Abstract
Background
The ability to identify bacterial pathogens that necessitate specific clinical management or public health action in children with acute gastroenteritis is crucial to patient care and public health. However, existing stool-testing guidelines offer inconsistent recommendations, and their performance characteristics are unknown. We evaluated 6 leading gastroenteritis guidelines (eg, those of the Centers for Disease Control and Prevention and Infectious Disease Society of America) that recommend when to test children’s stool for bacterial enteropathogens.
Methods
Via 2 emergency departments in Alberta, Canada, we enrolled 2447 children <18 years old who presented with ≥3 episodes of diarrhea and/or vomiting in a 24-hour period. All participants were tested for 9 bacterial enteropathogens: Aeromonas, Campylobacter, Escherichia coli O157, other Shiga toxin–producing E. coli, enterotoxigenic E. coli, Salmonella, Shigella, Vibrio, and Yersinia. Patient data gathered at the index visit were used to determine whether guidelines would recommend testing. Sensitivity and specificity to recommend testing for children with bacterial enteropathogens were calculated for each guideline.
Results
Outcome data were available for 2391 (97.7%) participants, and 6% (144/2391) of participants tested positive for a bacterial enteropathogen. Guideline sensitivity ranged from 25.8% (95% confidence interval [CI] 18.7–33.0%) to 66.9% (95% CI 59.3–74.6%), and varied for individual pathogens. Guideline specificity for all bacterial enteropathogens ranged from 63.6% (95% CI 61.6–65.6%) to 96.5% (95% CI 95.7–97.2%).
Conclusions
No guideline provided optimally balanced performance. The most sensitive guidelines missed one-third of cases and would drastically increase testing volumes. The most specific guidelines missed almost 75% of cases.
Keywords: acute gastroenteritis, enteric bacteria, stool culture, culture-independent diagnostic testing, laboratory utilization
The sensitivity and specificity of stool-testing guidelines regarding microbiologic testing for children with suspected bacterial enteropathogens vary widely. Sensitivity ranged from 25.8% to 66.9%, and specificity from 63.6% to 96.5%. No guideline optimally balanced sensitivity and laboratory utilization.
There are estimated to be 17 million annual episodes of acute gastroenteritis in the United Kingdom [1] and 179 million in the United States [2]. Routine performance of stool cultures to identify bacterial enteropathogens is not recommended [3, 4], because viral pathogens are the most common etiologic agents [5], illness is generally self-limited, and the aggregate cost of bacterial culture is high [6]. However, the identification of bacterial infections can guide therapy (eg, antibiotic avoidance in children infected with Shiga toxin-producing Escherichia coli [STEC] [7–9]) and public health investigations and interventions (eg, daycare/work exclusions, outbreak identification, and contact tracing) [10, 11]. Thus, guidelines that maximize the identification of bacterial illness while optimizing laboratory utilization are needed.
Several gastroenteritis guidelines provide recommendations regarding when to perform stool cultures or microbiologic testing targeting bacterial enteropathogens [4, 12–15], but they offer inconsistent criteria [3] and their performance for detecting pathogens of interest is unknown. Moreover, differences in guideline performance may exist between laboratory diagnostic approaches (eg, bacterial culture vs culture-independent diagnostic tests [CIDT]). To fill this knowledge gap, we compared the performance of 6 common pediatric stool-testing guidelines in a comprehensively tested cohort of children seeking care for acute gastroenteritis.
METHODS
Study Population
This study is a secondary analysis of data from the Alberta Provincial Pediatric EnTeric Infection TEam (APPETITE) study [16]. Children <18 years old with acute gastroenteritis were prospectively and consecutively enrolled between December 2014 and February 2018. Participants for this analysis were recruited in the emergency departments (EDs) of Alberta Children’s Hospital (Calgary, Alberta, Canada) and Stollery Children’s Hospital (Edmonton, Alberta, Canada). Alberta’s incidence of acute gastroenteritis is slightly higher than the average incidence throughout Canada [17], and the diarrhea prevalence in Canada is comparable to the United States and Australia [18].
Inclusion criteria required that participants had ≥3 episodes of vomiting and/or diarrhea in a 24-hour period, with <7 days of symptoms. Exclusion criteria included enrollment within the prior 14 days; an anticipated inability to complete the 14-day follow-up, including difficulty speaking and understanding English; neutropenia; emergent medical needs that preclude recruitment; and a visit related to coexisting mental health concerns. Study personnel obtained demographic and clinical data during the initial visit and 14 days after enrollment. Clinicians ordered stool testing as they routinely would (Supplementary Material). Details of clinical stool culture performance were obtained via chart review. APPETITE was approved by the Research Ethics Boards of both the University of Calgary and University of Alberta (REB14-1122). Informed consent was provided by caregivers; assent was obtained from the participants themselves when they were deemed mature enough to understand the study procedures and the potential benefits and harms.
Outcome Testing
We tested for 9 bacterial enteropathogens: Aeromonas, Campylobacter, Escherichia coli O157, other STEC, enterotoxigenic E. coli (ETEC), Salmonella, Shigella, Vibrio, and Yersinia. The primary outcome was the detection of any 1 of these agents from any of the tests. Secondary outcomes were (1) groupings of bacterial enteropathogens with high potential clinical or public health importance (Supplementary Material) and (2) specific bacterial enteropathogens (eg, Salmonella).
Full details of specimen retrieval, transport, storage, and testing have been described (Supplementary Material) [19]. Study personnel collected 2 rectal swabs and a stool sample, if available, from each child. All specimens were tested for 9 bacterial and 9 nonbacterial [20] enteropathogens (Supplementary Table S1). The Luminex xTAG Gastrointestinal Pathogen Panel (Luminex Molecular Diagnostics, Toronto, Canada) bacterial targets are Campylobacter spp., E. coli O157, STEC stx1/stx2, ETEC heat-labile (LT) and heat-stable (ST) toxin, Salmonella spp., Shigella spp., Vibrio cholerae, and Yersinia enterocolitica. Culture was used to detect Aeromonas spp., Campylobacter spp., E. coli O157, Salmonella spp., Shigella spp., Vibrio spp., and Yersinia spp. (except Y. pestis). A child was classified as positive for a bacterial enteropathogen if either a rectal swab or stool specimen yielded positive results using either test. The Gastrointestinal Pathogen Panel also detects Clostridioides [21] difficile toxin A/B, but this organism was not included in this analysis, because separate guidelines exist for C. difficile testing [22, 23]. Outcome testing was completed prior to and independently of stool-testing guideline application.
