Abstract
Prion diseases are caused by misfolded prion protein (PrPSc) and are accompanied by spongiform vacuolation of brain lesions. Approximately three centuries have passed since prion diseases were first discovered around the world; however, the exact role of certain factors affecting the causative agent of prion diseases is still debatable. In recent studies, somatic mutations were assumed to be cause of several diseases. Thus, we postulated that genetically unstable cancer tissue may cause somatic mutations in the prion protein gene (PRNP), which could trigger the onset of prion diseases. To identify somatic mutations in the PRNP gene in cancer tissues, we analyzed somatic mutations in the PRNP gene in cancer patients using the Cancer Genome Atlas (TCGA) database. In addition, to evaluate whether the somatic mutations in the PRNP gene in cancer patients had a damaging effect, we performed in silico analysis using PolyPhen-2, PANTHER, PROVEAN, and AMYCO. We identified a total of 48 somatic mutations in the PRNP gene, including 8 somatic mutations that are known pathogenic mutations of prion diseases. We identified significantly different distributions among the types of cancer, the mutation counts, and the ages of diagnosis between the total cancer patient population and cancer patients carrying somatic mutations in the PRNP gene. Strikingly, although invasive breast carcinoma and glioblastoma accounted for a high percentage of the total cancer patient population (9.9% and 5.4%, respectively), somatic mutations in the PRNP gene have not been identified in these two cancer types. We suggested the possibility that somatic mutations of the PRNP gene in glioblastoma can be masked by a diagnosis of prion disease. In addition, we found four aggregation-prone somatic mutations, these being L125F, E146Q, R151C, and K204N. To the best of our knowledge, this is the first specific analysis of the somatic mutations in the PRNP gene in cancer patients.
Keywords: cancer, prion, somatic mutation, aggregation, CJD, FFI, GSS, p53
1. Introduction
Prion diseases are fatal and irreversible neurodegenerative diseases caused by a deleterious form of the prion protein (PrPSc) derived from normal prion protein (PrPC) [1,2]. These diseases are accompanied by spongiform generation and astrocytosis in brain lesions. Prion diseases are the only zoonotic dementia with such a large host ranges, including scrapie in sheep and goats, bovine spongiform encephalopathy (BSE) in cattle, chronic wasting disease (CWD) in elk and deer, and Creutzfeldt–Jakob disease (CJD), fatal familial insomnia (FFI), and Gerstmann–Sträussler–Scheinker syndrome (GSS) in humans. In CJD, there are three representative types, including sporadic CJD, familial CJD, and iatrogenic CJD [3,4,5,6,7,8,9,10,11,12].
It has been approximately three centuries since scrapie in sheep was first discovered in 1732 [5]. However, the exact roles on certain factors influencing the causative agent of the disease have not been fully explained thus far. However, in recent studies, genetic variations including somatic mutations or postzygotic mutations have been shown to cause several diseases, including neurofibromatosis, McCune–Albright disease, paroxysmal nocturnal hemoglobinuria, incontinentia pigmenti, several types of cancer, and Alzheimer’s disease [13,14,15]. In human prion diseases, point mutations in the prion protein gene (PRNP), which encodes PrP, induce familial forms of human prion diseases, including familial CJD, GSS, and FFI [6,16,17]. In addition, spontaneous de novo mutations in the PRNP gene were detected in several types of prion diseases, including sporadic CJD and GSS [18,19,20,21]. In this regard, somatic mutations in the PRNP gene can be assumed to be a cause of prion diseases. In addition, previous studies have been reported that cancer tissues showed more genetic instability and elevated expression of the PRNP gene. Thus, we postulated that cancer tissue may induce somatic mutations in the PRNP gene, which can trigger the onset of prion diseases [22,23,24,25].
To find somatic mutations in the PRNP gene in cancer tissues, we searched the information on somatic mutations in the PRNP gene in cancer patients using the Cancer Genome Atlas (TCGA) database [26]. In addition, we compared the information on the somatic mutations in the PRNP gene in cancer patients with that of previously reported pathogenic mutations associated with prion diseases and with the total cancer patient population. Furthermore, to assess whether the somatic mutations in the PRNP gene in cancer patients are deleterious, we performed in silico annotation using PolyPhen-2, PANTHER, PROVEAN, and AMYCO [27,28,29,30,31,32].
2. Results
2.1. Previously Reported Prion Disease-Related Mutations
Genetic forms of human prion diseases include CJD, FFI, and GSS. Previous pathogenic mutations involved in genetic forms of prion diseases are described in Table 1. In detail, genetic CJD has been reported to carry G114V, D178N-129V, V180I, T183A, T188K, E196K, E196A, E200K, E200G, V203I, R208H, V210I, E211Q, I215V, M232R, and P238S mutations; double octapeptide deletions; and octapeptide insertions in the PRNP gene. In addition, FFI has been reported to carry D178N-129M mutations in the PRNP gene. Finally, GSS has been reported to carry P102L, P105L, P105T, P105S, A117V, G131V, Y145*, Q160*, V176G, H187R, F198S, D202N, Q212P, Q217R, Y226*, Q227*, and M232T mutations in the PRNP gene. Unique phenotypes of dementia have been reported to carry S17G, P39L, Y163*, D167N, D187fs, and R208C mutations in the PRNP gene.
Table 1.
Classification of pathogenic mutations of prion disease according to clinical phenotypes in previous studies.
| Clinical Phenotypes | Mutations |
|---|---|
| Creutzfeldt–Jakob Disease (CJD) | G114V, D178N-129V, V180I, T183A, T188K, E196K, E196A, E200K, E200G, V203I, R208H, V210I, E211Q, I215V, M232R, P238S, double octapeptide deletion, and octapeptide insertions |
| Fatal familial insomnia (FFI) | D178N-129M |
| Gerstmann–Sträussler–Scheinker syndrome (GSS) | P102L, P105L, P105T, P105S, A117V, G131V, Y145*, Q160*, V176G, H187R, F198S, D202N, Q212P, Q217R, Y226*, Q227*, and M232T |
| Others | S17G, P39L, Y163*, D167N, D187fs, and R208C |
Others: includes unique phenotypes of dementia. *: stop codon. fs: frame shift.
2.2. 48 Somatic Mutations in the PRNP Gene in Cancer Patients
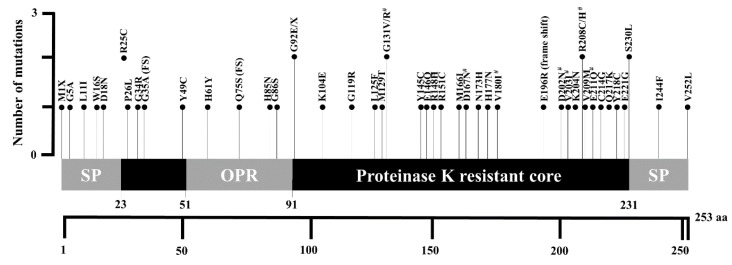
Using cBioPortal, we identified a total of 48 somatic mutations in the PRNP gene in 10,953 cancer patients. The detailed information is described in Table 2. Among the 48 mutations, 8 mutations, these being G131V, D167N, V180I, D202N, V203I, R208C, R208H, and E211Q (shaded boxes), were previously reported pathogenic mutations of prion diseases (Table 1). The locations of the identified somatic mutations are visualized in Figure 1. A total of five somatic mutations, these being M1X, G5A, L11I, W16S, and D18N, were located in the N-terminal signal peptide region. A total of five somatic mutations, including R25C, P26L, G34R, and G35A (fs), were located in the region of codons 23–51. A total of four somatic mutations, these being H61Y, Q75S (fs), H85N, and G86S, were located in an octapeptide repeat region. A total of 30 somatic mutations, these being G92E, G92X, K104E, G119R, L125F, M129T, G131V, G131R, Y145C, E146Q, R148H, R151C, M166I, D167N, N173H, H177N, V180I, E196R (FS), D202N, V203I, K204N, R208C, R208H, V209M, E211Q, C214G, Q217K, Y218C, E221G, and S230L, were located in the proteinase K resistant core region. Finally, two somatic mutations, I244F and V252L, were located in the C-terminal signal peptide region. Notably, of the 25 patients for whom we had access to cancer metastasis information, we observed no evidence of distant metastasis in 22 (92%) of them (Table 2).
