Abstract
Congenital hearing impairment is a sensory disorder that is genetically highly heterogeneous. By performing exome sequencing in two families with congenital nonsyndromic profound sensorineural hearing loss (SNHL), we identified autosomal dominantly inherited missense variants [p.(Asn283Ser); p.(Thr116Ile)] in GREB1L, a neural crest regulatory molecule. The p.(Thr116Ile) variant was also associated with bilateral cochlear aplasia and cochlear nerve aplasia upon temporal bone imaging, an ultra-rare phenotype previously seen in patients with de novo GREB1L variants. An important role of GREB1L in normal ear development has also been demonstrated by greb1l−/− zebrafish, which show an abnormal sensory epithelia innervation. Last, we performed a review of all disease-associated variation described in GREB1L, as it has also been implicated in renal, bladder and genital malformations. We show that the spectrum of features associated with GREB1L is broad, variable and with a high level of reduced penetrance, which is typically characteristic of neurocristopathies. So far, seven GREB1L variants (14%) have been associated with ear-related abnormalities. In conclusion, these results show that autosomal dominantly inherited variants in GREB1L cause profound SNHL. Furthermore, we provide an overview of the phenotypic spectrum associated with GREB1L variants and strengthen the evidence of the involvement of GREB1L in human hearing.
Keywords: autosomal dominant inheritance, exome sequencing, GREB1L, profound nonsyndromic hearing impairment, cochlear aplasia, cochlear nerve aplasia, neural crest, neurocristopathy
1. Introduction
Childhood hearing impairment (HI) is associated with impaired language acquisition, learning, speech development and affects 34 million children worldwide (World Health Organization). Approximately 1/1000 children are born with hearing loss, of which approximately 80% is genetic [1]. HI can be part of a syndrome with the presence of other medical anomalies, or it can be nonsyndromic. Currently, 120 nonsyndromic HI genes have been identified, with 59% having an autosomal recessive (AR), 37% an autosomal dominant (AD), and 5% an X-linked mode of inheritance (Hereditary hearing loss homepage). However, many genes remain to be identified due to the complexity of the hearing system and due to the understudy of some ancestries [2].
Nonsyndromic HI has no association with additional features or abnormalities. However, it can be associated with abnormalities of the middle ear and/or inner ear [1]. A large number of these abnormalities are mild, but bilateral cochlear aplasia, i.e., bilateral absence of the cochlea, is an ultra-rare and severe developmental abnormality of the inner ear. Approximately 0.3% of children with congenital sensorineural HI are estimated to have bilateral cochlear aplasia [3]. However, this estimate is predominately based on children who were candidates for a cochlear implant, and they usually present with severe-to-profound HI [3,4,5].
We previously identified de novo loss-of-function variants in GREB1L in two individuals with profound nonsyndromic HI with inner ear and cochleovestibular nerve (or 8th cranial) malformations (Table 1) [5,6]. Affected individuals had either absent cochleae bilaterally [p.(Glu1410fs)] or an absent cochlea on the right and incomplete partition type I on the left [p.(Arg328*)]. Both individuals also displayed abnormalities of their vestibules and absent 8th cranial nerves [6]. In addition, greb1l−/− zebrafish exhibit a loss of and/or abnormal sensory epithelia innervation, including a loss of the anterior cristae nerve and an abnormal innervation pathway from the occipital lateral line neuromast. These findings in humans and model organisms confirm the importance of GREB1L in sensory innervation [6]. Furthermore, Greb1l is widely expressed during craniofacial development, including the otic vesicle [6,7], and Greb1l−/− mice are embryological lethal and demonstrate severe abnormalities, including craniofacial and renal abnormalities [8]. Greb1l+/− mice show an abnormal embryo size, growth retardation [9] and mild abnormalities to their kidneys and ureters [8].
Table 1.
Variants identified in GREB1L associated with nonsyndromic hearing impairment.
| Family Type | Variant Segregation | Inheritance Model | Predicted Variant Effect | cDNA Change 1 | AA Change | gnomAD | CADD Score (v1.3) | GERP++RS | Splicing Effect Prediction 2 | Phenotype | ACMG 3 | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trio | de novo | AD | splicing | c.4368G>T | p.(Glu1410fs) | absent | 26 | 5.17 | splice site loss | profound bilateral SNHI; UCA (right); UIP-I (left); BVES + SCC; BCNA | P | [6] |
| Trio | de novo | AD | nonsense | c.982C>T | p.(Arg328*) | absent | 38 | 4.52 | NA | profound bilateral SNHI; BCA; BVES + SCC; BCNA | P | [6] |
| Family | inherited | AD | missense/splicing | c.848A>G | p.(Asn283Ser) | absent | 10 | 3.44 | ESE site loss | profound bilateral SNHI 4 | LP | This study (Family 1) |
| Trio | inherited 5 | AD | missense | c.347C>T | p.(Thr116Ile) | absent | 30 | 5.25 | NA | profound bilateral SNHI; BCA; BVES + SCC; BCNA | VUS | This study (Family 2) |
1 Based on NM_001142966.2; 2 Based on Human splicing finder (v.3.1), ESEfinder (v2.0) [10,11]. 3 Classified based on the American College of Medical Genetics (ACMG) guidelines: P, Pathogenic; LP, likely pathogenic; VUS, variant of unknown significance [12]. 4 Inner ear not evaluated via imaging. 5 Maternal reduced penetrance; AD, Autosomal Dominant; BCA, bilateral cochlear aplasia; BCNA, bilateral cochlear nerve aplasia; BVES + SCC: bilateral dysplastic vestibule and semicircular canals; ESE, exonic splicing enhancer; NA, Not applicable; SNHI, sensorineural hearing impairment; UCA, unilateral cochlear aplasia; UIP-I, unilateral incomplete partition type I.
In addition, de novo or autosomal dominantly inherited variants (often with reduced penetrance) have previously been implicated in individuals with renal, bladder and genital malformations [8,13,14]. Renal hypoplasia/aplasia 3 (RHDA3) is a severe developmental disorder characterized by abnormal kidney development and is caused by heterozygous GREB1L variants. Although the phenotype can be highly variable, the disorder falls within the most severe end of the spectrum of congenital anomalies of the kidney and urinary tract. In many of these cases, children were aborted or stillborn due to the severity of the malformations, such as bilateral renal aplasia [8,13,14].
