Abstract
Cyclin-dependent kinases (CDKs) are a group of serine/threonine protein kinases and play crucial roles in various cellular processes by regulating cell cycle and gene transcription. Cyclin-dependent kinase 12 (CDK12) is an important transcription-associated CDK. It shows versatile roles in regulating gene transcription, RNA splicing, translation, DNA damage response (DDR), cell cycle progression and cell proliferation. Recently, increasing evidence demonstrates the important role of CDK12 in various human cancers, illustrating it as both a biomarker of cancer and a potential target for cancer therapy. Here, we summarize the current knowledge of CDK12, and review the research advances of CDK12′s biological functions, especially its role in human cancers and as a potential target and biomarker for cancer therapy.
Keywords: cyclin-dependent kinase 12, gene transcription, cell cycle, cell proliferation, DNA damage response, cancer therapy
1. Introduction
Cyclin-dependent kinases (CDKs) are a group of serine/threonine protein kinases that are key regulators in various cellular processes [1,2,3]. CDK was first discovered as a cell division cycle (Cdc) gene in yeast [4]. The first cloned Cdc gene was Cdc2, which was named as CDK based on its kinase activity and its role in the cell cycle regulation [5]. With the successive discovery of CDK members, CDKs are divided into two subfamilies, including cell cycle-associated CDKs and transcription-associated CDKs. Cell cycle-associated CDKs mainly contain CDK1, CDK2, CDK4 and CDK6, which directly regulate the cell cycle progression. The transcription-associated CDKs, consisting of CDK7, CDK8, CDK9, CDK11, CDK12 and CDK13, control gene transcription [6,7]. The activity and substrate specificity of both cell cycle-associated CDKs and transcription-associated CDKs rely on a regulatory subunit known as cyclin. CDK binds a specific cyclin subunit to form a functional and active CDK/cyclin complex [7,8].
CDK12 is a transcription-associated CDK. It complexes with cyclin K to regulate gene transcription elongation via phosphorylating RNA polymerase II (RNAP II) [9,10,11,12,13] and also regulates translation [14]. Moreover, CDK12 plays a role in RNA splicing, cell cycle progression, cell proliferation, DNA damage response (DDR) and maintenance of genomic stability [2,9,10,13,14,15,16,17,18]. Since the mutation or amplification of CDK12 is closely related with tumorigenesis, CDK12 becomes an attractive therapeutic target for cancer treatment [7,19,20,21]. Here, we introduce the characteristics of CDK12, summarize the current advances of its biological functions and highlight its role in human cancer. Furthermore, we also discuss the future research direction of CDK12.
2. Cyclin Dependent Kinase 12 (CDK12): Gene, Structure and Expression
2.1. Gene and Isoforms of CDK12
CDK12, a ~164 kDa protein consisting of 1490 amino acids, is encoded by CDK12, located in human chromosome 17q12 and composed of 14 exons [22]. It was first identified as a novel human protein kinase by Ko et al. from the cDNA of HeLa cell in 2001 [22]. Because it is a Cdc2-related kinase with an arginine/serine-rich (RS) domain, it was named as CrkRS (Cdc2-related kinase with RS domain). Later in 2006, some researchers discovered that cyclin L1 and cyclin L2 were cyclins interacting with CrkRS, and thus CrkRS was renamed as CDK12 [23]. Chen et al. found that overexpressed CDK12 complexed with cyclin L via an immunoprecipitation experiment [23]. However, they did not point out the native interaction between CDK12 and cyclin L. To identify the associations between cyclin and endogenous CDK12, Bartkowiak et al. carried out a Mass Spectrometry (MS) analysis on co-immunopurified proteins and found that cyclin K was the only cyclin being identified, which demonstrates that CDK12 interacts with cyclin K [24]. Subsequent studies also confirmed that the cyclin combining with CDK12 is cyclin K [8,13,25]. Likewise, CDK13, the homologue of CDK12, has been proven to associate with cyclin K [13,25,26,27]. Moreover, cyclin K1 (a ~65 kDa isoform of cyclin K) has been demonstrated as the primary cyclin partner for CDK12 [8,13,25].
There are two isoforms of CDK12, which are identical at the 5′ end but different at the 3′ end [23] (Figure 1). According to the length of the open reading frame, the two CDK12 isoforms are named as CDK12S (the shorter isoform of CDK12) and CDK12L (the longer isoform of CDK12), respectively [23] (Figure 1).
Figure 1.
Genomic and messenger RNA (mRNA) structures of cyclin-dependent kinase 12 (CDK12). CDK12S: the shorter isoform of CDK12, CDK12L: the longer isoform of CDK12.
2.2. Structure of CDK12
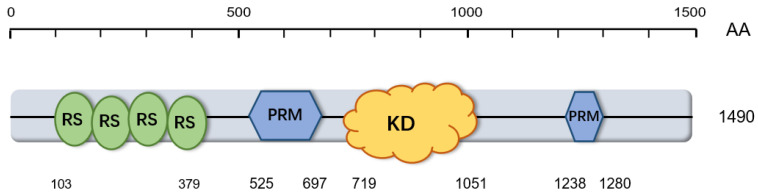
CDK12 is mainly composed of three domains: a central Cdc2-related protein kinase domain (KD), an N-terminal “arm”, about 700 amino acids, and a C-terminal “arm”, about 500 amino acids [22,28] (Figure 2). The central KD is composed of 300 amino acids and is located at the center of CDK12 [22]. Its main function is to mediate the phosphorylation of the C-terminal domain (CTD) of RNAP II. There are 21 RS motifs in the first 400 amino acids of CDK12, and only one RS motif in the rest of the approximately 1000 amino acids [22]. The RS domain, which is enriched arginine and serine, is considered as a prominent feature of CDK12 [22]. It was originally found in pre-messenger RNA (pre-mRNA) splicing factors that were important for spliceosome assembly and alternative splice-site selection [29]. In CDK12, the RS domain mainly functions to target CDK12 to the nuclear speckles [22]. The central KD and the RS domain endow CDK12 the capacity to directly link transcription with the splicing machinery. Proline-rich motifs (PRM) are located between the RS domain and the central KD and are also found in the C-terminal region [22]. The PRM contains the consensus binding sites for Src homology 3 (SH3) and tryptophan (WW) regions which can mediate protein–protein interactions by binding proline-rich modules in ligands [30,31,32,33,34,35]. The presence of the RS domain and PRM domain indicates that CDK12 is likely to take part in numerous protein–protein interactions [28]. Notably, the closest human homologue of CDK12 is CDK13. While their sequences of KD are highly homologous, their C- and N-terminal regions differ between CDK12 and CDK13 [26,28].
Figure 2.
Schematic diagram of CDK12 protein structure. AA: amino acid; RS: arginine/serine-rich domain; PRM: proline-rich motif; KD: kinase domain.
2.3. CDK12 Expression
As a transcription-associated CDK, CDK12 is ubiquitously expressed in mammalian tissues. The presence of CDK12 in all tissues has been determined via screening a panel of RNAs from specific human tissues [22]. CDK12 shows low tissue specificity according to The Human Protein Atlas (available online: https://www.proteinatlas.org/). Notably, high expression of CDK12 has been observed in bone marrow and testis compared with other tissues by The Human Protein Atlas. Besides, Castillo et al. have experimentally confirmed the high expression of CDK12 in human testis [36].
3. CDK12′s Biological Functions
3.1. CDK12 in Gene Transcription
In 2010, CDK12 was first demonstrated as a transcription-associated CTD kinase in Drosophila [24]. At present, CDK12 is regarded as a transcription-associated CDK, which phosphorylates the CTD of RNAP II [8,9,24,37]. RNAP II is responsible for RNA synthesis of eukaryotic genes. It directs the gene transcription process consisting of transcription initiation, elongation and termination [38]. The large subunit of RNAP II is RPB1 which contains a CTD. CTD contains repeats of the heptapeptide Y1S2P3T4S5P6S7, and single serine phosphorylation in these repeats is required for each step of the transcription cycle [39]. Phosphorylation of Ser2 is a hallmark of transcription elongation, and phosphorylation of Ser5 is required for proper transcription initiation, both of which are necessary for the transcription cycle [38,40]. Bartkowiak et al. have shown that treatment with RNA interference (RNAi) of CDK12 alters the phosphorylation state of the CTD and reduces the phosphorylation level of Ser2 [24]. Other findings have also found that CDK12 predominantly phosphorylates Ser2 [8,12,13,37,41,42]. Therefore, CDK12 is considered to phosphorylate Ser2 but not Ser5. In addition, CDK12 and cyclin K are considered to be proteins associated with RNAP II and transcription elongation [24,43]. CDK12 binds cyclin K to form a CDK12/cyclin K complex, which regulates phosphorylation of Ser2 in the CTD of RNAP II and expression of DDR genes, DNA replication genes and DNA repair genes [9,13].
