Abstract
Fusarium head blight (FHB) is one of the most devastating wheat disease due to its direct detrimental effects on grain-yield, quality and marketability. Resistant cultivars offer the most effective approach to manage FHB; however, the lack of different resistance resources is still a major bottleneck for wheat breeding programs. To identify and dissect FHB resistance, a doubled haploid wheat population produced from the Canadian spring wheat cvs AAC Innova and AAC Tenacious was phenotyped for FHB response variables incidence and severity, visual rating index (VRI), deoxynivalenol (DON) content, and agronomic traits days to anthesis (DTA) and plant height (PHT), followed by single nucleotide polymorphism (SNP) and simple sequence repeat (SSR) marker genotyping. A high-density map was constructed consisting of 10,328 markers, mapped on all 21 chromosomes with a map density of 0.35 cM/marker. Together, two major quantitative trait loci for FHB resistance were identified on chromosome 2D from AAC Tenacious; one of these loci on 2DS also colocated with loci for DTA and PHT. Another major locus for PHT, which cosegregates with locus for low DON, was also identified along with many minor and epistatic loci. QTL identified from AAC Tenacious may be useful to pyramid FHB resistance.
Keywords: QTL mapping, doubled haploid, disease resistance, Fusarium head blight, plant height, days to anthesis, epistasis
1. Introduction
Fusarium head blight (FHB), caused by Fusarium graminearum (Schwabe) (Fg) and another ~17 [1] species of the genus Fusarium, is one of the most devastating wheat diseases in the world due to its detrimental effects on grain-yield, quality and marketability. Moreover, contamination with Fusarium damaged kernels (FDK) and mycotoxins such as trichothecenes deoxynivalenol (DON), nivalenol and zearalenone [2] renders the grains unsuitable for food and feed. Recent FHB epidemics in the Canadian prairies resulted in significant economic impact on yield and grading losses to producers [3]. In western Canada alone, the eastern prairies have been most seriously affected with frequent outbreaks [3], but recently, concerns have arisen over the rapid spread of FHB in the western prairies [3]. FHB driven grade reductions caused significant economic losses in 2009, 2010 and 2012 (ranging from $2.9 to $8.7 million) in Alberta [4], especially in durum and highly susceptible soft white-spring wheat cultivars [3] but no serious outbreak was reported until 2016, when the downgrading of over 700,000 tons of wheat resulted in the loss of $12.8 million [5].
Although growing resistant cultivars has long been considered the most promising approach to alleviate the FHB associated risks [6], the development of FHB-resistant wheat cultivars (cvs) has been hindered due to a lack of knowledge, the complexities of resistance genetics and the absence of different resistance sources [7]. The complexities of the resistance genetics can be divided into two main categories: one which is characterized by different phenotypic responses or types of resistance within varietal resistance, and another which is associated with resistance but with unfavorable traits. Resistance to initial infection or disease incidence (DI, type I), the spread of symptoms within the head or disease severity (DS, type II) and DON accumulation (type III) are three well characterized phenotypic responses within complex varietal resistance, accompanied by three others: insensitivity to DON (type IV), resistance to FDK (type V) and tolerance or the ability to produce marketable grains in the presence of FHB (type VI) [7,8,9,10]. Associations between FHB resistance and a number of other traits such as days to anthesis (DTA) and plant height (PHT) [7,11,12,13,14,15,16] further complicate the genetics of FHB resistance which are controlled by polygenes/quantitative trait loci (QTL) that predominantly manifest in small additive effects and high genotype x environment interactions [17,18,19,20]. Venske et al. [21] identified as many as 65 meta-QTL for FHB resistance in wheat [21]. A few FHB-resistant QTL, such as Fhb1 [22], Fhb2 [23,24] and Fhb5 [25], respectively on chromosome arms 3BS, 6BS, and 5AS, are the most-characterized and used QTL in breeding programs. These loci possess relatively large additive effects, and are sometimes treated as discrete loci. All three loci were found in FHB-resistant Chinese wheat cv Sumai 3 and some of its derivatives. Fhb1 and Fhb5 have also been fine mapped [26,27,28,29]. Another reported prominent resistance QTL is on chromosome arm 2DL, initially detected from a Chinese breeding line ‘Wuhan-1′ [16]. Moreover, three resistance loci, i.e., Fhb3, Fhb6 and Fhb7, have been introgressed into wheat from its wild relatives Leymus racemosus [30], Elymus tsukushiensis [31] and Thinopyrum ponticum [32], respectively. A number of small-effect FHB resistance loci have also been detected from several north American and European wheat cvs and mapped to different chromosomes [19,33]. In addition to resistance QTL, a number of susceptibility factors have been identified from Sumai 3 [34,35,36] and Chinese Spring [37].
Recently, claims have been made that Fhb1 has been cloned [38,39,40], and candidate genes from the QTL regions on wheat chromosome arms 2AS [41], 2DS [42] and 2DL [43,44] have been identified; however, there is still no consensus among the breeding community regarding the real genes. PFT (pore-forming toxin-like protein) [38] and His (histidine-rich calcium-binding protein) [39,40] genes, singly or as a haplotype of both [45], have been reported to be pivotal genes in the Fhb1 region. Two recent studies [39,40] identified a critical deletion in the His gene as the causative mutation that gives rise to Fhb1-mediated resistance; however, these studies interpreted their findings differently. Su et al. [39] found that His is a susceptibility factor and the resistance is the result of a loss-of-function mutation, while Li et al. [40] observed that His is a resistance gene, and the same mutation results in a gain-of-function [46].
After decades of scientific research, our understanding about FHB resistance is still limited [6]. Although wheat cv Sumai 3 and its derived lines are seemingly the most studied so far, its host genetics are still a partial enigma. While efforts are still being made to understand Fhb1-mediated Sumai 3 resistance, there is increasing interest in understanding native FHB resistance in Canadian wheat cvs [47]. AAC Tenacious [48], an elite, highly FHB-resistant Canada Prairie Spring (CPS) wheat cv, is believed to carry its major resistance components from one of its progenitors, moderately resistant [49] Canadian heritage wheat cv Neepawa [50] (through male parent BW346) and FHB-resistant [48] female parent HY665. AAC Tenacious has excellent FHB resistance (likely better than Sumai 3). Despite its resistance response and parentage, it has not yet been explored for dissecting FHB resistance. The use of AAC Tenacious for genetic dissection of FHB resistance would be more beneficial for many breeding programs working on similar wheat classes/categories around the globe, since AAC Tenacious can be used directly for crossing due to its improved agronomic and quality traits compared to exotic resources and wild relatives of wheat. Therefore, a doubled haploid (DH) population developed from a cross of FHB susceptible Canada Western Special Purpose (CWSP) wheat cv AAC Innova [51] and AAC Tenacious was utilized to study FHB resistance. In this study, we identify major and stable FHB resistance from the Canadian wheat cultivar AAC Tenacious.
2. Results
2.1. Phenotypic Analysis: Varietal Resistance, Days to Anthesis and Plant Height
FHB nurseries have had good and homogeneous disease development over the years. The ANOVA showed significant differences among genotypes and environmental effect for DI, DS, visual rating index (VRI) and DON, and a genotype x environment effect for DON (Table 1). Among parents and common checks, while mean DI ranged from 2.5 to 9.1 in 2015, it was slightly higher in 2016 (ranged from 4.4 to 10.0), and very similar in 2017 (ranged from 2.2 to 10.0) (Supplementary Table S1). Similarly, DS was comparable over these years. Visual rating index (VRI) and DON response patterns were also quite comparable to DS over these years, except for a slightly higher index and DON accumulation of some checks and AAC Tenacious in 2016, an epidemic year (Supplementary Table S1). It seems that the higher index and DON in 2016 were largely due to the higher DI in 2016, as DS was similar over these years. Where both parents showed different phenotypes for DI, DS, VRI and DON in different years, AAC Tenacious showed the lowest DI, DS, VRI and DON among parents and checks. Mean DI, DS and VRI data showed that Sadash and CDC Teal were the most susceptible cvs among checks (Supplementary Table S1). DON data showed that while Sadash was one of two most susceptible cvs, AC Morse had highest DON accumulation. CDC Teal, AAC Indus and AAC Foray had similar DON accumulation (Supplementary Table S1).
Table 1.
Combined ANOVA table for disease incidence (DI), severity (DS), visual rating index (VRI = DI x DS) and deoxynivalenol content (DON) level of checks, parents and DH population AAC Innova x AAC Tenacious grown at Morden, Canada from 2015 to 2017.
| Source | d.f. | DI | DS | VRI | DON |
|---|---|---|---|---|---|
| Environment (E) | 1 | 20.25 ** | 175.80 ** | 967.00 * | 8135.00 ** |
| Block (within E) | 1 | 9.91 | 0.40 | 172.00 | 94.00 |
| Genotype (G) | 236 | 31.19 ** | 20.60 ** | 2864.00 ** | 1094.00 ** |
| (G х E) | 236 | 3.22 | 2.70 | 237.00 | 130.00 ** |
| Error | 875 | 2.93 | 2.30 | 250.00 | 83.00 |
| CV, % | 24.00 | 31.00 | 40.00 | 41.00 |
CV: coefficient of variation; d.f.: degrees of freedom; Signif. codes: p ** ≤ 0.01, p * ≤ 0.05.
Where the population frequency distribution of DI was skewed towards susceptibility, it was skewed towards resistance for DON. Distribution of DS and VRI were close to normal (Figure 1). Over the years, while population means of DI, DS, VRI and DON were within the range of the two parents (Supplementary Table S1), both upward and downward transgressive segregants were observed (Figure 1). Downward transgressive segregants indicate the presence of superior genes/QTL for FHB resistance in both parents [52].
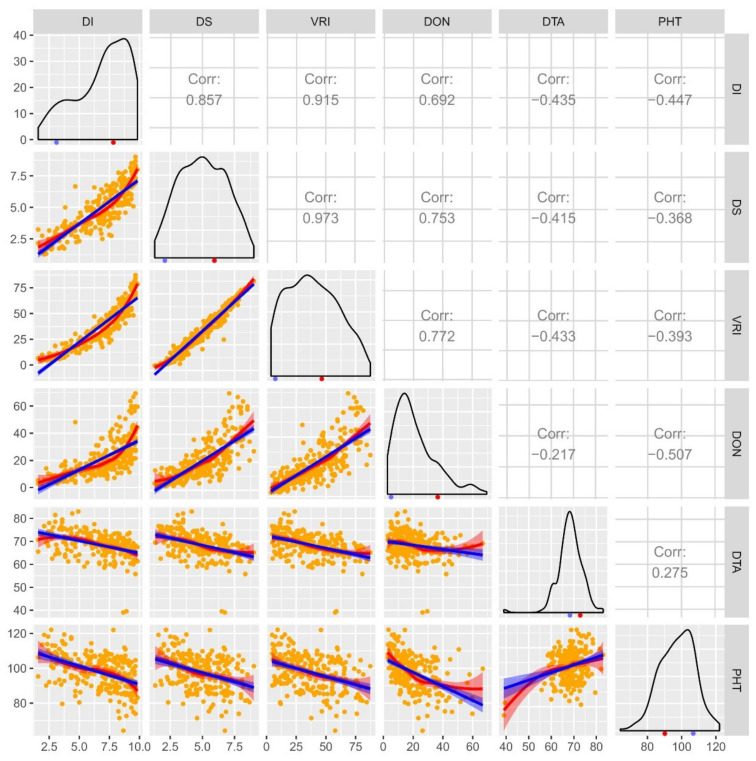
Figure 1.
A combined correlogram of doubled haploid (DH) population of cross AAC Innova x AAC Tenacious drawn using pooled phenotypic data (average of all environments) of four Fusarium head blight (FHB) traits disease incidence (DI; measured on a scale of 0-10), severity (DS; measured on a scale of 0-10), visual rating index (VRI; DI x DS) and deoxynivalenol (DON; measured in ppm) content, and days to anthesis (DTA; days) and plant height (PHT; measured in cm) evaluated at Lethbridge and/or Morden in Canada. Scatterplots with regression lines, linear (blue) and exponential (red), for each pair of traits are drawn on the lower-left part of the plot matrix. Orange dots in scatterplots represent DH lines. Pearson correlations are displayed on the upper-right part of the plot matrix. Frequency distribution plots for each trait are shown on the diagonal. The means of the parental genotypes AAC Innova and AAC Tenacious are indicated by red and blue dots, respectively, beneath frequency distribution plots.
ANOVA also showed significant differences among genotypes and environmental effect for both agronomic traits, i.e., DTA and PHT (Supplementary Tables S2 and S3), in addition to a genotype x environment interaction effect for DTA (Supplementary Table S2). Though both parents flower relatively late in the growing season, AAC Tenacious had anthesis 3.3 days earlier than AAC Innova (mean flowering in 70.1 days) (Supplementary Table S4). The population frequency distribution for DTA was near normal with a mean anthesis date within the range of two parents, but many upward and downward transgressive segregants were observed (Figure 1). AAC Innova had more desirable PHT (89.2 cm; 16.1 cm less) than AAC Tenacious (Supplementary Table S4). The DH population differed broadly for PHT, with the shortest and tallest DHs deviating by 56.7 cm where the mean of population was 98.0 cm. Similar to DTA, distribution was near normal for PHT, and both upward and downward transgressive segregants were observed (Figure 1).
A correlation analysis demonstrated positive relationships among DI, DS, VRI and DON, and between DTA and PHT, but a negative relationship between FHB related traits/variables and agronomic traits (DTA and PHT; Figure 1). Regression line analysis showed moderate to high contributions of DI and DS to DON accumulation (Figure 1), indicating the usefulness of DI and DS data for determining FHB resistance and DON accumulation.