In this article, we have, for the first time, identified a family with congenital profound HI that segregates a missense variant in GREB1L with an AD mode of inheritance and also report on an additional case with bilateral cochlear and cochlear nerve aplasia with a GREB1L variant.
2. Materials and Methods
2.1. Patient Recruitment and Clinical Assessment
The study was approved by the ethics committee of the Quaid-i-Azam University (IRB-QAU-153), University of Antwerp (B3002020000073) and the Institutional review board of Columbia University (IRB-AAAS2343). Informed consent and peripheral blood samples were obtained from all individuals of a non-consanguineous Pakistani family with deafness (Family 1 [4697]; Figure 1A) and a non-consanguineous Egyptian family with deafness (Family 2 [BAIE1]; Figure 1B). DNA was extracted using a phenol-chloroform procedure for the Pakistani family [15] and using magnetic beads with the chemagic™ blood DNA kit on a chemagic™ Prime™ instrument (PerkinElmer, Waltham, MA, USA) for the Egyptian family. The patient evaluation included a clinical history, physical, audiological and vestibular examination. Computed tomography (CT) and magnetic resonance imaging (MRI) of the temporal bone were performed in the Egyptian patient to identify the presence of cochleovestibular malformations (Family 2). Unfortunately, we were unable to perform CT or MRI on the Pakistani family (Family 1) due to the remote location of these individuals in the Khyber Pakhtunkhwa province, Pakistan.
Figure 1.
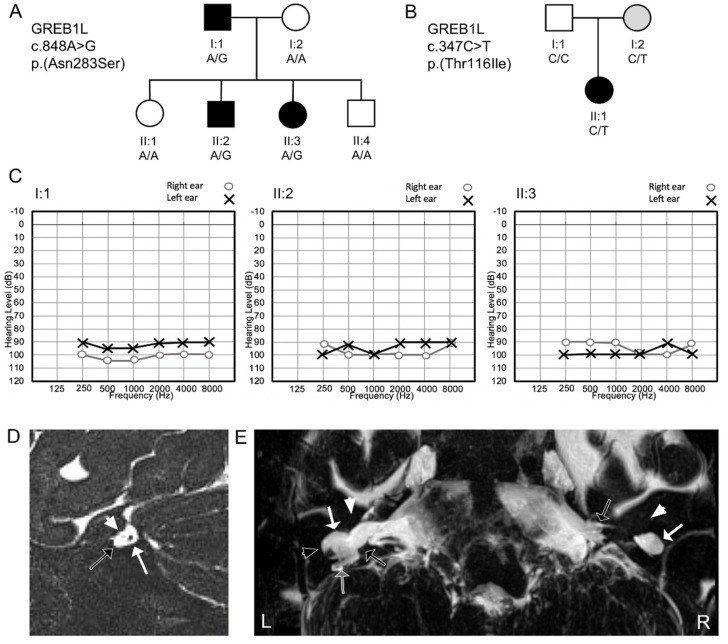
Segregation of the GREB1L missense variants in both families, audiological and imaging data. A/B. Segregation of the p.(Asn283Ser) GREB1L variant in family 1 (4697) (A) and p.(Thr116Ile) variant in family 2 (BAIE1) (B). Solid black symbols represent affected individuals and clear symbols unaffected family members. Grey symbols represent unaffected individuals that are also heterozygous for the variant (reduced penetrance). Females are represented by circles and males by squares. (C) Pure-tone audiograms of hearing-impaired family members of family 1 illustrate that each one presents with bilateral profound HI. (D) Oblique sagittal T2 sequence across the right internal auditory canal (IAC) of patient II:1 of family 2. The white arrow indicates the vestibular nerve (black dot). The arrowhead indicates a hypoplastic facial nerve (small grey dot). The black arrow indicates the area in the IAC where the cochlear nerve is expected but not observed. (E) Maximum intensity projection of a heavily T2 weighted sequence to the inner ear of the affected individual (II:1) of family 2. Bilateral cochlear aplasia and dysplasia of the vestibular system is visualized. The white arrowhead indicates the area where the cochlea is expected but not seen (bilaterally). The white arrow indicates a dysplastic cystic dilated vestibule on each side. The black arrow indicates a left narrow IAC and the right broad IAC with wide communication between the fundus of the IAC and the vestibule. The black arrowhead indicates a right dilated lateral semicircular canal. The grey arrow indicates a right rudimentary posterior semicircular canal. L, Left; R, right.
2.2. Exome Sequencing
For family 1, Sanger sequencing was performed to exclude coding variants in the HI gene GJB2 prior to exome sequencing. Additional variants were also excluded by Sanger sequencing that are common causes of HI in the Pakistani population: i.e., p.(Phe91Ser) and p.(Cys99Trp) within CIB2, two intronic variants in HGF (c.482+1986_1988delTGA and c.482+1991_2000delGATGATGAAA) and p.(Gln446Arg) and p.(Val239Asp) in SLC26A4 [16,17,18]. Next, a DNA sample from the affected member (II:2, family 1) underwent exome sequencing. From family 2, the affected patient (II:2) and both normal hearing parents (I:1; I:II) underwent exome sequencing. In short, exomic library preparation was performed using the SureSelect human all exon V6 kit (60.46 Mb target region) for family 1 and SeqCap EZ Exome Probes v3 (64 Mb target region) for family 2. Paired-end sequencing was performed on a HiSeq2500/4000 instrument (Illumina Inc, San Diego, CA, USA), with an average sequencing depth of on target regions of 61× for family 1 (II:2) and 119× (II:1), 97× (I:1), 106× (I:2) for family 2 and the fraction of targets covered >10× was 98.95% for family 1 and 96.80% for family 2. After removing low-quality reads, the filtered reads were aligned to the human reference genome (GRCh37/Hg19) using Burrows–Wheeler Aligner-MEM (BWAv0.7.15) [19]. Duplicate reads were marked using Picard-tools (v2.5.0). An insertions/deletion (Indel)-realignment and base quality score recalibration were performed with Genome Analysis Toolkit (GATK) (v3.7), and single nucleotide variants (SNVs) and InDels were called by the GATK HaplotypeCaller [20]. Variant annotation and filtering were performed using ANNOVAR [21]. In short for the analysis, (1) exonic and splice region variants +/− 12 bp from intron-exon boundary were retained; (2) An AD mode (including de novo for family 2) and AR mode of inheritance was considered for both families; (3) Variants with a predicted effect on protein function or pre-mRNA splicing (missense, nonsense, frameshift, start-loss, splice region, etc.) with a population-specific minor allele frequency (MAF) of <0.005 (for AR) and <0.0005 (for AD) in all populations of the Genome Aggregation Database (gnomAD) [22] and the Greater Middle East Variome Project (GME) [23] were retained to test for segregation; and (4) Bioinformatic prediction scores were annotated from dbnsfp35a and dbscSNV1.1 to evaluate missense and splice site variants respectively [24,25], including Combined Annotation Dependent Depletion (CADD) and Genomic Evolutionary Rate Profiling (GERP++) scores [26,27]. Genes previously involved in human/animal HI or genes expressed in the inner ear were prioritized [28,29]. Candidate variants obtained from filtering were visualized with the Integrative Genomics Viewer (IGV2.4.3). Sanger sequencing performed using an ABI3130XL Genetic Analyzer was used to validate the variants in both families and check the segregation of variants in additional family 1 members for which DNA was available.
