Abstract
Nucleophosmin (NPM1) is a ubiquitously expressed nucleolar protein involved in ribosome biogenesis, the maintenance of genomic integrity and the regulation of the ARF-p53 tumor-suppressor pathway among multiple other functions. Mutations in the corresponding gene cause a cytoplasmic dislocation of the NPM1 protein. These mutations are unique to acute myeloid leukemia (AML), a disease characterized by clonal expansion, impaired differentiation and the proliferation of myeloid cells in the bone marrow. Despite our improved understanding of NPM1 mutations and their consequences, the underlying leukemia pathogenesis is still unclear. Recent studies that focused on dysregulated gene expression in AML with mutated NPM1 have shed more light into these mechanisms. In this article, we review the current evidence on normal functions of NPM1 and aberrant functioning in AML, and highlight investigational strategies targeting these mutations.
Keywords: AML, nucleophosmin (NPM1), gene expression, targeted therapies
1. Introduction
Acute myeloid leukemia (AML) is a clinically and genomically heterogeneous malignancy despite having a relatively low number of genomic alterations compared to other cancers [1,2]. Mutations in bone marrow cells of the myeloid lineage or in myeloid progenitors lead to AML by hijacking properties from normal hematopoietic stem cells such as self-renewal, and via the recapitulation of the malignant progeny [3]. Treatment of AML had not changed in decades until recently, when several drugs received regulatory approval. This progress is largely due to an improved understanding of the biology and genomic landscape of this disease [4]. A mutation in the gene encoding nucleophosmin (NPM1) is one of the most commonly detected genomic alterations in AML. It is found in 20–30% of newly diagnosed AML and in 50% of those with a normal karyotype [1,5,6]. NPM1 is a chaperone protein that shuttles between the nucleus and cytoplasm with numerous functions. Multiple mechanisms through which mutations in NPM1 lead to leukemia have been described. However, despite an increased understanding of these mechanisms, there has not been to date any targeted therapy for NPM1-mutated AML. In this review, we summarize the current knowledge on the normal and aberrant functions of NPM1, highlight mechanisms of leukemogenesis and discuss rational therapeutic strategies.
2. Structure and Function of Wild-Type NPM1
2.1. NPM1 Protein Structure
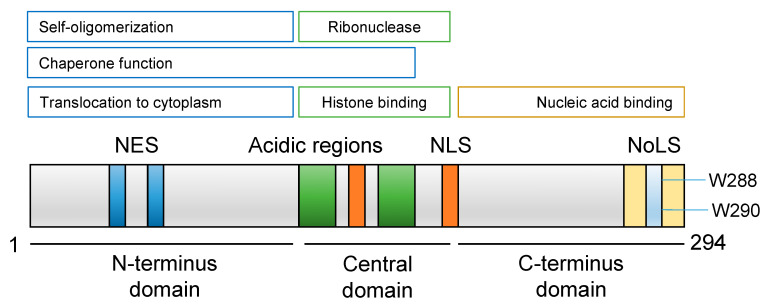
NPM1 is a ubiquitous and abundant protein that resides in the nucleoli under normal physiologic conditions but continuously shuttles between the nucleus and the cytoplasm. It is composed of 294 amino acids with a molecular weight of 37 kDa. It is divided into three structural and functional domains: N terminus, central and C terminus (Figure 1). The N terminus domain is highly conserved among proteins of the nucleophosmin family. Nuclear export signals (NES) which promote the translocation of NPM1 from the nucleus to the cytoplasm are found in the N terminus, with some contribution to this function by the central domain [7]. This translocation is dependent on the interaction of NES with the CRM1 export protein (Chromosomal Maintenance 1, also known as Exportin 1 or XPO1) [7,8]. NPM1 binds denatured proteins in vitro, a hallmark of chaperones [9]. This chaperone activity is mediated through the interaction of the N-terminus with numerous proteins [10]. The hydrophobic N terminus also promotes self-oligomerization, in which five monomers form a pentamer, driving NPM1 to the nucleolus [11,12]. The central domain contains two highly acidic regions necessary for binding histones, thus enabling the chromatin remodeling functions of NPM1 [13]. This domain also has a ribonuclease activity implicated in ribosome biogenesis and a nuclear localization signal (NLS) [11,14]. The C terminus domain has a highly conserved aromatic region, particularly at the Trp-288 and Trp-290 sites which form the nucleolar localization signal (NoLS) [10,15]. NoLS is critical for the localization of NPM1 to the nucleoplasm, with any alteration at this site being sufficient to delocalize NPM1 from the nucleolus [15].
Figure 1.
Nucleophosmin (NPM1) protein structure and functions. The nuclear export system (NES) (blue) located in the N-terminus domain promotes the translocation of NPM1 from the nucleus to the cytoplasm. The central domain contains the acidic regions (green) and a nuclear localization signal (NLS) (orange), and mediates histone binding and the ribonuclease activity. The C-terminus domain has a highly conserved aromatic region and the nucleolar localization signal (NoLS) (yellow) which contains Trp-288 and Trp-290.
The functions and localization of NPM1 are regulated by post-translational modifications [10,16]. NPM1 is phosphorylated by cyclin-dependent kinases at different sites affecting the various specific functions of this protein. The phosphorylation of Ser-227, Thr-234 and Thr-237 affects cell cycle progression [10], whereas the phosphorylation of Thr-199 regulates the duplication of cells by localizing NPM1 to the centrosome [17,18]. In addition, the phosphorylation of Thr-199, Thr-219, Thr-234 and Thr-237 inactivates the RNA binding capabilities, thus affecting the role of NPM1 in ribosomal biogenesis [19]. The acetylation of NPM1 lysine residues affects histone binding and chromatin transcription [20]. The acetyltransferase p300 modulates the subcellular localization of NPM1. Acetylated NPM1, predominantly localized in the nucleoplasm, associates with transcriptionally active RNA polymerase II [21]. Other post-translational modifications include sumoylation and ubiquitination, which affect NPM1 stabilization and degradation [10,22].
2.2. Physiologic NPM1 Functions
2.2.1. Ribosome Synthesis
The nucleolus—where NPM1 resides in physiologic states—is the site of the active transcription of the ribosomal ribonucleic acid (rRNA) and ribosome assembly [23]. Ribosomal biogenesis is the synthesis and processing of a precursor rRNA (pre-rRNA) and the assembly of ribosomal proteins on rRNAs to form premature ribosomes. NPM1 interacts with the ribosomal protein L5 (rpL5), a known chaperone for the 5S unit of rRNA [8]. The inhibition of NPM1 shuttling or loss of NPM1 blocks the nuclear export of rpL5 and 5S rRNA, resulting in cell cycle arrest [8]. This interaction of NPM1 with rpL5 allows NPM1 to access the maturing ribosome. As a chaperone, NPM1 prevents enzyme denaturation and aggregation [8,24,25]. The nucleic acid binding capacity of NPM1 and its ribonuclease activity further highlight its role in ribosome synthesis. Downregulation of NPM1 mRNA has been shown to inhibit the processing of pre-rRNA [26]. Thus, NPM1 supports cell growth by participating in the synthesis of ribosomes.
2.2.2. Genomic Stability and DNA Repair
NPM1 maintains genomic stability by binding to centrosomes and restricting their duplication to once per cell cycle [17]. The transcription of NPM1 is upregulated after UV irradiation in different cell lines [26,27]. Following DNA double stranded breaks, NPM1 functions as a chromatin binding factor [28]. While increased NPM1 levels have been associated with improved DNA repair, the loss of NPM1 leads to the increased phosphorylation of histone γ-H2AX, a target of the DNA repair kinases ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and RAD3-related (ATR) [29,30]. Heterozygous NPM1 knockout mice have uncontrolled centrosome duplication, which leads to supernumerary chromosomes and aneuploidy [31,32]. Finally, NPM1 functions may be regulated by BRCA1-mediated polyubiquitination, with BRCA1 inactivation leading to dysregulation in centrosome duplication [18].
2.2.3. Cellular Growth and Stress Response
NPM1 is involved in the regulation of proliferation and growth suppression pathways. NPM1 overexpression correlates with uncontrolled cell growth [33,34]. NPM1 contributes to the p53 stress response by modulating the activity and stability of p53 and the Arf tumor suppressor protein. NPM1 interacts directly with p53, regulating an increase in the stability and transcriptional activation of p53 after different types of stress [35]. It also interacts with the p53 negative regulator MDM2. Upon exposure to stress-inducing DNA damage, NPM1 translocates from the nucleoli to the nucleoplasm binding MDM2, thereby inhibiting p53 degradation by affecting the p53–MDM2 complex formation [36]. In addition, NPM1 regulates Arf, thus modulating growth suppression following cellular stress by a p53-independent mechanism [37,38].
