Abstract
The ability of Streptococcus mutans to persist in a variety of adverse environments and to emerge as a numerically dominant member of stable oral biofilm communities are essential elements for its cariogenicity. The S. mutans Cid/Lrg system has been studied as a key player in the integration of complex environmental signals into regulatory networks that modulate virulence and cell homeostasis. Cid/Lrg has also been shown to be closely associated with metabolic pathways of this organism, due to distinct patterns of cid and lrg expression in response to growth phase and glucose/oxygen levels. In this study, a comparison of cid and lrg promoter regions with conserved CodY (a regulator which responds to starvation stress)‐binding motifs revealed the presence of a potential CodY‐binding site, which is arranged similarly in both cid and lrg promoters. Electrophoretic mobility shift assays (EMSAs) and promoter reporter assays demonstrated that expression of the cid and lrg operons is directly mediated by the global transcriptional regulator CodY. DNase I footprinting analyses confirmed the predicted binding sequences for CodY in both the cid and the lrg promoter regions. Overexpression of CodY had no obvious effect on lrgAB expression, but deficiency of CodY still affected lrgAB expression in a lytST‐overexpressing strain, suggesting that CodY is required for the full regulation of lrgAB by LytST. We also demonstrated that both CodY and CcpA are involved in regulating pyruvate flux and utilization. Collectively, these data show that CodY directly regulates cid and lrg expression, and together with CcpA (previously shown to directly regulate cid and lrg promoters) contributes to coordinating pyruvate uptake and utilization in response to both the external environment and the cellular metabolic status.
Keywords: CcpA, Cid and Lrg, CodY, pyruvate, Streptococcus mutans
This article demonstrates the involvement of CodY as a direct regulator of cid and lrg operons, key players in the integration of complex environmental signals into regulatory networks that modulate virulence and cell homeostasis in Streptococcus mutans. It also improves our understanding of the functional links between stress response and cell death/lysis process.

1. INTRODUCTION
Streptococcus mutans is highly associated with the development of caries, a classic biofilm disease on the tooth surface (Loesche, 1986). The capacity to form biofilms is a major virulence trait of this organism and is closely related to its ability to persist in the oral cavity which experiences dynamic environmental changes such as nutrient limitation (Bowen, Burne, Wu, & Koo, 2018; Burne, 2018; Burne et al., 2012; Koo, Allan, Howlin, Stoodley, & Hall‐Stoodley, 2017; Lemos & Burne, 2008). Regulated cell death and lysis of a bacterial subpopulation which has been damaged by adverse environments have been suggested to be a survival strategy required for the formation of mature pathogenic biofilms (Bayles, 2003, 2007, 2014; Lewis, 2000; Rice & Bayles, 2008). In this respect, we have studied the S. mutans Cid/Lrg system, consisting of the two paralogously related dicistronic operons, lrgAB, and cidAB, encoding predicted membrane‐associated proteins (Ahn, Rice, Oleas, Bayles, & Burne, 2010). The CidA and LrgA proteins share predicted structural features with the bacteriophage‐encoded holin family of proteins, known to be involved in regulating the timing of host cell lysis during bacteriophage lytic infection (Bayles, 2003, 2007, 2014; Groicher, Firek, Fujimoto, & Bayles, 2000; Rice et al., 2003). Interestingly, mutation of the cid and lrg operons has demonstrated their impact on comprehensive S. mutans virulence traits, such as autolysis, biofilm development, oxidative and heat stress responses, antibiotic resistance, and genetic competence (Ahn, Gu, Koh, & Rice, 2017; Ahn et al., 2010; Rice, Turner, Carney, Gu, & Ahn, 2017). Most recently, it has been revealed that S. mutans LrgAB functions as a pyruvate uptake system that appears to function during stationary phase (when the primary carbon source is completely exhausted) (Ahn et al., 2019). Pyruvate is a central carbon metabolite and mediates several key pathways, such as the production of acetyl‐phosphate, organic acids and ATP, and the conversion of NAD/NADH. Given that pyruvate can also function as an effective scavenger of ROS, including hydrogen peroxide (H2O2) (Ahn et al., 2019), this molecule may serve as a central metabolic signal that coordinates the involvement of Lrg in coping with environmental stresses and modulating homeostasis, possibly initiating a metabolic response to induce cell death and lysis in a subpopulation.
Regulation of S. mutans cidAB and lrgAB expression is quite interesting, in that RNA levels and promoter activities of cidAB and lrgAB are counterbalanced throughout growth and in response to the availability of glucose and oxygen (Ahn et al., 2010). For example, in the early‐exponential phase, cid expression is dominant and lrg is repressed. As cells enter the stationary phase, lrg activated through the LytST complex, whereas cid expression diminishes (Ahn et al., 2010). Expression of the cid and lrg operons more sensitively responds to glucose levels. In particular, the lrg genes are only induced in cultures containing lower levels of glucose (≤15 mM) but are repressed in cultures containing glucose at concentrations of 20 mM and higher (Ahn et al., 2010). This expression pattern is correlated with the capacity of S. mutans to take up extracellular pyruvate into the cell at the stationary phase (Ahn et al., 2019). Oxygen levels also influence the magnitude of pyruvate flux during the growth of this organism (Ahn et al., 2019). Our previous study revealed that the expression of the cid and lrg operons is directly mediated by the global transcriptional regulator CcpA in response to glucose levels (Kim, Waters, Turner, Rice, & Ahn, 2019). Interestingly, two potential cre sites (for CcpA binding) in the cid promoter are arranged similarly to those in the lrg promoter region (Kim et al., 2019). CcpA also appeared to have the opposite effect on the regulation of cid (positive) and lrg (negative), suggesting that CcpA binding is an important mediator of cid and lrg transcriptional responses to glucose levels (Kim et al., 2019). Nevertheless, it does not appear that CcpA effects alone are sufficient to understand and explain the observed patterns of cid and lrg expression in response to glucose since the repression of lrgAB promoter activity was not relieved in the absence of CcpA when cells were cultivated in high‐glucose medium (Kim et al., 2019).
CcpA often works together with CodY (a global transcription regulator that participates in starvation adaptation) to sense changes in nutrient availability and coordinates, directly and indirectly, the expression of hundreds of genes involved in carbon and nitrogen metabolism of gram‐positive bacteria (Fujita, Satomura, Tojo, & Hirooka, 2014; Gorke & Stulke, 2008; Kim & Burne, 2017; Santiago, Marek, Faustoferri, & Quivey, 2013; Shivers, Dineen, & Sonenshein, 2006). CodY controls transcription by binding to a moderately conserved 15‐nucleotide (nt) consensus motif (AATTTTCNGAAAATT) in the vicinity of the promoter region of the target genes (Belitsky, 2011; Kim & Burne, 2017; Lemos, Nascimento, Lin, Abranches, & Burne, 2008), by competing with a positive regulator for binding or by serving as a roadblock to RNA polymerase (Belitsky, 2011). The DNA‐binding affinity of CodY is enhanced by its interaction with the branched‐chain amino acids (BCAAs; isoleucine, leucine, and valine) and GTP that act as effector molecules or signals of the nutritional status of the cell (Brinsmade & Sonenshein, 2011; Guedon, Serror, Ehrlich, Renault, & Delorme, 2001; Handke, Shivers, & Sonenshein, 2008; Ratnayake‐Lecamwasam, Serror, Wong, & Sonenshein, 2001; Shivers & Sonenshein, 2004; Sonenshein, 2005). CodY acts mainly as a repressor, and many of the genes which encode components of metabolic pathways are repressed during growth in the presence of excess nutrients and involved in adaptation to poor growth conditions (Sonenshein, 2007). In our previous microarray data, comparing RNA expression profiles of wild‐type and lytS‐deficient strains between early‐ and late‐exponential growth phases in BHI medium, it was shown that codY was markedly upregulated at early‐exponential phase by about 4.4‐fold and 2.4‐fold in both wild‐type and lytS‐deficient strains, respectively, compared to late‐exponential growth phase in both wild‐type and lytS‐deficient strains (Ahn, Qu, Roberts, Burne, & Rice, 2012). Thus, we assumed that this growth‐dependent expression of codY may be involved in the regulation of cid and lrg operons and may be, at least partly, responsible for the observed inversely correlated pattern of cid and lrg expression in response to the growth phase. In this study, we demonstrate that the cid and lrg promoter regions are direct targets of S. mutans CodY, using electrophoretic mobility shift assays (EMSAs). The potential binding sequences in each promoter region were also identified by DNase I footprinting analyses. We also show that lack of CodY influences pyruvate flux. The data presented clearly indicate that the balanced regulation between cid and lrg in response to carbohydrate availability and nutritional status of the organism is coordinated by the activities of CodY, together with CcpA, previously shown to directly regulate cid and lrg.