Table 2.
Detailed information on the 48 somatic mutations in cancer patients.
| Sample ID | Sex | Age | Cancer Type | AJCCMSC | AJCCTSC | Mutation | Mutation Type | Allele Frequency | # Mut |
|---|---|---|---|---|---|---|---|---|---|
| TCGA-4Z-AA7Q-01 | M | 79 | Bladder urothelial carcinoma | M0 | T3A | E146Q | Missense | 0.19 | 222 |
| TCGA-5N-A9KM-01 | F | 73 | Bladder urothelial carcinoma | MX | T4A | M1? | Nonstart | 0.36 | 189 |
| TCGA-IR-A3LK-01 | F | 69 | Cervical squamous cell carcinoma | M0 | T1B2 | G131V | Missense | 0.14 | 1189 |
| TCGA-KN-8428-01 | M | 71 | Chromophobe renal cell carcinoma | NA | T2 | V203I | Missense | 0.20 | 673 |
| TCGA-CK-5916-01 | F | 71 | Colon adenocarcinoma | M0 | T1 | V180I | Missense | 0.25 | 1407 |
| TCGA-EE-A29V-06 | M | 85 | Cutaneous melanoma | M0 | T3B | G92E | Missense | 0.42 | 1012 |
| TCGA-EB-A41A-01 | M | 90 | Cutaneous melanoma | M0 | T4B | L125F | Missense | 0.44 | 1533 |
| TCGA-D3-A2JC-06 | F | 53 | Cutaneous melanoma | M0 | T0 | M166I | Missense | 0.11 | 3148 |
| TCGA-D3-A8GM-06 | M | 73 | Cutaneous melanoma | M0 | T3B | G86S | Missense | 0.09 | 3288 |
| TCGA-YD-A9TA-06 | M | 75 | Cutaneous melanoma | NA | NA | H61Y | Missense | 0.29 | 2782 |
| TCGA-L5-A43J-01 | M | 90 | Esophageal squamous cell carcinoma | MX | T3 | R25C | Missense | 0.20 | 700 |
| TCGA-F7-A624-01 | M | 73 | Head and neck squamous cell carcinoma | M0 | T2 | R208H | Missense | 0.16 | 2744 |
| TCGA-VQ-A91D-01 | M | 70 | Intestinal type stomach adenocarcinoma | M0 | T4B | R208C | Missense | 0.25 | 2140 |
| TCGA-67-3771-01 | F | 77 | Lung adenocarcinoma | M0 | T1 | N173H | Missense | 0.12 | 951 |
| TCGA-44-5644-01 | F | 51 | Lung adenocarcinoma | NA | T2A | G5A | Missense | 0.34 | 908 |
| TCGA-69-A59K-01 | F | 60 | Lung adenocarcinoma | M0 | T3 | G119R | Missense | 0.21 | 438 |
| TCGA-17-Z023-01 | NA | NA | Lung adenocarcinoma | NA | NA | Y49C | Missense | 0.23 | 362 |
| TCGA-17-Z026-01 | NA | NA | Lung adenocarcinoma | NA | NA | E211Q | Missense | 0.19 | 873 |
| TCGA-22-5489-01 | M | 64 | Lung squamous cell carcinoma | M0 | T1B | Y145C | Missense | 0.21 | 227 |
| TCGA-63-A5MM-01 | F | 69 | Lung squamous cell carcinoma | M0 | T2 | H177N | Missense | 0.23 | 1070 |
| TCGA-NC-A5HH-01 | M | 53 | Lung squamous cell carcinoma | M0 | T1 | W16S | Missense | 0.27 | 314 |
| TCGA-G7-6790-01 | M | 57 | Papillary renal cell carcinoma | MX | T1A | Q217K | Missense | 0.09 | 71 |
| TCGA-F9-A97G-01 | M | 79 | Papillary renal cell carcinoma | M0 | T3 | V252L | Missense | 0.20 | 62 |
| TCGA-AG-A002-01 | M | 35 | Rectal adenocarcinoma | M0 | T2 | P26L | Missense | 0.45 | 11,438 |
| TCGA-B0-5713-01 | F | 75 | Renal clear cell carcinoma | M0 | T3B | E221G | Missense | 0.30 | 97 |
| TCGA-25-1313-01 | F | 62 | Serous ovarian cancer | NA | NA | G131R | Missense | 0.37 | 179 |
| TCGA-VQ-A8PO-01 | M | 74 | Signet ring cell carcinoma of the stomach | M0 | T4A | M129T | Missense | 0.23 | 751 |
| TCGA-BR-6452-01 | F | 78 | Stomach adenocarcinoma | M0 | T3 | I244F | Missense | 0.19 | 5050 |
| TCGA-CG-5717-01 | M | 58 | Stomach adenocarcinoma | M0 | T2B | R151C | Missense | 0.34 | 117 |
| TCGA-CG-5723-01 | M | 83 | Stomach adenocarcinoma | M0 | T2 | V209M | Missense | 0.08 | 1606 |
| TCGA-VQ-A8PP-01 | M | 76 | Tubular stomach adenocarcinoma | M0 | T4 | K104E | Missense | 0.36 | 1328 |
| TCGA-DX-AB2Z-01 | F | 87 | Undifferentiated pleomorphic sarcoma | NA | NA | R148H | Missense | 0.11 | 69 |
| TCGA-N7-A4Y0-01 | F | 65 | Uterine carcinosarcoma | NA | NA | C214G | Missense | 0.64 | 709 |
| TCGA-ND-A4WC-01 | NA | NA | Uterine carcinosarcoma | NA | NA | H85N | Missense | 0.20 | 3669 |
| TCGA-B5-A0JY-01 | F | 50 | Uterine endometrioid carcinoma | NA | NA | K204N | Missense | 0.34 | 9713 |
| TCGA-D1-A17Q-01 | F | 54 | Uterine endometrioid carcinoma | NA | NA | D167N | Missense | 0.40 | 5945 |
| TCGA-AP-A1DV-01 | F | 59 | Uterine endometrioid carcinoma | NA | NA | R25C | Missense | 0.52 | 12,071 |
| TCGA-FI-A2D5-01 | F | 56 | Uterine endometrioid carcinoma | NA | NA | D202N | Missense | 0.38 | 13,874 |
| TCGA-AJ-A3EL-01 | F | 47 | Uterine endometrioid carcinoma | NA | NA | L11I | Missense | 0.40 | 7391 |
| TCGA-AP-A0LT-01 | F | 57 | Uterine endometrioid carcinoma | NA | NA | E196Rfs*10 | Frame shift del | 0.28 | 638 |
| TCGA-AP-A1DV-01 | F | 59 | Uterine endometrioid carcinoma | NA | NA | Y218C | Missense | 0.42 | 12,071 |
| TCGA-AX-A3FT-01 | F | 64 | Uterine endometrioid carcinoma | NA | NA | Q75Sfs*35 | Frame shift del | 0.23 | 1281 |
| TCGA-B5-A1MX-01 | F | 47 | Uterine endometrioid carcinoma | NA | NA | G92* | Nonsense | 0.33 | 5699 |
| TCGA-BG-A222-01 | F | 49 | Uterine endometrioid carcinoma | NA | NA | G34R | Missense | 0.45 | 4276 |
| TCGA-DF-A2KU-01 | F | NA | Uterine endometrioid carcinoma | NA | NA | S230L | Missense | 0.30 | 10,058 |
| TCGA-EO-A22R- | F | 56 | Uterine endometrioid carcinoma | NA | NA | S230L | Missense | 0.30 | 12,783 |
| TCGA-EY-A1GK-01 | F | 74 | Uterine endometrioid carcinoma | NA | NA | G35Afs*75 | Frame shift del | 0.30 | 841 |
| TCGA-A5-A0G2-01 | F | 57 | Uterine serous carcinoma | NA | NA | D18N | Missense | 0.18 | 25,730 |
AJCCMSC: American Joint Committee on Cancer Metastasis Stage Code. AJCCTSC: American Joint Committee on Cancer Tumor Stage Code. # Mut: Total number of non-synonymous mutation in the sample. M0: No evidence of distant metastasis. MX: Distant metastasis cannot be assessed. NA: Not available. Shaded boxes indicate pathogenic somatic mutations of prion diseases. *: stop codon.