Interestingly, RNAP II transcription is not globally impaired in cells without CDK12/cyclin K complex [13]. Chirackal Manavalan et al. have found that the inhibition of CDK12 does not affect Ser2 phosphorylation level as well as global transcription but diminishes RNAP II processivity accompanied by transcript shortening of DNA replication genes, which is consistent with defective transcription elongation [9]. Moreover, CDK12 also plays a role in co-transcriptional processing of genes such as MYC, particularly at its 3′ end [41]. The Ser2 phosphorylation of CDK12 is important for the recruitment of 3′ end formation factors like cleavage stimulation factor 77 (CstF77). This mechanism involves RNAP II pausing that promotes Ser2 phosphorylation of CDK12, which serves to recruit CstF77 and is necessary for optimal 3′ end processing of the MYC gene [41]. Similarly, CDK12 is required for 3′ end processing of cellular oncogene fos (c-FOS) transcripts. Depletion of CDK12 leads to decreased levels of Ser2 phosphorylation, cleavage stimulation factor 64 (CstF64) and cleavage, and polyadenylation specificity factor 73 (CPSF73) at the c-FOS gene and attenuates the 3′ end formation of c-FOS transcripts [44]. In summary, CDK12 plays a key role in gene transcription.
3.2. CDK12 in RNA Splicing
CDK12 has been shown to play a role in RNA splicing. Rodrigues et al. have identified Drosophila CDK12 as a major determinant in regulating HOW (held out wings, a RNA-binding protein)-dependent splicing of Neurexin IV (a cell-adhesion molecule) [45]. Thus, they have demonstrated a mechanism in regulating timed splicing of newly synthesized mRNA molecules through phosphorylating of RNAP II CTD [45]. In addition, CDK12 is proven to alter splicing site selection of an E1a minigene [23]. Depletion of CDK12 shows diminished 3′ end processing of the activated c-FOS and c-MYC genes [41,44]. These findings demonstrate the importance of CDK12 in regulating and coordinating the transcription and pre-mRNA processing. Furthermore, CDK12 stabilizes serine-arginine splicing factor 1 (SRSF1) mRNA transcripts through skipping an alternative intron in the 3′ untranslated region (3′ UTR) [46]. Moreover, CDK12 associates with core spliceosome components and regulates alternative last exon (ALE) splicing of long transcripts in various cell types [20].
Recent studies have shown that minimal splicing alterations induced by the inhibition of CDK12 may be due to defective transcription elongation [9,10]. Treatment with THZ531 (inhibitor of CDK12) results in 13.4% intron retention, which is the largest proportion of splicing alteration [10]. Importantly, this phenomenon occurs primarily in long genes, such as DDR genes [10]. The apparent increased splicing efficiency in long genes may be due to defective transcription elongation accompanied by the reduction in the formation of such long transcripts, rather than a more efficient spliceosome [10]. Moreover, as DDR genes contain more intronic polyadenylation sites than other expressed genes, CDK12 can regulate DDR genes via suppressing the intronic polyadenylation [11]. It is worth noting that inhibition of CDK12 leads to transcript shortening of genes, which affects the expression of DNA replication genes and DNA repair genes [9]. In addition, the defective RNAP II processivity is usually accompanied by slower transcription elongation rates of CDK12-sensitive genes [9]. Thus, all these findings indicate that CDK12 indirectly regulates RNA splicing through regulating gene transcription.
3.3. CDK12 in Translation
Besides the regulatory role in mRNA biosynthesis, CDK12 also regulates the translation of mRNA [14]. Choi et al. found that CDK12 promoted translation of mRNAs via phosphorylating 4E-binding Protein 1 (4E-BP1), the mRNA 5′ cap-binding repressor [14]. More specifically, CDK12 cooperates with the mechanistic target of rapamycin (mTORC1) to affect the translation of mRNAs encoding DNA repair factors, ribosome and translation factors via phosphorylating 4E-BP1 at two Ser–Pro sites (S65, T70) that control the exchange of 4E-BP1, with eukaryotic initiation factor 4G (eIF4G) at the 5′ cap of target mRNAs. This finding reveals a new set of target genes (mTORC1-regulated genes) of CDK12. Therefore, CDK12 is important in the correct arrangement and progression of chromosomes through mitosis [14].
3.4. CDK12 in Cell Cycle
Normal cell cycle progression is of great significance for cell proliferation and the maintenance of genomic stability [9,47]. The dysregulation of cell cycle progression contributes to abnormal cell proliferation and oncogenesis [48,49]. The progression of the cell cycle relies on the periodic activity of the complex that binds CDKs and cyclins [50]. Different CDK/cyclin complexes exhibit different CDK kinase activity, thereby affecting different stages of cell cycle [50].
CDK12 plays a role in regulating cell cycle progression. Long-term depletion of CDK12 induces cell accumulation in G2/M phase [13,51]. Chen et al. conditionally deleted CDK12 in the neural progenitor cells (NPCs) of mice and found that the NPCs were accumulated at G2 and M phase [52]. There was a 1.3–4.6-fold increase of mitotic cells in the mutant mice compared with the control mice, suggesting that the cells lacking CDK12 had a longer cell cycle [52]. Therefore, deletion of CDK12 has been shown to prolong cell cycle, indicating the role of CDK12 in regulating cell cycle. A recent study has demonstrated that knockdown of CDK12 or cyclin K results in induction of mitotic catastrophe and decreased expression of Aurora B, a key regulator of mitosis [53]. The depletion of cyclin K induces inhibition of proliferation accompanied by G2/M arrest [53]. More recently, Chirackal Manavalan et al. have found that inhibition of CDK12 induces the G1/S cell cycle progression defect by using an analog-sensitive CDK12 cell line, in which CDK12 can be rapidly and specifically inhibited [9]. Inhibition of CDK12 induces the decreased expression of some crucial DNA replication genes (e.g., TOPBP1 (DNA topoisomerase II binding protein 1), CDC6 (cell division cycle 6) and CDT1 (Cdc10-dependent transcript 1)), which disrupts the formation of pre-replicative complex (pre-RC), thereby delaying G1/S progression [9]. This illustrates that CDK12 controls G1/S progression by regulating the expression of core DNA replication genes [9]. Moreover, the treatment of RNAi of CDK12 significantly increases the cell number of G0/G1 phase, indicating that CDK12 plays an important role in controlling the transition of G0/G1 phase to S phase [54].
3.5. CDK12 in Cell Proliferation
As cell cycle is closely related with cell proliferation, aberrant cell cycle progression may result in abnormal cell proliferation. CDK12 is involved in cell proliferation by regulating cell cycle. The assembly of pre-RC occurs during G1 phase, a process referred to as replication origin licensing, which is indispensable for sustaining cell proliferation [55]. Knockdown of cyclin K or its cognate kinase CDK12 prevents the assembly of pre-RC in G1 phase and inhibits cell proliferation [16], suggesting the involvement of CDK12 in cell proliferation. CDK12/cyclin K deficiency has been shown to inhibit cell proliferation and induce apoptosis via the induction of mitotic catastrophe [53]. Moreover, Choi et al. have found that CDK12/cyclin K complex is required for multiple steps in mitosis [14]. Cells deficient in CDK12 or cyclin K display profound mitotic defects [14]. Recently, it was found that a high level of CDK12 in various human cancers characterized by uncontrolled cell proliferation indicates the important regulatory role of CDK12 in cell proliferation [16]. Zhang et al. firstly used the CDK12 inhibitor, THZ531, to treat leukemia cells and found that THZ531 treatment caused an irreversible decrease in cell proliferation [12]. Subsequent studies also reported that suppression of CDK12 with either short hairpin RNAs (shRNAs) or THZ531 strongly inhibited cell proliferation and impaired the colony formation in cancer cells [51,56].