2.2. Genetic Map
A total of 10,328 high quality polymorphic single nucleotide polymorphism (SNP) markers from the wheat 90K SNP Infinium iSelect assay and simple sequence repeat (SSR) molecular markers were genetically mapped to 25 linkage groups (LGs) belonging to 21 wheat chromosomes in the AAC Innova x AAC Tenacious population (Table 2 and Supplementary Table S5). This map spanned a total of 3613.36 cM map distance (on average 172.06 cM per chromosome) with an average density of one marker every 0.35 cM (or 2.34 markers per cM) (Table 2). The highest number of markers were mapped to wheat homoeologous group 1 chromosomes (2565 markers; density = 6.29 markers/cM) and the lowest to group 4 chromosomes (760 markers; density = 1.72 markers/cM) (Table 2). On the other hand, where genome B showed the highest marker coverage (5077 markers; density = 3.96 markers/cM), i.e., slightly higher than the genome A (4112 markers; density = 2.75 markers/cM), genome D showed the lowest coverage (1139 markers; density = 1.36 markers/cM) (Table 2). Despite the mapping of vast number of markers and their high densities, the number of linkage bins spanned on all 21 chromosomes was 1543. This led to an average density of one linkage bin every 2.34 cM (ranging from 0.53 to 42.70) (Table 2; Supplementary Table S5). This showed that the distribution of loci along the chromosomal arms was uneven, and clusters were formed at certain regions; however, only 65 linkage bins exceeded 10.00 cM. A total of 438 and 263 (out of total 10,328) markers were newly mapped which were not present on any previously published individual wheat consensus maps [53,54,55,56,57] or integrated wheat consensus map [57], respectively. The order of the markers in the present study was largely in agreement (Supplementary Table S5) with previously published consensus maps [53,54,55,56,57].
Table 2.
Details of number of mapped molecular markers, map length and marker densities of individual linkage groups (LGs), seven homoeologous groups and three wheat genomes (A, B and D).
| Genome | LG/ Chromosome | No. of Markers | No. of Linkage Bins | Map Length (cM) | Marker Density | Map Density | ||
|---|---|---|---|---|---|---|---|---|
| Markers/cM | Linkage Bins/cM | cM/ Marker | cM/Linkage Bin | |||||
| A | 1 | 737.00 | 124.00 | 187.48 | 3.93 | 0.66 | 0.25 | 1.51 |
| 2 | 626.00 | 107.00 | 178.64 | 3.50 | 0.60 | 0.29 | 1.67 | |
| 3 | 430.00 | 87.00 | 237.23 | 1.81 | 0.37 | 0.55 | 2.73 | |
| 4 | 406.00 | 72.00 | 176.58 | 2.30 | 0.41 | 0.43 | 2.45 | |
| 5 | 760.00 | 116.00 | 232.93 | 3.26 | 0.50 | 0.31 | 2.01 | |
| 6 | 432.00 | 69.00 | 187.11 | 2.31 | 0.37 | 0.43 | 2.71 | |
| 7 | 721.00 | 138.00 | 297.25 | 2.43 | 0.46 | 0.41 | 2.15 | |
| Total | 4112.00 | 713.00 | 1497.23 | 2.75 | 0.48 | 0.36 | 2.10 | |
| % | 39.81 | 46.21 | 41.44 | |||||
| B | 1 | 1721.00 | 88.00 | 140.42 | 12.26 | 0.63 | 0.08 | 1.60 |
| 2 | 817.00 | 109.00 | 202.65 | 4.03 | 0.54 | 0.25 | 1.86 | |
| 3.1 | 89.00 | 28.00 | 38.94 | 2.29 | 0.72 | 0.44 | 1.39 | |
| 3.2 | 385.00 | 68.00 | 113.39 | 3.40 | 0.60 | 0.29 | 1.67 | |
| 4 | 302.00 | 60.00 | 119.83 | 2.52 | 0.50 | 0.40 | 2.00 | |
| 5 | 576.00 | 103.00 | 264.67 | 2.18 | 0.39 | 0.46 | 2.57 | |
| 6 | 693.00 | 87.00 | 206.49 | 3.36 | 0.42 | 0.30 | 2.37 | |
| 7 | 494.00 | 95.00 | 194.88 | 2.53 | 0.49 | 0.39 | 2.05 | |
| Total | 5077.00 | 638.00 | 1281.26 | 3.96 | 0.50 | 0.25 | 2.01 | |
| % | 49.16 | 41.35 | 35.46 | |||||
| D | 1 | 107.00 | 33.00 | 79.61 | 1.34 | 0.41 | 0.74 | 2.41 |
| 2 | 552.00 | 45.00 | 166.82 | 3.31 | 0.27 | 0.30 | 3.71 | |
| 3.1 | 25.00 | 6.00 | 18.14 | 1.38 | 0.33 | 0.73 | 3.02 | |
| 3.2 | 19.00 | 10.00 | 46.52 | 0.41 | 0.21 | 2.45 | 4.65 | |
| 3.3 | 87.00 | 7.00 | 26.64 | 3.27 | 0.26 | 0.31 | 3.81 | |
| 4 | 52.00 | 26.00 | 145.98 | 0.36 | 0.18 | 2.81 | 5.61 | |
| 5 | 156.00 | 30.00 | 181.74 | 0.86 | 0.17 | 1.16 | 6.06 | |
| 6 | 66.00 | 12.00 | 31.22 | 2.11 | 0.38 | 0.47 | 2.60 | |
| 7.1 | 45.00 | 9.00 | 23.12 | 1.95 | 0.39 | 0.51 | 2.57 | |
| 7.2 | 30.00 | 14.00 | 115.09 | 0.26 | 0.12 | 3.84 | 8.22 | |
| Total | 1139.00 | 192.00 | 834.87 | 1.36 | 0.23 | 0.73 | 4.35 | |
| % | 11.03 | 12.44 | 23.10 | |||||
| A+B+D | 1 | 2565.00 | 245.00 | 407.50 | 6.29 | 0.60 | 0.16 | 1.66 |
| 2 | 1995.00 | 261.00 | 548.11 | 3.64 | 0.48 | 0.27 | 2.10 | |
| 3 | 1035.00 | 206.00 | 480.86 | 2.15 | 0.43 | 0.46 | 2.33 | |
| 4 | 760.00 | 158.00 | 442.39 | 1.72 | 0.36 | 0.58 | 2.80 | |
| 5 | 1492.00 | 249.00 | 679.34 | 2.20 | 0.37 | 0.46 | 2.73 | |
| 6 | 1191.00 | 168.00 | 424.82 | 2.80 | 0.40 | 0.36 | 2.53 | |
| 7 | 1290.00 | 256.00 | 630.34 | 2.05 | 0.41 | 0.49 | 2.46 | |
| Total | 10328.00 | 1543.00 | 3613.36 | 2.86 | 0.43 | 0.35 | 2.34 | |
2.3. QTL Analysis
2.3.1. FHB Resistance and DON Content
QTL analyses were carried out separately for DI, DS, VRI and DON using phenotypic data of individual environments, as well as pooled (average of all environments) data. A total of seven main effect QTL for resistance to DI (type-I resistance), eleven to DS (type-II resistance), nine to index (VRI), and nine to DON (type-III resistance) were identified in one or more environments (Figure 2; Table 3 and Supplementary Table S6). Twenty of these QTL, along with two additional suggestive [58] QTL, one for DI on chromosome 7A and another for DON on chromosome 3B, were identified using pooled data (Table 3). While the phenotypic variation (R2) explained by these 22 QTL ranged from 2.10 (QFhi.lrdc-7A) to 34.50% (QDon.lrdc-2D.2) (Table 3), only 10 of these explained >10% R2 using pooled data, irrespective of their individual environment values. Notably, only one stable QTL for DI (QFhi.lrdc-2D) on chromosome 2DS, two for DS (QFhs.lrdc-2D.1 and QFhs.lrdc-4A) on chromosomes 2DS and 4A, two for VRI (QFhb.lrdc-2D.1 and QFhb.lrdc-4A) on chromosomes 2DS and 4A, and one for DON (QDon.lrdc-2D.2) on chromosome arm 2DL were detected across the tested environments (Supplementary Table S6); however, only those present on chromosome 2D were considered major QTL based on phenotypic variation explained.
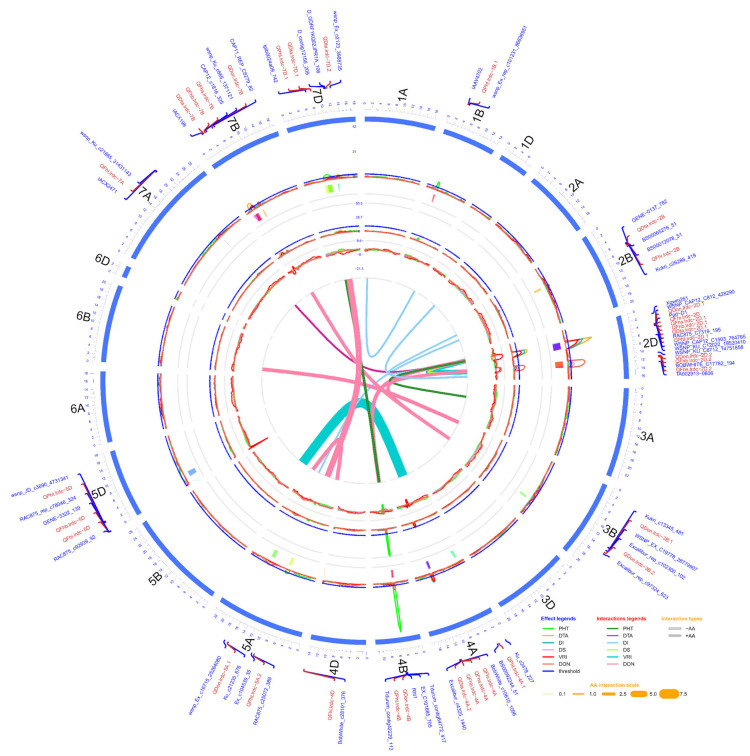
Figure 2.
Circos diagram of linkage and QTL map developed using pooled phenotypic data (average of all environments) of doubled haploid population produced from the cross AAC Innova x AAC Tenacious. The outermost circle/track shows the 21 chromosomes (1A -7D) arranged in a clockwise direction with selected QTL-linked/flanking markers in 0.1X scale (cM). Three inner circles and line connections in the middle represent the mean LOD score (second track from outside), % phenotypic variation (R2) explained (third track from outside), additive effect (AE) (fourth track from outside) of individual QTL and epistatic effect (AA) of digenic QTL x QTL interactions (line connections in the middle) for different measured traits. In the second and third tracks from outside, blue lines show a LOD threshold of 2.5 and R2 threshold of 10%, respectively. QTL confidence intervals are shown in different colors beneath the QTL scans in second track from outside. LOD, R2 and AE peaks of different traits are represented in different colors as shown in the effect legends in the lower-right corner of the diagram. Three most significant and stable QTL identified were on chromosomes 2D and 4B. AA interactions between QTL pairs of the same or different linkage groups for each trait are represented in middle of diagram by line connections of different colors, as shown in the interaction legend in the lower-right corner of the diagram. Negative and positive AA interactions are represented by hollow and solid lines, respectively. The width of the line connections represent the strength of AA effect, as shown in the AA interaction scale in the lower-right corner of diagram.
Table 3.
Details of quantitative trait loci identified for Fusarium head blight response variables/traits disease incidence (DI), severity (DS), visual rating index (VRI = DI x DS), deoxynivalenol content (DON), and days to anthesis (DTA) and plant height (PHT) on different wheat chromosomes in AAC Innova x AAC Tenacious doubled haploid population using pooled data of different environments.
