Abstract
A phenolic rich fraction purified from Simarouba glauca leaves was effective in alpha glucosidase inhibition. The purified fraction named ‘fraction-14’ had shown significant inhibition of yeast alpha glucosidase enzyme activity (IC50 = 2.4 ± 0.4 μg/mL) when compared to anti-diabetic drug acarbose (IC50 = 2450 ± 24 μg/mL). The purified fraction also had reasonable DPPH (IC50 = 14.4 ± 0.1 μg/mL) and ABTS (IC50 = 7.6 ± 0.5 μg/mL) free radical scavenging activity when compared to the standard ascorbic acid. The LC-MS analysis of bioactive ‘fraction-14’ revealed four compounds, eclalbasaponin-v (1), cyanidin-3-O-(2′galloyl)-galactoside (2), kaempferol-3-O-glucoside (3) and kaempferol-3-O-pentoside (4) for the first time in S. glauca in this study. The kinetic study of the ‘fraction-14’ indicates a mixed type of inhibition on the alpha glucosidase enzyme with Ki, 6.2 μg/mL. Docking studies showed promising binding energy for the compounds 2 (-7.769 kJ/mol), 3 (-7.04 kJ/mol) and 4 (-7.127 kJ/mol) against yeast alpha glucosidase which was better than acarbose (-6.867 kJ/mol). In conclusion, the phenolic rich fraction from S. glauca possessing good in-vitro antioxidant property and alpha glucosidase enzyme inhibition potential along with mixed inhibition kinetics. Also, better binding energy of compounds (1, 2 & 3) appears to contain potential lead-molecule for antidiabetic therapy.
Keywords: Food science, Food chemistry, Agriculture, Alpha glucosidas, Enzyme kinetics, Anti-diabetic, Molecular docking, Medicinal plant, Hypoglycaemic
Food Science; Food Chemistry; Agriculture; Alpha Glucosidas; Enzyme Kinetics; Anti-diabetic; Molecular Docking; Medicinal Plant; Hypoglycaemic.
1. Introduction
Diabetes Mellitus (DM) is known as one of the largest global threats of the 21st century in developed countries as well as in developing countries. Type 2 diabetes mellitus (T2DM) is characterized by constant hyperglycaemia due to the condition called insulin resistance in targeted organs and pancreatic β cell dysfunction, leading to deterioration in insulin secretions (Chatterjee et al., 2017). A survey from the International Diabetes Federation (IDF) in 2017 stated that globally 425 million people are suffering from DM in which more than 90 % of them have T2DM and, 629 million people may get diabetes by 2045 (Cho et al., 2018). Factors such as genetic susceptibility, environmental factors, obesity, physical inactivity, high sugar, fat-rich diets, etc. are believed to be responsible for the pathogenesis of T2DM. For the treatment of T2DM, several oral medications as well as injectable drugs have been available for decades, targeting a different kind of organ systems. Nevertheless, the main strategies of T2DM drugs are directed towards the control of hyperglycaemia and maintenance of the normal blood glucose homeostasis in the body (Kahn et al., 2014).
Amongst the many drug targets of T2DM, starch digesting enzymes such as alpha amylase and intestinal alpha glucosidase are targeted to achieve inhibition of these enzymes and hence to control the postprandial hyperglycaemia. Alpha glucosidases [EC 3.2.1.20] are digestive enzymes that belong to a family called ‘glycosides’, located in the brush border surface membrane of small intestinal cells that take part in the last step of carbohydrate digestion. These enzymes exclusively catalyse the hydrolysis of α-1,2, α-1,4 and α -1,6-glucosidic linkages in oligosaccharides and thereby release absorbable monosaccharides (Sim et al., 2008). Currently, acarbose, miglitol and voglibose are used as alpha glucosidase inhibitors (AGI) which are isolated from microbes. Unfortunately, severe adverse effects like vomiting, liver disorders, flatulence, abdominal discomfort, hypoglycaemia, urinary tract infection, etc. have been noticed in patients medicated with acarbose, miglitol and voglibose. Even though, the acarbose is a common drug prescribing as an AGI to treat T2DM (Dabhi et al., 2013; Mitrakou et al., 1998; Van De Laar et al., 2005).
Chronic hyperglycaemia is a condition observed in all forms of DM. According to the earlier reports, hyperglycaemia activates several biochemical pathways in the cells that are prone to the accumulation of reactive oxygen species (ROS). Because of dearth in the proper endogenous antioxidant defence mechanism, excessively accumulated ROS activates the stress-sensitive intracellular signalling pathways, which in turn, promote the cellular damage and play a role in the development of diabetic complications (Giacco and Brownlee, 2010; Vanessa Fiorentino et al., 2013). Among the several compounds that have been identified as AGI, generally phenolic compounds from a natural source are identified as the main phytochemicals with antioxidant properties. They work by reducing the accumulation of ROS in diabetic complications (Alam et al., 2017; Chen et al., 2019; Sarikurkcu et al., 2019). Also, flavonoids, phenylpropanoids, terpenes and quinines are known as good inhibitors of the alpha glucosidase enzyme as listed in a review (Yin et al., 2014). Even though myriad of phytochemical compounds have been reported from plant sources, there are no reports on the commercialization of phytochemicals as AGI so far. This lacuna was the drive for investigations to find a more potent natural AGI molecule from Simarouba glauca plant.
Simarouba glauca DC is rain-fed wasteland tree commonly called as ‘Laxmitaru’ or ‘Paradise tree’ belonging to the family Simaroubaceae. Different parts of the plant are being used traditionally as, antifungal, antibacterial, antioxidant (Osagie-Eweka et al., 2016; Santhosh et al., 2016; Umesh, 2015), antiparasitic, antipyretic and anticancer agents (Jose et al., 2019; Manasi and Gaikwad, 2011). The bark extract of this tree is commonly used for dysentery hence the bark is also called ‘dysentery-bark’ (Santhosh et al., 2016; Manasi and Gaikwad, 2011). The seed powder is being used to treat snakebites (Manasi and Gaikwad, 2011). Along with other species of Simaroubaceae family, S. glauca also contains a unique group of active compounds called quassinoids (ailanthinone, glaucarubinone, and holacanthone) which are shown to be responsible for the wide spectrum of biological activities of this plant (Alves et al., 2014). Hence, owing to the numerous biological activities reported in earlier studies and lack of comprehensive studies on phytochemical analysis, antioxidant and AGI activities from S. glauca, this plant was selected to explore the correlation between phytochemicals and different biological activities.
In this study, crude and purified ‘fraction-14’ from the ethanol extract of S. glauca leaves were analysed for phytochemicals and antioxidant assay followed by yeast alpha glucosidase enzyme inhibition assay. Further, the active ‘fraction-14’ was characterized by HPLC-Q–TOF-MS analysis and four compounds were tentatively identified for the first time in S. glauca leaf extract. The efficacy of alpha glucosidase enzyme inhibition was also studied by enzyme kinetics for purified ‘fraction-14’. The binding affinities of the identified compounds were analysed by in-silico docking of these compounds to alpha glucosidase enzyme. This study is expected to provide new insight into the potentiality of natural alpha glucosidase enzyme inhibitors as a alternative for the existing AGI to inhibit human alpha glucosidase.
2. Materials and methods
2.1. Chemicals and reagents
Alpha glucosidase from Saccharomyces cerevisiae (19.3 U/mg, Type1 lyophilized powder, G5003), p-Nitrophenyl-α-D-glucopyranoside (≥99%, N1377), 2, 2′-Azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS, ≥98%, A1888), quercetin (≥95% HPLC, Q4951), gallic acid (≥98% ACS reagent, 398225) and acarbose (PHR 1253) were procured from Sigma- Aldrich, India. 2, 2-diphenyl-1-picrylhydrazil (DPPH, 95%, Alfa Aesar-A Johnson Matthey Company) and acetonitrile HPLC grade (4400SP, Qualigens) obtained from Thermo Fisher Scientific India Pvt. Ltd. Vitamin C Purified (≥99%, QH1Q610760) and Methanol HPLC grade (SF8SF68931, 99.7%) were purchased from Merck Specialities Private Ltd, India. Chloroform analytical grade (2H2802300, 99.8%) purchased from SD Fine Chem. Ltd. India (SDFCL). Silica gel (Mesh size: 60–120) purchased from ACME Synthetic Chemicals, India.
2.2. Extraction and purification of active compounds
Fresh leaves were collected from S. glauca plants grown on the campus of Poornaprajna Institute of Scientific Research, Devanahalli, Bangalore Rural-562110, India (13.2884° N, 77.6713° E). Leaves were washed immediately in distilled water and air-dried at room temperature (RT), 28 ± 2 °C. Then, fine powder was prepared from the dried leaves using a mixer grinder and stored in polythene bags at 4 °C. The extract from the same powder was used for all the subsequent experiments.
Twenty grams of leaf powder was taken in a conical flask containing 150 mL of ethanol and the soluble content was extracted twice, using a rotary shaker at RT for 12 h. The extract in ethanol was filtered using a muslin cloth and the filtrate was centrifuged at 12,096 x g for 30 min (Eppendorf, 5810 R). The supernatant was collected and dried under vacuum using a rotary evaporator at 40 °C (IKA, RV 10 D S96). The dried extract was weighed and stored at -20 °C for further use.
The dried ethanol extract was re dissolved in absolute ethanol and was loaded on to a glass column (size of 50 × 4.5 cm) packed with the silica gel (60–120 mesh size). The column was equilibrated and eluted using a mobile phase containing chloroform, acetonitrile, and methanol at 5:4:1 ratio at a flow rate of 1 mL/min and collected 12 fractions of 10 mL each. Followed by, the same sample in the column was eluted with 50% aqueous methanol at the same flow rate and 4 fractions of 10 mL each were collected. All the 16 fractions were concentrated into 2 mL each using a rotary evaporator at 40 °C and further dried in a hot air oven at 40 °C and used for analysis.
2.3. Phytochemical assays
2.3.1. Estimation of total flavonoid
The total flavonoid content of ethanol extract and purified ‘fraction-14’ was determined using aluminum chloride assay (Medini et al., 2014) with slight modifications. Ethanol extract and purified ‘fraction-14’ were dissolved in Milli-Q water at 1 mg/ml concentration respectively, centrifuged at 16,873 x g for 10 min and filtered through 0.2 μm syringe filters before the analysis. The reaction mixture containing 150 μL of NaNO2 (0.72 M) and 100 μL of the sample in a 10 mL test tube. Then, the volume was made up to 2650 μL with Milli-Q water and incubated at RT (28 °C) for 5 min. Then 150 μL of AlCl3 (0.74 M) was added and incubated at RT for 1 min. Immediately after incubation, 1000 μL of NaOH (1 M) was added and the mixture was diluted to 5000 μL with Milli-Q water. The reaction mixture was mixed thoroughly and centrifuged at 16,873 x g for 5 min and absorbance was read at 510 nm using BioSpectrometer® kinetic (Eppendorf). Similarly, a standard curve was plotted for quercetin (Q) at different concentrations (200–1000 μg/mL) and used as flavonoid standard. The total flavonoid content in the sample extract was represented in milligram (mg) of quercetin equivalent/gram of the extract (mg Q/g extract).