2. METHODS
2.1. Bacterial strains and growth conditions
Streptococcus mutans UA159 and its derivatives were cultured in brain heart infusion (BHI) medium (Difco Laboratories, Detroit, MI) or chemically defined medium FMC (Terleckyj & Shockman, 1975) containing 11 mM (low‐glucose) or 45 mM (high‐glucose) at 37°C in a 5% CO2 incubator. The medium was supplemented by sodium pyruvate (Fisher Scientific) or β‐fluoropyruvic acid sodium salt monohydrate (3FP, Sigma‐Aldrich), as necessary. Antibiotics were used to supplement growth media in the following concentrations: kanamycin (1 mg/ml), spectinomycin (1 mg/ml), and tetracycline (10 μg/ml). For growth measurements, fresh medium was inoculated with 1:100 dilutions of overnight cultures of S. mutans. The optical density at 600 nm (OD600) was measured at 37°C at 30‐min intervals using a Bioscreen C growth curve analysis system. At least three independent experiments, each in quadruplicate, were performed. A representative result is presented in each corresponding figure.
2.2. Mutant construction
A strain constitutively expressing codY was constructed as previously described (Ahn & Rice, 2016). Briefly, the promoter region of codY (PcodY) was replaced by a fragment (ΩKm‐Pldh) containing a polar kanamycin resistance gene (ΩKm) and an ldh promoter region (Pldh). For this, two ~0.5‐kb fragments flanking the −35 and −10 sequences of the codY promoter were PCR‐amplified, ligated into the ΩKm‐Pldh cassette, and transformed into S. mutans. The lytST‐overexpressing strain (SAB163) was recently constructed (Ishkov, Ahn, Rice, & Hagen, 2020). To delete the codY gene in SAB163, we amplified a region containing the tetracycline gene (replacing codY) and its flanking arms (~0.5‐kb) in the ΔcodY strain by PCR and transformed into SAB163. Transformants were selected on BHI agar containing kanamycin (for a strain constitutively expressing codY) and tetracycline (for the deletion of codY in SAB163), and double‐crossover recombination into each gene was confirmed by PCR and sequencing to ensure that no mutations were introduced into flanking genes. The codY‐overexpressing (SAB399) and codY‐deficient strains in the lytST overexpression background (SAB392) were then each transformed by the PlrgA‐gfp construct for conducting the GFP promoter reporter assays described below.
2.3. Microplate promoter reporter assay
For measurement of GFP fluorescence of S. mutans strains harboring PlrgA‐gfp or PcidA‐gfp promoter fusion constructs (Kim et al., 2019), their overnight cultures were diluted 1:50 into 1.5 ml of FMC media and grown to an OD600 = 0.5. The cultures were diluted 1:50 into 175 μl FMC in individual wells of a 96‐well plate (black walls with clear bottoms; Corning Incorporated) and incubated at 37°C. The optical density at 600 nm (OD600) and green fluorescence (excitation 485/20 nm; emission 520/20 nm) were monitored at 30‐min intervals with a Synergy 2 multimode microplate reader (BioTek) controlled by Gen5 software. Fluorescence intensity was calculated by subtracting the fluorescence of wild‐type harboring plasmid without the reporter gene fusion from fluorescence readings of the S. mutans strains harboring the PlrgA‐gfp or PcidA‐gfp gene fusion.
2.4. Electrophoretic mobility shift assays (EMSAs) and DNase I footprinting assays
For EMSAs, DNA probes containing the promoter regions of cid or lrg were amplified by primers labeled on their 5′ end with biotinylated nucleotides. To delete each predicted cre site(s) or CodY‐box in cid or lrg promoter region, biotinylated DNA probes were amplified with primers with the desired base deletions in each promoter sequence. Four fmol of biotin‐labeled promoter regions of cid and lrg was incubated in combination with the increasing amount (0 and 25 pmol) of purified, recombinant His6‐tagged CodY protein (Kim & Burne, 2017) in a 10‐μl reaction mixture containing 10 mM HEPES (pH 7.8), 50 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol (DTT), 1 mM EDTA, 1 μg poly (dI‐dC), 1 μg bovine serum albumin (BSA), and 10% glycerol. The binding reaction, sample separation in a polyacrylamide gel, and DNA signal detection were performed as described previously (Kim et al., 2019). To map CodY‐box site(s) in the promoter regions of lrg and cid, DNase I footprinting assays were also carried out using a nonradiochemical capillary electrophoresis method (Yindeeyoungyeon & Schell, 2000), as described previously (Kim et al., 2019).
2.5. Measurement of extracellular pyruvate
Streptococcus mutans strains were grown in a low‐glucose (11 mM) FMC medium. Time‐course measurements of extracellular pyruvate during growth were measured as described previously (Ahn et al., 2019). Briefly, for monitoring growth, samples (200 μl) were taken at 1‐ to 2‐hr intervals and half of this volume (100 μl) was used to measure the optical density at 600 nm in a spectrophotometer. The other half (100 μl) was used to quantify extracellular pyruvate concentrations in the supernatant with an EnzyChrom™ Pyruvate Assay Kit (BioAssay Systems). The results are an average of two independent replicates, each performed in duplicate.
2.6. Quantitative real‐time PCR (qPCR) assay
To measure the expression of genes using qPCR, S. mutans UA159 and ΔcodY mutant strains were grown in BHI broth at 37°C in a 5% (vol/vol) CO2 atmosphere. To measure the growth‐dependent expression of cidA and lrgA genes, cells were harvested in early‐exponential (EE) and early‐stationary (ES) growth phases. Extraction of RNA, qPCR, and data analysis was performed as described elsewhere (Ahn et al., 2012; Ahn & Rice, 2016; Rice et al., 2017). Expression was normalized against an internal standard (gyrA), and data were presented as the relative copy number of mRNA (copies/μl). Statistical analyses were performed on data generated from n = 3 independent experiments using an unpaired t test.
3. RESULTS
3.1. CodY is involved in the regulation of cid and lrg expression
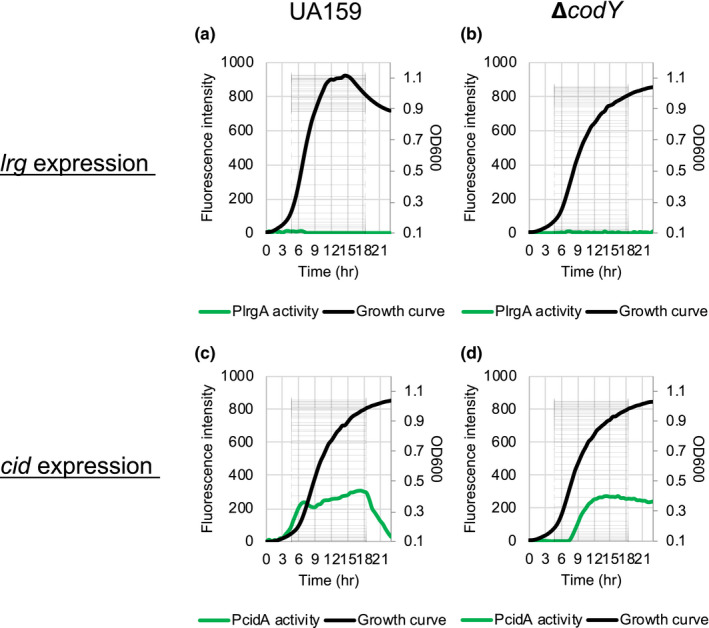
Given that lrgAB expression is induced in stationary phase and its expression is strongly repressed in the presence of excess glucose (Ahn et al., 2010, 2019), we assumed that the global stationary phase regulator CodY (Sonenshein, 2005, 2007) may be involved in the regulation of lrgAB and possibly cidAB. To assess whether CodY is involved in the regulation of lrgAB and cidAB, we transformed the PlrgA‐gfp and PcidA‐gfp reporter constructs, used in our previous study (Kim et al., 2019), into the ΔcodY mutant strain (Lemos et al., 2008), and monitored the expression of both lrgAB and cidAB in low‐ and high‐glucose cultures, allowing a strong stationary phase induction of lrgAB and cidAB, respectively (Ahn et al., 2010; Kim et al., 2019). We first confirmed that lrgAB promoter activity is sharply induced at the onset of the stationary phase in low‐glucose cultures (Figure 1a), similar to that reported in our previous studies with this transcriptional GFP reporter construct (Kim et al., 2019). Notably, the absence of the codY gene resulted in about a 40% reduction of lrgAB expression at the stationary phase (Figure 1b), compared to that of the wild‐type strain (Figure 1a). In high‐glucose cultures, no obvious induction of lrgAB was observed in both the absence and presence of CodY (Figure A1a,b in Appendix). In contrast, cid promoter (Pcid) activity was dramatically repressed at stationary phase in the low‐glucose culture of the wild‐type strain (Figure 1c), compared to Plrg activity in the same condition (Figure 1a), but cidAB showed higher promoter activity during exponential growth compared to stationary phase, as previously reported (Ahn et al., 2010). Notably, exponential phase cid induction was also markedly reduced in the absence of the codY gene in wild‐type low‐glucose cultures (Figure 1d). In high‐glucose cultures, cidAB promoter activity was comparable between wild type and the ΔcodY mutant strain (Figure A1c,d in Appendix 1). However, the absence of CodY resulted in repression of early‐exponential phase induction of cidAB during growth in high‐glucose cultures, compared to that in the wild type. These results suggest that CodY positively regulates the activity of both lrgAB and cidAB in low‐glucose culture conditions. To verify these read‐outs, we also monitored changes in the expression of lrg and cid during the transition to stationary phase in BHI, using quantitative real‐time PCR (qRT‐PCR), and observed a similar pattern in lrgAB and cidAB expression (Table A1 in Appendix 1).
