Abstract
Eosinophilic esophagitis (EoE) is a chronic, food antigen–driven, inflammatory disease of the esophagus and is associated with impaired barrier function. Evidence is emerging that loss of esophageal expression of the serine peptidase inhibitor, kazal type 7 (SPINK7), is an upstream event in EoE pathogenesis. Here, we provide evidence that loss of SPINK7 mediates its pro-EoE effects via kallikrein 5 (KLK5) and its substrate, protease-activated receptor 2 (PAR2). Overexpression of KLK5 in differentiated esophageal epithelial cells recapitulated the effect of SPINK7 gene silencing, including barrier impairment and loss of desmoglein-1 expression. Conversely, KLK5 deficiency attenuated allergen-induced esophageal protease activity, modified commensal microbiome composition, and attenuated eosinophilia in a murine model of EoE. Inhibition of PAR2 blunted the cytokine production associated with loss of SPINK7 in epithelial cells and attenuated the allergen-induced esophageal eosinophilia in vivo. Clinical samples substantiated dysregulated PAR2 expression in the esophagus of patients with EoE, and delivery of the clinically approved drug α1 antitrypsin (A1AT, a protease inhibitor) inhibited experimental EoE. These findings demonstrate a role for the balance between KLK5 and protease inhibitors in the esophagus and highlight EoE as a protease-mediated disease. We suggest that antagonizing KLK5 and/or PAR2 has potential to be therapeutic for EoE.
INTRODUCTION
Eosinophilic esophagitis (EoE) is an emerging, chronic, esophageal disease characterized by type 2 (allergic) immunologic responses to food, leading to esophageal-specific eosinophilic inflammation associated with a variety of symptoms, including failure to thrive, chest and abdominal pain, persistent heartburn, vomiting, dysphagia, and food impactions (1-3). EoE is currently treated with food elimination dietary therapy, proton pump inhibitor therapy, and topical glucocorticoids, but a substantial subset of patients have EoE that does not respond to these treatments, and EoE reoccurs nearly universally after cessation of any therapy (4).
We recently reported that the esophagus is a rich source of proteases and protease inhibitors, and we have shown that these molecules are part of a normal homeostatic surveillance mechanism (5-7). Furthermore, a local deficiency of the serine peptidase inhibitor, kazal type 7 (SPINK7) unleashes uncontrolled proteolytic activity and proinflammatory responses in esophageal epithelial cells; these responses are responsible for the propagation of EoE, at least in part. Here, we have uncovered that the extracellular serine protease kallikrein 5 (KLK5) is a direct target of SPINK7. In the skin, KLK5 has several roles, including the regulation of skin barrier function by processing mucins, antimicrobial peptides, and barrier proteins such as desmoglein-1 (DSG1) (8-10). Decreased expression of DSG1 occurs in EoE and is sufficient to induce barrier impairment and expression of proinflammatory genes in esophageal epithelial cells. In support of this central mechanism, rare, homozygous mutations in DSG1 are sufficient to develop EoE (11, 12).
KLK5 has the potential to serve as not only a degradative enzyme but also a proinflammatory signaling molecule by cleaving and activating the protease-activated receptor 2 (PAR2) (13, 14), which leads to the production of thymic stromal lymphopoietin (TSLP) (13, 15), a key immune checkpoint cytokine involved in stimulating dendritic cells to polarize the generation of type 2 T cells (TH2), key cells involved in EoE pathogenesis (16). TSLP is overproduced in EoE, and genetic variants in its gene modify EoE risk (17-19). In this study, we depleted KLK5 in vivo or inhibited KLK5 activity by delivering a protease inhibitor to the esophagus. We also used PAR2 antagonism to determine how these factors interact in EoE and how they could potentially be targeted for therapeutic benefit.
RESULTS
Expression, function, and regulation of KLKs
We recently reported that loss of SPINK7 in esophageal epithelial cells impairs barrier function and induces innate immune responses (6). We hypothesized that SPINK7 mediates its function, at least in part, by controlling the activity of serine proteases. An esophageal epithelial progenitor cell line (EPC2) and primary esophageal cells were stably transduced with a vector expressing either SPINK7-targeting short hairpin RNA (shRNA) or nonsilencing control shRNA to deplete SPINK7 expression. In addition, SPINK7 was depleted by generation of clustered regularly interspaced short palindromic repeats (CRISPR)–Cas9 genetic deletion of SPINK7 in EPC2 cells (6). The cells were grown at the air-liquid interface (ALI) to allow cellular differentiation (11). The trypsin-like activity of differentiated SPINK7-depleted cells was elevated by twofold (P ≤ 0.004; Fig. 1A). As a control, loss of SPINK7 did not modify expression of serine proteases like KLKs (fig. S1A), consistent with the effect being mediated by SPINK7 inhibition rather than changes in KLK expression. Because most of the members of the KLK family have trypsin-like activity and given their established role in skin homeostasis, we tested whether this family of proteins were SPINK7 targets. We first analyzed KLK expression in the esophagus and revealed that eight members of this family were readily detected (fig. S1B). KLK1, which was not expressed in non-EoE patients (control patients), was expressed in the esophagus of patients with EoE (fig. S1B); however, the overall expression of these eight KLKs was not significantly different in patients with EoE compared to control individuals (fig. S1B). We then performed a functional screen to reveal whether any of these KLKs were inhibited by SPINK7. Recombinant SPINK7 dose dependently inhibited the activity of KLK5 [inhibition constant (Ki) =130 nM], as well as KLK12, although less potently (Ki = 1500 nM) (Fig. 1B and fig. S1B). SPINK7 was 11.5-fold more potent toward KLK5 than KLK12 (Fig. 1B and fig. S1B). In contrast, SPINK7 did not inhibit KLK7, KLK11, or KLK13 (fig. S1, B and C). Structural analysis of SPINK7 and KLK5 using ClusPro predicted an interaction between the inhibitory loop of SPINK7 and the catalytic triad of KLK5 (Fig. 1C) by using an initial rotational screening of SPINK7 (resulting in ~109 conformations); the 1000 highest-scored conformations were clustered together (C-alpha root mean square deviation < 9 Å), and Fig. 1C shows the result of the largest cluster. Further, interaction of KLK5 and SPINK7 was analyzed using two other docking algorithms. These three computational applications all positioned the inhibitory loop of SPINK7 in close association to the serine residue of KLK5 in the catalytic triad (about 2.5 Å fig. S1D).
