Abstract
Plant peroxisomes are organelles enclosed by a single membrane whose biochemical composition has the capacity to adapt depending on the plant tissue, developmental stage, as well as internal and external cellular stimuli. Apart from the peroxisomal metabolism of reactive oxygen species (ROS), discovered several decades ago, new molecules with signaling potential, including nitric oxide (NO) and hydrogen sulfide (H2S), have been detected in these organelles in recent years. These molecules generate a family of derived molecules, called reactive nitrogen species (RNS) and reactive sulfur species (RSS), whose peroxisomal metabolism is autoregulated through posttranslational modifications (PTMs) such as S-nitrosation, nitration and persulfidation. The peroxisomal metabolism of these reactive species, which can be weaponized against pathogens, is susceptible to modification in response to external stimuli. This review aims to provide up-to-date information on crosstalk between these reactive species families and peroxisomes, as well as on their cellular environment in light of the well-recognized signaling properties of H2O2, NO and H2S.
Keywords: catalase, reactive oxygen, nitrogen and sulfur species, superoxide dismutase, nitric oxide, S-nitrosation, persulfidation
Introduction
For many years, peroxisomes in higher plants have been given different names, such as glyoxysomes during seed germination and leaf senescence, as well as leaf, root and fruit peroxisomes according to their presence in different organs and at different physiological stages (Tolbert and Essner, 1981; Palma et al., 2018). This is explained by the presence of metabolic pathways which appear to be specific to each type of peroxisome. However, peroxisomes, which share a number of metabolites and enzymes common to all types of peroxisome, is now the preferred term regardless of their specific metabolic characteristics (Pracharoenwattana and Smith, 2008). The most noteworthy metabolites and enzymes include H2O2 and catalase, which are directly involved in the metabolism of reactive oxygen species (ROS) (Su et al., 2018; Sousa et al., 2019).
Peroxisomes have a simple morphological constitution composed of a single membrane surrounding an amorphous matrix. Over the last 30 years, an increasing number of new and often unexpected components and processes in these organelles have been identified (Bueno and del Río, 1992; del Río et al., 1992; Corpas et al., 1994, 2001, 2017a, 2019a, Barroso et al., 1999; Reumann et al., 2009; Clastre et al., 2011; Simkin et al., 2011; Chowdhary et al., 2012; Guirimand et al., 2012; Oikawa et al., 2015; Reumann and Bartel, 2016; Kao et al., 2018; Pan et al., 2018, 2020; Borek et al., 2019), indicating that the plant peroxisomal metabolism and consequently peroxisomal enzymatic and non-enzymatic components are more diverse than previously predicted. The diverse complementary range of experimental approaches used to identify these new peroxisomal constituents includes: (i) the biochemical, proteomic and molecular analysis of purified peroxisomes combined with bioinformatics methodologies and (ii) cell biology studies of features such as immune localization with the aid of electron microscopy and specific fluorescent probes with appropriated controls. Although the model plant Arabidopsis thaliana has increased our knowledge of plant peroxisomes, it should be pointed out that studies of peroxisomes from other plant species have been essential, as the peroxisomal metabolism can be modulated depending on the plant organ, development time and plant species involved. Therefore, this review principally aims to provide an update of research on the metabolism of reactive species associated with oxygen, nitrogen and, more recently, sulfur, as well as to outline new challenges and possible future research perspectives regarding crosstalk between peroxisomes and other subcellular compartments such as oil bodies, mitochondria and plastids which are closely related both biochemically and structurally (Palma et al., 2006; Oikawa et al., 2019). Information on plant peroxisomes could also be useful in relation to peroxisome research into other organisms and vice versa.
Peroxisomal ROS Metabolism
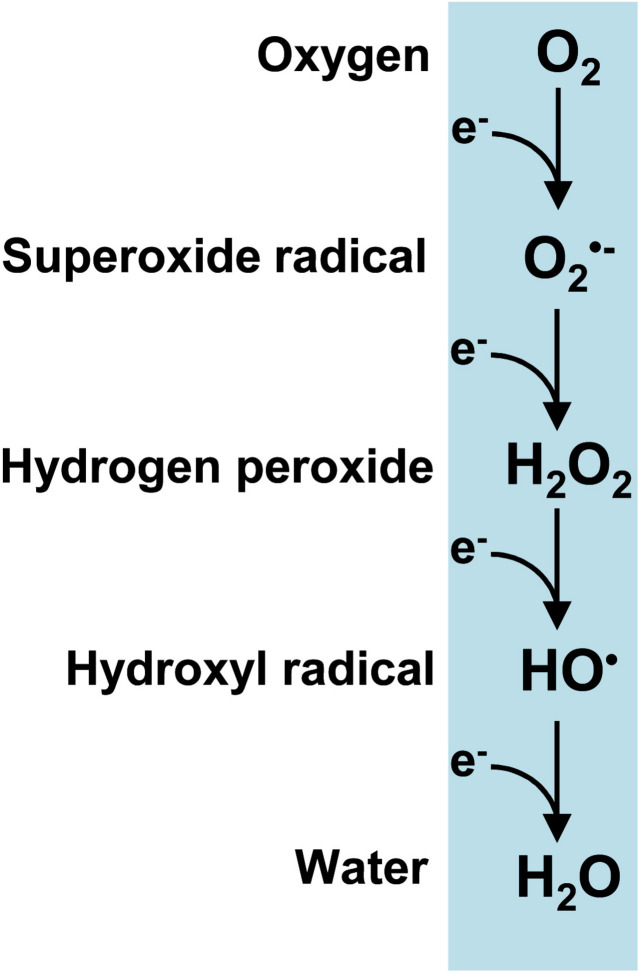
Reactive oxygen species (ROS) are produced by a series of single-electron reductions in molecular oxygen which sequentially form superoxide (O2•–), hydrogen peroxide (H2O2) and hydroxyl (HO•) radicals and ultimately ending in water (Figure 1). It is worth noting that the term peroxisomes, formerly known as microbodies, originates from their high H2O2 content (De Duve and Baudhuin, 1966; Corpas, 2015). Plant peroxisomes contain a significant number of enzymatic systems capable of generating H2O2 such as glycolate oxidase (GOX), acyl-CoA oxidase (AOX), urate oxidase (UO), polyamine oxidase, copper amine oxidase (CuAO), sulfite oxidase (SO), sarcosine oxidase (SOX), or superoxide dismutase (SOD) (Hauck et al., 2014; Corpas et al., 2017a and references therein). These H2O2-generating enzymes are involved in multiple biochemical pathways which are essential not only for the endogenous metabolism of plant peroxisomes but also for their interactions with other subcellular compartments such as plastids, mitochondria, cytosols, oil bodies and nuclei. In these subcellular interconnections, H2O2 itself plays a highly important role as a signal molecule in crosstalk between organelles in order to coordinate cell function.
FIGURE 1.
Reactive oxygen species (ROS) produced from a sequential one-electron reduction from oxygen.
Photorespiration has been estimated to be responsible for 70% of total H2O2 generated mainly from peroxisomal GOX in photosynthetic tissues (Noctor et al., 2002). Zhang et al. (2016) have described an elegant dynamic physical GOX-catalase association-dissociation mechanism that fine-tunes peroxisomal H2O2 in rice plants. Although peroxisomal H2O2 is kept under control when GOX and catalase are associated, under stress conditions and when mediated by salicylic acid (SA), this complex GOX-catalase dissociation mechanism inhibits catalase activity, leading to an increase in cellular H2O2 which acts as a signaling molecule (Zhang et al., 2016; Kohli et al., 2019). Another sophisticated mechanism, involving the interaction of the γb protein from the barley stripe mosaic virus with GOX, has been reported to inhibit GOX and to facilitate infection with the virus (Yang et al., 2018). More recently, Yamauchi et al. (2019) observed a connection between the H2O2-generating GOX and catalase, which is required in the stomatal movement. Thus, when there is an increase of oxidized peroxisomes they were removed by pexophagy allowing an increase in H2O2 in guard cells which mediated the stomatal closure. This mechanism of ROS homeostasis in guard cells seems to be relevant in response to environmental changes. On the other hand, the new peroxisomal small heat shock protein Hsp17.6CII, capable of increasing catalase activity especially under stress conditions, has been reported to be present in Arabidopsis plants (Li et al., 2017).
Acyl-CoA oxidase is another key peroxisomal H2O2-generating enzyme involved in fatty acid β-oxidation which, in collaboration with lipid bodies, enables triacylglyceride mobilization especially during seed germination and is also involved in the synthesis of signal molecules such as jasmonic acid (Baker et al., 2006; Chen et al., 2019b; Wang X. et al., 2019; Xin et al., 2019). However, under stress conditions such as salinity, ROS generated by peroxisomal fatty acid β-oxidation have a negative impact and contribute to oxidative damage (Yu et al., 2019).
Polyamines such as putrescine, spermidine and spermine are well known to be involved in multiple physiological processes, as well as mechanisms of response to various stress conditions (Wuddineh et al., 2018; Chen et al., 2019a; Wang W. et al., 2019). Several enzymes involved in the catabolism of polyamine, including H2O2-producing polyamine oxidase (PAO) and copper amino oxidase (CuAO), have been reported to be present in plant peroxisomes (Moschou et al., 2008; Kusano et al., 2015). These enzymes are also involved in the γ-aminobutyric acid (GABA) biosynthesis signaling pathway (Zarei et al., 2015; Corpas et al., 2009b).
In addition, peroxisomal xanthine oxidoreductase (XOR) and superoxide dismutase (SOD), key enzymes in O2•– and H2O2 metabolism, can be regulated by stress conditions such as salinity, heavy metal and ozone stress (Corpas et al., 1993, 2008; Ueda et al., 2013).