2.3.2. Estimation of total phenolics
The total phenolic content of the ethanol extract and purified ‘fraction-14’ was determined using the Folin-Ciocalteu (F–C) method (Medini et al., 2014) with slight modifications. Leaf crude extract and purified ‘fraction-14’ were dissolved in Milli-Q water at 1 mg/ml concentration respectively, centrifuged at 16,873 x g for 10 min and filtered through 0.2 μm syringe filters before the analysis. Briefly, 200 μL of each sample was taken in a test tube and diluted to 500 μL using Milli-Q water. Then 150 μL of F–C reagent was added, stirred and kept for incubation for 6 min at room temperature. Then 1250 μL of Na2CO3 (1 M) was mixed thoroughly with the reaction mixture and kept for incubation in dark for 2 h. Thereafter, the reaction mixture was made up to 3 mL by diluting with 1100 μL of Milli-Q water and absorbance was measured at 760 nm using Thermoscientific Multiscan® plate reader. A standard curve was plotted using gallic acid (GA) with a concentration range of 200–1600 μg/mL. The total phenolic content was represented as mg gallic acid equivalent per gram of the extract (mg GA/g extract).
2.4. Antioxidant assays
2.4.1. DPPH assay
Ethanol crude extract and purified ‘fraction-14’ were dissolved in methanol at 1 mg/ml concentration, centrifuged at 16,873 x g for 10 min and filtered through 0.2 μm filter before the analysis. The DPPH free radical scavenging activity of the extract was determined using the previously described method with slight modifications (Yan et al., 2018). The reaction mixture in a 96 well plate contained 50 μl of samples at different concentrations (1, 0.5, 0.1 μg/mL; diluted in Milli-Q water) and 100 μL DPPH free radicals (0.2 mM). The final volume was made up to 200 μL with methanol and incubated at RT (28 °C) in dark for 30 min. The control was prepared by replacing the sample with the methanol. Also, sample blanks were prepared for each sample by replacing DPPH free radicals with the methanol. The absorbance was measured at 517 nm using the Thermoscientific Multiscan® plate reader. Ascorbic acid was used as a positive control at different concentration range (1.76, 3.52, 5.28, 7.04 and 8.80 μg/mL) and the standard curve was plotted. DPPH radical scavenging ability of the samples and ascorbic acid was calculated using the formula (1).
| Inhibition (%) = [(Absorbance control– Absorbance sample)/Absorbance control] ×100 | (1) |
The absorbance values of control and sample represented are the net values obtained after subtracting the reaction blank and sample blank respectively. The DPPH free radical scavenging activity of the samples were represented in IC50 values and compared with ascorbic acid (standard antioxidant).
2.4.2. ABTS assay
Ethanol crude extract and purified ‘fraction-14’ were dissolved in 100% methanol at 1 mg/ml concentrations, centrifuged at 16,873 x g for 10 min and filtered through 0.2 μm filter before the analysis and ABTS free radical scavenging activity was determined using the protocol previously described with slight modifications (Yan et al., 2018). The ABTS free radical was generated by incubating an equal amount of 7 mM ABTS and 2.4 mM potassium persulphate for 16 h in dark. The absorbance of the free radical was adjusted to optical density of 0.7 ± 0.03 at 734 nm by diluting in methanol. In a 96 well plate, 50 μL of the sample at different concentrations (10, 5, 1, 0.1 and 0.05 μg/mL in Milli-Q water) was taken and 100 μL of ABTS free radicals was added. The final volume was made up to 200 μL with methanol. The reaction mixture was incubated at RT (28 °C) in dark condition for 6 min. The control was prepared by replacing the sample with the methanol. Also, the sample blank was prepared for each sample by replacing ABTS free radicals with the methanol. The absorbance was measured at 734 nm using the Thermoscientific Multiscan® plate reader. Ascorbic acid was used as a positive control and the standard curve was plotted for different concentrations (0.88, 1.76, 2.64, 3.52 and 4.40 μg/mL). ABTS radical scavenging ability of the sample and ascorbic acid was calculated using the formula (2).
| Inhibition (%) = [(Absorbance control– Absorbance sample)/Absorbance control] ×100 | (2) |
The absorbance values of control and sample represented are the net values obtained after subtracting the reaction blank and sample blank respectively. The ABTS free radical scavenging activity of the samples were represented in IC50 values and compared with ascorbic acid (standard antioxidant).
2.5. Yeast alpha glucosidase inhibition (AGI) assay
Ethanol extract and purified ‘fraction-14’ were dissolved in Milli-Q water at 10 mg/ml concentration, centrifuged at 16,873 x g for 10 min (Eppendorf, 5418) and filtered through 0.2 μm filters before the analysis. AGI assay was carried out using the previously described method with slight modifications (Bharadwaj et al., 2018). Briefly, in a 96 well microplate, the 50 μL of sample at different concentrations (10, 5, 1, 0.1, 0.01, 0.05 and 0.005 μg/mL) mixed with 40 μL (0.0015 units/mL) of yeast alpha glucosidase enzyme and 10 μL of phosphate buffer (100 mM) pH 7. The reaction mixture was incubated at 37 °C for 20 min. Then, 50 μL of the substrate (20 mM p-nitrophenyl-α-D-glucopyranoside (PNPG) was added to the reaction mixture and incubated at 37 °C for 30 min. Immediately, 100 μL of sodium carbonate (200 mM) solution was added to the reaction mixture to retard the enzyme activity. The blank for the reaction was prepared by replacing the sample and enzyme with buffer. Similarly, the control was prepared using a buffer in place of the sample. Also, blank solutions were prepared for each sample by replacing an enzyme with a buffer. The absorbance of p-nitrophenol released during the reaction was quantified by reading at 405 nm using the Thermoscientific Multiscan® plate reader (Multiscan Go 1510). Similarly, the anti-diabetic drug acarbose was used as a standard (10 mg/mL) for the assay. The percentage of alpha glucosidase enzyme inhibition was calculated using the formula (3).
| Inhibition (%) = [(Absorbance control– Absorbance sample)/Absorbance control] ×100 | (3) |
The absorbance values of control and sample represented are the net values obtained after subtracting the reaction blank and sample blank respectively. The alpha glucosidase enzyme inhibition activity of the samples was represented in IC50 values and compared with acarbose.
2.6. Characterization
2.6.1. UV-visible spectroscopy
The column purified ‘fraction-14’ was dissolved in Milli-Q water at 0.25 mg/mL concentration and UV-Visible spectra were measured. The absorbance spectrum was measured from 200-830 nm range using BioSpectrometer® kinetic.
2.6.2. Fourier transform infrared spectroscopy (FTIR)
The column purified ‘fraction-14’ was analysed using FTIR equipped with a DLATGS detector (BRUKER ALPHA). The dried sample was mixed with KBr at a ratio of 1:100 respectively and the mixture was ground into a fine powder using a mortar and pestle. The thin pellet was prepared and placed on the sample holder of the instrument and scanned at 4000 - 400 cm−1 range.
2.6.3. HPLC-Q–TOF-MS analysis
Column purified ‘fraction-14’ was further characterized using an HPLC equipped with Phenomenex C18 column (Aqua 5 μm, 200Å, 250 × 4.6 mm i.d.) to separate the analytes. Binary mobile phase system consisting solvent A: water +0.1% formic acid and solvent B: acetonitrile 90% + 0.1% formic acid with a gradient elution; 0–15 min (A:95% & B:5%), 15–17 min (A:65% & B:35%), 17–24 min (A:25% & B:75%), 24–30 min (A:95% & B 5%) was used for the elution. The separated analytes were detected from m/z 100–1000 using Accurate Mass Q-TOF- LC/MS (Agilent Technologies, 6520) equipped with dual electrospray ionization source (ESI). Analytes were identified in negative ionization mode using the following instrument conditions; gas flow, 8 L/min; gas temperature, 250 °C; nebulizer 35 psig; capillary voltage, 2750 V. The structures of identified molecules were drawn using the software ChemDraw Pro 8.0.
2.7. Enzyme inhibitory kinetics
Alpha glucosidase enzyme inhibition kinetic study was performed for column purified ‘fraction-14’ using a modified method previously explained elsewhere (Şöhretoğlu et al., 2018). P-nitrophenyl α-D-glucopyranoside (PNPG) was used as a substrate at different concentration range (0.01–0.8 mM). The enzyme concentration was maintained at 0.02 Units/mL for the experiment. The enzyme kinetic activity was studied in the absence as well as in the presence of inhibitors at different concentrations (0.83–4.15 μg/mL). The mode of enzyme inhibition, Vmax, and Km was determined using the following Lineweaver- Burk plot formula (4). The inhibitory constant Ki was determined from the graph slope versus inhibitor concentrations.
| (4) |
where Vo: Initial velocity; Km: Michaelis-Menten constant; Vmax: Maximum velocity; S: Substrate concentration.
2.8. Molecular docking
2.8.1. Homology model
The in-vitro studies are performed against the alpha glucosidase enzyme from Saccharomyces cerevisiae. The protein sequence of alpha glucosidase (MAL12-Yeast) in the FASTA format was retrieved from UniProt (access code P53341). Since the crystal structure of alpha glucosidase from Saccharomyces cerevisiae is not available, the 3D structure of the enzyme was generated using RaptorX homology modelling software (http://raptorx.uchicago.edu/). The quality of the homology model was authenticated by the Ramachandran plot using PROCHECK software (https://servicesn.mbi.ucla.edu/PROCHECK/).
2.8.2. Docking study
A docking study was performed against the homology model of Saccharomyces cerevisiae alpha glucosidase enzyme. The protein structure was prepared using the protein preparation module on Maestro with the default setting. The 2D structures of acarbose, kaempferol-3-O-pentoside and kaempferol-3-O-glucoside were retrieved from the PubChem database. Eclalbasaponin V and cyanidin-3-O- (2′galloyl)-galactoside were drawn using PubChem sketcher V2.4 software (https://pubchem.ncbi.nlm.nih.gov/edit2/index.html). The 3D conformations of compounds were generated by LigPrep (Schrodinger, LLC, New York, NY, 2015) with default settings using OPLS2005 force field. A docking grid of 20 Å was created around the enzyme active-site and the grid was identified as x: 11.33, y: -0.02 and z: 26.77. Docking was performed using Glide (Schrodinger, LLC, New York, NY, 2015) on Maestro version 10.1. The best docking poses for all the compounds were visually analysed using Pymol software (https://pymol.org/edu/?q=educational/).
2.9. Statistical analysis
The experiments were done in triplicates (n = 3) and verified as mean ± SD. The inhibition percentage and IC50 values were calculated by performing a regression analysis. Analysis of variance (ANOVA) was implemented and the differences between the values are considered to be statistically significant at p < 0.05 using Microsoft Excel 2010.
3. Results and discussion
3.1. Extraction and purification of the active compounds
The 20 g dried leaf of S. glauca was yielded 3.6 g of ethanol crude extract, which had AGI activity. One gram of the crude extract was purified on a silica gel column and 16 fractions were collected. Each fraction was dried and analysed for the AGI activity. Out of 16 fractions, ‘fraction-14’ (20 mg) was found to be highly active against alpha glucosidase enzyme as represented in Figure 1. Further, the ‘fraction-14’ was analysed for phytochemicals and antioxidant assays along with the crude extract.
Figure 1.
Silica gel column purified phenolic rich ‘fraction-14’. Out of 16 fractions colleced from the silica gel column chromatography of ethanol extract from S. glauca leaf, the ‘fraction-14’ had high phenolic content and found to be highly active against alpha glucosidase enzyme.