Figure 1.
Schematic map of human prion protein (PrP) with somatic mutations of the prion protein gene (PRNP) in cancer patients. Sharps indicate previous reported pathogenic mutations of prion diseases. SP: signal peptide; OPR: octapeptide repeat region.
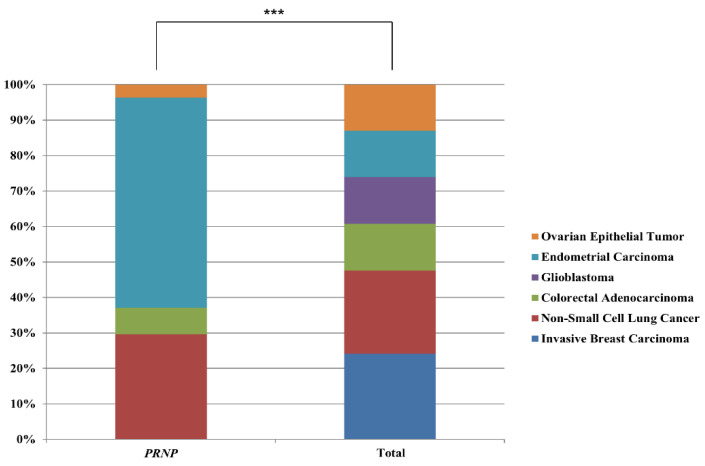
In addition, we compared the information on cancer patients who carried somatic mutations of the PRNP gene with that of the total cancer patient population. The detailed information is described in Table 3. In detail, we found 48 PRNP somatic mutations carriers (subgroup 1) and 8 PRNP pathogenic somatic mutation carriers of prion disease (subgroup 2). Subgroup 1 accounts for 0.43% of the total cancer patient population assessed, while subgroup 2 accounts for 0.07% of the total cancer patient population. Subgroup 2 accounts for 17.02% of subgroup 1. The age of diagnosis in the total cancer patient population was 59.1 ± 14.5 years, in subgroup 1 was 66 ± 12.8 years, and in subgroup 2 was 66.3 ± 7.8 years. Notably, compared to the age of diagnosis in the total cancer patient population, those of subgroups 1 showed significant differences with p-values of 0.000742. In addition, there were also significant differences in the mutation count distributions between the total cancer patient population and subgroups 1 and 2, with p-values < 0.00001 and 2.3 × 10−11, respectively. Specifically, a large number of the total cancer patient population displayed mutation counts <100 (67.3%). By contrast, subgroups 1 and 2 primarily displayed mutation counts >280 (83.3% and 100%, respectively). The distribution of cancer type between the total cancer patient population and subgroup 1 showed statistical significance. (p = 6 × 10−12). Specifically, the percentage of non-small cell lung cancer patients in subgroup 1 (16.7%) was increased by 1.7-fold compared to the total cancer patient population (9.6%). The percentage of endometrial carcinoma patients in subgroup 1 (33.3%) was increased by 6.3-fold compared to the total cancer patient population (5.3%). The percentage of non-small cell lung cancer patients in subgroup 2 (12.5%) was increased by 1.3-fold compared to the total cancer patient population (9.6%). The percentage of colorectal adenocarcinoma patients in subgroup 2 (12.5%) was increased by 2.3-fold compared to the total cancer patient population (5.4%). The percentage of endometrial carcinoma patients in subgroup 2 (25.0%) was increased by 4.7-fold compared to the total cancer patient population (5.3%). Notably, although invasive breast carcinoma and glioblastoma account for a high percentage of the total cancer patient population (9.9% and 5.4%, respectively), somatic mutations in the PRNP gene have not been identified in these two cancer types. Strikingly, distributions of six major cancer types (invasive breast cancer, non-small cell lung cancer, colorectal adenocarcinoma, glioblastoma, endometrial carcinoma, and ovarian epithelial tumor) of the patients with somatic mutations of the PRNP gene were shown to be significantly different from distributions of the six major cancer types of total patients (Figure 2).
Table 3.
Summary of information on the cancer patients carrying somatic mutations in the PRNP gene.
| Characteristics | All Cancer Patients | Subgroup 1 | Subgroup 2 | |
|---|---|---|---|---|
| Number of patients | 10,953 | 47 | 8 | |
| % compared to all cancer patients | - | 0.43% | 0.07% | |
| % compared to subgroup 1 | - | - | 17.02% | |
| Number of samples | 10,967 | 48 | 8 | |
| % compared to all cancer patients | 0.44% | 0.07% | ||
| % compared to subgroup 1 | - | - | 16.67% | |
| Diagnosis age (mean ± SD) | 59.1 ± 14.5 | 66 ± 12.8 | 66.3 ± 7.8 | |
| p-value compared to all cancer patients | 0.000742 | 0.093018 | ||
| p-value compared to subgroup 1 | - | - | 0.477389 | |
| Sex, N (%) | Male | 4866 (44.4%) | 18 (37.5%) | 3 (37.5%) |
| Female | 5315 (48.5%) | 27 (56.2%) | 4 (50.0%) | |
| NA | 772 (7.0%) | 3 (6.3%) | 1 (12.5%) | |
| Total | 10,953 | 48 | 8 | |
| p-value compared to all cancer patients | - | 0.30 | 0.79 | |
| p-value compared to subgroup 1 | - | - | 0.89 | |
| Mutation count | <100 | 6798 (67.3%) | 4 (8.3%) | 0 (0%) |
| 100 ≤ x < 200 | 1516 (15.0%) | 2 (4.2%) | 0 (0%) | |
| 200 ≤ x < 280 | 455 (4.5%) | 2 (4.2%) | 0 (0%) | |