Although catalase is the principal antioxidant enzyme in the matrix of all types of peroxisome (Mhamdi et al., 2010, 2012; Palma et al., 2020 and references therein), other enzymatic antioxidants are present in both the matrix and the membrane. It is also important to highlight the role of SOD isozymes, which differ according to peroxisomal origin (del Río et al., 2018). Thus, peroxisomes of watermelon cotyledons have two SOD isoenzymes, a CuZn-SOD located in the matrix and a Mn-SOD that is bound to the membrane (Bueno and del Río, 1992; Rodríguez-Serrano et al., 2007); pea leaf peroxisomes have a Mn-SOD present in the matrix; sunflower cotyledon peroxisomes have only a CuZn-SOD which is also located in the matrix (Corpas et al., 1998); carnation petal and pepper fruit peroxisomes have a Mn- and an Fe-SOD (Droillard and Paulin, 1990; Palma et al., 2018); and olive fruits peroxisomes contain four SOD isozymes, an Fe-SOD, two CuZn-SOD and a Mn-SOD (López-Huertas and del Río, 2014). Therefore, it could be hypothesized that the presence of two or more types of SOD in peroxisomes must have some physiological advantages. Thus, one of the SOD isozymes could be constitutive while the other one could be inducible under environmental or physiological stimuli such as seedling development, leaf senescence or fruit ripening.
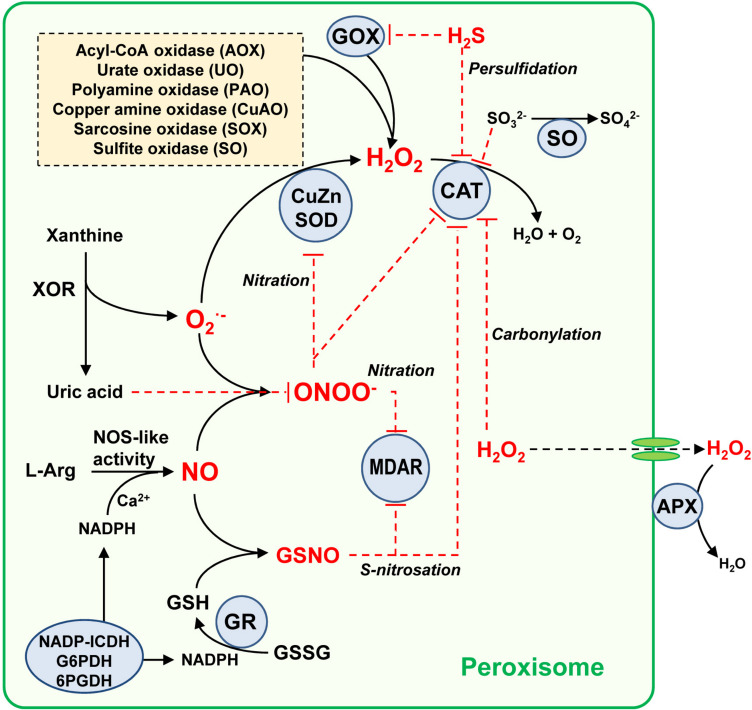
In addition, it is worth noting the role of ascorbate-glutathione cycle components, including ascorbate peroxidase (APX), monodehydroascorbate reductase (MDAR), dehydroasrcorbate reductase (DAR) and glutathione reductase (GR) (Jiménez et al., 1998; Romero-Puertas et al., 2006; López-Huertas and del Río, 2014; Corpas et al., 2017a). While MDAR is present in both matrix and membrane (Leterrier et al., 2005; Lisenbee et al., 2005; Eastmond, 2007), APX is exclusively located in the membrane (Corpas et al., 1994; Yamaguchi et al., 1995; Bunkelmann and Trelease, 1996). With its high affinity for H2O2 (low Km value around 74 μM), membrane-bound APX appears to have fine-tuned control of H2O2 (Ishikawa et al., 1998) as compared to catalase, which, with a Km value in the mM range, is less efficient at low concentrations of H2O2 (Huang et al., 1983; Mhamdi et al., 2010). The Km values for plant catalase are reported to vary quite considerably, with, for example, a Km of 50 mM in Beta vulgaris (Dinçer and Aydemir, 2001), 100 mM in rice (Ray et al., 2012) and 190 mM in pea (del Río et al., 1977). Peroxisomal APX appears to be critical in a diverse range of processes such as seedling development (Corpas and Trelease, 1998) and leaf senescence (Ribeiro et al., 2017). To maintain the ascorbate-glutathione cycle at the GR level, NADPH needs to be supplied by NADP-dependent endogenous dehydrogenases including glucose-6-phosphate dehydrogenase (G6PDH), 6-phosphogluconate dehydrogenase (6PGDH) and isocitrate dehydrogenase (NADP-ICDH) (Leterrier et al., 2016; Corpas and Barroso, 2018b and references therein). In addition, Corpas et al. (2017b) have reported the presence of a protein immunologically related to plant peroxiredoxins, whose expression is differentially modulated under oxidative stresses such as those induced by CdCl2 and the herbicide 2,4-dichlorophenoxyacetic acid (2,4-D); however, further research is necessary to clarify this phenomenon. Figure 2 shows a working model of the ROS metabolism and its interaction with other reactive species, including NO and H2S, which modulate the activity of peroxisomal enzymes through posttranslational modifications (PTMs), events which will be further discussed below.
FIGURE 2.
Simple model of the global metabolism of reactive oxygen/nitrogen/sulfur species in plant peroxisomes. Peroxisomes have an important battery of H2O2-generating enzymes, being the photorespiratory glycolate oxidase (GOX) one of the most relevant. Peroxisomal xanthine oxidoreductase (XOR) activity generates uric acid which the concomitant generation of superoxide radical (O2•–) which is dismutated to H2O2 by superoxide dismutase (SOD). All three SOD types have been described in plant peroxisomes from different origin, CuZ-SOD, Mn-SOD, and Fe-SOD. The H2O2 pool is mainly decomposed by catalase (CAT) but also by the membrane-bound ascorbate peroxidase (APX). An L-arginine (L-Arg) and Ca2+ dependent NOS-like activity generates NO which can react chemically with O2– to produce peroxynitrite (ONOO–), a nitrating molecule that facilitates PTMs such as tyrosine nitration. NO can also interact with reduced glutathione (GSH) to form S-nitrosoglutathione (GSNO), a NO donor which mediates S-nitrosation. GSH is regenerated by glutathione reductase (GR) which requires NADPH supplied by several NADPH-generating enzymes (NADPH-ICDH, G6PDH, and 6PGDH). Uric acid is a ONOO– scavenger, this being a mechanism of peroxisomal auto-regulation. With all these components, and according to reported data, the peroxisomal targets of NO-derived PTMs identified so far are CAT, CuZn-SOD, and monodehydroascorbate reductase (MDAR) which can undergo an inhibitory effect either by nitration or S-nitrosation. Additionally, CAT and GOX can be inhibited by hydrogen sulfide (H2S), and CAT is also inhibited by carbonylation. The H2O2-generating sulfite oxidase (SO) converts sulfite (SO32–) to sulfate (SO42–), which is a mechanism of protection because sulfite inhibits catalase activity. Red line denotes inhibition effect.
Given the capacity of ROS to mediate several PTMs, particularly carbonylation and S-sulfenylation, certain amino acid residues, especially arginine, lysine, threonine and proline, are carbonylated, which affects target protein function in many cases (Debska et al., 2012; Lounifi et al., 2013). Several studies have identified peroxisomal proteins, such as catalase, malate synthase and the fatty acid β-oxidation multifunctional protein AIM1, which undergo carbonylation (Nguyen and Donaldson, 2005; Anand et al., 2009; Mano et al., 2014; Rodríguez-Ruiz et al., 2019). On the other hand, H2O2 can oxidize specific protein cysteine thiols to sulfenic acid (SOH), a process known as S-sulfenylation, which usually results in enzymatic inactivation. Using proteomic techniques, approximately 2% of peroxisomal proteins have been reported to be susceptible to S-sulfenylation (Akter et al., 2017; Huang et al., 2019). This PTM has been observed to occur with respect to fatty acid β-oxidation acyl-coenzyme A oxidase 1, the multifunctional proteins MFP2, and AIM1, as well as amine oxidase, phosphomevalonate kinase, MDAR and NADP-ICDH. Table 1 shows a summary of peroxisomal enzymes targeted by carbonylation and S-sulfenylation, as well as other PTMs mediated by RNS and RSS, a subject which will be discussed below.
TABLE 1.
Peroxisomal enzymes target of diverse posttranslational modifications (PTMs) whose activities are affected by either ROS, RNS, or RSS.
| Peroxisomal enzyme | Pathway/Reaction | PTM | Effect on activity |
| Catalase (CAT) | H2O2 decomposition | Carbonylation | Inhibition |
| Tyr-nitration | Inhibition | ||
| S-nitrosation | Inhibition | ||
| Persufidation | Inhibition | ||
| Monodehydroascorbate reductase (MDAR) | Ascorbate-glutathione cycle | Tyr-nitration | Inhibition |
| S-nitrosation | Inhibition | ||
| S-sulfenylationa | Not reported | ||
| Hydroxypyruvate reductase (HPR) | Photorespiration | Tyr-nitration | Inhibition |
| S-nitrosation | Inhibition | ||
| Glycolate oxidase (GOX) | Photorespiration | S-nitrosation | Inhibition |
| Persufidation | Inhibition | ||
| CuZn-superoxide dismutase (CSD3) | O2•– dismutation | Tyr-nitration | Inhibition |
| Malate dehydrogenase (MDH) | Fatty acid β-oxidation | Tyr-nitration | Inhibition |
| S-nitrosation | Inhibition | ||
| Malate synthase (MS) | Glyoxylate cycle | Carbonylation | Inhibition |
| Isocitrate lyase (ICL) | Glyoxylate cycle | S-nitrosationa | Not reported |
| Acyl-coenzyme A oxidase 1 | Fatty acid β-oxidation | Persulfidationa | Not reported |
| S-sulfenylationa | Not reported | ||
| Multifunctional protein AIM1 isoform | Fatty acid β-oxidation | S-nitrosationa | Not reported |
| S-sulfenylationa | Not reported | ||
| Lon protease homolog 2 | Peroxisomal protein import | S-nitrosationa | Not reported |
| Phosphomevalonate kinase | Isoprenoid biosynthesis | S-sulfenylationa | Not reported |
| NADP-isocitrate dehydrogenase | NADPH supply | Tyr-nitration | Inhibition |
| S-nitrosation | Inhibition | ||
| Persufidation | Inhibition | ||
| S-sulfenylationa | Not reported |
aProteomic identification.