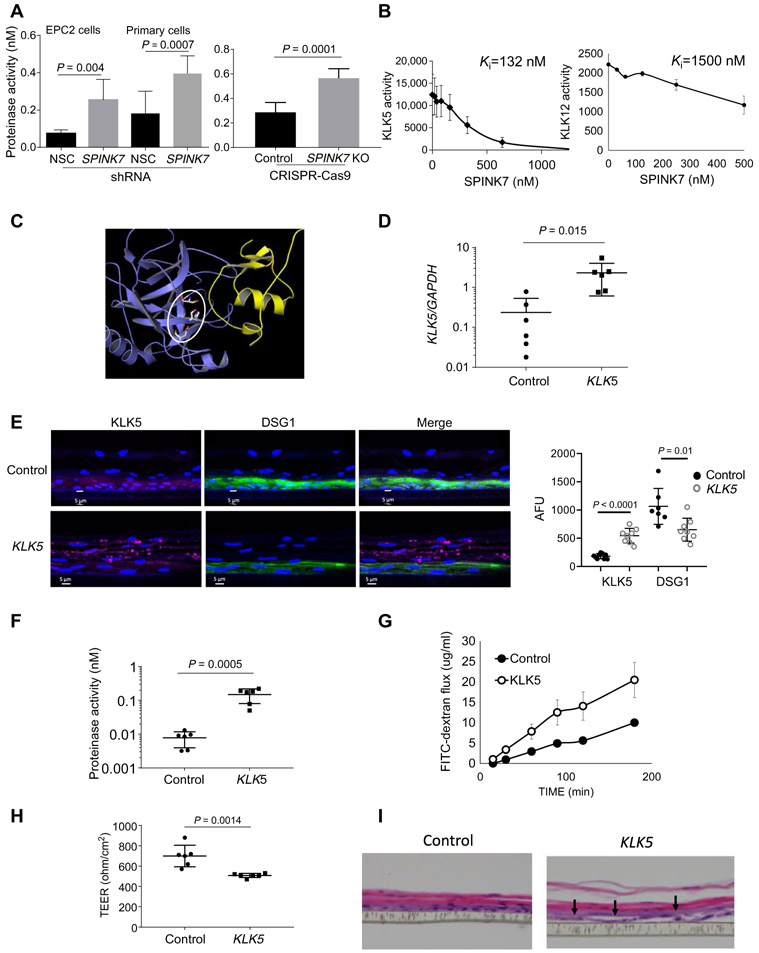
Fig. 1. Identification of SPINK7 targets and their function.
(A) Quantification of trypsin-like activity in supernatants derived from differentiated nonsilencing control (NSC) shRNA– or SPINK7 shRNA–treated EPC2 cells or primary esophageal cells or from control or CRISPR-Cas9 SPINK7 knockout (KO) EPC2 cells. Protease activity was quantitated by comparison to the activity of serial dilutions of KLK5. P values of SPINK7-depleted cells compared with control cells were calculated by t test (unpaired, two-tailed). Data shown from three independent experiments performed in triplicates. (B) KLK5 or KLK12 were incubated with their substrates and a wide range of doses of SPINK7. The proteolytic rate for each dose of SPINK7 was calculated. Results are the means ± SD. Values of inhibition constants (Ki) were calculated according to the Morrison equation (65). Data shown from three independent experiments performed in duplicates. (C) Docking results of SPINK7 (yellow), KLK5 (blue), and the catalytic triad of KLK5 in gray lines (circled) obtained from ClusPro (https://cluspro.bu.edu). The PDB files for SPINK7 (PDBID:2LEO chain A) and KLK5 (PDBID:2PSX chain A) were used with the default ClusPro settings. This represents the top result with balanced weighting. The images were generated with the program PyMOL (66). (D) Quantitative polymerase chain reaction analysis of KLK5 expression in the plasmid PLX304 (control) and KLK5-PLX304–transfected cells after lentivirus transduction. (E) Immunofluorescence of KLK5 (magenta) and desmoglein-1 (DSG1) (green) in PLX304 (control) and KLK5-PLX304–transfected EPC2 cells after differentiation. The graph on the right represents the mean fluorescent intensity of KLK5 and DSG1 in control and KLK5-overexpressing cells. Each dot represents the mean fluorescent intensity of one section. (F) Quantification of trypsin-like activity in supernatants derived from differentiated control or KLK5-overexpressing EPC2 cells. (G) A representative FITC-dextran flux that was measured at day 14 of air-liquid interface (ALI) differentiation from control and KLK5-overexpressing EPC2 cells at the indicated time points. (H) Transepithelial electrical resistance (TEER; ohm/cm2) measurement from control and KLK5-overexpressing EPC2 cells at day 9 of ALI differentiation. (I) Hematoxylin and eosin–stained sections of control or KLK5-overexpressing EPC2 cells after ALI differentiation (day 14). Arrows point to intercellular dilated spaces. Data in (A), (B), and (G) are the means ± SD (n ≥ 3). P values of KLK5 overexpressing cells compared with control cells in (D) to (H) were calculated by t test (unpaired, two-tailed). Data shown in (D) to (H) derived from three independent experiments performed in duplicates (D to H).
We investigated the effects of KLK5 overexpression in differentiated EPC2 cells. Lentivirus-mediated overexpression of KLK5 increased KLK5 mRNA expression by 10-fold compared to control cells (P = 0.015; Fig. 1D). KLK5 protein was also increased in KLK5-overexpressing cells compared to control cells (Fig. 1E). In addition, DSG1 protein expression was decreased by 33% in the KLK5-overexpressing cells compared to control cells (P = 0.01; Fig. 1E). The trypsin-like activity of the KLK5-overexpressing cells increased by 20-fold (P = 0.0005) compared to control cells (Fig. 1F). Overexpression of KLK5 impaired the barrier function as assessed by transcellular permeability, measured by the flux of macromolecules [fluorescein isothiocyanate (FITC)–dextran] and the transepithelial electrical resistance (Fig. 1, G and H). In addition, increased dilated intracellular spaces were observed in the differentiated, KLK5-overexpressing cells compared to control cells (Fig. 1I).
Impaired esophageal allergic inflammation in Klk5−/− mice
We generated Klk5−/− mice by CRISPR-Cas9–mediated genetic engineering as described in fig. S2 (A to C). Quantitative reverse transcription polymerase chain reaction demonstrated undetectable expression of Klk5 mRNA in the Klk5−/− esophagus (fig. S2D). The Klk5−/− mice were viable and fertile, consistent with previous strains of klk5-deficient mice (20, 21). Analysis of KLK5 expression revealed the highest expression of KLK5 was in the skin, pancreas, esophagus, tongue, uterus, and, to a lesser extent, in the brain and thymus (fig. S2E).