| >280 | 1328 (13.2%) | 40 (83.3%) | 8 (100%) | |
| Total | 10,097 (100%) | 48 | 8 | |
| p-value compared to all cancer patients | - | <0.00001 | 2.3 × 10−11 | |
| p-value compared to subgroup 1 | - | - | 0.67 | |
| Cancer type, N (%) | Invasive breast carcinoma | 1084 (9.9%) | 0 (0%) | 0 (0%) |
| Non-small cell lung cancer | 1053 (9.6%) | 8 (16.7%) | 1 (12.5%) | |
| Colorectal adenocarcinoma | 594 (5.4%) | 2 (4.2%) | 1 (12.5%) | |
| Glioblastoma | 592 (5.4%) | 0 (0%) | 0 (0%) | |
| Endometrial carcinoma | 586 (5.3%) | 16 (33.3%) | 2 (25.0%) | |
| Ovarian epithelial tumor | 585 (5.3%) | 1 (2.1%) | 0 | |
| Head and neck squamous cell carcinoma | 523 (4.8%) | 1 (2.1%) | 1 (12.5%) | |
| Esophagogastric adenocarcinoma | 514 (4.7%) | 5 (10.4%) | 1 (12.5%) | |
| Diffuse glioma | 513 (4.7%) | 0 (0%) | 0 (0%) | |
| Renal clear cell carcinoma | 512 (4.7%) | 1 (2.1%) | 0 (0%) | |
| Well-differentiated thyroid | 500 (4.6%) | 0 (0%) | 0 (0%) | |
| Prostate adenocarcinoma | 494 (4.5%) | 0 (0%) | 0 (0%) | |
| Melanoma | 448 (4.1%) | 5 (10.4%) | 0 (0%) | |
| Bladder urothelial carcinoma | 411 (3.7%) | 2 (4.2%) | 0 (0%) | |
| Hepatocellular carcinoma | 369 (3.4%) | 0 (0%) | 0 (0%) | |
| Renal non-clear cell carcinoma | 348 (3.2%) | 3 (6.3%) | 1 (12.5%) | |
| Sarcoma | 255 (2.3%) | 1 (2.1%) | 0 (0%) | |
| Cervical squamous cell carcinoma | 251 (2.3%) | 1 (2.1%) | 1 (12.5%) | |
| Leukemia | 200 (1.8%) | 0 (0%) | 0 (0%) | |
| Pancreatic adenocarcinoma | 184 (1.7%) | 0 (0%) | 0 (0%) | |
| Pheochromocytoma | 147 (1.3%) | 0 (0%) | 0 (0%) | |
| Thymic epithelial tumor | 123 (1.1%) | 0 (0%) | 0 (0%) | |
| Esophageal squamous cell carcinoma | 95 (0.9%) | 1 (2.1%) | 0 (0%) | |
| Adrenocortical carcinoma | 92 (0.8%) | 0 (0%) | 0 (0%) | |
| Pleural mesothelioma | 87 (0.8%) | 0 (0%) | 0 (0%) | |
| Non-seminomatous germ cell tumor | 86 (0.8%) | 0 (0%) | 0 (0%) | |
| Ocular melanoma | 80 (0.7%) | 0 (0%) | 0 (0%) | |
| Seminoma | 63 (0.6%) | 0 (0%) | 0 (0%) | |
| Mature B-cell neoplasms | 48 (0.4%) | 0 (0%) | 0 (0%) | |
| Cervical adenocarcinoma | 46 (0.4%) | 0 (0%) | 0 (0%) | |
| Cholangiocarcinoma | 36 (0.3%) | 0 (0%) | 0 (0%) | |
| Miscellaneous neuroepithelial tumor | 31 (0.3%) | 0 (0%) | 0 (0%) | |
| Undifferentiated stomach adenocarcinoma | 13 (0.1%) | 1 (2.1%) | 0 (0%) | |
| Fibrolamellar carcinoma | 3 (<0.1%) | 0 (0%) | 0 (0%) | |
| Encapsulated glioma | 1 (<0.1%) | 0 (0%) | 0 (0%) | |
| Total | 10,967 | 48 | 8 | |
| p-value compared to all cancer patients | - | 6 × 10−12 | 0.96 | |
| p-value compared to subgroup 1 | - | - | NA |
Subgroup 1: carriers of PRNP somatic mutations. Subgroup 2: carriers of PRNP pathogenic somatic mutations of prion disease. NA: not available. Shaded boxes indicate pathogenic somatic mutations of prion disease. Bold text indicates statistical significance (p < 0.05).
Figure 2.
Comparison of distributions of patients harboring somatic mutations of the PRNP gene according to six major cancer types. PRNP: distribution of cancer patients harboring somatic mutations of the PRNP gene in six major cancer types. Total: distribution of cancer patients harboring somatic mutations in six major cancer types. *** indicates p < 0.001.
2.3. In Silico Annotation of Mutations in the PRNP Gene
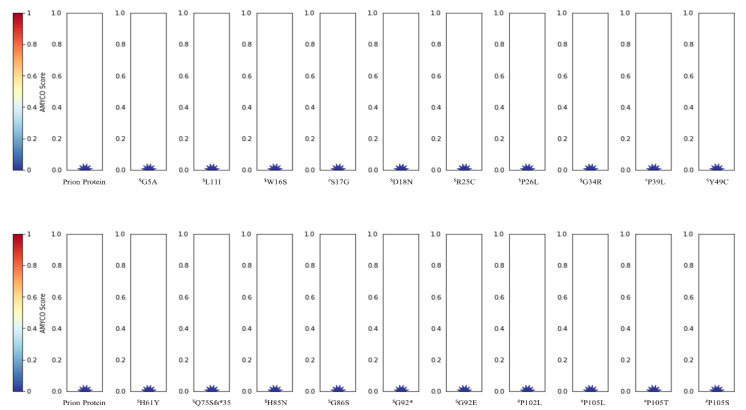
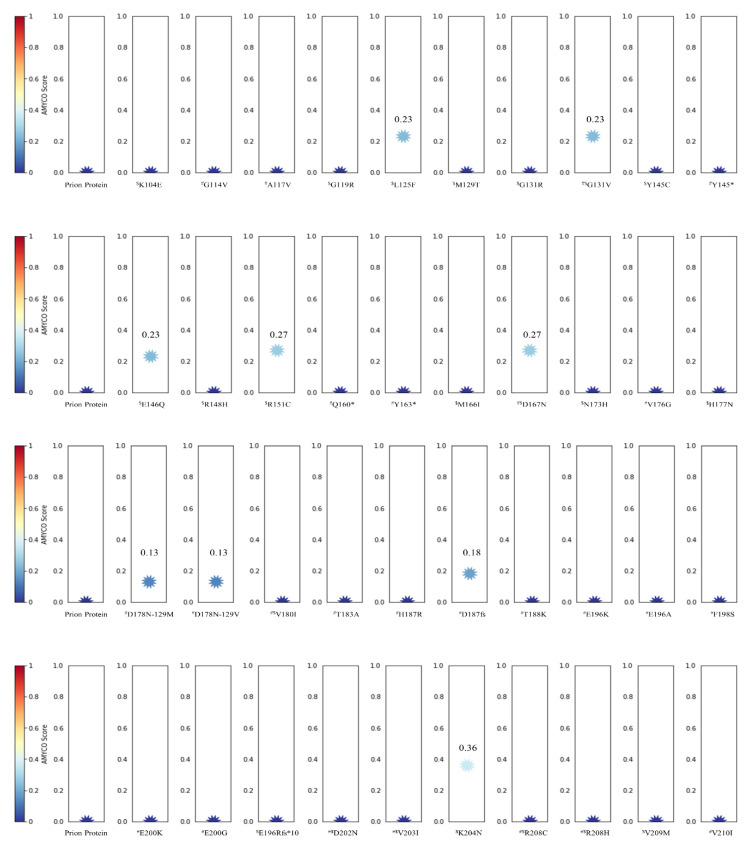
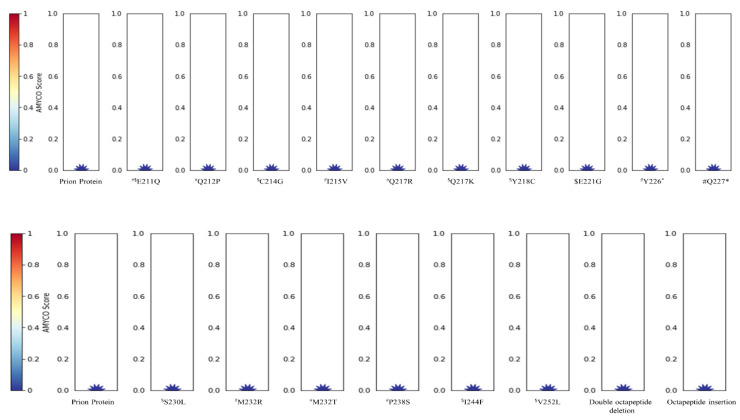
The impact of somatic mutations in PrP identified in this study and previously reported pathogenic mutations was analyzed by PolyPhen-2, PANTHER, and PROVEAN. The detailed information is described in Table 4. Notably, six somatic mutations, these being P26L, G34R, G119R, G131V, C214G, and Y218C, and five pathogenic mutations, these being P102L, P105L, G114V, H187R, and E200G were predicted to be deleterious by all three programs. We also evaluated the amyloid propensity of the mutations in the PRNP gene by AMYCO. The detailed information is described in Figure 3. Interestingly, nine mutations, these being L125F, G131V, E146Q, R151C, D167N, D178N-129M, D178N-129V, D187fs, and K204N, had an increased amyloid propensity with values of 0.23, 0.23, 0.23, 0.27, 0.27, 0.13, 0.13, 0.18, and 0.36, respectively. Among the six somatic mutations of cancer patients, four were newly identified amyloid-prone somatic mutations—L125F, E146Q, R151C, and K204N.