Given growing awareness of the important role of ROS peroxisomal metabolism in combating biotic stress, the expression of genes encoding for peroxisomal proteins involved in their biogenesis, fatty acid catabolism and the H2O2-generating glyoxylate cycle have been reported to increase during interactions between the pathogen Sclerotinia sclerotiorum and rapeseed (Brassica napus), thus facilitating pathogen cell wall degradation and metabolism detoxification (Chittem et al., 2020). On the other hand, using the Arabidopsis nca1 mutant with no catalase activity 1, containing residual activity of the three catalase isozymes, Hackenberg et al. (2013) identified a link between catalase and ROS production as autophagy-dependent cell death progresses. Table 2 shows some functional implications of peroxisomal H2O2 and other signal molecules generated in this organelle.
TABLE 2.
Signal molecules generated in plant peroxisomes during different processes and their functional implications.
| Peroxisomal signal | Functional implication | References |
| Hydrogen peroxide (H2O2) | Plant development and stress response | Zhang et al., 2016; Su et al., 2019 |
| Involved in peroxisome abundance under drought and heat stress | Hinojosa et al., 2019 | |
| Pexophagy | Yamauchi et al., 2019 | |
| Pathogen defense | Chittem et al., 2020 | |
| Nitric oxide (NO) | Pollen tube development | Prado et al., 2004 |
| Leaf senescence | Corpas et al., 2019 | |
| Lateral root formation | Schlicht et al., 2013 | |
| Heavy metal and root architecture | Piacentini et al., 2020 | |
| Hydrogen sulfide | Regulation of catalase | Corpas et al., 2019a |
| (H2S) | Herbicide glyphosate response | |
| Jasmonic acid (JA) | Plant growth | Wang X. et al., 2019 |
| Environmental stimuli | Xin et al., 2019 | |
| Insect defense | ||
| γ-aminobutyric acid (GABA) | Fruit flavor and flower fragrance | Zarei et al., 2015 |
| Abiotic stress tolerance | Shelp and Zarei, 2017 |
The generation of singlet oxygen (1O2) has always been associated with chloroplasts, particularly in photosystem II, responsible for various types of photo-damage which triggers distinct cellular responses (Wagner et al., 2004; Rosenwasser et al., 2011; Chen and Fluhr, 2018; Dogra et al., 2018). Using the green fluorescence probe to detect 1O2, peroxisomes, mitochondria and nuclei have been shown to be either the origin or target of 1O2, suggesting that this ROS is generated in a light-independent manner (Mor et al., 2014). These findings open up new questions about the importance of 1O2 in the mechanism of response to plant stress in which several subcellular compartments including peroxisomes are involved.
Peroxisomal Reactive Nitrogen Species (RNS)
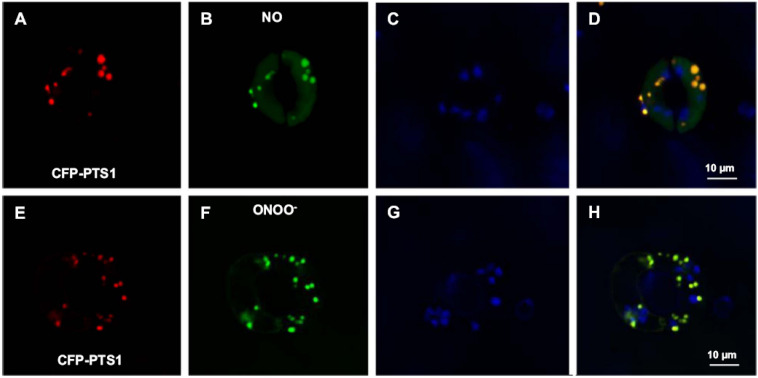
Nitric oxide (NO) metabolism has a significant impact on cellular metabolisms due to its involvement in the important plant physiological processes of seed and pollen germination, root development, stomatal closure, senescence and fruit ripening, as well as in the mechanism of response to many environmental stresses including salinity, drought, heavy metals and extreme temperature (Neill et al., 2008; León et al., 2014; Begara-Morales et al., 2018; Kolbert et al., 2019; Wei et al., 2020). NO belongs to a family of related molecules called reactive nitrogen species (RNS), with peroxynitrite (ONOO–) and S-nitrosogluthione (GSNO) being the most studied. Using various experimental approaches including electron paramagnetic resonance (EPR) spectroscopy, as well as biochemical and cellular biology, some RNS including NO, ONOO– and GSNO have been detected in plant peroxisomes (Barroso et al., 2013; Corpas and Barroso, 2014b; Corpas et al., 2019). Identification of peroxisomal proteins undergoing PTMs mediated by these NO-derived species is strong evidence of an active RNS metabolism in peroxisomes. Figures 3A–H shows in vivo images of NO and ONOO– in Arabidopsis guard cell peroxisomes detected by confocal laser scanning microscopy (CLSM) and specific fluorescent probes.
FIGURE 3.
Representative images illustrating the CLSM in vivo detection of nitric oxide (NO) and peroxynitrite (ONOO–), peroxisomes (red) and chloroplasts (blue) in guard cells of transgenic Arabidopsis seedlings expressing CFP-PTS1. (A,E) Fluorescence punctate (red) attributable to CFP-PTS1, indicating the localization of peroxisomes in guard cells. (B,F) Fluorescence punctate (green) attributable to the detection in the same guard cells of NO and ONOO–, respectively. (C,G) Chlorophyll autofluorescence (blue) attributable to the detection of chloroplasts. (D,H) Merged images for corresponding panels. Reproduced with permission from Corpas et al. (2017a) provided by Elsevier.
ONOO– results from a reaction between NO with O2•–, considered one of the fastest chemical reactions with a rate constant (k) of 1.9 × 1010 M–1 s–1 (Kissner et al., 1997). ONOO–, a strong oxidant and nitrating molecule involved in protein tyrosine nitration (NO2-Tyr), modifies protein function, mostly through inhibition (Corpas et al., 2009a; Mata-Pérez et al., 2016). This NO-derived PTM involves the covalent oxidative addition of a nitro group (-NO2) to tyrosine residues, a highly selective process which depends on factors such as the protein environment of the Tyr and the nitration mechanism (Bartesaghi and Radi, 2018). Table 1 shows some nitrated proteins identified in plant peroxisomes and how their function is affected. Interestingly, some of the proteins affected are directly involved in the ROS metabolism, indicating a close metabolic interconnection between both families of reactive species.
The antioxidant glutathione (GSH), a tripeptide (γ-Glu-Cys-Gly), undergoes S-nitrosation in order to generate GSNO, a low-molecular-weight NO reservoir, through a covalent addition of NO to the thiol group of Cys residues in order to form S-nitrosothiol (SNO) (Airaki et al., 2011). GSNO is a key molecule given its dynamic interaction with free cysteines, GSH and proteins through processes such as S-nitrosation, S-transnitrosation and S-glutathionylation (Broniowska et al., 2013; Corpas et al., 2013a, b). GSNO is enzymatically decomposed by GSNO reductase (GSNOR; Leterrier et al., 2011), an enzyme susceptible to S-nitrosation and consequently inhibition (Guerra et al., 2016). An increase in Tyr nitration, an irreversible process, is usually associated with nitro-oxidative stress; however, protein S-nitrosation, a reversible process, is a regulatory protein mechanism that occurs under physiological and stress conditions. Table 1 shows some peroxisomal proteins targeted by S-nitrosation, as well as proteins involved in ROS metabolism which are targeted by these NO-mediated PTMs.
The number of peroxisomal proteins targeted by NO-mediated PTMs is growing continuously. Using the biotin-switch technique and liquid chromatography/mass spectrometry/mass spectrometry (LC-MS/MS), several more S-nitrosated peroxisomal proteins have been identified during adventitious root growth induced by treatment with NO (Niu et al., 2019). These proteins include the peroxisomal LON2 protease, which is necessary for matrix protein import into peroxisomes (Lingard and Bartel, 2009); isocitrate lyase (ICL), involved in the glyoxylate cycle; and the multifunctional AIM1-like isoform, involved in fatty acid β-oxidation.
However, the source of enzymatic NO, as yet unelucidated, is currently the most controversial aspect of NO metabolism in higher plants (Kolbert et al., 2019). Two main candidates have been proposed: nitrate reductase (NR) (Mohn et al., 2019) and L-arginine-dependent NO synthase-like activity (Corpas et al., 2017a). Although no evidence of NR has been found in plant peroxisomes, NO synthase-like activity has been found and characterized in peroxisomes purified from pea leaves (Barroso et al., 1999). Though as yet unidentified, this protein is called NOS-like activity, as peroxisomal NO generation requires NOS proteins similar to those found in animals, including L-arginine, NADPH, FMN, FAD, tetrahydrobiopterin, calcium, and calmodulin (Corpas and Barroso, 2017b; Corpas et al., 2019). The protein responsible for NO generation is imported by a type 2 peroxisomal targeting signal involving a process dependent on calmodulin and calcium (Corpas and Barroso, 2014a, 2018a).
