthetic intermediates.
Deletion of gtsE leads to loss of SCWP β-galactosylation.
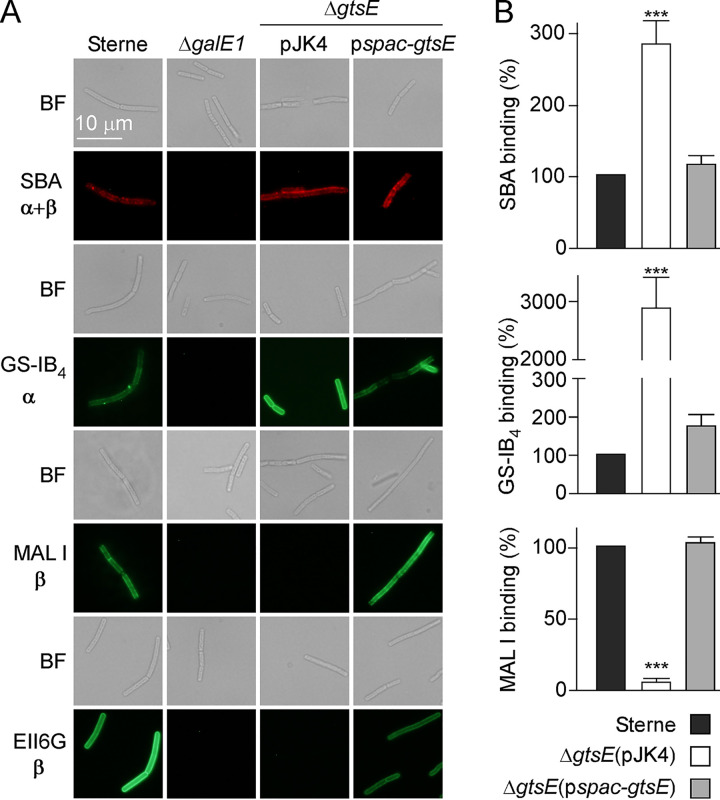
Since the deletion of gtsE did not result in a loss of viability, vegetative bacilli were derived from this mutant and subjected to binding with α- and β-Gal ligands modified with fluorophores for microscopy analysis. Soybean agglutinin (SBA) is a lectin that binds terminal α- and β-Gal linked to GalNAc (18, 25, 26). Isolectin GS-IB4 from Griffonia simplicifolia is selective for terminal α-d-Gal residues, and Maackia amurensis lectin I (MAL I) selectively binds the β (1,4) bond between Gal and GlcNAc (27). The monoclonal antibody (MAb) EAII6G6 was developed previously for the rapid identification of vegetative B. anthracis and shown to bind Gal-GlcNAc of SCWP (5, 18, 26, 28). As expected, vegetative bacilli of B. anthracis Sterne interacted with all four ligands, MAb EAII6G6, SBA, GS-IB4, and MAL I, and no immunofluorescence signal was detected with bacilli lacking galE1 (ΔgalE1) (Fig. 3A) (18). Bacilli lacking gtsE (ΔgtsE/pJK4) were stained with GS-IB4 and SBA lectins but not with MAb EAII6G6 and MAL I. Lectin binding was further quantified by recording fluorescence signals, revealing that, in fact, GS-IB4 and SBA binding increased in the absence of gtsE (Fig. 3B). All differences were restored upon complementation (ΔgtsE/pspac-gtsE) (Fig. 3A). Together, the data suggest that GtsE mediates β-galactosylation of SCWP in B. anthracis and that MAb EAII6G6, previously defined as a Gal-GlcNAc ligand (26), shares presumably the same stereospecificity with MAL I.
FIG 3.
gtsE deletion abolishes MAL I and EAII6G6 binding to B. anthracis. Vegetative bacilli were stripped of their S-layers by treatment with 3 M urea, fixed with 4% paraformaldehyde, and stained with monoclonal antibody EAII6G6-FITC, SBA lectin conjugated to Alexa Fluor 594, isolectin GS-IB4 conjugated to Alexa Fluor 488, or FITC-MAL I lectin. (A) Bright-field (BF) and fluorescence microscopy images were acquired. α and β indicate the configuration of ligands for lectins and antibodies. (B) Binding of fluorescent lectin was assessed by fluorescence measurements using a plate reader. Arbitrary units of fluorescence were converted to percentages of binding. Lectin binding to B. anthracis Sterne was set as 100%. Mean values and associated standard errors were derived from at least 3 independent experiments, and statistical analyses were performed by ANOVA and Tukey’s post hoc analysis. ***, P ≤ 0.001.
The C-terminal cell wall binding domain of PlyG (PlyGCBD) is sufficient to promote binding to B. anthracis SCWP (12, 13) and can be fused to mCherry without a loss of activity. S-layer-stripped vegetative forms of wild-type B. anthracis can be stained with PlyGCBD-mCherry, but staining is reduced for ΔgalE1 bacilli (18). When ΔgtsE bacilli were stripped of their S-layer by urea treatment, increased PlyGCBD-mCherry binding was observed compared to wild-type bacilli (Fig. 4). Plasmid complementation (ΔgtsE/pspac-gtsE) reduced PlyGCBD-mCherry binding to wild-type levels. As a control, PlyGCBD-mCherry did not bind B. cereus ATCC 14579, which assembles a distinct SCWP with a unique glycosylation pattern (Fig. 4).
FIG 4.
PlyGCBD-mCherry binding to B. anthracis. Vegetative bacilli of B. anthracis strains and B. cereus ATCC 14579 were stripped of their S-layers with 3 M urea and incubated with purified PlyGCBD-mCherry. (A) Representative bright-field (BF) and fluorescence microscopy images. (B) Binding of fluorescent protein was assessed by fluorescence measurements. Arbitrary units of fluorescence were converted to percentages of binding. PlyGCBD-mCherry binding to B. anthracis Sterne was set as 100%. Mean values and associated standard errors were derived from 5 independent experiments, and statistical analyses were performed by one-way ANOVA and Tukey’s post hoc analysis. **, P < 0.01; ***, P < 0.001.
Next, SCWP was extracted from wild-type B. anthracis Sterne and the isogenic ΔgtsE and complemented ΔgtsE/pspac-gtsE strains. The SCWP was purified by size exclusion chromatography, and fractions containing the polymer were analyzed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry. Ion signals at m/z 2,232.65 and m/z 3,329.62, corresponding to sodium adducts of two and three repeating units, respectively, were readily identified in SCWP preparations from the wild-type and complemented strains but missing from preparations of the ΔgtsE variant (Table 2). Instead, the ΔgtsE sample produced ions with signals at m/z 1,909.7 and m/z 2,843.1, which corresponded to two and three repeating units, respectively, lacking one galactose per repeating unit. These data are consistent with a model whereby gtsE is responsible for the incorporation of β-Gal residues in the SCWP.