Table 4.
In silico annotations of PRNP somatic mutations in cancer patients.
| Mutations | PolyPhen-2 | PANTHER | PROVEAN | |||
|---|---|---|---|---|---|---|
| Score | Prediction | Preservation Time | Prediction | Score | Prediction | |
| $ G5A | 0.729 | Possibly damaging | 176 | Probably benign | −1.238 | Neutral |
| $ L11I | 0.161 | Benign | 176 | Probably benign | −0.306 | Neutral |
| $ W16S | 0.546 | Possibly damaging | 176 | Probably benign | −1.237 | Neutral |
| # S17G | 0.528 | Possibly damaging | 176 | Probably benign | −0.239 | Neutral |
| $ D18N | 1.000 | Probably damaging | 176 | Probably benign | −0.104 | Neutral |
| $ R25C | 1.000 | Probably damaging | 176 | Probably benign | −1.801 | Neutral |
| $ P26L | 0.995 | Probably damaging | 324 | Possibly damaging | −3.263 | Deleterious |
| $ G34R | 1.000 | Probably damaging | 220 | Possibly damaging | −3.889 | Deleterious |
| # P39L | 1.000 | Probably damaging | 361 | Possibly damaging | −4.000 | Deleterious |
| $ Y49C | 1.000 | Probably damaging | 176 | Probably benign | −2.222 | Neutral |
| $ H61Y | 0.975 | Probably damaging | 176 | Probably benign | −1.021 | Neutral |
| $ H85N | 0.975 | Probably damaging | 176 | Probably benign | −0.833 | Neutral |
| $ G86S | 1.000 | Probably damaging | 324 | Possibly damaging | −0.995 | Neutral |
| $ G92E | 1.000 | Probably damaging | 176 | Probably benign | −1.253 | Neutral |
| # P102L | 1.000 | Probably damaging | 361 | Possibly damaging | −3.392 | Deleterious |
| # P105L | 1.000 | Probably damaging | 324 | Possibly damaging | −3.271 | Deleterious |
| # P105T | 0.998 | Probably damaging | 324 | Possibly damaging | −2.333 | Neutral |
| # P105S | 0.997 | Probably damaging | 324 | Possibly damaging | −1.496 | Neutral |
| $ K104E | 0.974 | Probably damaging | 324 | Possibly damaging | −1.208 | Neutral |
| # G114V | 1.000 | Probably damaging | 220 | Possibly damaging | −2.540 | Deleterious |
| # A117V | 0.999 | Probably damaging | 176 | Probably benign | −1.263 | Neutral |
| $ G119R | 1.000 | Probably damaging | 361 | Possibly damaging | −2.586 | Deleterious |
| $ L125F | 1.000 | Probably damaging | 324 | Possibly damaging | −0.398 | Neutral |
| $ M129T | 0.181 | Benign | 324 | Possibly damaging | −1.156 | Neutral |
| $ G131R | 1.000 | Probably damaging | 361 | Possibly damaging | −2.451 | Neutral |
| #,$ G131V | 1.000 | Probably damaging | 361 | Possibly damaging | −2.879 | Deleterious |
| $ Y145C | 0.997 | Probably damaging | 220 | Possibly damaging | −1.742 | Neutral |
| $ E146Q | 0.992 | Probably damaging | 361 | Possibly damaging | −0.985 | Neutral |
| $ R148H | 1.000 | Probably damaging | 361 | Possibly damaging | −1.735 | Neutral |
| $ R151C | 0.009 | Benign | 220 | Possibly damaging | −2.016 | Neutral |
| $ M166I | 0.000 | Benign | 30 | Probably benign | 0.254 | Neutral |
| #,$ D167N | 0.001 | Benign | 220 | Possibly damaging | −0.631 | Neutral |
| $ N173H | 0.952 | Probably damaging | 176 | Probably benign | −1.573 | Neutral |
| # V176G | 0.998 | Probably damaging | 361 | Possibly damaging | −2.253 | Neutral |
| $ H177N | 0.313 | Benign | 220 | Possibly damaging | −0.455 | Neutral |
| # D178N-129M | 1.000 | Probably damaging | 361 | Possibly damaging | −1.531 | Neutral |
| # D178N-129V | 1.000 | Probably damaging | 361 | Possibly damaging | −1.451 | Neutral |
| #,$ V180I | 0.009 | Benign | 220 | Possibly damaging | −0.11 | Neutral |
| # T183A | 0.978 | Probably damaging | 324 | Possibly damaging | −1.785 | Neutral |
| # H187R | 0.989 | Probably damaging | 220 | Possibly damaging | −2.607 | Deleterious |
| # T188K | 0.996 | Probably damaging | 324 | Possibly damaging | −0.550 | Neutral |
| # E196K | 0.624 | Possibly damaging | 220 | Possibly damaging | −0.641 | Neutral |
| # E196A | 0.472 | Possibly damaging | 220 | Possibly damaging | −1.206 | Neutral |
| # F198S | 0.994 | Probably damaging | 220 | Possibly damaging | −0.792 | Neutral |
| # E200K | 0.995 | Probably damaging | 361 | Possibly damaging | −1.478 | Neutral |
| # E200G | 0.994 | Probably damaging | 361 | Possibly damaging | −3.245 | Deleterious |
| #,$ D202N | 1.000 | Probably damaging | 220 | Possibly damaging | −1.118 | Neutral |
| #,$ V203I | 0.001 | Benign | 176 | Probably benign | −0.004 | Neutral |
| $ K204N | 0.898 | Possibly damaging | 220 | Possibly damaging | −1.815 | Neutral |
| #,$ R208C | 1.000 | Probably damaging | 220 | Possibly damaging | −2.324 | Neutral |
| #,$ R208H | 0.999 | Probably damaging | 220 | Possibly damaging | −0.855 | Neutral |
| $ V209M | 0.613 | Possibly damaging | 324 | Possibly damaging | −1.03 | Neutral |
| # V210I | 0.803 | Possibly damaging | 220 | Possibly damaging | 0.039 | Neutral |
| #,$ E211Q | 0.992 | Probably damaging | 220 | Possibly damaging | −0.36 | Neutral |
| # Q212P | 0.930 | Possibly damaging | 220 | Possibly damaging | −1.665 | Neutral |
| $ C214G | 0.975 | Probably damaging | 361 | Possibly damaging | −4.402 | Deleterious |
| # I215V | 0.000 | Benign | 220 | Possibly damaging | −0.099 | Neutral |
| # Q217R | 0.961 | Probably damaging | 220 | Possibly damaging | −1.306 | Neutral |
| $ Q217K | 0.942 | Probably damaging | 220 | Possibly damaging | −1.186 | Neutral |
| $ Y218C | 1.000 | Probably damaging | 361 | Possibly damaging | −3.626 | Deleterious |
| $ E221G | 0.651 | Possibly damaging | 220 | Possibly damaging | −1.244 | Neutral |
| $ S230L | 0.947 | Possibly damaging | 97 | Probably benign | −1.372 | Neutral |
| # M232R | 0.082 | Benign | 91 | Probably benign | −1.167 | Neutral |
| # M232T | 0.000 | Benign | 91 | Probably benign | −0.825 | Neutral |
| # P238S | 1.000 | Probably damaging | 361 | Possibly damaging | −1.189 | Neutral |
| $ I244F | 0.001 | Benign | 176 | Probably benign | −0.939 | Neutral |
| $ V252L | 0.990 | Probably damaging | 176 | Probably benign | −0.411 | Neutral |
$ indicates somatic mutations of the PRNP gene in cancer patients. # indicates previously reported pathogenic mutations of the PRNP gene in prion diseases. Shaded boxes indicate the mutations that were predicted to be deleterious by all three programs.