| Response Variable/ Trait |
Chr. (arm) | QTL Name a | QTL Position b | QTL Interval | LOD | Absolute Additive Effect | R 2 | Closest Marker | Individual Environment(s) | Favourable Donor Allele for Respective Trait | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker Name | Linkage Map Position | Physical Position (IWGSC RefSeq v2.0) c Start End |
|||||||||||
| DI | 2B | QFhi.lrdc-2B | 68.8 | 64.2–71.8 | 3.2 | 0.5 | 4.4 | BS00012078_51 | 66.73 | 265366661 | 265366634 | Mrd16, Mrd17 | T |
| 2DS | QFhi.lrdc-2D* | 40.9 | 40.2–63.7 | 15.5 | 1.0 | 25.0 | Ppd-D1 | 40.76 | 36205433 | 36204772 | Mrd15, Mrd16, Mrd17 | T | |
| 4A | QFhi.lrdc-4A | 118.0 | 114.8–121.6 | 8.0 | 0.7 | 11.9 | BobWhite_c10610_1096 | 117.82 | 688409042 | 688408942 | Mrd16, Mrd17 | I | |
| 4D | QFhi.lrdc-4D | 146.0 | 145.1–146.0 | 2.8 | 0.4 | 4.0 | BobWhite_c28101_376 | 145.98 | 515801764 | 515801671 | Mrd16, Mrd17 | I | |
| 5D | QFhi.lrdc-5D | 0.1 | 0.0–0.6 | 2.6 | 0.4 | 3.9 | RAC875_c92929_92 | 0.00 | 279491136 | 279491036 | Mrd17 | T | |
| 7A | QFhi.lrdc-7A | 206.4 | 205.9–206.8 | 2.6 | 0.4 | 2.1 | IACX2471 | 205.99 | 673715328 | 673715447 | -- | T | |
| DS | 2DS | QFhs.lrdc-2D.1* | 40.9 | 40.2–63.4 | 12.7 | 0.8 | 19.0 | Ppd-D1 | 40.76 | 36205433 | 36204772 | Mrd15, Mrd16, Mrd17 | T |
| 2DL | QFhs.lrdc-2D.2* | 115.6 | 105.2–126.4 | 5.4 | 0.5 | 15.2 | BobWhite_c17782_194 | 115.38 | 555098707 | 555098805 | Mrd15, Mrd16 | T | |
| 4A | QFhs.lrdc-4A | 118.0 | 114.8–121.3 | 5.4 | 0.5 | 10.1 | BobWhite_c10610_1096 | 117.82 | 688409042 | 688408942 | Mrd15, Mrd16, Mrd17 | I | |
| 7BS | QFhs.lrdc-7B | 32.3 | 24.3–35.5 | 3.6 | 0.4 | 6.3 | wsnp_CAP7_c44_26549 | 32.15 | 17091249 | 17091049 | Mrd16 | I | |
| VRI | 2DS | QFhb.lrdc-2D.1* | 40.9 | 40.2–63.4 | 12.4 | 9.4 | 18.0 | Ppd-D1 | 40.76 | 36205433 | 36204772 | Mrd15, Mrd16, Mrd17 | T |
| 2DL | QFhb.lrdc-2D.2* | 114.6 | 106.4–120.8 | 3.6 | 4.8 | 12.2 | BobWhite_c17782_194 | 115.38 | 555098707 | 555098805 | Mrd15 | T | |
| 4A | QFhb.lrdc-4A | 118.0 | 114.7–121.3 | 7.1 | 6.6 | 11.5 | BobWhite_c10610_1096 | 117.82 | 688409042 | 688408942 | Mrd15, Mrd16, Mrd17 | I | |
| 5D | QFhb.lrdc-5D | 0.2 | 0.0–0.4 | 2.5 | 3.4 | 3.0 | RAC875_c92929_92 | 0.00 | 279491136 | 279491036 | Mrd17 | T | |
| 7BS | QFhb.lrdc-7B | 31.6 | 24.1–35.2 | 3.9 | 4.9 | 7.3 | wsnp_CAP7_c44_26549 | 32.15 | 17091249 | 17091049 | Mrd16 | I | |
| DON | 2DS | QDon.lrdc-2D.1* | 40.6 | 37.9–41.9 | 3.9 | 3.0 | 12.3 | Ppd-D1 | 40.76 | 36205433 | 36204772 | Mrd16, Mrd17 | T |
| 2DL | QDon.lrdc-2D.2* | 112.6 | 105. 1–126.6 | 14.4 | 6.0 | 34.5 | BobWhite_c17782_194 | 115.38 | 555098707 | 555098805 | Mrd15, Mrd16, Mrd17 | T | |
| 3B | QDon.lrdc-3B.1* | 132.8 | 132.2–133.2 | 2.7 | 2.1 | 9.3 | Kukri_c13345_481 | 132.75 | 753508932 | 753508832 | Mrd15 | T | |
| 3B | QDon.lrdc-3B.2 | 147.3 | 144.5–150.3 | 2.7 | 2.3 | 4.0 | Excalibur_rep_c97324_623 | 151.49 | 771943609 | 771943709 | -- | T | |
| 4B | QDon.lrdc-4B* | 49.5 | 46.4–52.4 | 6.6 | 3.6 | 8.0 | Ex_c101685_705 | 49.48 | 31725418 | 31725518 | Mrd17 | T | |
| 5A | QDon.lrdc-5A.1 | 164.8 | 156.4–169.0 | 3.1 | 2.3 | 3.0 | IACX2540 | 164.62 | 621553818 | 621553699 | Mrd16, Mrd17 | T | |
| 7BS | QDon.lrdc-7B* | 52.9 | 52.2–57.7 | 3.4 | 2.7 | 5.0 | Kukri_c19823_491 | 52.69 | 60918162 | 60918262 | Mrd15 | I | |
| DTA | 2B | QDta.lrdc-2B | 65.9 | 64.9–66.7 | 2.6 | 0.6 | 3.0 | BS00065276_51 | 66.14 | 57526894 | 57526994 | Mrd17, Let18 | I |
| 2DS | QDta.lrdc-2D.1 | 41.9 | 40.2–63.5 | 22.9 | 2.1 | 30.0 | RAC875_c7319_195 | 41.83 | 36481727 | 36481627 | Mrd17, Let17, Let18, Let19 | I | |
| 4A | QDta.lrdc-4A.2 | 120.0 | 117.4–123.8 | 6.8 | 1.1 | 7.0 | Excalibur_c4325_1440 | 121.65 | 686650643 | 686650543 | Mrd17, Let18, Let19 | T | |
| 7BS | QDta.lrdc-7B | 27.3 | 18.1–31.9 | 6.6 | 1.1 | 8.0 | IACX198 | 24.26 | 10548520 | 10548633 | Mrd17, Let17, Let18, Let19 | T | |
| 7D | QDta.lrdc-7D.1 | 79.9 | 77.8–91.5 | 2.78 | 1.2 | 5.8 | D_GCE8AKX02ILA1U_88 | 79.8186 | 56638676 | 56638464 | Let17, Let18, Let19 | T | |
| 7D | QDta.lrdc-7D.2 | 117.7 | 113.0–118.0 | 3.4 | 0.8 | 3.5 | wsnp_Ex_c10430_17064001 | 118.58 | 114113970 | 114114170 | Mrd17 | T | |
| PHT | 1B | QPht.lrdc-1B.1 | 36.0 | 33.0–38.5 | 7.1 | 2.4 | 6.0 | IAAV4702 | 34.94 | 544295939 | 544296139 | Mrd17, LetPGH17 | I |
| 2DS | QPht.lrdc-2D.1 | 41.9 | 40.2–63.5 | 11.1 | 3.2 | 10.0 | RAC875_c7319_195 | 41.83 | 36481727 | 36481627 | Mrd17, Let17, Let18, Let19 | I | |
| 4A | QPht.lrdc-4A.1 | 22.6 | 17.8–26.8 | 2.6 | 1.5 | 2.0 | BS00092244_51 | 26.38 | 11756178 | 11756078 | Mrd17 | T | |
| 4B | QPht.lrdc-4B | 49.6 | 46.0–52.6 | 42.0 | 7.3 | 53.0 | Ex_c101685_705 | 49.48 | 31725418 | 31725518 | Mrd17, Let17, LetPGH17, Let18, Let19 | I | |
| 5A | QPht.lrdc-5A.2 | 97.8 | 91.9–103.0 | 3.7 | 1.7 | 3.0 | RAC875_c25072_389 | 97.75 | 524944276 | 524944376 | Let17, Let18 | T | |
| 5D | QPht.lrdc-5D | 26.5 | 24.7–42.5 | 2.9 | 1.7 | 3.0 | wsnp_JD_c3690_4731341 | 26.59 | 401527745 | 401527907 | -- | T | |
| 7D | QPht.lrdc-7D.1 | 79.9 | 67.9–95.2 | 5.0 | 2.0 | 4.0 | D_GCE8AKX02ILA1U_88 | 79.82 | 56638676 | 56638464 | Mrd17, Let17, Let18, Let19 | T | |
Note: Chr.: chromosome; QTL: quantitative trait loci; LOD: logarithm of the odds score; R2: percent of the phenotypic variance; Mrd: Morden; Let: Lethbridge; PGH: green house; 17: 2017; 18: 2018; 19: 2019; a QTL were named according to all loci identified using pooled (average of all environments) as well as individual environment phenotypic data; b QTL positions were determined by the highest LOD peak positions (cM) on linkage groups; c Physical positions were determined by SNP probe sequence mapped to reference genome; -- QTL detected using pooled data only; I = AAC Innova; T = AAC Tenacious; * loci also appeared during epistasis QTL analysis.
In addition to the aforementioned main effect QTL, 13 digenic epistasis interactions of 20 different loci were also identified (Figure 2; Table 4). All these epistasis interactions possessed a significant (p ≤ 0.05) additive x additive effect (Table 4). Notably, based on within 10.0 cM criteria, seven loci or chromosomal regions which appeared in digenic epistasis interactions were also identified during the main effect QTL analysis. These included QTL regions close to 72.6-82.5 cM on chromosome (arm) 2B (overlapping interval of QFhs.lrdc-2B and QFhb.lrdc-2B), 28.5-48.4 cM on 2DS (overlapping interval of QFhi.lrdc-2D, QFhs.lrdc-2D.1 and QFhb.lrdc-2D.1), 106.2-120.4 cM on 2DL (overlapping interval of QDon.lrdc-2D.2), 130.1-135.3 cM on 3B (overlapping interval of QDon.lrdc-3B.1), 47.7-50.0 cM on 4B (overlapping interval of QDon.lrdc-4B), 184.7-196.9 cM on 5A (overlapping interval of QDon.lrdc-5A.2) and 52.1-58.3 cM on 7BS (overlapping interval of QDon.lrdc-7B) (Table 3 and Table 4 and Supplementary Table S6). Out of all 20 epistasis loci, three QTL regions on chromosomes (or arms) 2DS, 2DL and 3B, as designated above with the respective QTL, were detected in interactions with more than one gene/QTL, either for the same or different response variable/trait (Table 4).
Table 4.
Details of epistasis quantitative trait loci (QTL) identified for disease incidence (DI), severity (DS), visual rating index (VRI = DI x DS), deoxynivalenol content (DON), and days to anthesis (DTA) and plant height (PHT) on different wheat chromosomes (chrs) in AAC Innova x AAC Tenacious doubled haploid population.
| Trait | QTL_i a | Chr | Interval_i b | Position_i c | Range_i d | QTL_j a | Chr | Interval_jb | Position_j c | Range_j d | AA e | SE f |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DI | 1–12 | 1A | BS00103478_51-BS00081682_51 | 17.7 | 12.7–23.2 | 2–86 | 1B | RAC875_c102886_73-Tdurum_contig9144_222 | 138.0 | 133.7–139.9 | 0.44 | 0.08 ** |
| 5–82 | 2B | IAAV1743-Jagger_c3435_145 | 118.4 | 115.5–126.3 | 13–83 | 5A | RAC875_rep_c109969_119-RFL_contig316_572 | 151.3 | 148.0–161.4 | −0.41 | 0.10 ** | |
| 6–11 # | 2D | Ppd-D1-wsnp_CAP12_c1503_764765 | 38.9 | 36–44.4 | 5–51 # | 2B | Tdurum_contig53156_111-Tdurum_contig1653_190 | 81.3 | 72.6–82.5 | −0.38 | 0.08 ** | |
| 6–42 | 2D | RAC875_c5998_1056-D_F5MV3MU01EDEO3_100 | 145.5 | 142.6–155.6 | 4–6 | 2A | Excalibur_rep_c106338_424-RAC875_c54668_102 | 36.5 | 25.5–49.0 | −0.51 | 0.09 ** | |
| DS | 6–10 # | 2D | wsnp_CAP12_c812_428290-RAC875_c7319_195 | 38.1 | 34.5–42.4 | 6–22 # | 2D | BobWhite_c17782_194-TA002913-0806 | 115.4 | 106.2–120.4 | −0.20 | 0.08 ** |
| VRI | 6–10 # | 2D | wsnp_CAP12_c812_428290-RAC875_c7319_195 | 38.1 | 36.1–39.4 | 6–22 # | 2D | BobWhite_c17782_194-TA002913-0806 | 112.4 | 108.2–119.4 | −2.90 | 0.98 ** |
| 10–41 | 4A | RAC875_c6075_214-BobWhite_c35402_66 | 84.8 | 84.2–87.8 | 14–7 | 5B | RAC875_c39204_91-TA015732-1144 | 20.6 | 12.6–27.6 | 6.40 | 0.89 ** | |
| DON | 6–10 # | 2D | wsnp_CAP12_c812_428290-RAC875_c7319_195 | 38.1 | 28.5–48.4 | 11–17 # | 4B | Ex_c101685_705-Tdurum_contig42229_113 | 49.5 | 47.7–50.0 | 2.02 | 0.41 ** |
| 6–21 # | 2D | wsnp_ku_c8712_14751858- BobWhite_c17782_194 | 107.2 | 104.2–110.2 | 8–69 # | 3B | Kukri_c13345_481-wsnp_Ex_c19778_28779907 | 132.7 | 130.1–135.3 | 1.52 | 0.57 ** | |
| 8–12 | 3B | RAC875_rep_C107068_182-RAC875_rep_C69171_241 | 6.7 | 0.0–18.4 | 17–12 | 6B | IAAV5385-Tdurum_contig61178_618 | 28.5 | 25.8–34.5 | 1.92 | 0.53 ** | |
| 8–69 # | 3B | Kukri_c13345_481-wsnp_Ex_c19778_28779907 | 132.7 | 130.1–135.3 | 20–16 # | 7B | Kukri_c19823_491-CAP11_rep_c8279_82 | 53.7 | 52.1–58.3 | −1.65 | 0.54 ** | |
| 13–9 | 5A | Ex_C95453_1499-Excalibur_c13536_202 | 20.3 | 16.3–33.3 | 13–98 # | 5A | Kukri_c108256_381-BS00090847_51 | 195.7 | 184.7–196.9 | −1.93 | 0.43 ** | |
| 13–52 | 5A | wsnp_Ku_c15816_24541162-GENE-3314_78 | 88.4 | 87.9–91.1 | 21–12 | 7D | D_GCE8AKX02ILA1U_88-D_CONTIG12156_209 | 103.8 | 94.8–111.9 | −3.62 | 0.46 ** | |
| DTA | 6–10 # | 2D | wsnp_CAP12_c812_428290-RAC875_c7319_195 | 38.1 | 36.1–40.4 | 19–123 | 7A | Wsnp_Ku_c8437_14341371-Excalibur_c3188_1352 | 234.8 | 228.3–238.5 | 0.55 | 0.20 ** |
| PHT | 6–12 # | 2D | wsnp_CAP12_c1503_764765-wsnp_Ku_c12022_19520410 | 39.4 | 36.1–45.4 | 7–35 # | 3A | Excalibur_c19671_139-wsnp_Ex_c9458_15679797 | 88.9 | 85.0–92.2 | −0.67 | 0.31 * |
| 11–17 # | 4B | Ex_c101685_705-Tdurum_contig42229_113 | 49.5 | 48.7–50.0 | 21–12 # | 7D | D_GCE8AKX02ILA1U_88-D_CONTIG12156_209 | 79.8 | 74.0–85.8 | −1.08 | 0.31 ** |
Note: a QTL_i and QTL_j: The two QTL involved in an epistatic interaction. QTL are named with the relevant chromosome and the marker intervals. The first (before ‘–’) and second (after ‘–’) part of QTL name represents chr number (1 to 21) and marker interval, respectively on relevant chromosome; b Interval_i and interval_j: The flanking markers of QTL_i and QTL_j, respectively; cPosition_i and position_j: The distance between QTL_i/QTL_j and the first marker of the relevant chromosome; d Range_i and range_j: The position support intervals of QTL_i and QTL_j, respectively; e AA: The estimated additive x additive (epistatic) effect. A positive epistasis value means that the phenotype is higher than expected, while a negative epistasis value means that the phenotype is lower than expected.; f SE: The standard error of estimated or predicted QTL effect; significance codes: p ** ≤0.01, p * ≤0.05; # loci also appeared during main effect QTL analysis.