TABLE 2.
MALDI-TOF mass spectrometry of hydrofluoric acid-released SCWPs from B. anthracis Sterne 34F2 (wild type) and isogenic ΔgtsE and ΔgtsE/pspac-gtsE variantsa
| Proposed composition of SCWPb | Observed m/z in: |
Theoretical m/z (monoisotopic) | ||
|---|---|---|---|---|
| Sterne | ΔgtsE strain | ΔgtsE/pspac-gtsE strain | ||
| HexNAc4-Gal2Na+ | 1,176.26 | 1,177.453 | ||
| HexNAc3-GlcN-Gal4Na+ | 1,458.41 | 1,459.52 | ||
| HexNAc5-GlcN-Gal32Na+ | 1,726.10 | 1,725.86 | ||
| HexNAc6-Gal32Na+ | 1,768.15 | 1,768.99 | ||
| HexNAc6-Gal4Na+ | 1,909.75 | 1,908.13 | ||
| HexNAc6-GlcN-Gal42Na+ | 2,091.76 | 2,092.11 | ||
| HexNAc6-Gal6Na+ | 2,232.65 | 2,233.64 | 2,231.79 | |
| HexNAc9-Gal6Na+ | 2,843.1 | 2,842 | ||
| HexNAc9-Gal9Na+ | 3,329.62 | 3,330.58 | 3,327.18 | |
Ion signals and proposed compositions of compounds from MALDI-TOF mass spectra of RP-HPLC-purified SCWPs from B. anthracis Sterne 34F2 and its ΔgtsE mutant and complemented variant ΔgtsE/pspac-gtsE.
The SCWPs isolated from wild-type Sterne and complemented ΔgtsE/pspac-gtsE strains generated nearly identical sets of ions, similar to the spectra characterizing the SCWP structure. Additional compositional explanations for the observed masses exist but are not listed here. HexNAc, ManNAc (N-acetylmannosaminyl) and GlcNAc (N-acetylglucosaminyl); GlcN, glucosaminyl; Gal, galactosyl.
Galactosylation of SCWP in permissive mutants lacking gtsA or gtsC.
Lectin staining was used to assess the contribution of gtsA, gtsC, and gtsD1 to the galactosylation of SCWP in a background lacking gtsE. The introduction of ΔgtsA::aad9, ΔgtsC::aad9, or the ΔgtsC-gtsD1::aad9 combination in the ΔgtsE background resulted in the loss of MAb EAII6G6 and MAL I binding, as expected, as well as the loss of GS-IB4 and SBA binding (Fig. 5). GS-IB4 and SBA binding defects could be reversed upon the addition of IPTG to cultures of ΔgtsA and ΔgtsC strains complemented with pspac-gtsA and pspac-gtsC (Fig. 5), suggesting that GtsA and GtsC may be involved in α-galactosylation of SCWP in B. anthracis. To verify this assumption, SCWP was extracted from conditional mutants bearing or not bearing the cognate complementing plasmids. Ion signals at m/z 1,136.31, m/z 3,327.78, and m/z 4,424.01, corresponding to sodium adducts of one, three, and four SCWP repeating units, respectively (18), were observed for wild-type or ΔgtsD1::aad9 samples, suggesting that GtsD1 does not contribute to galactosylation (Table 3). SCWP preparations isolated from the ΔgtsA::aad9 ΔgtsE and ΔgtsC-gtsD1::aad9 ΔgtsE strains generated fewer ion signals than the wild type, with a predominant signal of m/z 649.9. This ion corresponds to the sodium adduct of nongalactosylated trisaccharide [ManNAc-GlcNAc2] and was also the predominant ion observed previously in ΔgalE1 preparations (18). Ion signals at m/z 1,909.7 and m/z 2,841.7 were observed in preparations of ΔgtsA::aad9 ΔgtsE/pspac-gtsA and ΔgtsC-gtsD1::aad9 ΔgtsE/pspac-gtsC IPTG-treated cultures, respectively, and corresponded to two and three SCWP repeating units lacking one galactose per repeating unit, as observed for the ΔgtsE strain (Table 3). These data are consistent with a model whereby GtsA and GtsC support α-galactosylation of the SCWP.
FIG 5.
Deletion of gtsA or gtsC abolishes SBA and GS-IB4 lectin binding. Vegetative bacillus variants were prepared and stained with monoclonal antibody EAII6G6-FITC or lectin ligands as described in the legend to Fig. 3. Representative bright-field (BF) and fluorescence microscopy images are shown. α and β indicate the configuration of ligands for lectins and antibodies.
TABLE 3.
MALDI-TOF mass spectrometry of hydrofluoric acid-released SCWPs from B. anthracis variants lacking one or more predicted glycosyltransferasesa
| Proposed composition of SCWPb | Observed m/z in strain with genotype: |
Theoretical m/z (monoisotopic) | ||||
|---|---|---|---|---|---|---|
| ΔgtsD1::aad9 | ΔgtsC-gtsD1::aad9 ΔgtsE | ΔgtsC-gtsD1::aad9 ΔgtsE/pspac-gtsC | ΔgtsA::aad9 ΔgtsE | ΔgtsA::aad9 ΔgtsE/pspac-gtsA | ||
| HexNAc3Na+ | 649.85 | 649.82 | 649.80 | 649.81 | 649.83 | 649.97 |
| HexNAc3-Gal2Na+ | 976.34 | 974.26 | ||||
| HexNAc3-Gal3Na+ | 1,136.31 | 1,136.40 | ||||
| HexNAc4-Gal2Na+ | 1,175.30 | 1,175.40 | 1,177.45 | |||
| HexNAc3-GlcN-Gal4Na+ | 1,459.81 | 1,459.52 | ||||
| HexNAc5-GlcN-Gal32Na+ | 1,725.53 | 1,724.53 | 1,725.86 | |||
| HexNAc6-Gal4Na+ | 1,908.43 | 1,907.70 | 1,908.13 | |||
| HexNAc6-GlcN-Gal42Na+ | 2,090.60 | 2,091.63 | 2,092.11 | |||
| HexNAc9-Gal6Na+ | 2,841.73 | 2,842.00 | ||||
| HexNAc9-Gal9Na+ | 3,327.78 | 3,327.18 | ||||
| HexNAc12-Gal12Na+ | 4,424.01 | 4,422.57 | ||||
Ion signals and proposed compositions of compounds from MALDI-TOF mass spectra of RP-HPLC-purified SCWPs from various strains.