NPM1 is a transcriptional target of MYC [39,40]. MYC overexpression increases protein synthesis; therefore, NPM1 is a key transcriptional target, given its numerous functions. NPM1 controls Myc-induced proliferation and transformation by binding directly to Myc and regulating its binding to promotors of target genes [30,41].
2.3. NPM1 Gene Structure
The NPM1 gene is highly conserved between humans, rodents, chicken and fish. Human NPM1 is located on chromosome 5q35 and is composed of 12 exons with sizes ranging from 58 to 358 base pairs (bp) [42]. The regular-spliced NPM1 gene has 11 exons encoding 294 amino acids. There are two other isoforms in addition to the wild-type NPM1 gene (NPM1.2 and NPM1.3); however, less is known about their expression and function. The inactivation of NPM1 in the germ line is embryonically lethal. Npm1−/− mutant mice have aberrant organogenesis and die between embryonic day E11.5 and E16.5 because of severe anemia caused by defects in primitive hematopoiesis [31]. Other distinct developmental abnormalities in these mice include deficient anterior brain organogenesis, with complete absence of the eye [31].
3. NPM1 and Leukemia
3.1. NPM1 Mutations
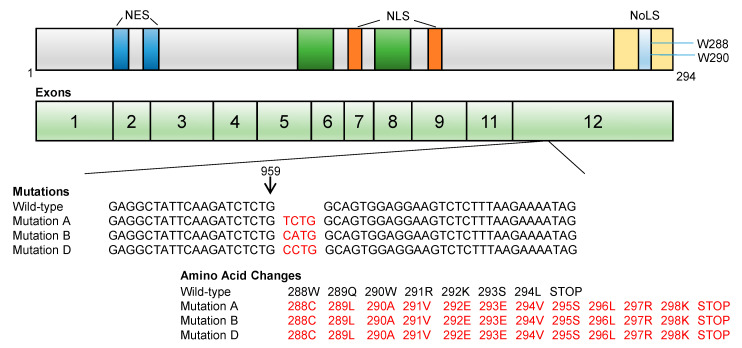
NPM1 is frequently overexpressed in multiple malignancies including gastric, colon, ovarian, bladder and prostate cancers, as reviewed by Grisendi and colleagues [33,34,43,44,45,46]. The NPM1 locus is involved in translocations causing hematologic malignancies such as acute promyelocytic leukemia where t(5;17)(q35;q12) leads to NPM1–RARα, anaplastic large cell lymphoma where t(2;5)(p23;q35) leads to NPM1-ALK, and myeloid neoplasms where t(3;5)(q25;q35) leads to NPM1-MLF1 [47,48,49]. In addition to the deletion of 5q35 in myelodysplastic syndrome, these abnormalities provided the rationale for the first studies that investigated alterations of NPM1 in AML. Subsequently, Falini and colleagues discovered that NPM1 mutations are relatively common in AML [50]. These mutations are found almost exclusively in exon 12, and to date have only been identified in myeloid malignancies but not in any other tumor. They often consist of 4 bp insertions or duplications between nucleotides 960 and 961. They cause the replacement of the last seven amino acids (WQWRKSL) with 11 different residues (Figure 2). Type A mutations, detected in 80% of cases, involve the duplication of “TCTG” (nucleotides 956–959), creating an insertion at position 960. Types B and D are the second and third most commonly occurring, followed by a few other rare mutations [50,51,52,53,54,55]. All of these mutations affect the Trp-289 and Trp-290 where the NoLS resides, leading instead to a cytoplasmic localization of the protein. Cytoplasmic NPM1 (NPM1c) is only detected in AML with the NPM1-mutated gene (NPM1c), and there are no NPM1 mutations with NPM1 remaining in the nucleolus. NPM1 mutations are exclusively heterozygous, which implies that NPM1c is able to form a dimer with wild-type NPM1, recruit it to the cytoplasm and perturb its normal function [31].
Figure 2.
NPM1 gene structure and common mutations in AML. Gene structure with exons 1–12 shown in addition to the sequence of the three most frequent exon 12 mutations/insertions in AML. These insertions lead to amino acid changes which affect the nucleolar localization signal (NoLS) (yellow) and cause the translocation of NPM1 from the nucleus to the cytoplasm. Nuclear export system (NES) (blue) and nuclear localization signal (NLS) (orange).
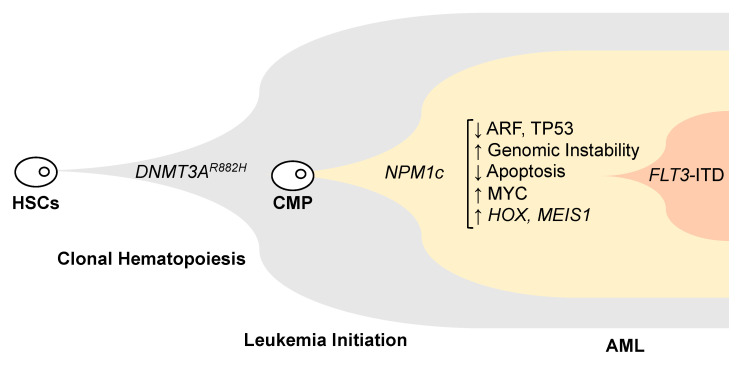
The lymphoid enzyme terminal deoxynucleotidyl transferase (TdT) has been implicated in the development of NPM1c through the addition of misprimed nucleotides causing these frameshift mutations [56]. The normal function of TdT is to increase the diversity of the immunoglobulin and T-cell receptor loci by adding non-templated nucleotides to their variable regions [57]. Interestingly, this off-target TdT activity has also been implicated in the development of internal tandem duplication mutations in the fms-like tyrosine kinase 3 gene (FLT3-ITD), which frequently co-occur with NPM1 mutations in AML [58]. FLT3-ITD mutations occur twice as often in AML with mutated NPM1 compared to AML with the wild-type NPM1 gene [50,51,52,53,54,55,59]. NPM1 mutations alone are not sufficient to cause AML and require cooperating mutations in other genes (Figure 3). A knock-in of NPM1c in mouse hematopoietic stem cells resulted in enhanced self-renewal and expanded myelopoeisis [60]. However, cooperating mutations were required for the development of overt leukemia [60,61]. Mutations in the gene DNA methyltransferase 3A (DNMT3A) also co-occur with NPM1c in 60% of AML cases [1]. DNMT3A mutations occur in hematopoietic stem cells and lead to a clonal expansion of these cells, a process known as clonal hematopoiesis (CH), which is an age-related process that predisposes to leukemia [62,63,64,65,66]. NPM1c mutations are only detected in overt myeloid malignancies and not in clonal hematopoiesis [67,68]. These mutations are restricted to the myeloid lineage and do not affect the lymphoid lineage, indicating that they probably arise in a common myeloid progenitor [62,69]. Using variant allele frequencies to infer the chronological sequence of these events in leukemia development, it was shown that mutations in DNMT3A likely develop first in hematopoietic stem cells (or other mutations causing CH), followed by NPM1c second in a CMP, and FLT3 mutations (or RAS) third (Figure 3) [1,54,70]. The co-occurrence of mutations in DNMT3A, NPM1 and FLT3-ITD is the most frequent combination of mutations found in AML, which highlights the synergy of this complex gene interaction in causing leukemia [1,61].
Figure 3.
Mechanisms of leukemogenesis in AML with mutated NPM1 gene. NPM1 mutations alone are not sufficient for causing leukemia. They arise from common progenitor cells (CMP) that have expanded following clonal hematopoiesis. This figure shows the most common combination of mutations, but other mutations causing clonal hematopoiesis could provide a similar background. NPM1 mutations are leukemia-initiating. They disrupt multiple mechanisms with a specific dependence on HOX genes and their DNA-binding cofactor MIES1.