2.3.2. Days to Anthesis and Plant Height
Similar to FHB response, QTL analyses were carried out separately for DTA and PHT using phenotypic data of individual environments as well as pooled data. A total of ten main effect QTL for DTA and fourteen for PHT were identified in one or more environments using data of individual environments (Figure 2; Table 3 and Supplementary Table S6). Twelve of these QTL along with one additional suggestive [58] QTL for PHT on chromosome 5D were identified using pooled data (Table 3). While these 13 QTL explained 2.00 (QPht.lrdc-4A.1) to 53.00% (QPht.lrdc-4B) R2 using pooled data (Table 3), only one DTA QTL (QDta.lrdc-2D.1) and two PHT QTL (QPht.lrdc-2D.1 and QPht.lrdc-4B) explained ≥10% of the R2 and were considered major QTLs. However, only two stable DTA QTL (QDta.lrdc-2D.1 and QDta.lrdc-7B) and one PHT QTL (QPht.lrdc-4B) were detected across the tested environments (Supplementary Table S6). Shorter DTA and reduced PHT alleles for different QTL were attributable to both parents (Table 3 and Supplementary Table S6).
In addition to the main effect QTL for DTA and PHT, three digenic epistasis interactions of six different loci were also identified (Figure 2; Table 4). All epistasis QTL also showed significant additive x additive effects (Table 4). Unlike FHB related QTL, the majority of epistasis loci (except one QTL close to 234.8 cM on 7A) overlapped with QTL regions identified during the main effect QTL analysis.
2.3.3. Pleiotropic QTL
Main effect QTL positions of FHB response variables/traits DI, DS, VRI and DON, and agronomic traits were compared. Chromosomal intervals which harboured two or more QTL within 10.0 cM were considered shared or common QTL regions, while the rest of the QTL intervals (>10.0 cM apart) were considered unique QTL regions. This analysis identified a total of 25 unique (10 for FHB variables or related traits on chromosomes 1A, 2A, 3B, 4B, 5A, 6A, 7A and 7B; 4 for DTA on 2D, 4A, 5B and 7D; and 11 for PHT on 1A, 1B, 2D, 3A, 4A, 5A, 5D, and 7D) and 11 shared QTL regions on different chromosomes identified from the QTL results that utilized phenotypic data belonging to all individual environments and pooled data (Supplementary Table S7). Notably, while all 11 shared regions harbouring QTL for FHB related variable(s)/traits, seven of these also harboured QTL for one of two agronomic traits, i.e., DTA (on 2B, 5A and 7B) and PHT (on 4B), or both (on 2DS, 4A and 7D). These seven were considered pleiotropic loci. On the other hand, only four shared regions harboured QTL solitarily for FHB related variables/traits, and were located on chromosomes 2DL, 3B, 4D and 5D (Supplementary Table S7). Only the 2DL region of these had a major effect QTL. Where all these regions harbouring FHB index QTL, 4D and 5D regions provided both type-I and -II resistance, the 2DL region provided both type-II and -III resistance and the 3B region provided only type-II resistance. Reduced FHB alleles at different loci in three (2DL, 3B and 5D) out of four regions were derived from AAC Tenacious. Among seven pleiotropic QTL regions, while one on chromosome arm 2DS harboured QTL for type-I, -II and -III resistances along with FHB index, the remaining six harboured QTL for ≤2 type of resistance with or without index QTL (Supplementary Table S7). Reduced FHB alleles at different loci in the 2B, 2DS, 4B and 5A regions were derived from AAC Tenacious, while in three other regions, i.e., 4A, 7BS and 7D, they were derived from AAC Innova (Supplementary Table S7).
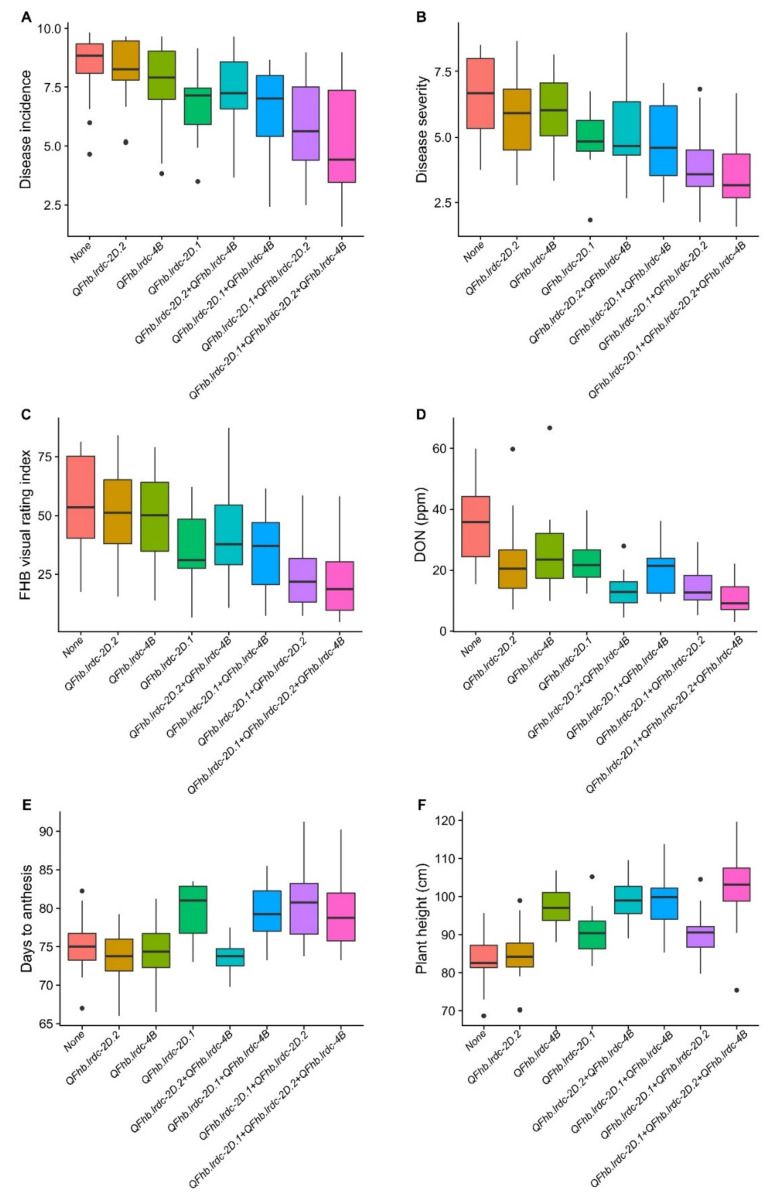
Three regions (solitarily FHB responsive 2DL and pleiotropic 2DS and 4B) which had QTL, explaining the largest phenotypic variation, and all derived from AAC Tenacious were assessed individually and in combination for their effect on different traits in DH population (Figure 3). It was observed that while individually, 2DS region contributed maximum to lower DI, DS and VRI, the 2DL region contributed towards a maximum reduction of DON content. Though the 4B region reduced DI, DS, VRI and DON, its effect was smaller on these response variables/traits compared to the other two regions (Figure 3). Similar results were observed in combinations (Figure 3). Interestingly, where low FHB 2DL and 4B alleles had no significant effect on days to anthesis, 2DS low FHB allele delayed anthesis time significantly (Figure 3). Conversely, except for the low FHB 2DL allele, both 2DS and 4B low FHB alleles increased plant height, with the effect of 4B being very prominent (Figure 3).
Figure 3.
Boxplot distributions of doubled haploid (DH) population produced from the cross AAC Innova x AAC Tenacious. Effects of positive alleles of single QTL (QFhb.lrdc-2D.1, QFhb.lrdc-2D.2 and QFhb.lrdc-4B) and their combinations on average Fusarium head blight incidence (A), severity (B), visual rating index (C), deoxynivalenol (DON) accumulation (D), days to anthesis (E) and plant height (F) are depicted alongside negative alleles at all three loci using pooled phenotypic data (average of all environments). Quartiles and medians are represented by boxes and continuous lines, respectively. Whiskers extend to the farthest points that are not outliers, whilst outliers are shown as black dots.
3. Discussion
There is increasing interest in identifying and characterizing native FHB resistance, as breeders have struggled to achieve sufficient progress in improving FHB resistance using exotic resources [47]. Although some attempts [15,47,59,60] have been made recently to genetically determine native FHB resistance in Europe and north America, there is an ever-increasing need to identify new resources, as FHB is spreading to previously unaffected areas [3]. In the present study, a spring wheat DH population, its parents and check cvs were subjected to extensive phenotyping for FHB-DI, -DS, -VRI and DON, as well as agronomic traits DTA and PHT, followed by high-throughput genotyping. The largely different phenotypic expression of both Canadian parents, i.e., AAC Innova, being susceptible, and AAC Tenacious, being highly resistant (Supplementary Table S1), and the broad genetic variation (Table 1) make this population ideal for dissecting FHB resistance. This study is the first effort to unravel the FHB resistance of AAC Tenacious, i.e., the first FHB-resistant wheat cultivar in Canada, which is supposed to carry a native or Canadian climate-adapted resistance component from its FHB-resistant female parent HY665 [48] and moderately FHB-resistant Canadian wheat cv Neepawa [49], a grandparent in lineage of its male parent BW346.
We utilized the Infinium iSelect 90K SNP assay [54] and a few other markers for genotyping, which resulted in the mapping of 10,328 markers representing 1543 linkage bins on all 21 wheat chromosomes. In the present study, the marker density of 2.34 markers per cM was comparable to the recently published individual wheat maps of two populations, ‘Doumai × Shi 4185′ and ‘Gaocheng 8901 × Zhoumai 16′, where marker densities were 3.85 and 3.0 markers/cM, respectively [57]. Conversely, the marker coverage of the present map is also consistent with previous studies, where B-genome had the highest marker coverage, followed by the A and D genomes [53,54,57,61,62]. Additionally, we identified at least 263 new markers that were not present on any individual or integrated previously published wheat consensus map [53,54,55,57]. These SNPs could be due to the use of the native germplasm, which was not present in previous studies [52] and helped us to explore the native FHB resistance in this population.
A total of 21 FHB responsive loci or chromosomal regions (15 from AAC Tenacious and 6 from AAC Innova), which involved both unique loci (10) and shared QTL regions (11; 4 solitarily FHB related and 7 most likely pleiotropic), were identified by this study (Supplementary Table S7). Notably, unique QTL, which were not detected in all environments, explained <10.0 % R2, and were thus considered small effect [63] suggestive [58] QTL. Instead, three shared QTL regions (i.e., 2DS, 2DL and 4B) harbored some major QTL (explained >10.0 % R2) for FHB response variables/traits which also showed high LOD score (>5.0) and additive effect, and were also detected over all tested environments and pooled data (Supplementary Table S7). These major and stable QTL in these shared regions include FHB-resistant QTL QFhi.lrdc-2D, QFhs.lrdc-2D.1, QFhb.lrdc-2D.1 and QDon.lrdc-2D.2, and DTA and PHT QTL QDta.lrdc-2D.1, QPht.lrdc-2D.1, and QPht.lrdc-4B. Broadly, the 2DL region of the three shared regions was the only specific form of FHB resistance; the rest appeared to result from the pleotropic effect of the loci also affecting either both DTA and PHT (2DS) or just PHT (4B). Interestingly, AAC Tenacious contributed to the resistance alleles at all major and stable loci in these regions. These QTL regions are discussed in greater detail below.
The 2DS FHB-resistant QTL had highest peaks, ranging from 40.6–40.9 cM, on the chromosome arm 2DS, which colocated with the photoperiod responsive gene Ppd-D1 (40.76 cM). The DTA QTL QDta.lrdc-2D.1 and PHT QTL QPht.lrdc-2D.1 confidence interval also overlapped 2DS FHB resistance-related QTL (40.6–40.9 cM) and the Ppd-D1 gene (40.76 cM). Associations among FHB, DTA and PHT are very common and have been thoroughly studied in wheat. FHB-resistant QTL have been shown to have overlapping anthesis time and plant height QTL with the European winter wheat population Renan/Recital [15].