Additional compositional explanations for the observed masses exist but are not listed here. HexNAc, ManNAc (N-acetylmannosaminyl) and GlcNAc (N-acetylglucosaminyl); GlcN, glucosaminyl; Gal, galactosyl.
GtsE is a cytosolic glycosyltransferase with a conserved DXD motif.
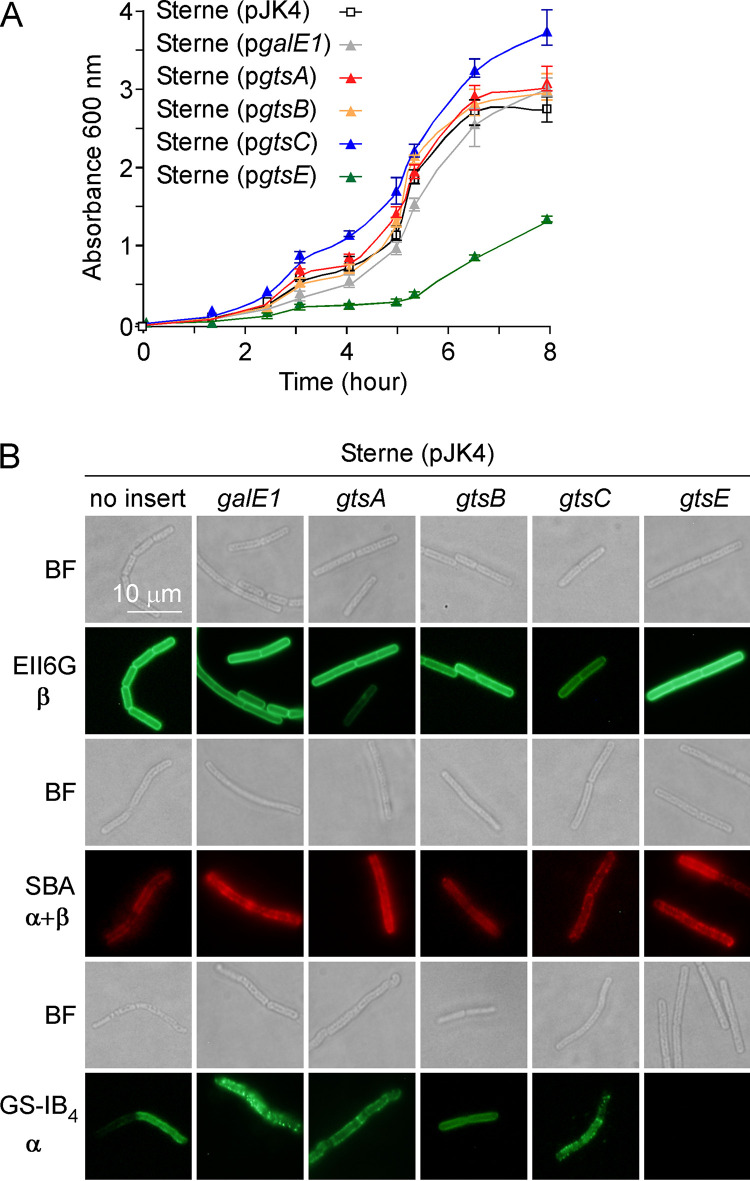
Our phenotypic analyses of gtsE variants suggest that GtsE carries out an inverting glycosyltransferase reaction. Sequence prediction tools identify GtsE as a soluble protein with no predicted signal sequence or transmembrane domain and as a member of the glycosyltransferase 2 family of proteins (PF00535) that clusters with the nucleotide–diphospho-sugar transferase superfamily (InterPro identifier IPR029044) (Table 1). InterPro analysis further identifies the conserved domain cd00761 of the glycosyltransferase family A (GT-A fold) enzymes. Many GT-A enzymes use a DXD motif to coordinate a divalent metal cation, which assists in the release of nucleoside diphosphate upon the transfer of the sugar from the nucleoside diphosphate sugar donor (e.g., UDP-Gal) to the substrate (e.g., repeating units of SCWP). In an attempt to further characterize GtsE, the protein was produced with an appended C-terminal histidine tag (GtsEHis). When used as a complementing plasmid, pgtsEHIS was found to restore MAb EAII6G6 and MAL I binding to bacilli lacking gtsE (Fig. 6A). Subcellular fractions of ΔgtsE/pgtsE and ΔgtsE/pgtsEHIS cultures were separated by SDS-PAGE and examined by Western blotting with an anti-His probe. This experiment revealed that GtsE is not found in the culture medium. The protein remains associated with intact cells. However, despite a lack of hydrophobic membrane segments, GtsE sediments with the membrane fraction along with the lipoprotein PrsA (Fig. 6B). Two putative DXD motifs, 86DDD88 and 162DED164, were identified in the sequence of GtsE. Variants with alanine substitutions were generated to yield plasmids pgtsED86A and pgtsED162A producing the proteins GtsED86A and GtsED162A, respectively. Plasmid pgtsED162A but not pgtsED86A restored MAb EAII6G6 and MAL I binding in the ΔgstE strain, suggesting that D86 is essential for enzymatic activity (Fig. 6C). Finally, we noted that the overproduction of GtsE in wild-type Sterne may also be toxic. Growth in the presence of IPTG was reduced in Sterne carrying pspac-gtsE compared to the vector control (Fig. 7A). gtsE overexpression also abolished GS-IB4 binding (Fig. 7B). Of note, the overexpression of gtsA, gtsB, gtsC, or galE1 in strain Sterne did not noticeably alter growth or GS-IB4 binding (Fig. 7A).
FIG 6.
Characterization of GtsE. (A) GtsE carrying a C-terminal six-histidine tag restores MAb EAII6G6 and MAL I binding to ΔgtsE bacilli. The experiment was performed as described in the legend to Fig. 3. (B) Cultures of ΔgtsE bacilli carrying pJK4 harboring either gtsE or gtsEHis were fractionated into culture medium (MD), total cell lysate (T), cytoplasm (C), and sedimented membrane (Mb) fractions. Proteins were separated on SDS-PAGE gels and either visualized by staining the gels with Coomassie (top) or transferred for immunoblotting with antibodies specific for the His tag, protective antigen (PA), Sap, and PrsA (bottom). A molecular ladder was loaded in the left lane of the top gel. Data are representative of results from two independent experiments. (C) Aspartic acid at position 86 is essential for GtsE activity. Vegetative ΔgtsE bacilli carrying pJK4 without an insert or with inserts harboring gtsE (wild type), gtsED86A, or gtsED162A were analyzed for MAb EAII6G6 and lectin binding. Representative bright-field (BF) and fluorescence microscopy images are shown. α and β indicate the configuration of ligands for lectins and antibodies.
FIG 7.