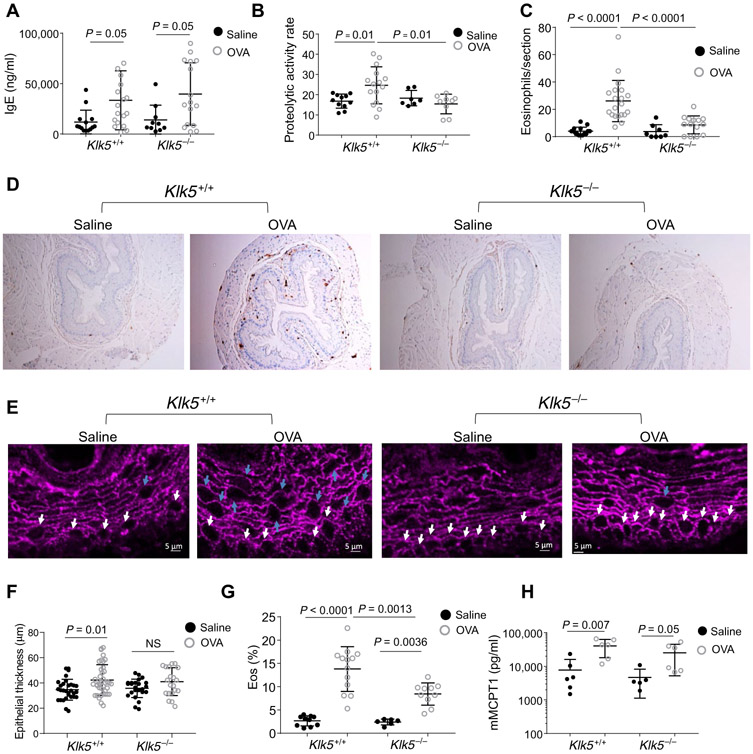
We induced allergic inflammation via a chicken egg albumin (OVA) model, as this is an established model and OVA does not contain proteolytic activity that may be inhibited by the protease inhibitors being examined. Mice were sensitized by two intraperitoneal injections of OVA and the adjuvant aluminum hydroxide. Intranasal allergen increased the expression of serum immunoglobulin E (IgE) in the OVA-treated mice compared to control saline-treated mice in both Klk5+/+ and Klk5−/− genotypes (Fig. 2A). Trypsin-like activity in the esophagus of the OVA-treated Klk5+/+ mice was increased by 50% compared to that of saline-treated wild-type mice but not OVA-treated Klk5−/− mice (P = 0.025; Fig. 2B), indicating a substantial contribution of KLK5 to the allergen-induced esophageal protease activity. Quantification of eosinophils, measured by major basic protein (MBP)+ cells demonstrated readily detectable eosinophils in the esophagus of the OVA-treated mice compared to saline-treated mice (P < 0.0001; Fig. 2, C and D). The number of eosinophils in the esophagus was decreased by threefold in OVA-treated Klk5−/− mice compared to OVA-treated Klk5+/+ mice (P = 0.0003; Fig. 2, C and D). Analysis of the esophageal epithelium using a confocal microscopy analysis revealed alterations in epithelial shape in the OVA-treated Klk5+/+ mice (Fig. 2E). Round (cuboidal) epithelial cells were localized to the basal epithelium in saline-treated Klk5+/+ mice but to the basal epithelium and suprabasal epithelium in OVA-treated Klk5+/+ mice (Fig. 2E). In addition, the epithelial thickness was increased in OVA-treated Klk5+/+ mice compared to control mice (Fig. 2F). In contrast, the epithelial thickness of the OVA-treated Klk5−/− mice was comparable to that of the saline-treated mice Klk5−/− mice and saline-treated Klk5+/+ mice (Fig. 2, E and F). OVA-treated Klk5−/− mice had a 40% decrease in the percent of eosinophils in the blood compared to that of OVA-treated Klk5+/+ mice (8.4 and 13.8%, respectively; P = 0.0013; Fig. 2G). The reduction of blood eosinophils was only seen after OVA challenge (Fig. 2G). In contrast to the decreased eosinophil number in the blood of the Klk5−/− mice, blood mast cell protease 1 (MCPT1, a marker of mast cell load and degranulation) was comparable between Klk5−/− and Klk5+/+ mice (Fig. 2H). Collectively, these findings demonstrate that esophageal proteolytic activity is induced after allergenic stimuli and that Klk5 depletion decreases the esophageal proteolytic activity and eosinophil accumulation in the esophagus and circulation.
Fig. 2. The effect of klk5 gene deletion on esophageal eosinophilia in murine experimental EoE.
(A) IgE concentration in the serum of the mice after induction of allergic inflammation. (B) Proteolytic activity in the esophagus after allergen challenge. (C) Quantification of eosinophils in the esophagus; data shown are the number of eosinophils per section. (D) Representative images of anti-MBP staining of esophageal sections after allergen challenge. (E) Immunofluorescence staining of murine esophageal sections after allergen challenge. For DSG1 (magenta), representative images of four sections from at least six different mice in each treatment group are presented. White arrows denote round (cuboidal) epithelial cells in the basal layer. Blue arrows denote round (cuboidal) epithelial cells in the suprabasal epithelium. (F) Quantification of esophageal epithelial thickness; data shown are the thickness of the epithelium from the basal cells to the lumen in different areas of the sections from at least six mice in each treatment from two independent experiments. NS, not significant. (G) Percent of eosinophils (Eos; % of circulating white blood cells) as determined by flow cytometry of SiglecF+ and CD11b+ cells in the blood after red blood cell lysis. (H) MCPT1 concentration in the serum of the mice after allergen challenge. Each data point represents a value from a single mouse from three independent experiments with at least three mice per group. Mean values are indicated by a horizontal line; error bars represent SD. P values in (A) to (C) and (F) to (H) were calculated by two-way ANOVA with multiple comparisons.
Delivery of a protease inhibitor to the esophagus
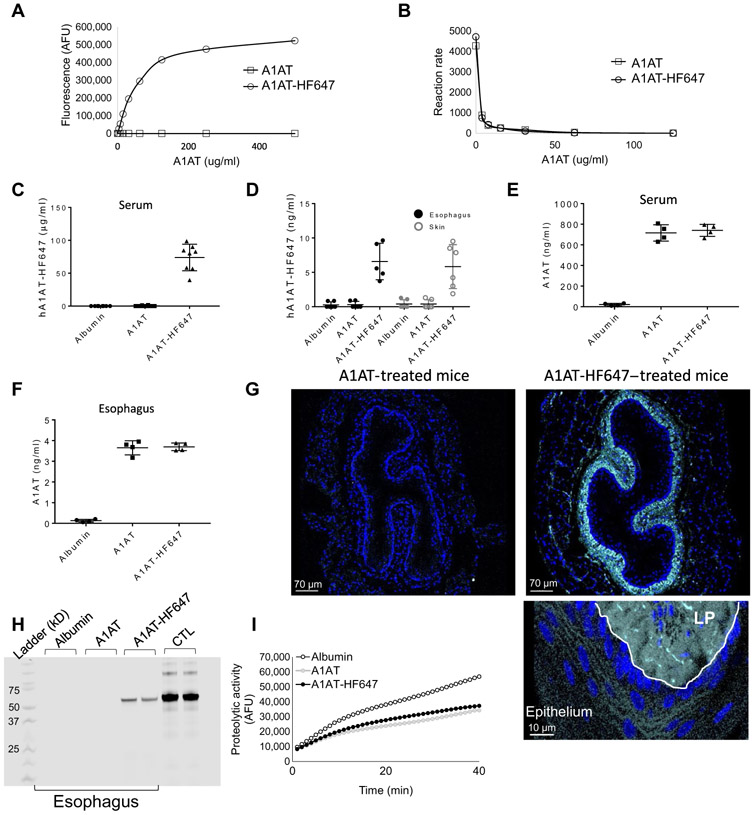
We have previously shown that α1 antitrypsin (A1AT), which is a broad serine protease inhibitor, was able to rescue the effects of SPINK7 deficiency in esophageal epithelial cells in vitro (6). Given that A1AT is a clinically approved drug and readily available, we focused on this antiprotease as a potential therapeutic. A1AT was able to block KLK5 activity in vitro (fig. S3A). We then aimed to establish an experimental framework for delivery of A1AT to the esophagus. To efficiently analyze the delivery of A1AT to various tissues including the esophagus, we conjugated A1AT with the fluorescent tag HiLyte Fluor 647, which produces a bright signal near the infrared spectrum, optimal for in vivo analyses because of low autofluorescence from live tissues. The fluorescent bioconjugates produced a high-fluorescent signal, and no autofluorescence was detected from unconjugated A1AT (Fig. 3A); in addition, the inhibitory activity of the bioconjugates was similar to the unconjugated A1AT (Fig. 3B). We then administered two intraperitoneal injections of 1 mg each of unlabeled A1AT, HiLyte Fluor 647–conjugated A1AT (A1AT-HF647), or human serum albumin as a control (in two consecutive days). Twenty-four hours after the last injection, no fluorescence signal was observed from the serum of albumin-treated mice or A1AT-treated mice (Fig. 3C). In contrast, a high-fluorescent signal was detected from the serum of the A1AT-HF647–treated mice with an estimated concentration of 70 μg/ml of A1AT (Fig. 3C). A1AT-HF647 produced a high-fluorescent signal also in esophageal and skin protein lysates, whereas albumin or nonlabeled A1AT-treated mice did not show any detectable fluorescent signal (Fig. 3D). A human A1AT enzyme-linked immunosorbent assay (ELISA) demonstrated that A1AT was present in the serum and esophagus of A1AT-treated mice but not in the albumin-treated mice (Fig. 3, E and F). Confocal microscopy analysis of the distal esophagus demonstrated a high, near-infrared signal from the A1AT-HF647–treated mice but not from A1AT-treated mice (Fig. 3G). A1AT-HF647 was detectable in all layers of the esophageal wall, including the epithelium, and was most abundant in the lamina propria (Fig. 3G). To validate that the fluorescent signal is from the A1AT bioconjugates and not from degrading products, we performed gel electrophoresis analysis of esophageal lysates, which revealed a fluorescent band in the protein lysates of the A1AT-HF647–treated mice that was the same molecular weight as purified A1AT conjugated to HF647 (A1AT, 55 kDa; HF-647, 1.2 kDa; Fig. 3H). Last, analysis of the trypsin-like activity in esophageal lysates revealed a decreased proteolytic activity in the A1AT and A1AT-HF647–treated mice compared to the albumin-treated mice (Fig. 3I). The trypsin-like proteolytic activity in serum samples was comparable between all groups (fig. S3B) likely because A1AT is normally at high concentrations in the circulation. These collective data indicate that intraperitoneal A1AT is effectively delivered to the esophagus, where it retains inhibitory activity.