Resistance loci on the chromosome arm 2DS were also identified in previous studies on wheat genotypes Alondra [64], Biscay [20], C615 [65], DH181 [66], Ernie [60], Furore [67], Gamenya [42,68], Jagger [69], Kenyon [47], Maringa [16], Massey [60], Naxos [11,70], Romanus [20], Shanghai-3 [11], Wangshuibai [71,72], Y1193-6 [36] and Yanzhan 1 [73]. The majority of previously published 2DS resistance loci mapped close to/overlap either one of/or two important markers loci, reduced height gene Rht8 [74] linked marker locus Xgwm261 in genotypes Furore, Gamenya, Kenyon, Maringa, Massey, Romanus, Wangshuibai, Y1193-6 and Yanzhan 1 [16,20,36,42,47,60,67,68,71,72,73] and photoperiod responsive gene Ppd-D1 in cvs/genotypes Ernie, Kenyon, Massey and Naxos [47,60,70]. Previously, it has been speculated that both the tall allele of Rht8 and the photoperiod-sensitive allele of Ppd-D1 may be causal genes for two different 2DS FHB-resistant QTL in cv Kenyon [47]. In another study, Liu et al. [60] reported that the photoperiod-sensitive allele Ppd-D1b pleiotropically reduced FHB susceptibility and increased plant height in Becker/Massey and Ernie/MO 94-317 populations. It was observed that previously mapped 2DS QTL from cvs/genotypes Alondra, Ernie, Furore, Gamenya, Jagger, Kenyon, Massey, Shanghai-3, Wangshuibai and Y1193-6 [11,36,47,60,64,67,68,69,71] not only overlap with the Ppd-D1 region, but also with each other when compared to the wheat reference genome using their linked marker sequences [75]. Since DTA and PHT loci, i.e., QDta.lrdc-2D.1 and QPht.lrdc-2D.1, were also identified in this 2DS region in present study, it became important to know whether the phenotypic effects of this interval were due to Rht8 or Ppd-D1. Thus, we genotyped the AAC Innova x AAC Tenacious population with Ppd-D1 and Xgwm261 markers. We observed that while AAC Innova carried a photoperiod-insensitive allele, i.e., Ppd-D1a, and a reduced brassinosteroid-sensitivity semidwarf allele, i.e., Rht8c, at Xgwm261 locus [76], AAC Tenacious carried the photoperiod-sensitive allele Ppd-D1b and the brassinosteroid-sensitive tall allele Rht8a [76]. These results seem obvious, as AAC Innova is a semidwarf cv, while AAC Tenacious is a tall cv, and do not reject the potential role of Rht8 affecting FHB response. However, the linkage and QTL mapping conducted using these markers eliminated the role of Rht8 in 2DS-mediated phenotypic effects. Our results showed the highest peak of all 2DS QTL (QFhi.lrdc-2D, QFhs.lrdc-2D.1, QFhb.lrdc-2D.1, QDon.lrdc-2D.1, QDta.lrdc-2D.1 and QPht.lrdc-2D.1) mapped close to/overlap Ppd-D1, while the Rht8 marker locus Xgwm261 was mapped at 27.2 and 24.4 cM distal to Ppd-D1 and 2DS QTL interval flanking marker wsnp_CAP12_c812_428290, respectively. These observations also indicate that AAC Innova and AAC Tenacious may be monomorphic at the Rht8 locus, despite the polymorphism at Xgwm261, which is perhaps due to historic recombination events between the two loci. Thus, the AAC Tenacious derived-photoperiod sensitive allele Ppd-D1b seems to be responsible for the phenotypic expression of 2DS QTL, particularly of QDta.lrdc-2D.1 and QPht.lrdc-2D.1, as Ppd-D1b is known to strongly regulate anthesis time [77], and has also been previously reported to pleiotropically control the height by shortening the plant life cycle [78]. Moreover, though all FHB, DTA and PHT QTL spanned the same interval, the highest peaks of disease-responsive (QFhi.lrdc-2D, QFhs.lrdc-2D.1, QFhb.lrdc-2D.1 and QDon.lrdc-2D.1) and agronomic (QDta.lrdc-2D.1 and QPht.lrdc-2D.1) QTL were separated by ~1.0 cM (Table 3), which is not statistically significant. Thus, the FHB resistance in this region may be due to either (i) Ppd-D1b, or (ii) a gene other than Ppd-D1, but with a linkage drag between the Ppd-D1b-sensitive allele and FHB resistance [1]. Further experiments involving a large fine mapping population are required to confirm the presence of two different loci in this region.
The second most important locus/chromosomal region identified during the present study was the solitarily FHB resistance-specific 2DL region. It harbored three important QTL, i.e., QFhs.lrdc-2D.2 (115.6 cM), QFhb.lrdc-2D.2 (114.6 cM) and QDon.lrdc-2D.2 (112.6 cM). While all of these QTL explained >10% phenotypic variation and are major loci, QDon.lrdc-2D.2 explained up to 34.5% phenotypic variation for DON, was stable over all of the tested environments and had no linkage drag. Interestingly, AAC Tenacious contributed resistance alleles at all these QTL in this region, and thus, all these QTL could be the result of a single gene. QTL on chromosome arm 2DL have been identified in previous individual QTL mapping studies utilizing cvs/lines of different origin, i.e., Canada (cv Kenyon), China (cvs Changjiang 9306 or CJ 9306, CS-SM3-7ADS, Shanghai-3, Wangshuibai, Wuhan-1), CIMMYT, Mexico (cvs C615, Soru#1 and SYN1), Swiss (cv Arina), and USA (cvs VA00W-38 and CASS94), [11,16,47,59,65,70,79,80,81,82,83,84,85]. Moreover, it appears that the 2DL QTL region from the present study overlapped with the 2DL QTL regions of at least Arina [83], CASS94 [82], CJ 9306 [79], CS-SM3-7ADS [85], Kenyon [47], Shanghai-3 [11], Soru#1 [70], SYN1 [80], Wangshuibai [81,84] and Wuhan-1 [16] when compared with the wheat reference genome using their linked marker sequences [75]. Interestingly, the 2DL QTL are reported to explain >10% (up to 60%) total phenotypic variation in the majority of the previous studies [47,70,79,80,81,82,84] with the exception of a few, such as QTL derived from Arina [83], Shanghai-3 [11] and Wuhan-1 [16]. This showed that QTL on the chromosome arm 2DL were not only detected repeatedly during different studies, but also that their expression was high, and they expressed in different genetic backgrounds, as witnessed by their widespread presence in global FHB-resistant wheat germplasm. With these qualities, in particular the absence of linkage drags or pleiotropic effect, and being a major contributor of DON reduction, it is considered a highly promising QTL for developing FHB-resistant cultivars.
Another important region identified during this study was on chromosome 4B. It harbored FHB-responsive QTL QFhi.lrdc-4B, QFhb.lrdc-4B and QDon.lrdc-4B, and PHT QTL QPht.lrdc-4B. Though QDon.lrdc-4B was also a major QTL in this region, QPht.lrdc-4B was the only stable QTL detected in all tested environments. QPht.lrdc-4B mapped to the same genomic interval as of Rht1 (Rht-B1) on chromosome 4B during synteny and collinearity analysis using QPht.lrdc-4B flanking marker sequences (Ex_c101685_705 and Tdurum_contig42229_113) and Rht-B1a gene sequence (GenBank Accession: FR719732) utilizing IWGSC RefSeq v2.0. AAC Tenacious contributed both increased height and DON reduction alleles along with DI and VRI reduction at this region. We found that AAC Tenacious possesses a gibberellic acid (GA)-sensitive, tall height allele of Rht1. Earlier studies reported an association between Rht-B1a (GA-sensitive allele) and low FHB as well as tall height [11,86]. A recent report showed decreased FHB spread in GA treated wheat plants [87]. Interestingly, despite the association of tall height with FHB resistance, we found five highly FHB-resistant DH lines carrying a GA-sensitive allele but with semidwarf statures in comparison to several DH lines carrying an insensitive allele, perhaps due to transgressive segregation at some other Rht loci. Rht24 is one such gene which exhibits reduced plant height without influencing FHB [88]. In addition to the direct effect of QPht.lrdc-4B, it also reduced DON in epistasis with Ppd-D1 during this study. Guo et al. [89] observed that the Ppd-D1b and Rht-D1a alleles affected the different floral developmental stages, extending the duration of stem elongation phase, floret primordium initiation and booting. It appears that sensitive alleles at the Ppd-D1 and Rht loci coordinate for different developmental activities and FHB resistance. Although the favorable alleles for DI, VRI and DON at this region were from AAC Tenacious, even in some semidwarf DHs with a GA-sensitive allele, its use in breeding may still result in a large trade-off, as the GA-sensitive allele of Rht1 is significantly associated with increased PHT. Thus, it would be more beneficial to deploy other resistance QTL to increase the resistance of genotypes carrying either Rht1 or other genes such as Rht24.
In addition to the aforementioned QTL, type-II and index QTL QFhs.lrdc-4A and QFhb.lrdc-4A, DTA QTL QDta.lrdc-7B and PHT QTL QPht.lrdc-7D.1, that belong to three shared regions on chromosomes 4A, 7B and 7D, were also detected in all environments and utilizing pooled data; however, these QTL generally explained <10.0% R2 in individual environments. In these regions, favourable alleles for FHB resistance-related QTL were contributed to by AAC Innova, while AAC Tenacious contributed favourable alleles for DTA and PHT. Based on comparative mapping, it seems that QFhs.lrdc-4A and QFhb.lrdc-4A mapped to the similar location as 4A QTL in 86ISMN 2137 [47], and QPht.lrdc-7D.1 to a similar location as 7D FHB-resistant QTL from Kenyon, Wesley-Fhb1-BC56 and AL-107-6106 [47,90].
The phenotypic effect of a gene or QTL allele at a locus may vary in different genetic backgrounds because of epistatic interaction with other gene loci. This means that an apparently “favorable” QTL allele at a locus may show a “neutral” or “unfavorable” phenotype effect in a new genetic background [91]. An understanding of epistatic loci is vital for breeding wheat, since epistasis not only complicates the genotype–phenotype relationship, but also affects the rate of genetic gain [91]. In this study, a number of epistasis loci involved in regulating FHB expression and other traits have been identified. Interestingly, seven FHB-influencing and five agronomic traits loci, involved in digenic epistatic interactions, were also identified during the main effect QTL analysis, clearly suggesting the influence of epistasis on the additive effect of the main effect QTL. Despite their importance and possible roles in crop improvement, epistatic loci influencing FHB are seldom reported, except in a few studies on wheat [47,85] and triticale [52]. This could be due to many factors, including the complexity of FHB phenotypic response and the less attractive minor effects resulting from individual interactions.
In summary, the results of the present study add to our knowledge of the unique, pleotropic and epistatic FHB-resistance QTL derived from Canadian wheat cvs AAC Innova and AAC Tenacious. More particularly, the three major FHB-resistance QTL identified during present study were derived from AAC Tenacious. These QTL will not only be valuable for Canadian wheat breeders, but may be useful for breeders in other growing regions. These QTL, along with the resources generated in this study, including the high-density SNP map and the DH lines with different gene/QTL combinations, will facilitate the development of FHB-resistant cvs using marker-assisted breeding, and will also help in map-based cloning.
4. Materials and Methods
4.1. Plant Material
A doubled haploid mapping population (224 lines) was generated from a biparental cross AAC Innova x AAC Tenacious using the wheat-maize pollination technique [92] at the Lethbridge Research and Development Centre, Lethbridge, AB to genetically examine FHB resistance and associated traits. AAC Innova is an FHB-susceptible, semidwarf cultivar, while AAC Tenacious is a resistant, tall cultivar. The cv AAC Innova was developed from the cross AC Andrew/N9195 (L01164) [51]. The resistant cv AAC Tenacious was derived from the cross HY665/BW346 [48]. The female parent, HY665, which was supposed to be a FHB-resistant source, was derived from the cross 96W137//Howell/HY617BSR [48]. The male parent, BW346, is believed to carry FHB resistance from Canadian wheat cv Neepawa, and had the pedigree RL4802//(96MHN5295-1)BW174*2/Clark [48].
A number of Canadian wheat cvs/breeding lines developed and maintained at Agriculture and Agri-Food Canada with different levels of resistance were also used as checks for comparisons in nurseries; these included the FHB-resistant breeding line FHB 37, the moderately resistant cv AAC Penhold, the intermediate cv AC Cora, the moderately susceptible cvs AAC Foray, Carberry, AAC Indus and 5602 HR and the susceptible cvs AC Morse, CDC Teal and Sadash.
4.2. Fusarium Head Blight (FHB) Phenotyping and Deoxynivalenol (DON) Assessment
The doubled haploid lines, their parents and checks were screened in replicated trials in the FHB nurseries at Morden, MB, Canada in 2015, 2016 and 2017. Trial entries and checks were replicated in a randomized block design. The entries were planted in a single 1-m-long row using a six-row cassette Row XL, Wintersteiger planter. The rows were spaced 30 cm apart with passes of 0.5 m between each set of six rows. Fg corn kernel inoculum was prepared using four Fg isolates HSW-15-39 (3-ADON), HSW-15-87 (3-ADON), HSW-15-27 (15-ADON) and HSW-15-57 (15-ADON). The inoculum was dispersed at a rate of 8 g per row on biweekly intervals, starting when earliest lines reached a 4–5 leaf stage. The application of the inoculum was followed by irrigation three times a week using Cadman Irrigations Travellers with Briggs booms. Visual observations were taken at 18 to 21 days postinoculation for infected heads (disease incidence; DI) and spikelets (disease severity; DS) on a scale of 0 to 10, and used to calculate the FHB visual rating index (VRI: DI × DS) [93]. Matured wheat heads were harvested from the center of each plot, dried and threshed using an Almaco (Nevada, IA) stationary thresher without the use of a fan, followed by cleaning the seed samples by hand to prevent the loss of Fusarium-damaged kernels. One 10 g aliquot from the two replications of each line/cultivar were ground using a Romermill (Model 2A, Romer Labs Inc., Union, MO, USA) to make a whole-grain flour. Quantification of DON was carried out using two 1-g aliquots sampled from each ground sample using EZ-Quant® Vomitoxin ELISA kit (Diagnostix, Ltd., Mississauga, ON, Canada) with an accuracy of 0.5 ppm [94].