Overexpression of gtsE slows the growth of B. anthracis. (A) Growth curves of strain Sterne carrying empty vector pJK4 or plasmids harboring galE1, gtsA, gtsB, gtsC, and gtsE grown in BHI broth with IPTG at 37°C were obtained by monitoring the absorbance of cultures over time at 600 nm. Bars indicate standard deviations (n = 3). (B) Vegetative bacillus variants were prepared and stained with monoclonal antibody EAII6G6-FITC or lectin ligands as described in the legend to Fig. 3. Representative bright-field (BF) and fluorescence microscopy images are shown. α and β indicate the configuration of ligands for lectins and antibodies.
DISCUSSION
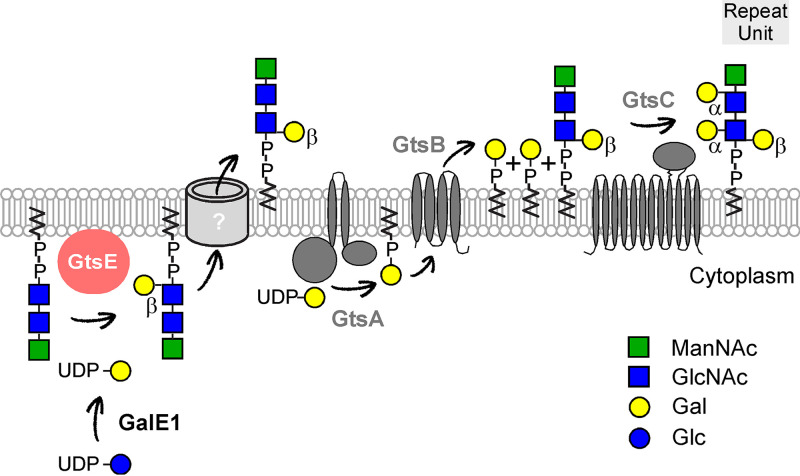
Unlike Bacillus subtilis and Staphylococcus aureus, B. anthracis produces SCWP instead of wall teichoic acid (WTA); the trisaccharide repeats that constitute the SCWP are not found in B. subtilis and S. aureus. In these species, WTA is made of simple glycerol or ribitol repeats (20, 29). Nonetheless, B. anthracis encodes TagO- and TagA-like enzymes. TagO initiates WTA synthesis in S. aureus and B. subtilis by transferring GlcNAc-1-phosphate from UDP-GlcNAc onto the undecaprenol-phosphate carrier C55-(PO4) to generate C55-(PO4)2-GlcNAc (lipid III) (29). TagA transfers ManNAc from UDP-ManNAc onto O-4 of GlcNAc within lipid III to yield C55-(PO4)2-GlcNAc-ManNAc (29). In B. anthracis, the genetic repression of tagO inhibits SCWP synthesis and causes expansion in the size and spherical shapes of vegetative forms that can no longer divide (3, 30). Like with WTA, B. anthracis TagO and TagA may synthesize linkage units of SCWP; alternatively, TagO and TagA could support the synthesis of trisaccharide units. Bioinformatics and experimental inquiries have defined four gene clusters that contribute to the assembly of SCWP (20). The current model favors the synthesis of trisaccharide repeat units onto undecaprenol-phosphate on the cis side of the membrane followed by flipping across the membrane and polymerization of repeat units on the trans side of the membrane in a reaction mediated by WpaA and WpaB (11, 20). Presumably, polymerization is terminated by LytR, CspA, and Psr (LCP) enzymes that transfer polymerized repeats from undecaprenol onto peptidoglycan (31, 32). Here, we identify the glycosyltransferases that add galactosyl residues to trisaccharide subunits and propose a slightly updated model. Using lectins and MALDI-TOF analyses, we show that the deletion of bas5280 results in the assembly of SCWP lacking β-Gal modifications. We name this gene gtsE for glycosyltransferase E. gtsE is harbored in the variable region of the scwp2 gene cluster encountered in strains of the B. cereus sensu lato group that elaborate SCWP (Fig. 1). gtsE is not conserved in the scwp2 gene clusters of B. cereus ATCC 10987 and B. cereus ATCC 14579, which lack β-Gal in the SCWP. Lectin accessibility to α-Gal is increased in the envelope of ΔgtsE bacilli, suggesting steric hindrance by β-Gal. The galactosylation of SCWP also influences the binding of the phage murein hydrolases PlyG and PlyL. Previous work using chemically synthesized trisaccharides suggested a preferred binding of these hydrolases to either α-GlcNAc modified with β-Gal at O-4 or β-GlcNAc modified with α-Gal at O-3 (15). Using the PlyGCBD-mCherry fusion protein, we find that binding is increased in the gtsE mutant, suggesting that β-Gal modification may hinder PlyG and PlyL binding or that PlyG and PlyL preferentially bind repeats with α-Gal modification in the SCWP. Our in vivo data agree with a model whereby gtsE catalyzes the transfer of UDP-Gal synthesized by GalE1 onto GlcNAc. However, attempts to demonstrate such an activity using UDP-Gal as the donor and C55-(PO4)2-GlcNAc or the simpler GlcNAc or UDP-GlcNAc as the acceptor substrates failed (data not shown). We propose that GtsE transfers Gal residues onto the lipid-bound trisaccharide repeats C55-(PO4)2-GlcNAc-GlcNAc-ManNAc on the cis side of the plasma membrane. This activity may account for GtsE sedimentation with the subcellular membrane fraction.
B. anthracis Sterne appears to encode a truncated GtsD enzyme that is otherwise intact in B. cereus G9241 (Fig. 1B). No changes were observed upon the deletion of gtsD1. Thus, we speculate that GtsD may account for α-Gal substitution at O-3 of ManNAc residues of B. cereus G9241 SCWP.