3.2. Potential Leukemogenic Mechanisms of NPM1 Mutations
Given all the various important functions of NPM1 in maintaining homeostasis, any disruption of these functions could perturb numerous mechanisms and lead to malignancy (Figure 3). However, the striking specificity of findings associated with NPM1c possibly offer the best clues on the mechanisms underlying their neoplastic effect: (1) NPM1c mutations are only detected in myeloid malignancies and not in any other cancer despite the ubiquitous expression of NPM1 in cells from various tissues; (2) All NPM1c mutations lead to a translocation of NPM1 from the nucleus to the cytoplasm; (3) All NPM1c mutations in AML are heterozygous; therefore, they likely act as a dominant negative mutant leading to the retention of wild-type NPM1 in the cytoplasm; (4) NPM1c alone without co-operating mutations is not sufficient for leukemogenesis. Therefore, it is not clear based on current knowledge whether NPM1c mutations drive leukemia predominantly through loss of function, gain of function or both.
3.2.1. Genomic Instability
NPM1 regulates centrosome duplication, and NPM1 loss affects genomic stability. However, it is unclear whether the disruption of this function following mutations in NPM1 is a central mechanism contributing to leukemogenesis. Npm1+/− mice have a significant degree of genomic instability that increased their susceptibility to oncogenic transformation [31]. On the other hand, NPM1c are most common in normal karyotype AML, indicating no evidence of genomic instability at least at the cytogenetic analysis level. It is possible that NPM1c cells retain their capacity to control centrosome duplication in those cases. Additional chromosomal abnormalities are detected in 15% of AML with an NPM1 mutation. In a recent analysis, the presence of a complex karyotype in AML with an NPM1-mutated gene, with negative or low burden FLT-ITD, was associated with lower response rates and shorter survival despite earlier reports showing no impact on prognosis [71,72,73].
3.2.2. Loss of ARF and/or p53 Functions
The tumor suppressor Arf residing in the nucleolus is stabilized by NPM1. Arf is delocalized to the cytoplasm by NPM1c, which in turn exerts a dominant negative effect on wild-type NPM1, and it limits the inhibitory effect of NPM1 on this protein’s turnover, thus decreasing the half-life of Arf [74]. However, it appears that the perturbation of Arf functions is not enough to explain the oncogenic effects of NPM1c [75].
NPM1 stabilizes both TP53 and MDM2, the TP53 negative regulator, hence modulating cellular growth suppression through p53-dependent and independent mechanisms. Loss of NPM1 could affect levels of TP53, leading to oncogenic transformation. However, mouse cells and embryos that are Npm1 null have stabilized p53, indicating that NPM1 is not critical for p53 induction [30,31].
3.2.3. Effect on Apoptosis
NPM1c binds to cleaved caspase-6 and 8, which are the active forms of these proteins, thereby exerting a gain of function by inhibiting their proteolytic activity [76]. This affects apoptosis by offering protection from death ligand-induced cell apoptosis, in addition to suppressing the effect of caspase-6/-8 on myeloid differentiation [76]. In addition, NPM1c could affect the migration of the proapoptotic protein GADD45A. This protein interacts with p53 in the nucleus to enhance cell cycle arrest in response to stress. However, it lacks a nuclear localization signal and relies on NPM1 for this function [77,78].
3.2.4. Increased MYC
Cells with NPM1c have decreased the degradation of Myc by acting on the F-box protein Fbw7gamma, a component of the E3 ligase complex involved in the ubiquitination and proteasome degradation of this oncogene [79]. Myc protein expression by immunohistochemistry in the bone marrow of patients with AML was found to be relatively higher in AML patients with a mutated NPM1 gene compared to other subtypes [80]. This perhaps highlights the role of this oncogene in the neoplastic process triggered by mutations in NPM1.
3.2.5. HOX Genes Expression
AML with NPM1c is associated with the upregulation of the homeobox cluster or HOX genes, which are genes that are important for hematopoietic development. Specifically, members of this distinctive stem cell signature include the genes HOXA, HOXB and their DNA-binding cofactor MEIS1 [5,60,61,81]. However, the link between this signature and NPM1c had not been clear until recently. To assess whether the presence of NPM1c, its cytoplasmic location and the overexpression of HOX genes are just an association or a series of events related to the mutation, Brunetti and colleagues employed a series of experiments modulating these factors genetically or pharmacologically [82]. They found that NPM1c mutant-specific degradation or re-localization to the nucleus led to a loss of this stem cell signature and induced the differentiation of these myeloid cells by disrupting the oncogenic program. This highlights that the cytoplasmic location of NPM1c is critical for the pathogenesis of the disease and not just the decrease of NPM1 in the nucleus. HOX genes are also upregulated in leukemias with a rearrangement of the Mixed Lineage Leukemia (MLL) gene (also known as Lysine Methyltransferase 2A or KMT2A) [83]. MLL1 is a histone methyltransferase with multiple functions that affect transcription. Drawing on the gene expression similarity with MLL-rearranged leukemia, and the dependence of MLL rearrangements on their DNA-binding cofactor menin to exert an oncogenic effect, recent studies have shed more light into how NPM1c induces the aberrant expression of HOX genes [83,84]. Using CRISPR/Cas9 editing, AML with the mutated NPM1 gene was found to be dependent on menin and on MEIS1 [85,86]. Though this series of investigations established the central role of the MLL complex and HOX genes in causing this leukemia, the mechanism by which NPM1c interacts with these epigenetic regulators is still unknown.
3.3. Phenotype and Clinical Implications of NPM1 Mutations
AML with the mutated NPM1 gene is a distinct subtype according the 2016 World Health Organization (WHO) classification, due to its specific mutational profile, immunophenotype, clinical behavior and mutual exclusiveness to other recurring genomic alterations [87]. NPM1c mutations are associated with the female sex, even though AML is more common in men, in those with a higher median age (age 39–47 years), and in those with higher bone marrow blast, white blood cell and platelet counts compared to other subtypes of AML [1,50,51,54,55,59,88,89]. NPM1c AML is most commonly classified under the M4 (Acute myelomonocytic leukemia) or M5 (Acute monocytic leukemia) subtypes of the French–American–British (FAB) classification [52]. The blasts in NPM1-mutated AML typically have a distinctive fish-mouth appearance. It has a relatively better prognosis with higher rates of remission and longer survival compared to other subtypes of AML [51,52,53,54,55,88,90]. However, prognosis depends strongly on the presence of co-occurring mutations. FLT3-ITD mutations confer a worse prognosis, especially when their allelic burden is high [1,91]. Furthermore, this adverse effect of FLT3-ITD is most pronounced when present in combination with mutations in NPM1 and DNMT3A [2]. Numerous studies have shown the benefit of an allogeneic stem cell transplant in patients with concomitant NPM1 and FLT3-ITD mutations, especially when the FLT3-ITD allelic burden is high [92,93].
NPM1c mutations are persistent throughout the course of AML and disappear with remission. This finding highlights their clinical significance and use in the monitoring of minimal or measurable residual disease (MRD) following treatment. MRD provides powerful prognostic information and is increasingly incorporated in the routine management of AML. Detectable MRD is consistently associated with an increased risk of relapse and worse long-term outcomes [94]. Using a reverse-transcriptase quantitative polymerase-chain-reaction assay to detect NPM1c transcripts, Ivey and colleagues established the importance of NPM1c as a prognostic MRD marker in AML [95]. These mutations detected at the time of relapse provided a better marker of the disease status, and the persistence of NPM1c following treatment could be considered an indication for an allogeneic stem cell transplant [96].
3.4. Therapeutic Strategies for Targeting AML with Mutated NPM1 Gene
An improved understanding of the mechanisms underlying the oncogenic effect of NPM1c has resulted in multiple innovative and promising therapeutic strategies. The current treatment of AML with the NPM1-mutated gene consists of combinations of chemotherapeutic agents with the backbone of an anthracycline and cytarabine [4]. These combinations result in high remission rates in NPM1c AML, where the FLT3 mutational burden is low, leading to prolonged survival compared to other subtypes [4]. However, relapses and resistance still occur in AML with NPM1c, and the current treatment can be associated with a significant morbidity, especially in older patients. Therefore, novel therapeutic strategies can further improve outcomes in this entity, especially when specifically targeting NPM1c—a leukemia-initiating event (Table 1).
Table 1.