3.2. Phytochemical assays
3.2.1. Estimation of total phenolics and flavonoids
The total phenolic and flavonoid compounds of ethanol extract and purified ‘fraction-14’ are represented in Table 1. The total phenolic compounds in crude extract and purified ‘fraction-14’ were determined to be 414 ± 11 and 178 ± 7 mg GA/g of extract respectively. The total flavonoid content in crude ethanol extract was found to be 166 ± 29 mg Q/g of extract. However, flavonoid content was not detected in purified ‘fraction-14’ up to a concentration of 1 mg/mL. Earlier studies on S. glauca, Umesh et al. reported the phenolic contents of about 200 GA/g and flavonoids as 14.98 mg Q/g in the crude ethanol extract (Umesh, 2015). Another report confirmed the presence of phenolic compounds in ethanol extract as 84.55 ± 0.59 mg GA/g of extract (Puranik et al., 2017). Another group reported a very low level of phenolic and flavonoid compounds in the crude ethanol extract of S. glauca leaves as 0.12 ± 0.01 mg GA/g and 0.14 ± 0.02 mg Q/g of extract respectively (Osagie-Eweka et al., 2016). However, all the earlier studies are on crude ethanol extract alone.
Table 1.
Alpha-glucosidase inhibitory (AGI) activity, phytochemical analysis, antioxidant activity of ethanol crude extract and column purified ‘fraction-14’ of S. glauca leaf.
| Sample | AGI activity (IC50 μg/mL) | Phytochemicals |
Antioxidant activity (IC50 in μg/mL) |
||
|---|---|---|---|---|---|
| Total phenolics (mg GA/g) | Total flavonoids (mg Q/g) | DPPH Assay | ABTS Assay | ||
| Ethanol crude extract | 0.5 ± 0.04 | 414 ± 11 | 166 ± 29 | 12.5 ± 0.6 | 4.0 ± 0.5 |
| Purified fraction-14 | 2.4 ± 0.40 | 178 ± 70 | - | 14.4 ± 0.1 | 7.6 ± 0.5 |
| Ascorbic acid | - | - | - | 5.1 ± 0.1 | 3.9 ± 0.2 |
| Acarbose | 2450 ± 24 | - | - | - | - |
Note: AGI activity and antioxidant activity are represented by IC50 values. Acarbose was used as a standard for AGI activity. Total phenolics are represented in mg GA per gram of extract. Total flavonoids are represented in mg Q per gram of extract. Ascorbic acid was used as standard for antioxidant activities. All the experiments are expressed as mean ± SD (n = 3). The values are statistically significant at p < 0.05.
Q: Quercetin, GA: Gallic acid, SD: Standard Deviation, “-“: Not determined.
Even though the phenolic compounds are ubiquitous in plants, their recovery greatly depends on their extraction method, solubility in the solvents, the extent of polymerization of phenols, the interference of other metabolites, the formation of insoluble complexes, etc (Medini et al., 2014). In this study, the S. glauca leaf extract showed a high amount of total phenolic and flavonoid compounds than the previous reports. This observation is probably due to factors such as different geographical origin of the plant, environmental conditions, seasonal differences, solvents, etc (Medini et al., 2014). Most of the earlier reports on plant extracts which had high phenolic compounds, also showed good antioxidant activity and AGI activity (Chen et al., 2019; Jiang et al., 2017; Sarikurkcu et al., 2019; Tan and Chang, 2017; Yan et al., 2018). So, the present study was focused on the antioxidant property and identification of AGI from phenolic rich purified ‘fraction-14’ of S. glauca.
3.3. Antioxidant assays
DPPH and ABTS free radical scavenging activity of ethanol extract purified ‘fraction-14’ and a standard were determined and expressed their IC50 values as shown in Table 1. The DPPH free radical scavenging activity (IC50) of the ethanol extract and purified ‘fraction-14’ were 12.5 ± 0.6 and 14.4 ± 0.1 μg/mL respectively. Whereas, the ABTS free radical scavenging activity (IC50) for ethanol extract and purified ‘fraction-14’ were 4 ± 0.5 and 7.6 ± 0.5 μg/mL respectively. For both the assays, ascorbic acid was used as a positive control. DPPH and ABTS free radical scavenging ability of ascorbic acid were found to be 5.1 ± 0.1 and 3.9 ± 0.2 μg/mL respectively. Both the ethanol extract and purified ‘fraction-14’ had lower antioxidant activity than ascorbic acid.
The antioxidant properties of the phenolic and flavonoid compounds have been studied extensively for decades. It has been reported that the antioxidant properties of the phenolics and flavonoids are due to the presence of aromatic rings containing hydroxyl groups. These hydroxyl moieties associated with an aromatic ring can donate a hydrogen atom or an electron, chelate metals, activate antioxidant enzymes or inhibit oxidases (Amarowicz et al., 2004). Crude ethanol extract from S. glauca leaves containing phenolic and flavonoid were shown to possess good DPPH activity (IC50) of 13.12 μg/mL (Umesh, 2015) and 4.91 μg/mL (Osagie-Eweka et al., 2016). The earlier report on ABTS free radical scavenging activity for the crude ethanol extract of S. glauca was 45.20 μg/mL (Osagie-Eweka et al., 2016). In the present study, DPPH (IC50 = 12.5 ± 0.6 μg/mL) and ABTS (IC50 = 4 ± 0.5 μg/mL) values of crude ethanol extract (which had phenolics and flavanoids) are comparable to the antioxidant values of earlier studies. Similarly, another study on S. glauca leaves reported very low DPPH scavenging activity in ethyl acetate (IC50 = 1200 μg/mL) and petroleum ether (IC50 = 1280 μg/mL) extracts (Santhosh et al., 2016). However, in the present study along with alpha glucosidase inhibition, the column purified ‘fraction-14’ of S. glauca also showed the presence of phenolics, as well as DPPH and ABTS free radical scavenging activity for the first time.
3.4. Yeast alpha glucosidase inhibition (AGI) assay
The AGI activities of ethanol extract and purified ‘fraction-14’ were represented in Table1. The ethanol extract exhibited good activity with an IC50 value of 0.5 ± 0.04 μg/mL. The AGI activity of ‘fraction-14’ showed the highest activity than the other fractions with an IC50 value of 2.4 ± 0.4 μg/mL as represented in Table 1. Also, Table 2 represents the AGI activities of the remaining purified fractions of ethanol extract. Further, the crude ethanol extract and ‘fraction-14’ have shown very good inhibition of alpha glucosidase enzyme which are significantly better than the standard acarbose (IC50 = 2450 ± 24 μg/mL).
Table 2.
Alpha-glucosidase inhibitory (AGI) activity of column purified fractions of ethanol extract from S. glauca leaf.
| S.N. | Purified fractions | AGI activity (%Inhibition) |
|---|---|---|
| 1 | 1–12 | - |
| 2 | 13 | 24.5 |
| 3 | 14 | 78.7 |
| 4 | 15 | 26.1 |
| 5 | 16 | 39.1 |
Note: AGI values are represented in percentage inhibition at the sample concentration of 0.05 mg/mL “-“: Not observed.
There are several reports on the AGI activity of the phenolic extracts from other plants. The phenolic extract from Panicum sumatrense has shown AGI activity with an IC50 value of 18.97 ± 0.43 μg/mL against yeast enzyme (Pradeep and Sreerama, 2018). The extractable and non-extractable polyphenols from Camellia sinensis exhibited yeast AGI activity with IC50 values of 0.77 ± 0.02 and 1.96 ± 0.21 μg/mL respectively (Yan et al., 2018). The semi-purified phenolic rich extract of Glycine max L showed AGI activity against yeast enzyme with an IC50 value of 13.81 ± 0.081 μg/mL (Tan and Chang, 2017). Also, the phenolic rich ethyl acetate extract of Eucalyptus grandis inhibited yeast AG enzyme with an IC50 value of 1.40 ± 0.18 μg/mL (Jiang et al., 2017). In another report, the phenolic extract of Juglans regia leaf showed AGI activity in-vitro and anti-hyperglycaemic effect against streptozotocin-induced rat models (Mollica et al., 2017).
However, there are few in-vivo studies on the anti-diabetic properties of the plant extracts from Simaroubaceae family. One study shows, the compounds bruceine E and D isolated from Brucea javanica lowers the blood glucose concentration by 73.57 ± 13.64% and 87.99 ± 2.91% respectively in streptozotocin-induced rat models, when compared to the non-treated streptozotocin-induced rats (NoorShahida et al., 2009). In another study, the methanolic extract of Quassia amara at 200 mg/kg dose reduces the elevated blood glucose level near to the normal in a streptozotocin-induced rat model (Husain et al., 2011). But there are no available reports on the in-vitro or in-vivo analysis of S. glauca extracts for AGI activity. Hence, based on the available reports on the biological properties of different parts of S. glauca, the AGI activity is reported for the first time in this study.
It was interesting to observe that the in-vitro analysis of the purified ‘fraction-14’ showed about 1000-fold better AGI activity (IC50 = 2.4 ± 0.4 μg/mL) when compared to the standard acarbose (IC50 = 2450 ± 24 μg/mL), indicating the presence of highly potent inhibitor of AG in S. galuca leaf extract, which may be a potential lead molecule to treat T2DM.There was a marginal decrease in AGI activity of purified ‘fraction-14’ when compared to the crude extract, which could be due to the synergistic effect of phenolic compounds present in the crude extract (Şöhretoğlu et al., 2018). The loss of activity in the purified fraction was also reported in earlier studies (Tan and Chang, 2017; Yin et al., 2014). Most of the reports on different plant extracts indicate the AGI activity of the phenolic extracts. The current study also showed the correlation between high phenolics of ‘fraction-14’ and its AGI activity. Hence, to identify the possible compounds responsible for AGI activity, the ‘fraction-14’ was characterized using UV-Visible, FTIR and LC-MS techniques.
3.5. Characterization
3.5.1. UV-visible spectroscopy and FTIR analysis
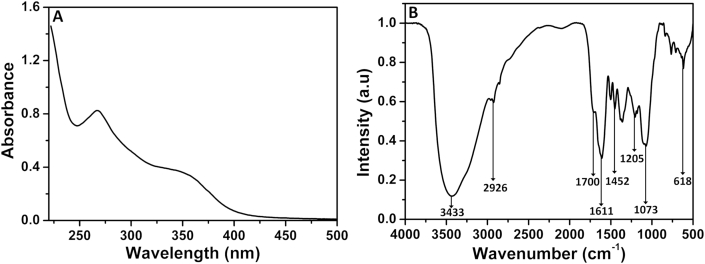
The UV-visible spectroscopic analysis of the column purified ‘fraction-14’ showed two absorption peaks at 267 nm and 350 nm with the absorbance of 0.82 and 0.36 respectively indicating the presence of phenolic compounds as represented in Figure 2A. The above results correlate with the earlier studies, where they illustrate the presence of two absorption peaks for phenolic compounds in the range between 320 to 380 and 250–285 nm range in the UV region (Matthäus, 2002).
Figure 2.
UV-Visible spectroscopic analysis (A) and FTIR spectra (B) of column purified ‘fraction-14’ of ethanol extract from S. glauca leaf. For UV-Visible spectroscopy, the sample was dissolved in Milli-Q water at 0.25 mg/mL concentration. For FTIR, the sample was mixed with KBr at a ratio of 1:100, ground into a fine powder and a thin pellet was made.
