Abstract
Dendroctonus valens is the main pest of the genus Pinus. To facilitate gene expression analyses, suitable reference genes for adults and mature larvae of D. valens under different temperature conditions were determined. In particular, we obtained the sequences of candidate reference genes, ACT, TUB, SHDA, PRS18, 18S rRNA, and CYP4G55, from transcriptome data. Real-time quantitative PCR was used to analyze gene expression, and geNorm, NormFinder, and BestKeeper were used to evaluate expression stability. Under different temperature conditions, the expression levels of 18S rRNA, PRS18, and TUB were stable in adults, in which 18S rRNA > PRS18 > TUB. In mature larvae, the expression levels of TUB, 18S rRNA, and SDHA were stable, in which TUB > 18S rRNA > SDHA. The combination of 18S rRNA and PRS18 is recommended for studies of gene expression in adults and the combination of 18S rRNA and TUB is effective for studies of gene expression in mature larvae of D. valens under different temperature conditions.
Keywords: Dendroctonus valens, qRT-PCR, reference genes
1. Introduction
Dendroctonus valens (Coleoptera: Scolytidae) is a major pest of the genus Pinus, native to Canada and North America. In China, abundant food resources, the lack of interspecific competition, and the lack of effective natural enemies have enabled the spread of D. valens [1]. Since its discovery in Shanxi Province in the late 1990s, the area of infestation has expanded rapidly, reaching Beijing, Hebei, Henan, Inner Mongolia, and Liaoning. The pest has caused serious harm to Pinus tabulaeformis and P. Sylvestris and has become one of the main invasive pests in China.
Precise analyses of gene expression levels are important for molecular biology research. Real-time quantitative PCR (qRT-PCR) has become the “gold standard” for transcript-level expression analyses, owing to its high accuracy, specificity, sensitivity, and rapidity [2,3]. However, in the implementation of qRT-PCR, RNA extraction, polymerase amplification, and cDNA synthesis can all lead to systematic errors [2,4]. To eliminate these sources of error, genes with constant expression levels across conditions are usually selected as references for normalization. Under different experimental treatments, the expression level of an ideal reference gene remains constant [5]. However, the wide application of qRT-PCR has revealed that a single gene is not expressed stably under complex experimental treatments [6,7]. Appropriate reference genes may differ depending on the insect species, experimental treatment, development stage, tissue type, and temperature [8,9,10,11,12]. Common reference genes for insect studies include beta-actin, beta-tubulin, alpha-tubulin, ribosomal protein S18 (PRS18), 18S ribosomal RNA (18S rRNA), and succinate dehydrogenase complex subunit A (SDHA) [13]. These genes are involved in normal metabolic processes in cells. Reference gene screening has been performed in Coleoptera, including Leptinotarsa decemlineata, Diabrotica virgifera virgifera, Colaphellus bowringi, and Harmonia axyridis [14,15,16,17].
Few studies have evaluated D. valens at the molecular level. Cano-Ramirez studied P450 expression in the antennae and intestines of D. valens exposed to monoterpenes; the cytochrome P450 gene CYP4G55 was stably expressed under these conditions and therefore was identified as a suitable reference gene [18]. However, it is not clear whether this reference gene exhibits stable expression under other experimental treatments. In this study, we further screened reference genes to obtain a reliable internal reference gene for studying the gene expression pattern of D. valens.
2. Materials and Methods
2.1. Insects
Mature larvae and adults of D. valens were collected from Chifeng (Inner Mongolia, China). Live insects were brought to the laboratory in dark conditions. All samples were maintained for 2 h at 25 °C; some were placed in liquid nitrogen for quick-freezing and the rest were maintained at −10 °C, −5 °C, 0 °C, 5 °C and 10 °C for 1 h, immediately frozen in liquid nitrogen, and stored in a refrigerator at −80 °C.
2.2. RNA Isolation and cDNA Synthesis
TRIzol (No. 15596018; Invitrogen, Carlsbad, CA, USA) and the RNeasy Plus Mini Kit (No. 74134; Qiagen, Hilden, Germany) were used for RNA extraction from mature larvae and adults of D. valens according to the manufacturers’ instructions. The micro-ultraviolet/visible spectrophotometer NanoDrop 8000 (Thermo, Waltham, MA, USA) was used to determine the A260/A280 ratio for evaluations of the quality and concentration of extracted RNA. Single stranded cDNA was synthesized from 1.0 μg total RNA using the Prime Script RT Reagent Kit with gDNA Eraser Kit (TaKaRa, Shiga, Japan).