GtsA, GtsB, and GtsC are reminiscent of the three-component glycosylation systems described previously in temperate phages of Shigella flexneri or Salmonella spp. (33) and now found to be widely distributed in bacteria (34). In SfV, SfX, and SfI lysogenic strains of Shigella, a three-gene operon unit (gtrA, gtrB, and gtrV, -X, or -I, respectively) is required for O-antigen glycosylation accounting for serotype conversion (35–37). This led to a model whereby the glycosyltransferase GtrA transfers a carbohydrate from its nucleoside diphosphate (NDP) donor onto undecaprenol phosphate, GtrB flips undecaprenol-bound sugars to the trans side of the membrane, and the third integral membrane glycosyltransferase, named for the prophage, e.g., GtrV (SfV), transfers the carbohydrate onto O antigen (Table 4) (33, 34). This model was based on biochemical evidence that preceded the identification of gtr genes. Using radiolabeled glucose, Nikaido and colleagues isolated a [14C]Glc-lipid intermediate that served as the donor for the transfer of glucosyl residues onto the O antigen of Salmonella enterica serovar Typhimurium lipopolysaccharide (38). Incubation of UDP-[14C]Gal or UDP-[14C]GlcNAc with membranes of Bacillus coagulans or B. subtilis, respectively, was shown to result in the formation of radioactive undecaprenol intermediates and radioactive lipoteichoic acid (LTA) (39, 40). Genes for LTA-modifying enzymes were recently described for Listeria monocytogenes, B. subtilis, and S. aureus, and their identification is in agreement with three-component glycosylation systems (Table 4) (34, 41, 42). In L. monocytogenes, a second three-component glycosylation system modifies WTA (41) and appears to share the GtcA flippase of the LTA-modifying enzymes (43) (Table 4). Unlike with Shigella and B. anthracis, genes of three-component glycosylation systems do not often cluster in operons, and the cognate terminal glycosyltransferases that govern substrate specificity are weakly related (Table 4). Genes encoding three-component glycosylation systems could readily be deleted in L. monocytogenes, B. subtilis, and S. aureus (41, 42). This is in contrast to B. anthracis, where such deletions are tolerated only in a ΔgalE1 background. Presumably, trisaccharide units lacking all galactosyl modifications (ΔgalE1) remain substrates for polymerization, whereas in mutants lacking gtsA, gtsB, or gtsC, trisaccharide units bearing β-Gal residues cannot be polymerized and instead accumulate on the trans side of the membrane (Fig. 8). This accumulation depletes the pool of undecaprenol and kills the cell. Accordingly, ΔgtsA ΔgtsE and ΔgtsC ΔgtsE double mutants are also viable, yet ΔgtsB ΔgtsE double mutants are nonviable. Perhaps toxicity results from the irreversible conversion of UDP-Gal to C55-(PO4)-Gal by GtsA. This toxicity is alleviated by deleting galE1, i.e., by eliminating the UDP-Gal substrate. When gtsE is expressed on a plasmid, bacterial replication slows. This is in agreement with the notion that the transient accumulation of trisaccharide units bearing β-Gal residues limits the pool of undecaprenol. Alternatively, the overproduction of GtsE may lead to the aberrant glycosylation of repeat units and prevent further modification by GtsA, GtsB, and GtsC. Together, our findings support a model whereby the addition of β-Gal occurs intracellularly, whereas α-Gal addition occurs extracellularly (Fig. 8). Further biochemical characterization is needed to support this model and to establish whether α-Gal modification represents a key limiting step for SCWP polymerization and length in B. anthracis.
TABLE 4.
Three-component glycosylation proteins from various bacteria compared to B. anthracis GtsA, GtsB, and GtsC
| Bacterial species/substrates | Dolichol-phosphate-mannose synthetase |
Flippase |
Dolichyl-phosphate-mannose-protein mannosyltransferasea
|
|||
|---|---|---|---|---|---|---|
| Name | % homology | Name | % homology | Name | % homology | |
| B. anthracis/Gal and SCWP | GtsA (BAS5287) | GtsB (BAS5286) | GtsC (BAS5285) | |||
| B. subtilis/GlcNAc and LTA | CsbB (BSU08600) | 62 | GtcA (BSU38210) | 28 | YfhO (BSU08610) | No significant similarity |
| L. monocytogenes/Gal and LTA | GtlA (Lmo0933) | 71 | GtcA (Lmo2549) | 57 | GtlB (Lmo0626) | No significant similarity |
| L. monocytogenes/GlcNAc and WTA | Lmo2550 | 64 | GtcA (Lmo2549) | 57 | Lmo1079 | No significant similarity |
| S. aureus/GlcNAc and LTA | CsbB-like SAUSA300_0689 | 59 | GtrA-like SAUSA300_2376 | 58 | YfhO-like SAUSA300_1135 | No significant similarity |
| S. flexneri/Glu and O antigen | GtrB | 61 | GtrA | No significant similarity | GtrI | No significant similarity |
Dolichyl-phosphate-mannose-protein mannosyltransferases share no sequence similarity among bacterial species, as they catalyze sugar transfer on species-specific substrates.
FIG 8.
Revised model for SCWP synthesis. Trisaccharide repeats are synthesized on undecaprenol-P on the cis side of the membrane and are substrates for β-Gal modification by GtsE. Precursor units are flipped to the trans side of the membrane by an as-yet-uncharacterized enzyme. GtsA, GtsB, and GtsC add α-Gal residues to the newly translocated subunit. Presumably, the subunits are further polymerized by wall polysaccharide assembly factors (Wpa), and the new polymer is transferred onto peptidoglycan by LCP proteins (20). GtsA, GtsB, and GtsC are represented with 2, 4, and 12 predicted membrane-spanning segments, respectively. GtsA and GtsC are endowed with large cytoplasmic and extracytoplasmic domains, respectively.
MATERIALS AND METHODS
Bacterial growth and reagents.
The B. anthracis Sterne 34F2 strain and its variants were grown in brain heart infusion (BHI) broth or agar at temperatures ranging between 30°C and 40°C. E. coli was grown in lysogeny broth (LB) or agar at 37°C. Where necessary, kanamycin (Kan), chloramphenicol (Cam), and spectinomycin (Spec) (Fisher Scientific) were added at concentrations of 20, 10, and 200 μg ml−1, respectively, for B. anthracis. Ampicillin (Amp) and Kan were added at concentrations of 100 and 50 μg ml−1, respectively, for E. coli. Isopropyl-β-d-thiogalactopyranoside (IPTG) was used at a 0.1 mM final concentration. Culture media were obtained from BD. SBA conjugated to Alexa Fluor 594 and GS-IB4 conjugated to Alexa Fluor 488 were obtained from Thermo Fisher Scientific. Fluorescein-labeled MAL I was obtained from Vector Laboratories. EAII6G6 antibody was a previous gift from Teresa Abshire and Arthur Friedlander. Unless otherwise indicated, all other reagents were purchased from Sigma.
B. anthracis strains and plasmids.
The temperature-sensitive vector pLM4 carrying the Kan resistance gene was used for the allelic replacement of gts genes as previously described (44). Briefly, upstream and downstream DNA sequences flanking the target gene were amplified using specific primers and PCR, digested with restriction enzymes, and ligated into pLM4 digested with the same enzymes. In all cases but gtsE, the aad9 gene encoding spectinomycin resistance was cloned at the location of the missing gene. Complementing plasmids for various gts genes were generated by amplifying the minimum coding sequence of each gene using PCR and cloning the DNA fragments into pWWW412 (45), pJK4 (3), or pLM5 (44). Bacteriophage CP-51 was used for transduction experiments as described previously (11, 46). pJK4-derived plasmids pgtsED86A and pgtsED162A were generated by PCR with oligonucleotides harboring either a D86A or D162A mutation and the wild-type plasmid pspac-gtsE as a template. Following PCR, products were treated with the DpnI enzyme prior to transformation into E. coli. All recombinant constructs were verified by DNA sequencing, and all oligonucleotides used in this work are listed in Table S1 in the supplemental material.