Fig. 3. A1AT delivery to the esophagus.
(A) Arbitrary fluorescence unit (AFU) measurements of different concentrations of A1AT and A1AT-HF647. Fluorescent signal was measured by the far-red filter (excitation, 625 nm; emission, 660 to 720 nm) that corresponds to the wavelength of HF647. (B) Measurements of trypsin activity using FITC-casein substrate at the indicated concentrations of A1AT or A1AT-HF647. The proteolytic activity for each concentration of A1AT or A1AT-HF647 was calculated by the average increase in fluorescent intensity per 1 min after serial of 30 measurements in 1-min intervals for a total time of 30 min. (C to I) Mice were intraperitoneally injected twice with 1 mg of human serum albumin or A1AT or A1AT-HF647 for two consecutive days. Twenty-four hours after the last injection, fluorescence in the serum (C) or protein lysates from the esophagus and skin (D) were measured. Concentrations of A1AT were calculated per 1 ml of serum or 1 mg of tissue using serial dilutions of A1AT-HF647. A1AT concentration in the serum (E) and esophageal protein lysates (F) were measured by a human A1AT ELISA. (G) Representative images of esophageal sections from mice that were injected with A1AT or A1AT-HF647. HF647 is shown in cyan, and nuclei are marked in blue by 4′,6-diamidino-2-phenylindole (DAPI). White line separates the LP from the epithelium. (H) After two intraperitoneal injections (1 mg) of albumin, A1AT, or AIAT-HF647, esophageal lysates were electrophoresed in a polyacrylamide gel; 100 ng of A1AT-HF647 was a positive control (CTL). HF647 fluorescence was visualized using ImageStudio software. (I) Proteolytic activity measurement of esophageal protein lysates after two intraperitoneal injections of albumin, A1AT, or AIAT-HF647 (1 mg each) using the substrate BOC-VPR-AMC and 5 nM recombinant human KLK5. Data in (C) to (F) are presented as means ± SD, with data points representing individual mice from three independent experiments with at least two mice per group.
A1AT delivery inhibits eosinophilic responses in a murine EoE model
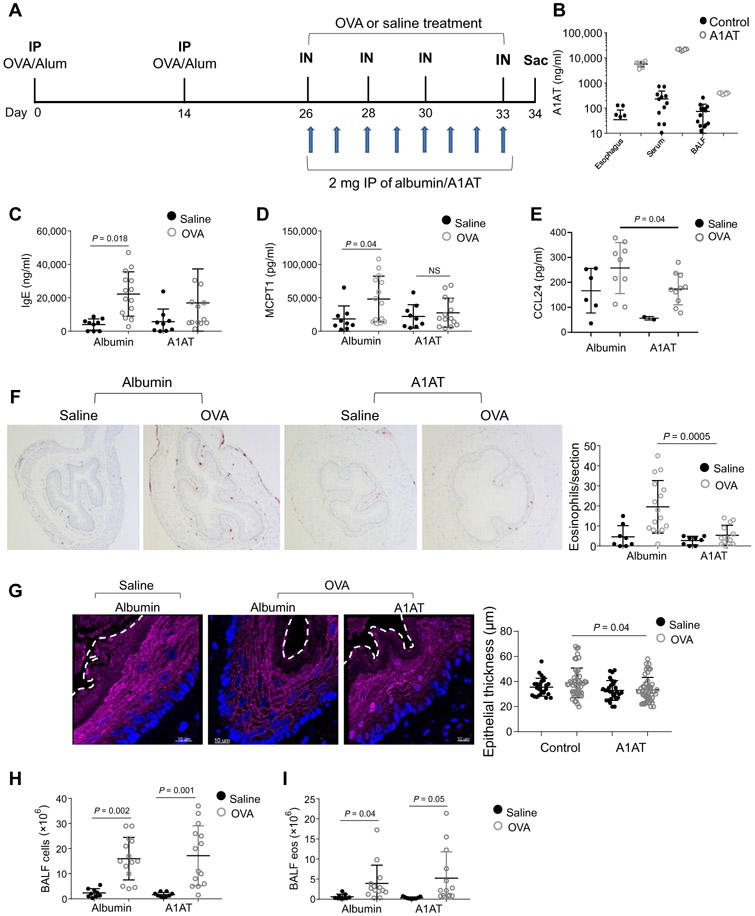
We treated mice with A1AT or control human albumin daily for the last 8 days of the allergen-induced model of esophageal eosinophilia (Fig. 4A). After the OVA challenges (day 34), A1AT concentration was elevated in the A1AT-treated group compared to the control group (albumin-treated mice) in the blood, esophagus, and broncho-alveolar lavage fluid (BALF) (Fig. 4B). Serum IgE concentration was not modified in the A1AT-treated mice compared to the control groups (Fig. 4C). MCPT1 concentrations in the A1AT group were not significantly increased after OVA challenge compared with saline challenge (Fig. 4D). The eosinophil chemoattractants CCL11 and CCL24 concentrations were increased after OVA challenge (Fig. 4E and fig. S3C). CCL24 concentrations in the A1AT group were decreased after OVA challenge (P = 0.04; Fig. 4E), whereas the concentration of CCL11 was unaffected by A1AT (fig. S3C). Consistent with the CCL24 increase, analysis of allergen-induced eosinophil accumulation in the esophagus revealed a 3.6-fold decrease in the A1AT group compared to the control group (P = 0.0005; Fig. 4F). Confocal microscopy analysis of the epithelium revealed that the number of round (cuboidal) epithelial cells and the epithelial thickness of the A1AT-treated group after OVA challenges was decreased compared to the albumin-treated group after OVA challenges (Fig. 4G). Analysis of the BALF revealed that the beneficial effect of A1AT was not seen in the lung (Fig. 4, H and I).