2.3. Selection of Candidate Reference Genes
Based on transcriptome data obtained by high-throughput sequencing in our laboratory, gene sequences were compared with sequences of related species in GenBank. Finally, six common housekeeping genes were selected as candidate reference genes (ACT, TUB, SHDA, PRS18, 18S rRNA, and CYP4G55). The sequences of the candidate reference genes were verified by RT-PCR. Total RNA was extracted and cDNA was obtained by reverse transcription as described above. Primers for RT-PCR were designed, and cDNA was used as the template for PCR amplification. The PCR system (25 μL) included cDNA template (1 μL), 2× PrimeSTAR Max Premix (12.5 μL), forward and reverse primers (10 μmol/L; 0.5 μL), and ddH2O (10.5 μL). PCR conditions were as follows: 94 °C for 2 min; 35 cycles of 94 °C for 30 s, 58 °C for 30 s, 72 °C for 30 s; 72 °C for 3 min.
2.4. Primer Design
Based on the nucleotide sequences of candidate reference genes, Primer3Plus was used to design primers (Figure S1) and the primer sequences were synthesized by Beijing Ruiboxingke Biotechnology Co., Ltd. (Beijing, China). The specificity of the primers was determined by the confirmation of a single peak in the melting curve. A 5× gradient dilution of the cDNA of adults and mature larvae was used as a template to draw the standard curve to determine the amplification efficiency of primers.
2.5. qRT-PCR Analysis
cDNA was used as a template for qRT-PCR using the newly designed primers. The analysis was repeated three times for each sample with three technical repetitions for each group. The reaction system (12.5 μL) was as follows: cDNA template (1 μL), forward and reverse primers (10 μmol/L; 2 μL), SYBRPremix Ex Taq II (6.25 μL), and ddH2O (4.25 μL). The reaction conditions were as follows: 95 °C for 3 min; 40 cycles of 95 °C for 10 s, 58 °C for 30 s; 5 s at 95 °C, 65 °C–95 °C, 0.5 °C increase/cycle. After the reaction, the amplification curve and melting curve were confirmed.
2.6. Analysis and Verification of the Stability of Reference Genes
The experimental data obtained by qRT-PCR were analyzed by four methods: ΔCt, geNorm, BestKeeper, and NormFinder. The standard procedures for each method were followed. Based on the expression level of each candidate reference gene and results for each algorithm, stable expression across different temperatures was evaluated.
For verification, HSP21, which encodes a heat shock protein in D. valens, was used as a target gene. With different candidate reference genes, the 2−ΔΔCt method was used to calculate the relative expression of HSP21 under different temperature conditions [19] to verify the stability of reference genes.
3. Results
3.1. Selection of Candidate Reference Genes and Primer Design
ACT, SDHA, 18S rRNA, RPS18, CYP4G55, and TUB were selected as candidate reference genes based on transcriptome sequencing data. An alignment showed high sequence similarity with Dendroctonus ponderosae loci of over 90%, and the sequences determined by RT-PCR were similar to those obtained by transcriptome sequencing (over 99% similarity). Specific primers for qPCR were designed based on gene sequences. A standard curve analysis indicated that the amplification efficiency of the qPCR primers was 91.3–109.8% with high correlation coefficients (R2 > 99%), indicating that the standard curve showed a good linear relationship (Table 1). In addition, the melting curves for all candidate genes exhibited single peaks, indicating high primer specificity (Figure S2).
Table 1.
Primer sequences, product sizes, and PCR efficiencies for the assessed genes.
| Gene | RT-qPCR | ||
|---|---|---|---|
| Primer Sequences | Amplification Efficiency | R 2 | |
| CYP4G55 | F: AGCCAACGAGTTCGGAAGAG | 98.7 | 0.994 |
| R: TCAGAAGACCATCGCCCAAC | |||
| 18S rRNA | F: TGGAGGAAAACGGGCACTAC | 91.3 | 0.998 |
| R: GACTTGTCTGCGTTGCACAG | |||
| SDHA | F: CTGGTGCCGATACGCAAATG | 92.1 | 1.000 |
| R: TAAGGCTGTTGTCGGACACC | |||
| TUB | F: CTTACCACCCCCACATACGG | 97.8 | 1.000 |
| R: ATTGCTGACTGCCTCTGGAC | |||
| ACT | F: TGCTGCAGGAAGATCCACTG | 109.8 | 0.995 |
| R: GCACTGTCCCTGTCAGGTAC | |||
| PRS18 | F: CATCGCTCTGTCCTCGGTAC | 102.3 | 0.999 |
| R: TCGGTGTGCTTGACATCCAA | |||
| HSP21 | F: TGGATGTGGAGGGCTTCAAG | 107.1 | 0.997 |
| R: TGTTAGAACGCCGTCCTCAC | |||
3.2. Stability Analysis of Candidate Reference Genes
3.2.1. Expression Levels of the Candidate Reference Genes
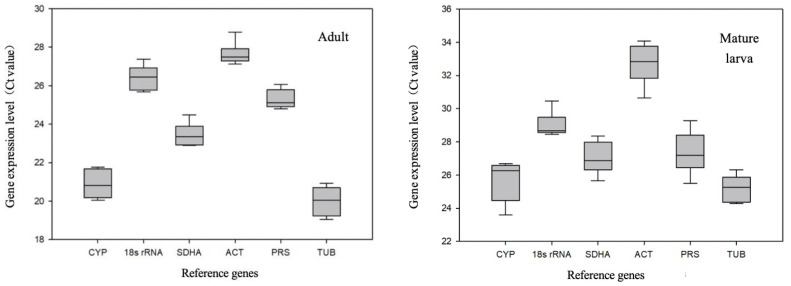
Transcript abundance and cycle threshold (Ct) variation are important parameters for screening reference genes. The stability of Ct values is an important criterion for reference gene selection. The smaller the Ct value of the gene, the higher its expression level; conversely, the larger the Ct value, the lower its expression level. As shown in Figure 1, the average Ct values for the six candidate reference genes were between 19 and 35 at different temperatures, and transcript abundances in adults were higher than those in mature larvae. The expression patterns of the six candidate reference genes were similar at the two developmental stages. The transcript abundances of TUB and CYP4G55 were higher than those of other genes, while the abundance of ACT was the lowest. In addition, the average standard deviation of each gene set negatively correlated with its stability. Ct values for CYP4G55 and TUB exhibited high variation among adults, while Ct values for CYP4G55 and ACT exhibited substantial variation among mature larvae.