Peroxisomal NO metabolism is involved in processes such as pollen tube germination (Prado et al., 2004), lateral root formation (Schlicht et al., 2013), and leaf senescence (Corpas et al., 2019), as well as in responses to environmental and heavy metal stresses such as salinity (Corpas et al., 2009b), lead (Corpas and Barroso, 2017a), and cadmium (Corpas and Barroso, 2014b; Piacentini et al., 2020).
Reactive Sulfur Species (RSS) in Plant Peroxisomes
Reactive sulfur species (RSS) are chemically comparable to ROS (Olson, 2019) and can be generated from hydrogen sulfide (H2S), some of these species are thiyl radical (HS•), hydrogen persulfide (H2S2), persulfide radical (HS2•–), sulfite (SO32–) or sulfate (SO42–) among others (Gruhlke and Slusarenko, 2012; Ono et al., 2014; Mishanina et al., 2015; Park et al., 2015; Schöneich, 2016). However, the biochemistry of H2S in cells, given its multiple interactions with other reactive species, is more complex than previously thought (see Filipovic et al., 2018 for a more in-depth review); for example, protein thiyl radicals are generated during the reaction of H2O2 with heme proteins, possibly inducing protein degradation (Schöneich, 2016).
Different molecules and enzymes, such as GSH (Müller et al., 2004), glutathione reductase (Romero-Puertas et al., 2006), and sulfite oxidase (Nowak et al., 2004; Hänsch and Mendel, 2005), involved in sulfur metabolism, are present in plant peroxisomes. Sulfite oxidase (SO) catalyzes the conversion of sulfite to sulfate by producing H2O2. The functional relevance of this enzyme is that it can protect catalase activity since sulfite, at low concentration, has the capacity to inhibit catalase activity (Veljovic-Jovanovic et al., 1998). Nevertheless, despite the greater importance attributed to peroxisomal SO in a recent study, mitochondrial SO in animal cells has the capacity to generate NO from nitrite (Bender et al., 2019), while NO enzymatic generation from SO in plant peroxisomes remains to be proven. An earlier study confirmed the important role played by the peroxisomal RSS metabolism (Corpas and Barroso, 2015).
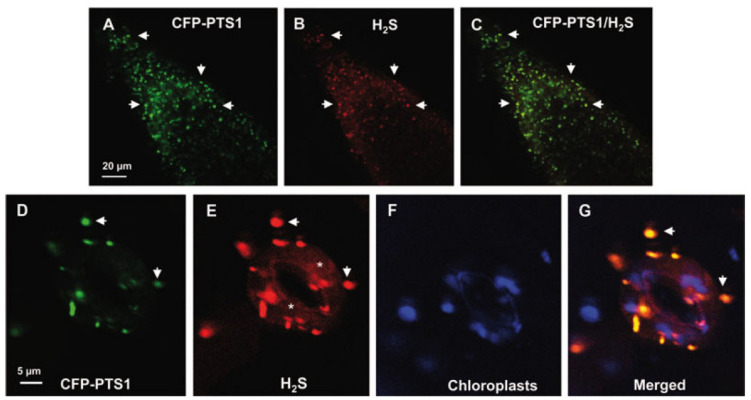
H2S has recently been proven to be present in plant peroxisomes (Corpas et al., 2019a). Figures 4A–G shows representative images of H2S in peroxisomes from the root tips and guard cells of Arabidopsis seedlings detected by in vivo CLSM and a specific fluorescent probe. Using proteomic techniques, some peroxisomal enzymes have been identified as targets of persulfidation (Aroca et al., 2015, 2017). On the other hand, in vitro analysis shows that catalase activity from Arabidopsis and sweet pepper fruits is inhibited in the presence of H2S (Corpas et al., 2019a). Although, to our knowledge, the enzymatic source of peroxisomal H2S remains unknown, previous studies have proposed some potential candidates. For example, catalase, which functions as a sulfide oxidase or sulfur reductase, is capable of oxidizing or generating H2S (Olson et al., 2017). SOD has also been reported to have the capacity to catalyze the reaction between O2 and H2S to generate persulfide (Olson et al., 2018). In a previous study by Corpas and Barroso (2015), the presence of these enzymatic and non-enzymatic components in plant peroxisomes indicates that, in addition to ROS and RNS, these organelles also have an active RSS metabolism.
FIGURE 4.
Representative images illustrating the CLSM in vivo detection of H2S (red color) and peroxisomes (green color) in root tips (A–C) and guard cells (D–G) of 10 days old Arabidopsis seedlings expressing CFP-PTS1. (A,D) Fluorescence puncta (green) attributable to CFP-PTS1, indicating the localization of peroxisomes. (B,E) Fluorescence punctate (red) attributable to H2S detection in the same area. (C) Merged image of (A,B) showing colocalized fluorescence punctate (yellow). (F) Chlorophyll autofluorescence (blue) demonstrating location of chloroplasts. (G) Merged images of (D–F). H2S (red color) was detected by using 5 mM WSP-5, a fluorescence probe for H2S. Arrows indicate representative punctate spots corresponding to the colocalization of H2S with peroxisomes. Asterisks indicate localization of H2S in the cytosol. Reproduced with permission from Corpas et al. (2019a) provided by John Wiley and Sons.
Crosstalk Between Peroxisomal Reactive Species
Functional interactions and inter-regulation through PTMs in these families of reactive species are shown in Figure 2. In this working model, under physiological conditions, catalase, the main antioxidant enzyme, regulates levels of H2O2 generated by different pathways, principally photorespiratory glycolate oxidase (GOX) (Noctor et al., 2002). On the other hand, peroxisomal xanthine oxidoreductase (XOR) activity involved in purine catabolism generates uric acid, with the concomitant formation of the O2•– (Corpas et al., 1997, 2008; Zarepour et al., 2010), which, in turn, is dismutated to H2O2 by SOD. The pool of H2O2 is mainly decomposed by catalase (CAT) and also by membrane-bound ascorbate peroxidase (APX). L-Arg-dependent NOS-like activity generates NO (Corpas et al., 2019) which chemically reacts with O2•– to produce peroxynitrite (ONOO–), a nitrating molecule that facilitates PTMs such as tyrosine nitration. NO also interacts with reduced glutathione (GSH) to form S-nitrosoglutathione (GSNO), a NO donor that mediates S-nitrosation. Uric acid is a physiological ONOO– scavenger (Alamillo and García-Olmedo, 2001) involved in endogenous peroxisomal auto-regulation. Thus, the peroxisomal enzymes targeted by NO-derived PTMs, catalase (CAT), CuZn-SOD, and monodehydroascorbate reductase (MDAR), are inhibited by nitration and S-nitrosation. Both CAT, and GOX are inhibited by H2S; the former is also inhibited by carbonylation when H2O2 is overproduced. In addition, H2O2-generating sulfite oxidase (SO) is involved in the conversion of sulfite to sulfate which, given sulfite’s ability to inhibit SO, has been reported to be a catalase protection mechanism. These interconnections highlight the biochemical complexity of this self-regulated plant peroxisome network, in which the antioxidant catalase is one of the most regulated peroxisomal enzymes (Palma et al., 2020).
Conclusion
Much of our knowledge of reactive species metabolism in plant peroxisomes is now well established. The three molecular families ROS, RNS, and RSS are present in plant peroxisomes, which are considered to be potential producers of reactive species and to play an important role in the cell signaling network. However, our limited knowledge of reactive species families needs to be expanded by identifying new peroxisomal protein targets. We also need to determine the effect of the different PTMs, carbonylation, S-sulfenylation, S-nitrosation, tyrosine nitration, and persulfidation, on target protein function and peroxisomal metabolism. In addition, interactions with other subcellular compartments which share biochemical pathways such as photorespiration, fatty acid β-oxidation, isoprenoid biosynthesis and purine and polyamine metabolism (Clastre et al., 2011; Guirimand et al., 2012; Corpas et al., 2019b) should be investigated. Similarly, the relationship between reactive species and complex peroxisomal biogenesis, division and matrix/membrane protein import mechanisms (Reumann and Bartel, 2016; Kao et al., 2018) has been underexplored (López-Huertas et al., 2000). Further research should also be carried out to identify the proteins responsible for endogenous peroxisomal generation of NO and H2S. This would increase our knowledge of how organelle biochemistry is modulated within the framework of the whole cell metabolism. This research could lead to biotechnological applications given the important role of peroxisomes in many physiological processes and in responses to biotic and abiotic stresses. Furthermore, in addition to harboring reactive species, with their known signaling properties, peroxisomes are a source of other signaling molecules such as jasmonic acid and γ-aminibutic acid (GABA), which extends the functional role of plant peroxisomes. Table 2 shows signaling molecules generated in the plant peroxisomal metabolism and some examples of their role in various plant processes.
Author Contributions
Authors have made a collaborative, direct and intellectual contribution to the work, and have approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
SG-G acknowledges a “Formación de Personal Investigador” contract (BES-2016-078368) from the Ministry of Economy and Competitiveness, Spain.
Footnotes
Funding. This work was supported by the European Regional Development Fund co-financed grant from the Ministry of Economy and Competitiveness (AGL2015-65104-P and PID2019-103924GB-I00), the Plan Andaluz de Investigación, Desarrollo e Innovación (P18-FR-1359), and the Junta de Andalucía (group BIO192), Spain.