Investigational strategies for targeting AML with mutated NPM1.
| Drug Class | Target | Mechanism | References | |
|---|---|---|---|---|
| ATRA + ATO | Antineoplastic agents | ? | Proteasomal degradation of NPM1 | Martelli et al., 2015; El Hajj et al., 2015 |
| Dactinomycin | Chemotherapy | RNA-Pol 1 | Unclear in this setting | Falini et al., 2015 |
| GO | ADC | CD33 | Induces DSBs in DNA, cell cycle Arrest and apoptosis | Lambert et al., 2014; Olombel et al., 2016 |
| Venetoclax | Small molecule | Bcl-2 | Induces apoptosis | Lachowiez et al., 2020 |
| Selinexor | Small molecule | XPO1 | Inhibits translocation of NPM1 to the cytoplasm | Brunetti et al., 2019 |
| EAPB0503 | Small molecule | ? | Proteasomal degradation of NPM1 | Nabbouh et al., 2017 |
| Modified T cells | Immunotherapy | Neoantigen | T-cell activation | Van der Lee et al., 2019 |
| VTP-50469 | Small molecule | Menin | Disrupts MLL chromatin complex Preferential effect on MEIS1 (not HOX) | Uckelman et al., 2020 |
| MI-2, MI-503, MI-463, MI-3454 | Small molecule | Menin | Inhibits HOX genes and MEIS1 | Borkin et al., 2015; Grembecka et al., 2012; Krivtsov et al. 2019 |
| Deguelin | Rotenoid | ? | Induces apoptosis in NPM1c cell lines | Yi et al., 2015 |
| NSC348884 | Small molecule | NPM1 Oligomerization domain | Inhibits oligomerization, leading to apoptosis | Balusa et al., 2011 |
Abbreviations: ATRA + ATO, all-trans retinoic acid with arsenic trioxide; GO, Gemtuzumab ozogamicin; ADC, antibody-drug conjugate. GO is an IgG4 κ antibody bound to calicheamicin. DSBs: double-stranded breaks. Targets of EAPB0503 and Deguelin are unknown.
The combination of all-trans-retinoic acid (ATRA) and arsenic trioxide (ATO), a curative treatment for acute promyelocytic leukemia, leads to the degradation of NPM1c, inducing apoptosis and cell differentiation in NPM1-mutated cells [97,98]. Given the high expression levels of CD33 in NPM1 mutant AML, the antibody-drug conjugate gemtuzumab ozogamicin (GO) was identified as an effective addition to combination strategies [99,100]. Bcl-2 inhibition in combination with chemotherapy or hypomethylating agents is emerging as an attractive strategy for AML patients who are unable to tolerate conventional chemotherapy [101,102]. The Bcl-2 inhibitor drug venetoclax shows an excellent activity in patients with NPM1c, resulting in higher response rates in this entity compared to other mutational profiles [103]. Dactinomycin is a chemotherapeutic agent used in the treatment of malignancies such as Wilms tumors or gestational trophoblastic tumors. With the hypothesis that it induces nucleolar stress, dactinomycin was used to successfully treat a 60-year-old patient who was not a candidate for conventional chemotherapy and induced a durable complete remission, including for two other patients with refractory NPM1c AML (among six patients with refractory NPM1c AML) [104].
The cytoplasmic location of NPM1c is a key event in leukemia development. The translocation of NPM1 to the cytoplasm depends on the interaction with the CRM1 export protein, also known as Exportin 1 or XPO1. Selinexor is an orally bioavailable selective XPO1 inhibitor approved for the treatment of advanced multiple myeloma and is under investigation for the treatment of lymphoma and AML. Brunetti and colleagues targeted XPO1 with selinexor in an NPM1c model, which led to the relocalization of NPM1 to the nucleus, inhibition of the HOX genes and an anti-leukemia effect [82]. However, a clinical trial investigating selinexor in AML showed modest efficacy and concerns about tolerability because of anorexia attributed to CNS penetration [105]. The low number of patients included in this trial did not allow for the correlation of activity with the mutational status [105].
Given the growing knowledge on the central role of HOX genes in NPM1c pathogenesis and the interaction with the MLL chromatin complex, recent studies have leveraged the use of small molecule inhibitors of the MLL co-factor menin to target NPM1-mutated AML [106,107,108]. Using an inducible Npm1 single mutant and Npm1/Dnmt3a double mutant mouse model in addition to patient-derived xenograft models, Uckelmann and colleagues tested the effect of VTP-50469, a small molecule inhibitor of menin [86]. They demonstrated that menin inhibition induced the loss of MEIS1 expression and led to the significant differentiation of Npm1c leukemic cells in vitro, with a potent anti-leukemic activity on NPM1c in pre-leukemia and in overt leukemia. Interestingly, the expression levels of HOX genes were not affected by this drug despite the dramatic anti-tumor effect, indicating maybe that MEIS1 could be the shared vulnerability for AML with NPM1c. Similarly, Klossowski and colleagues used the menin inhibitor MI-3454 in a mouse model of NPM1-mutated AML and in patient-derived xenograft models, and showed remarkable anti-leukemic activity, this time, however, downregulating HOX genes and MEIS1 [109]. Both of these inhibitors have analogs currently under investigation in clinical trials which just started (NCT04065399, NCT04067336). One question, among many others, that could be answered by these trials is whether targeting HOX genes and MEIS1 versus MEIS1 alone would add to efficacy or affect tolerability.
NPM1c mutations produce the same alternative reading frame in the C-terminus domain. Therefore, this can be considered a shared neoantigen with the NPM1-mutated gene in AML, which can be targeted with immunotherapy. Neoantigens arise from tumor-specific mutations and are limited to malignant cells [110,111,112]. Van der Lee and colleagues developed genetically engineered T cells after identifying one peptide (HLA-A*02:01-binding CLAVEEVSL) as immunogenic [112]. They subsequently generated T cell clones and retrovirally transduced T cell populations that kill NPM1-mutated AML [113].
Other investigational strategies to target NPM1c, including proteasomal degradation or inhibiting NPM1 oligomerization, are summarized In Table 1 [114,115,116].
4. Conclusions and Future Directions
NPM1 is involved in critical mechanisms that maintain cellular homeostasis. Our knowledge on the aberrant function of NPM1 mutations which lead to a unique leukemia has expanded. The most important next steps are to translate these discoveries in the clinical setting and improve therapy for patients with this illness. NPM1 mutations can be used as a robust biomarker indicating MRD. Therefore, future studies should focus on how to incorporate this biomarker into clinical decision making. The obvious consequence of NPM1 persistence following standard treatment would be the use of allogeneic stem cell transplants to eradicate MRD. However, all targeted therapies under development for NPM1-mutated AML should leverage this disease, indicating characteristics for the monitoring of therapy. There are likely multiple mechanisms leading to leukemia development triggered by NPM1c, but the critical question is which mechanism would be considered a shared therapeutic vulnerability. Ultimately, similar to what is seen with targeted therapies in multiple cancers, a resistance caused by clonal heterogeneity or escape could emerge; therefore, safe combination strategies will be key for achieving cures for all AML patients with a mutated NPM1 gene.
Acknowledgments
G.C.I. received funding through the K12 Paul Calabresi Clinical Scholarship Award (NIH/NCI K12 CA088084).