Binding assays and subcellular fractionation of bacilli.
Binding assays were performed using vegetative bacilli stripped of S-layers as described previously (18). Briefly, bacterial colonies were scraped off agar plates, suspended and washed once in phosphate-buffered saline (PBS), and boiled at 95°C for 10 min in PBS containing 3 M urea. Cells were sedimented by centrifugation for 1 min at 16,000 × g to remove SCWP-bound proteins in the supernatant. For lectin binding, cells were washed twice with PBS, fixed with formalin, and washed 3 more times with PBS before staining. For binding with PlyGCBD-mCherry, cells were washed twice with PBS and normalized by an optical density at 600 nm (OD600) of 1.0. Bacterial cell suspensions (100 μl) were incubated either for 1 h with SBA-Alexa Fluor 594 (50 μg/ml), GS-IB4–Alexa Fluor 488 (25 μg/ml), and MAL I-fluorescein (20 to 40 μg/ml) or overnight at 4°C with purified PlyGCBD-mCherry as previously described (30). Only 4 μl of the bacterial suspension was used for a 30-min incubation with 1 μl of fluorescein isothiocyanate (FITC)-labeled monoclonal antibody EAII6G6. To analyze binding, bacteria were washed twice with PBS to remove the unbound ligand. Next, the cells were imaged using a charge-coupled-device (CCD) camera on an Olympus IX2-UCB microscope with a 100× objective or transferred to 96-well plates for quantification of fluorescence signals using a BioTek Synergy HT microplate reader. The excitation and emission wavelengths were set at 590 ± 20 nm and 645 ± 40 nm, respectively. Fluorescence measurements were converted to percent binding compared to that of the wild-type B. anthracis strain 34F2, for which protein binding, i.e., fluorescence units, was arbitrarily set as 100%. Statistical analysis was performed by one-way analysis of variance (ANOVA) and Tukey’s post hoc analysis.
For subcellular fractionation experiments, bacterial cultures were grown to an OD600 of 1.0 and sedimented at 16,000 × g for 10 min to recover the medium (MD) fraction. Cells in the pellet were washed, suspended in cytoplasmic buffer (50 mM HEPES, 66 mM potassium acetate, 10 mM magnesium acetate [pH 7.5]), and lysed by bead beating for 10 min at 4.5 m/s to yield the total cell lysate (T). Lysates were subjected to ultracentrifugation at 100,000 × g for 1 h. Soluble proteins from the cytoplasm (C) were removed, and pellets were suspended in 1 M Tris-HCl (pH 8.0)–4% SDS to extract membrane proteins (Mb). Proteins in all samples were precipitated with trichloroacetic acid, washed with acetone, and solubilized in sample buffer (4% SDS, 1% β-mercaptoethanol, 10% glycerol, 50 mM Tris-HCl [pH 7.5], 0.2% bromophenol blue) prior to separation by SDS-PAGE. Proteins in gels were stained with Coomassie brilliant blue or electrotransferred to a polyvinylidene difluoride (PVDF) membrane for immunoblot analysis using rabbit antisera raised against purified antigens or a HisProbe horseradish peroxidase (HRP) conjugate (Promega). Immune complexes were revealed by chemiluminescence using HRP-conjugated secondary antibody (Cell Signaling Technology).
Purification of SCWP and MALDI-TOF mass spectrometry.
Bacteria were scraped off agar plates, suspended in 25 ml water, sedimented by centrifugation for 10 min at 6,000 × g, and washed once in water. Next, cells were suspended in 400 ml 4% SDS, boiled for 30 min, washed, suspended in water, and mechanically lysed with 0.1-mm glass beads. The resulting murein sacculi were sedimented at 17,000 × g for 15 min, suspended in 100 mM Tris-HCl (pH 7.5), and incubated for 4 h at 37°C with 10 μg/ml DNase and 10 μg/ml RNase supplemented with 20 mM MgSO4. Samples were incubated for 16 h at 37°C with 10 μM trypsin supplemented with 10 mM CaCl2. Enzymes were inactivated by boiling for 30 min in a water bath in 1% SDS. The SDS was removed by 5 cycles of centrifugation and washing in water. Murein sacculi were washed with water, 100 mM Tris-HCl (pH 8.0), water, 0.1 M EDTA (pH 8.0), water, and acetone, and twice more with water, before suspension in 5 ml of water and the addition of 25 ml of 48% hydrofluoric acid (HF). Samples were incubated overnight on ice with shaking and centrifuged at 17,000 × g for 15 min. The SCWP-containing supernatant was mixed with ice-cold ethanol in a 1:5 ratio, causing SCWP precipitation. The polysaccharide was washed extensively with ice-cold ethanol, recovered by centrifugation at 17,000 × g at 4°C for 15 min, and suspended in water to a concentration of 100 mg/ml. One hundred microliters of this preparation was subjected to reversed-phase high-performance liquid chromatography (RP-HPLC) analysis as described previously (18, 47). RP-HPLC fractions containing SCWP were spotted onto a prespotted AnchorChip II (PAC II) plate (Bruker) that contained an α-cyano-4-hydroxycinnamic acid (HCCA) matrix and subjected to MALDI-TOF mass spectrometry using a Bruker autoflex speed MALDI instrument in positive-reflectron mode. Calibrants were used directly from the PAC II plate. Predicted molecular weights were calculated using the following average incremental values based on the atomic weights of the elements: 162.142 for hexose, 203.195 for 2-N-acetamido-2-deoxyhexose, and 18.0153 for the free reducing end.
Supplementary Material
ACKNOWLEDGMENTS
EAII6G6 antibody was a previous gift from Teresa Abshire and Arthur Friedlander. We thank Stephanie Willing and members of our laboratory for experimental advice and discussion.
This research was supported by grant AI069227 from the National Institute of Allergy and Infectious Diseases Infectious Disease Branch.