References
- Airaki M., Sánchez-Moreno L., Leterrier M., Barroso J. B., Palma J. M., Corpas F. J. (2011). Detection and quantification of S-nitrosoglutathione (GSNO) in pepper (Capsicum annuum L.) plant organs by LC-ES/MS. Plant Cell Physiol. 52 2006–2015. 10.1093/pcp/pcr133 [DOI] [PubMed] [Google Scholar]
- Akter S., Carpentier S., Van Breusegem F., Messens J. (2017). Identification of dimedone-trapped sulfenylated proteins in plants under stress. Biochem. Biophys. Rep. 9 106–113. 10.1016/j.bbrep.2016.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamillo J. M., García-Olmedo F. (2001). Effects of urate, a natural inhibitor of peroxynitrite-mediated toxicity, in the response of Arabidopsis thaliana to the bacterial pathogen Pseudomonas syringae. Plant J. 25 529–540. 10.1046/j.1365-313x.2001.00984.x [DOI] [PubMed] [Google Scholar]
- Anand P., Kwak Y., Simha R., Donaldson R. P. (2009). Hydrogen peroxide induced oxidation of peroxisomal malate synthase and catalase. Arch. Biochem. Biophys. 491 25–31. 10.1016/j.abb.2009.09.019 [DOI] [PubMed] [Google Scholar]
- Aroca A., Benito J. M., Gotor C., Romero L. C. (2017). Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis. J. Exp. Bot. 68 4915–4927. 10.1093/jxb/erx294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca Á, Serna A., Gotor C., Romero L. C. (2015). S-sulfhydration: a cysteine posttranslational modification in plant systems. Plant Physiol. 168 334–342. 10.1104/pp.15.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A., Graham I. A., Holdsworth M., Smith S. M., Theodoulou F. L. (2006). Chewing the fat: beta-oxidation in signalling and development. Trends Plant Sci. 11 124–132. 10.1016/j.tplants.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Barroso J. B., Corpas F. J., Carreras A., Sandalio L. M., Valderrama R., Palma J. M., et al. (1999). Localization of nitric-oxide synthase in plant peroxisomes. J. Biol. Chem. 274 36729–36733. [DOI] [PubMed] [Google Scholar]
- Barroso J. B., Valderrama R., Corpas F. J. (2013). Immunolocalization of S-nitrosoglutathione, S-nitrosoglutathione reductase and tyrosine nitration in pea leaf organelles. Acta Physiol. Plant 35 2635–2640. 10.1007/s11738-013-1291-0 [DOI] [Google Scholar]
- Bartesaghi S., Radi R. (2018). Fundamentals on the biochemistry of peroxynitrite and protein tyrosine nitration. Redox Biol. 14 618–625. 10.1016/j.redox.2017.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begara-Morales J. C., Chaki M., Valderrama R., Sánchez-Calvo B., Mata-Pérez C., Padilla M. N., et al. (2018). Nitric oxide buffering and conditional nitric oxide release in stress response. J. Exp. Bot. 69 3425–3438. 10.1093/jxb/ery072 [DOI] [PubMed] [Google Scholar]
- Bender D., Tobias Kaczmarek A., Niks D., Hille R., Schwarz G. (2019). Mechanism of nitrite-dependent NO synthesis by human sulfite oxidase. Biochem. J. 476 1805–1815. 10.1042/bcj20190143 [DOI] [PubMed] [Google Scholar]
- Borek S., Stefaniak S., Śliwiñski J., Garnczarska M., Pietrowska-Borek M. (2019). Autophagic machinery of plant peroxisomes. Int. J. Mol. Sci. 20:4754. 10.3390/ijms20194754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broniowska K. A., Diers A. R., Hogg N. (2013). S-nitrosoglutathione. Biochim. Biophys. Acta 1830 3173–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno P., del Río L. A. (1992). Purification and properties of glyoxysomal cuprozinc superoxide dismutase from watermelon cotyledons (Citrullus vulgaris Schrad). Plant Physiol. 98 331–336. 10.1104/pp.98.1.331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunkelmann J. R., Trelease R. N. (1996). Ascorbate peroxidase. A prominent membrane protein in oilseed glyoxysomes. Plant Physiol. 110 589–598. 10.1104/pp.110.2.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Shao Q., Yin L., Younis A., Zheng B. (2019a). Polyamine function in plants: metabolism, regulation on development, and roles in abiotic stress responses. Front. Plant Sci. 9:1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Lu X., Ge L., Sun X., Xin Z. (2019b). Wound- and pathogen-activated de novo JA synthesis using different ACX isozymes in tea plant (Camellia sinensis). J. Plant Physiol. 243:153047. 10.1016/j.jplph.2019.153047 [DOI] [PubMed] [Google Scholar]
- Chen T., Fluhr R. (2018). Singlet oxygen plays an essential role in the root’s response to osmotic stress. Plant Physiol. 177 1717–1727. 10.1104/pp.18.00634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittem K., Yajima W. R., Goswami R. S., Del Río Mendoza L. E. (2020). Transcriptome analysis of the plant pathogen Sclerotinia sclerotiorum interaction with resistant and susceptible canola (Brassica napus) lines. PLoS One 15:e0229844. 10.1371/journal.pone.0229844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary G., Kataya A. R., Lingner T., Reumann S. (2012). Non-canonical peroxisome targeting signals: identification of novel PTS1 tripeptides and characterization of enhancer elements by computational permutation analysis. BMC Plant Biol. 12:142. 10.1186/1471-2229-12-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clastre M., Papon N., Courdavault V., Giglioli-Guivarc’h N., St-Pierre B., Simkin A. J. (2011). Subcellular evidence for the involvement of peroxisomes in plant isoprenoid biosynthesis. Plant Signal. Behav. 6 2044–2046. 10.4161/psb.6.12.18173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas F. J. (2015). What is the role of hydrogen peroxide in plant peroxisomes? Plant Biol. 17 1099–1103. 10.1111/plb.12376 [DOI] [PubMed] [Google Scholar]
- Corpas F. J., Sandalio L. M., del Río L. A., Trelease R. N. (1998). Copper–zinc superoxide dismutase is a constituent enzyme of the matrix of peroxisomes in the cotyledons of oilseed plants. New Phytol. 138 307–314. 10.1046/j.1469-8137.1998.00899.x [DOI] [PubMed] [Google Scholar]
- Corpas F J., Barroso J B., del Río L A. (2001). Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant. Sci. 6 145–50. 10.1016/s1360-1385(01)01898-2 [DOI] [PubMed] [Google Scholar]
- Corpas F. J., Alché J. D., Barroso J. B. (2013a). Current overview of S-nitrosoglutathione (GSNO) in higher plants. Front. Plant Sci. 4:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas F. J., Leterrier M., Begara-Morales J. C., Valderrama R., Chaki M., López-Jaramillo J., et al. (2013b). Inhibition of peroxisomal hydroxypyruvate reductase (HPR1) by tyrosine nitration. Biochim. Biophys. Acta 1830 4981–4989. 10.1016/j.bbagen.2013.07.002 [DOI] [PubMed] [Google Scholar]
- Corpas F. J., Barroso J. B. (2014a). Peroxisomal plant nitric oxide synthase (NOS) protein is imported by peroxisomal targeting signal type 2 (PTS2) in a process that depends on the cytosolic receptor PEX7 and calmodulin. FEBS Lett. 588 2049–2054. 10.1016/j.febslet.2014.04.034 [DOI] [PubMed] [Google Scholar]
- Corpas F. J., Barroso J. B. (2014b). Peroxynitrite (ONOO-) is endogenously produced in Arabidopsis peroxisomes and is overproduced under cadmium stress. Ann. Bot. 113 87–96. 10.1093/aob/mct260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas F. J., Barroso J. B. (2015). Reactive sulfur species (RSS): possible new players in the oxidative metabolism of plant peroxisomes. Front Plant Sci. 6:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas F. J., Barroso J. B. (2017a). Lead-induced stress, which triggers the production of nitric oxide (NO) and superoxide anion (O2⋅-) in Arabidopsis peroxisomes, affects catalase activity. Nitric Oxide 68 103–110. 10.1016/j.niox.2016.12.010 [DOI] [PubMed] [Google Scholar]
- Corpas F. J., Barroso J. B. (2017b). Nitric oxide synthase-like activity in higher plants. Nitric Oxide 68 5–6. 10.1016/j.niox.2016.10.009 [DOI] [PubMed] [Google Scholar]
- Corpas F. J., Barroso J. B. (2018a). Calmodulin antagonist affects peroxisomal functionality by disrupting both peroxisomal Ca2+ and protein import. J. Cell Sci. 131:jcs201467. 10.1242/jcs.201467 [DOI] [PubMed] [Google Scholar]
- Corpas F. J., Barroso J. B. (2018b). Peroxisomal plant metabolism - an update on nitric oxide, Ca2+ and the NADPH recycling network. J. Cell Sci. 131:jcs202978. 10.1242/jcs.202978 [DOI] [PubMed] [Google Scholar]
- Corpas F. J., Barroso J. B., González-Gordo S., Muñoz-Vargas M. A., Palma J. M. (2019a). Hydrogen sulfide: a novel component in Arabidopsis peroxisomes which triggers catalase inhibition. J. Integr. Plant Biol. 61 871–883. [DOI] [PubMed] [Google Scholar]
- Corpas F. J., del Río L. A., Palma J. M. (2019b). Plant peroxisomes at the crossroad of NO and H2O2 metabolism. J. Integr. Plant Biol. 61 803–816. [DOI] [PubMed] [Google Scholar]