Stool-testing Guidelines
Guidelines were chosen for evaluation based on literature review and expert consensus opinion (Supplementary Material). There were 4 guidelines from leading organizations (Table 1): the 2003 US Centers for Disease Control and Prevention (CDC) guideline [12]; the 2009 (updated in 2014) British National Institute for Health and Care Excellence (NICE) guideline [4], which apply to children <5 years of age; the 2014 European Society for Pediatric Gastroenterology, Hepatology, and Nutrition (ESPGHAN) guideline [14]; and the 2017 Infectious Diseases Society of America (IDSA) guideline [15]. Criteria for culture proposed by Klein et al in 2006 [24] after a single-center analysis of children with diarrhea and a guideline published by Hatchette and Farina in 2011 [13] were also included (Table 1).
Table 1.
Case Characteristics and Symptoms Referenced in Acute Gastroenteritis Guidelines, Algorithms, and Published Reports
| CDC [12] | Klein et ala [24] | NICEb [4] | Hatchette and Farina [13] | ESPGHAN [14] | IDSA [15] | |
|---|---|---|---|---|---|---|
| Applies to | Children with ≥3 diarrhea events in 24 hours | Children and young adults with diarrhea | Children <5yo with ≤14 days diarrhea | Children and adults with ≥1 day diarrhea | Children with decrease in stool consistency and/or increase in frequency | Children and adults with diarrhea |
| Recent antibiotics use | - | absent | - | - | - | - |
| International travel | - | history | recent history | - | history | recent history if diarrhea lasts ≥14 days |
| Daycare attendancec | - | - | - | present | - | - |
| Underlying chronic conditiond | - | - | - | present | present | - |
| Diarrhea events in 24 hours | - | >10 | - | - | >10e | - |
| Diarrhea duration | - | ≤10 days | ≥7 | - | >14 daysf | - |
| Vomiting events in 24 hours | - | <1 | - | - | - | - |
| Blood in stool | present | present | present | present | present | present |
| Fever | - | present | - | present | - | present |
| Dehydration score | - | - | - | present | (severe)e | - |
| Maximal pain level | - | - | - | - | - | (severe) |
| Other criteriag | X | - | X | X | X | X |
Values in parentheses were used for the sensitivity analysis only.
Abbreviations: APPETITE, Alberta Provincial Pediatric Enteric Infection Team; CDC, US Centers for Disease Control and Prevention; ESPGHAN, European Society for Pediatric Gastroenterology, Hepatology, and Nutrition; IDSA, Infectious Disease Society of America; NICE, National Institute for Health and Care Excellence; yo, years old
aBlood in the stool or the combination of all other criteria indicated testing.
bReviewed by NICE in 2012 and 2014.
cGuideline indicated testing in daycare workers; to adapt this for a pediatric population, we converted the criterion to daycare attendance.
dGuidelines did not enumerate qualifying conditions, and APPETITE was similarly unrestrictive in defining chronic conditions.
eGuideline indicated “extremely severe conditions”; we applied this as a high number of diarrheal episodes and, for the sensitivity analysis, severe dehydration.
fGuideline indicated “prolonged” illness; we applied this as >14 days.
gCriteria not available or not applicable to APPETITE population, including immune status, doubt about gastroenteritis diagnosis, septicemia, particular exposures, and inclusion in an outbreak.
For each guideline, criteria were reviewed, and corresponding data collected by the APPETITE study were identified without reference to bacterial enteropathogen positivity associated with particular fields. All guidelines were written for individuals with diarrhea (Table 1). The only common criterion to test was blood in stool. Several criteria lacked details; to include them, we established a priori definitions (Supplementary Material). A test/don’t test decision for each child by each guideline was determined independently of bacterial enteropathogen positivity. Although multiple guidelines recommended testing individuals who appear septic or are immunocompromised, these criteria could not be assessed in our study, because such children were excluded.
Statistical Analysis
We summarized the frequency of pathogens detected in the cohort by primary symptoms at enrollment and 14-day follow-up as having vomiting (ie, ≥3 episodes of vomiting in a 24-hour period), diarrhea (ie, ≥3 episodes of diarrhea in a 24-hour period), or both (Table 2). We described the frequency of bacterial enteropathogens detected according to important demographic and clinical variables at enrollment (Table 3).
Table 2.
Pathogen Detection by Symptom Complex at Index Presentation and at the Completion of 14-Day Follow-up
| Pathogen | Total Casesa | Symptom Complex Index Presentation n (%) |
Symptom Complex 14-Day Follow-up n (%) |
||||
|---|---|---|---|---|---|---|---|
| Isolated Diarrhea | Diarrhea + Vomiting | Isolated Vomiting | Isolated Diarrhea | Diarrhea + Vomiting | Isolated Vomiting | ||
| Bacteria | 144 | 59 (41.0) | 49 (34.0) | 36 (25.0) | 55 (38.2) | 64 (44.4) | 25 (17.4) |
| Salmonella spp. | 54 | 32 (59) | 12 (22) | 10 (19) | 29 (54) | 18 (33) | 7 (13) |
| Aeromonas spp. | 26 | 5 (19) | 11 (42) | 10 (38) | 4 (15) | 14 (54) | 8 (31) |
| Campylobacter spp. | 18 | 8 (44) | 8 (44) | 2 (11) | 8 (44) | 9 (50) | 1 (6) |
| STEC, non-O157 | 17 | 4 (24) | 8 (47) | 5 (29) | 4 (24) | 9 (53) | 4 (24) |
| Escherichia coli O157 | 10 | 4 (40) | 3 (30) | 3 (30)b | 4 (40) | 5 (50) | 1 (10) |
| Shigella spp. | 8 | 3 (38) | 4 (50) | 1 (13) | 3 (38) | 4 (50) | 1 (13) |
| ETEC | 6 | 2 (33) | 2 (33) | 2 (33) | 2 (33) | 4 (67) | 0 (0) |
| Yersinia spp. | 5 | 1 (20) | 1 (20) | 3 (60) | 1 (20) | 1 (20) | 3 (60) |
| Vibrio spp. | 0 | - | - | - | - | - | - |
| Clostridioides difficile c | 46 | 4 (9) | 11 (24) | 31 (67) | 4 (9) | 20 (43) | 22 (48) |
| Virus(es) | 1520 | 212 (13.9) | 585 (38.5) | 723 (47.6) | 188 (12.4) | 893 (58.8) | 439 (28.9) |
| Parasite(s) | 11 | 1 (9) | 5 (45) | 5 (45) | 0 (0) | 7 (64) | 4 (36) |
| No pathogen detected | 769 | 153 (19.9) | 135 (17.6) | 481 (62.5) | 147 (19.1) | 234 (30.4) | 388 (50.5) |
Symptom complex refers to the combination of ≥3 diarrhea episodes or ≥3 vomiting episodes in a 24-hour period, necessary to meet the definition of acute gastroenteritis. Percentiles for pathogen categories with >100 cases were rounded to the first decimal place, and others were rounded to the nearest whole number.