Copy number variants (CNVs) were called in exome data from both families using CONiFER (v0.2.2) [30]. Gene annotation was done using the BioMart Database [31] and variant frequency was assessed using the Database of Genomic Variants [32] and gnomAD [22] using the same frequency cut-offs as above for SNV/InDels.
3. Results
3.1. Clinical Findings
In the Pakistani family (Family 1), hearing impairment was prelingual for the three affected family members, and pure-tone audiometry revealed bilateral profound sensorineural HI (Figure 1). No gross vestibular dysfunction was observed via a tandem gait test, and Romberg test in affected individuals I:1, II:2, and II:3. Clinical histories were obtained, and the patients underwent a physical exam at the ages of 45 years of age (y) (I:2), 15y (II:2), and 17y (II:3) with no other health problems reported, including no kidney or bladder issues, however asymptomatic kidney disease could not be excluded. History of head trauma, severe infections or ototoxic treatment was not present. None of the other family members displayed HI or any other clinical features, and the parents have no reported consanguinity.
In the Egyptian patient (Family 2), auditory brainstem responses and cochlear microphonic potentials were bilaterally absent. Vestibular testing showed bilateral aberrant head impulse test, minimal nystagmi on the rotational chair test, and no nystagmus response on the caloric test (with water 44 °C), suggesting a reduced canalar function. C-Vemp (cervical-vestibular evoked myogenic potentials) were bilaterally present at 130 dBSPL, implying a functioning vestibule. MRI and CT imaging showed bilateral cochlear aplasia, aplasia of the cochlear nerve and dysplasia of the vestibule and semicircular canals (Figure 1). The vestibular nerve was present on both sides. On the left side, a hypoplastic narrow internal auditory canal (IAC) was found. On the right side, the IAC was wide with deficient fundus and wide communication between the IAC and vestibule. The facial nerve had a hypoplastic aspect bilaterally. The patient did not have any other known health issues, however, mild kidney disease could not be excluded. The parents have normal hearing, reported no health issues and are non-consanguineous. There is no family history of congenital or progressive HI.
3.2. Exome Sequencing
In family 1, exome sequencing and variant filtering identified variants in MYO15A, POLE and GREB1L as candidates and were validated and tested for segregation (Table S1). Only a missense [(NM_001142966.2:c.848A>G:p.(Asn283Ser)] variant in GREB1L, a gene previously associated with HI, segregated with HI in pedigree 4697 with an AD mode of inheritance (Table 1, Figure 1, Figure S1) [6]. The variant is absent from gnomAD and GME. It is located at a conserved position amongst species (GERP++ RS: 3.44; phastCons20way_mammalian: 1.00). The variant has a CADD score = 10 and is predicted damaging by fathmm-MKL. Based on ESEfinder (v2.0) [10], the variant is located in an exonic splicing enhancer motif (ACAGTAG; score 2.74; threshold >2.67) predicted responsive to Pre-MRNA-Splicing Factor SRp40, which is lost due to the variant (GCAGTAG; score 1.28; threshold >2.67). Therefore, the variant might impact normal protein functioning through various mechanisms, however, we do not know its exact effect in vivo as we were unable to obtain RNA from the patients. GREB1L is intolerant to loss-of-function (LoF) variants and is likely under selection against them (pLI = 1; o/e = 0.02 [0.01–0.07]) [22], with only 2% of the expected LoF variants observed. In addition, only 52% of the expected missense variants are observed in GREB1L (z score = 5.37; o/e = 0.52 [0.49–0.56]) [22]. The p.(Asn283Ser) missense variant is located in a position and region with a gene-specific missense tolerance ratio (MTR) percentile of <25 [33], which signifies that this region of the protein is also less likely to tolerate missense variants. Pathogenic missense variants are enriched within the 25th percentile of the intolerant region of the gene’s MTR distribution [33].
In family 2, all family members were sequenced via exome sequencing. We also identified a variant in GREB1L [(NM_001142966.2:c.347C>T:p.(Thr116Ile)], which was verified with Sanger sequencing (Figure S1). None of the other identified variants were likely to be related to HI (Table S1). The p.(Thr116Ile) variant in GREB1L was inherited from the unaffected mother, and the variant is absent from gnomAD and GME. It is located at a conserved position amongst species (GERP++ RS: 5.25; phastCons20way_mammalian: 0.935), has a CADD score = 30, and is predicted damaging by fathmm-MKL. The temporal bone imaging phenotype of the patient in this family is remarkably similar to patients previously described with de novo GREB1L variants (Table 1), a phenotype that is ultra-rare [6].
Based on the ACMG guidelines for variant classification, p.(Asn283Ser) was classified as likely pathogenic (PM2, PP1-M [Bayes Factor = 16 [34]], PP2 and PP3) and p.(Thr116Ile) was classified as a variant of unknown significance (PM2, PP2, PP3 and PP4) [12]. Finally, no CNVs were identified in either family that were likely to be involved in disease etiology.