Fig. 4. The effect of A1AT delivery on experimental EoE.
(A) Schematic representations of the EoE model and A1AT or control albumin administration. IP, intraperitoneal injections; IN, intranasal challenges. (B) A1AT concentration in the esophagus, skin, and BALF. A1AT concentrations were determined using a human A1AT ELISA of 1 mg of tissue or 1 ml of BALF. (C) IgE concentration in the serum. (D) MCPT1 concentration in the serum. (E) CCL24 concentration in the esophagus of mice after induction of allergic inflammation. (F) Representative images of anti-MBP staining in esophageal sections after induction of allergic inflammation. In the right graph, a quantification of eosinophils per section of the esophagus is given. (G) Immunofluorescence staining of murine esophageal sections after allergen-induced EoE model for DSG1 (magenta) and DAPI-stained nuclei (blue); representative images of four sections from six different mice in each treatment group are presented. The white dashed line highlights the boundary between the epithelium and the lumen. In the right graph, each dot represents the thickness of the epithelium in a specific section of the esophagus. (H) Quantification of cell number in the BALF after induction of allergic inflammation. (I) Quantification of eosinophil (eos) number in the BALF after induction of allergic inflammation. In (B) to (H), each data point represents a value from a single mouse from three independent experiments, and means ± SD are indicated by horizontal lines. P values in (C) to (I) were calculated by two-way ANOVA with multiple comparisons. Data in (B) to (I) derived from three independent experiments with at least three mice per group.
We then speculated that patients with A1AT deficiency (A1AD) may be enriched for EoE. Accordingly, we probed the electronic medical record at our hospital for the overlap of both diseases and found a ~7.5-fold enrichment (P < 0.0001) (table S1).
KLK5 modifies essential components of the esophagus including the microbiome
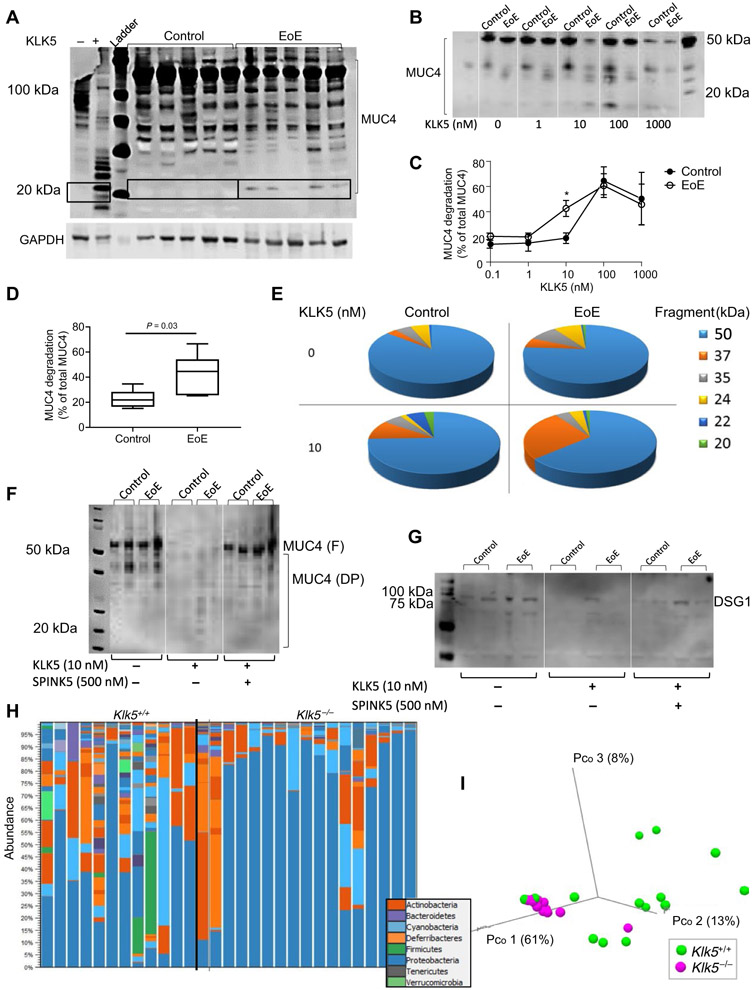
We first tested whether KLK5 had the capacity to digest esophageal proteins that are implicated in barrier maintenance. We focused on mucin proteins, as KLK5 has been shown to be essential for mucin processing (10), although not addressed in the esophagus. In protein lysates from esophageal biopsies, anti-MUC4 antibody stained multiple bands between 50 and 150 kDa that correspond to multiple posttranslational modifications of MUC4. Consistent with our hypothesis that there is a deficiency of KLK5 inhibitors in EoE, analysis of MUC4 revealed a cleavage band of MUC4 that was ~20 kDa and was only present in protein lysates derived from patients with EoE; this cleavage product was missing in all of the control biopsies (Fig. 5A). When protein lysates were incubated at 37°C for 20 hours in the absence of protease inhibitors, anti-MUC4 antibody stained multiple bands between 50 and 100 kDa. In contrast, when protein lysates were incubated at 37°C for 20 hours in the absence of protease inhibitors and with the presence of recombinant human KLK5, anti-MUC4 antibody stained multiple bands that were primarily below 50 kDa. The most notable cleavage product of MUC4 after KLK5 incubation was ~20 kDa (Fig. 5A), which is likely a MUC4 cleavage product that was cleaved by KLK5 as expected (10).
Fig. 5. The effect of KLK5 activity on mucin and DSG1 processing and microbiome homeostasis.
(A) MUC4 protein expression in esophageal biopsies. A representative image of 5 EoE samples and 5 control samples of 11 EoE samples and 10 control samples. The first two lanes from the left represent MUC4 expression from an EoE biopsy that was not treated with protease inhibitor cocktail and incubated at 37°C for 20 hours with recombinant KLK5 (1 μm) (+) or control buffer (−). The right lanes represent MUC4 protein expression in EoE biopsies compared to control biopsies that were treated with protease inhibitor cocktail during protein extraction. (B) Representative image of MUC4 expression in EoE and control biopsies after 20 hours of incubation with the indicated concentrations of recombinant KLK5 at 37°C and in the absence of protease inhibitor cocktail. (C) Quantitative analysis of MUC4 degradation in esophageal biopsies with the indicated concentrations of recombinant KLK5. Results are the means ± SD of three control and three EoE biopsies. *P < 0.05 according to t test (unpaired, two-tailed). (D) Quantitative analysis of the percent of MUC4 degradation from total MUC4. P value was calculated according to t test (unpaired, two-tailed). (E) Percentage of all cleavage bands after 20 hours of incubation at 37°C with or without recombinant KLK5 (10 nM) in the absence of protease inhibitor cocktail from seven control and seven EoE esophageal biopsies. (F) Western blot analysis of MUC4 or (G) DSG1 degradation in esophageal biopsies after 20 hours of incubation at 37°C with or without recombinant KLK5 (10 nM) and with or without SPINK5 (500 nM) in the absence of protease inhibitor cocktail. Degradation products are marked (DP) and full-length protein are marked (F). (H) 16S sequence analysis of phylum abundance of Klk5+/+ and Klk5−/− in the esophagus. (I) Variability analysis of the microbiome of Klk5+/+ and Klk5−/− in the esophagus according to a PERMANOVA weighted test. Each data point represents one mouse. Results are from three independent experiments with at least four mice in each group.