4.3. Days to Anthesis and Plant Height Phenotyping
Days to anthesis (DTA; Z00-61) was measured as the number of days from planting to Zadoks growth stage 61, when a few anthers started to emerge at the middle of spikes [95]. DTA was recorded in green house experiments in 2017, field experiments in 2018 and 2019 at Lethbridge, AB and field experiments in 2017 at Morden, MB.
Plant height (PHT; cm) was measured in green house experiments in 2017, field experiments in 2017, 2018 and 2019 at Lethbridge, AB and field experiments at Morden, MB in 2017. All measurements were taken on at least three plants in green house conditions and five plants per line/plot in field conditions.
4.4. Phenotypic Data Evaluation
The phenotypic data was subjected to ANOVA using the agricolae (version 1.2–4) package of the R (R version 3.2.3) software [96]. For the ANOVA model, DHs, their parents and checks were considered fixed effects, while environments and blocks were considered random effects. ANOVA was conducted both within and across environments. A combined ANOVA table for all FHB related traits was generated. Pearson correlations and regression between traits and scatterplots were calculated using the R package GGally software [97].
4.5. Genotyping and Linkage and Quantitative Trait Loci (QTL) Mapping
A total of 188 DH lines and their parents (AAC Innova and AAC Tenacious) were used for genotyping. Two seeds from each line were seeded in 2 x 96 cell seed planting trays (parents repeated over both trays) in a soil mixture of Turface (9.07 kg), Peat Moss (0.907 kg) and Vermiculite (0.06 m3). Leaf tissue samples were collected from 10 days old seedlings followed by isolation of DNA using DNeasy 96 Plant Kit (Qiagen Inc., Valencia, CA, USA) following Dhariwal et al. [52]. Quant-iT™ PicoGreen® dsDNA Assay Kit (Thermo Fisher Scientific Inc., Bartlesville, OK, USA) was used to quantify DNA samples, followed by dilutions of DNA samples to 50 ng/µl. Marker genotyping was carried out using wheat 90K Infinium iSelect SNP assay, functional marker Ppd-D1, and simple sequence repeat (SSR) marker Xgwm261. SNP genotyping data was analyzed using the Genotyping module of the GenomeStudio software package (Illumina Inc., San Diego, CA, USA), as described previously [52]. Briefly, high-quality SNPs were selected from the list of all SNPs evaluated for genotyping using the following logical expression filter features in GenomeStudio (i) Call Freq: >0.50, (ii) Minor Allele Freq: >0.03, (iii) AA Freq: !=1, (iv) AB Freq: !=1, and (v) BB Freq: !=1. The parental profiles of SNPs were compared, followed by the selection of identical SNPs to remove monomorphic SNPs together. SNPs that showed segregation distortion were removed using a ‘two-step’ strategy: In the first step, SNPs with allele frequencies <0.4 and >0.6 were removed; In the second step, SNPs that deviated significantly from 1:1 ratio on the bases of χ2 values, were removed. Only high-quality polymorphic markers clearly segregating for a 1:1 ratio were used for genetic linkage mapping.
Initially, markers were assigned to linkage groups (LGs) using the minimum spanning tree algorithm implemented in the MSTMAP software (version 2.0) [98] (available freely at GitHub). Distance and objective functions, i.e., Kosambi [99] and maximum likelihood (ML) [100], respectively, were utilized to calculate the recombination frequencies and order of LGs. Double recombinants were corrected after rescoring, and individual LGs were refined at a threshold of LOD = 3.0 and recombination frequency = 0.35 using the MapDisto software (version 1.7.7.011) [101]. LGs were assigned to wheat chromosomes based on the wheat 90K consensus SNP map [54]. Different LGs generated from the same chromosome were merged to a single LG using gradually less stringent cut off values, followed by recalculating the map length using MapDisto.
The QTL Cartographer (version 1.6) software package [102,103] was used to map main effect QTL (M-QTL) using a composite interval mapping (CIM) method with a regression method, i.e., forwards and backwards cofactor (p = 0.05). QTL significance thresholds were determined by 1000 permutations at a significance level of p = 0.05 to declare putative QTL. QTL (2.5 < LOD < threshold) were also reported when detected in at least one environment or pooled data reaching the significance level following the method described by Yi et al. [65]. QTL intervals were calculated using the entire CIM interval above the threshold/selected LOD score. QTLNetwork (version 2.0) [104] was used for two-locus analyses to identify additive (A), epistatic and QTL × environment interaction (A*E) effects of QTL. A Circos diagram representing the AAC Innova x AAC Tenacious linkage and QTL map with epistatic interactions in a circular network plot was generated using the R package OmicCircos (version 1.14.0) software [105].
5. Conclusions
This study provides new insights into the genetic basis of the FHB resistance of wheat cv AAC Tenacious, which is the first Canadian spring wheat cv to obtain a FHB ‘resistant’ rating. In this study, we identified stable as well as environment-specific, pleotropic and epistatic FHB-resistance QTL. Notably, two important major, stable FHB-resistance QTL, mapped on wheat chromosome arms 2DS and 2DL, were derived from AAC Tenacious. While 2DS QTL is colocated with DTA and PHT QTL, 2DL is a solitary FHB responsive QTL. The latter is very important and may be useful to pyramid FHB resistance from sources such as Sumai 3 and others. The QTL identified in the present study will be valuable resources, not only for Canadian wheat breeders, but also for breeders working on similar wheat classes around the globe. However, environment-specific QTL should be subject to further investigation using a suitable approach such as meta-analysis to see if they possess any significant effect on FHB, for example FHB susceptibility.
Acknowledgments
We gratefully acknowledged the financial support provided by Alberta Wheat Commission and AAFC Canadian National Wheat Cluster Program. We greatly acknowledged the technical support provided by Leslie Bihari and Zafrin Akter in producing of doubled haploid population, and Mark Virginallo and Kelly Ryan in the field experiments.
Abbreviations
| AA | Additive × additive (epistatic) effect |
| AE | Additive effect |
| A*E | QTL × environment interaction |
| ANOVA | Analysis of variance |
| Chr | Chromosome |
| CIM | Composite interval mapping |
| cM | centi-Morgan |
| cm | Centimetre |
| CPS | Canada Prairie Spring wheat class |
| CWSP | Canada Western Special Purpose wheat class |
| cv | Cultivar |
| CV | Coefficient of variation |
| df | Degrees of freedom |
| DH | Doubled haploid |
| DI | Disease incidence |
| DON | Deoxynivalenol |
| DS | Disease severity |
| DTA | Days to anthesis |
| E | Environment |
| FDK | Fusarium damaged kernels |
| Fg | Fusarium graminearum (Schwabe) |
| FHB | Fusarium head blight |
| G | Genotype |
| GA | Gibberellic acid |
| GDD | Growing degree days |
| His | Histidine-rich calcium-binding protein |
| I | Intermediate |
| JD | Julian calendar days |
| LG | Linkage group |
| LOD | Logarithm of the odds |
| MR | Moderately resistant |
| MS | Moderately susceptible |
| PFT | Pore-forming toxin-like protein |
| PHT | Plant height |
| QTL | Quantitative trait loci |
| R | Resistant |
| R2 | Phenotypic variation |
| Rht | Reduced height |
| S | Susceptible |
| SNP | Single nucleotide polymorphism |
| SSR | Simple sequence repeat |
| VRI | Visual rating index |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/12/4497/s1.
Author Contributions
Conceptualization, R.D. and H.S.R.; methodology, investigation and data curation, R.D., M.A.H., C.H., C.A.M. and H.S.R.; formal analysis, R.D., C.A.M.; resources, M.A.H., C.H. and H.S.R.; writing—original draft preparation, R.D.; writing—review and editing, R.D., C.A.M., M.A.H., C.H. and H.S.R.; visualization, R.D.; supervision, H.S.R; project administration, R.D. and H.S.R.; funding acquisition, H.S.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Alberta Wheat Commission and AAFC Canadian National Wheat Cluster Program.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Niwa S., Kazama Y., Abe T., Ban T. Tracking haplotype for QTLs associated with Fusarium head blight resistance in Japanese wheat (Triticum aestivum L.) lineage. Agric. Food Secur. 2018;7:4. doi: 10.1186/s40066-017-0139-z. [DOI] [Google Scholar]
- 2.Gilbert J., Tekauz A. Review: Recent developments in research on Fusarium head blight of wheat in Canada. Can. J. Plant. Pathol. 2000;22:1–8. doi: 10.1080/07060660009501155. [DOI] [Google Scholar]
- 3.Sieusahai G. Alberta Fusarium Graminearum Management Plan. [(accessed on 8 November 2019)]; Available online: https://www.alberta.ca/alberta-fusarium-graminearum-management-plan.aspx.
- 4.Heikkila R. Economic Cost of Fusarium: Farm-level and Regional Economic Impact of Fusarium in Alberta. (External Release) Alberta Agriculture and Forestry; Edmonton, AB, Canada: 2015. p. 6. [Google Scholar]
- 5.Komirenko Z. Economic Cost of Fusarium: Farm-Level and Regional Economic Impact of Fusarium in Alberta. (External Release) Alberta Agriculture and Forestry; Edmonton, AB, Canada: 2018. p. 8. [Google Scholar]
- 6.Jia H.Y., Zhou J.Y., Xue S.L., Li G.Q., Yan H.S., Ran C.F., Zhang Y.D., Shi J.X., Jia L., Wang X., et al. A journey to understand wheat Fusarium head blight resistance in the Chinese wheat landrace Wangshuibai. Crop. J. 2018;6:48–59. doi: 10.1016/j.cj.2017.09.006. [DOI] [Google Scholar]
- 7.Mesterhazy A. Types and components of resistance to Fusarium head blight of wheat. Plant. Breed. 1995;114:377–386. doi: 10.1111/j.1439-0523.1995.tb00816.x. [DOI] [Google Scholar]
- 8.Mesterhazy A., Bartok T., Mirocha C.G., Komoroczy R. Nature of wheat resistance to Fusarium head blight and the role of deoxynivalenol for breeding. Plant. Breed. 1999;118:97–110. doi: 10.1046/j.1439-0523.1999.118002097.x. [DOI] [Google Scholar]
- 9.Bai G.H., Shaner G. Scab of wheat-prospects for control. Plant. Dis. 1994;78:760–766. [Google Scholar]
- 10.McMullen M., Jones R., Gallenberg D. Scab of wheat and barley: A re-emerging disease of devastating impact. Plant. Dis. 1997;81:1340–1348. doi: 10.1094/PDIS.1997.81.12.1340. [DOI] [PubMed] [Google Scholar]
- 11.Lu Q.X., Lillemo M., Skinnes H., He X.Y., Shi J.R., Ji F., Dong Y.H., Bjornstad A. Anther extrusion and plant height are associated with type I resistance to Fusarium head blight in bread wheat line ‘Shanghai-3/Catbird’. Theor. Appl. Genet. 2013;126:317–334. doi: 10.1007/s00122-012-1981-9. [DOI] [PubMed] [Google Scholar]
- 12.Miedaner T., Voss H.-H. Effect of dwarfing Rht genes on Fusarium head blight resistance in two sets of near-isogenic lines of wheat and check cultivars. Crop. Sci. 2008;48:2115–2122. doi: 10.2135/cropsci2008.02.0107. [DOI] [Google Scholar]
- 13.Voss H.H., Holzapfel J., Hartl L., Korzun V., Rabenstein F., Ebmeyer E., Coester H., Kempf H., Miedaner T. Effect of the Rht-D1 dwarfing locus on Fusarium head blight rating in three segregating populations of winter wheat. Plant. Breed. 2008;127:333–339. doi: 10.1111/j.1439-0523.2008.01518.x. [DOI] [Google Scholar]
- 14.McCartney C.A., Somers D.J., Fedak G., DePauw R.M., Thomas J., Fox S.L., Humphreys D.G., Lukow O., Savard M.E., McCallum B.D., et al. The evaluation of FHB resistance QTLs introgressed into elite Canadian spring wheat germplasm. Mol. Breed. 2007;20:209–221. doi: 10.1007/s11032-007-9084-z. [DOI] [Google Scholar]
- 15.Gervais L., Dedryver F., Morlais J.Y., Bodusseau V., Negre S., Bilous M., Groos C., Trottet M. Mapping of quantitative trait loci for field resistance to Fusarium head blight in an European winter wheat. Theor. Appl. Genet. 2003;106:961–970. doi: 10.1007/s00122-002-1160-5. [DOI] [PubMed] [Google Scholar]
- 16.Somers D.J., Fedak G., Savard M. Molecular mapping of novel genes controlling Fusarium head blight resistance and deoxynivalenol accumulation in spring wheat. Genome. 2003;46:555–564. doi: 10.1139/g03-033. [DOI] [PubMed] [Google Scholar]