Author Contributions
J.Z. and G.C.I. designed and wrote the paper; N.J.S. and R.K.-S. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Disclosures
G.C.I. received research funding from Celgene, Syndax, and served on an advisory board for Novartis. N.J.S. received consulting fees from Takeda Oncology and AstraZeneca, research funding from Takeda Oncology and Astellas Pharma Inc., and honoraria from Amgen.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Papaemmanuil E., Gerstung M., Bullinger L., Gaidzik V.I., Paschka P., Roberts N.D., Potter N.E., Heuser M., Thol F., Bolli N., et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A.J.R., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.L., et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dombret H., Gardin C. An update of current treatments for adult acute myeloid leukemia. Blood. 2016;127:53–61. doi: 10.1182/blood-2015-08-604520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Short N.J., Rytting M.E., Cortes J.E. Acute myeloid leukaemia. Lancet. 2018;392:593–606. doi: 10.1016/S0140-6736(18)31041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia. N. Engl. J. Med. 2013;369:98. doi: 10.1056/NEJMx130028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel J.P., Gönen M., Figueroa M.E., Fernandez H., Sun Z., Racevskis J., Van Vlierberghe P., Dolgalev I., Thomas S., Aminova O., et al. Prognostic Relevance of Integrated Genetic Profiling in Acute Myeloid Leukemia. N. Engl. J. Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W., Budhu A., Forgues M., Wang X.W. Temporal and spatial control of nucleophosmin by the Ran-Crm1 complex in centrosome duplication. Nat. Cell Biol. 2005;7:823–830. doi: 10.1038/ncb1282. [DOI] [PubMed] [Google Scholar]
- 8.Yu Y., Maggi L.B., Jr., Brady S.N., Apicelli A.J., Dai M.S., Lu H., Weber J.D. Nucleophosmin is essential for ribosomal protein L5 nuclear export. Mol. Cell. Biol. 2006;26:3798–3809. doi: 10.1128/MCB.26.10.3798-3809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szebeni A., Olson M.O. Nucleolar protein B23 has molecular chaperone activities. Protein Sci. 1999;8:905–912. doi: 10.1110/ps.8.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Federici L., Falini B. Nucleophosmin mutations in acute myeloid leukemia: A tale of protein unfolding and mislocalization. Protein Sci. 2013;22:545–556. doi: 10.1002/pro.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolli N., De Marco M.F., Martelli M.P., Bigerna B., Pucciarini A., Rossi R., Mannucci R., Manes N., Pettirossi V., Pileri S.A., et al. A dose-dependent tug of war involving the NPM1 leukaemic mutant, nucleophosmin, and ARF. Leukemia. 2009;23:501–509. doi: 10.1038/leu.2008.326. [DOI] [PubMed] [Google Scholar]
- 12.Jian Y., Gao Z., Sun J., Shen Q., Feng F., Jing Y., Yang C. RNA aptamers interfering with nucleophosmin oligomerization induce apoptosis of cancer cells. Oncogene. 2009;28:4201–4211. doi: 10.1038/onc.2009.275. [DOI] [PubMed] [Google Scholar]
- 13.Dutta S., Akey I.V., Dingwall C., Hartman K.L., Laue T., Nolte R.T., Head J.F., Akey C.W. The crystal structure of nucleoplasmin-core: Implications for histone binding and nucleosome assembly. Mol. Cell. 2001;8:841–853. doi: 10.1016/S1097-2765(01)00354-9. [DOI] [PubMed] [Google Scholar]
- 14.Dingwall C., Dilworth S.M., Black S.J., Kearsey S.E., Cox L.S., Laskey R.A. Nucleoplasmin cDNA sequence reveals polyglutamic acid tracts and a cluster of sequences homologous to putative nuclear localization signals. EMBO J. 1987;6:69–74. doi: 10.1002/j.1460-2075.1987.tb04720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishimura Y., Ohkubo T., Furuichi Y., Umekawa H. Tryptophans 286 and 288 in the C-terminal region of protein B23.1 are important for its nucleolar localization. Biosci. Biotechnol. Biochem. 2002;66:2239–2242. doi: 10.1271/bbb.66.2239. [DOI] [PubMed] [Google Scholar]
- 16.Mitrea D.M., Grace C.R., Buljan M., Yun M.K., Pytel N.J., Satumba J., Nourse A., Park C.G., Madan Babu M., White S.W., et al. Structural polymorphism in the N-terminal oligomerization domain of NPM1. Proc. Natl. Acad. Sci. USA. 2014;111:4466–4471. doi: 10.1073/pnas.1321007111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okuda M., Horn H.F., Tarapore P., Tokuyama Y., Smulian A.G., Chan P.K., Knudsen E.S., Hofmann I.A., Snyder J.D., Bove K.E., et al. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103:127–140. doi: 10.1016/S0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 18.Okuwaki M. The structure and functions of NPM1/Nucleophsmin/B23, a multifunctional nucleolar acidic protein. J. Biochem. 2008;143:441–448. doi: 10.1093/jb/mvm222. [DOI] [PubMed] [Google Scholar]
- 19.Okuwaki M., Tsujimoto M., Nagata K. The RNA binding activity of a ribosome biogenesis factor, nucleophosmin/B23, is modulated by phosphorylation with a cell cycle-dependent kinase and by association with its subtype. Mol. Biol. Cell. 2002;13:2016–2030. doi: 10.1091/mbc.02-03-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swaminathan V., Kishore A.H., Febitha K.K., Kundu T.K. Human histone chaperone nucleophosmin enhances acetylation-dependent chromatin transcription. Mol. Cell. Biol. 2005;25:7534–7545. doi: 10.1128/MCB.25.17.7534-7545.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shandilya J., Swaminathan V., Gadad S.S., Choudhari R., Kodaganur G.S., Kundu T.K. Acetylated NPM1 localizes in the nucleoplasm and regulates transcriptional activation of genes implicated in oral cancer manifestation. Mol. Cell. Biol. 2009;29:5115–5127. doi: 10.1128/MCB.01969-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heath E.M., Chan S.M., Minden M.D., Murphy T., Shlush L.I., Schimmer A.D. Biological and clinical consequences of NPM1 mutations in AML. Leukemia. 2017;31:798–807. doi: 10.1038/leu.2017.30. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez-Verdun D., Junera H.R. Chapter 4 The nucleolus. In: Bittar E.E., Bittar N., editors. Principles of Medical Biology. Volume 2. Elsevier; Amsterdam, The Netherlands: 1995. pp. 73–92. [Google Scholar]
- 24.Hingorani K., Szebeni A., Olson M.O. Mapping the functional domains of nucleolar protein B23. J. Biol. Chem. 2000;275:24451–24457. doi: 10.1074/jbc.M003278200. [DOI] [PubMed] [Google Scholar]
- 25.Schnittger S., Bacher U., Haferlach C., Alpermann T., Dicker F., Sundermann J., Kern W., Haferlach T. Characterization of NPM1-mutated AML with a history of myelodysplastic syndromes or myeloproliferative neoplasms. Leukemia. 2011;25:615–621. doi: 10.1038/leu.2010.299. [DOI] [PubMed] [Google Scholar]
- 26.Itahana K., Bhat K.P., Jin A., Itahana Y., Hawke D., Kobayashi R., Zhang Y. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell. 2003;12:1151–1164. doi: 10.1016/S1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- 27.Wu M.H., Chang J.H., Chou C.C., Yung B.Y. Involvement of nucleophosmin/B23 in the response of HeLa cells to UV irradiation. Int. J. Cancer. 2002;97:297–305. doi: 10.1002/ijc.1606. [DOI] [PubMed] [Google Scholar]
- 28.Lee S.Y., Park J.H., Kim S., Park E.J., Yun Y., Kwon J. A proteomics approach for the identification of nucleophosmin and heterogeneous nuclear ribonucleoprotein C1/C2 as chromatin-binding proteins in response to DNA double-strand breaks. Biochem J. 2005;388:7–15. doi: 10.1042/BJ20042033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu M.H., Chang J.H., Yung B.Y. Resistance to UV-induced cell-killing in nucleophosmin/B23 over-expressed NIH 3T3 fibroblasts: Enhancement of DNA repair and up-regulation of PCNA in association with nucleophosmin/B23 over-expression. Carcinogenesis. 2002;23:93–100. doi: 10.1093/carcin/23.1.93. [DOI] [PubMed] [Google Scholar]