The FTIR analysis of purified ‘fraction-14’ shows several bands at different intensities as represented in Figure 2B. The band at 3433 cm−1 and 618 cm−1 may be due to stretching and bending vibration of -O-H groups respectively. The band at 2926 cm−1 could be attributed to the -C-H stretching of aromatics. The stretching vibration of –C=C- of aromatic rings could appear at 1611 cm−1. The band appeared at 1452 cm−1 could be attributed to the presence of aromatic –C=C- bond. The band appeared at 1700 cm−1 may be due to the stretching vibration of –C=O bond. These are the common bands present in the identified compound 1 (eclalbasaponin V), 2 (cyanidin-3-O-(2′galloyl)-galactoside), 3 (kaempferol-3-O-glucoside) and 4 (kaempferol-3-O-pentoside). However, two bands appeared at 1073 and 1205cm−1 which may be attributed to the symmetric and asymmetric vibrations of the O=S=O group of compounds 1. The description of FTIR bands to the respective functional groups in this study are explained in comparison with the earlier reports (González-Cabrera et al., 2018).
3.5.2. LC-MS analysis
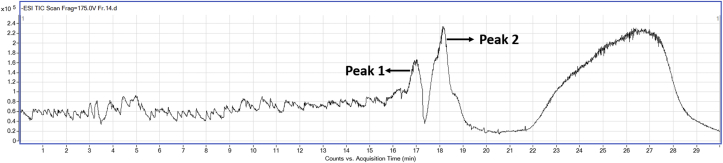
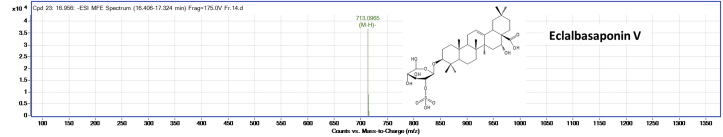
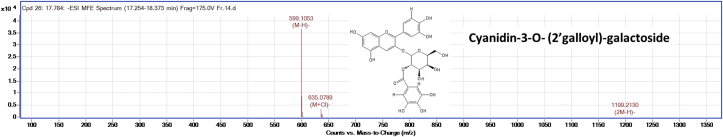
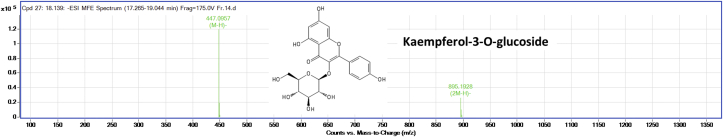
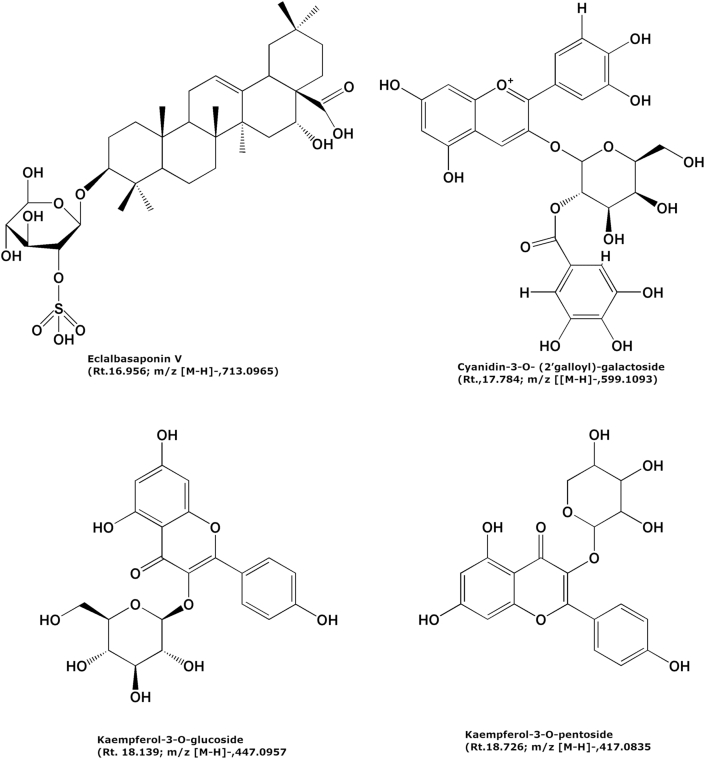
The silica gel column purified ‘fraction-14’ was subjected to HPLC-Q–TOF-MS analysis to identify the possible compound responsible for the AGI activity. The LC-MS analysis of the compound showed two major peaks as represented in Figure 3. The LC-MS analysis of all the peaks of ‘fraction-14’ was represented in Table 3. The peaks at retention time between 16.4 - 18.8 min which had high intensity was annotated for m/z values. These m/z values were compared with the m/z values of the known phytochemical compounds, and four compounds were tentatively identified in negative ion mode. Figures 4, 5, 6, and 7 represents the Mass spectra of tentatively identified compounds. The structures of all four compounds are represented in Figure 8. The list of compounds with their m/z values along with the retention time and other data is represented in Table 4. The compound 1 with m/z 713.0965 [M-H]¯ is identified as eclalbasaponin V; a triterpenoid saponin has been identified previously in Eclipta prostrata L. The compound 2; an anthocyanin, identified as cyanidin-3-O-(2′galloyl)-galactoside with m/z 599.1053 [M-H]¯ also reported previously from Rhus coriaria L (Abu-Reidah et al., 2015; Han et al., 2015) and Victoria amazonica (Han et al., 2015; Strack et al., 1992). These compounds are rarely reported in green leaves. However, there are some reports on the presence of anthocyanins in early as well as later stages of the vegetative part including leaves and petioles (Hatier and Gould, 2008; Lee, 2002). Study conducted by Manetas and group observed the presence of anthocyanins in the mature green leaves of Rosa sp. and Ricinus communis L (Manetas et al., 2002). Further, the presence of anthocyanins were documented in the leaves of tropical plants during their different stages of the development (Lee and Collins, 2001). In the present study extract was from the leaves of different age groups along with petiole, might have contained anthocyanins. The compound 3 with m/z 447.0957 [M-H]¯ identified as kaempferol-3-O-glucoside has been reported previously in Pollen Typhae, Phaseolus vulgaris L and Flaveria bidentis (L.) Kuntze (Chen et al., 2015; Pitura and Arntfield, 2019; Wei et al., 2011). The compound 4 identified as kaempferol-3-O-pentoside showed molecular ion peak at m/z 417.0835 [M-H] ¯ has been identified previously in Bauhinia species and Rubus grandifolius Lowe. The compounds 3 and 4 belong to the flavonoid group (Farag et al., 2015; Gouveia-Figueira and Castilho, 2015). All the four compounds are reported in different plant species but, reported for the first time from S. glauca.
Figure 3.
ESI-TIC spectra of column purified ‘fraction-14’. The LC-MS peaks at retention time between 16.4-18.8 min which has high intensity was analysed for mass spectroscopy.
Table 3.
LC-MS analysis of purified ‘fraction-14’. The peak at retention time between 16.4-18.8 min which had high intensity was annotated for m/z values.
| Show/Hide | Cpd | File | Saturated | RT | Precursor | Mass | Ion Polarity | Ions | Height | Volume | Algorithm | Quality Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TRUE | 27 | Fr.14.d | - | 18.139 | 447.0957 | 448.1026 | Negative | 8 | 119599 | 7445123 | Find by Molecular Feature | 100 |
| TRUE | 23 | Fr.14.d | - | 16.956 | 713.0965 | 714.1038 | Negative | 4 | 36790 | 1299330 | Find by Molecular Feature | 100 |
| TRUE | 26 | Fr.14.d | - | 17.784 | 599.1053 | 600.1123 | Negative | 9 | 36451 | 1476837 | Find by Molecular Feature | 100 |
| TRUE | 10 | Fr.14.d | - | 9.625 | 353.088 | 354.0953 | Negative | 3 | 27554 | 784165 | Find by Molecular Feature | 100 |
| TRUE | 30 | Fr.14.d | - | 18.726 | 417.0835 | 418.0908 | Negative | 3 | 23452 | 856989 | Find by Molecular Feature | 100 |
| TRUE | 22 | Fr.14.d | - | 16.949 | 577.0812 | 578.0884 | Negative | 3 | 20970 | 777064 | Find by Molecular Feature | 100 |
| TRUE | 2 | Fr.14.d | - | 3.233 | 362.9402 | 363.9474 | Negative | 3 | 20155 | 165424 | Find by Molecular Feature | 81.8 |
| TRUE | 1 | Fr.14.d | - | 3.039 | 362.9401 | 363.9474 | Negative | 3 | 18606 | 214072 | Find by Molecular Feature | 80 |
| TRUE | 18 | Fr.14.d | - | 16.215 | 729.0911 | 730.0983 | Negative | 4 | 18606 | 740919 | Find by Molecular | 100 |
| TRUE | 7 | Fr.14.d | - | 5.027 | 265.0929 | 266.1 | Negative | 5 | 15749 | 430217 | Find by Molecular Feature | 100 |
| TRUE | 11 | Fr.14.d | - | 10.571 | 183.03 | 184.0373 | Negative | 2 | 14872 | 541952 | Find by Molecular Feature | 100 |
| TRUE | 20 | Fr.14.d | - | 16.94 | 463.0883 | 464.0952 | Negative | 7 | 13827 | 571896 | Find by Molecular Feature | 91.3 |
| TRUE | 5 | Fr.14.d | - | 3.889 | 248.9602 | 249.9675 | Negative | 2 | 13548 | 449609 | Find by Molecular Feature | 100 |
| TRUE | 25 | Fr.14.d | - | 17.778 | 713.0958 | 714.1031 | Negative | 3 | 11945 | 395509 | Find by Molecular Feature | 100 |
| TRUE | 3 | Fr.14.d | - | 3.345 | 248.9601 | 249.9674 | Negative | 2 | 11416 | 106904 | Find by Molecular Feature | 100 |
| TRUE | 4 | Fr.14.d | - | 3.854 | 520.9083 | 521.9156 | Negative | 2 | 10581 | 175521 | Find by Molecular Feature | 100 |
| TRUE | 19 | Fr.14.d | - | 16.221 | 615.0978 | 616.105 | Negative | 6 | 9636 | 377572 | Find by Molecular Feature | 100 |
| TRUE | 6 | Fr.14.d | - | 3.928 | 154.9736 | 155.9809 | Negative | 2 | 9430 | 276406 | Find by Molecular Feature | 86.5 |
| TRUE | 16 | Fr.14.d | - | 12.999 | 633.0717 | 634.079 | Negative | 5 | 9163 | 226214 | Find by Molecular Feature | 100 |
| TRUE | 28 | Fr.14.d | - | 18.165 | 561.0843 | 562.0916 | Negative | 3 | 8935 | 408128 | Find by Molecular | 100 |
| TRUE | 9 | Fr.14.d | - | 6.291 | 169.0141 | 170.0214 | Negative | 2 | 8767 | 274588 | Find by Molecular | 100 |
| TRUE | 29 | Fr.14.d | - | 18.718 | 599.1034 | 600.1103 | Negative | 6 | 8624 | 495158 | Find by Molecular Feature | 82.1 |
| TRUE | 13 | Fr.14.d | - | 11.465 | 531.0956 | 532.1029 | Negative | 3 | 8351 | 248916 | Find by Molecular Feature | 100 |
| TRUE | 8 | Fr.14.d | - | 5.294 | 331.0664 | 332.0737 | Negative | 3 | 7720 | 221418 | Find by Molecular Feature | 100 |
| TRUE | 12 | Fr.14.d | - | 11.455 | 417.1032 | 418.1104 | Negative | 5 | 7289 | 230316 | Find by Molecular Feature | 87.1 |
| TRUE | 24 | Fr.14.d | - | 17.763 | 433.0768 | 434.0841 | Negative | 4 | 6128 | 231414 | Find by Molecular Feature | 100 |
| TRUE | 21 | Fr.14.d | - | 16.949 | 599.1024 | 600.109 | Negative | 6 | 5973 | 278252 | Find by Molecular Feature | 100 |
| TRUE | 17 | Fr.14.d | - | 15 | 651.1877 | 652.195 | Negative | 3 | 5747 | 180282 | Find by Molecular Feature | 80 |
| TRUE | 14 | Fr.14.d | - | 12.015 | 467.0797 | 468.0869 | Negative | 3 | 5336 | 198210 | Find by Molecular Feature | 100 |
| TRUE | 15 | Fr.14.d | - | 12.023 | 353.0873 | 354.0946 | Negative | 2 | 5135 | 154844 | Find by Molecular Feature | 100 |
Figure 4.