Abbreviations: ETEC, enterotoxigenic Escherichia coli; PCR, polymerase chain reaction; spp, species; STEC, Shiga toxin-producing Escherichia coli.
aTotal does not equal the 2391 sample size because of coinfections, including viral coinfections among 54 children with a bacterial enteropathogen. No children were infected with >1 bacterial enteropathogen. See Supplementary Table S2 for additional information on coinfection.
bAll E. coli O157 cases who presented with vomiting but did not meet the definition of diarrhea were PCR-positive for E. coli O157, PCR-negative for Shiga toxin genes, and culture-negative. There was 1 case with diarrhea but no vomiting that also had this combination of laboratory results.
cIncludes only cases ≥2 years old.
Table 3.
Frequency of Bacterial Enteropathogen Detection by Demographic Characteristics and Acute Gastroenteritis Event Data at Index Presentation
| Na | Bacteria Detected (%) | |
|---|---|---|
| Total | 2391 | 144 (6.0) |
| Case characteristics | ||
| Age, years | ||
| <1 | 714 | 39 (5.5) |
| 1 to <2 | 628 | 32 (5.1) |
| 2 to <3 | 294 | 16 (5.4) |
| 3 to <4 | 191 | 10 (5.2) |
| 4 to <5 | 150 | 11 (7.3) |
| 5 to <11 | 345 | 31 (9.0) |
| 11 to <18 | 69 | 5 (7.2) |
| Antibiotics, previous 60 days | ||
| Yes | 396 | 19 (4.8) |
| No | 1954 | 120 (6.1) |
| International travel,b previous 12 months | ||
| Yes | 442 | 40 (9.0) |
| No | 1943 | 103 (5.3) |
| Daycare attendance | ||
| Yes | 739 | 32 (4.3) |
| No | 1650 | 112 (6.8) |
| Chronic medical condition | ||
| Yes | 263 | 9 (3.4) |
| No | 2128 | 135 (6.3) |
| Season | ||
| Winter (December–February) | 543 | 23 (4.2) |
| Spring (March–May) | 641 | 27 (4.2) |
| Summer (June–August) | 630 | 49 (7.8) |
| Fall (September–November) | 577 | 45 (7.8) |
| Clinical event data | ||
| Diarrhea events in 24 hours | ||
| 0 | 957 | 27 (2.8) |
| 1–2 | 275 | 9 (3.3) |
| 3–5 | 476 | 21 (4.4) |
| 6–10 | 411 | 42 (10) |
| >10 | 272 | 45 (17) |
| Any | 1434 | 117 (8.2) |
| Diarrhea duration, days | ||
| <3.0 | 873 | 67 (7.7) |
| 3.0 to <5.0 | 401 | 37 (9.2) |
| ≥5 | 160 | 13 (8.1) |
| Vomiting events in 24 hours | ||
| 0 | 268 | 43 (16) |
| 1–2 | 145 | 16 (11) |
| 3–5 | 839 | 37 (4.4) |
| 6–10 | 702 | 30 (4.3) |
| >10 | 437 | 18 (4.1) |
| Any | 2123 | 101 (4.8) |
| Vomiting duration, days | ||
| <3.0 | 1571 | 74 (4.7) |
| 3.0 to <5.0 | 406 | 20 (4.9) |
| ≥5 | 147 | 7 (4.8) |
| Fever (subjective) | ||
| Yes | 1034 | 87 (8.4) |
| No | 1225 | 47 (3.8) |
| Highest temperature, °C | ||
| <38.0 | 168 | 10 (6.0) |
| 38.0 to <39.0 | 356 | 31 (8.7) |
| ≥39.0 | 407 | 33 (8.1) |
| Bloody diarrhea | ||
| Yes | 99 | 31 (31) |
| No | 1826 | 77 (4.2) |
| Dehydration score | ||
| None (0) | 224 | 17 (7.6) |
| Some (1–4) | 1310 | 74 (5.6) |
| Moderate/ severe (5–8) | 852 | 53 (6.2) |
| Maximal pain level | ||
| None (0) | 261 | 7 (2.7) |
| Mild (1–3) | 347 | 14 (4.0) |
| Moderate (4–6) | 698 | 44 (6.3) |
| Severe (7–10) | 1076 | 79 (7.3) |
| Stool culture ordered during emergency department visit | ||
| Yes | 165 | 45 (27) |
| No | 2226 | 99 (4.4) |
aIncludes only participants whose samples were tested for bacterial enteropathogens.
bOutside Canada and the United States.
The presence of bloody diarrhea was not recorded for participants enrolled between 1 December 2014 and 29 October 2015. We imputed missing data using multiple imputation by chained equations, as implemented in the mice [25] analysis package for the R [26] statistical computing environment (Supplementary Material). 5 datasets were imputed using 50 iterations each. Descriptive statistics were based on reported, not imputed, data.
For the primary outcome of positivity for any bacterial enteropathogen, we calculated sensitivity and specificity for the identification of children with bacterial enteropathogens. Using symptoms reported at the enrollment visit, we applied the guidelines to each of the 5 multiply imputed datasets to determine which children the guidelines would recommend to have stool testing. We calculated sensitivity and specificity in each dataset, and pooled them with their variances using Rubin’s rules [27]. We calculated asymptotic 95% confidence intervals (CIs) from the pooled variance of each measure. The confusion matrix for each guideline was back-calculated by applying the pooled sensitivity and specificity estimates to the total of bacterial enteropathogen–positive and –negative children, respectively.
We calculated positive and negative predictive values of the guidelines for a range of bacterial enteropathogen prevalences among children with acute gastroenteritis by applying Bayes Theorem to the pooled sensitivity and specificity estimates (Supplementary Material).
For secondary outcomes, we calculated the sensitivity and specificity of the guidelines to recommend testing for children with combinations of bacterial enteropathogens that clinicians or public health practitioners may identify as particularly important. We calculated bacterial enteropathogen–specific case ascertainment of the guidelines by pooling the pathogen-specific sensitivity measures from each of the multiply imputed datasets, as in the primary analysis, with the exception that exact CIs were calculated using the Clopper-Pearson method to account for small cell sizes [28].
We determined the sensitivity of our results to changes in 3 aspects of the analysis: modified guideline criteria definitions, weaker assumptions for missing data patterns, and separate analyses of culture and CIDT positives (Supplementary Material).