3.3. Phenotypic Spectrum of GREB1L Variation
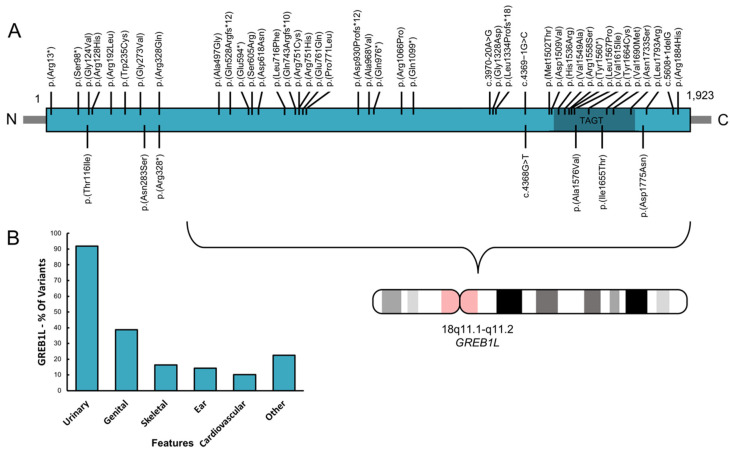
We performed a detailed literature search of all disease-associated variation (N = 49) reported in GREB1L, which are listed in Table 2 and displayed in Figure 2. This illustrates that variants are present over the entire length of the gene, with some clustered in or near the TAGT domain. Although previous studies were mostly focused on renal malformations, this table shows that a variety of malformations can be present in affected individuals, including renal, bladder, uterus, ear and other issues such as skeletal abnormalities. Reduced penetrance was observed in 50% of the reported variants in which parents or unaffected siblings were also assessed. In addition, variants were inherited maternally in a large majority of cases (71%). Three patients previously studied for renal malformations also showed ear-related issues. Therefore, of the total of 49 variants that have been reported, 7 variants (14%) have been associated with a hearing or an ear abnormality.
Table 2.
All variants reported in GREB1L and their associated phenotypic features.
| cDNA Variant 1 | Amino Acid Variant | Urinary Phenotype | Genital Phenotype | Ear Phenotype 2 | Other Phenotypes | Inheritance | Reduced Penetrance | Reference |
|---|---|---|---|---|---|---|---|---|
| c.37C>T | p.(Arg13*) | unilateral MCD, congenital megaureter | – | – | – | NA | NA | [14] |
| c.293C>G | p.(Ser98*) | BKA | – | – | – | de novo | no | [13] |
| c.347C>T | p.(Thr116Ile) | – | – | profound bilateral SNHI, BCA; BVES+SCC; BCNA | – | mat | yes (mat) | This study |
| c.371G>T | p.(Gly124Val) | BKA, UKA, bladder hypoplasia | – | – | Potter sequence | suspected pat 4 | NA | [35] |
| c.383G>A | p.(Arg128His) | UKA | unicornate uterus, agenesis of left ovary | – | – | NA | NA | [14] |
| c.575G>T | p.(Arg192Leu) | BKA, UKA | unique fallopian trump and ovary | – | insulin-dependent diabetes | mat | no | [8] |
| c.705G>T | p.(Trp235Cys) | BKA, UKA, renal cysts, clear cell renal carcinoma | MRKH, arcuate uterus | – | – | AD family (2 mat, 2 pat) | yes (2 mat, 1 unaffected female sib) | [36] |
| c.818G>T | p.(Gly273Val) | UKA | – | – | – | NA | NA | [14] |
| c.848A>G | p.(Asn283Ser) + splicing | – | – | profound bilateral SNHI 3 | – | AD family (1 pat) | no | This study |
| c.982C>T | p.(Arg328*) | – | – | profound bilateral SNHI, BCA; BVES+SCC; BCNA | – | de novo | no | [6] |
| c.983G>A | p.(Arg328Gln) | pelvic kidney, MCD, VUR | – | – | mat | no | [8] | |
| c.1490C>G | p.(Ala497Gly) | UKA | – | – | – | NA | NA | [14] |
| c.1582delC | p.(Gln528Argfs*12) | BKA, UKA | UA, unicornuated uterus | – | clinodactyly | mat | yes (mat) | [8] |
| c.1780G>T | p.(Glu594*) | BKA, UKA, VUR | UA, fallopian trumps absence, ovarian hernia, uterine left artery absent | – | – | mat | no | [8] |
| c.1813A>C | p.(Ser605Arg) | UKA, multilocular cyst | blind ending hemi-vagina and bicornuated uterus | – | – | pat | yes (pat) | [8] |
| c.1852G>A | p.(Asp618Asn) | Ectopic kidney, VUR, duplicated ureter | MRKH type 2 | – | unilateral polydactyly, facial asymmetry | NA | NA | [37] |
| c.2148G>T | p.(Leu716Phe) | VUR | – | – | iris anomaly | NA | NA | [8] |
| c.2227del | p.(Gln743Argfs*10) | UKA, MCD | MRKH type 2, UA | – | scoliosis | AD family (1 mat) | yes (mat) | [37] |
| c.2251C>T | p.(Arg751Cys) | BKA, unilateral hypoplasia | unicornuated uterus | – | – | mat | no | [8] |
| c.2252G>A | p.(Arg751His) | UKA, MCD, megaurethra | – | – | hepatic portal fibrosis | mat 5 | NA | [8] |
| c.2281G>C | p.(Glu761Gln) | UKA, duplication of the ureter, unilateral MCD, congenital megaureter | – | – | – | pat | yes (pat) | [14] |
| c.2312C>T | p.(Pro771Leu) | UKA, MCD | UA, streak ovaries, rudimentary follopian tubes | – | Right unique umbilical artery, 11 pairs of ribs, 6 cervical hemivertebrae with 1 hemivertebrae | NA | NA | [37] |
| c.2787_2788del | p.(Asp930Profs*12) | BKA, UKA, MCD | UA | – | – | AD family (1 mat; 1pat) | yes (mat) | [37] |
| c.2903C>T | p.(Ala968Val) | BKA, with agenesis of ureters, bladder hypoplasia | – | – | – | de novo | no | [13] |
| c.2926C>T | p.(Gln976*) | BKA, UKA | – | – | – | mat | no | [8] |
| c.3197G>C | p.(Arg1066Pro) | UKA | – | – | – | NA | NA | [14] |
| c.3295C>T | p.(Gln1099*) | unilateral MCD | – | – | – | mat | no | [14] |
| c.3970-20A>G | splicing | BKA, ureter and bladder aplasia | UA | – | Unilateral hexadactyly | pat | yes (pat) | [37] |
| c.3983G>A | p.(Gly1328Asp) | UKA | MRKH type 2 | – | – | NA | NA | [37] |
| c.3998_3999insC | p.(Leu1334Profs*18) | UKA | – | – | – | mat | no | [14] |