- 30.Colombo E., Bonetti P., Lazzerini Denchi E., Martinelli P., Zamponi R., Marine J.C., Helin K., Falini B., Pelicci P.G. Nucleophosmin is required for DNA integrity and p19Arf protein stability. Mol. Cell. Biol. 2005;25:8874–8886. doi: 10.1128/MCB.25.20.8874-8886.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grisendi S., Bernardi R., Rossi M., Cheng K., Khandker L., Manova K., Pandolfi P.P. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437:147–153. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 32.Sportoletti P., Grisendi S., Majid S.M., Cheng K., Clohessy J.G., Viale A., Teruya-Feldstein J., Pandolfi P.P. Npm1 is a haploinsufficient suppressor of myeloid and lymphoid malignancies in the mouse. Blood. 2008;111:3859–3862. doi: 10.1182/blood-2007-06-098251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsui K.H., Cheng A.J., Chang P., Pan T.L., Yung B.Y. Association of nucleophosmin/B23 mRNA expression with clinical outcome in patients with bladder carcinoma. Urology. 2004;64:839–844. doi: 10.1016/j.urology.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 34.Grisendi S., Mecucci C., Falini B., Pandolfi P.P. Nucleophosmin and cancer. Nat. Rev. Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 35.Colombo E., Marine J.C., Danovi D., Falini B., Pelicci P.G. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat. Cell Biol. 2002;4:529–533. doi: 10.1038/ncb814. [DOI] [PubMed] [Google Scholar]
- 36.Kurki S., Peltonen K., Latonen L., Kiviharju T.M., Ojala P.M., Meek D., Laiho M. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell. 2004;5:465–475. doi: 10.1016/S1535-6108(04)00110-2. [DOI] [PubMed] [Google Scholar]
- 37.Kuo M.L., den Besten W., Bertwistle D., Roussel M.F., Sherr C.J. N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev. 2004;18:1862–1874. doi: 10.1101/gad.1213904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertwistle D., Sugimoto M., Sherr C.J. Physical and Functional Interactions of the Arf Tumor Suppressor Protein with Nucleophosmin/B23. Mol. Cell. Biol. 2004;24:985–996. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeller K.I., Haggerty T.J., Barrett J.F., Guo Q., Wonsey D.R., Dang C.V. Characterization of nucleophosmin (B23) as a Myc target by scanning chromatin immunoprecipitation. J. Biol. Chem. 2001;276:48285–48291. doi: 10.1074/jbc.M108506200. [DOI] [PubMed] [Google Scholar]
- 40.Boon K., Caron H.N., van Asperen R., Valentijn L., Hermus M.C., van Sluis P., Roobeek I., Weis I., Voûte P.A., Schwab M., et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001;20:1383–1393. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z., Boone D., Hann S.R. Nucleophosmin interacts directly with c-Myc and controls c-Myc-induced hyperproliferation and transformation. Proc. Natl. Acad. Sci. USA. 2008;105:18794–18799. doi: 10.1073/pnas.0806879105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim M.J., Wang X.W. Nucleophosmin and human cancer. Cancer Detect. Prev. 2006;30:481–490. doi: 10.1016/j.cdp.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nozawa Y., Van Belzen N., Van der Made A.C., Dinjens W.N., Bosman F.T. Expression of nucleophosmin/B23 in normal and neoplastic colorectal mucosa. J. Pathol. 1996;178:48–52. doi: 10.1002/(SICI)1096-9896(199601)178:1<48::AID-PATH432>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 44.Shields L.B., Gercel-Taylor C., Yashar C.M., Wan T.C., Katsanis W.A., Spinnato J.A., Taylor D.D. Induction of immune responses to ovarian tumor antigens by multiparity. J. Soc. Gynecol. Investig. 1997;4:298–304. doi: 10.1016/S1071-5576(97)00068-3. [DOI] [PubMed] [Google Scholar]
- 45.Subong E.N., Shue M.J., Epstein J.I., Briggman J.V., Chan P.K., Partin A.W. Monoclonal antibody to prostate cancer nuclear matrix protein (PRO:4-216) recognizes nucleophosmin/B23. Prostate. 1999;39:298–304. doi: 10.1002/(SICI)1097-0045(19990601)39:4<298::AID-PROS11>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 46.Skaar T.C., Prasad S.C., Sharareh S., Lippman M.E., Brunner N., Clarke R. Two-dimensional gel electrophoresis analyses identify nucleophosmin as an estrogen regulated protein associated with acquired estrogen-independence in human breast cancer cells. J. Steroid Biochem. Mol. Biol. 1998;67:391–402. doi: 10.1016/S0960-0760(98)00142-3. [DOI] [PubMed] [Google Scholar]
- 47.Redner R.L., Rush E.A., Faas S., Rudert W.A., Corey S.J. The t(5;17) variant of acute promyelocytic leukemia expresses a nucleophosmin-retinoic acid receptor fusion. Blood. 1996;87:882–886. doi: 10.1182/blood.V87.3.882.bloodjournal873882. [DOI] [PubMed] [Google Scholar]
- 48.Morris S.W., Kirstein M.N., Valentine M.B., Dittmer K.G., Shapiro D.N., Saltman D.L., Look A.T. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 49.Raimondi S.C., Dube I.D., Valentine M.B., Mirro J., Jr., Watt H.J., Larson R.A., Bitter M.A., Le Beau M.M., Rowley J.D. Clinicopathologic manifestations and breakpoints of the t(3;5) in patients with acute nonlymphocytic leukemia. Leukemia. 1989;3:42–47. [PubMed] [Google Scholar]
- 50.Falini B., Mecucci C., Tiacci E., Alcalay M., Rosati R., Pasqualucci L., La Starza R., Diverio D., Colombo E., Santucci A., et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki T., Kiyoi H., Ozeki K., Tomita A., Yamaji S., Suzuki R., Kodera Y., Miyawaki S., Asou N., Kuriyama K., et al. Clinical characteristics and prognostic implications of NPM1 mutations in acute myeloid leukemia. Blood. 2005;106:2854–2861. doi: 10.1182/blood-2005-04-1733. [DOI] [PubMed] [Google Scholar]
- 52.Dohner K., Schlenk R.F., Habdank M., Scholl C., Rucker F.G., Corbacioglu A., Bullinger L., Frohling S., Dohner H. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: Interaction with other gene mutations. Blood. 2005;106:3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 53.Schnittger S., Schoch C., Kern W., Mecucci C., Tschulik C., Martelli M.F., Haferlach T., Hiddemann W., Falini B. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106:3733–3739. doi: 10.1182/blood-2005-06-2248. [DOI] [PubMed] [Google Scholar]
- 54.Thiede C., Koch S., Creutzig E., Steudel C., Illmer T., Schaich M., Ehninger G., Leukämie F.T.D.S. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML) Blood. 2006;107:4011–4020. doi: 10.1182/blood-2005-08-3167. [DOI] [PubMed] [Google Scholar]
- 55.Verhaak R.G.W., Goudswaard C.S., van Putten W., Bijl M.A., Sanders M.A., Hugens W., Uitterlinden A.G., Erpelinck C.A.J., Delwel R., Löwenberg B., et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): Association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106:3747–3754. doi: 10.1182/blood-2005-05-2168. [DOI] [PubMed] [Google Scholar]
- 56.Borrow J., Dyer S.A., Akiki S., Griffiths M.J. Molecular roulette: Nucleophosmin mutations in AML are orchestrated through N-nucleotide addition by TdT. Blood. 2019;134:2291–2303. doi: 10.1182/blood.2019001240. [DOI] [PubMed] [Google Scholar]
- 57.Vassiliou G.S. The curious incident of TdT-mediated mutations in AML. Blood. 2019;134:2229–2231. doi: 10.1182/blood.2019003619. [DOI] [PubMed] [Google Scholar]
- 58.Borrow J., Dyer S.A., Akiki S., Griffiths M.J. Terminal deoxynucleotidyl transferase promotes acute myeloid leukemia by priming FLT3-ITD replication slippage. Blood. 2019;134:2281–2290. doi: 10.1182/blood.2019001238. [DOI] [PubMed] [Google Scholar]
- 59.Chou W.C., Tang J.L., Lin L.I., Yao M., Tsay W., Chen C.Y., Wu S.J., Huang C.F., Chiou R.J., Tseng M.H., et al. Nucleophosmin Mutations in De novo Acute Myeloid Leukemia: The Age-Dependent Incidences and the Stability during Disease Evolution. Cancer Res. 2006;66:3310–3316. doi: 10.1158/0008-5472.CAN-05-4316. [DOI] [PubMed] [Google Scholar]