Mass spectra of eclalbasaponin V (compound 1). The m/z value was obtained in negative ionization mode and compared with m/z value of previously identified compounds in literature.
Figure 5.
Mass spectra of cyanidin-3-O- (2′galloyl)-galactoside (compound 2). The m/z value was obtained in negative ionization mode and compared with m/z value of previously identified compounds in literature.
Figure 6.
Mass spectra of kaempferol-3-O-glucoside (compound 3). The m/z value was obtained in negative ionization mode and compared with m/z value of previously identified compounds in literature.
Figure 7.
Mass spectra of kaempferol-3-O-pentoside (compound 4). The m/z value was obtained in negative ionization mode and compared with m/z value of previously identified compounds in literature.
Figure 8.
Chemical structures of tentatively identified compounds from column purified ‘fraction-14’ of ethanol extract from S. glauca leaf using LC-MS analysis. The compounds were identified based on the comparison of m/z values with m/z values of previously identified compounds in literature.
Table 4.
Identified compounds from column purified ‘fraction-14’ using HPLC-Q–TOF-MS analysis.
| S.N. | Rt (min) | Observed m/z [M-H] ¯ | Other fragments | m/z [M-H] ¯ from literature | Tentative identification |
|---|---|---|---|---|---|
| 01 | 16.956 | 713.0965 | - | 713.3582 | Eclalbasaponin V |
| 02 | 17.784 | 599.1053 | 635.0789 [M + Cl] ¯, 1199.2130[2M-H] ¯ | 599.1039 | Cyanidin-3-O- (2′galloyl)-galactoside |
| 03 | 18.139 | 447.0957 | 895.1928 [2M-H] ¯ | 447.0957 | Kaempferol-3-O-glucoside |
| 04 | 18.726 | 417.0835 | - | 417.0845 | Kaempferol-3-O-pentoside |
Note: The m/z values were obtained in negative ionization mode. Compounds (S.N. 1,2, 3 & 4)were identified based on comparison of m/z values with the previous reports. “-“: Not observed.
Although these four compounds are reported by others, there is no available literature on the biological activities of these compounds. However, biological activity was analysed for the compounds eclalbasaponin, eclalbasaponin I and eclalbasaponin II which are closely related to the compound 1 (Cho et al., 2016; Ray et al., 2013; Wang et al., 2018). Many anthocyanins, other than compound 2, showed antioxidant activity and anti-inflammatory activities in an earlier report (Blando et al., 2018; Kong et al., 2003). Compound 3 was previously identified in Helichrysum compactum extract, which showed antioxidant activity (Süzgeç et al., 2005). In another study, the semi-purified fraction of legumes showed AGI activity and later on one of the compounds from this fraction was identified as kaempferol-3-O-glucoside (Tan and Chang, 2017). However, antioxidant activity, AGI activity, and antiproliferation effect have been determined in the crude extracts of Bauhinia species and Euphorbia supina, where later characterization of the extracts confirms the presence of compound 4 which is also identified in the present study as kaempferol-3-O-pentoside (Farag et al., 2015; Song et al., 2014).
Recently, many purified compounds like rhinacanthin C (IC50 = 22.6 ± 0.6 μg/mL) (Shah et al., 2017), norathyriol (IC50 = 4.22 ± 0.19 μg/mL) (Gu et al., 2019), mangiferin (IC50 = 36.84 μg/mL) (Sekar et al., 2019), etc. have been reported as AGI from different medicinal plants. The purified ‘fraction-14’ in this study showed better AGI activity (IC50 = 2.4 ± 0.4 μg/mL) than the above-mentioned compounds. Compounds 3 and 4 identified in this study may contribute to the AGI activity and antioxidant activity observed in ‘fraction-14’, as these compounds are already known for alpha glucosidase inhibition and antioxidant properties according to the earlier reports (Farag et al., 2015; Song et al., 2014; Tan and Chang, 2017). So, the purified ‘fraction-14’ is expected to have a higher potential inhibitor for an alpha glucosidase enzyme and can be used for T2DM treatment after a detailed toxicological, in-vivo and clinical studies.
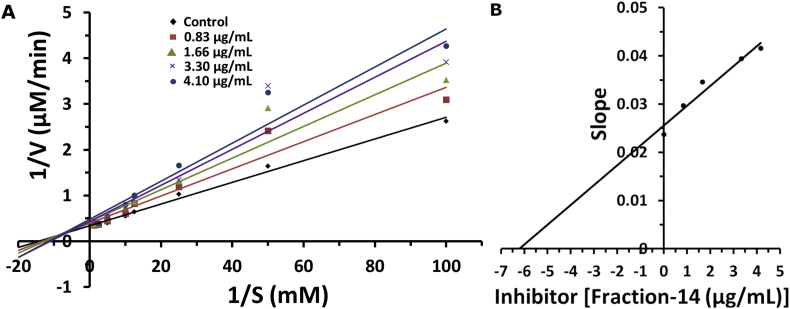
3.6. Enzyme inhibitory kinetics
The double reciprocal Lineweaver-Burk plot was used to determine the mode of inhibition and kinetic parameters for the column purified ‘fraction-14’. Figure 9A represents the Lineweaver-Burk plot for different concentrations of inhibitor (purified ‘fraction-14’) analysed for different concentrations of the substrate (PNPG). In this plot all lines drawn were intersected at the second quadrant, which indicates, the purified ‘fraction-14’ exhibited the mixed type of inhibition on the alpha glucosidase enzyme. Also, the apparent Vmax was slightly changed and Michaelis-Menten constant, Km values were increasing with the increasing concentration of inhibitor as shown in Table 5. This behaviour indicate that the inhibitor prefers binding to the free enzyme rather than the enzyme-substrate complex. Precisely, Figure 9B represents the slope versus inhibitor was a linear fit, which shows, the purified ‘fraction-14’ has a single inhibition site or a single class of inhibition on the alpha glucosidase enzyme. Our findings are in line with previous reports where kaempferol and hesperetin show the mixed type inhibition on yeast alpha glucosidase enzyme. Interestingly, unlike acarbose (a competitive inhibitor), where its effectiveness is found to be hindered by the high concentration of carbohydrate intake, the mixed inhibitors would still be effective at lower concentration (Gong et al., 2017; Peng et al., 2016; Rouzbehan et al., 2017). Furthermore, the inhibition constant, Ki for purified ‘fraction-14’ is 6.2 μg/mL which is better than the acarbose (Ki = 49 mg/mL) reported in the earlier study (Kim et al., 1999).
Figure 9.
Lineweaver- Burk plots (A) and Secondary plot of slope versus inhibitor (‘fraction-14’) (B). The inhibitor concentrations were used as 0.83, 1.66, 3.32 and 4.15 μg/mL. Alpha glucosidase concentration was 0.02 units/mL. Substrate, PNPG concentrations were 0.01, 0.02, 0.04, 0.08, 0.1, 0.2, 0.4 and 0.8 mM.
Table 5.
Maximum velocity; Vmax and Michaelis-Menten constant; Km for different concentrations of column purified ‘fraction-14’.
| S.N. | Fraction-14 (μg/mL) | Vmax (μM/min.) | Km (μM) |
|---|---|---|---|
| 1 | 0 | 2.95 | 70 |
| 2 | 0.83 | 2.50 | 74 |
| 3 | 1.66 | 2.33 | 80 |
| 4 | 3.32 | 2.28 | 90 |
| 5 | 4.15 | 2.13 | 90 |
Note: The alpha glucosidase concentration was 0.02 units/mL. The substrate, PNPG concentration was 0.01, 0.02, 0.04, 0.08, 0.1, 0.2, 0.4 and 0.8 mM. The differences among the values are considered to be statistically significant at p < 0.05. PNPG: p-nitrophenyl-α-D-glucopyranoside.
3.7. Homology model
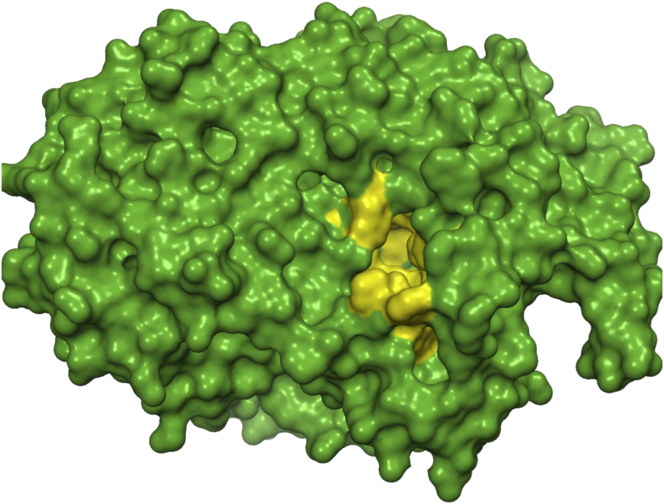
The homology model of S. cerevisiae alpha glucosidase was generated using S. cerevisiae isomaltase (PDB:3A47; 71.8% sequence identity and 84.9% similarity with S. cerevisiae alpha glucosidase) and MalL mutant enzyme from Bacillus subtilis (PDB:4MB1; 41% sequence identity and 58.8% similarity with S. cerevisiae alpha glucosidase) as templates. Figure 10 represents the structure of S. cerevisiae alpha glucosidase in a surface view generated using RaptorX software. The active site of the enzyme is represented in the yellow-coloured patch where all the identified compounds are perfectly stacked during the docking study. Figure 11 represents the Ramachandran plot where 90.5% residues reside in the most favoured region, 8.5% residues reside in additional allowed regions, 0.8% residues located in generously allowed regions and 0.2% residues located in disallowed regions. Analysis of the Ramachandran plot further strengthens the quality of the S. cerevisiae alpha glucosidase homology model. So, the validated and analysed homology model was used for the docking study.
Figure 10.
Surface representation of modelled structure of the yeast alpha glucosidase. The structure was generated using RaptorX software. The active site of the enzyme is represented in a yellow-colored patch where the docked compounds interacts with enzyme which involves both hydrogen bonds and hydrophobic interactions.
Figure 11.
Ramachandran plot for homology model of yeast alpha glucosidase. The homology model was built using RaptorX software. The model was validated and authenticated by Ramachandran plot using PROCHECK software.