We then hypothesized that the EoE esophageal lysates are more sensitive to KLK5 proteolysis than control esophageal lysates. Protein lysates were incubated in 37°C for 20 hours in the absence of protease inhibitors and with the presence of recombinant human KLK5. Anti-MUC4 antibody stained multiple bands, and the most notable band was 50 kDa. After incubation of 10 nM recombinant human KLK5, the 50 kDa band of MUC4 was decreased in EoE biopsies but not in control biopsies (Fig. 5B). Instead, lower molecular species of MUC4 appeared in EoE biopsies that were absent in control biopsies (Fig. 5B). When biopsies were incubated with 1000 nM recombinant KLK5, all MUC4 species were reduced in control and EoE biopsies (Fig. 5B), indicating that KLK5 mediates degradation of MUC4, which is no longer detectable by the anti-MUC4 antibody. Quantification of MUC4 degradation products revealed a significant increase (P = 0.03) in degrading products in EoE biopsies compared to control biopsies when biopsies were incubated with 10 nM recombinant human KLK5; differences at other concentrations were not statistically significant (Fig. 5C). Incubation of EoE biopsy lysates with 10 nM KLK5 resulted in a twofold increase in the percent of degradation of MUC4 products compared to control biopsies (20 versus 40%, respectively; P = 0.03; Fig. 5D). Figure 5E shows the average percent of each MUC4 degradation product in EoE and control biopsies with or without recombinant KLK5. Conversely, we hypothesized that addition of SPINK would reduce generation of the MUC4-reactive bands. We used recombinant human SPINK5 given its availability, our finding that it efficiently inhibited KLK5 activity in vitro (fig. S4) (22) and its coreduction along with SPINK7 in the esophagus of patients with EoE (6). SPINK5 abolished KLK5-mediated MUC4 degradation (Fig. 5F).
We also analyzed another KLK5 target, DSG1 (9, 11, 12). DSG1 expression was decreased in EoE biopsies compared to control (Fig. 5G). Incubation of esophageal lysates from patients with EoE with KLK5 resulted in cleavage of DSG1, whereas adding SPINK5 abolished this KLK5-mediated DSG1 degradation (Fig. 5G). Collectively, these findings substantiate a role for the KLK5/SPINK axis in regulating essential components of the esophagus including processing of mucus (MUC4) and barrier integrity (DSG1).
Having identified that KLK5 likely regulates esophageal barrier integrity and mucin expression, we hypothesized that the local microbiome in the esophagus would be regulated by KLK5. Analysis of 16S sequencing revealed that the commensal microbiome in the esophagus in the Klk5−/− mice was modified compared to Klk5+/+ mice, with an increase in the portion of Proteobacteria and a decrease in Firmicutes, Bacteroidetes, and Cyanobacteria (Fig. 5H). The abundance of Ruminococcaceae and Lachnospiraceae bacteria, which are increased in the intestine of patients with milk allergy (23), was decreased in the Klk5−/− mice compared to Klk5+/+ mice (table S2). Principal components analysis using a permutational multivariate analysis of variance (PERMANOVA) weighted test revealed distinct clustering of the Klk5−/− mice and Klk5+/+ mice (P = 0.005; Fig. 5I), suggesting that KLK5 regulates host-microbe homeostasis.
KLK5 and PAR2 expression and function in vitro and ex vivo
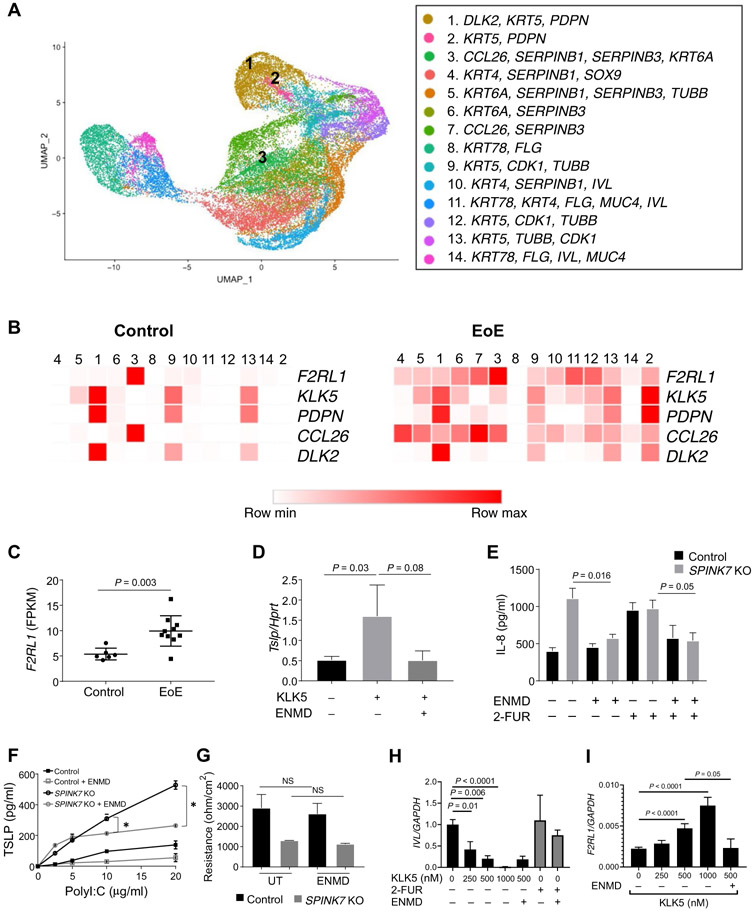
Single-cell RNA sequencing analysis in human esophageal tissue revealed 14 epithelial populations based on principal components analysis (Fig. 6A). Analysis of human esophageal tissue from control patients revealed that the highest expression of KLK5 was in a distinct cell population designated population 1 (Fig. 6, A and B) and that lesser expression was in populations 9 and 13 (Fig. 6B). In patients with EoE, KLK5 expression was dispersed between several cell populations, with the highest expression being in populations 1 and 2 (Fig. 6B and fig. S5A). KLK5+ cells were expressed by clusters that were classified as basal and undifferentiated epithelial cells as defined by DLK2 expression, an inhibitor of NOTCH1 signaling, which regulates esophageal cell growth (5, 24, 25). KLK5 expression was associated with PDPN expression, which is also expressed by basal and undifferentiated epithelial cells (Fig. 6B) (26). The expression of KLK5 was distinct from that of other epithelial KLKs (fig. S5A). In patients with EoE, KLK5 was expressed in additional clusters that were enriched with F2RL1+ cells (cluster 3). F2RL1 (encoding for the KLK5 substrate, PAR2) expression was increased in patients with EoE compared to control individuals (fig. S5B). Cluster 3 was enriched with cells that were actively expressing the eosinophil chemoattractant CCL26 (Fig. 6B). Analysis of the genes that are most enriched in clusters 1, 2, and 3 revealed that these cells are actively expressing mRNAs and proteins that are specialized in transcription regulation, adhesion molecule binding, translation of proteins, and unfolded protein responses (fig. S5C and table S3). Consistent with this, F2RL1 mRNA was overexpressed in RNA isolated from whole biopsies from patients with EoE compared to control individuals (1.85-fold increase, P = 0.0035; Fig. 6C). Collectively, these data support a functional circuit involving KLK5 and PAR2 in the same cellular units, although not exclusively.