Figure 1.
Expression levels of candidate reference genes in adults and mature larvae of D. valens. Expression levels are displayed as raw cycle threshold (Ct) values of the candidate reference genes of D. valens at different temperatures. The boxes represent the values between the 25th and 75th percentiles, the lines in the boxes indicate the median values, and the whisker caps denote the minimal to maximal values.
3.2.2. geNorm Analysis
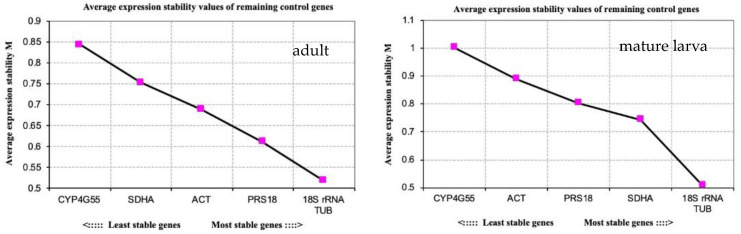
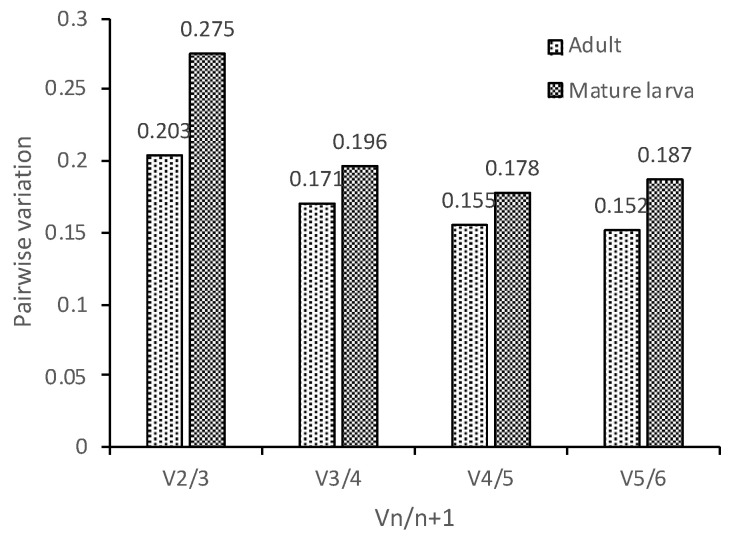
geNorm was used to analyze the expression stability of the candidate reference genes at different temperatures. Stability values (M) were obtained. A lower value indicates more stable gene expression. As shown in Figure 2, TUB and 18S rRNA were the most stable and CYP4G55 was the most unstable gene in both adults and mature larvae exposed to different temperatures. In Figure 3, we found that the pairwise variation Vn/(Vn + 1) was greater than 0.15 for samples at different temperatures. Therefore, to reduce experimental error, it is necessary to use multiple reference genes to analyze target gene expression in D. valens under different temperature treatments.
Figure 2.
Expression stability of the candidate reference genes in adults and mature larvae of D. valens evaluated using geNorm.
Figure 3.
Pairwise variation (Vn/Vn + 1) analysis of the number of candidate reference genes in D. valens. According to the paired difference analysis of the standardized factor of the internal reference gene, the mutation value V was obtained for pairwise comparison, so as to determine the optimal number of the internal reference gene. When the Vn/n + 1 value was less than the threshold value of 0.15, the optimal number of the internal reference gene was n.