3.8. Docking study
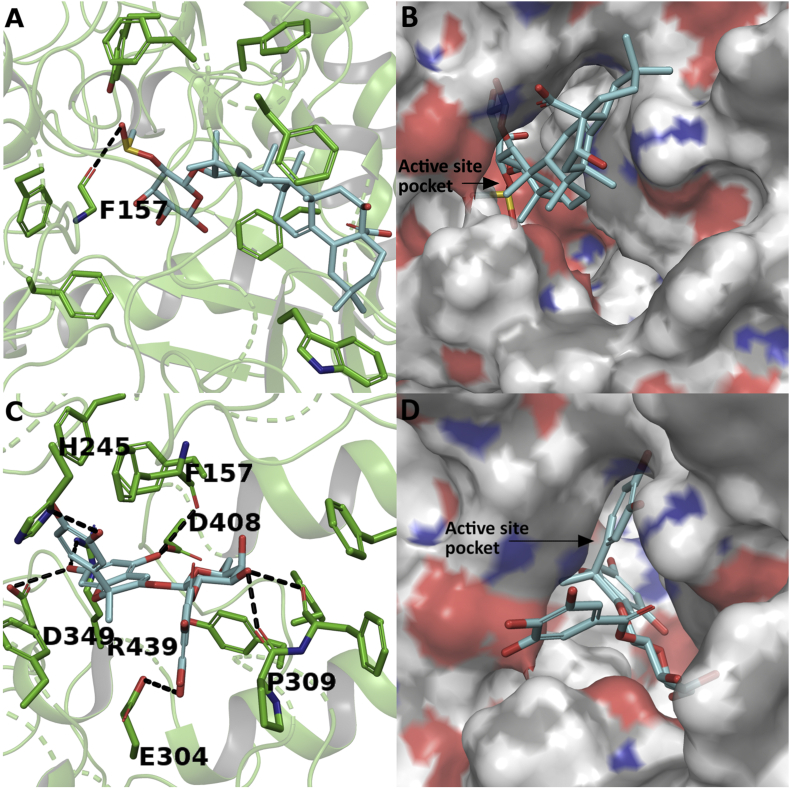
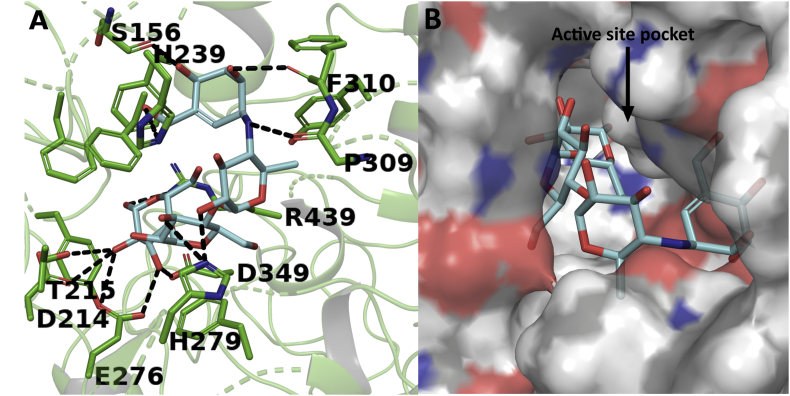
A docking study was performed to predict the mode of interaction between the identified compounds and the alpha glucosidase enzyme. The homology modelling of S. cerevisiae alpha glucosidase and docking studies have been reported in earlier studies where several compounds were docked with the alpha glucosidase enzyme (Gollapalli et al., 2019; Picot et al., 2017; Wang et al., 2017). The docking study of the identified compounds are represented in Figure 12 and Figure 13. The binding energy of the compounds and active site residues of S. cerevisiae alpha glucosidase are represented in Table 6. Among the docked compounds, compound 2 showed highest binding energy as -7.769 kJ/mol followed by compound 3, compound 4, acarbose and compound 1 at binding energy -7.127, -7.04, -6.867 and -4.409 kJ/mol respectively. The compound 1 showed only one hydrogen bond interaction with the main-chain of residue PHE157 as shown in Figure 12A and stabilized in the active site pocket with hydrophobic interactions with residues PHE310, 231, 300, 311, 158, 177, TRP242 and TYR313 as represented in Figure 12B and Table 6. Figure 12C represents the mode of interaction between compound 2 and enzyme in a stick view. The cyanidin moiety of compound 2 showed five hydrogen bond interactions with the residues HIS245, ASP408, 349, PHE157 and ARG439. The galactoside moiety showed hydrogen bond interaction with PRO309 while GLU304 involved in the hydrogen bonding interaction with the galloyl moiety of compound 2. Further, surface characteristic of compound 2 is perfectly complementary to the active site and stacks deep inside the active site pocket of the enzyme with several hydrophobic interactions such as PHE177, 310, 231, 311, 158, 300 and TYR313 as represented in Figure 12D and Table 6.
Figure 12.
Docking study of compounds 1 and 2 with yeast alpha glucosidase. Interaction of compound 1 in stick representation (A) and surface representation form (B); and interaction of compound 2 in stick representation (C) and surface representation form (D) with active site residues of alpha glucosidase. The compounds were perfectly stacked inside the active site pocket of the enzyme with several hydrogen bonds and hydrophobic interactions. The hydrogen bond interactions were represented in black coloured broken lines.
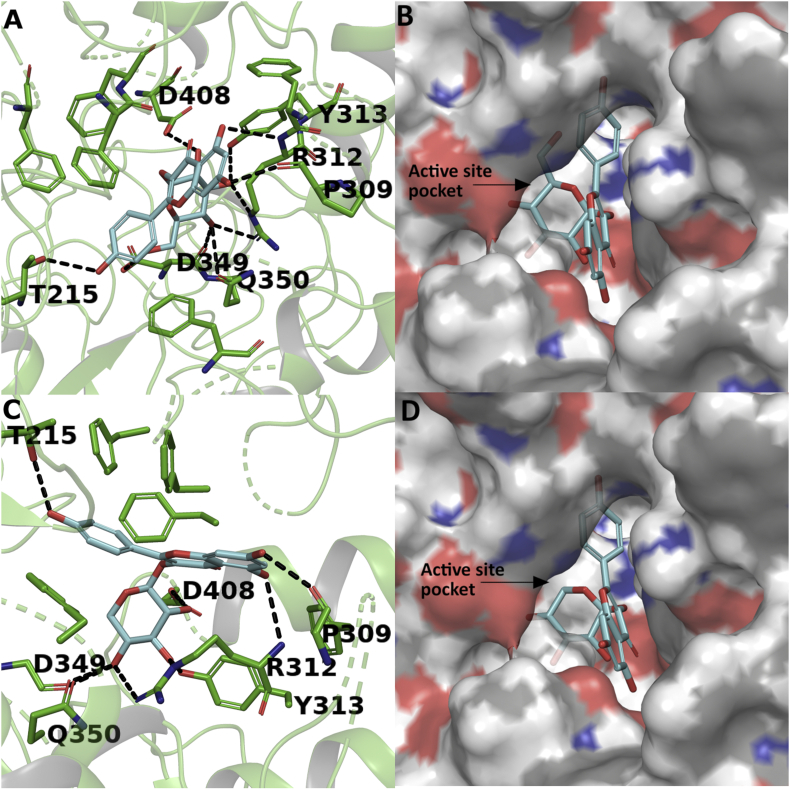
Figure 13.
Docking study of compounds 3 and 4 with yeast alpha glucosidase. Interaction of compound 3 in stick representation (A) and surface representation form (B); and interaction of compound 4 in stick representation (C) and surface representation form (D) with active site residues of alpha glucosidase. The compounds were perfectly stacked inside the active site pocket of the enzyme with several hydrogen bonds and hydrophobic interactions. The hydrogen bond interactions were represented in black coloured broken lines.
Table 6.
Binding energy of the compounds and interacting active site residues of Saccharomyces cerevisiae alpha-glucosidase.
| S.N. | Compound | Binding energy (KJ/mol) | Interacting residues of the active site |
|---|---|---|---|
| 1 | Eclalbasaponin V | -4.409 | PHE157, 310, 231, 300, 311, 158, 177, TRP242, TYR313 |
| 2 | Cyanidin-3-O- (2′galloyl)-galactoside | -7.769 | HIS245, PRO309, ASP408, 349, GLU304, ARG439, PHE157, 177, 310, 231, 311, 158, 300, TYR313 |
| 3 | Kaempferol-3-O-glucoside | -7.127 | PRO309, ARG312, THR215, ASP349, 408, TYR313, GLN350, PHE157, 158, 300, 177, 311 |
| 4 | Kaempferol-3-O-pentoside | -7.04 | THR215, ASP349, 408, GLN350, TYR313, ARG312, PRO309, PHE300, 157, 177, 158, 311 |
| 5 | Acarbose | -6.867 | PRO309, HIS239,279, ASP349, SER156, ARG439, THR215, GLU276, ASP214, PHE310, 300, 157, 177, 311, 158, TYR17, 313 |
Note: The hydrogen bond interactions and hydrophobic interctions were observed during the docking study.
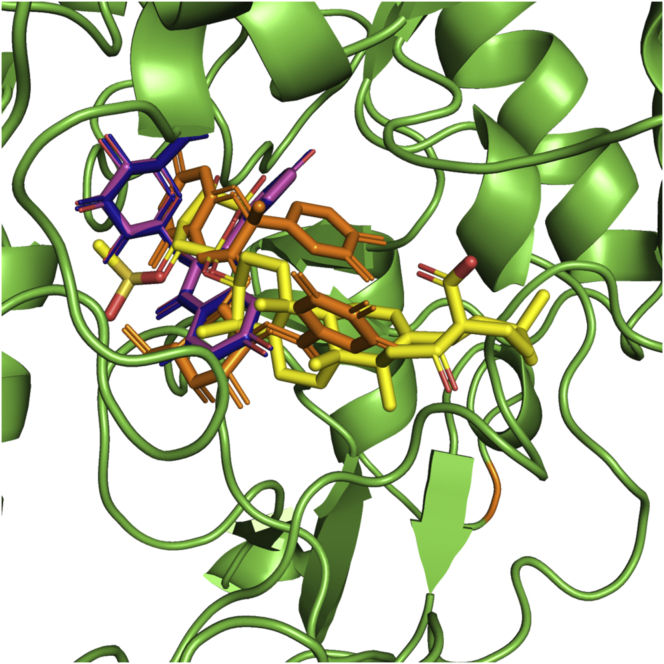
Similarly, the kaempferol moieties of the compounds 3 and 4 make hydrogen bond interactions with the residues THR215, PRO309, ARG312. The glucoside and pentoside moieties of both the compound are involved in hydrogenbond interaction with ARG312, ASP349, 408, TYR313 and GLN350 as represented in Figure 13A and Figure 13C. Further, both the compounds were stabilized in the active site pocket due to hydrophobic interactions with residues PHE157, 158, 300, 177, 311 as represented in Figure 13B, Figure13D and Table 6. Figure 14 represents the superimposition of the docked compounds in the active site pocket of the enzyme. Kaempferol, glucoside moieties of the compounds 3 and pentoside moiety of compound 4 were perfectly superimposed to each other, which indicates the similar kind of interactions of both the compounds with the enzyme. Also, the docking study was performed with antidiabetic drug acarbose represented in Figure 15 and Table 6. The acarvosine ring showed hydrogen bond interactions with residues PHE310, PRO309, HIS239 and SER156. The maltose unit occupied the active site pocket with hydrogen bond interaction with residues ARG439, GLU276, THR215, ASP349, HIS279 and ASP214 as represented in Figure 15A. Further, the acarbose was stabilized in the active site pocket with residues PHE310, 300, 157, 177, 311, 158, TYR71 and 313 as represented in Figure 15B and Table 6.