FIGURE 1.
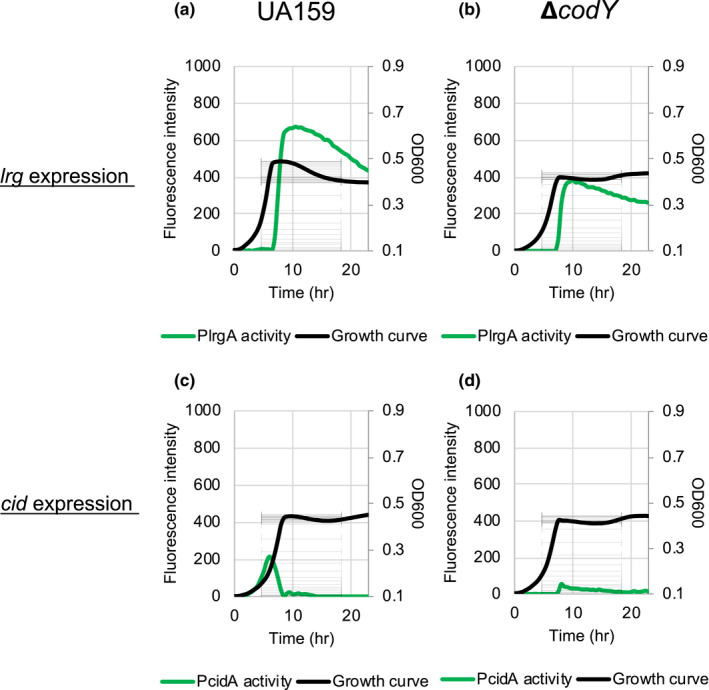
Effect of CodY on Streptococcus mutans lrg and cid promoter activity over the growth in the low‐glucose cultures. Strains harboring PlrgA‐gfp (a and b) and PcidA‐gfp (c and d) constructs in pDL278 in the S. mutans UA159 wild‐type (a and c) and ∆codY (b and d) backgrounds were grown in a chemically defined FMC medium, supplemented by 11 mM glucose. Relative gfp expression (colored lines) and OD600 (black lines) were monitored on a plate reader (see Materials and Methods for details). The results are representative of three independent experiments
3.2. Both lrgAB and cidAB are regulated by direct binding with CodY
Given the potential involvement of CodY in the regulation of both cidAB and lrgAB, we further inspected the promoter region of each operon to identify potential CodY‐binding site(s). In the cid promoter region, we discovered a potential CodY‐binding site (AAcTTTCTGAAAAgT) with two mismatches to the consensus sequence, AATTTTCWGAAAATT (Belitsky & Sonenshein, 2008), at positions −30 to −16 with respect to the transcription initiation start (TIS) site of cidA, partially overlapped with cid‐cre2, recently identified for CcpA binding (Kim et al., 2019), and predicted −35 region of the promoter (Figure 2a). Although we were unable to identify a well‐conserved CodY‐binding site in the lrg promoter region, a candidate CodY‐binding site (gtTTTaCAcAAAATg, at positions −30 to −16 with respect to TIS of lrgA) was recognized, which contains 5 mismatches to the consensus sequence (Figure 2b). This CodY‐binding site also partially overlaps the lrg‐cre2 for CcpA binding (Kim et al., 2019) and is located immediately downstream of the predicted −35 region of the promoter (Figure 2b). Next, to determine whether CodY is capable of binding to the lrg and/or cid promoter regions, including the predicted CodY‐binding site, electrophoretic mobility shift assays (EMSAs) were performed with increasing amounts of purified His‐CodY and biotin‐labeled cid or lrg promoter regions. The CodY protein bound to both promoter regions of cid (Figure 2c, lane 2) and lrg (Figure 2d, lane 2), so migration of labeled cid and lrg promoter fragments was hindered in the presence of CodY. Unlabeled promoter regions of lrg and cid inhibited the binding of labeled DNA probes to CodY (Figure 2c,d, lane 3, respectively), indicating that the binding interactions are specific. Branched‐chain amino acids (BCAAs; leucine, isoleucine, valine) have been known to be effector molecules for CodY binding to DNA promoter regions in S. mutans (Santiago et al., 2013). When BCAAs were used in the binding assays for CodY with the promoter regions of cid or lrg, 10 mM BCAAs substantially enhanced CodY binding to both cid (Figure 2c, lane 4) and lrg (Figure 2d, lane 4) promoter regions. These data suggest that interactions between CodY and the lrg (or cid) promoter occur more effectively in the presence of BCAAs and implicate CodY as another major regulator of lrg and cid expression, together with CcpA, recently shown to directly bind to the cid and lrg promoter regions (Kim et al., 2019).
FIGURE 2.
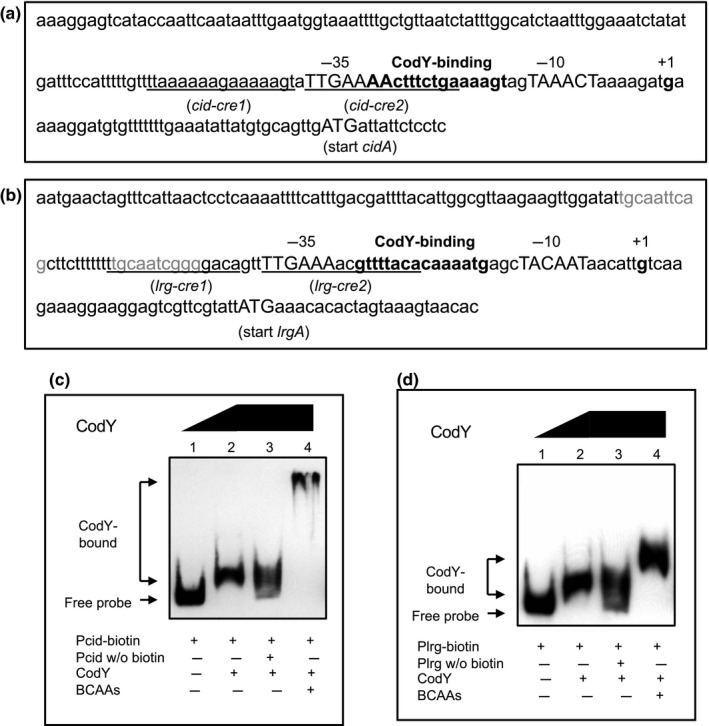
Potential binding site and activity of CodY in the cid and lrg promoters. a, b; The promoter regions of cid (a) and lrg (b). The potential CodY‐binding sites are displayed in bold font. The transcriptional start sites (TSSs) are numerically denoted (+1) in bold font, and −10/−35 sequences indicated by capital letters. The hypothetical cre sites are underlined. Predicted LytT DNA‐binding motif in the lrg promoter region is indicated in gray font. c, d; Electrophoretic mobility shift assays (EMSAs) of CodY binding with the promoter region of cid (c) or lrg (d). Biotin‐labeled promoter regions (4 fmol) of cid (a) and lrg (b) were incubated with the increasing amount of purified His‐CodY protein (0 and 25 pmol). Unlabeled promoter regions (0.3 pmol; lane 3) or BCAAs (10 mM; lane 4) were added in the binding reaction mixture, respectively. The reactions were run on a non‐denaturing polyacrylamide gel and the signal observed via chemiluminescence
3.3. Identification of binding sites for CodY in the cid and lrg promoter regions
EMSAs showed that CodY could directly bind to the promoter region of cid and lrg (Figure 2c,d, respectively). First, to identify the precise location of binding by CodY on the cid promoter region, DNase I footprinting experiments using capillary electrophoresis (fragment analysis) were conducted using a PCR‐amplified 6‐carboxyfluorescein (6‐FAM)‐labeled DNA probe of the cid promoter region (201‐bp) bound to CodY under the same conditions as used for EMSAs. Sites protected from DNase I digestion by DNA binding by CodY were visualized as regions lacking discernable peaks compared to a control reaction mixture containing BSA (bovine serum albumin). Reaction mixtures containing CodY yielded only a single region of protection spanning from −77 to −54, with respect to the ATG start site of the cidA gene. This protected region corresponded to the predicted cid‐cre2 (nt −73/−59) and CodY‐binding sites (nt −68/−54) (Figure 3a), showing that two predicted cre sites are involved in binding with CcpA, as well as CodY, and provide further support that both CcpA and CodY proteins may work