R (base 3.4.3) [26] with packages plyr [29], epiR [30], ggplot2 [31], and binom, [32] was used for statistical analyses.
RESULTS
During the study period, APPETITE enrolled 2447 children, among whom 2391 (97.7%) provided stool specimens and/or rectal swabs that underwent microbiologic testing: bacterial enteropathogens were detected in 144 (6.0%). Salmonella (n = 54), Aeromonas (n = 26), and Campylobacter (n = 18) were the most common bacterial enteropathogens (Table 2; Supplementary Table S2). Vomiting was present without diarrhea at enrollment in 36 (25.0%) of the 144 children with a detected bacterial enteropathogen; at the 14-day follow-up, 25 (17.4%) children with a detected bacterial enteropathogen still had no diarrhea (Table 2; Supplementary Table S3).
The proportion of children with a bacterial enteropathogen detected was highest for children with bloody diarrhea (31%, 31/99; Table 3). During the ED visit, stool cultures were ordered for 165 children by the attending physician (separate from study testing).
Guideline Performance
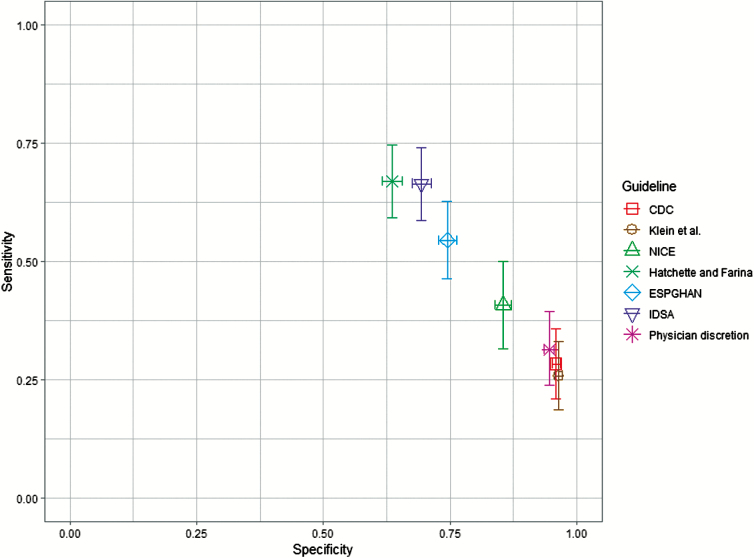
Sensitivity to detect bacterial enteropathogens ranged from 25.8% (95% CI 18.7–33.0%) for the Klein et al [24] criteria to 66.9% (95% CI 59.3–74.6%) for the Hatchette and Farina [13] guideline (Figure 1; Supplementary Table S4). Specificity ranged from 63.6% (95% CI 61.6–65.6%) for the Hatchette and Farina [13] guideline to 96.5% (95% CI 95.7–97.2%) for the Klein et al criteria [24]. In comparison, stool cultures ordered during the ED visit by attending physicians as part of routine care (n = 165) had a sensitivity of 31.3% (45/144; 95% CI 23.8–39.5%) and specificity of 94.7% (95% CI 93.6–95.6%) (Figure 1).
Figure 1.
Sensitivity and specificity of 6 stool-testing guidelines for identifying children with bacterial enteropathogens. Guidelines were applied to a cohort of children <18 years old with acute gastroenteritis (n = 2391) and were evaluated for their ability to recommend testing for children with bacterial enteropathogens (n = 144). Sensitivity and specificity were calculated separately for each multiply imputed dataset and were pooled, with their variances, using Rubin’s rules. Error bars indicate 95% confidence intervals (bars did not extend past the width of the point for the specificity of the CDC[12] and Klein et al [24]guidelines). The NICE[4] guidelines applied only to children <5 years old (n = 1977; bacterial enteropathogens n = 108). Physician discretion refers to laboratory tests ordered as part of routine care by the attending physician, separate from the study. Abbreviations: CDC, US Centers for Disease Control and Prevention; ESPGHAN, European Society for Pediatric Gastroenterology, Hepatology, and Nutrition; IDSA, Infectious Disease Society of America; NICE, National Institute for Health and Care Excellence.
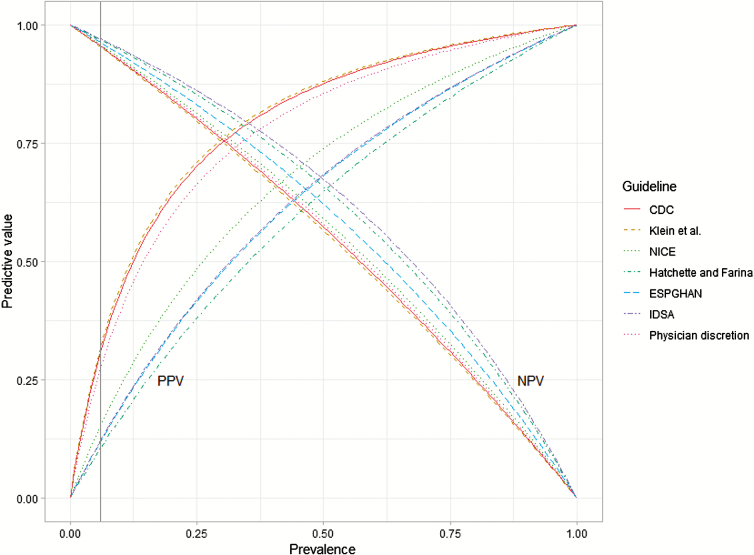
Guidelines recommended testing for 116 (Klein et al [24]) to 914 (Hatchette and Farina [13]) total children (Supplementary Table S4), the latter of which constitutes 38% (914/2391) of all study participants and 79% (914/1159) of those meeting the definition of diarrhea. Relative to the number of bacterial cultures ordered by ED physicians, this means application of the least specific guidelines would increase testing volumes 2- to 5-fold. The CDC [12] guideline and Klein et al [24] criteria had the highest positive predictive value (Figure 2; Supplementary Table S5). At low prevalence, there was little difference in the negative predictive value across guidelines.
Figure 2.
Predictive values of 6 stool-testing guidelines for identifying children with bacterial enteropathogens. Using the pooled sensitivity and specificity for each guideline, Bayes Theorem was used to calculate positive (from 0.0 to 1.1) and negative (from 0.1 to 1.0) predictive values. Prevalence refers to the proportion of acute gastroenteritis cases who would test positive for a bacterial enteropathogen. Physician discretion refers to laboratory tests ordered as part of routine care by the attending physician, separate from the study. Prevalence in the study cohort was 6.0% (vertical line). Abbreviations: CDC, US Centers for Disease Control and Prevention; ESPGHAN, European Society for Pediatric Gastroenterology, Hepatology, and Nutrition; IDSA, Infectious Disease Society of America; NICE, National Institute for Health and Care Excellence; NPV, negative predictive value; PPV, positive predictive value.