Author order was determined on the basis of contribution to the experimental design and its execution.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Choudhury B, Leoff C, Saile E, Wilkins P, Quinn CP, Kannenberg EL, Carlson RW. 2006. The structure of the major cell wall polysaccharide of Bacillus anthracis is species-specific. J Biol Chem 281:27932–27941. doi: 10.1074/jbc.M605768200. [DOI] [PubMed] [Google Scholar]
- 2.Mesnage S, Fontaine T, Mignot T, Delepierre M, Mock M, Fouet A. 2000. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J 19:4473–4484. doi: 10.1093/emboj/19.17.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kern J, Ryan C, Faull K, Schneewind O. 2010. Bacillus anthracis surface-layer proteins assemble by binding to the secondary cell wall polysaccharide in a manner that requires csaB and tagO. J Mol Biol 401:757–775. doi: 10.1016/j.jmb.2010.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang YT, Oh SY, Hendrickx AP, Lunderberg JM, Schneewind O. 2013. Bacillus cereus G9241 S-layer assembly contributes to the pathogenesis of anthrax-like disease in mice. J Bacteriol 195:596–605. doi: 10.1128/JB.02005-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forsberg LS, Abshire TG, Friedlander A, Quinn CP, Kannenberg EL, Carlson RW. 2012. Localization and structural analysis of a conserved pyruvylated epitope in Bacillus anthracis secondary cell wall polysaccharides and characterization of the galactose-deficient wall polysaccharide from avirulent B. anthracis CDC 684. Glycobiology 22:1103–1117. doi: 10.1093/glycob/cws080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lunderberg JM, Nguyen-Mau SM, Richter GS, Wang YT, Dworkin J, Missiakas DM, Schneewind O. 2013. Bacillus anthracis acetyltransferases PatA1 and PatA2 modify the secondary cell wall polysaccharide and affect the assembly of S-layer proteins. J Bacteriol 195:977–989. doi: 10.1128/JB.01274-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couture-Tosi E, Delacroix H, Mignot T, Mesnage S, Chami M, Fouet A, Mosser G. 2002. Structural analysis and evidence for dynamic emergence of Bacillus anthracis S-layer networks. J Bacteriol 184:6448–6456. doi: 10.1128/jb.184.23.6448-6456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mignot T, Mesnage S, Couture-Tosi E, Mock M, Fouet A. 2002. Developmental switch of S-layer protein synthesis in Bacillus anthracis. Mol Microbiol 43:1615–1627. doi: 10.1046/j.1365-2958.2002.02852.x. [DOI] [PubMed] [Google Scholar]
- 9.Kern VJ, Kern JW, Theriot JA, Schneewind O, Missiakas D. 2012. Surface-layer (S-layer) proteins Sap and EA1 govern the binding of the S-layer-associated protein BslO at the cell septa of Bacillus anthracis. J Bacteriol 194:3833–3840. doi: 10.1128/JB.00402-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson VJ, Kern JW, McCool JW, Schneewind O, Missiakas D. 2011. The SLH-domain protein BslO is a determinant of Bacillus anthracis chain length. Mol Microbiol 81:192–205. doi: 10.1111/j.1365-2958.2011.07688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh SY, Lunderberg JM, Chateau A, Schneewind O, Missiakas D. 2017. Genes required for Bacillus anthracis secondary cell wall polysaccharide synthesis. J Bacteriol 199:e00613-16. doi: 10.1128/JB.00613-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Low LY, Yang C, Perego M, Osterman A, Liddington RC. 2005. Structure and lytic activity of a Bacillus anthracis prophage endolysin. J Biol Chem 280:35433–35439. doi: 10.1074/jbc.M502723200. [DOI] [PubMed] [Google Scholar]
- 13.Schuch R, Nelson D, Fischetti VA. 2002. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 418:884–889. doi: 10.1038/nature01026. [DOI] [PubMed] [Google Scholar]
- 14.Ganguly J, Low LY, Kamal N, Saile E, Forsberg LS, Gutierrez-Sanchez G, Hoffmaster AR, Liddington R, Quinn CP, Carlson RW, Kannenberg EL. 2013. The secondary cell wall polysaccharide of Bacillus anthracis provides the specific binding ligand for the C-terminal cell wall-binding domain of two phage endolysins, PlyL and PlyG. Glycobiology 23:820–832. doi: 10.1093/glycob/cwt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mo KF, Li X, Li H, Low LY, Quinn CP, Boons GJ. 2012. Endolysins of Bacillus anthracis bacteriophages recognize unique carbohydrate epitopes of vegetative cell wall polysaccharides with high affinity and selectivity. J Am Chem Soc 134:15556–15562. doi: 10.1021/ja3069962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forsberg LS, Choudhury B, Leoff C, Marston CK, Hoffmaster AR, Saile E, Quinn CP, Kannenberg EL, Carlson RW. 2011. Secondary cell wall polysaccharides from Bacillus cereus strains G9241, 03BB87 and 03BB102 causing fatal pneumonia share similar glycosyl structures with the polysaccharides from Bacillus anthracis. Glycobiology 21:934–948. doi: 10.1093/glycob/cwr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okinaka RT, Price EP, Wolken SR, Gruendike JM, Chung WK, Pearson T, Xie G, Munk C, Hill KK, Challacombe J, Ivins BE, Schupp JM, Beckstrom-Sternberg SM, Friedlander A, Keim P. 2011. An attenuated strain of Bacillus anthracis (CDC 684) has a large chromosomal inversion and altered growth kinetics. BMC Genomics 12:477. doi: 10.1186/1471-2164-12-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chateau A, Lunderberg JM, Oh SY, Abshire T, Friedlander A, Quinn CP, Missiakas DM, Schneewind O. 2018. Galactosylation of the secondary cell wall polysaccharide of Bacillus anthracis and its contribution to anthrax pathogenesis. J Bacteriol 200:e00562-17. doi: 10.1128/JB.00562-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leoff C, Saile E, Sue D, Wilkins P, Quinn CP, Carlson RW, Kannenberg EL. 2008. Cell wall carbohydrate compositions of strains from the Bacillus cereus group of species correlate with phylogenetic relatedness. J Bacteriol 190:112–121. doi: 10.1128/JB.01292-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Missiakas D, Schneewind O. 2017. Assembly and function of the Bacillus anthracis S-layer. Annu Rev Microbiol 71:79–98. doi: 10.1146/annurev-micro-090816-093512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuch R, Pelzek AJ, Raz A, Euler CW, Ryan PA, Winer BY, Farnsworth A, Bhaskaran SS, Stebbins CE, Xu Y, Clifford A, Bearss DJ, Vankayalapati H, Goldberg AR, Fischetti VA. 2013. Use of a bacteriophage lysin to identify a novel target for antimicrobial development. PLoS One 8:e60754. doi: 10.1371/journal.pone.0060754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang YT, Missiakas D, Schneewind O. 2014. GneZ, a UDP-GlcNAc 2-epimerase, is required for S-layer assembly and vegetative growth of Bacillus anthracis. J Bacteriol 196:2969–2978. doi: 10.1128/JB.01829-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy M, Gowrishankar J. 1997. Identification and characterization of ssb and uup mutants with increased frequency of precise excision of transposon Tn10 derivatives: nucleotide sequence of uup in Escherichia coli. J Bacteriol 179:2892–2899. doi: 10.1128/jb.179.9.2892-2899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlier L, Haase AS, Burgos Zepeda MY, Dassa E, Lequin O. 2012. The C-terminal domain of the Uup protein is a DNA-binding coiled coil motif. J Struct Biol 180:577–584. doi: 10.1016/j.jsb.