3.6. CDK12 in DNA Damage Response (DDR)
DDR is biologically significant because it is responsible for detecting the DNA damage and repairing it to maintain normal cellular processes [57]. The CDK12/cyclin K complex plays an important role in regulating the expression of DDR genes by phosphorylating RNAP II CTD [9,13]. Inhibition of CDK12/cyclin K results in decreased expression of DDR genes, such as BRCA1 (breast and ovarian cancer type 1 susceptibility protein 1), ATR (ataxia telangiectasia and Rad3-related), FANCI (Fanconi anemia complementation groups - I) and FANCD2 (Fanconi anemia complementation groups - D2) [13], which are important for maintaining genome stability. Moreover, cells without CDK12/cyclin K are sensitive to DNA damage agents and develop spontaneous DNA damage signaling [13]. Therefore, CDK12 is important for maintaining genomic stability by interacting with cyclin K to regulate the expression of DDR genes [13]. In addition, CDK12 is proven to regulate pre-RC assembly during G1 phase as well as the expression of DNA replication genes and homologous recombination (HR) DNA repair genes to protect cells from genomic instability [9,16]. Zhang et al. have shown that after the cells are treated with THZ531 (CDK12 inhibitor), the expression of core DDR genes (BRCA1, FANCF (Fanconi anemia complementation group F) and ERCC4 (excision repair cross-complementing group 4)) is decreased [12].
The molecular basis for the effect of CDK12 on DDR genes has been further studied [10]. It has been indicated that inhibition of CDK12 leads to a gene length-dependent elongation defect associated with early termination through premature cleavage and polyadenylation (PCPA). Thus, the expression of DDR genes is affected by CDK12, primarily due to their relatively longer length and lower ratio of U1 small nuclear ribonucleoprotein (U1 snRNP) binding to intronic polyadenylation site [10]. Chirackal et al. have also confirmed that CDK12 is essential for optimal RNAP II processivity at longer genes, such as genes involved in DNA replication and DNA repair [9]. Recently, it has been reported that CDK12 responds to DNA damage through regulating the translation of mTORC1-dependent mRNAs [14]. To be more specific, CDK12 regulates translation of the DNA damage response checkpoint kinase 1 (CHK1). Therefore, CDK12 acts indirectly to control p53 stability in response to DNA damage through regulating the translation of CHK1 [14]. In addition, CDK12 selectively regulates the translation of many critical mitotic regulatory complexes. Loss of CDK12 results in defective DNA repair, mitotic catastrophe and profound genome instability [14,53].
4. CDK12 and Human Cancer
Recently, more and more evidence demonstrates the involvement of CDK12 in cancer (Table 1) [7,10,15]. This may be due to the key role of CDK12 in regulating transcription elongation and the expression of genes involved in DDR, DNA replication and mRNA processing [9,10,12,24]. Abnormal expression or mutation of CDK12 is detected in various cancers, such as breast cancer, ovarian cancer, prostate cancer and gastric cancer [7,15,17,18]. Moreover, CDK12 is also indirectly implicated in esophageal, endometrial, uterine, bladder, colorectal and pancreatic ductal carcinomas [15]. Interestingly, CDK12 shows both tumorigenic and tumor-suppressive effects in different cancer types, which will be introduced in detail in the following.
Table 1.
The role of cyclin-dependent kinase 12 (CDK12) in various cancers and the associated mechanism.
| Cancer Type | CDK12′s Function | Mechanism | References |
|---|---|---|---|
| Breast cancer (HER2 1 -positive breast cancer) | Tumor promoter | Overexpression of CDK12 regulates the splicing of ATM 5 and DNAJB6-L 6 and actives WNT 7 and IRS1-ErbB-PI3K 8 signaling | [20,59] |
| Breast cancer (TNBC 2) | Tumor suppressor | Loss of CDK12 leads to downregulation of DDR 9 genes | [61,76] |
| Ovarian cancer (HGSOC 3) | Tumor suppressor | Loss of CDK12 leads to downregulation of DDR 9 genes | [21,63,65] |
| Prostate cancer (mCRPC 4) | Tumor suppressor | Loss of CDK12 leads to downregulation of DDR 9 genes | [68,69] |
| Gastric cancer | Tumor promoter | Overexpression of CDK12 actives the CDK12/CCL21 10 pathway | [75] |
1 HER2: human epidermal growth factor receptor 2 2 TNBC: triple-negative breast cancer 3 HGSOC: high-grade serous ovarian cancer 4 mCRPC: metastatic castration-resistant prostate cancer 5 ATM: ataxia telangiectasia-mutated 6 DNAJB6-L: the long isoform of DNAJB6 (DnaJ homolog subfamily B member 6, MRJ) 7 WNT: Wingless-Integrated 8 IRS1-ErbB-PI3K: IRS1 (insulin receptor substrate-1)-ErbB (epidermal growth factor receptor)-PI3K (phosphatidylinositol-3-kinase) 9 DDR: DNA damage response 10 CCL21: CC-chemokine ligand 21.
4.1. CDK12 in Breast Cancer
Increasing evidence shows that CDK12 is closely linked with breast cancer. Interestingly, CDK12 plays distinguishing roles among various subtypes of breast cancer, especially for HER2 (human epidermal growth factor receptor 2)-positive breast cancer and triple-negative breast cancer (TNBC). In HER2-positive breast cancer, CDK12 acts as a tumor promoter, while in TNBC, CDK12 acts as a tumor suppressor.
HER2-positive breast cancer is a subtype of breast cancer and presents an amplification pattern of oncogene HER2 (ERBB2). It is shown that CDK12 and HER2 oncogenes are co-amplified in breast cancer [20]. CDK12 promotes migration and invasion of HER2-positive breast tumor cells through regulating the ALE splicing of DDR activator ATM (ataxia telangiectasia-mutated) and DNAJB6 (DnaJ homolog subfamily B member 6, MRJ)-L [20]. In addition, Chen et al. have demonstrated that mutations in CDK12, TP53 (tumor suppressor p53) and PIK3CA are the most frequent in 107 HER2-positive breast cancer patients [58]. Choi et al. have indicated that CDK12 drives the development of HER2-positive breast cancer via affecting WNT (Wingless-Integrated) and IRS1 (insulin receptor substrate-1)-ErbB (epidermal growth factor receptor)-PI3K (phosphatidylinositol-3-kinase) signaling [59]. CDK12 promotes tumor initiation through regulating cancer stem cells (CSCs) or affecting the genes which are necessary to activate downstream pathways such as ErbB-PI3K-AKT (Protein Kinase B) or WNT-signaling cascades [59]. In addition, inhibition of CDK12 facilitates anticancer efficacy of trastuzumab in HER2-positive tumors [59]. Another subtype of breast cancer is TNBC, which can be characterized by the low expression of estrogen receptor (ER), progesterone receptor (PR) and HER2 [60]. High expression of CDK12 is associated with HER2 status and plays important roles during the tumorigenesis and development of breast cancer [61]. However, CDK12 is not an independent predictor of breast cancer-specific survival [61]. Notably, absent CDK12 is associated with a triple-negative phenotype (ER-, PR-, HER2-) [61]. There is a small proportion of HER2-positive patients that show absent CDK12 protein expression but a large proportion of absent CDK12 protein expression in TNBC patients [61]. In addition, absence of CDK12 protein is often accompanied by downregulation of DDR proteins (ATR (ataxia-telangiectasia and Rad3-related), Ku70/Ku80 (the classical non-homologous end joining (cNHEJ) factors), PARP1 (poly ADP-ribose polymerase 1), DNA-PK (DNA-dependent protein kinase) and γH2AX (phosphorylated histone H2AX)), suggesting a novel mechanism of CDK12-associated DDR dysregulation in breast cancer [61]. In summary, CDK12 acts as tumor promoter in HER2-positive breast cancer, but as a tumor suppressor in TNBC.