- Corpas F. J., Barroso J. B., Palma J. M., Rodríguez-Ruiz M. (2017a). Plant peroxisomes: a nitro-oxidative cocktail. Redox Biol. 11 535–542. 10.1016/j.redox.2016.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas F. J., Pedrajas J. R., Palma J. M., Valderrama R., Rodríguez-Ruiz M., Chaki M., et al. (2017b). Immunological evidence for the presence of peroxiredoxin in pea leaf peroxisomes and response to oxidative stress conditions. Acta Physiol. Plant 39:57. [Google Scholar]
- Corpas F. J., Bunkelmann J., Trelease R. N. (1994). Identification and immunochemical characterization of a family of peroxisome membrane proteins (PMPs) in oilseed glyoxysomes. Eur. J. Cell Biol. 65 280–290. [PubMed] [Google Scholar]
- Corpas F. J., Chaki M., Leterrier M., Barroso J. B. (2009a). Protein tyrosine nitration: a new challenge in plants. Plant Signal. Behav. 4 920–923. 10.4161/psb.4.10.9466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas F. J., Hayashi M., Mano S., Nishimura M., Barroso J. B. (2009b). Peroxisomes are required for in vivo nitric oxide accumulation in the cytosol following salinity stress of Arabidopsis plants. Plant Physiol. 151 2083–2094. 10.1104/pp.109.146100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas F. J., de la Colina C., Sánchez-Rasero F., del Río L. A. (1997). A role for leaf peroxisomes in the catabolism of purines. J. Plant Physiol. 151 246–250. 10.1016/s0176-1617(97)80161-7 [DOI] [Google Scholar]
- Corpas F. J., del Río L. A., Palma J. M. (2019). Impact of nitric oxide (NO) on the ROS metabolism of peroxisomes. Plants 8:E37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas F. J., Gómez M., Hernández J. A., del Rio L. A. (1993). Metabolism of activated oxygen in peroxisomes from two Pisum salivum L. cultivars with different sensitivity to sodium chloride. J. Plant Physiol. 141 160–165. 10.1016/s0176-1617(11)80753-4 [DOI] [Google Scholar]
- Corpas F. J., Palma J. M., Sandalio L. M., Valderrama R., Barroso J. B., del Río L. A. (2008). Peroxisomal xanthine oxidoreductase: characterization of the enzyme from pea (Pisum sativum L.) leaves. J. Plant Physiol. 165 1319–1330. 10.1016/j.jplph.2008.04.004 [DOI] [PubMed] [Google Scholar]
- Corpas F. J., Trelease R. N. (1998). Differential expression of ascorbate peroxidase and a putative molecular chaperone in the boundary membrane of differentiating cucumber seedling peroxisomes. J. Plant Physiol. 153 332–338. 10.1016/s0176-1617(98)80159-4 [DOI] [Google Scholar]
- De Duve C., Baudhuin P. (1966). Peroxisomes (microbodies and related particles). Physiol. Rev. 46 323–357. 10.1152/physrev.1966.46.2.323 [DOI] [PubMed] [Google Scholar]
- Debska K., Bogatek R., Gniazdowska A. (2012). Protein carbonylation and its role in physiological processes in plants. Postepy Biochem. 58 34–43. [PubMed] [Google Scholar]
- del Río L. A., Ortega M. G., López A. L., Gorgé J. L. (1977). A more sensitive modification of the catalase assay with the Clark oxygen electrode. Application to the kinetic study of the pea leaf enzyme. Anal Biochem. 80 409–415. 10.1016/0003-2697(77)90662-5 [DOI] [PubMed] [Google Scholar]
- del Río L. A., Sandalio L. M., Palma J. M., Bueno P., Corpas F. J. (1992). Metabolism of oxygen radicals in peroxisomes and cellular implications. Free Radic. Biol. Med. 13 557–580. 10.1016/0891-5849(92)90150-f [DOI] [PubMed] [Google Scholar]
- del Río L. A., Corpas F. J., López-Huertas E., Palma J. M. (2018). “Plant superoxide dismutases: function under abiotic stress conditions,” in Antioxidants and Antioxidant Enzymes in Higher Plants, (Cham: Springer International Publishing; ), 1–26. 10.1007/978?3?319?75088-0_1 [DOI] [Google Scholar]
- Dinçer A., Aydemir T. (2001). Purification and characterization of catalase from chard (Beta vulgaris var. cicla). J. Enzyme Inhib. 16 165–175. 10.1080/14756360109162366 [DOI] [PubMed] [Google Scholar]
- Dogra V., Rochaix J. D., Kim C. (2018). Singlet oxygen-triggered chloroplast-to-nucleus retrograde signalling pathways: an emerging perspective. Plant Cell Environ. 41 1727–1738. 10.1111/pce.13332 [DOI] [PubMed] [Google Scholar]
- Droillard M. J., Paulin A. (1990). Isozymes of superoxide dismutase in mitochondria and peroxisomes isolated from petals of carnation (Dianthus caryophyllus) during senescence. Plant Physiol. 94 1187–1192. 10.1104/pp.94.3.1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond P. J. (2007). MONODEHYROASCORBATE REDUCTASE4 is required for seed storage oil hydrolysis and postgerminative growth in Arabidopsis. Plant Cell 19 1376–1387. 10.1105/tpc.106.043992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovic M. R., Zivanovic J., Alvarez B., Banerjee R. (2018). Chemical biology of H2S signaling through persulfidation. Chem Rev. 118 1253–1337. 10.1021/acs.chemrev.7b00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhlke M. C., Slusarenko A. J. (2012). The biology of reactive sulfur species (RSS). Plant Physiol. Biochem. 59 98–107. 10.1016/j.plaphy.2012.03.016 [DOI] [PubMed] [Google Scholar]
- Guerra D., Ballard K., Truebridge I., Vierling E. (2016). S-Nitrosation of conserved cysteines modulates activity and stability of S-nitrosoglutathione reductase (GSNOR). Biochemistry 55 2452–2464. 10.1021/acs.biochem.5b01373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirimand G., Simkin A. J., Papon N., Besseau S., Burlat V., St-Pierre B., et al. (2012). Cycloheximide as a tool to investigate protein import in peroxisomes: a case study of the subcellular localization of isoprenoid biosynthetic enzymes. J. Plant Physiol. 169 825–829. 10.1016/j.jplph.2012.01.020 [DOI] [PubMed] [Google Scholar]
- Hackenberg T., Juul T., Auzina A., Gwizdz S., Malolepszy A., Van Der Kelen K., et al. (2013). Catalase and NO CATALASE ACTIVITY1 promote autophagy-dependent cell death in Arabidopsis. Plant Cell 25 4616–4626. 10.1105/tpc.113.117192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänsch R., Mendel R. R. (2005). Sulfite oxidation in plant peroxisomes. Photosynth Res. 86 337–343. 10.1007/s11120-005-5221-x [DOI] [PubMed] [Google Scholar]
- Hauck O. K., Scharnberg J., Escobar N. M., Wanner G., Giavalisco P., Witte C. P. (2014). Uric acid accumulation in an Arabidopsis urate oxidase mutant impairs seedling establishment by blocking peroxisome maintenance. Plant Cell 26 3090–3100. 10.1105/tpc.114.124008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinojosa L., Sanad M. N. M. E., Jarvis D. E., Steel P., Murphy K., Smertenko A. (2019). Impact of heat and drought stress on peroxisome proliferation in quinoa. Plant J. 99 1144–1158. 10.1111/tpj.14411 [DOI] [PubMed] [Google Scholar]
- Huang A. H. C., Moore T. S., Trelease R. N. (1983). Plant Peroxisomes. New York, NY: Academic Press. [Google Scholar]
- Huang J., Willems P., Wei B., Tian C., Ferreira R. B., Bodra N., et al. (2019). Mining for protein S-sulfenylation in Arabidopsis uncovers redox-sensitive sites. Proc. Natl. Acad. Sci. U.S.A. 116 21256–21261. 10.1073/pnas.1906768116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T., Yoshimura K., Sakai K., Tamoi M., Takeda T., Shigeoka S. (1998). Molecular characterization and physiological role of a glyoxysome-bound ascorbate peroxidase from spinach. Plant Cell Physiol. 39 23–34. 10.1093/oxfordjournals.pcp.a029285 [DOI] [PubMed] [Google Scholar]
- Jiménez A., Hernández J. A., Pastori G., del Río L. A., Sevilla F. (1998). Role of the ascorbate-glutathione cycle of mitochondria and peroxisomes in the senescence of pea leaves. Plant Physiol. 118 1327–35. 10.1104/pp.118.4.1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao Y. T., Gonzalez K. L., Bartel B. (2018). Peroxisome function, biogenesis, and dynamics in plants. Plant Physiol. 176 162–177. 10.1104/pp.17.01050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissner R., Nauser T., Bugnon P., Lye P. G., Koppenol W. H. (1997). Formation and properties of peroxynitrite as studied by laser flash photolysis, high-pressure stopped-flow technique, and pulse radiolysis. Chem. Res. Toxicol. 10 1285–1292. 10.1021/tx970160x [DOI] [PubMed] [Google Scholar]
- Kohli S. K., Khanna K., Bhardwaj R., Abd Allah E. F., Ahmad P., Corpas F. J. (2019). Assessment of subcellular ROS and NO metabolism in higher plants: multifunctional signaling molecules. Antioxidants 8:641. 10.3390/antiox8120641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbert Z., Barroso J. B., Brouquisse R., Corpas F. J., Gupta K. J., Lindermayr C., et al. (2019). A forty year journey: the generation and roles of NO in plants. Nitric. Oxide 93 53–70. [DOI] [PubMed] [Google Scholar]
- Kusano T., Kim D. W., Liu T., Berberich T. (2015). “Polyamine catabolism in plants,” in Polyamines, eds Kusano T., Suzuki H., (Tokyo: Springer; ), 77–88. 10.1007/978-4-431-55212-3_6 [DOI] [Google Scholar]
- León J., Castillo M. C., Coego A., Lozano-Juste J., Mir R. (2014). Diverse functional interactions between nitric oxide and abscisic acid in plant development and responses to stress. J. Exp. Bot. 65 907–921. 10.1093/jxb/ert454 [DOI] [PubMed] [Google Scholar]