- 60.Vassiliou G.S., Cooper J.L., Rad R., Li J., Rice S., Uren A., Rad L., Ellis P., Andrews R., Banerjee R., et al. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat. Genet. 2011;43:470–475. doi: 10.1038/ng.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dovey O.M., Cooper J.L., Mupo A., Grove C.S., Lynn C., Conte N., Andrews R.M., Pacharne S., Tzelepis K., Vijayabaskar M.S., et al. Molecular synergy underlies the co-occurrence patterns and phenotype of NPM1-mutant acute myeloid leukemia. Blood. 2017;130:1911–1922. doi: 10.1182/blood-2017-01-760595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shlush L.I., Zandi S., Mitchell A., Chen W.C., Brandwein J.M., Gupta V., Kennedy J.A., Schimmer A.D., Schuh A.C., Yee K.W., et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Corces-Zimmerman M.R., Hong W.J., Weissman I.L., Medeiros B.C., Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc. Natl. Acad. Sci. USA. 2014;111:2548–2553. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Genovese G., Kahler A.K., Handsaker R.E., Lindberg J., Rose S.A., Bakhoum S.F., Chambert K., Mick E., Neale B.M., Fromer M., et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xie M., Lu C., Wang J., McLellan M.D., Johnson K.J., Wendl M.C., McMichael J.F., Schmidt H.K., Yellapantula V., Miller C.A., et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKerrell T., Park N., Moreno T., Grove C.S., Ponstingl H., Stephens J., Crawley C., Craig J., Scott M.A., Hodkinson C., et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 2015;10:1239–1245. doi: 10.1016/j.celrep.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Steensma D.P., Bejar R., Jaiswal S., Lindsley R.C., Sekeres M.A., Hasserjian R.P., Ebert B.L. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P.V., Mar B.G., Lindsley R.C., Mermel C.H., Burtt N., Chavez A., et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martelli M.P., Manes N., Pettirossi V., Liso A., Pacini R., Mannucci R., Zei T., Bolli N., di Raimondo F., Specchia G., et al. Absence of nucleophosmin leukaemic mutants in B and T cells from AML with NPM1 mutations: Implications for the cell of origin of NPMc+ AML. Leukemia. 2008;22:195–198. doi: 10.1038/sj.leu.2404857. [DOI] [PubMed] [Google Scholar]
- 70.Metzeler K.H., Herold T., Rothenberg-Thurley M., Amler S., Sauerland M.C., Görlich D., Schneider S., Konstandin N.P., Dufour A., Bräundl K., et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood. 2016;128:686–698. doi: 10.1182/blood-2016-01-693879. [DOI] [PubMed] [Google Scholar]
- 71.Haferlach C., Mecucci C., Schnittger S., Kohlmann A., Mancini M., Cuneo A., Testoni N., Rege-Cambrin G., Santucci A., Vignetti M., et al. AML with mutated NPM1 carrying a normal or aberrant karyotype show overlapping biologic, pathologic, immunophenotypic, and prognostic features. Blood. 2009;114:3024–3032. doi: 10.1182/blood-2009-01-197871. [DOI] [PubMed] [Google Scholar]
- 72.Micol J.B., Boissel N., Renneville A., Castaigne S., Gardin C., Preudhomme C., Dombret H. The role of cytogenetic abnormalities in acute myeloid leukemia with NPM1 mutations and no FLT3 internal tandem duplication. Blood. 2009;114:4601–4602. doi: 10.1182/blood-2009-07-230995. [DOI] [PubMed] [Google Scholar]
- 73.Angenendt L., Röllig C., Montesinos P., Martínez-Cuadrón D., Barragan E., García R., Botella C., Martínez P., Ravandi F., Kadia T., et al. Chromosomal Abnormalities and Prognosis in NPM1-Mutated Acute Myeloid Leukemia: A Pooled Analysis of Individual Patient Data From Nine International Cohorts. J. Clin. Oncol. 2019;37:2632–2642. doi: 10.1200/JCO.19.00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Colombo E., Martinelli P., Zamponi R., Shing D.C., Bonetti P., Luzi L., Volorio S., Bernard L., Pruneri G., Alcalay M., et al. Delocalization and Destabilization of the Arf Tumor Suppressor by the Leukemia-Associated NPM Mutant. Cancer Res. 2006;66:3044–3050. doi: 10.1158/0008-5472.CAN-05-2378. [DOI] [PubMed] [Google Scholar]
- 75.Besten W.D., Kuo M.L., Williams R.T., Sherr C.J. Myeloid Leukemia-Associated Nucleophosmin Mutants Perturb p53-Dependent and Independent Activities of the Arf Tumor Suppressor Protein. Cell Cycle. 2005;4:1593–1598. doi: 10.4161/cc.4.11.2174. [DOI] [PubMed] [Google Scholar]
- 76.Leong S.M., Tan B.X., Bte Ahmad B., Yan T., Chee L.Y., Ang S.T., Tay K.G., Koh L.P., Yeoh A.E.J., Koay E.S.C., et al. Mutant nucleophosmin deregulates cell death and myeloid differentiation through excessive caspase-6 and -8 inhibition. Blood. 2010;116:3286–3296. doi: 10.1182/blood-2009-12-256149. [DOI] [PubMed] [Google Scholar]
- 77.Gao H., Jin S., Song Y., Fu M., Wang M., Liu Z., Wu M., Zhan Q. B23 regulates GADD45a nuclear translocation and contributes to GADD45a-induced cell cycle G2-M arrest. J. Biol. Chem. 2005;280:10988–10996. doi: 10.1074/jbc.M412720200. [DOI] [PubMed] [Google Scholar]
- 78.Wang X.W., Zhan Q., Coursen J.D., Khan M.A., Kontny H.U., Yu L., Hollander M.C., O’Connor P.M., Fornace A.J., Harris C.C. GADD45 induction of a G2/M cell cycle checkpoint. Proc. Natl. Acad. Sci. USA. 1999;96:3706–3711. doi: 10.1073/pnas.96.7.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bonetti P., Davoli T., Sironi C., Amati B., Pelicci P.G., Colombo E. Nucleophosmin and its AML-associated mutant regulate c-Myc turnover through Fbw7γ. J. Cell Biol. 2008;182:19–26. doi: 10.1083/jcb.200711040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ohanian M., Rozovski U., Kanagal-Shamanna R., Abruzzo L.V., Loghavi S., Kadia T., Futreal A., Bhalla K., Zuo Z., Huh Y.O., et al. MYC protein expression is an important prognostic factor in acute myeloid leukemia. Leuk. Lymphoma. 2019;60:37–48. doi: 10.1080/10428194.2018.1464158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alcalay M., Tiacci E., Bergomas R., Bigerna B., Venturini E., Minardi S.P., Meani N., Diverio D., Bernard L., Tizzoni L., et al. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood. 2005;106:899–902. doi: 10.1182/blood-2005-02-0560. [DOI] [PubMed] [Google Scholar]
- 82.Brunetti L., Gundry M.C., Sorcini D., Guzman A.G., Huang Y.H., Ramabadran R., Gionfriddo I., Mezzasoma F., Milano F., Nabet B., et al. Mutant NPM1 Maintains the Leukemic State through HOX Expression. Cancer Cell. 2018;34:499–512. doi: 10.1016/j.ccell.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Armstrong S.A., Staunton J.E., Silverman L.B., Pieters R., den Boer M.L., Minden M.D., Sallan S.E., Lander E.S., Golub T.R., Korsmeyer S.J. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 84.Yokoyama A., Somervaille T.C., Smith K.S., Rozenblatt-Rosen O., Meyerson M., Cleary M.L. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 85.Kuhn M.W., Song E., Feng Z., Sinha A., Chen C.W., Deshpande A.J., Cusan M., Farnoud N., Mupo A., Grove C., et al. Targeting Chromatin Regulators Inhibits Leukemogenic Gene Expression in NPM1 Mutant Leukemia. Cancer Discov. 2016;6:1166–1181. doi: 10.1158/2159-8290.CD-16-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Uckelmann H.J., Kim S.M., Wong E.M., Hatton C., Giovinazzo H., Gadrey J.Y., Krivtsov A.V., Rucker F.G., Dohner K., McGeehan G.M., et al. Therapeutic targeting of preleukemia cells in a mouse model of NPM1 mutant acute myeloid leukemia. Science. 2020;367:586–590. doi: 10.1126/science.aax5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arber D.A., Orazi A., Hasserjian R., Thiele J., Borowitz M.J., Le Beau M.M., Bloomfield C.D., Cazzola M., Vardiman J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 88.Gale R.E., Green C., Allen C., Mead A.J., Burnett A.K., Hills R.K., Linch D.C. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood. 2008;111:2776–2784. doi: 10.1182/blood-2007-08-109090. [DOI] [PubMed] [Google Scholar]
- 89.Boissel N., Renneville A., Biggio V., Philippe N., Thomas X., Cayuela J.M., Terre C., Tigaud I., Castaigne S., Raffoux E., et al. Prevalence, clinical profile, and prognosis of NPM mutations in AML with normal karyotype. Blood. 2005;106:3618–3620. doi: 10.1182/blood-2005-05-2174. [DOI] [PubMed] [Google Scholar]