| c.4368G>T | splicing | – | – | profound bilateral SNHI, UCA (right); UIP-I (left); BVES+SCC; BCNA | – | de novo | no | [37] |
| c.4369−1G>C | splicing | BKA | UA | – | thickened left ventricular wall, 10 pairs of ribs | mat | yes (mat) | [8] |
| c.4505T>C | p.(Met1502Thr) | BKA | – | – | retro-esophageal subclavian artery, adrenal gland hypoplasia, enlarged thymus, one pair of cervical ribs | mat 5 | NA | [8] |
| c.4526A>T | p.(Asp1509Val) | BKA | – | – | adrenal cytomegaly | NA | NA | [8] |
| c.4607A>G | p.(His1536Arg) | BKA, UKA, MCD, horseshoe kidney | UA | – | 11 pairs of ribs | mat | no | [8] |
| c.4646T>C | p.(Val1549Ala) | UKA | UA | – | Henoch Schönlein Purpura | NA | NA | [14] |
| c.4672C>A | p.(Arg1558Ser) | BKA | – | – | – | mat | yes (mat) | [8] |
| c.4680C>A | p.(Tyr1560*) | BKA, bladder agenesis, VUR | uterus anomaly | – | Potter sequence | AD family (1 mat) | no | [14] |
| c.4700T>C | p.(Leu1567Pro) | UKA | – | – | – | de novo | no | [14] |
| c.4727C>T | p.(Ala1576Val) | BKA | – | auricular tag | hypertrophic left ventricle, aortic stenosis | NA | NA | [8] |
| c.4843G>A | p.(Val1615Ile) | BRHD, congenital hydronephrosis | – | – | – | mat | yes (mat) | [14] |
| c.4964T>C | p.(Ile1655Thr) | UKA | – | unilateral SNHI | genu valgum, flat feet | pat | yes (pat) | [14] |
| c.4991A>C | p.(Tyr1664Cys) | UKA | – | – | – | NA | NA | [14] |
| c.5068G>A | p.(Val1690Met) | UKA, VUR | – | – | – | pat 5 | NA | [14] |
| c.5198A>G | p.(Asn1733Ser) | BKA, UKA, ureter and bladder aplasia | UA, hemi-uterus, streak ovaries | – | – | AD family (1 mat, 1 pat) | yes (mat; unaffected female sib) | [37] |
| c.5323G>A | p.(Asp1775Asn) | BKA | – | preauricular tag, lop ear | – | NA | NA | [8] |
| c.5378T>G | p.(Leu1793Arg) | BKA, RKA, hypertrophy of the kidney | – | – | – | AD family (2 mat) | yes (1 mat) | [38] |
| c.5608+1delG | splicing | BKA, UKA, bladder agenesis | undifferentiated external female genitalia | – | Potter sequence | de novo, mat | yes (2 male siblings) | [35,38] |
| c.5651G>A | p.(Arg1884His) | URHD | – | – | – | NA | NA | [14] |
1 Based on NM_001142966.2. 2 Many of the children with renal malformations listed here were aborted/stillborn, in which hearing could not have been assessed. 3 Inner ear not evaluated via imaging. 4 No genetic evaluation. 5 Parent not evaluated via renal ultrasound. AD, Autosomal Dominant; AD family, family with multiple affected (>2) showing autosomal dominant inheritance; BCA, bilateral cochlear aplasia; BCNA, bilateral cochlear nerve aplasia; BKA, bilateral kidney agenesis; BVES + SCC, bilateral dysplastic vestibule and semicircular canals; ESE, exonic splicing enhancer; mat, maternal inheritance; MCD, multi-cystic dysplasia; MRKH, Mayer-Rokitansky-Küster-Hauser syndrome; NA, Not assessed; pat, paternal inheritance; SNHI, sensorineural hearing impairment; UCA, unilateral cochlear aplasia; UIP-I, unilateral incomplete partition type I; UKA, unilateral kidney agenesis; UA, uterovaginal aplasia or uterus aplasia; VUR, vesicoureteral reflux.
Figure 2.
All variants reported in GREB1L and their associated phenotypic features. A. GREB1L protein structure with all variants indicated. The seven bottom variants are associated with ear-related abnormalities. Dark green, TAGT or Ten-eleven translocation/J binding protein (TET/JBP)-associated glycosyltrasferase domain [39]. B. The percentage (%) of variants associated with the most prevalent phenotypic features seen in affected individuals.
4. Discussion
HI in children is both genetically and phenotypically heterogeneous. Identification of novel genes implicated in congenital HI is important to understand normal hearing and ear development, for patient management and intervention and for the development of novel therapeutic strategies.
We identified two families with congenital profound nonsyndromic sensorineural HI that segregate missense variants [p.(Asn283Ser) and p.(Thr116Ile)] in GREB1L (Figure 1). GREB1L is a premigratory neural crest (NC) regulatory molecule implicated in the embryonic development of many tissues [40]. The cranial NC is important in the development of the peripheral nervous system and non-neural tissues, including craniofacial connective and skeletal tissues [41]. In addition, it also gives rise to the stria vascularis of the inner ear and the glia cells of the cochleovestibular nerve and inner ear ganglion [42]. greb1l has also been implicated in Hoxb1 and Shha signaling in zebrafish [14], important pathways in the inner ear and cranial nerve development [43,44,45].