Figure 3.
Prediction on amyloid propensity of prion protein (PrP) according to somatic mutations of the prion protein gene (PRNP) gene in cancer patients. $ indicates somatic mutations of the PRNP gene in cancer patients. # indicates previously reported pathogenic mutations of the PRNP gene in prion diseases.
3. Discussion
In the present study, we identified a total of 48 somatic mutations in the PRNP gene in 10,967 cancer patients. Prion diseases are considered to be monogenic diseases, and the association of the PRNP gene with the onset of prion diseases was confirmed by genome-wide association studies (GWAS) [33,34]. Thus, the somatic mutations in the PRNP gene in cancer tissues are very important. Of the 48 somatic mutations, 8 were previously confirmed pathogenic mutations of prion diseases. Although we cited pathogenic mutations of the PRNP gene from previous studies, the penetrance on the pathogenic mutations of the PRNP gene was still debatable. In a previous study, several mutations including V180I, V210I, and M232R were found in healthy controls and these mutations showed low penetrance under 10%. Thus, these results suggest that these mutations were benign variants or low-risk variants [35]. In the present study, we further analyzed previously reported pathogenic mutations using in silico programs. Notably, V180I, V210I, and M232R mutations were predicted to be low amyloid propensity and benign by some programs (Figure 3, Table 4). Further study to confirm the deleterious effects of the mutations is needed in the future. In addition, cancer patients carrying these somatic mutations showed no evidence of distant metastasis (Table 2). Since PrP contributes to metastasis in several types of cancers, somatic mutations of the PRNP gene, which can affect normal function of PrP, may impact the property of distant metastasis [36,37]. Further validation of distant metastatic property in cancer cells carrying PRNP mutations is highly desirable in the future. Interestingly, the age of patients carrying somatic mutations (66 ± 12.8 years) was much older than that of the total cancer patient population (59.1 ± 14.5 years) (Table 2). Since sporadic prion disease has been reported to develop around the ages of 60–70, this was a very interesting finding [38]. In addition, since PrP contributes to several properties of cancer including tumorigenesis, protection of apoptotic stress, and metastasis, further study of the relationship between age and several characteristics of cancers mediated by somatic mutations of the PRNP gene is needed in the future [36,37,39,40]. Furthermore, 83.3% of patients carrying somatic mutations of the PRNP gene showed over 280 mutation counts. This distribution is significantly different from that of the total cancer patient population. In addition, the cancer type of the patients carrying somatic mutations was also significantly different from the total cancer patient population. Notably, although invasive breast carcinoma (9.9%) and glioblastoma (5.4%) account for high percentage of the total cancer patient population, somatic mutation of the PRNP gene has not been identified in these two cancer types. Specifically, because somatic mutations of the PRNP gene in glioblastoma, which occurs in the brain, can be masked by a diagnosis of dementia, the absence of somatic mutations in the PRNP gene in glioblastoma can be explained. Previous study has been reported that transgenic mice carrying a pathogenic mutation of the PRNP gene can lead to spontaneous prion disease [41,42]. In addition, a recent study has been reported that PrPSc were also detected in healthy people, who have not been diagnosed with prion diseases [43]. On the basis of these studies, we postulated that cancer patients carrying pathogenic somatic mutation of the PRNP gene may produce a basal level of PrPSc in peripheral tissues and may not be diagnosed with prion disease. However, to date, there is no clinical evidence for this hypothesis. Since PrP protects cancer cells against apoptotic stress and contributes to tumorigenesis, the difference on distribution of somatic mutation of the PRNP gene may be induced depending on the degree of contribution of PrP to different cancer types [39,40]. Further study to verify functional role of prion protein in various cancer types is needed in the future.
We also analyzed somatic mutations of the PRNP gene in cancer patients by PolyPhen-2, PANTHER, and PROVEAN. Notably, six somatic mutations—P26L, G34R, G119R, G131V, C214G, and Y218C—and five pathogenic mutations—P102L, P105L, G114V, H187R, and E200G—were considered to be damaging to PrP function by all three programs. Since PrPC has also been reported to be related to the survival signaling pathway and copper ion-associated enzyme activity, these somatic mutations may impact the normal function of PrP [44]. In addition, we predicted the amyloid propensity of the somatic mutations in the PRNP gene in cancer patients. Notably, four somatic mutations—L125F, E146Q, R151C, and K204N—were newly identified amyloid-prone somatic mutations in the PRNP gene in cancer patients. In addition, we found several nonsense somatic mutations. These nonsense mutations induced PrP cleavage, which can affect the survival signaling pathway or the aggregation of PrP [6,17,45]. Thus, further investigation of the mutation counts, the absence of somatic mutations in glioblastoma, and these nonsense mutations are highly desirable in the future.
In recent cancer studies, p53 has been demonstrated to play a pivotal role in several types of cancer and showed prion-like aggregation. Aggregation of p53 causes additional deleterious effects in cancer patients [46,47,48]. In addition, although cross-seeding activity between aggregated p53 and PrPSc has not been investigated thus far, the similarity of cross-seeding activities in other misfolded proteins suggests that aggregated p53 may act as a seed to induce the aggregation of PrP [49]. Thus, the detection of PrPSc in cancer patients using Western blot or protein misfolded cyclic amplification (PMCA) is highly desirable in the future.
4. Material and Methods
4.1. Information on the Somatic Mutations in the PRNP Gene in Cancer Patients
The protein sequence of human PrP in Figure 1 was obtained from GenBank at the National Center for Biotechnology Information (NCBI, AAH22532.1). Previously reported pathogenic mutations associated with prion disease were collected from previous studies [6,16,17]. The data on somatic mutations of the PRNP gene in cancer patients were extracted from TCGA database using cBioPortal (http://www.cbioportal.org/) [26]. Among total mutations of the PRNP gene, germline mutations were excluded using the filtering option. Somatic mutations were analyzed using reference sequence from GenBank at the NCBI (NM_000311.5). Clinical data including somatic mutations, age of diagnosis, sex, mutation count, cancer type, and allele frequencies were retrieved from cBioPortal and the data were read by R (https://www.r-project.org/).
4.2. Statistical Analysis
Statistical analyses were performed using SAS version 9.4 (SAS Institute Inc., Cary, NC, USA). The differences in the distributions of sex, mutation count, and cancer type between the total cancer patient population and the subgroups were compared using the χ2 test. The age of diagnosis was reported as the mean values ± standard deviation (SD). Statistical significance based on p-values was calculated with a one-tailed Student’s t-test for single comparison.