3.2.3. NormFinder Analysis
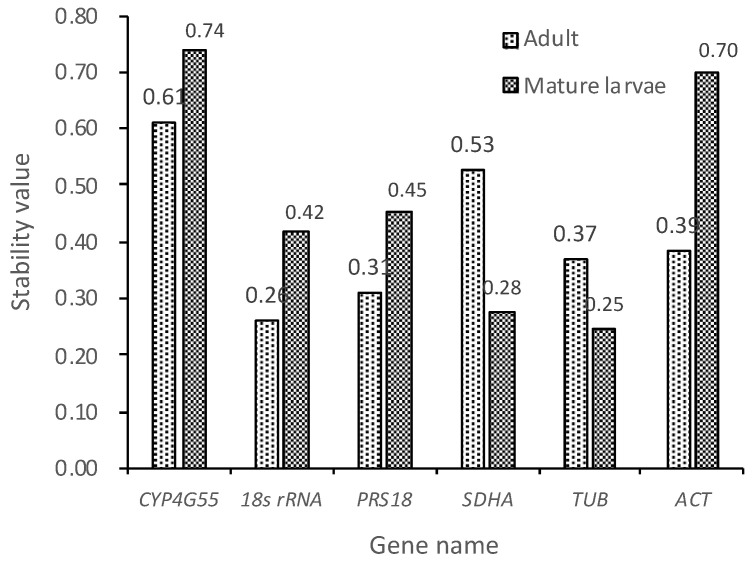
The results of a NormFinder analysis differed slightly from those of the geNorm analysis, as shown in Figure 4. Using NormFinder, under the same experimental conditions, the most stable reference gene in adults was 18S rRNA with a stability value of 0.26, followed by PRS18 and TUB with stability values of 0.31 and 0.37. In mature larvae, TUB was the most stable gene (M = 0.25), followed by SDHA and 18S rRNA, with stability values of 0.28 and 0.45. However, the most unstable reference gene for both adults and mature larvae was CYP4G55.
Figure 4.
Expression stability of candidate reference genes in adults and mature larvae of D. valens evaluated using NormFinder. A lower stability value indicates a more stable expression.
3.2.4. BestKeeper Analysis
In a BestKeeper analysis, a higher correlation coefficient (r) indicates a lower standard deviation (SD) and coefficient of variation (CV) and higher reference gene stability. In this analysis, SDHA and PRS18 were the most stable genes in adults, and TUB and 18S rRNA were the most stable genes in mature larvae (Table 2 and Table 3). Moreover, the p-values for the candidate reference genes were all less than 0.05, indicating that these genes could be used in combination as co-reference genes.
Table 2.
Expression stability of the candidate reference genes in adult D. valens evaluated using BestKeeper.
| Gene | n | geo Mean | AR Mean | min | max | SD | CV [%] | min [x-fold] |
max [x-fold] |
SD [±x-fold] |
CC [r] | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CYP4G55 | 18 | 20.83 | 20.88 | 19.22 | 24.21 | 1.20 | 5.77 | −3.06 | 10.35 | 2.31 | 0.765 | 0.001 |
| 18S rRNA | 18 | 26.39 | 26.42 | 24.07 | 28.70 | 1.01 | 3.81 | −5.00 | 4.98 | 2.01 | 0.915 | 0.001 |
| SDHA | 18 | 23.42 | 23.45 | 21.52 | 26.15 | 0.87 | 3.72 | −3.73 | 6.64 | 1.83 | 0.761 | 0.001 |
| ACT | 18 | 27.62 | 27.65 | 25.99 | 30.94 | 1.17 | 4.22 | −3.09 | 10.00 | 2.24 | 0.878 | 0.001 |
| PRS18 | 18 | 25.26 | 25.29 | 23.14 | 27.16 | 0.88 | 3.50 | −4.35 | 3.73 | 1.85 | 0.876 | 0.001 |
| TUB | 18 | 19.90 | 19.99 | 16.40 | 23.09 | 1.65 | 8.26 | −11.32 | 9.14 | 3.14 | 0.908 | 0.001 |
Table 3.
Expression stability of the candidate reference genes in mature larval D. valens evaluated using BestKeeper.