together to coordinate cid expression. To investigate the requirement of the protected regions in DNA binding by CodY, the entire cid‐cre1 or cid‐cre2 site was deleted, and the binding affinity of CodY to these deletion constructs was measured by EMSA. The binding of the deleted cid‐cre1 probe by CodY was weak, showing a slight smear of the probe on the gel (Figure 4a, lane 6). When we tested the ability of CodY to bind the probe in which cid‐cre2 (including part of predicted CodY‐binding site) was deleted, the probe was less bound by CodY, resulting to more smears on the gel (Figure 4a, lane 9), supporting the contribution of the predicted CodY‐binding sequence (Figure 2a). The data show that the cid‐cre2 site seems to be more important for CodY binding relative to the cid‐cre1 site and also suggest that CodY and CcpA may have some competitive interactions on the cid promoter. In contrast, in the lrg promoter region, CodY binding occurred at two separate sites, as revealed by the DNase I footprinting experiment (Figure 3b). The upstream site, designated lrg‐CodY1 (AgTTggatAttgca; mismatches are indicated by lowercase letters), mapped to the positions −114 to −101 with respect to the ATG start site of the lrgA gene, but mismatched to the consensus sequence (AATTTTCWGAAAATT) (Belitsky & Sonenshein, 2008). The second site, designated lrg‐CodY2 (AGTTTTGAAAACGTTT), extended from −70 to −55 with respect to the ATG start site of the gene included a 15‐bp CodY motif (gtTTTaCAcAAAATg; mismatches are indicated by lowercase letters) with five mismatches to the consensus. Since two potential CodY‐binding sites (lrg‐CodY1 and lrg‐CodY2) were identified in the lrg promoter region (Figure 3b), we tested the ability of CodY to bind the probe in which either lrg‐CodY1 or lrg‐CodY2 was deleted. As shown in Figure 4b, both deletions impeded the full migration of labeled lrg promoter region, showing a strong smear of the probe on the gel, although they could not completely interfere with CodY binding. These data suggest that the regions identified by DNase I footprinting (Figure 3) may contribute to the CodY binding to the lrg promoter.
FIGURE 3.
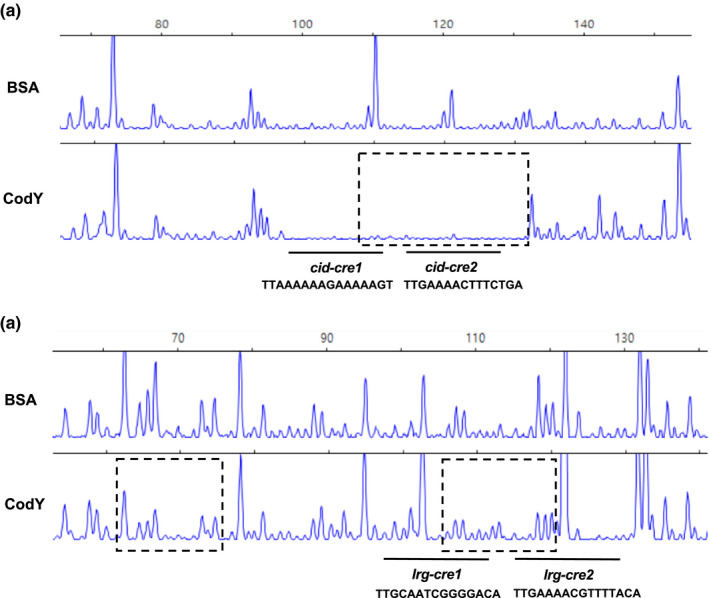
DNase I footprinting assays of CodY binding to the cid and lrg promoter regions. Dotted boxes depict the regions protected from DNase I digestion upstream of cid (a) and lrg (b) by CodY. The electropherograms represent control DNA with BSA (bovine serum albumin) on top and footprints with of CodY in the bottom of each panel
FIGURE 4.
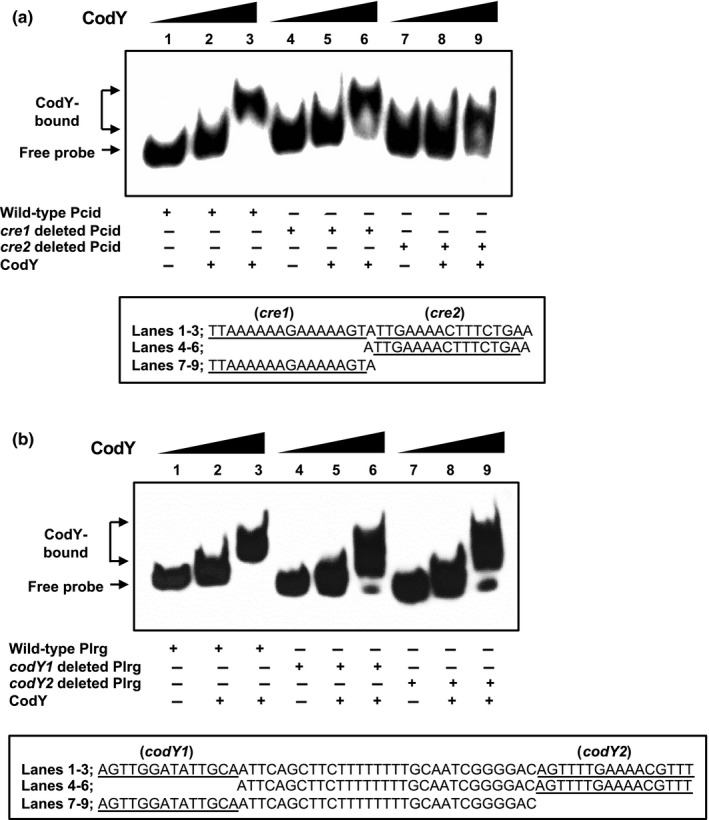
Effects of deleting predicted cre and CodY‐box on CodY binding to the cid and lrg promoter regions. Underlined and letters indicate putative CcpA‐binding (cre1 and cre2) and CodY‐binding (CodY1 and CodY2) sequences in the cid (a) and lrg (b) promoter regions, respectively. EMSAs were performed with 1 fmol biotinylated cid (Pcid) and lrg (Plrg) regions with a deletion in the putative cre and CodY‐box sequences and various amounts of purified His‐CodY. Each binding reaction mixture contains 0, 6.25, or 12.5 pmol of His‐CodY protein. (a) Lanes 1–3 contain biotinylated Pcid wild‐type DNA probes; lanes 4–6 contain biotinylated cre1 deleted DNA probes; and lanes 7–9 contain biotinylated cre2 deleted DNA probes. (b) Lanes 1–3 contain biotinylated Plrg wild‐type DNA probes; lanes 4–6 contain biotinylated CodY1 deleted DNA probes; and lanes 7–9 contain biotinylated CodY2 deleted DNA probes. The reactions were run on a non‐denaturing polyacrylamide gel and the signal observed via chemiluminescence. Presented results are representative of two independent experiments
3.4. Lack of CodY affects stationary phase pyruvate uptake through LrgAB
While the role of CidAB remains enigmatic, we recently reported that LrgAB is responsible for reuptake of pyruvate, excreted during growth as an overflow metabolite (Ahn et al., 2019). Thus, we evaluated whether the transcriptional change of lrgAB by lack of CodY affects the capacity of S. mutans to take up pyruvate. For this, we monitored the level of extracellular pyruvate during growth in a low‐glucose FMC medium using a Pyruvate Assay Kit (Ahn et al., 2019). In wild‐type culture, pyruvate reuptake initiated at the onset of the stationary phase (Figure 5a), as reported previously (Ahn et al., 2019). In the ΔcodY strain, however, reuptake of pyruvate was delayed relative to wild type, initiating after about an hour delay and reuptake was overall reduced, with considerable amounts of pyruvate left in the medium (Figure 5b), consistent with the observation that stationary phase lrgAB expression is reduced in the ΔcodY mutant (Figure 1b). It is also noteworthy that the maximum level (approx. 200 μM) of pyruvate excreted during the growth of the ΔcodY strain was about 45% reduced compared to wild type, suggesting that lack of CodY may accelerate pyruvate metabolism in the cell, consequently decreasing the amount of overflow pyruvate excretion. Given that the level of accumulated pyruvate is the primary stimulus for lrgAB induction and subsequent pyruvate uptake (Ahn et al., 2019), reduced pyruvate excretion (Figure 5b) may be responsible, at least in part, for the reduced stationary phase lrgAB expression (Figure 1b) and pyruvate uptake (Figure 5b) in the ΔcodY strain.