4.2. CDK12 in Ovarian Cancer
Ovarian cancer is one of the most common malignant tumors for women. High-grade serous ovarian cancer (HGSOC) has a higher mortality rate [62]. The mutations of TP53 play a dominant role in HGSOC and mutated BRCA1/BRCA2 are found in 22% of tumors [63]. Significantly, the mutation of CDK12 is detected, which is mainly nonsense or indel, suggesting the potential loss of function [63]. Loss-of-function (LOF) mutations of CDK12 contribute to genomic instability, underlying the genesis of the cancer by causing defects in multiple DNA repair signaling pathways [21]. More importantly, CDK12 LOF genomic alterations are associated with focal tandem duplications (FTDs) in ovarian cancer [64]. In addition, BRCA1 promoter hypermethylation or mutational inactivation of CDK12 can downregulate transcription of BRCA1, thereby disrupting HR DNA repair in ovarian cancer, and then leading to metabolic reprogramming of ovarian cancer cells [65]. A c.1047-2A>G splice site variant of the CDK12 gene was recently reported to be strongly associated with hereditary ovarian cancer [66]. These results demonstrate that CDK12 is a tumor suppressor in ovarian cancer. Moreover, recent reports have shown that suppression of MYC via inhibition of CDK7, CDK12 and CDK13 may be an effective treatment for MYC-dependent ovarian cancer [67].
4.3. CDK12 in Prostate Cancer
Prostate cancer (PCa) is the second most frequently diagnosed cancer in men. It was recently demonstrated that CDK12 is associated with PCa [68,69]. It is considered that loss or mutation of CDK12 leads to genomic instability, which contributes to metastatic prostate cancer [68]. In addition, TP53, PTEN (phosphatase and tensin homolog) and CDK12 defects are commonly detected in metastatic castration-resistant prostate cancer (mCRPC) patients [70]. It has been shown that inactivation of CDK12, TP53 and BRCA2 affects distinct classes of structural variation in mCRPC based on a whole-genome analysis from 101 mCRPC patients [71]. Specifically, CDK12 mutation is related to tandem duplications [71]. Recent studies also show that inactivation of CDK12 (biallelic inactivation) is associated with a global tandem duplication phenotype [72,73]. Wu et al. have identified a novel subtype of prostate cancer characterized by biallelic loss of CDK12 [69]. They detected the aberrations of CDK12 in 25/360 mCRPC patients (6.9%). In addition, the CDK12 mutant is often accompanied by FTDs [64,69]. The presence of FTDs in CDK12-mutated cancers may result in highly recurrent gains of genes involved in cell cycle and DNA replication [69]. In addition, CDK12-mutant prostate cancers are characterized by increased gene fusions, fusion-induced neoantigen open reading frames and high immune infiltration [69]. CDK12-mutant prostate cancer patients have a higher likelihood of response to immunotherapy than an unselected metastatic prostate cancer population from the pilot clinical study [69]. Therefore, inhibition of CDK12 may sensitize tumors to checkpoint inhibitor-based immunotherapies [69].
4.4. CDK12 in Gastric Cancer
CDK12 is also involved in gastric cancer. According to the different status of HER2, gastric cancer is divided into two subtypes, including HER2-positive gastric cancer and HER2-negative gastric cancer [74]. Among these, CDK12 amplification is mainly detected in HER2-positive gastric cancer [74]. Ji et al. have shown that high-level expression of CDK12 is detected in gastric tumor samples compared with normal samples [75]. Moreover, patients with high expression of CDK12 show lower overall survival rates than patients with low expression of CDK12 [75]. They have identified positive correlations of CD8+ cell number and CCL21 (CC-chemokine ligand 21) mRNA expression with CDK12 level [75]. These evidences indicate the involvement of CDK12 in gastric cancer.
5. CDK12 as a Potential Target and Biomarker for Cancer Therapy
Evidence shows that CDK12 is not only a biomarker but also a potential therapeutic target of cancer (Table 2). CDK12 mutation or deficiency sensitizes cells to PARP (poly ADP-ribose polymerase) inhibitors and agents that target cell-cycle checkpoints, such as CHK1 [77]. PARP is a nuclear enzyme that modifies the substrates by poly(ADP-ribose)ylation (PARylation) [78]. PARP inhibitors are Food and Drug Administration (FDA)-approved drugs that target cancers with defects in HR, including those with BRCA1 or BRCA2 mutations [79,80]. Cancers with a BRCA1 mutation, such as TNBC and ovarian cancer, are usually treated with PARP inhibitors as targeted drugs [79]. Johnson et al. have indicated that loss or inhibition of CDK12 sensitizes cells to PARP inhibitors and helps patients overcome the resistance of PARP inhibitors [81]. CHK1 is a cellular factor that targets tumor cells with genomic instability [82]. CHK1 inhibitors have been tested as anti-tumor agents and are used in treating a variety of cancers [79]. Loss of CDK12 enhances the anti-proliferative effect of CHK1 inhibitors [79]. Previous studies have shown that the anti-tumor effect of CHK1 inhibitors is determined by p53 status, while other findings have illustrated that CHK1 inhibitors decrease cellular viability irrespective of p53 status [83,84,85]. However, Paculova et al. have indicated that the anti-proliferative effect of CHK1 inhibitor combined with loss of CDK12 is comparable in cell lines regardless of p53 status [79]. CHK1 is important in the effective repair of endogenous DNA damage, especially in cells lacking CDK12 or BRCA1 [79]. Thus, CDK12 deficiency should be considered as a CHK1 sensitivity biomarker candidate [79].
Table 2.
CDK12 as potential target for cancer therapy.
| Treatment | Function | Cancer Type | References |
|---|---|---|---|
| Dinaciclib | Inhibition of multiple CDKs including CDK12 | Breast cancer and metastatic osteosarcoma | [59,96] |
| THZ1 | Inhibition of CDK7/12 | Ovarian cancer and neuroblastoma | [67,87] |
| THZ531 | Inhibition of CDK12/13 | Breast cancer, hepatocellular carcinoma and metastatic osteosarcoma | [56,59,96] |
| SR-4835 | Inhibition of CDK12/13 | TNBC 3 (use with PARP inhibitors) | [76] |
| PARP 1 inhibitors + CDK12 inhibitors | Synthetic lethality | TNBC 3, ovarian cancer and Ewing sarcoma | [76,81,86] |
| CHK1 2 inhibitors | Synthetic lethality | Ovarian cancer | [79] |
1 PARP: poly ADP-ribose polymerase 2 CHK1: checkpoint kinases 1 3 TNBC: triple-negative breast cancer.
CDK12 plays an important role in promoting cancer cell growth, especially in cancers driven by dysregulated transcription factors, such as cancers dependent on MYC (neuroblastoma) and the EWS–FLI1 fusion oncoprotein (Ewing sarcoma) [77]. Neuroblastoma is a cancer highly dependent on transcriptional programs [86]. MYC is a proto-oncogene and a major driver of many human cancers. Amplification of n-MYC (MYCN) leads to neuroblastoma [87]. Studies have indicated that THZ1, a CDK12 inhibitor, inhibits MYC expression and tumor growth [67,87]. In addition, CDK12 plays an important role in the processing of MYC precursor mRNA. Ewing sarcoma is characterized by chromosome rearrangement which fuses the strong transactivation domain of EWS protein with the DNA binding domain of FLI1 protein [86]. EWS/FLI acts as both a transcriptional activator and a transcriptional repressor [88]. Currently, treatment of Ewing sarcoma mainly uses CDK12 inhibitors THZ1 and THZ531, which impair DNA damage repair in an EWS/FLI-dependent manner [86]. The combination of CDK12 and PARP inhibitors is highly active in Ewing Sarcoma [86]. Taken together, targeting CDK12 may be a viable treatment strategy for cancers driven by dysregulated transcription factors.