| Gene | n | geo Mean | AR Mean | min | max | SD | CV [%] | Min [x-fold] |
max [x-fold] |
SD [±x-fold] |
CC [r] | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CYP4G55 | 18 | 25.63 | 25.68 | 22.93 | 28.27 | 1.32 | 5.15 | −6.50 | 6.26 | 2.50 | 0.773 | 0.001 |
| 18S rRNA | 18 | 28.96 | 29.00 | 26.88 | 32.40 | 1.16 | 3.98 | −4.24 | 10.82 | 2.23 | 0.836 | 0.001 |
| SDHA | 18 | 26.95 | 27.02 | 24.53 | 32.14 | 1.53 | 5.67 | −5.35 | 36.53 | 2.89 | 0.905 | 0.001 |
| ACT | 18 | 32.66 | 32.71 | 29.05 | 35.18 | 1.62 | 4.95 | −12.23 | 5.71 | 3.07 | 0.696 | 0.001 |
| PRS18 | 18 | 27.28 | 27.33 | 24.44 | 32.01 | 1.36 | 4.97 | −7.13 | 26.67 | 2.56 | 0.946 | 0.001 |
| TUB | 18 | 25.17 | 25.20 | 23.51 | 28.25 | 1.01 | 4.01 | −3.17 | 8.46 | 2.01 | 0.931 | 0.001 |
3.3. Comprehensive Ranking of Candidate Reference Genes
According to the results obtained using the three algorithms, reference gene sequences were obtained. The geometric mean parameter values for each reference gene sequence were calculated as the final ranking [20]. As shown in Table 4, under different temperature treatments, the order of reference genes ranked by stability in adults was 18S rRNA > PRS18 > TUB > SDHA > ACT > CYP4G55 and in mature larvae was TUB > 18S rRNA > SDHA > PRS18 > CYP4G55 > ACT.
Table 4.
Reference genes ranks by geNorm, NormFinder, BestKeeper, and overall rank.
| RANK | geNorm | NormFinder | BestKeeper | OVERALL | |
|---|---|---|---|---|---|
| Adults | 1 | 18S rRNA/TUB | 18S rRNA | SDHA | 18S rRNA |
| 2 | PRS18 | PRS18 | PRS18 | ||
| 3 | PRS18 | TUB | 18S rRNA | TUB | |
| 4 | ACT | ACT | ACT | SDHA | |
| 5 | SDHA | SDHA | CYP4G55 | ACT | |
| 6 | CYP4G55 | CYP4G55 | TUB | CYP4G55 | |
| Mature larvae | 1 | 18S rRNA /TUB | TUB | TUB | TUB |
| 2 | SDHA | 18S rRNA | 18S rRNA | ||
| 3 | SDHA | 18S rRNA | CYP4G55 | SDHA | |
| 4 | PRS18 | PRS18 | PRS18 | PRS18 | |
| 5 | ACT | ACT | SDHA | CYP4G55 | |
| 6 | CYP4G55 | CYP4G55 | ACT | ACT |
3.4. Verification of Reference Genes
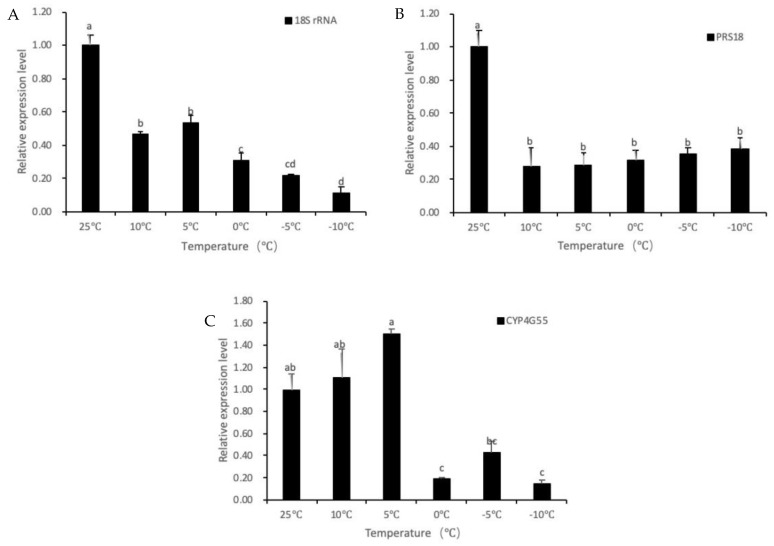
To verify the stability of the reference genes, 18S rRNA (the gene with the highest stability in comprehensive analyses in adults), PRS18 (with slightly less stability), and CYP4G55 (with the lowest stability) were selected to determine the relative expression level of HSP21 in adults under different low-temperature conditions. As shown in Figure 5, using 18S rRNA and PRS18 as reference genes, levels of gene expression were similar. When 18S rRNA was used as the reference gene, the expression level of HSP21 under low temperatures was significantly lower than that in the 25 °C. When PRS18 was used as the reference gene, the expression level of HSP21 under temperature stress was slightly different from that of 18S rRNA but was also lower than that in the 25 °C. However, when CYP4G55 was used as the reference gene, the expression levels of the target gene HSP21 at 10 °C and 5 °C were not significantly lower than those in the 25 °C, in contrast to the results obtained using 18S rRNA or PRS18 as the reference gene. This indicates that an unstable reference gene may affect the accuracy of qPCR results and even lead to incorrect conclusions.
Figure 5.