4.3. In Silico Evaluation of Somatic Mutations in the PRNP Gene in Cancer Patients
Somatic mutations in the PRNP gene in cancer patients were evaluated by PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), PANTHER (http://www.pantherdb.org/), PROVEAN (http://provean.jcvi.org/index.php), and AMYCO (http://bioinf.uab.es/amycov04/) [27,28,29,30]. The effects of sequence variation were evaluated on the basis of protein structure by PolyPhen-2. The PolyPhen-2 score corresponds to the substitution effect and ranges from 0.0 to 1.0. The prediction outcome can be presented as “Benign”, “Possibly damaging”, or “Probably damaging” on the basis of the score. PROVEAN assessed amino acid substitution by building up and comparing the clusters of related sequences and calculating the score. Scores below −2.5 are predicted as “Deleterious” and scores above −2.5 are predicted as “Neutral”. PANTHER analyzes data in groups on the basis of similarities in molecular function and biological processes. PANTHER analyzed preservation time to estimate the substitution of amino acids. The interpretation of preservation time is described as follows: “Probably damaging” is greater than 450; “Possibly damaging” is between 200 and 450; “Probably benign” is less than 200. AMYCO evaluates the impact of variations on the aggregation propensity of prion-like proteins. The AMYCO score corresponds to the substitution effect and ranges from 0.0 to 1.0.
5. Conclusions
In conclusion, we identified 48 somatic mutations in the PRNP gene in 10,967 cancer patients. Among them, eight somatic mutations are pathogenic for prion diseases. We identified significantly different distributions of cancer type, mutation count, and the age of diagnosis between the total cancer patient population and cancer patients carrying PRNP somatic mutations. We found six somatic mutations, these being P26L, G34R, G119R, G131V, C214G, and Y218C, and five pathogenic mutations, these being P102L, P105L, G114V, H187R, and E200G, which can affect PrPC function, along with four aggregation-prone somatic mutations—L125F, E146Q, R151C, and K204N. To the best of our knowledge, this is the first specific analysis on the somatic mutations of the PRNP gene in cancer patients.
Acknowledgments
The results obtained in this study are partially based on data produced by the TCGA Research Network (http://cancergenome.nih.gov/).
Abbreviations
| PrPSc | Misfolded prion protein |
| PrPC | Normal prion protein |
| PrP | Prion protein |
| TCGA | The Cancer Genome Atlas |
| BSE | Bovine spongiform encephalopathy |
| CJD | Creutzfeldt–Jakob disease |
| CWD | Chronic wasting disease |
| FFI | Fatal familial insomnia |
| GSS | Gerstmann–Sträussler–Scheinker syndrome |
| PRNP | Prion protein gene |
Author Contributions
Y.-C.K. and B.-H.J. conceived and designed the experiments. Y.-C.K., S.-Y.W., and B.-H.J. analyzed the data. Y.-C.K. and B.-H.J. wrote the paper. All authors read and approved the final manuscript.
Funding
This research was supported by the Basic Science Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (2018R1D1A1B07048711). This research was also supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2017R1A6A1A03015876). This work was supported by the NRF (National Research Foundation of Korea) grant funded by the Korean Government (NRF-2019—Fostering Core Leaders of the Future Basic Science Program/Global Ph.D. Fellowship Program).
Conflicts of Interest
The authors declare no conflict of interest, financial or otherwise.
References
- 1.Prusiner S.B. Prions. Proc. Natl. Acad. Sci. USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prusiner S.B. The prion diseases. Brain Pathol. 1998;8:499–513. doi: 10.1111/j.1750-3639.1998.tb00171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murdoch B.M., Murdoch G.K. Genetics of Prion Disease in Cattle. Bioinform. Biol. Insights. 2015;9:1–10. doi: 10.4137/BBI.S29678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurt T.D., Sigurdson C.J. Cross-species transmission of CWD prions. Prion. 2016;10:83–91. doi: 10.1080/19336896.2015.1118603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenlee J.J. Review: Update on Classical and Atypical Scrapie in Sheep and Goats. Vet. Pathol. 2019;56:6–16. doi: 10.1177/0300985818794247. [DOI] [PubMed] [Google Scholar]
- 6.Jeong B.H., Kim Y.S. Genetic studies in human prion diseases. J. Korean Med. Sci. 2014;29:623–632. doi: 10.3346/jkms.2014.29.5.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaccari G., Panagiotidis C.H., Acin C., Peletto S., Barillet F., Acutis P., Bossers A., Langeveld J., van Keulen L., Sklaviadis T., et al. State-of-the-art review of goat TSE in the European Union, with special emphasis on PRNP genetics and epidemiology. Vet. Res. 2009;40:48. doi: 10.1051/vetres/2009031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim Y.C., Jeong B.H. The first report of prion-related protein gene (PRNT) polymorphisms in goat. Acta. Vet. Hung. 2017;65:291–300. doi: 10.1556/004.2017.028. [DOI] [PubMed] [Google Scholar]
- 9.Kim Y.C., Jeong B.H. Bovine spongiform encephalopathy (BSE) associated polymorphisms of the prion-like protein gene (PRND) in Korean dairy cattle and Hanwoo. J. Dairy Res. 2018;85:7–11. doi: 10.1017/S0022029917000814. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y.C., Jeong B.H. First report of prion-related protein gene (PRNT) polymorphisms in cattle. Vet. Rec. 2018;182:717. doi: 10.1136/vr.104123. [DOI] [PubMed] [Google Scholar]
- 11.Kim Y.C., Kim S.K., Jeong B.H. Scrapie susceptibility-associated indel polymorphism of shadow of prion protein gene (SPRN) in Korean native black goats. Sci. Rep. 2019;9:15261. doi: 10.1038/s41598-019-51625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S.K., Kim Y.C., Won S.Y., Jeong B.H. Potential scrapie-associated polymorphisms of the prion protein gene (PRNP) in Korean native black goats. Sci. Rep. 2019;9:15293. doi: 10.1038/s41598-019-51621-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erickson R.P. Somatic gene mutation and human disease other than cancer. Mutat. Res. 2003;543:125–136. doi: 10.1016/S1383-5742(03)00010-3. [DOI] [PubMed] [Google Scholar]
- 14.Leija-Salazar M., Piette C., Proukakis C. Review: Somatic mutations in neurodegeneration. Neuropathol. Appl. Neurobiol. 2018;44:267–285. doi: 10.1111/nan.12465. [DOI] [PubMed] [Google Scholar]
- 15.Kim Y.C., Jeong M.J., Jeong B.H. Strong association of regulatory single nucleotide polymorphisms (SNPs) of the IFITM3 gene with influenza H1N1 2009 pandemic virus infection. Cell. Mol. Immunol. 2019 doi: 10.1038/s41423-019-0322-1. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bagyinszky E., Giau V.V., Youn Y.C., An S.S.A., Kim S. Characterization of mutations in PRNP (prion) gene and their possible roles in neurodegenerative diseases. Neuropsychiatr. Dis. Treat. 2018;14:2067–2085. doi: 10.2147/NDT.S165445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lloyd S., Mead S., Collinge J. Genetics of prion disease. Top. Curr. Chem. 2011;305:1–22. doi: 10.1007/128_2011_157. [DOI] [PubMed] [Google Scholar]
- 18.Nicolas G., Veltman J.A. The role of de novo mutations in adult-onset neurodegenerative disorders. Acta. Neuropathol. 2019;137:183–207. doi: 10.1007/s00401-018-1939-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alzualde A., Moreno F., Martinez-Lage P., Ferrer I., Gorostidi A., Otaegui D., Blazquez L., Atares B., Cardoso S., Martinez de Pancorbo M., et al. Somatic mosaicism in a case of apparently sporadic Creutzfeldt-Jakob disease carrying a de novo D178N mutation in the PRNP gene. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010;153B:1283–1291. doi: 10.1002/ajmg.b.31099. [DOI] [PubMed] [Google Scholar]