Secondary Outcomes
For the most restrictive bacterial enteropathogen set, including only Shigella spp., STEC, and Salmonella spp., sensitivity ranged from 29% (Klein et al [24]) to 73% (Hatchette and Farina [13]; Table 4). For the set including all bacterial enteropathogens except Aeromonas spp., sensitivity ranged from 31% (Klein et al [24]) to 71% (Hatchette and Farina [13]).
Table 4.
Sensitivity and Specificity of 6 Common Stool-testing Guidelines for Subsets of Bacterial Enteropathogens
| CDC [12] % (95% CI) |
Klein et al [24] % (95% CI) |
NICE [4] % (95% CI) |
Hatchette and Farina [13] % (95% CI) |
ESPGHAN [14] % (95% CI) |
IDSA [15] % (95% CI) |
|
|---|---|---|---|---|---|---|
| Shigella spp. + STEC + Salmonella spp. (n = 89, n<5 = 63) | ||||||
| Sensitivity | 32 (23–42) | 29 (20–39) | 48 (36–61) | 73 (64–82) | 60 (50–70) | 72 (63–81) |
| Specificity | 95 (95–96) | 96 (95–97) | 85 (83–87) | 63 (61–65) | 74 (72–76) | 68 (66–70) |
| Shigella spp. + STEC + Salmonella spp. + Campylobacter spp. (n = 107, n<5 = 72) | ||||||
| Sensitivity | 37 (28–46) | 33 (24–42) | 49 (38–61) | 74 (66–82) | 62 (53–72) | 73 (64–81) |
| Specificity | 96 (95–97) | 96 (96–97) | 85 (84–87) | 63 (61–65) | 74 (72–76) | 69 (67–70) |
| Shigella spp. + STEC + Salmonella spp. + Campylobacter spp. + Yersinia spp. (n = 112, n<5 = 77) | ||||||
| Sensitivity | 36 (27–45) | 33 (24–42) | 49 (38–60) | 72 (64–81) | 62 (53–71) | 71 (63–80) |
| Specificity | 96 (95–97) | 96 (96–97) | 85 (84–87) | 63 (61–65) | 74 (72–76) | 69 (67–70) |
| Shigella spp. + STEC + Salmonella spp. + Campylobacter spp. + Yersinia spp. + ETEC (n = 118, n<5 = 83) | ||||||
| Sensitivity | 34 (26–43) | 31 (23–40) | 48 (37–59) | 71 (63–80) | 60 (52–69) | 70 (62–78) |
| Specificity | 96 (95–97) | 96 (96–97) | 85 (84–87) | 63 (61–65) | 74 (72–76) | 69 (67–70) |
Guidelines were evaluated vs combinations of bacterial enteropathogens that had a high likelihood of necessitating specific clinical management or public health action. Sensitivity and specificity were calculated separately for each multiply-imputed dataset and pooled, with their variances, using Rubin’s rules. The 95% CIs were calculated using the normal approximation. NICE guidelines applied only to children <5 years old.
Abbreviations: CDC, US Centers for Disease Control and Prevention; CI, confidence interval; ESPGHAN, European Society for Pediatric Gastroenterology, Hepatology, and Nutrition; ETEC, enterotoxigenic Escherichia coli; IDSA, Infectious Disease Society of America; NICE, National Institute for Health and Care Excellence; spp., species; STEC, Shiga toxin-producing Escherichia coli.
For individual bacterial enteropathogens, case ascertainment was greatest for Shigella spp., ranging from 56% (NICE [4]) to 88% (IDSA [15]), and Campylobacter spp., ranging from 52% (Klein et al [24]) to 78% (Hatchette and Farina [13]; Supplementary Table S6). Sensitivity was variable for Salmonella spp. and STEC. Testing was recommended for ≤50% of patients with detected Aeromonas spp., ETEC, and Yersinia spp. cases.
Sensitivity of Results to Analysis Design
Including criteria for dehydration for ESPGHAN and pain for IDSA guidelines, increased sensitivity and decreased specificity by ~10%, relative to the primary analysis (Supplementary Table S7). The guideline performance changed little when testing weaker assumptions regarding the pattern of missing data (Supplementary Table S8). Sensitivity was higher for both culture and CIDT when results were considered separately; there was no meaningful change in specificity (Supplementary Table S8).
DISCUSSION
Bacterial enteropathogens are most important to detect in children with acute gastroenteritis, and the importance of their identification is determined by the clinical and public health implications of the specific pathogens. Treating all bacterial enteropathogens as important, we found that existing stool-testing guidelines show sub-optimal performance. Sensitivity ranged from 25.8% to 66.9%, and specificity from 63.6% to 96.5%. Use of the guidelines would have resulted in large numbers of missed cases, requiring specific clinical management, public health notification, and/or substantially elevated testing volumes. For alternate combinations of pathogens that may be particularly important, guideline sensitivity was higher, but it generally increased by less than 10%, and specificity was unchanged. For individual bacterial enteropathogens, sensitivity was highest for Shigella spp., Campylobacter spp., and Salmonella spp., but all guidelines would have missed cases, and sensitivity for non-O157 STEC was only 15–56%.
Our findings support and extend the evidence of inconsistencies among guideline recommendations for stool testing [3]. Although the quality of evidence for testing recommendations has been assessed as very low [14], low [14, 15], or moderate [15], the inconsistency and suboptimal sensitivity we report is alarming. The sensitivity of clinical testing by attending physicians was slightly higher than those of the least sensitive/most specific guidelines, but did not adhere to any single guideline. This finding is consistent with a recent study showing clinicians are more likely to adhere to guidelines based on high-quality evidence [33].
Our study highlights 3 primary concerns when evaluating guidelines for use. The first is the need to use a highly sensitive guideline that recommends testing when bacterial enteropathogens of high clinical or public health importance are present. Salmonella spp., Campylobacter spp., and Shigella spp. are recognized for their high disease severity [34–36], and missed cases present a risk of transmission to others or delayed outbreak detection. Public health control measures, such as daycare/work exclusions, and clinical management [37] are also crucial. In our study, sensitivity was particularly low for STEC. Both O157 [38–41] and non-O157 [42–46] STEC serogroups have been linked to outbreaks and severe outcomes. STEC cases may benefit from early-in-illness fluid administration [47, 48], and antibiotics should be avoided [7, 8], making them model candidates for rapid clinical action upon detection.