Relative expression levels of HSP21 under various temperatures in adult D. valens using different reference genes. (A–C) 18S rRNA, RPS18 and CYP4G55 were used as reference genes, respectively. Data are mean ± SE. Different letters above bars mean significant differences (p < 0.05, Duncan’s test).
4. Discussion
Our results showed that 18S rRNA and TUB were stable reference genes in adults and mature larvae at different temperatures, respectively. 18S rRNA is a commonly used reference gene. In Coccinella septempunctata, 18S rRNA is considered the best reference gene for analyses of gene expression in different tissue types; in H. axyridis, it is considered the best reference gene for analyses of gene expression at different temperatures [17,21]. Tubulin is a cytoskeleton component. It functions in the maintenance cell shape, mitosis, cell movement, intracellular transport, and organelle composition. Genes encoding tubulin are often used as reference genes. TUB is expressed stably in various tissues of D. virgifera virgifera [15]. It is also stably expressed under different temperature treatments in Nilaparvata lugens [7], but is not stable in second-instar larvae of Galeruca daurica under different temperature treatments [22].
Few studies have focused on reference genes in D. valens. Cano-Ramirez preliminarily screened reference genes and found that CYP4G55 is stable and could be used as a reference for studies of P450 in the antennae and intestines of D. valens exposed to monoterpenes [18]. However, in this study, CYP4G55 stability was low relative to those of the other candidates in both adults and mature larvae in low temperatures. Actin (ACT) is the main structural protein of the cytoskeleton with an important role in cell functions. However, we found that this gene, which is commonly used as a reference in other insects, is far less stable than other candidate reference genes in adults and mature larvae. Similarly, in different tissues of C. septempunctata and at different temperatures in Phenacoccus solenopsis, Henosepilachna vigintioctomaculata, and Hippodamia convergens, ACT has low stability [21,23,24,25]. It is a relatively stable reference gene in Myzus persicae under temperature stress [26]. These results prove that reference gene screening results depend on the species, sample type, and experimental conditions, further supporting the importance of screening.
Research has shown that none of the genes exhibited constant expression levels. Therefore, the use of two or more reference genes may yield more accurate results. Under different temperatures, GAPDH and EF-1α are stably expressed in Spodoptera litura [27], and Actin, Mnf, and α-TUB are stably expressed in Drosophila melanogaster [28]. In addition, for the same insect taxa and experimental conditions, different candidate genes can be identified. For example, a study has shown that RPS15 and RPL27 are stably expressed in Helicoverpa armigera [29], while another study has shown that the combination of RPL28 and RPS15 is the most suitable reference under the same conditions [30].
In this study, geNorm, NormFinder, and BestKeeper were used to evaluate and verify the stability of reference genes. The results obtained using the three algorithms were not completely consistent. Under different temperatures, geNorm, NormFinder, and BestKeeper all showed that TUB is the most stable reference genes in mature larvae. However, in adults exposed to different temperatures, geNorm and NormFinder both showed that 18S rRNA and PRS18 were relatively stable reference genes, while BestKeeper showed that SDHA and PRS18 were relatively stable. Similar differences have been reported in studies of Lipaphis erysimi and Anastrepha obliqua; differences in software and statistical methods may influence stable gene identification [31,32]. Finally, the geometric mean value for the stability of each reference gene was used for a comprehensive analysis. Under different temperature conditions, the expression levels of 18S rRNA, PRS18, and TUB (in order) were most stable in adults, and the expression levels of TUB, 18S rRNA, and SDHA (in order) were most stable in larvae. These results not only provide an experimental basis for the selection of reference genes for studies of the cold tolerance of D. valens, but also provide a reference for screening studies focused on other insects. The application of these loci as reference genes under other physiological or experimental conditions remains to be determined.
5. Conclusions
In this study, six candidate reference genes were screened from transcriptome data for D. valens. The expression stability of the candidate reference genes in adults and mature larvae under temperature stress was evaluated. Based on our results, the combination of 18S rRNA and PRS18 is recommended for studies of gene expression in adult D. valens at different temperatures, and the combination of 18S rRNA and TUB is effective for studies of gene expression in mature larvae. These results contribute to studies of reference genes in Coleoptera and provides a basis for molecular studies of D. valens.
Acknowledgments
We thank Bioedit (www.bioedit.com) for linguistic assistance during manuscript preparation.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4450/11/6/328/s1, Figure S1: The design strategy of Primer3Plus, Figure S2: Specificity of real-time qRT-PCR amplification in Dendroctonus valens. Melting curves of six genes [ACT (A), SDHA (B), 18S rRNA (C), RPS18 (D), CYP4G55 (E) and TUB (F)] reveal single peaks.