- 20.Dagvadorj A., Petersen R.B., Lee H.S., Cervenakova L., Shatunov A., Budka H., Brown P., Gambetti P., Goldfarb L.G. Spontaneous mutations in the prion protein gene causing transmissible spongiform encephalopathy. Ann. Neurol. 2002;52:355–359. doi: 10.1002/ana.10267. [DOI] [PubMed] [Google Scholar]
- 21.Webb T.E., Poulter M., Beck J., Uphill J., Adamson G., Campbell T., Linehan J., Powell C., Brandner S., Pal S., et al. Phenotypic heterogeneity and genetic modification of P102L inherited prion disease in an international series. Brain. 2008;131:2632–2646. doi: 10.1093/brain/awn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santos T.G., Lopes M.H., Martins V.R. Targeting prion protein interactions in cancer. Prion. 2015;9:165–173. doi: 10.1080/19336896.2015.1027855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinton C., Antony H., Hashimi S.M., Munn A., Wei M.Q. Significance of prion and prion-like proteins in cancer development, progression and multi-drug resistance. Curr. Cancer. Drug Targets. 2013;13:895–904. doi: 10.2174/156800961131300092. [DOI] [PubMed] [Google Scholar]
- 24.Negrini S., Gorgoulis V.G., Halazonetis T.D. Genomic instability—an evolving hallmark of cancer. Nat. Rev. Mol. Cell. Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 25.Shen Z. Genomic instability and cancer: An introduction. J. Mol. Cell. Biol. 2011;3:1–3. doi: 10.1093/jmcb/mjq057. [DOI] [PubMed] [Google Scholar]
- 26.Tomczak K., Czerwinska P., Wiznerowicz M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. (Pozn) 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013;76:7–20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi Y., Chan A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iglesias V., Conchillo-Sole O., Batlle C., Ventura S. AMYCO: Evaluation of mutational impact on prion-like proteins aggregation propensity. BMC Bioinform. 2019;20:24. doi: 10.1186/s12859-019-2601-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang H., Thomas P.D. PANTHER-PSEP: Predicting disease-causing genetic variants using position-specific evolutionary preservation. Bioinformatics. 2016;32:2230–2232. doi: 10.1093/bioinformatics/btw222. [DOI] [PubMed] [Google Scholar]
- 31.Won S.Y., Kim Y.C., Kim K., Kim A.D., Jeong B.H. The First Report of Polymorphisms and Genetic Features of the prion-like Protein Gene (PRND) in a Prion Disease-Resistant Animal, Dog. Int. J. Mol. Sci. 2019;20:1404. doi: 10.3390/ijms20061404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim Y.C., Jeong B.H. In Silico Evaluation of Acetylation Mimics in the 27 Lysine Residues of Human Tau Protein. Curr. Alzheimer. Res. 2019;16:379–387. doi: 10.2174/1567205016666190321161032. [DOI] [PubMed] [Google Scholar]
- 33.Mead S., Poulter M., Uphill J., Beck J., Whitfield J., Webb T.E., Campbell T., Adamson G., Deriziotis P., Tabrizi S.J., et al. Genetic risk factors for variant Creutzfeldt-Jakob disease: A genome-wide association study. Lancet Neurol. 2009;8:57–66. doi: 10.1016/S1474-4422(08)70265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mead S., Uphill J., Beck J., Poulter M., Campbell T., Lowe J., Adamson G., Hummerich H., Klopp N., Ruckert I.M., et al. Genome-wide association study in multiple human prion diseases suggests genetic risk factors additional to PRNP. Hum. Mol. Genet. 2012;21:1897–1906. doi: 10.1093/hmg/ddr607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minikel E.V., Vallabh S.M., Lek M., Estrada K., Samocha K.E., Sathirapongsasuti J.F., McLean C.Y., Tung J.Y., Yu L.P., Gambetti P., et al. Quantifying prion disease penetrance using large population control cohorts. Sci. Transl. Med. 2016;8:322ra329. doi: 10.1126/scitranslmed.aad5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan Y., Zhao L., Liang J., Liu J., Shi Y., Liu N., Zhang G., Jin H., Gao J., Xie H., et al. Cellular prion protein promotes invasion and metastasis of gastric cancer. FASEB J. 2006;20:1886–1888. doi: 10.1096/fj.06-6138fje. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q., Qian J., Wang F., Ma Z. Cellular prion protein accelerates colorectal cancer metastasis via the Fyn-SP1-SATB1 axis. Oncol. Rep. 2012;28:2029–2034. doi: 10.3892/or.2012.2025. [DOI] [PubMed] [Google Scholar]
- 38.Manix M., Kalakoti P., Henry M., Thakur J., Menger R., Guthikonda B., Nanda A. Creutzfeldt-Jakob disease: Updated diagnostic criteria, treatment algorithm, and the utility of brain biopsy. Neurosurg. Focus. 2015;39:E2. doi: 10.3171/2015.8.FOCUS15328. [DOI] [PubMed] [Google Scholar]
- 39.Gao Z., Peng M., Chen L., Yang X., Li H., Shi R., Wu G., Cai L., Song Q., Li C. Prion Protein Protects Cancer Cells against Endoplasmic Reticulum Stress Induced Apoptosis. Virol. Sin. 2019;34:222–234. doi: 10.1007/s12250-019-00107-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang X., Cheng Z., Zhang L., Wu G., Shi R., Gao Z., Li C. Prion Protein Family Contributes to Tumorigenesis via Multiple Pathways. Adv. Exp. Med. Biol. 2017;1018:207–224. doi: 10.1007/978-981-10-5765-6_13. [DOI] [PubMed] [Google Scholar]
- 41.Watts J.C., Giles K., Bourkas M.E., Patel S., Oehler A., Gavidia M., Bhardwaj S., Lee J., Prusiner S.B. Towards authentic transgenic mouse models of heritable PrP prion diseases. Acta. Neuropathol. 2016;132:593–610. doi: 10.1007/s00401-016-1585-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sigurdson C.J., Joshi-Barr S., Bett C., Winson O., Manco G., Schwarz P., Rulicke T., Nilsson K.P., Margalith I., Raeber A., et al. Spongiform encephalopathy in transgenic mice expressing a point mutation in the beta2-alpha2 loop of the prion protein. J. Neurosci. 2011;31:13840–13847. doi: 10.1523/JNEUROSCI.3504-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gill O.N., Spencer Y., Richard-Loendt A., Kelly C., Brown D., Sinka K., Andrews N., Dabaghian R., Simmons M., Edwards P., et al. Prevalence in Britain of abnormal prion protein in human appendices before and after exposure to the cattle BSE epizootic. Acta Neuropathol. 2020 doi: 10.1007/s00401-020-02153-7. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wulf M.A., Senatore A., Aguzzi A. The biological function of the cellular prion protein: An update. BMC Biol. 2017;15:34. doi: 10.1186/s12915-017-0375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altmeppen H.C., Puig B., Dohler F., Thurm D.K., Falker C., Krasemann S., Glatzel M. Proteolytic processing of the prion protein in health and disease. Am. J. Neurodegener. Dis. 2012;1:15–31. [PMC free article] [PubMed] [Google Scholar]
- 46.Rangel L.P., Costa D.C., Vieira T.C., Silva J.L. The aggregation of mutant p53 produces prion-like properties in cancer. Prion. 2014;8:75–84. doi: 10.4161/pri.27776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheckel C., Aguzzi A. Prions, prionoids and protein misfolding disorders. Nat. Rev. Genet. 2018;19:405–418. doi: 10.1038/s41576-018-0011-4. [DOI] [PubMed] [Google Scholar]
- 48.Silva J.L., Rangel L.P., Costa D.C., Cordeiro Y., De Moura Gallo C.V. Expanding the prion concept to cancer biology: Dominant-negative effect of aggregates of mutant p53 tumour suppressor. Biosci. Rep. 2013;33:e00054. doi: 10.1042/BSR20130065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morales R., Moreno-Gonzalez I., Soto C. Cross-seeding of misfolded proteins: Implications for etiology and pathogenesis of protein misfolding diseases. PLoS Pathog. 2013;9:e1003537. doi: 10.1371/journal.ppat.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]