Second, laboratory utilization can be dramatically impacted by stool-testing guidelines. The CDC [12] guidelines and Klein et al [24] criteria would maintain current testing volumes, but the more sensitive and less specific guidelines would increase testing volumes up to 5-fold.
Third, all guidelines were limited to patients with diarrhea, but our findings show that lack of diarrhea at the initial healthcare encounter does not rule out the possibility of a bacterial etiology. Excluding children with isolated vomiting at the index visit would give the appearance of increased sensitivity (by decreasing the denominator, not by increasing the number of cases ascertained) and decreased specificity, with no change in the number of tests, but it would also miss up to 25% of children in whom bacterial enteropathogens were identified (some of whom developed diarrhea after the index visit). Under current guidelines, caregivers should be encouraged to return for further assessment if their child subsequently experiences diarrhea with other predictors of a bacterial etiology, and new criteria should be explored for the microbiologic evaluation of children with isolated vomiting.
Because guideline performance varies by pathogen, factors that affect pathogen distribution might also affect the generalizability of our results. Study participants were recruited from 2 Eds, located in tertiary-care pediatric hospitals. However, referrals constituted a minority of participants, and the ED was the site of primary presentation for almost all participants. Thus, our results may generalize to primary-care settings, but differences in severity between settings should be investigated further. Geographically, there may be differences within North America and abroad in the patterns of ED use and pathogen distribution, particularly relative to low-resource settings [49]. Moreover, secular trends in pathogen recovery rates [24, 50] and seasonal variation must be considered. The study period encompassed several small, community outbreaks, but it was not dominated by any particular outbreak. Our findings should not be applied to outbreak settings in which a large portion of cases are due to a single pathogen, but our secondary analysis of individual pathogens may be helpful in such contexts.
A limitation of our study is that CIDT detection of an enteropathogen might reflect colonization or shedding from a past infection or non-viable organism. Conversely, bacterial viability could be compromised during transport, yielding falsely negative culture results. In sensitivity analyses we found that, relative to the primary analysis, in which a positive result from culture or CIDT was considered a true positive, both culture and CIDT individually yielded higher guideline sensitivity (Supplementary Table S8). Our testing did not include enteropathogenic E. coli, enteroinvasive E. coli, enteroaggregative E. coli, or Plesiomonas shigelloides, which are included in some commercial testing platforms. Enteroinvasive E. coli is similar in presentation to Shigella and enteropathogenic E. coli, and enteroaggregative E. coli is similar to ETEC, with enteropathogenic E. coli generally somewhat more severe [51]. Given the low sensitivity of most guidelines for identifying ETEC, testing for these pathogens may have further lowered the sensitivity we observed. The prevalence of enteroinvasive E. coli and P. shigelloides in Canada is low and would likely not have altered the findings significantly.
We used multiple imputation to address the issue of missing data and to avoid the bias inherent in a complete case approach [52], but it is possible that our data violate the missing at random assumption required by this method. Therefore, we tested alternate assumptions in sensitivity analyses, including limiting the analysis to only those cases recruited after bloody diarrhea was added to the questionnaire. We observed very little change in guideline performance in any of these sensitivity analyses. This included an analysis of only data after the introduction of the bloody diarrhea question. Some guideline criteria were non-specific (eg, “severe” disease), preventing objective assessment. We tested multiple definitions and observed minimal variation in sensitivity. Lastly, APPETITE excluded immunocompromised children and those needing emergent care; however, <1% of children excluded met these criteria, suggesting that their inclusion would have little effect on our findings. Children whose caregivers could not complete a 14-day follow-up because of insufficient ability to communicate in English were excluded. These patients may have been more likely to have recent travel histories, exposing them to the pathogens for which the guidelines had lowest sensitivity. Thus, their exclusion may have falsely increased sensitivity of the guidelines.
Our study points to the need for updated stool-testing recommendations that are based on strong evidence and that balance testing volumes with the identification of bacterial enteropathogens that may necessitate specific clinical management or public health action. Such recommendations should consider all pertinent information from the patient at presentation, recommend testing for children with pathogens of high clinical or public health importance, and include criteria for children with isolated vomiting at initial presentation.
Modern molecular testing provides opportunities to detect more enteric pathogens than ever before [53], but how that testing should be employed is unclear based on our data. Given deficiencies in sensitivity or drastic increases in testing volume, we cannot recommend the use of existing guidelines to identify children for bacterial enteropathogen testing. Updated, validated recommendations are urgently needed to guide responsible, evidence-informed testing for enteropathogens in children with acute gastroenteritis.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. G. A. M. T. conceived and planned the analysis, cleaned and analyzed the data, and drafted and revised the paper. L. C., B. E. L., X.-L. P., and S. B. F. conceived and planned Alberta Provincial Pediatric Enteric Infection Team (APPETITE). A. N.-A., O. G. V., J. D., P. I. T., S. D., and J. M. advised on the design and implementation of APPETITE. B. E. L. and S. A. implemented APPETITE at the Stollery Children’s Hospital site. S. B. F. oversaw APPETITE, conceived the analysis, and implemented APPETITE at the Alberta Children’s Hospital site. A. N.-A. contributed to the analysis plan. B. M. B. interpreted results of the analysis. K. K. designed data collection tools, coordinated data collection at the Alberta Children’s Hospital site, and assessed the raw data for quality. L. C. led the bacterial testing. X.-L. P. led the virologic testing. All authors revised the paper and approved the final version.
Acknowledgments. The authors thank the patients and their families for cooperating with their study and the APPETITE for their contributions. They also thank the Department of Laboratory Medicine and Pathology, University of Alberta, DynaLIFE Diagnostic Laboratory Services, community laboratories, and the Provincial Laboratory for Public Health (ProvLab), and especially the bacteriology staff, for their assistance with receiving, handling, and processing specimens. The authors thank the emergency department research nurses and Pediatric Emergency Medicine Research Associate Program at the Alberta Children’s Hospital for recruiting study participants; the emergency department bedside nurses for assisting with rectal swab performance, and the research assistants, research nurses, and the Little Bit of Help research volunteer program for their assistance with participant recruitment at the Stollery Children’s Hospital.
Financial support. This work was supported by the APPETITE, which is funded by a Team Collaborative Research Innovation Opportunity grant from Alberta Innovates. APPETITE is also supported by the Alberta Children’s Hospital Research Institute (Calgary, Alberta, Canada) and the Women and Children’s Health Research Institute (Edmonton, Alberta, Canada), through a Partnership Award. The Pediatric Emergency Medicine Research Associate Program is supported by a grant from the Alberta Children’s Hospital Foundation.