CDK inhibitors have been studied and applied to cancer treatment. Considering that CDK12 plays an important role in regulating transcription elongation and maintenance of genome stability, CDK12 aberrations are found in various types of cancer [59,61,63,69]. Inhibition of CDK12 is considered a favorable strategy for cancer treatment [24,81,89,90,91]. Dinaciclib is a multi-specific CDK inhibitor that exhibits potent antiproliferative effects on various cancers [92]. It was initially found to inhibit CDK1, CDK2, CDK5 and CDK9, and was recently reported to have an inhibitory effect on CDK12 [81,93]. Dinaciclib inhibits phosphorylation of Ser2 of RNAP II CTD and downregulates HR DNA repair genes [81]. Moreover, dinaciclib can reverse the resistance of PARP inhibitor, converting tumor growth inhibition to durable regression [81]. This suggests that combined inhibition of CDK12 and PARP may be a good therapeutic strategy. THZ1 is a CDK7 inhibitor and has therapeutic effects on both breast and lung cancer [94,95]. Currently, it has been demonstrated that high concentrations of THZ1 can also be used as an inhibitor of CDK12 [90]. Based on the study of THZ1, THZ531 (a novel CDK12 inhibitor) is developed [12]. In 2016, Zhang et al. designed a covalent inhibitor, THZ531, which can inhibit both CDK12 and CDK13 [12]. Studies have indicated that THZ531 inhibits cell proliferation via preferentially suppressing the expression of DNA repair-related genes and inducing strong DDR in cancer cells [56]. Another novel CDK12/13 inhibitor, SR-4835, has been developed by Quereda’s group [76]. It has potential for treatment of TNBC through the downregulation of core DDR genes and upregulation of genes involved in cell apoptosis. Accordingly, SR-4835 is synergized with PARP inhibitors to inhibit cancer cell proliferation [76]. Clinical trials of CDK12 inhibitors combined with PARP inhibitors treatment are currently under way. One clinical trial in Phase I adopts dinaciclib (CDK inhibitor SCH 727965) and veliparib (PARP-1 inhibitor ABT-888) for treatment in patients with advanced solid tumors (available online: http://clinicaltrials.gov, NCT01434316). This clinical trial is still in the process of recruiting and is estimated to be completed in December 2020. Once the recommended phase 2 dose for ABT-888 in combination with SCH727965 is established, the trial will be included in an extended cohort to assess preliminary activity in both BRCA wild-type and BRCA-mutated TNBCs. In summary, CDK12 inhibitors may become good candidates for anticancer drugs.
6. Conclusion and Perspectives
Here, we summarized the current knowledge and research advances of CDK12 and its biological functions, and highlighted the role of CDK12 in human cancers, demonstrating that it is a potential target for cancer therapy. By regulating transcription elongation and the expression of genes involved in DDR, DNA replication and mRNA processing, CDK12 participates in various cellular processes such as DDR, RNA splicing, cell cycle progression and cell proliferation. As all of these cellular processes are closely related to cancer development, CDK12 has been demonstrated as an important molecule involved in cancer development, such as breast cancer, ovarian cancer, prostate cancer and gastric cancer. This suggests that CDK12 is an important biomarker and may serve as a potential therapeutic target. More recent studies have verified the therapeutic effects by targeting CDK12 with CDK12 inhibitors during the treatment of cancers. The CDK12 inhibitors not only inhibit the transcription and proliferation of cancer cells but also enhance the sensitivity of tumor cells to drugs and overcome drug resistance. Moreover, suppression of CDK12 has a good therapeutic effect on cancers, especially those driven by dysregulated transcription factors. Recently, CDK12 inhibitors have been applied for clinical trials for cancer treatment. All these demonstrate CDK12 as a biomarker and target for cancer diagnosis and therapy.
In summary, CDK12, a transcription-associated CDK, shows versatility in regulating gene transcription, RNA splicing, translation, cell cycle, cell proliferation and DDR, alteration of which contributes to cancer development. Thus, the alteration of CDK12 drives tumorigenesis. Given their biological functions, important roles among various cancers, and the therapeutic effects for cancer cells by targeting CDK12, CDK12 may become a novel potential biomarker and target for human cancer diagnosis and therapy in the future. However, there are still some concerns. As CDK12 is important for normal cell cycle progress and cell proliferation, will the modulation of CDK12 when treating cancer induce other disease? How can we specifically target CDK12 in the cancer cells? Considering the complicated role of CDK12 acting both as a tumor suppressor and promoter, especially for different subtypes of breast cancer, what kind of strategy should be adopted to target CDK12 for each subtype (e.g., triple-negative breast cancer)? As some CDK12 inhibitors also target CDK13, how should these inhibitors be used for clinical application and how do we develop the inhibitor specifically targeting CDK12? Answering these questions will make CDK12 a potential target for cancer therapy.
Acknowledgments
The authors would like to thank Yu Li (Institute of Medical Research, Northwestern Polytechnical University) and Xiaohui Zhan (Department of Medicine, Indiana University School of Medicine) for their help to improve the manuscript, including the English writing.
Author Contributions
Paper design: A.Q. and L.H. Literature collection and summary: S.L., L.H., Z.W., Z.C., S.L., and X.X. Drafting manuscript: S.L., L.H., Z.W., and Z.C. Figure drawing: S.L., and Z.W. Table: S.L., S.L., and X.X. Revising manuscript: L.H., and A.Q. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China, grant number 81772017, Young Talent Fund of University Association for Science and Technology in Shaanxi, grant number 20170401, and The Project Supported by Natural Science Basic Research Plan in Shaanxi Province of China, grant number 2018JM3040, the grant BKJ17J004.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Lim S., Kaldis P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 2.Malumbres M., Barbacid M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 3.Malumbres M. Cyclin-dependent kinases. Genome Biol. 2014;15:122. doi: 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nurse P. Genetic control of cell size at cell division in yeast. Nature. 1975;256:547–551. doi: 10.1038/256547a0. [DOI] [PubMed] [Google Scholar]