- Leterrier M., Barroso J. B., Valderrama R., Begara-Morales J. C., Sánchez-Calvo B., Chaki M., et al. (2016). Peroxisomal NADP-isocitrate dehydrogenase is required for Arabidopsis stomatal movement. Protoplasma 253 403–415. 10.1007/s00709-015-0819-0 [DOI] [PubMed] [Google Scholar]
- Leterrier M., Chaki M., Airaki M., Valderrama R., Palma J. M., Barroso J. B., et al. (2011). Function of S-nitrosoglutathione reductase (GSNOR) in plant development and under biotic/abiotic stress. Plant Signal. Behav. 6 789–793. 10.4161/psb.6.6.15161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leterrier M., Corpas F. J., Barroso J. B., Sandalio L. M., del Río L. A. (2005). Peroxisomal monodehydroascorbate reductase. Genomic clone characterization and functional analysis under environmental stress conditions. Plant Physiol. 138 2111–2123. 10.1104/pp.105.066225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Li J., Hao R., Guo Y. (2017). Activation of catalase activity by a peroxisome-localized small heat shock protein Hsp17.6CII. J. Genet. Genomics 44 395–404. 10.1016/j.jgg.2017.03.009 [DOI] [PubMed] [Google Scholar]
- Lingard M. J., Bartel B. (2009). Arabidopsis LON2 is necessary for peroxisomal function and sustained matrix protein import. Plant Physiol. 151 1354–1365. 10.1104/pp.109.142505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisenbee C. S., Lingard M. J., Trelease R. N. (2005). Arabidopsis peroxisomes possess functionally redundant membrane and matrix isoforms of monodehydroascorbate reductase. Plant J. 43 900–914. 10.1111/j.1365-313x.2005.02503.x [DOI] [PubMed] [Google Scholar]
- López-Huertas E., Charlton W. L., Johnson B., Graham I. A., Baker A. (2000). Stress induces peroxisome biogenesis genes. EMBO J. 19 6770–6777. 10.1093/emboj/19.24.6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Huertas E., del Río L. A. (2014). Characterization of antioxidant enzymes and peroxisomes of olive (Olea europaea L.) fruits. J. Plant Physiol. 171 1463–1471. 10.1016/j.jplph.2014.06.014 [DOI] [PubMed] [Google Scholar]
- Lounifi I., Arc E., Molassiotis A., Job D., Rajjou L., Tanou G. (2013). Interplay between protein carbonylation and nitrosylation in plants. Proteomics 13 568–578. 10.1002/pmic.201200304 [DOI] [PubMed] [Google Scholar]
- Mano J., Nagata M., Okamura S., Shiraya T., Mitsui T. (2014). Identification of oxidatively modified proteins in salt-stressed Arabidopsis: a carbonyl-targeted proteomics approach. Plant Cell Physiol. 55 1233–1244. 10.1093/pcp/pcu072 [DOI] [PubMed] [Google Scholar]
- Mata-Pérez C., Begara-Morales J. C., Chaki M., Sánchez-Calvo B., Valderrama R., Padilla M. N., et al. (2016). Protein tyrosine nitration during development and abiotic stress response in plants. Front. Plant Sci. 7:1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A., Noctor G., Baker A. (2012). Plant catalases: peroxisomal redox guardians. Arch. Biochem. Biophys. 525 181–194. 10.1016/j.abb.2012.04.015 [DOI] [PubMed] [Google Scholar]
- Mhamdi A., Queval G., Chaouch S., Vanderauwera S., Van Breusegem F. (2010). Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 61 4198–4220. [DOI] [PubMed] [Google Scholar]
- Mishanina T. V., Libiad M., Banerjee R. (2015). Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways. Nat. Chem. Biol. 11 457–464. 10.1038/nchembio.1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn M. A., Thaqi B., Fischer-Schrader K. (2019). Isoform-specific NO synthesis by Arabidopsis thaliana nitrate reductase. Plants 8:67. 10.3390/plants8030067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor A., Koh E., Weiner L., Rosenwasser S., Sibony-Benyamini H., Fluhr R. (2014). Singlet oxygen signatures are detected independent of light or chloroplasts in response to multiple stresses. Plant Physiol. 165 249–261. 10.1104/pp.114.236380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschou P. N., Sanmartin M., Andriopoulou A. H., Rojo E., Sanchez-Serrano J. J., Roubelakis-Angelakis K. A. (2008). Bridging the gap between plant and mammalian polyamine catabolism: a novel peroxisomal polyamine oxidase responsible for a full back-conversion pathway in Arabidopsis. Plant Physiol. 147 1845–1857. 10.1104/pp.108.123802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Zechmann B., Zellnig G. (2004). Ultrastructural localization of glutathione in cucurbita pepo plants. Protoplasma 223 213–219. [DOI] [PubMed] [Google Scholar]
- Neill S., Barros R., Bright J., Desikan R., Hancock J., Harrison J., et al. (2008). Nitric oxide, stomatal closure, and abiotic stress. J. Exp. Bot. 59 165–176. [DOI] [PubMed] [Google Scholar]
- Nguyen A. T., Donaldson R. P. (2005). Metal-catalyzed oxidation induces carbonylation of peroxisomal proteins and loss of enzymatic activities. Arch. Biochem. Biophys. 2005 25–31. 10.1016/j.abb.2005.04.018 [DOI] [PubMed] [Google Scholar]
- Niu L., Yu J., Liao W., Xie J., Yu J., Lv J., et al. (2019). Proteomic investigation of S-nitrosylated proteins during NO-induced adventitious rooting of cucumber. Int. J. Mol. Sci. 20:5363. 10.3390/ijms20215363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G., Veljovic-Jovanovic S., Driscoll S., Novitskaya L., Foyer C H. (2002). Drought and oxidative load in the leaves of C3 plants: a predominant role for photorespiration? Ann. Bot. 89 841–50. 10.1093/aob/mcf096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak K., Luniak N., Witt C., Wüstefeld Y., Wachter A., Mendel R. R., et al. (2004). Peroxisomal localization of sulfite oxidase separates it from chloroplast-based sulfur assimilation. Plant Cell Physiol. 45 1889–1894. 10.1093/pcp/pch212 [DOI] [PubMed] [Google Scholar]
- Oikawa K., Hayashi M., Hayashi Y., Nishimura M. (2019). Re-evaluation of physical interaction between plant peroxisomes and other organelles using live-cell imaging techniques. J. Integr. Plant Biol. 61 836–852. [DOI] [PubMed] [Google Scholar]
- Oikawa K., Matsunaga S., Mano S., Kondo M., Yamada K., Hayashi M., et al. (2015). Physical interaction between peroxisomes and chloroplasts elucidated by in situ laser analysis. Nat. Plants. 1:15035. [DOI] [PubMed] [Google Scholar]
- Olson K. R. (2019). Hydrogen sulfide, reactive sulfur species and coping with reactive oxygen species. Free Radic. Biol. Med. 140 74–83. 10.1016/j.freeradbiomed.2019.01.020 [DOI] [PubMed] [Google Scholar]
- Olson K. R., Gao Y., Arif F., Arora K., Patel S., DeLeon E. R., et al. (2018). Metabolism of hydrogen sulfide (H2S) and production of reactive sulfur species (RSS) by superoxide dismutase. Redox Biol. 15 74–85. 10.1016/j.redox.2017.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson K. R., Gao Y., DeLeon E. R., Arif M., Arif F., Arora N., et al. (2017). Catalase as a sulfide-sulfur oxido-reductase: an ancient (and modern?) regulator of reactive sulfur species (RSS). Redox Biol. 12 325–339. 10.1016/j.redox.2017.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K., Akaike T., Sawa T., Kumagai Y., Wink D. A., Tantillo D. J., et al. (2014). Redox chemistry and chemical biology of H2S, hydropersulfides, and derived species: implications of their possible biological activity and utility. Free Radic. Biol. Med. 77 82–94. 10.1016/j.freeradbiomed.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma J. M., de Morales P. Á, del Río L. A., Corpas F. J. (2018). The proteome of fruit peroxisomes: sweet pepper (Capsicum annuum L.) as a model. Subcell Biochem. 89 323–341. 10.1007/978-981-13-2233-4_14 [DOI] [PubMed] [Google Scholar]
- Palma J. M., Jiménez A., Sandalio L. M., Corpas F. J., Lundqvist M., Gómez M., et al. (2006). Antioxidative enzymes from chloroplasts, mitochondria, and peroxisomes during leaf senescence of nodulated pea plants. J. Exp. Bot. 57 1747–1758. 10.1093/jxb/erj191 [DOI] [PubMed] [Google Scholar]
- Palma J. M., Mateos R. M., López-Jaramillo J., Rodríguez-Ruiz M., González-Gordo S., Lechuga-Sancho A. M., et al. (2020). Plant catalases as NO and H2S targets. Redox Biol. 24 131–137. 10.1016/j.redox.2020.101525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan R., Liu J., Wang S., Hu J. (2020). Peroxisomes: versatile organelles with diverse roles in plants. New Phytol. 225 1410–1427. 10.1111/nph.16134 [DOI] [PubMed] [Google Scholar]
- Pan R., Reumann S., Lisik P., Tietz S., Olsen L. J., Hu J. (2018). Proteome analysis of peroxisomes from dark-treated senescent Arabidopsis leaves. J. Integr. Plant Biol. 60 1028–1050. 10.1111/jipb.12670 [DOI] [PubMed] [Google Scholar]