FIGURE 5.
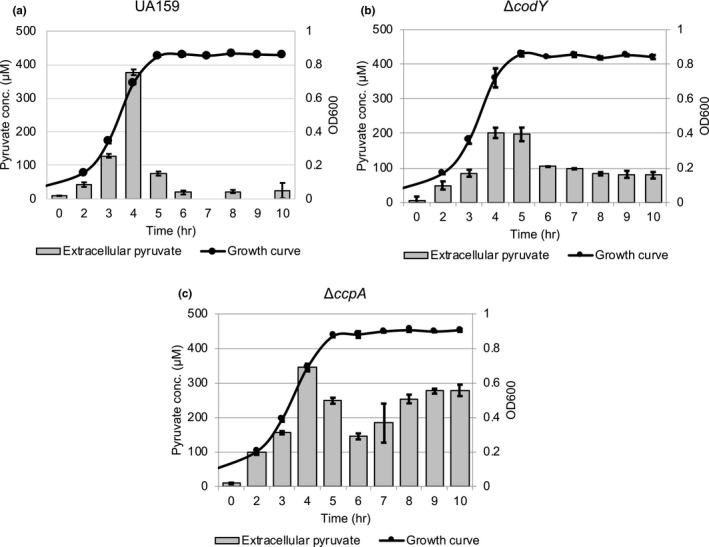
Measurement of extracellular pyruvate during the growth of Streptococcus mutans wild‐type,ΔcodY, ΔccpA strains. The wild‐type UA159 (a), ΔcodY (b), and ΔccpA (c) strains were grown in the FMC medium supplemented by 11 mM glucose. For time‐course measurements of extracellular pyruvate and growth, samples were taken at 1‐ or 2‐hr intervals (see Materials and Methods for details). The concentration of pyruvate was determined using an EnzyChrom™ Pyruvate Assay Kit and growth was measured by the optical density at 600 nm (OD600). Bars indicate the concentration of extracellular pyruvate; and solid line with circles indicates the growth curve. The results are an average of two independent experiments
3.5. Lack of CodY affects pyruvate flux during exponential growth
We previously showed that supplemented pyruvate had no impact on the growth rate of wild‐type cells but delayed the onset of the stationary phase (Ahn et al., 2019). Therefore, we wondered whether the lack of CodY affects the ability of S. mutans to utilize pyruvate during the stationary phase. A similar growth trend to that observed in the wild‐type strain (Figure 6a) was observed in the ΔcodY strain (Figure 6b), suggesting that the imported pyruvate could be normally utilized for further cell growth when the primary carbon source is exhausted. To further evaluate the effect of codY deficiency on pyruvate flux and utilization, we also monitored the growth of wild‐type and codY mutant strains in the presence of increasing amounts (0, 0.1, 1, and 10 mM) of 3‐fluoropyruvate (3FP), previously shown to compete with pyruvate and inhibit cell growth by binding to PDH complex in the cell (Apfel, Ikeda, Speckhard, & Frey, 1984; Flournoy & Frey, 1989; Lang, Leystra‐Lantz, & Cook, 1987). As shown in Figure 7a, wild‐type cells displayed normal growth in the presence of 0.1 and 1 mM 3FP, but demonstrated a growth defect in the presence of 10 mM 3FP, as previously observed (Ahn et al., 2019). In contrast, the ΔcodY mutant was unable to normally grow in the presence of 1 mM 3FP and no growth was observed in the presence of 10 mM 3FP (Figure 7b), suggesting that lack of CodY affected exponential phase pyruvate uptake or utilization, likely in an Lrg‐independent manner. We also cultivated the PlrgA‐gfp reporter strain in this same condition to measure lrgAB promoter activity. In the wild‐type background, stationary phase expression of lrgAB was markedly reduced in the presence of 0.1 mM 3FP (Figure A2a,b in Appendix 1) and almost completely inhibited at concentrations of 1 mM and 10 mM 3FP (Figure A2c,d, respectively, as previously observed (Ahn et al., 2019). Somewhat surprisingly, a similar trend of lrgAB expression in response to 3FP was also observed in the ΔcodY background, although the overall levels of lrgAB expression were lower in the ΔcodY background than in the wild‐type background (Figure A3e–h). Therefore, these results suggest that the growth defect of the ΔcodY strain by 3FP may be independent of lrgAB expression, possibly due to the perturbation of pyruvate transport and metabolism during exponential growth by lack of CodY.
FIGURE 6.
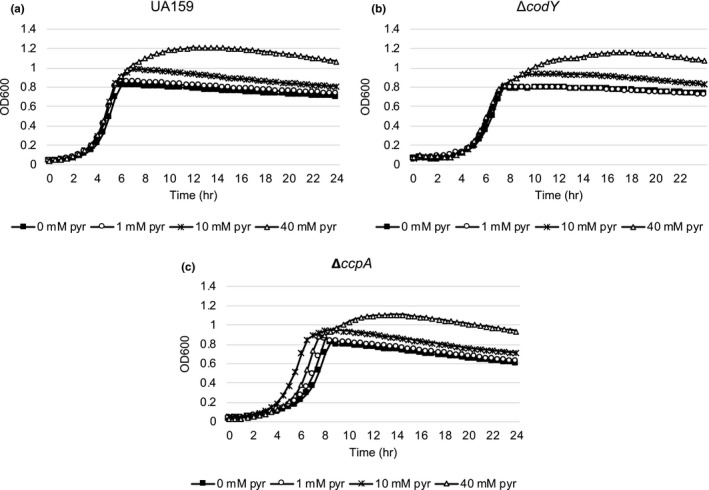
The effect of exogenously added pyruvate on the growth of Streptococcus mutans wild‐type,ΔcodY, and ΔccpA strains. The wild‐type UA159 (a), ΔcodY (b), and ΔccpA (c) strains were each grown in a low‐glucose (11 mM) FMC medium supplemented by different concentrations of pyruvate (0, 1, 10, and 40 mM). Optical density at 600 nm was monitored every 30 min at 37°C using the Bioscreen C lab system. The results are representative of three independent experiments
FIGURE 7.
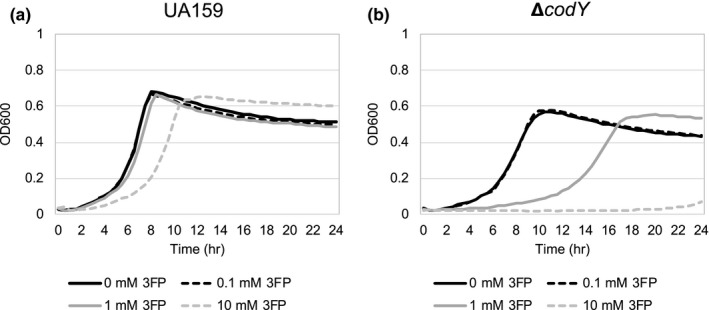
The effect of exogenously added 3‐fluoropyruvate (3FP; pyruvate analog) on the growth of Streptococcus mutans wild‐type and ΔcodY strains. Wild type (a) and ΔcodY mutant (b) were each grown in a low‐glucose (11 mM) FMC medium supplemented by different concentrations of 3FP (0, 0.1, 1, and 10 mM). Optical density at 600 nm was monitored every 30 min at 37°C using the Bioscreen C lab system. The results are representative of three independent experiments
3.6. Overexpression of codY does not affect lrgAB expression
The finding that an identified lrg‐CodY1 site (Figure 2b) partially overlaps a predicted LytT‐binding motif in the lrg promoter (Kim et al., 2019) suggests a possible interplay between LytT and CodY in activating the lrgAB promoter, in agreement with the role of CodY as a positive regulator (Figure 1a,b). To evaluate the role of CodY in regulating lrgAB, we created a strain (SAB399), constitutively expressing codY from ldh promoter in the wild‐type background, as previously described (Ahn & Rice, 2016). Then, we transformed the PlrgA‐gfp reporter constructs (Kim et al., 2019) into SAB399 and monitored the expression of lrg in the low‐glucose condition. However, overexpression of codY had no obvious effect on lrgAB expression (Figure A3a,b in Appendix 1), suggesting that the accessibility of LytT to the lrg promoter is not fully controlled by CodY. In contrast, overexpression of lytST (SAB163) in a wild‐type background led to a dramatic increase of stationary phase lrgAB expression (Figure A3c in Appendix 1), as recently reported (Ishkov et al., 2020). To evaluate the effect of CodY on the potential interaction with LytT, we also inactivated codY in the lytST overexpression background (SAB392), followed by transformation with the PlrgA‐gfp reporter construct, and monitored the expression of lrg in low‐glucose cultures. As shown in Figure A3d in Appendix 1, deficiency of CodY still reduced the stationary phase expression of lrgAB in the lytST overexpression strain, an expression pattern similar to that observed in the ΔcodY strain (Figure 1b). These results suggest that the effect of CodY on lrgAB promoter activity is independent of LytST.