- 17.Snijders C.H.A., Perkowski J. Effects of head blight caused by Fusarium culmorum on toxin content and weight of wheat kernels. Phytopathology. 1990;80:566–570. doi: 10.1094/Phyto-80-566. [DOI] [Google Scholar]
- 18.Ban T., Suenaga K. Genetic analysis of resistance to Fusarium head blight caused by Fusarium graminearum in Chinese wheat cultivar Sumai 3 and the Japanese cultivar Saikai 165. Euphytica. 2000;113:87–99. doi: 10.1023/A:1003951509797. [DOI] [Google Scholar]
- 19.Buerstmayr H., Ban T., Anderson J.A. QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: A review. Plant. Breed. 2009;128:1–26. doi: 10.1111/j.1439-0523.2008.01550.x. [DOI] [Google Scholar]
- 20.Holzapfel J., Voss H.H., Miedaner T., Korzun V., Haberle J., Schweizer G., Mohler V., Zimmermann G., Hartl L. Inheritance of resistance to Fusarium head blight in three European winter wheat populations. Theor. Appl. Genet. 2008;117:1119–1128. doi: 10.1007/s00122-008-0850-z. [DOI] [PubMed] [Google Scholar]
- 21.Venske E., dos Santos R.S., da Rosa F.D., Rother V., da Maia L.C., Pegoraro C., Costa de Oliveira A. Meta-analysis of the QTLome of Fusarium head blight resistance in bread wheat: Refining the current puzzle. Front. Plant. Sci. 2019:10. doi: 10.3389/fpls.2019.00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waldron B.L., Moreno-Sevilla B., Anderson J.A., Stack R.W., Frohberg R.C. RFLP mapping of QTL for Fusarium head blight resistance in wheat. Crop. Sci. 1999;39:805–811. doi: 10.2135/cropsci1999.0011183X003900030032x. [DOI] [Google Scholar]
- 23.Anderson J.A., Stack R.W., Liu S., Waldron B.L., Fjeld A.D., Coyne C., Moreno-Sevilla B., Fetch J.M., Song Q.J., Cregan P.B., et al. DNA markers for Fusarium head blight resistance QTLs in two wheat populations. Theor. Appl. Genet. 2001;102:1164–1168. doi: 10.1007/s001220000509. [DOI] [Google Scholar]
- 24.Yang Z.P., Gilbert J., Somers D.J., Fedak G., Procunier J.D., McKenzie I.H. Marker assisted selection of Fusarium head blight resistance genes in two doubled haploid populations of wheat. Mol. Breed. 2003;12:309–317. doi: 10.1023/B:MOLB.0000006834.44201.48. [DOI] [Google Scholar]
- 25.Buerstmayr H., Steiner B., Hartl L., Griesser M., Angerer N., Lengauer D., Miedaner T., Schneider B., Lemmens M. Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. II. Resistance to fungal penetration and spread. Theor. Appl. Genet. 2003;107:503–508. doi: 10.1007/s00122-003-1272-6. [DOI] [PubMed] [Google Scholar]
- 26.Cuthbert P.A., Somers D.J., Thomas J., Cloutier S., Brule-Babel A. Fine mapping Fhb1, a major gene controlling Fusarium head blight resistance in bread wheat (Triticum aestivum L.) Theor. Appl. Genet. 2006;112:1465–1472. doi: 10.1007/s00122-006-0249-7. [DOI] [PubMed] [Google Scholar]
- 27.Liu S., Zhang X., Pumphrey M.O., Stack R.W., Gill B.S., Anderson J.A. Complex microcolinearity among wheat, rice, and barley revealed by fine mapping of the genomic region harboring a major QTL for resistance to Fusarium head blight in wheat. Funct. Integr. Genom. 2006;6:83–89. doi: 10.1007/s10142-005-0007-y. [DOI] [PubMed] [Google Scholar]
- 28.Xue S., Xu F., Tang M., Zhou Y., Li G., An X., Lin F., Xu H., Jia H., Zhang L., et al. Precise mapping Fhb5, a major QTL conditioning resistance to Fusarium infection in bread wheat (Triticum aestivum L.) Theor. Appl. Genet. 2011;123:1055–1063. doi: 10.1007/s00122-011-1647-z. [DOI] [PubMed] [Google Scholar]
- 29.Buerstmayr M., Steiner B., Wagner C., Schwarz P., Brugger K., Barabaschi D., Volante A., Vale G., Cattivelli L., Buerstmayr H. High-resolution mapping of the pericentromeric region on wheat chromosome arm 5AS harbouring the Fusarium head blight resistance QTL Qfhs.ifa-5a. Plant. Biotechnol. J. 2018;16:1046–1056. doi: 10.1111/pbi.12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi L.L., Pumphrey M.O., Friebe B., Chen P.D., Gill B.S. Molecular cytogenetic characterization of alien introgressions with gene Fhb3 for resistance to Fusarium head blight disease of wheat. Theor. Appl. Genet. 2008;117:1155–1166. doi: 10.1007/s00122-008-0853-9. [DOI] [PubMed] [Google Scholar]
- 31.Cainong J.C., Bockus W.W., Feng Y., Chen P., Qi L., Sehgal S.K., Danilova T.V., Koo D.-H., Friebe B., Gill B.S. Chromosome engineering, mapping, and transferring of resistance to Fusarium head blight disease from Elymus tsukushiensis into wheat. Theor. Appl. Genet. 2015;128:1019–1027. doi: 10.1007/s00122-015-2485-1. [DOI] [PubMed] [Google Scholar]
- 32.Guo J., Zhang X., Hou Y., Cai J., Shen X., Zhou T., Xu H., Ohm H.W., Wang H., Li A., et al. High-density mapping of the major FHB resistance gene Fhb7 derived from Thinopyrum ponticum and its pyramiding with Fhb1 by marker-assisted selection. Theor. Appl. Genet. 2015;128:2301–2316. doi: 10.1007/s00122-015-2586-x. [DOI] [PubMed] [Google Scholar]
- 33.Buerstmayr H., Buerstmayr M., Schweiger W., Steiner B. Genomics-assisted breeding for Fusarium head blight resistance in wheat. In: Varshney R.K., Tuberosa R., editors. Translational Genomics for Crop Breeding: Biotic Stress. Volume 1. John Wiley & Sons; Hoboken, NJ, USA: 2013. pp. 45–61. [Google Scholar]
- 34.Yao J., Ge Y., Wang S., Yao G., Zhou C., Qian C. Chromosomal location of genes for scab resistance in wheat cultivar Sumai 3. Zuo Wu Xue Bao. 1997;23:450–453. [Google Scholar]
- 35.Zhou W.-C., Kolb F.L., Bai G.-H., Domier L.L., Yao J.-B. Effect of individual Sumai 3 chromosomes on resistance to scab spread within spikes and deoxynivalenol accumulation within kernels in wheat. Hereditas. 2002;137:81–89. doi: 10.1034/j.1601-5223.2002.01674.x. [DOI] [PubMed] [Google Scholar]
- 36.Basnet B.R., Glover K.D., Ibrahim A.M.H., Yen Y., Chao S. A QTL on chromosome 2DS of ‘Sumai 3’ increases susceptibility to Fusarium head blight in wheat. Euphytica. 2012;186:91–101. doi: 10.1007/s10681-011-0495-x. [DOI] [Google Scholar]
- 37.Ma H.-X., Bai G.-H., Gill B.S., Hart L.P. Deletion of a chromosome arm altered wheat resistance to Fusarium head blight and deoxynivalenol accumulation in Chinese Spring. Plant. Dis. 2006;90:1545–1549. doi: 10.1094/PD-90-1545. [DOI] [PubMed] [Google Scholar]
- 38.Rawat N., Pumphrey M.O., Liu S., Zhang X., Tiwari V.K., Ando K., Trick H.N., Bockus W.W., Akhunov E., Anderson J.A., et al. Wheat Fhb1 encodes a chimeric lectin with agglutinin domains and a pore-forming toxin-like domain conferring resistance to Fusarium head blight. Nat. Genet. 2016;48:1576. doi: 10.1038/ng.3706. [DOI] [PubMed] [Google Scholar]
- 39.Su Z., Bernardo A., Tian B., Chen H., Wang S., Ma H., Cai S., Liu D., Zhang D., Li T., et al. A deletion mutation in TaHRC confers Fhb1 resistance to Fusarium head blight in wheat. Nat. Genet. 2019;51:1099–1105. doi: 10.1038/s41588-019-0425-8. [DOI] [PubMed] [Google Scholar]
- 40.Li G., Zhou J., Jia H., Gao Z., Fan M., Luo Y., Zhao P., Xue S., Li N., Yuan Y., et al. Mutation of a histidine-rich calcium-binding-protein gene in wheat confers resistance to Fusarium head blight. Nat. Genet. 2019;51:1106–1112. doi: 10.1038/s41588-019-0426-7. [DOI] [PubMed] [Google Scholar]
- 41.Gadaleta A., Colasuonno P., Giove S.L., Blanco A., Giancaspro A. Map-based cloning of Qfhb.mgb-2A identifies a WAK2 gene responsible for Fusarium head blight resistance in wheat. Sci. Rep. 2019;9:6929. doi: 10.1038/s41598-019-43334-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Handa H., Namiki N., Xu D., Ban T. Dissecting of the FHB resistance QTL on the short arm of wheat chromosome 2D using a comparative genomic approach: From QTL to candidate gene. Mol. Breed. 2008;22:71–84. doi: 10.1007/s11032-008-9157-7. [DOI] [Google Scholar]
- 43.Kage U., Karre S., Kushalappa A.C., McCartney C. Identification and characterization of a Fusarium head blight resistance gene TaACT in wheat QTL-2DL. Plant. Biotechnol. J. 2017;15:447–457. doi: 10.1111/pbi.12641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kage U., Yogendra K.N., Kushalappa A.C. TaWRKY70 transcription factor in wheat QTL-2DL regulates downstream metabolite biosynthetic genes to resist Fusarium graminearum infection spread within spike. Sci. Rep. 2017;7:42596. doi: 10.1038/srep42596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hao Y., Zhu Z., Gao C., Cheng S., Xia X., He C. The complexity of Fhb1, is it a gene or a gene cluster?; Proceedings of the 9th Canadian Workshop on Fusarium Head Blight 4th Canadian Wheat Symposium; Winnipeg, Manitoba, ON, Canada. 19–22 November 2018; p. 28. [Google Scholar]
- 46.Lagudah E.S., Krattinger S.G. A new player contributing to durable Fusarium resistance. Nat. Genet. 2019;51:1070–1071. doi: 10.1038/s41588-019-0454-3. [DOI] [PubMed] [Google Scholar]
- 47.McCartney C.A., Brûlé-Babel A.L., Fedak G., Martin R.A., McCallum B.D., Gilbert J., Hiebert C.W., Pozniak C.J. Fusarium head blight resistance QTL in the spring wheat cross Kenyon/86ISMN 2137. Front. Microbiol. 2016;7:1542. doi: 10.3389/fmicb.2016.01542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown P.D., Randhawa H.S., Mitchell Fetch J., Meiklejohn M., Fox S.L., Humphreys D.G., Green D., Wise I., Fetch T., Gilbert J., et al. AAC Tenacious red spring wheat. Can. J. Plant. Sci. 2015;95:805–810. doi: 10.4141/cjps-2015-011. [DOI] [Google Scholar]
- 49.Tekauz A., Gilbert J., Abramson D. Reaction to Fusarium head blight of spring wheats based on symptoms, Fusarium spp., and mycotoxins. Cereal Res. Commun. 1997;25:821–822. doi: 10.1007/BF03543862. [DOI] [Google Scholar]
- 50.Campbell A.B. Neepawa hard red spring wheat. Can. J. Plant. Sci. 1970;50:752–753. doi: 10.4141/cjps70-143. [DOI] [Google Scholar]
- 51.Randhawa H.S., Graf R.J., Sadasivaiah R.S. AAC Innova general purpose spring wheat. Can. J. Plant. Sci. 2015;95:787–791. doi: 10.4141/cjps-2015-024. [DOI] [Google Scholar]
- 52.Dhariwal R., Fedak G., Dion Y., Pozniak C., Laroche A., Eudes F., Randhawa H. High density single nucleotide polymorphism (SNP) mapping and quantitative trait loci (QTL) analysis in a biparental spring triticale population localized major and minor effect Fusarium head blight resistance and associated traits QTL. Genes. 2018;9:19. doi: 10.3390/genes9010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cavanagh C.R., Chao S., Wang S., Huang B.E., Stephen S., Kiani S., Forrest K., Saintenac C., Brown-Guedira G.L., Akhunova A., et al. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. USA. 2013;110:8057–8062. doi: 10.1073/pnas.1217133110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S., Wong D., Forrest K., Allen A., Chao S., Huang B.E., Maccaferri M., Salvi S., Milner S.G., Cattivelli L., et al. Characterization of polyploid wheat genomic diversity using a high-density 90000 single nucleotide polymorphism array. Plant. Biotechnol. J. 2014;12:787–796. doi: 10.1111/pbi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu L.-X., Barbier H., Rouse M.N., Singh S., Singh R.P., Bhavani S., Huerta-Espino J., Sorrells M.E. A consensus map for UG99 stem rust resistance loci in wheat. Theor. Appl. Genet. 2014;127:1561–1581. doi: 10.1007/s00122-014-2326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li C., Bai G., Chao S., Wang Z. A high-density SNP and SSR consensus map reveals segregation distortion regions in wheat. Biomed. Res. Int. 2015;2015:1–10. doi: 10.1155/2015/830618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wen W., He Z., Gao F., Liu J., Jin H., Zhai S., Qu Y., Xia X. A high-density consensus map of common wheat integrating four mapping populations scanned by the 90k snp array. Front. Plant. Sci. 2017;8:1389. doi: 10.3389/fpls.2017.01389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lander E., Kruglyak L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat. Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 59.Liu S., Christopher M.D., Griffey C.A., Hall M.D., Gundrum P.G., Brooks W.S. Molecular characterization of resistance to Fusarium head blight in U.S. soft red winter wheat breeding line VA00W-38. Crop. Sci. 2012;52:2283–2292. doi: 10.2135/cropsci2012.03.0144. [DOI] [Google Scholar]