Potential conflicts of interest. G. A. M. T. is supported by the University of Calgary Eyes High Postdoctoral Scholar program. P. I. T. is supported by the Biobank Core of the Washington University Digestive Diseases Research Core Center (grant number P30DK052574) and was a coauthor of 2 of the assessed guidelines. S. B. F. is supported by the Alberta Children’s Hospital Foundation Professorship in Child Health and Wellness and reports grants from Alberta Innovates, Alberta Children’s Hospital Foundation, and the University of Calgary during the conduct of the study. J. D. received a recurrent grant from Alberta Health for the Alberta Community Influenza Surveillance Program, outside the submitted work but during the conduct of the study. J. M. is employed by Alberta Health Services as a Medical Officer of Health with responsibility for Communicable Disease Control, which includes certain notifiable enteric diseases. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Tam CC, Rodrigues LC, Viviani L, et al. Longitudinal study of infectious intestinal disease in the UK (IID2 study): incidence in the community and presenting to general practice. Gut 2012; 61:69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scallan E, Griffin PM, Angulo FJ, Tauxe RV, Hoekstra RM. Foodborne illness acquired in the United States–unspecified agents. Emerg Infect Dis 2011; 17:16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van den Berg J, Berger MY. Guidelines on acute gastroenteritis in children: a critical appraisal of their quality and applicability in primary care. BMC Fam Pract 2011; 12:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. National Collaborating Centre for Women’s and Children’s Health. Diarrhoea and vomiting caused by gastroenteritis: diagnosis, assessment and management in children younger than 5 years. London, United Kingdom: National Institute for Health and Clinical Excellence, 2009. [Google Scholar]
- 5. Payne DC, Vinjé J, Szilagyi PG, et al. Norovirus and medically attended gastroenteritis in U.S. children. N Engl J Med 2013; 368:1121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schmutz C, Mäusezahl D, Bless PJ, Hatz C, Schwenkglenks M, Urbinello D. Estimating healthcare costs of acute gastroenteritis and human campylobacteriosis in Switzerland. Epidemiol Infect 2017; 145:627–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wong CS, Mooney JC, Brandt JR, et al. Risk factors for the hemolytic uremic syndrome in children infected with Escherichia coli O157:H7: a multivariable analysis. Clin Infect Dis 2012; 55:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smith KE, Wilker PR, Reiter PL, Hedican EB, Bender JB, Hedberg CW. Antibiotic treatment of Escherichia coli O157 infection and the risk of hemolytic uremic syndrome, Minnesota. Pediatr Infect Dis J 2012; 31:37–41. [DOI] [PubMed] [Google Scholar]
- 9. Freedman SB, Xie J, Neufeld MS, et al. ; Alberta Provincial Pediatric Enteric Infection Team (APPETITE) Shiga toxin-producing Escherichia coli infection, antibiotics, and risk of developing hemolytic uremic syndrome: a meta-analysis. Clin Infect Dis 2016; 62:1251–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Atkinson R, Maguire H, Gerner-Smidt P. A challenge and an opportunity to improve patient management and public health surveillance for food-borne infections through culture-independent diagnostics. J Clin Microbiol 2013; 51:2479–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Humphries RM, Linscott AJ. Laboratory diagnosis of bacterial gastroenteritis. Clin Microbiol Rev 2015; 28:3–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. King CK, Glass R, Bresee JS, Duggan C; Centers for Disease Control and Prevention Managing acute gastroenteritis among children: oral rehydration, maintenance, and nutritional therapy. MMWR Recomm Rep 2003; 52:1–16. [PubMed] [Google Scholar]
- 13. Hatchette TF, Farina D. Infectious diarrhea: when to test and when to treat. Can Med Assoc J 2011; 183:339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guarino A, Ashkenazi S, Gendrel D, Lo Vecchio A, Shamir R, Szajewska H; European Society for Pediatric Gastroenterology, Hepatology, and Nutrition; European Society for Pediatric Infectious Diseases European Society for Pediatric Gastroenterology, Hepatology, and Nutrition/European Society for Pediatric Infectious Diseases evidence-based guidelines for the management of acute gastroenteritis in children in Europe: update 2014. J Pediatr Gastroenterol Nutr 2014; 59:132–52. [DOI] [PubMed] [Google Scholar]
- 15. Shane AL, Mody RK, Crump JA, et al. 2017 Infectious Diseases Society of America Clinical Practice Guidelines for the diagnosis and management of infectious diarrhea. Clin Infect Dis 2017; 65:e45–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Freedman SB, Lee BE, Louie M, et al. Alberta Provincial Pediatric EnTeric Infection TEam (APPETITE): epidemiology, emerging organisms, and economics. BMC Pediatr 2015; 15:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thomas MK, Murray R, Nesbitt A, Pollari F. The incidence of acute gastrointestinal illness in Canada, Foodbook Survey 2014–2015. Can J Infect Dis Med Microbiol 2017; 2017:11. Article ID 5956148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scallan E, Majowicz SE, Hall G, et al. Prevalence of diarrhoea in the community in Australia, Canada, Ireland, and the United States. Int J Epidemiol 2005; 34:454–60. [DOI] [PubMed] [Google Scholar]
- 19. Freedman SB, Xie J, Nettel-Aguirre A, et al. ; Alberta Provincial Pediatric EnTeric Infection TEam (APPETITE) Enteropathogen detection in children with diarrhoea, or vomiting, or both, comparing rectal flocked swabs with stool specimens: an outpatient cohort study. Lancet Gastroenterol Hepatol 2017; 2:662–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pang XL, Preiksaitis JK, Lee BE. Enhanced enteric virus detection in sporadic gastroenteritis using a multi-target real-time PCR panel: a one-year study. J Med Virol 2014; 86:1594–601. [DOI] [PubMed] [Google Scholar]
- 21. Lawson PA, Citron DM, Tyrrell KL, Finegold SM. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) Prevot 1938. Anaerobe 2016; 40:95–9. [DOI] [PubMed] [Google Scholar]
- 22. McDonald LC, Gerding DN, Johnson S, et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 2018; 66:987–94. [DOI] [PubMed] [Google Scholar]