Fig. 6. KLK5 and PAR2 expression and function.
(A) Uniform manifold approximation and projection (UMAP) plot displaying single cells, colored by shared nearest neighbor clusters and cell types. (B) Heat maps represent the expression of the indicated genes in epithelial clusters based on single-cell RNA sequencing data of dispersed cells from esophageal EoE biopsies and control biopsies. Data derived from two control and five EoE biopsies and total of 47,141 cells. (C) Fragments per kilobase of transcript per million (FPKM) values for F2LR1 determined from bulk RNA sequencing of esophageal biopsies [n = 6 control patients (Control) and n = 10 patients with active EoE (EoE)]. Mean values are indicated by a horizontal line; error bars represent SD, with each circle or square representing an individual. (D) Tslp expression was examined in murine esophageal explants that were either untreated or treated with KLK5 (800 nM) in the presence or absence of ENMD-1068 (1 mM). Data represent as the means ± SD from three independent experiments. (E) IL-8 protein expression in supernatants from control or SPINK7-KO EPC2 cells that were differentiated in ALI culture (day 14). Cells were treated with either ENMD-1068 (500 μM), 2-FUR (1 μM), or ENMD-1068 (500 μM) 30 min before 2-FUR (1 μM) on day 9 and day 12 of differentiation. P values were calculated by two-way ANOVA with multiple comparisons from four independent experiments performed in duplicates. (F) TSLP release from control or SPINK7-KO EPC2 cells that were grown in high-calcium media and high confluency for 64 hours and then stimulated for 8 hours with the indicated concentrations of polyI:C with or without ENMD (1 mM). Cell supernatants were assessed for TSLP concentrations from three independent experiments performed in duplicates. Data are means ± SD. (G) TEER (ohm/cm2) measurement from EPC2 cells at day 12 of ALI differentiation. After the final 48 hours, cells were either left untreated (UT) or treated with ENMD-1068 (500 μM). Data from three independent experiments performed in triplicates. (H) IVL mRNA expression in EPC2 cells that were grown in high-calcium media and high confluency (250,000 cells per well) for 48 hours and then stimulated for 18 hours with the indicated concentrations of KLK5 or 2-FUR (1 μM) with or without ENMD-1068 (500 μM). (I) F2RL1 mRNA expression in EPC2 cells that were grown in high-calcium media and high confluency (250,000 cells per well) for 48 hours and then stimulated for 18 hours with the indicated concentrations of KLK5 with or without ENMD-1068 (500 μM). Data in (H) and (I) are from three independent experiments performed in triplicates. (D) to (I) represent the mean values, and error bars represent SD. P values in (C) and (F) to (I) were calculated by t test (unpaired, two-tailed).
We subsequently tested the role of KLK5 and PAR2 in murine esophageal explants ex vivo by inhibiting KLK5-stimulated PAR2 activation with a selective PAR2 antagonist, 6-amino-1-[4-(3-methyl-1-oxobutyl)-1-piperazinyl]-1-hexanone hydrochloride (ENMD-1068). KLK5 stimulated TSLP production, whereas ENMD-1068 inhibited the KLK5-induced TSLP production from the esophageal explants (Fig. 6D).
Because SPINK7 is an upstream regulator of KLK5, we investigated whether PAR2 activation had a role in the phenotypes induced by loss of SPINK7. We investigated the effects of ENMD-1068 in a SPINK7 CRISPR-Cas9 gene-deleted human epithelial cell line. ENMD-1068 inhibited interleukin-8 (IL-8) release from SPINK7 knockout cells and the release of IL-8 after PAR2 activation using the PAR2 agonist 2-Furoyl-LIGRLO-amide (2-FUR) (Fig. 6E). ENMD-1068 inhibited polyI:C-induced TSLP production by SPINK7 knockout EPC2 cells (Fig. 6F). ENMD-1068 did not affect the barrier function of differentiated SPINK7 knockout nor control EPC2 cells, indicating roles for PAR2 in regulating distinct SPINK7 responses (Fig. 6G). We analyzed the effect of KLK5 and PAR2 activation and inhibition on epithelial differentiation by analyzing the expression of the differentiation marker IVL (involucrin-encoding gene). We found that KLK5 dose dependently inhibited IVL expression, whereas 2-FUR and ENMD-1068 did not affect IVL expression (Fig. 6H). We suggest that KLK5 activity can inhibit epithelial cell differentiation, and this may occur independent of PAR2 activation consistent with the lack of effect of PAR2 inhibition on epithelial barrier function (Fig. 6G). KLK5 stimulation dose dependently increased F2RL1 expression and ENMD-1068 inhibited the KLK5-mediated F2RL1 increase (Fig. 6I).
Inhibition of PAR2 attenuates allergen-induced esophageal eosinophilia
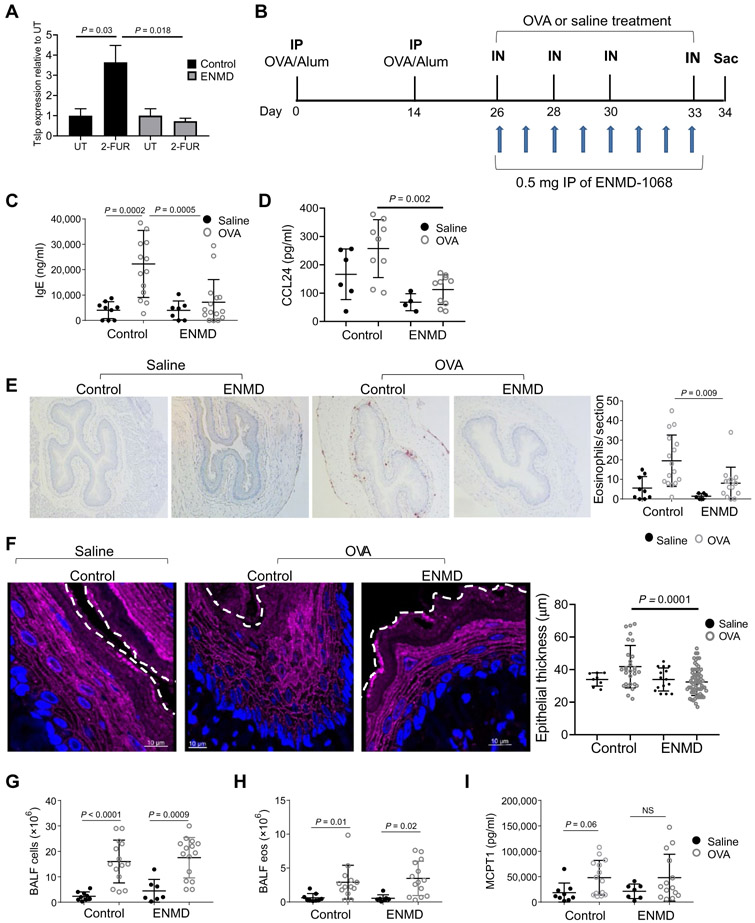
We administered two intraperitoneal injections of 0.5 mg of ENMD-1068 or saline as a control (on two consecutive days) to mice. Twenty-four hours after the last injection, esophageal explants were either left untreated or treated with 2-FUR. Stimulation with 2-FUR increased Tslp production compared to untreated esophageal explants of saline-treated mice (Fig. 7A). In contrast, stimulation with 2-FUR did not induce Tslp production in the esophageal explants of ENMD-1068–treated mice (Fig. 7A). We next blocked PAR2 in vivo using ENMD-1068 (Fig. 7B). ENMD-1068 inhibited IgE production by threefold in OVA-treated compared to control mice (P = 0.0005; Fig. 7C). ENMD-1068 also inhibited esophageal CCL24 production by >2-fold in OVA-treated compared to control mice (P = 0.002; Fig. 7D), whereas CCL11 production was unaffected by ENMD-1068 (fig. S6). Allergen-induced eosinophilia was 2.4-fold less abundant in the esophagus of the ENMD-1068–treated compared to control-treated mice (P = 0.009; Fig. 7E). Analysis of the esophageal epithelium revealed that the epithelial thickness of the ENMD-1068–treated group after OVA challenges was decreased compared to the control group after OVA challenges (Fig. 7F). ENMD-1068 did not affect the accumulation of BALF total cells (Fig. 7G), accumulation of eosinophils (Fig. 7H), nor MCPT1 concentrations in the serum (Fig. 7I).