Previous reports on disease-related GREB1L variants showed that a variable phenotype is present, including within families segregating the same variant (e.g., left vs. right ear) [36,37]. In addition, a high level of reduced penetrance has been reported, including in family 2 of this study. There is no evidence that variants cluster within specific domains of the protein (Table 2; Figure 2). This finding is similar to what was observed for EYA1, an NC regulatory molecule which is involved branchio-oto-renal (BOR)/branchio-otic (BO) syndrome etiology [46]. EYA1 is also characterized by a high level of phenotypic variation between patients, even within the same family, and the severity of the phenotype does not correlate with the type of variant nor with the domain involved. In BOR patients with EYA1 variants, which presents with both ear and renal abnormalities, normal kidneys were often observed in family members with BOR while other family members had renal abnormalities [46]. Many neurocristopathies typically show this variable phenotypic profile amongst patients, even within families or within the same individual (left vs. right) [47], and multiple hypotheses have been suggested to explain this phenomenon, such as environmental factors and genetic modifiers [6,46,48]. However, as the NC is a transient and migratory cell population during development, there are also complex micro-regulations that could disturb NC migration during development. Because of this, the path of NC migration that ends up affected due to GREB1L dysfunction could perhaps be attributed to chance. An example of this can be found in knockout (Wv/Wv) mice. These mice have a defect in c-kit, a NC migration regulatory molecule involved in the migration and proliferation of melanocytes in the inner ear. Wv/Wv mice show uni- or bilateral inner ear issues with variable hearing levels, and this variability in inner-ear phenotype was found to be reflected by the number of melanocytes present and how far they migrated along each cochlea during development [49].
The particular link between renal and ear abnormalities has previously been demonstrated [47], including in neurocristopathies. Several neural crest regulatory molecules are known to cause ear/kidney syndromes with variable expression of both ear and kidney phenotypes (e.g., EYA1, SIX1, SIX5, CHD7, MASP1, TBX1), involved in BOR/BO syndrome, CHARGE syndrome, 3MC syndrome and DiGeorge syndrome [5,47]. In addition to these, there are also several other disorders with a specific renal/ear link, such as Alport syndrome and Bartter syndrome [50,51]. Interestingly, when reviewing all variants reported in GREB1L to date, we also demonstrate that 14% of GREB1L variants (N = 7) have been associated with ear-related issues. It is to be noted however, that many of the previous reports (focused on renal malformations) included aborted/stillborn fetuses, in which hearing could not have been assessed. In addition, inner ear and cochlear nerve malformations cannot be assessed via prenatal ultrasound and if an autopsy was performed and would usually not be detected on routine autopsy. Therefore, the number of ear malformations associated with GREB1L variants is likely under-reported. Last, this renal/ear link is also seen in Mayer-Rokitansky-Kuster-Hauser (MRKH) syndrome, characterized by abnormal development of the internal reproductive system in females, and is also caused by GREB1L variants (Table 2). Interestingly, HI is reported in 10–25% of individuals with MRKH syndrome [37].
We also detected a maternal bias in the inheritance of GREB1L variants (Table 2). This maternal bias has previously observed and two mechanisms have been suggested: (1) imprinting [8,36] (2) or GREB1L variants could affect male fertility resulting in a low rate of paternal inheritance [8]. Genital issues, including uterus aplasia, are common and have been reported in many females (Table 2), but the presence in males may be underestimated as the defect might not be a gross morphological abnormality that causes infertility.
De novo GREB1L variants have been previously implicated in a phenotype which consists of profound HI and inner ear and cochleovestibular nerve malformations [6]. The inner ear malformation seen in family 2 is remarkably similar to the patients previously reported with de novo GREB1L variants (Table 1) [6], and includes cochlear aplasia, cochlear nerve aplasia and bilateral dysplastic vestibules and semicircular canals (Figure 1), an ultra-rare phenotype. The finding of multiple independent cases with GREB1L variants and this exact ultra-rare phenotype is significant [6]. In addition, greb1l−/− zebrafish (p.Gln408Ter) exhibit a loss of or abnormal sensory epithelia innervation [6], supporting the importance of GREB1L in the inner ear and nerve development.
Unfortunately, we were unable to perform temporal bone imaging in the affected members of family 1 since they are located in a remote village in Pakistan. The profound bilateral congenital HI phenotype observed for affected members of this family suggests that it may also be due to inner ear/cochleovestibular nerve malformations. Since sample collection for DNA extraction and genetic screening is easier to implement in areas with limited access to modern healthcare systems than temporal bone imaging, we believe including GREB1L in diagnostic screening for nonsyndromic HI is valuable.
In conclusion, we demonstrate that autosomal dominantly inherited variants in GREB1L are involved in profound sensorineural HI etiology and show that GREB1L behaves with a similar phenotypic variance compared to other neurocristopathies. In addition, we recommend including GREB1L in diagnostic screening panels for nonsyndromic HI.
Acknowledgments
We would like to thank the families for their participation in this study.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/6/687/s1, Table S1: Candidate variants identified in both families, genotypes and annotations. Figure S1: Sanger traces of the GREB1L variants in both families.
Author Contributions
Conceptualization, methodology, I.S. (Isabelle Schrauwen), W.A., G.V.C. and S.M.L.; formal analysis, I.S. (Isabelle Schrauwen) T.B., and A.N.; investigation: K.L., I.S. (Isabelle Schatteman) and A.A.; data curation, K.L. and I.S. (Isabelle Schatteman); writing—original draft preparation, I.S. (Isabelle Schrauwen); writing—review and editing, all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the American Hearing Research Foundation to I.S., the Higher Education Commission of Pakistan (to W.A.) and the National Institute of Deafness and other Communication Disorders grants R01 DC011651 and DC003594 to S.M.L.
Conflicts of Interest
The authors have no conflicts of interest related to the work in this manuscript.