- Park C. M., Weerasinghe L., Day J. J., Fukuto J. M., Xian M. (2015). Persulfides: current knowledge and challenges in chemistry and chemical biology. Mol. Biosyst. 11 1775–1785. 10.1039/c5mb00216h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacentini D., Corpas F. J., D’Angeli S., Altamura M. M., Falasca G. (2020). Cadmium and arsenic-induced-stress differentially modulates Arabidopsis root architecture, peroxisome distribution, enzymatic activities and their nitric oxide content. Plant Physiol. Biochem. 148 312–323. 10.1016/j.plaphy.2020.01.026 [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I., Smith S. M. (2008). When is a peroxisome not a peroxisome? Trends Plant Sci. 13 522–525. 10.1016/j.tplants.2008.07.003 [DOI] [PubMed] [Google Scholar]
- Prado A. M., Porterfield D. M., Feijó J. A. (2004). Nitric oxide is involved in growth regulation and re-orientation of pollen tubes. Development 131 2707–2714. 10.1242/dev.01153 [DOI] [PubMed] [Google Scholar]
- Ray M., Mishra P., Das P., Sabat S. C. (2012). Expression and purification of soluble bio-active rice plant catalase-A from recombinant Escherichia coli. J. Biotechnol. 157 12–19. 10.1016/j.jbiotec.2011.09.022 [DOI] [PubMed] [Google Scholar]
- Reumann S., Bartel B. (2016). Plant peroxisomes: recent discoveries in functional complexity, organelle homeostasis, and morphological dynamics. Curr. Opt. Plant Biol. 34 17–26. 10.1016/j.pbi.2016.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Quan S., Aung K., Yang P., Manandhar-Shrestha K., Holbrook D., et al. (2009). In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol. 150 125–143. 10.1104/pp.109.137703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro C. W., Korbes A. P., Garighan J. A., Jardim-Messeder D., Carvalho F. E. L., Sousa R. H. V., et al. (2017). Rice peroxisomal ascorbate peroxidase knockdown affects ROS signaling and triggers early leaf senescence. Plant Sci. 263 55–65. 10.1016/j.plantsci.2017.07.009 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ruiz M., González-Gordo S., Cañas A., Campos M. J., Paradela A., Corpas F. J., et al. (2019). Sweet pepper (Capsicum annuum L.) fruits contain an atypical peroxisomal catalase that is modulated by reactive oxygen and nitrogen species. Antioxidants 8:374. 10.3390/antiox8090374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Serrano M., Romero-Puertas M. C., Pastori G. M., Corpas F. J., Sandalio L. M., del Río L. A., et al. (2007). Peroxisomal membrane manganese superoxide dismutase: characterization of the isozyme from watermelon (Citrullus lanatus Schrad.) cotyledons. J. Exp. Bot. 58 2417–2427. 10.1093/jxb/erm095 [DOI] [PubMed] [Google Scholar]
- Romero-Puertas M. C., Corpas F. J., Sandalio L. M., Leterrier M., Rodríguez-Serrano M., del Río L. A., et al. (2006). Glutathione reductase from pea leaves: response to abiotic stress and characterization of the peroxisomal isozyme. New Phytol. 170 43–52. 10.1111/j.1469-8137.2006.01643.x [DOI] [PubMed] [Google Scholar]
- Rosenwasser S., Rot I., Sollner E., Meyer A. J., Smith Y., Leviatan N., et al. (2011). Organelles contribute differentially to reactive oxygen species-related events during extended darkness. Plant Physiol. 156 185–201. 10.1104/pp.110.169797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicht M., Ludwig-Müller J., Burbach C., Volkmann D., Baluska F. (2013). Indole-3-butyric acid induces lateral root formation via peroxisome-derived indole-3-acetic acid and nitric oxide. New Phytol. 200 473–482. 10.1111/nph.12377 [DOI] [PubMed] [Google Scholar]
- Schöneich C. (2016). Thiyl radicals and induction of protein degradation. Free Radic. Res. 50 143–149. 10.3109/10715762.2015.1077385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelp B. J., Zarei A. (2017). Subcellular compartmentation of 4-aminobutyrate (GABA) metabolism in Arabidopsis: an update. Plant Signal. Behav. 12:e1322244. 10.1080/15592324.2017.1322244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin A. J., Guirimand G., Papon N., Courdavault V., Thabet I., Ginis O., et al. (2011). Peroxisomal localisation of the final steps of the mevalonic acid pathway in planta. Planta 234 903–914. 10.1007/s00425-011-1444-6 [DOI] [PubMed] [Google Scholar]
- Sousa R. H. V., Carvalho F. E. L., Lima-Melo Y., Alencar V. T. C. B., Daloso D. M., Margis-Pinheiro M., et al. (2019). Impairment of peroxisomal APX and CAT activities increases protection of photosynthesis under oxidative stress. J. Exp. Bot. 70 627–639. 10.1093/jxb/ery354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T., Li W., Wang P., Ma C. (2019). Dynamics of peroxisome homeostasis and its role in stress response and signaling in Plants. Front. Plant Sci. 10:705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T., Wang P., Li H., Zhao Y., Lu Y., Dai P., et al. (2018). The Arabidopsis catalase triple mutant reveals important roles of catalases and peroxisome-derived signaling in plant development. J. Integr. Plant Biol. 60 591–607. 10.1111/jipb.12649 [DOI] [PubMed] [Google Scholar]
- Tolbert N. E., Essner E. (1981). Microbodies: peroxisomes and glyoxysomes. J. Cell Biol. 91 271s–283s. 10.1083/jcb.91.3.271s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y., Uehara N., Sasaki H., Kobayashi K., Yamakawa T. (2013). Impacts of acute ozone stress on superoxide dismutase (SOD) expression and reactive oxygen species (ROS) formation in rice leaves. Plant Physiol. Biochem. 70 396–402. 10.1016/j.plaphy.2013.06.009 [DOI] [PubMed] [Google Scholar]
- Veljovic-Jovanovic S., Oniki T., Takahama U. (1998). Detection of monodehydroascorbic acid radical in sulfite-treated leaves and mechanism of its formation. Plant Cell Physiol. 39 1203–1208. 10.1093/oxfordjournals.pcp.a029321 [DOI] [Google Scholar]
- Wagner D., Przybyla D., Op den Camp R., Kim C., Landgraf F., Lee K. P., et al. (2004). The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306 1183–1185. 10.1126/science.1103178 [DOI] [PubMed] [Google Scholar]
- Wang X., Zhu B., Jiang Z., Wang S. (2019). Calcium-mediation of jasmonate biosynthesis and signaling in plants. Plant Sci. 287:110192. 10.1016/j.plantsci.2019.110192 [DOI] [PubMed] [Google Scholar]
- Wang W., Paschalidis K., Feng J. C., Song J., Liu J. H. (2019). Polyamine catabolism in plants: a universal process with diverse functions. Front. Plant Sci. 10:561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Zhang M., Wei S., Zhang J., Wang C., Liao W. (2020). Roles of nitric oxide in heavy metal stress in plants: cross-talk with phytohormones and protein S-nitrosylation. Environ. Pollut. 259:113943. 10.1016/j.envpol.2020.113943 [DOI] [PubMed] [Google Scholar]
- Wuddineh W., Minocha R., Minocha S. C. (2018). Polyamines in the context of metabolic networks. Methods Mol. Biol. 1694 1–23. 10.1007/978-1-4939-7398-9_1 [DOI] [PubMed] [Google Scholar]
- Xin Z., Chen S., Ge L., Li X., Sun X. (2019). The involvement of a herbivore-induced acyl-CoA oxidase gene, CsACX1, in the synthesis of jasmonic acid and its expression in flower opening in tea plant (Camellia sinensis). Plant Physiol. Biochem. 135 132–140. 10.1016/j.plaphy.2018.11.035 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Mori H., Nishimura M. (1995). A novel isoenzyme of ascorbate peroxidase localized on glyoxysomal and leaf peroxisomal membranes in pumpkin. Plant Cell Physiol. 36 1157–1162. 10.1093/oxfordjournals.pcp.a078862 [DOI] [PubMed] [Google Scholar]
- Yamauchi S., Mano S., Oikawa K., Hikino K., Teshima K. M., Kimori Y., et al. (2019). Autophagy controls reactive oxygen species homeostasis in guard cells that is essential for stomatal opening. Proc. Natl. Acad. Sci. U.S.A. 116 19187–19192. 10.1073/pnas.1910886116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Li Z., Zhang K., Zhang X., Zhang Y., Wang X., et al. (2018). Barley stripe mosaic virus γb interacts with glycolate oxidase and inhibits peroxisomal ROS production to facilitate virus infection. Mol. Plant 11 338–341. 10.1016/j.molp.2017.10.007 [DOI] [PubMed] [Google Scholar]
- Yu L., Fan J., Xu C. (2019). Peroxisomal fatty acid β-oxidation negatively impacts plant survival under salt stress. Plant Signal. Behav. 14:1561121. 10.1080/15592324.2018.1561121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Xu Y., Xie Z., Li X., He Z H., Peng X X. (2016). Association-dissociation of glycolate oxidase with catalase in rice: a potential switch to modulate intracellular H2O2 levels. Mol. Plant. 9 737–748. 10.1016/j.molp.2016.02.002 [DOI] [PubMed] [Google Scholar]
- Zarei A., Trobacher C. P., Cooke A. R., Meyers A. J., Hall J. C., Shelp B. J. (2015). Apple fruit copper amine oxidase isoforms: peroxisomal MdAO1 prefers diamines as substrates, whereas extracellular MdAO2 exclusively utilizes monoamines. Plant Cell Physiol. 56 137–147. 10.1093/pcp/pcu155 [DOI] [PubMed] [Google Scholar]
- Zarepour M., Kaspari K., Stagge S., Rethmeier R., Mendel R. R., Bittner F. (2010). Xanthine dehydrogenase AtXDH1 from Arabidopsis thaliana is a potent producer of superoxide anions via its NADH oxidase activity. Plant Mol. Biol. 72 301–310. 10.1007/s11103-009-9570-2 [DOI] [PubMed] [Google Scholar]