Figure 14.
Superimposition of the docked compounds in the active site of the enzyme. Compounds 1, 2, 3 and 4 were represented in yellow, orange, blue and purple coloured sticks respectively.
Figure 15.
Docking study of acarbose with yeast alpha glucosidase. (A) Stick representation and (B) Surface representation. The hydrogen bond interactions were represented in black coloured broken lines.
In this study docking of the identified compounds reported for the first time. Similar to the in-vitro AGI activities, identified compounds from the ‘fraction-14’ have shown better binding activities when compared to the standard drug acarbose. Though, there are no literature available on docking studies of these compounds, the binding energy of compounds 2, 3 and 4 with the yeast alpha glucosdiase are very promising. If the identified compounds preserve an effective binding affinity against human alpha glucosidase enzyme similar to this study, there would be an opportunity to develop these molecules into a new alpha glucosidase inhibitor.
Quassinoids and other compounds like kaempferol, amarolide, saponin, etc. have been isolated and reported in Simaroubaceae family for different biological activities including antidiabetic activity (Alves et al., 2014). But, the detailed study on antidiabetic properties of isolated compounds from Simaroubaceae family are lacking except for few studies on Quassia amara (Husain et al., 2011) and Brucea javanica plants (NoorShahida et al., 2009). In the present study, alpha glucosidase inhibition activity of the purified ‘fraction-14’ was analysed using in-vitro, in-silico as well as kinetic study indicates that there are effective inhibitors present in the S. glauca leaf extract. This study would be a inspiration for further detailed studies on these identified compounds to develop them as a novel natural alpha glucosidase inhibitor.
4. Conclusion
To the best of our knowledge, the detailed study on phytochemical analysis, antioxidant activity, AGI activity, kinetics and docking studies of purified phenolic rich fraction from S. glauca leaf was conducted for the first time in this report. The column purified ‘fraction-14’ of S. glauca possess high AGI activity when compared to commercial drug acarbose. The antioxidant activity and high alpha glucosidase inhibition activity of ‘fraction-14’ is comparable to the earlier studies and, the presence of phenolics was confirmed by phytochemical study. The strong AGI activity was proved by the inhibition kinetic study which shows the mixed type of inhibition on the alpha glucosidase enzyme. The superior Ki value than the acarbose suggests that the compounds present in the ‘fraction -14’ would have an effective AGI for the management of T2DM. The LC-MS analysis of purified ‘fraction-14’ lead to the identification of four compounds (compounds 1, 2, 3 and 4), out of them, two compounds are already known for AGI activity and antioxidant activity. The docking study also showed that compounds 2, 3 and 4 have higher binding energy than the known drug acarbose against alpha glucosidase, which indicates the identified compounds are better AGI than the acarbose. The phenolic rich and antioxidant fraction with significant AGI activity is a promising result for its application as a therapeutic agent to treat T2DM. Even though the in-vitro analysis exhibits excellent AGI activity, the implication of in-vivo studies would reveal the potentiality of these molecules before considering for the T2DM treatment.
Declarations
Author contribution statement
Kirana P Mugaranja: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Ananda Kulal: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by Vision Group on Science and Technology (VGST), Govt. of Karnataka, India (CISEE/GRD No. 535/2016-17).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
KMP would like to thank Admar Mutt Education Foundation, Udupi, India for the fellowship and research facility.
References
- Abu-Reidah I.M., Ali-Shtayeh M.S., Jamous R.M., Arráez-Román D., Segura-Carretero A. HPLC–DAD–ESI-MS/MS screening of bioactive components from Rhus coriaria L.(Sumac) fruits. Food Chem. 2015;166:179–191. doi: 10.1016/j.foodchem.2014.06.011. [DOI] [PubMed] [Google Scholar]
- Alam M.A., Zaidul I., Ghafoor K., Sahena F., Hakim M., Rafii M., Abir H., Bostanudin M., Perumal V., Khatib A. In vitro antioxidant and, α-glucosidase inhibitory activities and comprehensive metabolite profiling of methanol extract and its fractions from Clinacanthus nutans. BMC Compl. Alternative Med. 2017;17(181):1–10. doi: 10.1186/s12906-017-1684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves I.A., Miranda H.M., Soares L.A., Randau K.P. Simaroubaceae family: botany, chemical composition and biological activities. Revista Brasileira de Farmacognosia. 2014;24(4):481–501. [Google Scholar]
- Amarowicz R., Pegg R., Rahimi-Moghaddam P., Barl B., Weil J. Free-radical scavenging capacity and antioxidant activity of selected plant species from the Canadian prairies. Food Chem. 2004;84(4):551–562. [Google Scholar]
- Blando F., Calabriso N., Berland H., Maiorano G., Gerardi C., Carluccio M.A., Andersen Ø.M. Radical scavenging and anti-inflammatory activities of representative anthocyanin groupings from pigment-rich fruits and vegetables. Int. J. Mol. Sci. 2018;19(169):1–15. doi: 10.3390/ijms19010169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj S.S., Poojary B., Nandish S.K.M., Kengaiah J., Kirana M.P., Shankar M.K., Das A.J., Kulal A., Sannaningaiah D. Efficient synthesis and in silico studies of the benzimidazole hybrid scaffold with the quinolinyloxadiazole skeleton with potential α-glucosidase inhibitory, anticoagulant, and antiplatelet activities for type-II diabetes mellitus management and treating thrombotic disorders. ACS Omega. 2018;3(10):12562–12574. doi: 10.1021/acsomega.8b01476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S., Khunti K., Davies M.J. Type 2 diabetes. Lancet. 2017;389(10085):2239–2251. doi: 10.1016/S0140-6736(17)30058-2. [DOI] [PubMed] [Google Scholar]
- Chen Y., Wang E., Wei Z., Zheng Y., Yan R., Ma X. Phytochemical analysis, cellular antioxidant, α-glucosidase inhibitory activities of various herb plant organs. Ind. Crop. Prod. 2019;141:111771. [Google Scholar]
- Chen Y., Yu H., Wu H., Pan Y., Wang K., Jin Y., Zhang C. Characterization and quantification by LC-MS/MS of the chemical components of the heating products of the flavonoids extract in pollen typhae for transformation rule exploration. Molecules. 2015;20(10):18352–18366. doi: 10.3390/molecules201018352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho N.H., Shaw J.E., Karuranga S., Huang Y., da Rocha Fernandes J.D., Ohlrogge A.W., Malanda B. IDF Diabetes Atlas: global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res. Clin. Pract. 2018;138:271–281. doi: 10.1016/j.diabres.2018.02.023. [DOI] [PubMed] [Google Scholar]
- Cho Y.J., Woo J.-H., Lee J.-S., Jang D.S., Lee K.-T., Choi J.-H. Eclalbasaponin II induces autophagic and apoptotic cell death in human ovarian cancer cells. J. Pharmacol. Sci. 2016;132(1):6–14. doi: 10.1016/j.jphs.2016.02.006. [DOI] [PubMed] [Google Scholar]
- Dabhi A.S., Bhatt N.R., Shah M.J. Voglibose: an alpha glucosidase inhibitor. J. Clin. Diagn. Res.: J. Clin. Diagn. Res. 2013;7(12):3023–3027. doi: 10.7860/JCDR/2013/6373.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag M.A., Sakna S.T., El-fiky N.M., Shabana M.M., Wessjohann L.A. Phytochemical, antioxidant and antidiabetic evaluation of eight Bauhinia L. species from Egypt using UHPLC–PDA–qTOF-MS and chemometrics. Phytochemistry. 2015;119:41–50. doi: 10.1016/j.phytochem.2015.09.004. [DOI] [PubMed] [Google Scholar]
- Giacco F., Brownlee M. Oxidative stress and diabetic complications. Circ. Res. 2010;107(9):1058–1070. doi: 10.1161/CIRCRESAHA.110.223545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollapalli M., Taha M., Javid M.T., Almandil N.B., Rahim F., Wadood A., Mosaddik A., Ibrahim M., Alqahtani M.A., Bamarouf Y.A. Synthesis of benzothiazole derivatives as a potent α-glucosidase inhibitor. Bioorg. Chem. 2019;85:33–48. doi: 10.1016/j.bioorg.2018.12.021. [DOI] [PubMed] [Google Scholar]
- Gong Y., Qin X.-Y., Zhai Y.-Y., Hao H., Lee J., Park Y.-D. Inhibitory effect of hesperetin on α-glucosidase: molecular dynamics simulation integrating inhibition kinetics. Int. J. Biol. Macromol. 2017;101:32–39. doi: 10.1016/j.ijbiomac.2017.03.072. [DOI] [PubMed] [Google Scholar]
- González-Cabrera M., Domínguez-Vidal A., Ayora-Cañada M. Hyperspectral FTIR imaging of olive fruit for understanding ripening processes. Postharvest Biol. Technol. 2018;145:74–82. [Google Scholar]
- Gouveia-Figueira S.C., Castilho P.C. Phenolic screening by HPLC–DAD–ESI/MSn and antioxidant capacity of leaves, flowers and berries of Rubus grandifolius Lowe. Ind. Crop. Prod. 2015;73:28–40. [Google Scholar]
- Gu C., Yang M., Zhou Z., Khan A., Cao J., Cheng G. Purification and characterization of four benzophenone derivatives from Mangifera indica L. leaves and their antioxidant, immunosuppressive and α-glucosidase inhibitory activities. Journal of Functional Foods. 2019;52:709–714. [Google Scholar]
- Han L., Liu E., Kojo A., Zhao J., Li W., Zhang Y., Wang T., Gao X. Qualitative and quantitative analysis of Eclipta prostrata L. by LC/MS. Sci. World J. 2015;2015:1–15. doi: 10.1155/2015/980890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatier J.-H.B., Gould K.S. Anthocyanins, Springer. 2008. Anthocyanin function in vegetative organs; pp. 1–19. [Google Scholar]
- Husain G.M., Singh P.N., Singh R.K., Kumar V. Antidiabetic activity of standardized extract of Quassia amara in nicotinamide–streptozotocin-induced diabetic rats. Phytother Res. 2011;25(12):1806–1812. doi: 10.1002/ptr.3491. [DOI] [PubMed] [Google Scholar]
- Jiang P., Xiong J., Wang F., Grace M.H., Lila M.A., Xu R. α-Amylase and α-glucosidase inhibitory activities of phenolic extracts from Eucalyptus grandis × E. urophylla bark. J. Chem. 2017;2:1–7. [Google Scholar]