Web Resources
References
- 1.Shearer A.E., Hildebrand M.S., Smith R.J. Hereditary hearing loss and deafness overview. In: Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J., Stephens K., Amemiya A., editors. GeneReviews®. University of Washington; Seattle, WA, USA: 1993. [Google Scholar]
- 2.Chakchouk I., Zhang D., Zhang Z., Francioli L.C., Santos-Cortez R.L.P., Schrauwen I., Leal S.M. Disparities in discovery of pathogenic variants for autosomal recessive non-syndromic hearing impairment by ancestry. Eur. J. Hum. Genet. 2019;27:1456–1465. doi: 10.1038/s41431-019-0417-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaplan A.B., Kozin E.D., Puram S.V., Owoc M.S., Shah P.V., Hight A.E., Sethi R.K.V., Remenschneider A.K., Lee D.J. Auditory brainstem implant candidacy in the United States in children 0–17 years old. Int. J. Pediatr. Otorhinolaryngol. 2015;79:310–315. doi: 10.1016/j.ijporl.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sennaroglu L., Saatci I. A new classification for cochleovestibular malformations. Laryngoscope. 2002;112:2230–2241. doi: 10.1097/00005537-200212000-00019. [DOI] [PubMed] [Google Scholar]
- 5.Kari E., Llaci L., Go J.L., Naymik M., Knowles J.A., Leal S.M., Rangasamy S., Huentelman M.J., Liang W., Friedman R.A., et al. Genes Implicated in Rare Congenital Inner Ear and Cochleovestibular Nerve Malformations. Ear Hear. 2020 doi: 10.1097/AUD.0000000000000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schrauwen I., Kari E., Mattox J., Llaci L., Smeeton J., Naymik M., Raible D.W., Knowles J.A., Crump J.G., Huentelman M.J., et al. De novo variants in GREB1L are associated with non-syndromic inner ear malformations and deafness. Hum. Genet. 2018;137:459–470. doi: 10.1007/s00439-018-1898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunskill E.W., Potter A.S., Distasio A., Dexheimer P., Plassard A., Aronow B.J., Potter S.S. A gene expression atlas of early craniofacial development. Dev. Biol. 2014;391:133–146. doi: 10.1016/j.ydbio.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Tomasi L., David P., Humbert C., Silbermann F., Arrondel C., Tores F., Fouquet S., Desgrange A., Niel O., Bole-Feysot C., et al. Mutations in GREB1L cause bilateral kidney agenesis in humans and mice. Am. J. Hum. Genet. 2017;101:803–814. doi: 10.1016/j.ajhg.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cacheiro P., Haendel M.A., Smedley D. International Mouse Phenotyping Consortium and the Monarch Initiative New models for human disease from the International Mouse Phenotyping Consortium. Mamm. Genome. 2019;30:143–150. doi: 10.1007/s00335-019-09804-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cartegni L., Wang J., Zhu Z., Zhang M.Q., Krainer A.R. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desmet F.-O., Hamroun D., Lalande M., Collod-Béroud G., Claustres M., Béroud C. Human splicing finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boissel S., Fallet-Bianco C., Chitayat D., Kremer V., Nassif C., Rypens F., Delrue M.-A., Soglio D.D., Oligny L.L., Patey N., et al. Genomic study of severe fetal anomalies and discovery of GREB1L mutations in renal agenesis. Genet. Med. 2018;20:745–753. doi: 10.1038/gim.2017.173. [DOI] [PubMed] [Google Scholar]
- 14.Sanna-Cherchi S., Khan K., Westland R., Krithivasan P., Fievet L., Rasouly H.M., Ionita-Laza I., Capone V.P., Fasel D.A., Kiryluk K., et al. Exome-wide Association Study Identifies GREB1L Mutations in Congenital Kidney Malformations. Am. J. Hum. Genet. 2017;101:789–802. doi: 10.1016/j.ajhg.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J., Russell D.W. Purification of nucleic acids by extraction with phenol: Chloroform. CSH Protoc. 2006;2006 doi: 10.1101/pdb.prot4455. [DOI] [PubMed] [Google Scholar]
- 16.Schultz J.M., Khan S.N., Ahmed Z.M., Riazuddin S., Waryah A.M., Chhatre D., Starost M.F., Ploplis B., Buckley S., Velásquez D., et al. Noncoding mutations of HGF are associated with nonsyndromic hearing loss, DFNB39. Am. J. Hum. Genet. 2009;85:25–39. doi: 10.1016/j.ajhg.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riazuddin S., Belyantseva I.A., Giese A.P.J., Lee K., Indzhykulian A.A., Nandamuri S.P., Yousaf R., Sinha G.P., Lee S., Terrell D., et al. Alterations of the CIB2 calcium- and integrin-binding protein cause Usher syndrome type 1J and nonsyndromic deafness DFNB48. Nat. Genet. 2012;44:1265–1271. doi: 10.1038/ng.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shahzad M., Sivakumaran T.A., Qaiser T.A., Schultz J.M., Hussain Z., Flanagan M., Bhinder M.A., Kissell D., Greinwald J.H., Khan S.N., et al. Genetic Analysis through OtoSeq of Pakistani Families Segregating Prelingual Hearing Loss. Otolaryngol. -Head Neck Surg. 2013;149:478–487. doi: 10.1177/0194599813493075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H., Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 2015;10:1556–1566. doi: 10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott E.M., Halees A., Itan Y., Spencer E.G., He Y., Azab M.A., Gabriel S.B., Belkadi A., Boisson B., Abel L., et al. Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat. Genet. 2016;48:1071–1076. doi: 10.1038/ng.3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X., Wu C., Li C., Boerwinkle E. dbNSFP v3.0: A one-stop database of functional predictions and annotations for human Nonsynonymous and splice-site SNVs. Hum. Mutat. 2016;37:235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jian X., Boerwinkle E., Liu X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014;42:13534–13544. doi: 10.1093/nar/gku1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S. Identifying a High Fraction of the Human Genome to be under Selective Constraint Using GERP++ PLoS Comput. Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen J., Scheffer D.I., Kwan K.Y., Corey D.P. SHIELD: An integrative gene expression database for inner ear research. Database (Oxford) 2015;2015 doi: 10.1093/database/bav071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrauwen I., Hasin-Brumshtein Y., Corneveaux J.J., Ohmen J., White C., Allen A.N., Lusis A.J., Van Camp G., Huentelman M.J., Friedman R.A. A comprehensive catalogue of the coding and non-coding transcripts of the human inner ear. Hear. Res. 2016;333:266–274. doi: 10.1016/j.heares.