- 23. Surawicz CM, Brandt LJ, Binion DG, et al. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol 2013; 108:478–98; quiz 99. [DOI] [PubMed] [Google Scholar]
- 24. Klein EJ, Boster DR, Stapp JR, et al. Diarrhea etiology in a Children’s Hospital Emergency Department: a prospective cohort study. Clin Infect Dis 2006; 43:807–13. [DOI] [PubMed] [Google Scholar]
- 25. van Buuren S, Groothuis-Oudshoorn K. Mice: multivariate imputation by chained equations in R. J Stat Soft 2011; 45:1–67. [Google Scholar]
- 26. R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2017. [Google Scholar]
- 27. Rubin DB. Multiple imputation for nonresponse in surveys. New York, NY: John Wiley & Sons, 1987. [Google Scholar]
- 28. Clopper CJ, Pearson ES. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika 1934; 26:404–13. [Google Scholar]
- 29. Wickham H. The split-apply-combine strategy for data analysis. J Stat Soft 2011; 40:1–29. [Google Scholar]
- 30.Stevenson M with contributions from Nunes T, Heuer C, Marshall J, et al. epiR: Tools for the analysis of epidemiological data, 2018. R package version 0.9-99. Available at: https://CRAN.R-project.org/package=epiR [Google Scholar]
- 31. Wickham H. Ggplot2: elegant graphics for data analysis. New York, NY: Springer-Verlag, 2009. [Google Scholar]
- 32.Dorai-Raj S. binom: Binomial Confidence Intervals For Several Parameterizations, 2014. R package version 1.1-1. Available at: https://CRAN.R-project.org/package=binom [Google Scholar]
- 33. O’Sullivan JW, Albasri A, Koshiaris C, Aronson JK, Heneghan C, Perera R. Diagnostic test guidelines based on high-quality evidence had greater rates of adherence: a meta-epidemiological study. J Clin Epidemiol 2018; 103:40–50. [DOI] [PubMed] [Google Scholar]
- 34. Scallan E, Hoekstra RM, Angulo FJ, et al. Foodborne illness acquired in the United States–major pathogens. Emerg Infect Dis 2011; 17:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adams DA, Thomas KR, Jajosky RA, et al. Summary of notifiable infectious diseases and conditions - United States, 2015. MMWR Morb Mortal Wkly Rep 2017; 64:1–143. [DOI] [PubMed] [Google Scholar]
- 36. Xie J, Nettel-Aguirre A, Lee BE, et al. Relationship between enteric pathogens and acute gastroenteritis disease severity: a prospective cohort study. Clin Microbiol Infect. 2018. pii:S1198-743X(18)30479-8. [DOI] [PubMed] [Google Scholar]
- 37. Christopher PR, David KV, John SM, Sankarapandian V. Antibiotic therapy for Shigella dysentery. Cochrane Database Syst Rev. 2009;(4):CD006784. [DOI] [PubMed] [Google Scholar]
- 38. Bell BP, Goldoft M, Griffin PM, et al. A multistate outbreak of Escherichia coli O157:H7-associated bloody diarrhea and hemolytic uremic syndrome from hamburgers. The Washington experience. JAMA 1994; 272:1349–53. [PubMed] [Google Scholar]
- 39. Carter AO, Borczyk AA, Carlson JA, et al. A severe outbreak of Escherichia coli O157:H7–associated hemorrhagic colitis in a nursing home. N Engl J Med 1987; 317:1496–500. [DOI] [PubMed] [Google Scholar]
- 40. Neill MA, Tarr PI, Clausen CR, Christie DL, Hickman RO. Escherichia coli O157:H7 as the predominant pathogen associated with the hemolytic uremic syndrome: a prospective study in the Pacific Northwest. Pediatrics 1987; 80:37–40. [PubMed] [Google Scholar]
- 41. Mody RK, Gu W, Griffin PM, et al. Postdiarrheal hemolytic uremic syndrome in United States children: clinical spectrum and predictors of in-hospital death. J Pediatr 2015; 166:1022–9. [DOI] [PubMed] [Google Scholar]
- 42. Luna-Gierke RE, Griffin PM, Gould LH, et al. Outbreaks of non-O157 Shiga toxin-producing Escherichia coli infection: USA. Epidemiol Infect 2014; 142:2270–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Preußel K, Höhle M, Stark K, Werber D. Shiga toxin-producing Escherichia coli O157 is more likely to lead to hospitalization and death than non-O157 serogroups–except O104. PLOS One 2013; 8:e78180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morton V, Cheng JM, Sharma D, Kearney A. Notes from the field: an outbreak of shiga toxin-producing Escherichia coli O121 infections associated with flour - Canada, 2016-2017. MMWR Morb Mortal Wkly Rep 2017; 66:705–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brooks JT, Sowers EG, Wells JG, et al. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983–2002. J Infect Dis 2005; 192:1422–9. [DOI] [PubMed] [Google Scholar]
- 46. Scavia G, Gianviti A, Labriola V, et al. A case of haemolytic uraemic syndrome (HUS) revealed an outbreak of Shiga toxin-2-producing Escherichia coli O26:H11 infection in a nursery, with long-lasting shedders and person-to-person transmission, Italy 2015. J Med Microbiol 2018; 67:775–82. [DOI] [PubMed] [Google Scholar]
- 47. Ake JA, Jelacic S, Ciol MA, et al. Relative nephroprotection during Escherichia coli O157:H7 infections: association with intravenous volume expansion. Pediatrics 2005; 115:e673–80. [DOI] [PubMed] [Google Scholar]
- 48. Hickey CA, Beattie TJ, Cowieson J, et al. Early volume expansion during diarrhea and relative nephroprotection during subsequent hemolytic uremic syndrome. Arch Pediatr Adolesc Med 2011; 165:884–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kotloff KL, Nataro JP, Blackwelder WC, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 2013; 382:209–22. [DOI] [PubMed] [Google Scholar]
- 50. Denno DM, Shaikh N, Stapp JR, et al. Diarrhea etiology in a pediatric emergency department: a case control study. Clin Infect Dis 2012; 55:897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. O’Reilly CE, Iwamoto M, Griffin PM. Escherichia coli, diarrheagenic. In: Centers for Disease Control and Prevention. CDC Yellow Book 2018: health information for international travel. New York, NY: Oxford University Press, 2017. Available at: https://wwwnc.cdc.gov/travel/yellowbook/2018/infectious-diseases-related-to-travel/escherichia-coli-diarrheagenic [Google Scholar]
- 52. Sterne JA, White IR, Carlin JB, et al. Multiple imputation for missing data in epidemiological and clinical research: potential and pitfalls. BMJ 2009; 338:b2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Anderson N, Tarr PI. Multiplex nucleic acid amplification technology and gut infection diagnosis: challenges, opportunities, and result interpretation. Gastroenterol Clin North Am. 2018; 47:793-812. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.