- 90.Schlenk R.F., Dohner K., Krauter J., Frohling S., Corbacioglu A., Bullinger L., Habdank M., Spath D., Morgan M., Benner A., et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N. Engl. J. Med. 2008;358:1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 91.Döhner H., Estey E., Grimwade D., Amadori S., Appelbaum F.R., Büchner T., Dombret H., Ebert B.L., Fenaux P., Larson R.A., et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meleveedu K.S., Sidiqi M.H., Nadiminti K., Hefazi M., Alkhateeb H.B., Shah M.V., Phelps A., He R., Viswanatha D.S., Dingli D., et al. Impact of Allogeneic Stem Cell Transplant on Outcomes of Patients with Acute Myeloid Leukemia Based on NPM1 and FLT3 Mutational Status. Biol. Blood Marrow Transplant. 2019;25:S113–S114. doi: 10.1016/j.bbmt.2018.12.390. [DOI] [Google Scholar]
- 93.Hemmati P., Terwey T.H., Vuong L.G., le Coutre P.D., Dörken B., Arnold R. Allogeneic Stem Cell Transplantation For Cytogenetically Normal Acute Myeloid Leukemia: Impact Of FLT3 and NPM1 Mutational Status. Blood. 2013;122:2104. doi: 10.1182/blood.V122.21.2104.2104. [DOI] [Google Scholar]
- 94.Short N.J., Ravandi F. How close are we to incorporating measurable residual disease into clinical practice for acute myeloid leukemia? Haematologica. 2019;104:1532–1541. doi: 10.3324/haematol.2018.208454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ivey A., Hills R.K., Simpson M.A., Jovanovic J.V., Gilkes A., Grech A., Patel Y., Bhudia N., Farah H., Mason J., et al. Assessment of Minimal Residual Disease in Standard-Risk AML. N. Engl. J. Med. 2016;374:422–433. doi: 10.1056/NEJMoa1507471. [DOI] [PubMed] [Google Scholar]
- 96.Grimwade D., Freeman S.D. Defining minimal residual disease in acute myeloid leukemia: Which platforms are ready for “prime time”? Blood. 2014;124:3345–3355. doi: 10.1182/blood-2014-05-577593. [DOI] [PubMed] [Google Scholar]
- 97.Martelli M.P., Gionfriddo I., Mezzasoma F., Milano F., Pierangeli S., Mulas F., Pacini R., Tabarrini A., Pettirossi V., Rossi R., et al. Arsenic trioxide and all-trans retinoic acid target NPM1 mutant oncoprotein levels and induce apoptosis in NPM1-mutated AML cells. Blood. 2015;125:3455–3465. doi: 10.1182/blood-2014-11-611459. [DOI] [PubMed] [Google Scholar]
- 98.El Hajj H., Dassouki Z., Berthier C., Raffoux E., Ades L., Legrand O., Hleihel R., Sahin U., Tawil N., Salameh A., et al. Retinoic acid and arsenic trioxide trigger degradation of mutated NPM1, resulting in apoptosis of AML cells. Blood. 2015;125:3447–3454. doi: 10.1182/blood-2014-11-612416. [DOI] [PubMed] [Google Scholar]
- 99.Lambert J., Lambert J., Nibourel O., Pautas C., Hayette S., Cayuela J.M., Terre C., Rousselot P., Dombret H., Chevret S., et al. MRD assessed by WT1 and NPM1 transcript levels identifies distinct outcomes in AML patients and is influenced by gemtuzumab ozogamicin. Oncotarget. 2014;5:6280–6288. doi: 10.18632/oncotarget.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Olombel G., Guerin E., Guy J., Perrot J.Y., Dumezy F., de Labarthe A., Bastie J.N., Legrand O., Raffoux E., Plesa A., et al. The level of blast CD33 expression positively impacts the effect of gemtuzumab ozogamicin in patients with acute myeloid leukemia. Blood. 2016;127:2157–2160. doi: 10.1182/blood-2016-01-689976. [DOI] [PubMed] [Google Scholar]
- 101.DiNardo C.D., Pratz K., Pullarkat V., Jonas B.A., Arellano M., Becker P.S., Frankfurt O., Konopleva M., Wei A.H., Kantarjian H.M., et al. Venetoclax combined with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood. 2019;133:7–17. doi: 10.1182/blood-2018-08-868752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wei A.H., Strickland S.A., Hou J.Z., Fiedler W., Lin T.L., Walter R.B., Enjeti A., Tiong I.S., Savona M., Lee S., et al. Venetoclax Combined With Low-Dose Cytarabine for Previously Untreated Patients With Acute Myeloid Leukemia: Results From a Phase Ib/II Study. J. Clin. Oncol. 2019;37:1277–1284. doi: 10.1200/JCO.18.01600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lachowiez C.A., Loghavi S., Kadia T.M., Daver N., Borthakur G., Pemmaraju N., Naqvi K., Alvarado Y., Yilmaz M., Short N., et al. Outcomes of older patients with NPM1-mutated AML: Current treatments and the promise of venetoclax-based regimens. Blood Adv. 2020;4:1311–1320. doi: 10.1182/bloodadvances.2019001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Falini B., Brunetti L., Martelli M.P. Dactinomycin in NPM1-Mutated Acute Myeloid Leukemia. N. Engl. J. Med. 2015;373:1180–1182. doi: 10.1056/NEJMc1509584. [DOI] [PubMed] [Google Scholar]
- 105.Garzon R., Savona M., Baz R., Andreeff M., Gabrail N., Gutierrez M., Savoie L., Mau-Sorensen P.M., Wagner-Johnston N., Yee K., et al. A phase 1 clinical trial of single-agent selinexor in acute myeloid leukemia. Blood. 2017;129:3165–3174. doi: 10.1182/blood-2016-11-750158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Borkin D., He S., Miao H., Kempinska K., Pollock J., Chase J., Purohit T., Malik B., Zhao T., Wang J., et al. Pharmacologic Inhibition of the Menin-MLL Interaction Blocks Progression of MLL Leukemia In Vivo. Cancer Cell. 2015;27:589–602. doi: 10.1016/j.ccell.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Grembecka J., He S., Shi A., Purohit T., Muntean A.G., Sorenson R.J., Showalter H.D., Murai M.J., Belcher A.M., Hartley T., et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat. Chem. Biol. 2012;8:277–284. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Krivtsov A.V., Evans K., Gadrey J.Y., Eschle B.K., Hatton C., Uckelmann H.J., Ross K.N., Perner F., Olsen S.N., Pritchard T., et al. A Menin-MLL Inhibitor Induces Specific Chromatin Changes and Eradicates Disease in Models of MLL-Rearranged Leukemia. Cancer Cell. 2019;36:660–673. doi: 10.1016/j.ccell.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Klossowski S., Miao H., Kempinska K., Wu T., Purohit T., Kim E., Linhares B.M., Chen D., Jih G., Perkey E., et al. Menin inhibitor MI-3454 induces remission in MLL1-rearranged and NPM1-mutated models of leukemia. J. Clin. Investig. 2020;130:981–997. doi: 10.1172/JCI129126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Blankenstein T., Leisegang M., Uckert W., Schreiber H. Targeting cancer-specific mutations by T cell receptor gene therapy. Curr. Opin. Immunol. 2015;33:112–119. doi: 10.1016/j.coi.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schumacher T.N., Schreiber R.D. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 112.Tran E., Robbins P.F., Rosenberg S.A. ‘Final common pathway’ of human cancer immunotherapy: Targeting random somatic mutations. Nat. Immunol. 2017;18:255–262. doi: 10.1038/ni.3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Van der Lee D.I., Reijmers R.M., Honders M.W., Hagedoorn R.S., de Jong R.C., Kester M.G., van der Steen D.M., de Ru A.H., Kweekel C., Bijen H.M., et al. Mutated nucleophosmin 1 as immunotherapy target in acute myeloid leukemia. J. Clin. Investig. 2019;129:774–785. doi: 10.1172/JCI97482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nabbouh A.I., Hleihel R.S., Saliba J.L., Karam M.M., Hamie M.H., Wu H.J.M., Berthier C.P., Tawil N.M., Bonnet P.A., Deleuze-Masquefa C., et al. Imidazoquinoxaline derivative EAPB0503: A promising drug targeting mutant nucleophosmin 1 in acute myeloid leukemia. Cancer. 2017;123:1662–1673. doi: 10.1002/cncr.30515. [DOI] [PubMed] [Google Scholar]
- 115.Balusu R., Fiskus W., Rao R., Chong D.G., Nalluri S., Mudunuru U., Ma H., Chen L., Venkannagari S., Ha K., et al. Targeting levels or oligomerization of nucleophosmin 1 induces differentiation and loss of survival of human AML cells with mutant NPM1. Blood. 2011;118:3096–3106. doi: 10.1182/blood-2010-09-309674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yi S., Wen L., He J., Wang Y., Zhao F., Zhao J., Zhao Z., Cui G., Chen Y. Deguelin, a selective silencer of the NPM1 mutant, potentiates apoptosis and induces differentiation in AML cells carrying the NPM1 mutation. Ann. Hematol. 2015;94:201–210. doi: 10.1007/s00277-014-2206-x. [DOI] [PubMed] [Google Scholar]