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krumm N., Sudmant P.H., Ko A., O’Roak B.J., Malig M., Coe B.P., Quinlan A.R., Nickerson D.A., Eichler E.E. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–1532. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J., Haider S., Baran J., Cros A., Guberman J.M., Hsu J., Liang Y., Yao L., Kasprzyk A. BioMart: A data federation framework for large collaborative projects. Database (Oxford) 2011;2011 doi: 10.1093/database/bar038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacDonald J.R., Ziman R., Yuen R.K.C., Feuk L., Scherer S.W. The Database of Genomic Variants: A curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Optimizing Genomic Medicine in Epilepsy Through a Gene-Customized Approach to Missense Variant Interpretation. [(accessed on 6 March 2020)]; doi: 10.1101/gr.226589.117. Available online: https://genome.cshlp.org/content/27/10/1715/F1.expansion.html. [DOI] [PMC free article] [PubMed]
- 34.Jarvik G.P., Browning B.L. Consideration of Cosegregation in the Pathogenicity Classification of Genomic Variants. Am. J. Hum. Genet. 2016;98:1077–1081. doi: 10.1016/j.ajhg.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasmussen M., Sunde L., Nielsen M.L., Ramsing M., Petersen A., Hjortshøj T.D., Olsen T.E., Tabor A., Hertz J.M., Johnsen I., et al. Targeted gene sequencing and whole-exome sequencing in autopsied fetuses with prenatally diagnosed kidney anomalies. Clin. Genet. 2018;93:860–869. doi: 10.1111/cge.13185. [DOI] [PubMed] [Google Scholar]
- 36.Herlin M.K., Le V.Q., Højland A.T., Ernst A., Okkels H., Petersen A.C., Petersen M.B., Pedersen I.S. Whole-exome sequencing identifies a GREB1L variant in a three-generation family with Müllerian and renal agenesis: A novel candidate gene in Mayer-Rokitansky-Küster-Hauser (MRKH) syndrome. A case report. Hum. Reprod. 2019;34:1838–1846. doi: 10.1093/humrep/dez126. [DOI] [PubMed] [Google Scholar]
- 37.Adeline J., Bouchra B., Corinne F., Jerôme T., Claire J., Aimé L., Elise B.-B., Christèle D., Véronique D., Laurent P., et al. GREB1L variants in familial and sporadic hereditary urogenital adysplasia and Mayer-Rokitansky-Kuster-Hauser syndrome. Clin. Genet. 2020 doi: 10.1111/cge.13769. [DOI] [PubMed] [Google Scholar]
- 38.Brophy P.D., Rasmussen M., Parida M., Bonde G., Darbro B.W., Hong X., Clarke J.C., Peterson K.A., Denegre J., Schneider M., et al. A Gene Implicated in Activation of Retinoic Acid Receptor Targets Is a Novel Renal Agenesis Gene in Humans. Genetics. 2017;207:215–228. doi: 10.1534/genetics.117.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iyer L.M., Zhang D., Burroughs A.M., Aravind L. Computational identification of novel biochemical systems involved in oxidation, glycosylation and other complex modifications of bases in DNA. Nucleic Acids Res. 2013;41:7635–7655. doi: 10.1093/nar/gkt573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plouhinec J.-L., Roche D.D., Pegoraro C., Figueiredo A.-L., Maczkowiak F., Brunet L.J., Milet C., Vert J.-P., Pollet N., Harland R.M., et al. Pax3 and Zic1 trigger the early neural crest gene regulatory network by the direct activation of multiple key neural crest specifiers. Dev. Biol. 2014;386:461–472. doi: 10.1016/j.ydbio.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shakhova O., Sommer L. StemBook. Harvard Stem Cell Institute; Cambridge, MA, USA: 2008. Neural crest-derived stem cells. [PubMed] [Google Scholar]
- 42.Whitfield T.T. Development of the inner ear. Curr. Opin. Genet. Dev. 2015;32:112–118. doi: 10.1016/j.gde.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 43.Webb B.D., Shaaban S., Gaspar H., Cunha L.F., Schubert C.R., Hao K., Robson C.D., Chan W.-M., Andrews C., MacKinnon S., et al. HOXB1 founder mutation in humans recapitulates the phenotype of Hoxb1-/- mice. Am. J. Hum. Genet. 2012;91:171–179. doi: 10.1016/j.ajhg.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vogel M., Velleuer E., Schmidt-Jiménez L.F., Mayatepek E., Borkhardt A., Alawi M., Kutsche K., Kortüm F. Homozygous HOXB1 loss-of-function mutation in a large family with hereditary congenital facial paresis. Am. J. Med. Genet. A. 2016;170:1813–1819. doi: 10.1002/ajmg.a.37682. [DOI] [PubMed] [Google Scholar]
- 45.Brown A.S., Epstein D.J. Otic ablation of smoothened reveals direct and indirect requirements for Hedgehog signaling in inner ear development. Development. 2011;138:3967–3976. doi: 10.1242/dev.066126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orten D.J., Fischer S.M., Sorensen J.L., Radhakrishna U., Cremers C.W.R.J., Marres H.A.M., Van Camp G., Welch K.O., Smith R.J.H., Kimberling W.J. Branchio-oto-renal syndrome (BOR): Novel mutations in the EYA1 gene, and a review of the mutational genetics of BOR. Hum. Mutat. 2008;29:537–544. doi: 10.1002/humu.20691. [DOI] [PubMed] [Google Scholar]
- 47.Vega-Lopez G.A., Cerrizuela S., Tribulo C., Aybar M.J. Neurocristopathies: New insights 150 years after the neural crest discovery. Dev. Biol. 2018;444(Suppl 1):S110–S143. doi: 10.1016/j.ydbio.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 48.Marques A.H., O’Connor T.G., Roth C., Susser E., Bjørke-Monsen A.-L. The influence of maternal prenatal and early childhood nutrition and maternal prenatal stress on offspring immune system development and neurodevelopmental disorders. Front. Neurosci. 2013;7:120. doi: 10.3389/fnins.2013.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cable J., Barkway C., Steel K.P. Characteristics of stria vascularis melanocytes of viable dominant spotting (Wv/Wv) mouse mutants. Hear. Res. 1992;64:6–20. doi: 10.1016/0378-5955(92)90164-I. [DOI] [PubMed] [Google Scholar]
- 50.Mochizuki T., Lemmink H.H., Mariyama M., Antignac C., Gubler M.C., Pirson Y., Verellen-Dumoulin C., Chan B., Schröder C.H., Smeets H.J. Identification of mutations in the alpha 3(IV) and alpha 4(IV) collagen genes in autosomal recessive Alport syndrome. Nat. Genet. 1994;8:77–81. doi: 10.1038/ng0994-77. [DOI] [PubMed] [Google Scholar]
- 51.Birkenhäger R., Otto E., Schürmann M.J., Vollmer M., Ruf E.M., Maier-Lutz I., Beekmann F., Fekete A., Omran H., Feldmann D., et al. Mutation of BSND causes Bartter syndrome with sensorineural deafness and kidney failure. Nat. Genet. 2001;29:310–314. doi: 10.1038/ng752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.