3.7. CcpA is also involved in pyruvate flux and utilization
Since the predicted LytT‐binding motif overlaps predicted CcpA‐binding site (lrg‐cre1) in the lrg promoter (Figure 2b), we assumed that together with CodY, CcpA may be also involved in the ability of S. mutans to take up and utilize extracellular pyruvate. For this, we monitored the level of extracellular pyruvate during the growth of the ΔccpA strain in the same condition used above (Figure 5a,b). As shown in Figure 5c, pyruvate excretion in the ccpA mutant was similar to that observed in the wild‐type strain, but reuptake of pyruvate at the stationary phase was marked decelerated in the ccpA mutant, with more than 75% of excreted pyruvate left in the medium in stationary phase. This result was quite surprising, because stationary phase lrgAB expression was elevated in the ΔccpA strain, relative to the wild‐type strain (Kim et al., 2019). We also evaluated the ability of the ΔccpA strain to utilize pyruvate in the stationary phase by growing S. mutans in low‐glucose FMC medium, supplemented by different concentrations of pyruvate, as described above (Figure 6a,b). As shown in Figure 6c, the addition of external pyruvate to the medium effectively prolonged exponential growth of the ΔccpA strain but interestingly, it also enhanced the growth rate of the organism. Taken together, these data suggest that although CcpA regulates stationary phase lrgAB expression (Kim et al., 2019), it also appears to affect exponential growth by rerouting pyruvate metabolism.
4. DISCUSSION
The ability of S. mutans to maintain balanced carbon metabolism in response to environmental conditions, particularly in the transition from conditions that allow exponential growth, to conditions that limit growth, is an important virulence attribute of this organism that promotes its survival and persistence during periods of “feast or famine” in oral cavity. Through our previous studies, we have demonstrated that the Cid/Lrg system is regulated in a growth phase‐dependent manner and by glucose and oxygen levels (Ahn et al., 2010, 2017; Ahn & Rice, 2016). In particular, the findings that the lrg genes are specifically induced at stationary phase (Ahn et al., 2010; Kim et al., 2019), and that their gene products are required for reuptake of pyruvate (Ahn et al., 2019), further support the importance of the Cid/Lrg system in preserving cellular homeostasis when exogenous carbohydrate has been depleted in the environment. The findings presented here reveal that the global transcriptional regulator CodY directly regulates transcription of the cid and lrg genes in S. mutans. CodY is known to repress stationary phase gene expression during exponential phase in many bacteria (Sonenshein, 2005). In our previous microarray data for comparison of early‐ versus late‐exponential growth phases in the wild type, it was shown that the codY gene was >4‐fold upregulated at early‐exponential phase, compared to that of late‐exponential growth phase (Ahn et al., 2012; Kim et al., 2019), indicating that CodY may actively repress the stationary phase induced genes, including lrg, during exponential growth. Given that LytT is required for expression of lrgAB and that lrgAB expression is reduced in the ΔcodY mutant strain, we assumed that CodY may be required for full activity of LytT. This idea may be supported by the finding that one of predicted CodY‐binding sites (lrg‐CodY1) partially overlaps a predicted LytT‐binding motif in the lrg promoter. However, the finding that the overexpression of codY had no evident effect on the stationary phase expression of lrgAB suggests that the abundance of CodY alone does not affect the accessibility of LytT to the promoter region. Nevertheless, based on the EMSA and DNases I footprinting data, it is likely that CodY may interact with LytT on the lrg promoter. Notably, unlike the codY gene, the lytT gene was >14‐fold downregulated at early‐exponential phase, compared to that of late‐exponential growth phase (Ahn et al., 2012; Kim et al., 2019). Thus, a possible scenario for the interplay between CodY and LytT may be that expression of lrg is strongly repressed by CodY during exponential growth, and as the cells approach stationary phase, CodY may interact with LytT in order to activate the lrgAB promoter. This regulatory switch for lrg expression may be affected by intracellular metabolic status. In this study, we showed that BCAA enhanced the binding of CodY to lrg (and cid) without a further requirement for GTP, suggesting that BCAAs are predominant co‐effectors in mediating an interaction between CodY and lrg (or cid), and the cid and lrg operons respond to changes in amino acid availability. Therefore, it is possible that CodY loses its repressing activity as a result of the decrease in the intracellular levels of BCAAs and/or GTP that occurs at the onset of stationary phase, or by interacting with LytT. The levels of BCAAs may reflect the metabolic/nutritional status of cell, allowing CodY to coordinate a variety of genes that are induced when S. mutans cells make the transition from rapid exponential growth to stationary phase. However, it does not appear that CodY competes with LytT for the potential binding site on the lrgAB promoter, because (a) lack of CodY does not release the repression of lrgAB during exponential growth and (b) it still markedly reduces the stationary phase expression of lrgAB in the strain overexpressing lytST, showing dramatic elevation of lrgAB expression at stationary phase. Thus, the access of LytT to the lrgAB promoter or its full function as an activator may require CodY at stationary phase. To address this idea, we are currently investigating the potential interaction between CodY and LytT in the regulation of lrgAB. Given that CodY is a major repressor of stationary phase gene expression during exponential phase, and deficiency of CodY renders the cell more sensitive to 3FP during growth, it is also possible that CodY may be involved in both carbohydrate and pyruvate metabolism, related to cell growth. In fact, the S. mutans ΔcodY strain grows slowly (Lemos et al., 2008), compared to the wild‐type strain.
More interestingly, the predicted LytT‐binding motif, partially overlapping with the lrg‐CodY site, also overlaps the predicted cre1 site (for CcpA binding) in the lrg promoter, indicating that activation of lrg by LytT is influenced by both CcpA and CodY. In our previous EMSAs, it is quite apparent that CcpA has a higher binding affinity to the cid and particularly lrg promoter regions than CodY, unless the level of BCAAs is increased in the culture (Kim et al., 2019). The addition of BCAAs to the EMSA experiments was more effective at promoting CodY binding to the cid promoter than to the lrg promoter, suggesting that regulation by CodY may overcome regulation by CcpA or RNA polymerase for cid expression in the presence of BCAAs. Based on the data so far, it is still premature to speculate on how CodY and CcpA contribute to the expression and function of cid and lrg, even though it is clear that CodY and CcpA are involved in the regulation of cid and lrg at a molecular level. Given that CcpA would be activated by an elevated level of glycolytic intermediates (Deutscher, Kuster, Bergstedt, Charrier, & Hillen, 1995; Saier, 1996), competition or cooperation among these regulators may depend on the cellular metabolic status and its surrounding environment. An interesting finding is that supplemented pyruvate can be efficiently utilized even during exponential growth in the ΔccpA strain, although no lrgAB induction was observed (Kim et al., 2019), suggesting that CcpA plays an important role in coordinating the utilization of glucose, as well as pyruvate during cell growth. On the contrary, stationary phase pyruvate uptake does not seem to efficiently occur in the absence of CcpA. As well, the overlapped positioning of the cre (CcpA binding) and the CodY‐binding sites further support the idea that CcpA and CodY work together in regulating both cid and lrg expression, and changes in glycolytic metabolite and amino acid pools act as signals that alter the balance between the effects these two regulatory proteins have on cid and lrg expression. This current study offers an intriguing potential insight toward understanding metabolic linkages with cid and lrg operons.