- 5.Hartwell L.H. Three additional genes required for deoxyribonucleic acid synthesis in Saccharomyces cerevisiae. J. Bacteriol. 1973;115:966–974. doi: 10.1128/JB.115.3.966-974.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loyer P., Trembley J.H., Katona R., Kidd V.J., Lahti J.M. Role of CDK/cyclin complexes in transcription and RNA splicing. Cell. Signal. 2005;17:1033–1051. doi: 10.1016/j.cellsig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Paculova H., Kohoutek J. The emerging roles of CDK12 in tumorigenesis. Cell Div. 2017;12:7. doi: 10.1186/s13008-017-0033-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng S.W., Kuzyk M.A., Moradian A., Ichu T.A., Chang V.C., Tien J.F., Vollett S.E., Griffith M., Marra M.A., Morin G.B. Interaction of cyclin-dependent kinase 12/CrkRS with cyclin K1 is required for the phosphorylation of the C-terminal domain of RNA polymerase II. Mol. Cell. Biol. 2012;32:4691–4704. doi: 10.1128/mcb.06267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chirackal Manavalan A.P., Pilarova K., Kluge M., Bartholomeeusen K., Rajecky M., Oppelt J., Khirsariya P., Paruch K., Krejci L., Friedel C.C., et al. CDK12 controls G1/S progression by regulating RNAPII processivity at core DNA replication genes. EMBO Rep. 2019;20:e47592. doi: 10.15252/embr.201847592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krajewska M., Dries R., Grassetti A.V., Dust S., Gao Y., Huang H., Sharma B., Day D.S., Kwiatkowski N., Pomaville M., et al. CDK12 loss in cancer cells affects DNA damage response genes through premature cleavage and polyadenylation. Nat. Commun. 2019;10:1757. doi: 10.1038/s41467-019-09703-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubbury S.J., Boutz P.L., Sharp P.A. CDK12 regulates DNA repair genes by suppressing intronic polyadenylation. Nature. 2018;564:141–145. doi: 10.1038/s41586-018-0758-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang T., Kwiatkowski N., Olson C.M., Dixon-Clarke S.E., Abraham B.J., Greifenberg A.K., Ficarro S.B., Elkins J.M., Liang Y., Hannett N.M., et al. Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat. Chem. Biol. 2016;12:876–884. doi: 10.1038/nchembio.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blazek D., Kohoutek J., Bartholomeeusen K., Johansen E., Hulinkova P., Luo Z., Cimermancic P., Ule J., Peterlin B.M. The Cyclin K/CDK12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011;25:2158–2172. doi: 10.1101/gad.16962311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi S.H., Martinez T.F., Kim S., Donaldson C., Shokhirev M.N., Saghatelian A., Jones K.A. CDK12 phosphorylates 4E-BP1 to enable mTORC1-dependent translation and mitotic genome stability. Genes Dev. 2019;33:418–435. doi: 10.1101/gad.322339.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lui G.Y.L., Grandori C., Kemp C.J. CDK12: An emerging therapeutic target for cancer. J. Clin. Pathol. 2018;71:957–962. doi: 10.1136/jclinpath-2018-205356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei T., Zhang P., Zhang X., Xiao X., Zhang J., Qiu T., Dai Q., Zhang Y., Min L., Li Q., et al. Cyclin K regulates prereplicative complex assembly to promote mammalian cell proliferation. Nat. Commun. 2018;9:1876. doi: 10.1038/s41467-018-04258-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chila R., Guffanti F., Damia G. Role and therapeutic potential of CDK12 in human cancers. Cancer Treat. Rev. 2016;50:83–88. doi: 10.1016/j.ctrv.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Pilarova K., Herudek J., Blazek D. CDK12: Cellular functions and therapeutic potential of versatile player in cancer. NAR Cancer. 2020;2 doi: 10.1093/narcan/zcaa003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng F., Yang C., Kong Y., Huang X., Chen Y., Zhou Y., Xie X., Liu P. CDK12 Promotes Breast Cancer Progression and Maintains Stemness by Activating c-myc/beta -catenin Signaling. Curr. Cancer Drug Targets. 2020;20:156–165. doi: 10.2174/1568009619666191118113220. [DOI] [PubMed] [Google Scholar]
- 20.Tien J.F., Mazloomian A., Cheng S.G., Hughes C.S., Chow C.C.T., Canapi L.T., Oloumi A., Trigo-Gonzalez G., Bashashati A., Xu J., et al. CDK12 regulates alternative last exon mRNA splicing and promotes breast cancer cell invasion. Nucleic Acids Res. 2017;45:6698–6716. doi: 10.1093/nar/gkx187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ekumi K.M., Paculova H., Lenasi T., Pospichalova V., Bosken C.A., Rybarikova J., Bryja V., Geyer M., Blazek D., Barboric M. Ovarian carcinoma CDK12 mutations misregulate expression of DNA repair genes via deficient formation and function of the CDK12/CycK complex. Nucleic Acids Res. 2015;43:2575–2589. doi: 10.1093/nar/gkv101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ko T.K., Kelly E., Pines J. CrkRS: A novel conserved Cdc2-related protein kinase that colocalises with SC35 speckles. J. Cell Sci. 2001;114:2591–2603. doi: 10.1242/jcs.114.14.2591. [DOI] [PubMed] [Google Scholar]
- 23.Chen H.H., Wang Y.C., Fann M.J. Identification and characterization of the CDK12/cyclin L1 complex involved in alternative splicing regulation. Mol. Cel. Biol. 2006;26:2736–2745. doi: 10.1128/MCB.26.7.2736-2745.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartkowiak B., Liu P., Phatnani H.P., Fuda N.J., Cooper J.J., Price D.H., Adelman K., Lis J.T., Greenleaf A.L. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010;24:2303–2316. doi: 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai Q., Lei T., Zhao C., Zhong J., Tang Y.Z., Chen B., Yang J., Li C., Wang S., Song X., et al. Cyclin K-containing kinase complexes maintain self-renewal in murine embryonic stem cells. J. Biol. Chem. 2012;287:25344–25352. doi: 10.1074/jbc.M111.321760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohoutek J., Blazek D. Cyclin K goes with CDK12 and Cdk13. Cell Div. 2012;7:12. doi: 10.1186/1747-1028-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greifenberg A.K., Hönig D., Pilarova K., Düster R., Bartholomeeusen K., Bösken C.A., Anand K., Blazek D., Geyer M. Structural and Functional Analysis of the Cdk13/Cyclin K Complex. Cell Rep. 2016;14:320–331. doi: 10.1016/j.celrep.2015.12.025. [DOI] [PubMed] [Google Scholar]
- 28.Greenleaf A.L. Human CDK12 and CDK13, multi-tasking CTD kinases for the new millenium. Transcription. 2019;10:91–110. doi: 10.1080/21541264.2018.1535211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valcárcel J., Green M.R. The SR protein family: Pleiotropic functions in pre-mRNA splicing. Trends Biochem. Sci. 1996;21:296–301. doi: 10.1016/S0968-0004(96)10039-6. [DOI] [PubMed] [Google Scholar]
- 30.Kay B.K., Williamson M.P., Sudol M. The importance of being proline: The interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2000;14:231–241. doi: 10.1096/fasebj.14.2.231. [DOI] [PubMed] [Google Scholar]
- 31.Ball L.J., Kuhne R., Schneider-Mergener J., Oschkinat H. Recognition of proline-rich motifs by protein-protein-interaction domains. Angewandte Chem. 2005;44:2852–2869. doi: 10.1002/anie.200400618. [DOI] [PubMed] [Google Scholar]
- 32.Kaneko T., Li L., Li S.S. The SH3 domain—A family of versatile peptide- and protein-recognition module. Front. Biosci. J. Virtual Library. 2008;13:4938–4952. doi: 10.2741/3053. [DOI] [PubMed] [Google Scholar]
- 33.Mayer B.J. SH3 domains: Complexity in moderation. J. Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 34.Ponna S.K., Myllykoski M., Boeckers T.M., Kursula P. Structure of an unconventional SH3 domain from the postsynaptic density protein Shank3 at ultrahigh resolution. Biochem. Biophys. Res. Commun. 2017;490:806–812. doi: 10.1016/j.bbrc.2017.06.121. [DOI] [PubMed] [Google Scholar]
- 35.Bedford M.T., Chan D.C., Leder P. FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J. 1997;16:2376–2383. doi: 10.1093/emboj/16.9.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castillo J., Knol J.C., Korver C.M., Piersma S.R., Pham T.V., de Goeij-de Haas R.R., van Pelt A.M.M., Jimenez C.R., Jansen B.J.H. Human Testis Phosphoproteome Reveals Kinases as Potential Targets in Spermatogenesis and Testicular Cancer. Mol. Cell. Proteom. MCP. 2019;18:S132–S144. doi: 10.1074/mcp.RA118.001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bosken C.A., Farnung L., Hintermair C., Merzel Schachter M., Vogel-Bachmayr K., Blazek D., Anand K., Fisher R.P., Eick D., Geyer M. The structure and substrate specificity of human CDK12/Cyclin K. Nat. Commun. 2014;5:3505. doi: 10.1038/ncomms4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuda N.J., Ardehali M.B., Lis J.T. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egloff S., Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. TIG. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 40.Eick D., Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 2013;113:8456–8490. doi: 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- 41.Davidson L., Muniz L., West S. 3’ end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes Dev. 2014;28:342–356. doi: 10.1101/gad.231274.