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Goldstein IJ, Hayes CE. 1978. The lectins: carbohydrate-binding proteins of plants and animals. Adv Carbohydr Chem Biochem 35:127–340. doi: 10.1016/s0065-2318(08)60220-6. [DOI] [PubMed] [Google Scholar]
- 26.Ezzell JW Jr, Abshire TG, Little SF, Lidgerding BC, Brown C. 1990. Identification of Bacillus anthracis by using monoclonal antibody to cell wall galactose-N-acetylglucosamine polysaccharide. J Clin Microbiol 28:223–231. doi: 10.1128/JCM.28.2.223-231.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knibbs RN, Goldstein IJ, Ratcliffe RM, Shibuya N. 1991. Characterization of the carbohydrate binding specificity of the leukoagglutinating lectin from Maackia amurensis. Comparison with other sialic acid-specific lectins. J Biol Chem 266:83–88. [PubMed] [Google Scholar]
- 28.Kamal N, Ganguly J, Saile E, Klee SR, Hoffmaster A, Carlson RW, Forsberg LS, Kannenberg EL, Quinn CP. 2017. Structural and immunochemical relatedness suggests a conserved pathogenicity motif for secondary cell wall polysaccharides in Bacillus anthracis and infection-associated Bacillus cereus. PLoS One 12:e0183115. doi: 10.1371/journal.pone.0183115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown S, Santa Maria JP Jr, Walker S. 2013. Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol 67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lunderberg JM, Liszewski Zilla M, Missiakas D, Schneewind O. 2015. Bacillus anthracis tagO is required for vegetative growth and secondary cell wall polysaccharide synthesis. J Bacteriol 197:3511–3520. doi: 10.1128/JB.00494-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liszewski Zilla M, Chan YG, Lunderberg JM, Schneewind O, Missiakas D. 2015. LytR-CpsA-Psr enzymes as determinants of Bacillus anthracis secondary cell wall polysaccharide assembly. J Bacteriol 197:343–353. doi: 10.1128/JB.02364-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liszewski Zilla M, Lunderberg JM, Schneewind O, Missiakas D. 2015. Bacillus anthracis lcp genes support vegetative growth, envelope assembly, and spore formation. J Bacteriol 197:3731–3741. doi: 10.1128/JB.00656-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allison GE, Verma NK. 2000. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol 8:17–23. doi: 10.1016/s0966-842x(99)01646-7. [DOI] [PubMed] [Google Scholar]
- 34.Mann E, Whitfield C. 2016. A widespread three-component mechanism for the periplasmic modification of bacterial glycoconjugates. Can J Chem 94:883–893. doi: 10.1139/cjc-2015-0594. [DOI] [Google Scholar]
- 35.Huan PT, Bastin DA, Whittle BL, Lindberg AA, Verma NK. 1997. Molecular characterization of the genes involved in O-antigen modification, attachment, integration and excision in Shigella flexneri bacteriophage SfV. Gene 195:217–227. doi: 10.1016/s0378-1119(97)00143-1. [DOI] [PubMed] [Google Scholar]
- 36.Huan PT, Whittle BL, Bastin DA, Lindberg AA, Verma NK. 1997. Shigella flexneri type-specific antigen V: cloning, sequencing and characterization of the glucosyl transferase gene of temperate bacteriophage SfV. Gene 195:207–216. doi: 10.1016/s0378-1119(97)00144-3. [DOI] [PubMed] [Google Scholar]
- 37.Guan S, Bastin DA, Verma NK. 1999. Functional analysis of the O antigen glucosylation gene cluster of Shigella flexneri bacteriophage SfX. Microbiology 145(Part 5):1263–1273. doi: 10.1099/13500872-145-5-1263. [DOI] [PubMed] [Google Scholar]
- 38.Nikaido H, Nikaido K, Nakae T, Makela PH. 1971. Glucosylation of lipopolysaccharide in Salmonella: biosynthesis of O antigen factor 12 2. I. Over-all reaction. J Biol Chem 246:3902–3911. [PubMed] [Google Scholar]
- 39.Mancuso DJ, Chiu TH. 1982. Biosynthesis of glucosyl monophosphoryl undecaprenol and its role in lipoteichoic acid biosynthesis. J Bacteriol 152:616–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwasaki H, Shimada A, Yokoyama K, Ito E. 1989. Structure and glycosylation of lipoteichoic acids in Bacillus strains. J Bacteriol 171:424–429. doi: 10.1128/jb.171.1.424-429.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rismondo J, Percy MG, Grundling A. 2018. Discovery of genes required for lipoteichoic acid glycosylation predicts two distinct mechanisms for wall teichoic acid glycosylation. J Biol Chem 293:3293–3306. doi: 10.1074/jbc.RA117.001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kho K, Meredith TC. 2018. Salt-induced stress stimulates a lipoteichoic acid-specific three-component glycosylation system in Staphylococcus aureus. J Bacteriol 200:e00017-18. doi: 10.1128/JB.00017-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rismondo J, Haddad TFM, Shen Y, Loessner MJ, Grundling A. 2020. GtcA is required for LTA glycosylation in Listeria monocytogenes serovar 1/2a and Bacillus subtilis. Cell Surf 6:100038. doi: 10.1016/j.tcsw.2020.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marraffini LA, Schneewind O. 2006. Targeting proteins to the cell wall of sporulating Bacillus anthracis. Mol Microbiol 62:1402–1417. doi: 10.1111/j.1365-2958.2006.05469.x. [DOI] [PubMed] [Google Scholar]
- 45.Bubeck Wardenburg J, Williams WA, Missiakas D. 2006. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc Natl Acad Sci U S A 103:13831–13836. doi: 10.1073/pnas.0603072103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Green BD, Battisti L, Koehler TM, Thorne CB, Ivins BE. 1985. Demonstration of a capsule plasmid in Bacillus anthracis. Infect Immun 49:291–297. doi: 10.1128/IAI.49.2.291-297.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chateau A, Schneewind O, Missiakas D. 2019. Extraction and purification of wall-bound polymers of Gram-positive bacteria. Methods Mol Biol 1954:47–57. doi: 10.1007/978-1-4939-9154-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.