- 60.Liu S., Griffey C.A., Hall M.D., McKendry A.L., Chen J., Brooks W.S., Brown-Guedira G., Van Sanford D., Schmale D.G. Molecular characterization of field resistance to Fusarium head blight in two US soft red winter wheat cultivars. Theor. Appl. Genet. 2013;126:2485–2498. doi: 10.1007/s00122-013-2149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Su Q., Zhang X., Zhang W., Zhang N., Song L., Liu L., Xue X., Liu G., Liu J., Meng D., et al. QTL detection for kernel size and weight in bread wheat (Triticum aestivum L.) using a high-density SNP and SSR-based linkage map. Front. Plant. Sci. 2018;9:1484. doi: 10.3389/fpls.2018.01484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qi L.L., Echalier B., Chao S., Lazo G.R., Butler G.E., Anderson O.D., Akhunov E.D., Dvorák J., Linkiewicz A.M., Ratnasiri A., et al. A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics. 2004;168:701–712. doi: 10.1534/genetics.104.034868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Collard B.C.Y., Jahufer M.Z.Z., Brouwer J.B., Pang E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica. 2005;142:169–196. doi: 10.1007/s10681-005-1681-5. [DOI] [Google Scholar]
- 64.Shen X., Zhou M., Lu W., Ohm H. Detection of Fusarium head blight resistance QTL in a wheat population using bulked segregant analysis. Theor. Appl. Genet. 2003;106:1041–1047. doi: 10.1007/s00122-002-1133-8. [DOI] [PubMed] [Google Scholar]
- 65.Yi X., Cheng J., Jiang Z., Hu W., Bie T., Gao D., Li D., Wu R., Li Y., Chen S., et al. Genetic analysis of Fusarium head blight resistance in CIMMYT bread wheat line C615 using traditional and conditional QTL mapping. Front. Plant. Sci. 2018;9:573. doi: 10.3389/fpls.2018.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Z., Gilbert J., Fedak G., Somers D.J. Genetic characterization of QTL associated with resistance to Fusarium head blight in a doubled-haploid spring wheat population. Genome. 2005;48:187–196. doi: 10.1139/g04-104. [DOI] [PubMed] [Google Scholar]
- 67.Buerstmayr M., Lemmens M., Steiner B., Buerstmayr H. Advanced backcross QTL mapping of resistance to Fusarium head blight and plant morphological traits in a Triticum macha × T. aestivum population. Theor. Appl. Genet. 2011;123:293. doi: 10.1007/s00122-011-1584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu D.H., Juan H.F., Nohda M., Ban T. QTLs mapping of type I and type II resistance to FHB in wheat. In: Canty S.M., Lewis J., Siler L., Ward R.W., editors. 2001 National Fusarium Head Blight Forum Proceedings Erlanger, KY, USA. Kinko’s, FEdex; Okemos, MI, USA: 2001. pp. 40–42. [Google Scholar]
- 69.Cai J., Bai G. Quantitative trait loci for Fusarium head blight resistance in Huangcandou × ‘Jagger’ wheat population. Crop. Sci. 2014;54:2520–2528. doi: 10.2135/cropsci2013.12.0835. [DOI] [Google Scholar]
- 70.He X., Lillemo M., Shi J., Wu J., Bjørnstad Å., Belova T., Dreisigacker S., Duveiller E., Singh P. QTL characterization of Fusarium head blight resistance in CIMMYT bread wheat line Soru#1. PLoS ONE. 2016;11:e0158052. doi: 10.1371/journal.pone.0158052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jia G., Chen P., Qin G., Bai G., Wang X., Wang S., Zhou B., Zhang S., Liu D. QTLs for Fusarium head blight response in a wheat DH population of Wangshuibai/Alondra‘s’. Euphytica. 2005;146:183–191. doi: 10.1007/s10681-005-9001-7. [DOI] [Google Scholar]
- 72.Zhang M., Zhang R., Yang J., Luo P. Identification of a new QTL for Fusarium head blight resistance in the wheat genotype “Wang shui-bai”. Mol. Biol. Rep. 2010;37:1031–1035. doi: 10.1007/s11033-009-9809-7. [DOI] [PubMed] [Google Scholar]
- 73.Lv C., Song Y., Gao L., Yao Q., Zhou R., Xu R., Jia J. Integration of QTL detection and marker assisted selection for improving resistance to Fusarium head blight and important agronomic traits in wheat. Crop. J. 2014;2:70–78. doi: 10.1016/j.cj.2013.10.004. [DOI] [Google Scholar]
- 74.Korzun V., Röder M.S., Ganal M.W., Worland A.J., Law C.N. Genetic analysis of the dwarfing gene (Rht8) in wheat. Part, I. Molecular mapping of Rht8 on the short arm of chromosome 2D of bread wheat (Triticum aestivum L.) Theor. Appl. Genet. 1998;96:1104–1109. doi: 10.1007/s001220050845. [DOI] [Google Scholar]
- 75.Dhariwal R., Graf J.R., Randhawa H.S. Validation and origin of FHB resistance QTL identified from Canadian wheat cv AAC Tenacious. (unpublished) [Google Scholar]
- 76.Gasperini D., Greenland A., Hedden P., Dreos R., Harwood W., Griffiths S. Genetic and physiological analysis of Rht8 in bread wheat: An alternative source of semi-dwarfism with a reduced sensitivity to brassinosteroids. J. Exp. Bot. 2012;63:4419–4436. doi: 10.1093/jxb/ers138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Law C.N., Sutka J., Worland A.J. A genetic study of day-length response in wheat. Heredity. 1978;41:185–191. doi: 10.1038/hdy.1978.87. [DOI] [Google Scholar]
- 78.Worland A.J., Law C.N. Genetic analysis of chromosome 2D of wheat. I. The location of genes affecting height, daylength insensitivity, hybrid dwarfism and yellow-rust resistance. Zeitschrift für Pflanzenzüchtung. 1986;96:331–345. [Google Scholar]
- 79.Jiang G.-L., Shi J., Ward R.W. QTL analysis of resistance to Fusarium head blight in the novel wheat germplasm CJ 9306. I. Resistance to fungal spread. Theor. Appl. Genet. 2007;116:3–13. doi: 10.1007/s00122-007-0641-y. [DOI] [PubMed] [Google Scholar]
- 80.Zhu Z., Bonnett D., Ellis M., He X., Heslot N., Dreisigacker S., Gao C., Singh P. Characterization of Fusarium head blight resistance in a CIMMYT synthetic-derived bread wheat line. Euphytica. 2016;208:367–375. doi: 10.1007/s10681-015-1612-z. [DOI] [Google Scholar]
- 81.Lin F., Xue S.L., Zhang Z.Z., Zhang C.Q., Kong Z.X., Yao G.Q., Tian D.G., Zhu H.L., Li C.J., Cao Y., et al. Mapping QTL associated with resistance to Fusarium head blight in the Nanda2419 × Wangshuibai population. II: Type I resistance. Theor. Appl. Genet. 2006;112:528–535. doi: 10.1007/s00122-005-0156-3. [DOI] [PubMed] [Google Scholar]
- 82.Lewis J.M., Suenaga K., Ginkel M.V., Gilchrist L., Shi J.R., Jiang G.L., Kravchenko S., Mujeeb-Kazi A., Ward R.W. Identification and mapping of a QTL for type II resistance to Fusarium head blight on chromosome arm 2DL of wheat. In: Canty S.M., Boring T., Wardwell J., Ward R.W., editors. Proceedings of the 2nd International Symposium on Fusarium Head Blight Incorporating the 8th European Fusarium Seminar. Volume 1. Michigan State University; Orlando, FL, USA: 2004. pp. 89–92. [Google Scholar]
- 83.Paillard S., Schnurbusch T., Tiwari R., Messmer M., Winzeler M., Keller B., Schachermayr G. QTL analysis of resistance to Fusarium head blight in Swiss winter wheat (Triticum aestivum L.) Theor. Appl. Genet. 2004;109:323–332. doi: 10.1007/s00122-004-1628-6. [DOI] [PubMed] [Google Scholar]
- 84.Mardi M., Buerstmayr H., Ghareyazie B., Lemmens M., Mohammadi S.A., Nolz R., Ruckenbauer P. QTL analysis of resistance to Fusarium head blight in wheat using a ‘Wangshuibai’-derived population. Plant. Breed. 2005;124:329–333. doi: 10.1111/j.1439-0523.2005.01103.x. [DOI] [Google Scholar]
- 85.Ma H.-X., Bai G.-H., Zhang X., Lu W.-Z. Main effects, epistasis, and environmental interactions of quantitative trait loci for Fusarium head blight resistance in a recombinant inbred population. Phytopathology. 2006;96:534–541. doi: 10.1094/PHYTO-96-0534. [DOI] [PubMed] [Google Scholar]
- 86.He X., Singh P.K., Dreisigacker S., Singh S., Lillemo M., Duveiller E. Dwarfing genes Rht-B1b and Rht-D1b are associated with both type I FHB susceptibility and low anther extrusion in two bread wheat populations. PLoS ONE. 2016;11:e0162499. doi: 10.1371/journal.pone.0162499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Buhrow L.M., Cram D., Tulpan D., Foroud N.A., Loewen M.C. Exogenous abscisic acid and gibberellic acid elicit opposing effects on Fusarium graminearum infection in wheat. Phytopathology. 2016;106:986–996. doi: 10.1094/PHYTO-01-16-0033-R. [DOI] [PubMed] [Google Scholar]
- 88.Herter C.P., Ebmeyer E., Kollers S., Korzun V., Leiser W.L., Würschum T., Miedaner T. Rht24 reduces height in the winter wheat population ‘Solitär × Bussard’ without adverse effects on Fusarium head blight infection. Theor. Appl. Genet. 2018;131:1263–1272. doi: 10.1007/s00122-018-3076-8. [DOI] [PubMed] [Google Scholar]
- 89.Guo Z., Chen D., Roeder M., Ganal M., Schnurbusch T. Genetic dissection of pre-anthesis sub-phase durations during the reproductive spike development of wheat. Plant. J. 2018;95:909–918. doi: 10.1111/tpj.13998. [DOI] [PubMed] [Google Scholar]
- 90.Eckard J.T., Gonzalez-Hernandez J.L., Caffe M., Berzonsky W., Bockus W.W., Marais G.F., Baenziger P.S. Native Fusarium head blight resistance from winter wheat cultivars ‘Lyman,’ ‘Overland,’ ‘Ernie,’ and ‘Freedom’ mapped and pyramided onto ‘Wesley’-Fhb1 backgrounds. Mol. Breed. 2015;35:6. doi: 10.1007/s11032-015-0200-1. [DOI] [Google Scholar]
- 91.Kumar A., Mantovani E.E., Simsek S., Jain S., Elias E.M., Mergoum M. Genome wide genetic dissection of wheat quality and yield related traits and their relationship with grain shape and size traits in an elite × non-adapted bread wheat cross. PLoS ONE. 2019;14:e0221826. doi: 10.1371/journal.pone.0221826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Suenaga K., Nakajima K. Efficient production of haploid wheat (Triticum aestivum) through crosses between Japanese wheat and maize (Zea mays) Plant Cell Rep. 1989;8:263–266. doi: 10.1007/BF00274125. [DOI] [PubMed] [Google Scholar]
- 93.Gilbert J., Morgan K. Field-based rating of spring wheat infected with Fusarium graminearum, cause of Fusarium head blight; Proceedings of the 6th European Seminar on Fusarium—Mycotoxins, Taxonomy and Pathogenicity; Berlin, Germany. 11–16 September 2000; p. 73. [Google Scholar]
- 94.Sinha R.C., Savard M.E. Comparison of immunoassay and gas chromatography methods for the detection of the mycotoxin deoxynivalenol in grain samples. Can. J. Plant Pathol. 1996;18:233–236. doi: 10.1080/07060669609500617. [DOI] [Google Scholar]
- 95.Zadoks J.C., Chang T.T., Konzak C.F. A decimal code for the growth stages of cereals. Weed Res. 1974;14:415–421. doi: 10.1111/j.1365-3180.1974.tb01084.x. [DOI] [Google Scholar]
- 96.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
- 97.Schloerke B., Crowley J., Cook D., Briatte F., Marbach M., Thoen E., Elberg A., Larmarange J. Ggally: Extension to ‘Ggplot2’, R package version 1.4.0. R Foundation for Statistical Computing; Vienna, Austria: 2018. [Google Scholar]
- 98.Wu Y., Bhat P.R., Close T.J., Lonardi S. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. 2008;4:e1000212. doi: 10.1371/journal.pgen.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kosambi D.D. The estimation of map distances from recombination values. Ann. Eugen. 1943;12:172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x. [DOI] [Google Scholar]
- 100.Jansen J., de Jong A.G., van Ooijen J.W. Constructing dense genetic linkage maps. Theor. Appl. Genet. 2001;102:1113–1122. doi: 10.1007/s001220000489. [DOI] [Google Scholar]
- 101.Lorieux M. Mapdisto: Fast and efficient computation of genetic linkage maps. Mol. Breed. 2012;30:1231–1235. doi: 10.1007/s11032-012-9706-y. [DOI] [Google Scholar]
- 102.Zeng Z.B. Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc. Natl Acad. Sci. USA. 1993;90:10972–10976. doi: 10.1073/pnas.90.23.10972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zeng Z.B. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang J., Hu C., Hu H., Yu R., Xia Z., Ye X., Zhu J. QTLNetwork: Mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics. 2008;24:721–723. doi: 10.1093/bioinformatics/btm494. [DOI] [PubMed] [Google Scholar]
- 105.Hu Y., Yan C. Omiccircos: High-Quality Circular Visualization of Omics Data, R Package Version 1.16.0. R Foundation for Statistical Computing; Vienna, Austria: 2015. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.