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu M., Yang W., Ni T., Tang Z., Nakadai T., Zhu J., Roeder R.G. RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science. 2015;350:1383–1386. doi: 10.1126/science.aad2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edwards M.C., Wong C., Elledge S.J. Human cyclin K, a novel RNA polymerase II-associated cyclin possessing both carboxy-terminal domain kinase and Cdk-activating kinase activity. Mol. Cell. Biol. 1998;18:4291–4300. doi: 10.1128/MCB.18.7.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eifler T.T., Shao W., Bartholomeeusen K., Fujinaga K., Jäger S., Johnson J.R., Luo Z., Krogan N.J., Peterlin B.M. Cyclin-dependent kinase 12 increases 3’ end processing of growth factor-induced c-FOS transcripts. Mol. Cell. Biol. 2015;35:468–478. doi: 10.1128/MCB.01157-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodrigues F., Thuma L., Klambt C. The regulation of glial-specific splicing of Neurexin IV requires HOW and CDK12 activity. Development. 2012;139:1765–1776. doi: 10.1242/dev.074070. [DOI] [PubMed] [Google Scholar]
- 46.Liang K., Gao X., Gilmore J.M., Florens L., Washburn M.P., Smith E., Shilatifard A. Characterization of human cyclin-dependent kinase 12 (CDK12) and CDK13 complexes in C-terminal domain phosphorylation, gene transcription, and RNA processing. Mol. Cell. Biol. 2015;35:928–938. doi: 10.1128/MCB.01426-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertoli C., Skotheim J.M., de Bruin R.A. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013;14:518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gutgemann I., Lehman N.L., Jackson P.K., Longacre T.A. Emi1 protein accumulation implicates misregulation of the anaphase promoting complex/cyclosome pathway in ovarian clear cell carcinoma. Mod. Pathol. 2008;21:445–454. doi: 10.1038/modpathol.3801022. [DOI] [PubMed] [Google Scholar]
- 49.Messner D.J., Kowdley K.V. Neoplastic transformation of rat liver epithelial cells is enhanced by non-transferrin-bound iron. BMC Gastroenterol. 2008;8:2. doi: 10.1186/1471-230X-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perez-Posada A., Dudin O., Ocana-Pallares E., Ruiz-Trillo I., Ondracka A. Cell cycle transcriptomics of Capsaspora provides insights into the evolution of cyclin-CDK machinery. PLoS Genet. 2020;16:e1008584. doi: 10.1371/journal.pgen.1008584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geng M., Yang Y., Cao X., Dang L., Zhang T., Zhang L. Targeting CDK12-mediated transcription regulation in anaplastic thyroid carcinoma. Biochem. Biophys. Res. Commun. 2019;520:544–550. doi: 10.1016/j.bbrc.2019.10.052. [DOI] [PubMed] [Google Scholar]
- 52.Chen H.R., Juan H.C., Wong Y.H., Tsai J.W., Fann M.J. CDK12 Regulates Neurogenesis and Late-Arising Neuronal Migration in the Developing Cerebral Cortex. Cerebral Cortex. 2017;27:2289–2302. doi: 10.1093/cercor/bhw081. [DOI] [PubMed] [Google Scholar]
- 53.Schecher S., Walter B., Falkenstein M., Macher-Goeppinger S., Stenzel P., Krümpelmann K., Hadaschik B., Perner S., Kristiansen G., Duensing S., et al. Cyclin K dependent regulation of Aurora B affects apoptosis and proliferation by induction of mitotic catastrophe in prostate cancer. Int. J. Cancer. 2017;141:1643–1653. doi: 10.1002/ijc.30864. [DOI] [PubMed] [Google Scholar]
- 54.Li Y., Jiang F., Shi X., Liu X., Yang H., Zhang Z. Identification and Characterization of the Cyclin-Dependent Kinases Gene Family in Silkworm, Bombyx mori. DNA Cell Biol. 2016;35:13–23. doi: 10.1089/dna.2015.3049. [DOI] [PubMed] [Google Scholar]
- 55.Blow J.J., Gillespie P.J. Replication licensing and cancer--a fatal entanglement? Nat. Rev. Cancer. 2008;8:799–806. doi: 10.1038/nrc2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang C., Wang H., Lieftink C., du Chatinier A., Gao D., Jin G., Jin H., Beijersbergen R.L., Qin W., Bernards R. CDK12 inhibition mediates DNA damage and is synergistic with sorafenib treatment in hepatocellular carcinoma. Gut. 2019 doi: 10.1136/gutjnl-2019-318506. [DOI] [PubMed] [Google Scholar]
- 57.Jackson S.P., Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen B., Zhang G., Wei G., Wang Y., Guo L., Lin J., Li K., Mok H., Cao L., Ren C., et al. Heterogeneity of genomic profile in patients with HER2-positive breast cancer. Endocr. Related Cancer. 2020;27:153–162. doi: 10.1530/ERC-19-0414. [DOI] [PubMed] [Google Scholar]
- 59.Choi H.J., Jin S., Cho H., Won H.Y., An H.W., Jeong G.Y., Park Y.U., Kim H.Y., Park M.K., Son T., et al. CDK12 drives breast tumor initiation and trastuzumab resistance via WNT and IRS1-ErbB-PI3K signaling. EMBO Rep. 2019;20:e48058. doi: 10.15252/embr.201948058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Al-Mahmood S., Sapiezynski J., Garbuzenko O.B., Minko T. Metastatic and triple-negative breast cancer: Challenges and treatment options. Drug Deliv. Transl. Res. 2018;8:1483–1507. doi: 10.1007/s13346-018-0551-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Naidoo K., Wai P.T., Maguire S.L., Daley F., Haider S., Kriplani D., Campbell J., Mirza H., Grigoriadis A., Tutt A., et al. Evaluation of CDK12 Protein Expression as a Potential Novel Biomarker for DNA Damage Response-Targeted Therapies in Breast Cancer. Mol. Cancer Ther. 2018;17:306–315. doi: 10.1158/1535-7163.MCT-17-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koonings P.P., Campbell K., Mishell D.R., Jr., Grimes D.A. Relative frequency of primary ovarian neoplasms: A 10-year review. Obstet. Gynecol. 1989;74:921–926. doi: 10.1016/0020-7292(90)90378-X. [DOI] [PubMed] [Google Scholar]
- 63.Bell D., Berchuck A., Birrer M., Chien J., Cramer D.W., Dao F., Dhir R., DiSaia P., Gabra H., Glenn P., et al. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sokol E.S., Pavlick D., Frampton G.M., Ross J.S., Miller V.A., Ali S.M., Lotan T.L., Pardoll D.M., Chung J.H., Antonarakis E.S. Pan-Cancer Analysis of CDK12 Loss-of-Function Alterations and Their Association with the Focal Tandem-Duplicator Phenotype. Oncologist. 2019;24:1526–1533. doi: 10.1634/theoncologist.2019-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kanakkanthara A., Kurmi K., Ekstrom T.L., Hou X., Purfeerst E.R., Heinzen E.P., Correia C., Huntoon C.J., O’Brien D., Wahner Hendrickson A.E., et al. BRCA1 Deficiency Upregulates NNMT, Which Reprograms Metabolism and Sensitizes Ovarian Cancer Cells to Mitochondrial Metabolic Targeting Agents. Cancer Res. 2019;79:5920–5929. doi: 10.1158/0008-5472.CAN-19-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bogdanova N.V., Schurmann P., Valova Y., Dubrowinskaja N., Turmanov N., Yugay T., Essimsiitova Z., Mingazheva E., Prokofyeva D., Bermisheva M., et al. A Splice Site Variant of CDK12 and Breast Cancer in Three Eurasian Populations. Front. Oncol. 2019;9:493. doi: 10.3389/fonc.2019.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zeng M., Kwiatkowski N.P., Zhang T., Nabet B., Xu M., Liang Y., Quan C., Wang J., Hao M., Palakurthi S., et al. Targeting MYC dependency in ovarian cancer through inhibition of CDK7 and CDK12/13. eLife. 2018;7 doi: 10.7554/eLife.39030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reimers M.A., Yip S.M., Zhang L., Cieslik M., Dhawan M., Montgomery B., Wyatt A.W., Chi K.N., Small E.J., Chinnaiyan A.M., et al. Clinical Outcomes in Cyclin-dependent Kinase 12 Mutant Advanced Prostate Cancer. Eur. Urol. 2020;77:333–341. doi: 10.1016/j.eururo.2019.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu Y.M., Cieślik M., Lonigro R.J., Vats P., Reimers M.A., Cao X., Ning Y., Wang L., Kunju L.P., de Sarkar N., et al. Inactivation of CDK12 Delineates a Distinct Immunogenic Class of Advanced Prostate Cancer. Cell. 2018;173:1770–1782. doi: 10.1016/j.cell.2018.