- Jose A., Kannan E., Kumar P.R.A.V., Madhunapantula S.V. Therapeutic potential of phytochemicals isolated from Simarouba glauca for inhibiting cancers: a review. Sys. Rev. Pharm. 2019;10(1):73–80. [Google Scholar]
- Kahn S.E., Cooper M.E., Del Prato S. Pathophysiology and treatment of type 2 diabetes: perspectives on the past, present, and future. Lancet. 2014;383(9922):1068–1083. doi: 10.1016/S0140-6736(13)62154-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.-J., Lee S.-B., Lee H.-S., Lee S.-Y., Baek J.-S., Kim D., Moon T.-W., Robyt J.F., Park K.-H. Comparative study of the inhibition of α-glucosidase, α-amylase, and cyclomaltodextrin glucanosyltransferase by acarbose, isoacarbose, and acarviosine–glucose. Arch. Biochem. Biophys. 1999;371(2):277–283. doi: 10.1006/abbi.1999.1423. [DOI] [PubMed] [Google Scholar]
- Kong J.-M., Chia L.-S., Goh N.-K., Chia T.-F., Brouillard R. Analysis and biological activities of anthocyanins. Phytochemistry. 2003;64(5):923–933. doi: 10.1016/s0031-9422(03)00438-2. [DOI] [PubMed] [Google Scholar]
- Lee D.W. Anthocyanins in leaves: distribution, phylogeny and development. Adv. Bot. Res. 2002;37:37–53. [Google Scholar]
- Lee D.W., Collins T.M. Phylogenetic and ontogenetic influences on the distribution of anthocyanins and betacyanins in leaves of tropical plants. Int. J. Plant Sci. 2001;162(5):1141–1153. [Google Scholar]
- Manasi P.S., Gaikwad D. A critical review on medicinally important oil yielding plant laxmitaru (Simarouba glauca DC.) J. Pharmaceut. Sci. Res. 2011;3(4):1195–1213. [Google Scholar]
- Manetas Y., Drinia A., Petropoulou Y. High contents of anthocyanins in young leaves are correlated with low pools of xanthophyll cycle components and low risk of photoinhibition. Photosynthetica. 2002;40(3):349–354. [Google Scholar]
- Matthäus B. Antioxidant activity of extracts obtained from residues of different oilseeds. J. Agric. Food Chem. 2002;50(12):3444–3452. doi: 10.1021/jf011440s. [DOI] [PubMed] [Google Scholar]
- Medini F., Fellah H., Ksouri R., Abdelly C. Total phenolic, flavonoid and tannin contents and antioxidant and antimicrobial activities of organic extracts of shoots of the plant Limonium delicatulum. Journal of Taibah University for science. 2014;8(3):216–224. [Google Scholar]
- Mitrakou A., Tountas N., Raptis A., Bauer R., Schulz H., Raptis S. Long-term effectiveness of a new α-glucosidase inhibitor (BAY m1099-miglitol) in insulin-treated Type 2 diabetes mellitus. Diabet. Med. 1998;15(8):657–660. doi: 10.1002/(SICI)1096-9136(199808)15:8<657::AID-DIA652>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Mollica A., Zengin G., Locatelli M., Stefanucci A., Macedonio G., Bellagamba G., Onaolapo O., Onaolapo A., Azeez F., Ayileka A. An assessment of the nutraceutical potential of Juglans regia L. leaf powder in diabetic rats. Food Chem. Toxicol. 2017;107:554–564. doi: 10.1016/j.fct.2017.03.056. [DOI] [PubMed] [Google Scholar]
- NoorShahida A., Wong T.W., Choo C.Y. Hypoglycemic effect of quassinoids from Brucea javanica (L.) Merr (Simaroubaceae) seeds. J. Ethnopharmacol. 2009;124(3):586–591. doi: 10.1016/j.jep.2009.04.058. [DOI] [PubMed] [Google Scholar]
- Osagie-Eweka S.D.E., Orhue N.J., Ekhaguosa D.O. Comparative phytochemical analyses and in-vitro antioxidant activity of aqueous and ethanol extracts of Simarouba glauca (paradise tree) Eur. J. Med. Plants. 2016;13(3) [Google Scholar]
- Peng X., Zhang G., Liao Y., Gong D. Inhibitory kinetics and mechanism of kaempferol on α-glucosidase. Food Chem. 2016;190:207–215. doi: 10.1016/j.foodchem.2015.05.088. [DOI] [PubMed] [Google Scholar]
- Picot M.C., Zengin G., Mollica A., Stefanucci A., Carradori S., Mahomoodally M. In vitro and in silico studies of mangiferin from Aphloia theiformis on key enzymes linked to diabetes type 2 and associated complications. Med. Chem. 2017;13(7):633–640. doi: 10.2174/1573406413666170307163929. [DOI] [PubMed] [Google Scholar]
- Pitura K., Arntfield S.D. Characteristics of flavonol glycosides in bean (Phaseolus vulgaris L.) seed coats. Food Chem. 2019;272:26–32. doi: 10.1016/j.foodchem.2018.07.220. [DOI] [PubMed] [Google Scholar]
- Pradeep P., Sreerama Y.N. Phenolic antioxidants of foxtail and little millet cultivars and their inhibitory effects on α-amylase and α-glucosidase activities. Food Chem. 2018;247:46–55. doi: 10.1016/j.foodchem.2017.11.103. [DOI] [PubMed] [Google Scholar]
- Puranik S.I., Ghagane S.C., Nerli R.B., Jalalpure S.S., Hiremath M.B. Evaluation of in vitro antioxidant and anticancer activity of Simarouba glauca leaf extracts on T-24 bladder cancer cell line. Phcog. J. 2017;9(6):906–912. [Google Scholar]
- Ray A., Bharali P., Konwar B. Mode of antibacterial activity of eclalbasaponin isolated from Eclipta alba. Appl. Biochem. Biotechnol. 2013;171(8):2003–2019. doi: 10.1007/s12010-013-0452-3. [DOI] [PubMed] [Google Scholar]
- Rouzbehan S., Moein S., Homaei A., Moein M.R. Kinetics of α-glucosidase inhibition by different fractions of three species of Labiatae extracts: a new diabetes treatment model. Pharmaceut. Biol. 2017;55(1):1483–1488. doi: 10.1080/13880209.2017.1306569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhosh S.K., Venugopal A., Radhakrishnan M.C. Study on the phytochemical, antibacterial and antioxidant activities of Simarouba glauca. South Indian Journal of Biological Sciences. 2016;2(1):119-124. [Google Scholar]
- Sarikurkcu C., Eskici M., Karanfil A., Tepe B. Phenolic profile, enzyme inhibitory and antioxidant activities of two endemic Nepeta species: Nepeta nuda subsp. glandulifera and N. cadmea. South Afr. J. Bot. 2019;120:298–301. [Google Scholar]
- Sekar V., Chakraborty S., Mani S., Sali V., Vasanthi H. Mangiferin from Mangifera indica fruits reduces post-prandial glucose level by inhibiting α-glucosidase and α-amylase activity. South Afr. J. Bot. 2019;120:129–134. [Google Scholar]
- Shah M.A., Khalil R., Ul-Haq Z., Panichayupakaranant P. α-Glucosidase inhibitory effect of rhinacanthins-rich extract from Rhinacanthus nasutus leaf and synergistic effect in combination with acarbose. Journal of Functional Foods. 2017;36:325–331. [Google Scholar]
- Sim L., Quezada-Calvillo R., Sterchi E.E., Nichols B.L., Rose D.R. Human intestinal maltase–glucoamylase: crystal structure of the N-terminal catalytic subunit and basis of inhibition and substrate specificity. J. Mol. Biol. 2008;375(3):782–792. doi: 10.1016/j.jmb.2007.10.069. [DOI] [PubMed] [Google Scholar]
- Şöhretoğlu D., Sari S., Šoral M., Barut B., Özel A., Liptaj T. Potential of Potentilla inclinata and its polyphenolic compounds in α-glucosidase inhibition: kinetics and interaction mechanism merged with docking simulations. Int. J. Biol. Macromol. 2018;108:81–87. doi: 10.1016/j.ijbiomac.2017.11.151. [DOI] [PubMed] [Google Scholar]
- Song Y., Jeong S.W., Lee W.S., Park S., Kim Y.-H., Kim G.-S., Lee S.J., Jin J.S., Kim C.-Y., Lee J.E. Determination of polyphenol components of Korean prostrate spurge (Euphorbia supina) by using liquid chromatography—tandem mass spectrometry: overall contribution to antioxidant activity. Journal of analytical methods in chemistry. 2014;418690:1–8. doi: 10.1155/2014/418690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack D., Wray V., Metzger J.W., Grosse W. Two anthocyanins acylated with gallic acid from the leaves of Victoria amazonica. Phytochemistry. 1992;31(3):989–991. [Google Scholar]
- Süzgeç S., Meriçli A.H., Houghton P.J., Çubukçu B. Flavonoids of Helichrysum compactum and their antioxidant and antibacterial activity. Fitoterapia. 2005;76(2):269–272. doi: 10.1016/j.fitote.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Tan Y., Chang S.K. Digestive enzyme inhibition activity of the phenolic substances in selected fruits, vegetables and tea as compared to black legumes. Journal of functional foods. 2017;38:644–655. [Google Scholar]
- Umesh T. In-vitro antioxidant potential, free radical scavenging and cytotoxic activity of Simarouba gluaca leaves. Int. J. Pharm. Pharmaceut. Sci. 2015;7(2):411–416. [Google Scholar]
- Van De Laar F.A., Lucassen P.L., Akkermans R.P., Van De Lisdonk E.H., Rutten G.E., Van Weel C. α-Glucosidase inhibitors for patients with type 2 diabetes: results from a Cochrane systematic review and meta-analysis. Diabetes Care. 2005;28(1):154–163. doi: 10.2337/diacare.28.1.154. [DOI] [PubMed] [Google Scholar]
- Vanessa Fiorentino T., Prioletta A., Zuo P., Folli F. Hyperglycemia-induced oxidative stress and its role in diabetes mellitus related cardiovascular diseases. Curr. Pharmaceut. Des. 2013;19(32):5695–5703. doi: 10.2174/1381612811319320005. [DOI] [PubMed] [Google Scholar]
- Wang G., Peng Z., Wang J., Li X., Li J. Synthesis, in vitro evaluation and molecular docking studies of novel triazine-triazole derivatives as potential α-glucosidase inhibitors. Eur. J. Med. Chem. 2017;125:423–429. doi: 10.1016/j.ejmech.2016.09.067. [DOI] [PubMed] [Google Scholar]
- Wang W., Yao G.-D., Shang X.-Y., Gao J.-C., Zhang Y., Song S.-J. Eclalbasaponin I from Aralia elata (Miq.) Seem. reduces oxidative stress-induced neural cell death by autophagy activation. Biomed. Pharmacother. 2018;97:152–161. doi: 10.1016/j.biopha.2017.10.106. [DOI] [PubMed] [Google Scholar]
- Wei Y., Xie Q., Fisher D., Sutherland I.A. Separation of patuletin-3-O-glucoside, astragalin, quercetin, kaempferol and isorhamnetin from Flaveria bidentis (L.) Kuntze by elution-pump-out high-performance counter-current chromatography. J. Chromatogr. A. 2011;1218(36):6206–6211. doi: 10.1016/j.chroma.2011.01.058. [DOI] [PubMed] [Google Scholar]
- Yan S., Shao H., Zhou Z., Wang Q., Zhao L., Yang X. Non-extractable polyphenols of green tea and their antioxidant, anti-α-glucosidase capacity, and release during in vitro digestion. Journal of Functional Foods. 2018;42:129–136. [Google Scholar]
- Yin Z., Zhang W., Feng F., Zhang Y., Kang W. α-Glucosidase inhibitors isolated from medicinal plants. Food Science and Human Wellness. 2014;3(3):136–174. [Google Scholar]