In conclusion, our data show that the Cid/Lrg system, presumably integrating signals associated with cellular metabolic status and its surrounding environment, is coordinated through direct interaction with global regulators CodY and CcpA, the latter which has previously been shown to regulate cid and lrg expression (Kim et al., 2019). Given that lack of CodY affects lrgAB expression and excreted pyruvate flux, CodY seems to be closely linked to the uptake and utilization of pyruvate, a key intermediate in various metabolic pathways and rerouted to the synthesis of BCAAs by the action of the ilv biosynthetic pathways that are also under the control of CcpA and CodY (Fujita et al., 2014; Santiago, Macgilvray, Faustoferri, & Quivey, 2012; Santiago et al., 2013; Shivers & Sonenshein, 2004, 2005). Given the hypothesized role of Cid and Lrg in inducing cell death and lysis, pyruvate may serve as a metabolic determinant that influences the fate of cells within a population to lyse under unfavorable conditions, which seems to be regulated by LytT and at least two global regulators, CodY and CcpA. From a regulatory standpoint, it is also interesting to determine how cid and lrg regulation is influenced by competitive interactions among those regulatory proteins. Taken together, this study provides new insights into how the regulation of cid and lrg expression mediates the response of S. mutans to the unpredictable changing oral cavity environment and improves our understanding of the functional links between stress response and cell death/lysis process.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
Sang‐Joon Ahn: Conceptualization (lead); formal analysis (supporting); funding acquisition (lead); investigation (supporting); methodology (lead); supervision (lead); validation (supporting); writing‐original draft (lead); writing‐review and editing (lead). Hey‐Min Kim: Formal analysis (lead); investigation (supporting); methodology (supporting); visualization (lead); writing‐original draft (supporting); writing–review and editing (supporting). Shailja Desai: Formal analysis (supporting); investigation (supporting); methodology (supporting); visualization (supporting). Kamal Deep: Formal analysis (supporting); investigation (supporting); methodology (supporting); visualization (supporting); writing–review and editing (supporting). Kelly C Rice: Conceptualization (supporting); funding acquisition (supporting); writing‐original draft (supporting); writing–review and editing (supporting).
ETHICS STATEMENT
None required.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (NIH)‐National Institute of Dental and Craniofacial Research (NIDCR) grant R01 DE025237 (SJA). We thank Professor Robert A. Burne (Department of Oral Biology, University of Florida) for providing all the resources needed.
Appendix 1.
FIGURE A1.
Effect of CodY on Streptococcus mutans lrg and cid promoter activity during growth in high‐glucose cultures. The strains harboring PlrgA‐gfp (a and b) and PcidA‐gfp (c and d) constructs in pDL278 in the S. mutans UA159 wild‐type (a and c) and ∆codY (b and d) backgrounds were grown in a chemically defined FMC medium, supplemented by 45 mM glucose. Relative gfp expression (colored lines) and OD600 (black lines) were monitored on a plate reader (see Materials and Methods for details). The results are representative of three independent experiments
FIGURE A2.
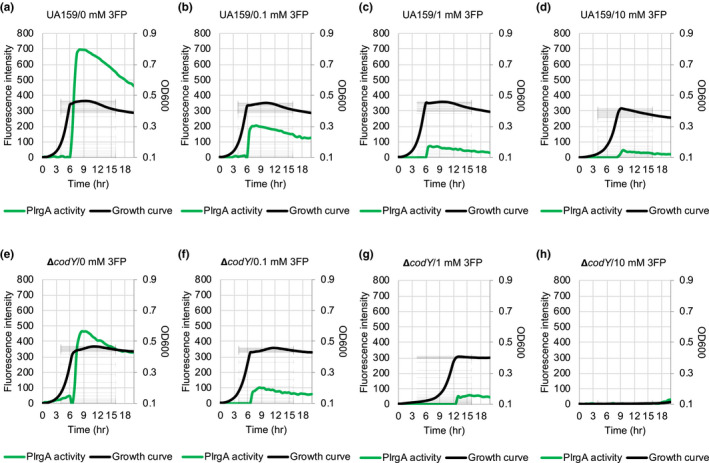
The effect of 3‐fluoropyruvate (3FP; toxic pyruvate analog) on lrg promoter (PlrgA) activity and cell growth in wild‐type and ΔcodY strains. The PlrgA‐gfp reporter strains in wild‐type (a–d) and ΔcodY (E‐H) backgrounds were grown in low‐glucose (11 mM) FMC medium supplemented by 0 (a and e), 0.1 (b and f), 1 (c and g), or 10 mM (d and h) 3FP. Relative gfp expression (green line) and OD600 (black line) were monitored on a plate reader (see Materials and Methods for details). The results are representative of two independent experiments
FIGURE A3.
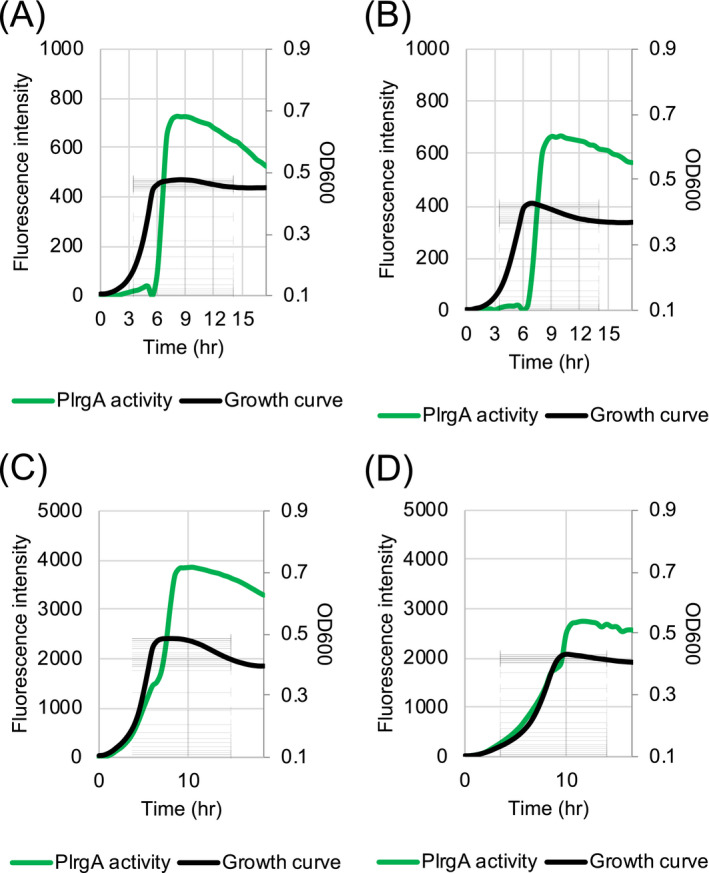
Effect of codY or lytST overexpression on Streptococcus mutans lrg promoter activity during growth in low‐glucose cultures. Strains harboring PlrgA‐gfp constructs in pDL278 in the S. mutans UA159 wild‐type (a), SAB399 (overexpressing codY; b), SAB163 (overexpressing lytST; c), and SAB392 (overexpressing lytST and lacking codY; d) backgrounds were grown in a chemically defined FMC medium, supplemented by 11 mM glucose. Relative gfp expression (colored lines) and OD600 (black lines) were monitored on a plate reader (see Materials and Methods for details). The results are representative of two independent experiments
TABLE A1.
The comparison of pyruvate metabolism‐related gene expression in early‐exponential (EE) versus early‐stationary (ES) growth phases of Streptococcus mutans wild‐type and ΔcodY strains by real‐time qPCR
| Wild type | Fold change (ES/EE) | ΔcodY | Fold change (ES/EE) | |||
|---|---|---|---|---|---|---|
| EE | ES | EE | ES | |||
| lrgA |
1.28E+03 a (9.39E±02) |
4.09E+07 (2.34E±07) |
32,953 |
2.27E+03 (1.78E±0.3) |
3.06E+06 (2.33E±05) |
1,348 |
| cidA |
1.89E+03 (7.70E±02) |
1.55E+02 (8.54E±01) |
0.08 |
1.16E+03 (1.32E±02) |
9.41E+01 (6.31E±01) |
0.08 |
Relative copy number of mRNA (copies/μl); standard deviation in parentheses.
Ahn S‐J, Kim H‐M, Desai S, Deep K, Rice KC. Regulation of cid and lrg expression by CodY in Streptococcus mutans . MicrobiologyOpen. 2020;9:e1040 10.1002/mbo3.1040
DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in this published article.