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mateo J., Seed G., Bertan C., Rescigno P., Dolling D., Figueiredo I., Miranda S., Nava Rodrigues D., Gurel B., Clarke M., et al. Genomics of lethal prostate cancer at diagnosis and castration resistance. J. Clin. Investig. 2020 doi: 10.1172/JCI132031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quigley D.A., Dang H.X., Zhao S.G., Lloyd P., Aggarwal R., Alumkal J.J., Foye A., Kothari V., Perry M.D., Bailey A.M., et al. Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. Cell. 2018;174:758–769. doi: 10.1016/j.cell.2018.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Viswanathan S.R., Ha G., Hoff A.M., Wala J.A., Carrot-Zhang J., Whelan C.W., Haradhvala N.J., Freeman S.S., Reed S.C., Rhoades J., et al. Structural Alterations Driving Castration-Resistant Prostate Cancer Revealed by Linked-Read Genome Sequencing. Cell. 2018;174:433–447. doi: 10.1016/j.cell.2018.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liang C., Niu L., Xiao Z., Zheng C., Shen Y., Shi Y., Han X. Whole-genome sequencing of prostate cancer reveals novel mutation-driven processes and molecular subgroups. Life Sci. 2019 doi: 10.1016/j.lfs.2019.117218. [DOI] [PubMed] [Google Scholar]
- 74.Zhou C., Feng X., Yuan F., Ji J., Shi M., Yu Y., Zhu Z., Zhang J. Difference of molecular alterations in HER2-positive and HER2-negative gastric cancers by whole-genome sequencing analysis. Cancer Manag. Res. 2018;10:3945–3954. doi: 10.2147/CMAR.S172710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ji J., Zhou C., Wu J., Cai Q., Shi M., Zhang H., Yu Y., Zhu Z., Zhang J. Expression pattern of CDK12 protein in gastric cancer and its positive correlation with CD8(+) cell density and CCL12 expression. Int. J. Med. Sci. 2019;16:1142–1148. doi: 10.7150/ijms.34541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Quereda V., Bayle S., Vena F., Frydman S.M., Monastyrskyi A., Roush W.R., Duckett D.R. Therapeutic Targeting of CDK12/CDK13 in Triple-Negative Breast Cancer. Cancer Cell. 2019;36:545–558. doi: 10.1016/j.ccell.2019.09.004. [DOI] [PubMed] [Google Scholar]
- 77.Chou J., Quigley D.A., Robinson T.M., Feng F.Y., Ashworth A. Transcription-Associated Cyclin-Dependent Kinases as Targets and Biomarkers for Cancer Therapy. Cancer Discov. 2020;10:351–370. doi: 10.1158/2159-8290.CD-19-0528. [DOI] [PubMed] [Google Scholar]
- 78.Schiewer M.J., Goodwin J.F., Han S., Brenner J.C., Augello M.A., Dean J.L., Liu F., Planck J.L., Ravindranathan P., Chinnaiyan A.M., et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2:1134–1149. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paculova H., Kramara J., Simeckova S., Fedr R., Soucek K., Hylse O., Paruch K., Svoboda M., Mistrik M., Kohoutek J. BRCA1 or CDK12 loss sensitizes cells to CHK1 inhibitors. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2017;39:1010428317727479. doi: 10.1177/1010428317727479. [DOI] [PubMed] [Google Scholar]
- 80.Lord C.J., Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnson S.F., Cruz C., Greifenberg A.K., Dust S., Stover D.G., Chi D., Primack B., Cao S., Bernhardy A.J., Coulson R., et al. CDK12 Inhibition Reverses De Novo and Acquired PARP Inhibitor Resistance in BRCA Wild-Type and Mutated Models of Triple-Negative Breast Cancer. Cell Rep. 2016;17:2367–2381. doi: 10.1016/j.celrep.2016.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thompson R., Eastman A. The cancer therapeutic potential of Chk1 inhibitors: How mechanistic studies impact on clinical trial design. Br. J. Clin. Pharmacol. 2013;76:358–369. doi: 10.1111/bcp.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ma C.X., Janetka J.W., Piwnica-Worms H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol. Med. 2011;17:88–96. doi: 10.1016/j.molmed.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ma C.X., Cai S., Li S., Ryan C.E., Guo Z., Schaiff W.T., Lin L., Hoog J., Goiffon R.J., Prat A., et al. Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. J. Clin. Investig. 2012;122:1541–1552. doi: 10.1172/JCI58765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guzi T.J., Paruch K., Dwyer M.P., Labroli M., Shanahan F., Davis N., Taricani L., Wiswell D., Seghezzi W., Penaflor E., et al. Targeting the replication checkpoint using SCH 900776, a potent and functionally selective CHK1 inhibitor identified via high content screening. Mol. Cancer Ther. 2011;10:591–602. doi: 10.1158/1535-7163.MCT-10-0928. [DOI] [PubMed] [Google Scholar]
- 86.Iniguez A.B., Stolte B., Wang E.J., Conway A.S., Alexe G., Dharia N.V., Kwiatkowski N., Zhang T., Abraham B.J., Mora J., et al. EWS/FLI Confers Tumor Cell Synthetic Lethality to CDK12 Inhibition in Ewing Sarcoma. Cancer cell. 2018;33:202–216. doi: 10.1016/j.ccell.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Delehouzé C., Godl K., Loaëc N., Bruyère C., Desban N., Oumata N., Galons H., Roumeliotis T.I., Giannopoulou E.G., Grenet J., et al. CDK/CK1 inhibitors roscovitine and CR8 downregulate amplified MYCN in neuroblastoma cells. Oncogene. 2014;33:5675–5687. doi: 10.1038/onc.2013.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Riggi N., Knoechel B., Gillespie S.M., Rheinbay E., Boulay G., Suva M.L., Rossetti N.E., Boonseng W.E., Oksuz O., Cook E.B., et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell. 2014;26:668–681. doi: 10.1016/j.ccell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Menghi F., Barthel F.P., Yadav V., Tang M., Ji B., Tang Z., Carter G.W., Ruan Y., Scully R., Verhaak R.G.W., et al. The Tandem Duplicator Phenotype Is a Prevalent Genome-Wide Cancer Configuration Driven by Distinct Gene Mutations. Cancer Cell. 2018;34:197–210. doi: 10.1016/j.ccell.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kwiatkowski N., Zhang T., Rahl P.B., Abraham B.J., Reddy J., Ficarro S.B., Dastur A., Amzallag A., Ramaswamy S., Tesar B., et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014;511:616–620. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Asghar U., Witkiewicz A.K., Turner N.C., Knudsen E.S. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat. Rev. Drug Discov. 2015;14:130–146. doi: 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Novotna E., Bukum N., Hofman J., Flaxova M., Kouklikova E., Louvarova D., Wsol V. Aldo-keto reductase 1C3 (AKR1C3): A missing piece of the puzzle in the dinaciclib interaction profile. Arch. Toxicol. 2018;92:2845–2857. doi: 10.1007/s00204-018-2258-0. [DOI] [PubMed] [Google Scholar]
- 93.Parry D., Guzi T., Shanahan F., Davis N., Prabhavalkar D., Wiswell D., Seghezzi W., Paruch K., Dwyer M.P., Doll R., et al. Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Mol. Cancer Ther. 2010;9:2344–2353. doi: 10.1158/1535-7163.MCT-10-0324. [DOI] [PubMed] [Google Scholar]
- 94.McDermott M.S.J., Sharko A.C., Munie J., Kassler S., Melendez T., Lim C.U., Broude E.V. CDK7 Inhibition is Effective in all the Subtypes of Breast Cancer: Determinants of Response and Synergy with EGFR Inhibition. Cells. 2020;9:638. doi: 10.3390/cells9030638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Christensen C.L., Kwiatkowski N., Abraham B.J., Carretero J., Al-Shahrour F., Zhang T., Chipumuro E., Herter-Sprie G.S., Akbay E.A., Altabef A., et al. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell. 2014;26:909–922. doi: 10.1016/j.ccell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bayles I., Krajewska M., Pontius W.D., Saiakhova A., Morrow J.J., Bartels C., Lu J., Faber Z.J., Fedorov Y., Hong E.S., et al. Ex vivo screen identifies CDK12 as a metastatic vulnerability in osteosarcoma. J. Clin. Investig. 2019;129:4377–4392. doi: 10.1172/JCI127718. [DOI] [PMC free article] [PubMed] [Google Scholar]