Author Contributions
Conceptualization, C.Z.; methodology, C.Z. and J.T.; validation, C.Z., J.T., D.Z., Y.X., F.S. and S.Z.; formal analysis, C.Z.; resources, J.T. and S.Z.; data curation, C.Z. and J.T.; writing—original draft preparation, C.Z.; writing—review and editing, C.Z.; project administration, J.T. and S.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by “Chinese National Natural Science Foundation” (NO. 31870642).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Yan Z., Sun J., Don O., Zhang Z. The red turpentine beetle, Dendroctonus valens LeConte (Scolytidae): An exotic invasive pest of pine in China. Biodivers. Conserv. 2005;14:1735–1760. doi: 10.1007/s10531-004-0697-9. [DOI] [Google Scholar]
- 2.Bustin S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- 3.Valasek M.A., Repa J.J. The power of real-time PCR. Adv. Physiol. Educ. 2005;29:151–159. doi: 10.1152/advan.00019.2005. [DOI] [PubMed] [Google Scholar]
- 4.Vandesompele J., De Preter K., Pattyn F., Poppe B., Roy N.V., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:1–11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su Y.L., He W.B., Wang J., Li J.M., Liu S.S., Wang X.W. Selection of endogenous reference genes for gene expression analysis in the Mediterranean species of the whitefly Bemisia tabaci (Hemiptera: Aleyrodidae) complex. J. Econ. Entomol. 2013;106:1446–1455. doi: 10.1603/EC12459. [DOI] [PubMed] [Google Scholar]
- 6.Omondi B.A., Latorre-Estivalis J.M., Oliveira I.H.R., Ignell R., Lorenzo M.G. Evaluation of reference genes for insect olfaction studies. Parasit. Vector. 2015;8:243. doi: 10.1186/s13071-015-0862-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan M., Lu Y., Zhu X., Wan H., Shakeel M., Zhan S., Jin B.R., Li J.H. Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PLoS ONE. 2014;9:e86503. doi: 10.1371/journal.pone.0086503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basu S., Pereira A.E., Pinheiro D.H., Wang H.C., Valencia-Jimenez A. Evaluation of reference genes for real-time quantitative PCR analysis in southern corn rootworm, Diabrotica undecimpunctata howardi (Barber) Sci. Rep. 2019;9:10703. doi: 10.1038/s41598-019-47020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qu C., Wang R., Che W.N., Zhu X., Li F.Q., Luo C. Selection and evaluation of reference genes for expression analysis using quantitative real-time PCR in the Asian Ladybird Harmonia axyridis (Coleoptera: Coccinellidae) PLoS ONE. 2018;13:e0192521. doi: 10.1371/journal.pone.0192521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajarapu S.P., Mamidala P., Mittapalli O. Validation of reference genes for gene expression studies in the emerald ash borer (Agrilus planipennis) Insect Sci. 2012;19:41–46. doi: 10.1111/j.1744-7917.2011.01447.x. [DOI] [Google Scholar]
- 11.Yu S.H., Yang P., Sun T., Qi Q., Wang X.Q., Xu D.L., Chen X.M. Identification and evaluation of reference genes in the Chinese white wax scale insect Ericerus pela. SpringerPlus. 2016;5:791. doi: 10.1186/s40064-016-2548-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi C.H., Yang F.S., Zhu X., Du E., Yang Y.T., Wang S.L., Wu Q.J., Zhang Y.J. Evaluation of Housekeeping Genes for Quantitative Real-Time PCR Analysis of Bradysia odoriphaga (Diptera: Sciaridae) Int. J. Mol. Sci. 2016;17:1034. doi: 10.3390/ijms17071034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekalska B., Ciechanowicz A., Dolegowska B., Naruszewicz M. Optimized RT-PCR Method for Assaying Expression of Monocyte Chemotactic Protein Type 1 (MCP-1) in Rabbit Aorta. Biochem. Genet. 2006;44:129–139. doi: 10.1007/s10528-006-9015-4. [DOI] [PubMed] [Google Scholar]
- 14.Shi X.Q., Guo W.C., Wan P.J., Zhou L.T., Ren X.L., Ahmat T., Fu K.Y., Li G.Q. Validation of reference genes for expression analysis by quantitative real-time PCR in Leptinotarsa decemlineata (Say) BMC Res. Notes. 2013;6:93. doi: 10.1186/1756-0500-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodrigues T.B., Khajuria C., Wang H., Matz N., Cunha Cardoso D., Valicente F.H., Zhou X., Siegfried B. Validation of Reference Housekeeping Genes for Gene Expression Studies in Western Corn Rootworm (Diabrotica virgifera virgifera) PLoS ONE. 2014;9:e109825. doi: 10.1371/journal.pone.0109825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan Q.Q., Zhu L., Li Y., Liu W., Wang X.P. A De Novo Transcriptome and Valid Reference Genes for Quantitative Real-Time PCR in Colaphellus bowringi. PLoS ONE. 2015;10:e0118693. doi: 10.1371/journal.pone.0118693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X., Pan H., Yuan L., Zhou X. Reference gene selection for RT-qPCR analysis in Harmonia axyridis, a global invasive lady beetle. Sci. Rep. 2018;8:2689. doi: 10.1038/s41598-018-20612-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cano-Ramírez C., López M.F., Cesar-Ayala A.K., Pineda-Martíneza V., Sullivanb B.T., Zúñigaa G. Isolation and expression of cytochrome P450 genes in the antennae and gut of pine beetle Dendroctonus rhizophagus (Curculionidae: Scolytinae) following exposure to host monoterpenes. Gene. 2012;520:47–63. doi: 10.1016/j.gene.2012.11.059. [DOI] [PubMed] [Google Scholar]