REFERENCES
- Ahn, S. J. , Deep, K. , Turner, M. E. , Ishkov, I. , Waters, A. , Hagen, S. J. , & Rice, K. C. (2019). Characterization of LrgAB as a stationary phase‐specific pyruvate uptake system in Streptococcus mutans . BMC Microbiology, 19, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, S.‐J. , Gu, T. , Koh, J. , & Rice, K. C. (2017). Remodeling of the Strpetococcus mutans proteome in response to LrgAB and external stresses. Scientific Reports, 7, 14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, S. J. , Qu, M. D. , Roberts, E. , Burne, R. A. , & Rice, K. C. (2012). Identification of the Streptococcus mutans LytST two‐component regulon reveals its contribution to oxidative stress tolerance. BMC Microbiology, 12, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, S. J. , & Rice, K. C. (2016). Understanding the Streptococcus mutans Cid/Lrg System through CidB Function. Applied and Environment Microbiology, 82, 6189–6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, S. J. , Rice, K. C. , Oleas, J. , Bayles, K. W. , & Burne, R. A. (2010). The Streptococcus mutans Cid and Lrg systems modulate virulence traits in response to multiple environmental signals. Microbiology, 156, 3136–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfel, M. A. , Ikeda, B. H. , Speckhard, D. C. , & Frey, P. A. (1984). Escherichia coli pyruvate dehydrogenase complex. Thiamin pyrophosphate‐dependent inactivation by 3‐bromopyruvate. Journal of Biological Chemistry, 259, 2905–2909. [PubMed] [Google Scholar]
- Bayles, K. W. (2003). Are the molecular strategies that control apoptosis conserved in bacteria? Trends in Microbiology, 11, 306–311. [DOI] [PubMed] [Google Scholar]
- Bayles, K. W. (2007). The biological role of death and lysis in biofilm development. Nature Reviews Microbiology, 5, 721–726. [DOI] [PubMed] [Google Scholar]
- Bayles, K. W. (2014). Bacterial programmed cell death: Making sense of a paradox. Nature Reviews Microbiology, 12, 63–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky, B. R. (2011). Indirect repression by Bacillus subtilis CodY via displacement of the activator of the proline utilization operon. Journal of Molecular Biology, 413, 321–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky, B. R. , & Sonenshein, A. L. (2008). Genetic and biochemical analysis of CodY‐binding sites in Bacillus subtilis . Journal of Bacteriology, 190, 1224–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen, W. H. , Burne, R. A. , Wu, H. , & Koo, H. (2018). Oral biofilms: Pathogens, matrix, and polymicrobial interactions in microenvironments. Trends in Microbiology, 26, 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinsmade, S. R. , & Sonenshein, A. L. (2011). Dissecting complex metabolic integration provides direct genetic evidence for CodY activation by guanine nucleotides. Journal of Bacteriology, 193, 5637–5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne, R. A. (2018). Getting to know "The Known Unknowns": Heterogeneity in the oral microbiome. Advances in Dental Research, 29, 66–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne, R. A. , Zeng, L. , Ahn, S. J. , Palmer, S. R. , Liu, Y. , Lefebure, T. , … Nascimento, M. M. (2012). Progress dissecting the oral microbiome in caries and health. Advances in Dental Research, 24, 77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher, J. , Kuster, E. , Bergstedt, U. , Charrier, V. , & Hillen, W. (1995). Protein kinase‐dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in gram‐positive bacteria. Molecular Microbiology, 15, 1049–1053. [DOI] [PubMed] [Google Scholar]
- Flournoy, D. S. , & Frey, P. A. (1989). Inactivation of the pyruvate dehydrogenase complex of Escherichia coli by fluoropyruvate. Biochemistry, 28, 9594–9602. [DOI] [PubMed] [Google Scholar]
- Fujita, Y. , Satomura, T. , Tojo, S. , & Hirooka, K. (2014). CcpA‐mediated catabolite activation of the Bacillus subtilis ilv‐leu operon and its negation by either CodY‐ or TnrA‐mediated negative regulation. Journal of Bacteriology, 196, 3793–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorke, B. , & Stulke, J. (2008). Carbon catabolite repression in bacteria: Many ways to make the most out of nutrients. Nature Reviews Microbiology, 6, 613–624. [DOI] [PubMed] [Google Scholar]
- Groicher, K. H. , Firek, B. A. , Fujimoto, D. F. , & Bayles, K. W. (2000). The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. Journal of Bacteriology, 182, 1794–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedon, E. , Serror, P. , Ehrlich, S. D. , Renault, P. , & Delorme, C. (2001). Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched‐chain amino acids in Lactococcus lactis . Molecular Microbiology, 40, 1227–1239. [DOI] [PubMed] [Google Scholar]
- Handke, L. D. , Shivers, R. P. , & Sonenshein, A. L. (2008). Interaction of Bacillus subtilis CodY with GTP. Journal of Bacteriology, 190, 798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishkov, I. , Ahn, S. J. , Rice, K. C. , & Hagen, S. J. (2020). Environmental triggers of lrgA expression in Streptococcus mutans . Frontiers in Microbiology, 11 Article 18. 10.3389/fmicb.2020.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. M. , Waters, A. , Turner, M. E. , Rice, K. C. , & Ahn, S. J. (2019). Regulation of cid and lrg expression by CcpA in Streptococcus mutans . Microbiology, 165, 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. N. , & Burne, R. A. (2017). CcpA and CodY coordinate acetate metabolism in Streptococcus mutans . Applied and Environmental Microbiology, 83, e03274‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo, H. , Allan, R. N. , Howlin, R. P. , Stoodley, P. , & Hall‐Stoodley, L. (2017). Targeting microbial biofilms: Current and prospective therapeutic strategies. Nature Reviews Microbiology, 15, 740–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, V. J. , Leystra‐Lantz, C. , & Cook, R. A. (1987). Characterization of the specific pyruvate transport system in Escherichia coli K‐12. Journal of Bacteriology, 169, 380–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos, J. A. , & Burne, R. A. (2008). A model of efficiency: Stress tolerance by Streptococcus mutans . Microbiology, 154, 3247–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos, J. A. , Nascimento, M. M. , Lin, V. K. , Abranches, J. , & Burne, R. A. (2008). Global regulation by (p)ppGpp and CodY in Streptococcus mutans . Journal of Bacteriology, 190, 5291–5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, K. (2000). Programmed death in bacteria. Microbiology and Molecular Biology Reviews, 64, 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loesche, W. J. (1986). Role of Streptococcus mutans in human dental decay. Microbiological Reviews, 50, 353–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnayake‐Lecamwasam, M. , Serror, P. , Wong, K. W. , & Sonenshein, A. L. (2001). Bacillus subtilis CodY represses early‐stationary‐phase genes by sensing GTP levels. Genes & Development, 15, 1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, K. C. , & Bayles, K. W. (2008). Molecular control of bacterial death and lysis. Microbiology and Molecular Biology Reviews, 72(1), 85–109. 10.1128/MMBR.00030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, K. C. , Firek, B. A. , Nelson, J. B. , Yang, S. J. , Patton, T. G. , & Bayles, K. W. (2003). The Staphylococcus aureus cidAB operon: Evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. Journal of Bacteriology, 185, 2635–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, K. C. , Turner, M. E. , Carney, O. V. , Gu, T. , & Ahn, S. J. (2017). Modification of the Streptococcus mutans transcriptome by LrgAB and environmental stressors. Microbial Genomics, 3, e000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier, M. H., Jr. (1996). Cyclic AMP‐independent catabolite repression in bacteria. FEMS Microbiology Letters, 138, 97–103. [DOI] [PubMed] [Google Scholar]
- Santiago, B. , Macgilvray, M. , Faustoferri, R. C. , & Quivey, R. G., Jr. (2012). The branched‐chain amino acid aminotransferase encoded by ilvE is involved in acid tolerance in Streptococcus mutans . Journal of Bacteriology, 194, 2010–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago, B. , Marek, M. , Faustoferri, R. C. , & Quivey, R. G., Jr. (2013). The Streptococcus mutans aminotransferase encoded by ilvE is regulated by CodY and CcpA. Journal of Bacteriology, 195, 3552–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivers, R. P. , Dineen, S. S. , & Sonenshein, A. L. (2006). Positive regulation of Bacillus subtilis ackA by CodY and CcpA: Establishing a potential hierarchy in carbon flow. Molecular Microbiology, 62, 811–822. [DOI] [PubMed] [Google Scholar]
- Shivers, R. P. , & Sonenshein, A. L. (2004). Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched‐chain amino acids. Molecular Microbiology, 53, 599–611. [DOI] [PubMed] [Google Scholar]
- Shivers, R. P. , & Sonenshein, A. L. (2005). Bacillus subtilis ilvB operon: An intersection of global regulons. Molecular Microbiology, 56, 1549–1559. [DOI] [PubMed] [Google Scholar]
- Sonenshein, A. L. (2005). CodY, a global regulator of stationary phase and virulence in Gram‐positive bacteria. Current Opinion in Microbiology, 8, 203–207. [DOI] [PubMed] [Google Scholar]
- Sonenshein, A. L. (2007). Control of key metabolic intersections in Bacillus subtilis . Nature Reviews Microbiology, 5, 917–927. [DOI] [PubMed] [Google Scholar]
- Terleckyj, B. , & Shockman, G. D. (1975). Amino acid requirements of Streptococcus mutans and other oral streptococci. Infection and Immunity, 11, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yindeeyoungyeon, W. , & Schell, M. A. (2000). Footprinting with an automated capillary DNA sequencer. BioTechniques, 29, 1034–6, 1038, 1040–1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