- 19.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Wang H.L., Wen H.S., Li Y., Zhang K., Liu Y. Evaluation of potential reference genes for quantitative RT-PCR analysis in spotted sea bass (Lateolabrax maculatus) under normal and salinity stress conditions. PeerJ. 2018;6:e5631. doi: 10.7717/peerj.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang C.X., Preisser E.L., Zhang H.J., Liu Y., Dai L.Y., Pan H.P., Zhou X.G. Selection of Reference Genes for RT-qPCR Analysis in Coccinella septempunctata to Assess Un-intended Effects of RNAi Transgenic Plants. Front. Plant. Sci. 2016;7:1672. doi: 10.3389/fpls.2016.01672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan Y., Zhou X.R., Pang B.P. Reference gene selection and evaluation for expression analysis using qRT-PCR in Galeruca daurica (Joannis) Bull. Entomol. Res. 2017;107:359–368. doi: 10.1017/S0007485316000948. [DOI] [PubMed] [Google Scholar]
- 23.Arya S.K., Jain G., Upadhyay S.K., Yadav S., Singh H., Dixit S., Verma P.C. Reference genes validation in Phenacoccus solenopsis under various biotic and abiotic stress conditions. Sci. Rep. 2017;7:13520. doi: 10.1038/s41598-017-13925-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu J., Chen S.M., Guo M.J., Ye C.Y., Qiu B.L., Wu J.H., Yang C.X., Pan H.P. Selection and Validation of Reference Genes for RT-qPCR Analysis of the Ladybird Beetle Henosepilachna vigintioctomaculata. Front. Physiol. 2018;9:1614. doi: 10.3389/fphys.2018.01614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan H.P., Yang X.W., Siegfried B.D., Zhou X.G. A Comprehensive Selection of Reference Genes for RT-qPCR Analysis in a Predatory Lady Beetle, Hippodamia convergens (Coleoptera: Coccinellidae) PLoS ONE. 2015;10:e0125868. doi: 10.1371/journal.pone.0125868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang Z.W., Liu F.H., Tian H.G., Zhang M., Guo S.S., Liu T.X. Evaluation of the reference genes for expression analysis using quantitative real-time polymerase chain reaction in the green peach aphid, Myzus persicae. Insect Sci. 2017;24:222–234. doi: 10.1111/1744-7917.12310. [DOI] [PubMed] [Google Scholar]
- 27.Lu Y., Yuan M., Gao X., Kang T., Zhan S., Wan H. Identification and validation of reference genes for gene expression analysis using quantitative PCR in Spodoptera litura (Lepidoptera:Noctuidae) PLoS ONE. 2013;8:e68059. doi: 10.1371/journal.pone.0068059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ponton F., Chapuis M.P., Pernice M., Sword G.A., Simpson S.J. Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J. Insect Physiol. 2011;57:840–850. doi: 10.1016/j.jinsphys.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Zhang S.D., An S.H., Li Z., Wu F.M., Yang Q.P., Liu Y.C., Cao J.J., Zhang H.J., Zhang Q.W., Liu X.X. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae) Gene. 2015;555:393–402. doi: 10.1016/j.gene.2014.11.038. [DOI] [PubMed] [Google Scholar]
- 30.Shakeel M., Zhu X., Kang T.H., Wan H., Li J.H. Selection and evaluation of reference genes for quantitative gene expression studies in cotton bollworm, Helicoverpa armigera (Lepidoptera: Noctuidae) J. Asia Pac. Entomol. 2015;18:123–130. doi: 10.1016/j.aspen.2015.01.001. [DOI] [Google Scholar]
- 31.Koramutla M.K., Aminedi R., Bhattacharya R. Comprehensive evaluation of candidate reference genes for qRT-PCR studies of gene expression in mustard aphid, Lipaphis erysimi (Kalt) Sci. Rep. 2016;6:25883. doi: 10.1038/srep25883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura A.M., Chahad-Ehlers S., Lima A.L.A., Taniguti C.H., Sobrinho I., Torres F.R., Brito R.A. Reference genes for accessing differential expression among developmental stages and analysis of differential expression of OBP genes in Anastrepha obliqua. Sci. Rep. 2015;6:17480. doi: 10.1038/srep17480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.