Fig. 7. The effect of selective inhibition of PAR2 on experimental EoE.
(A) Tslp mRNA expression was examined in murine esophageal explants that were taken from mice that were intraperitoneally injected twice with 0.5 mg of ENMD-1068 or saline. Esophageal explants were either untreated or treated with 2-FUR (2 μM) for 6 hours at 37°C. Data represented as the mean values of three independent experiments; error bars represent SD. P values was calculated by t test (unpaired, two-tailed). (B) Schematic representation of the EoE model and PAR2 antagonist administration. (C) IgE concentration in the serum and (D) CCL24 concentration in esophageal protein lysates after induction of allergic inflammation. (E) Representative image of anti-MBP staining in esophageal sections after induction of allergic inflammation. The graph on the right shows the quantification of MBP+ eosinophils per field in the esophagus. (F) Immunofluorescence staining of murine esophageal sections after induction of allergic inflammation for DSG1 (magenta) and DAPI-stained nuclei (blue); representative images of four sections from six different mice in each treatment group are presented. The white dashed line represents the boundary between the epithelium and the lumen. In the right graph, each dot in the graph represents the thickness of the epithelium in a specific section of the esophagus. (G) Quantification of total cell number in the BALF after induction of allergic inflammation. (H) Quantification of eosinophil numbers in the BALF after induction of allergic inflammation. (I) MCPT1 concentration in the serum of the mice after allergen and saline challenge. In (C) to (I), mean values are indicated by a horizontal line when A each circle represents data point from one mouse; error bars represent SD. P values in (A) and (C) to (I) were calculated by two-way ANOVA with multiple comparisons. Data derived from three independent experiments with at least three mice in each group.
DISCUSSION
The data presented herein identify a dysregulated proteolytic pathway in EoE. We demonstrate that the serine protease KLK5 is a target of SPINK7, a protease inhibitor whose deficiency has a causal role in EoE (6). Overexpression of KLK5 in differentiated esophageal epithelial cells induced epithelial barrier impairment reminiscent to the effect of SPINK7 depletion in differentiated esophageal epithelial cells (6). Here, we show that overexpression of KLK5 impaired the barrier as measured by increased transcellular permeability and decreased expression of the barrier protein DSG1. In addition, we show that A1AT inhibits KLK5 activity in vitro and allergen-induced esophageal eosinophilia in vivo. These findings extend our previous observation that A1AT treatment of SPINK7-deficient esophageal epithelial cells blocks proteolytic activity, restores barrier function, and abolishes cytokine production (6). Therefore, we suggest that loss of SPINKs unleashes uncontrolled KLK5 activity, which is causal, at least in part, in the development of the epithelial barrier dysfunction and the proinflammatory state in EoE (fig. S7). We suggest that A1AT has the potential to be therapeutic in EoE by restricting KLK5 activity in the esophagus.
We focused on KLKs as candidate substrates of SPINK7 based on previous publications that report inhibition of members of KLK family by SPINK proteins (22, 27-31). This is due to the presence of the consensus Kazal domain in the SPINK proteins that is importantly present in SPINK7 (6). We then tested the role of Klk5 in a murine EoE-like model. Repeated intranasal OVA challenge failed to increase esophageal eosinophilia in the Klk5−/− mice, although the mice produced IgE normally and had unabated numbers of eosinophils in the blood at baseline. The proteolytic activity in the esophagus was increased after OVA challenges but was abrogated in the Klk5−/− mice. These data place KLK5 as a nonredundant regulator of esophageal eosinophilia, at least in the setting of this experimental EoE murine model. We showed that KLK5 mRNA expression was not changed between EoE and control esophageal biopsies. However, in EoE biopsies, KLK5 expression was more diffuse and expressed in several epithelial clusters compared with control biopsies, where KLK5 expression was restricted to only three epithelial cell clusters. We suggest that KLK5 is more active in EoE biopsies compared to control biopsies because of loss of its natural inhibitors (i.e., SPINK7 and SPINK5). In support of this hypothesis, we demonstrated that the KLK5 substrate MUC4 is overprocessed in biopsies from patients with EoE compared with control biopsies.
We used two different strategies for intervention in the KLK5 pathway to potentially attenuate EoE in vivo by (i) inhibiting the downstream signaling events of KLK5 activation (i.e., inhibiting PAR2 activation by administering PAR2 antagonist) and (ii) by upstream inhibition of KLK5 activity by delivery of protease inhibitors (i.e., A1AT delivery). Both of these strategies attenuated esophageal eosinophilia. Although A1AT is a nonspecific anti-serine protease, we show that A1AT inhibits KLK5 activity in vitro. The findings are consistent with an effect through KLK5. In addition, we show that A1AT inhibits KLK5 activity in vitro and allergen-induced esophageal eosinophilia in vivo. These findings extend our previous observation that A1AT treatment of SPINK7-deficient esophageal epithelial cells blocks proteolytic activity, restores barrier function, and abolishes cytokine production (6). The A1AT findings provide preclinical proof of concept that protease inhibitors may be therapeutic for EoE. We suggest that A1AT has the potential to be therapeutic in EoE by restricting KLK5 activity in the esophagus.
Our findings show that pharmacological delivery of A1AT is sufficient to correct SPINK7 deficiency in vitro (6) and attenuate experimental EoE in vivo. Our data revealed a ~7.5-fold enrichment of EoE in patients with A1AD. These preliminary findings are consistent with a recent clinical report (32) and call attention to examining this association more deeply in future studies.
KLK5 is implicated in skin desquamation, as it cleaves junctional proteins including DSG1, desmocollin-1, and corneodesmosin (33). DSG1 is down-regulated in EoE, and IL-13 down-regulates DSG1 expression. DSG1 has been shown to be important in the esophageal epithelial barrier (11). Our data reveal that in addition to the IL-13–dependent DSG1 inhibitory pathway, there is a KLK5-dependent pathway that regulates DSG1 posttranslationally by proteolytic degradation in the esophagus. We suggest that KLK5 and possibly other proteases are partially responsible for the decreased DSG1 protein expression in the esophagus.
The importance of KLK5 in barrier integrity and eliciting TH2 responses is illustrated by predisposition to atopy in individuals harboring loss-of-function mutations in the protease inhibitor SPINK5 (termed Netherthon’s syndrome). Homozygous loss of SPINK5 results in uncontrolled KLK5, KLK7, and KLK14 proteolytic activities in the skin, which leads to barrier defects and severe atopy, including EoE (8, 34, 35). Murine modeling of overexpressing human KLK5 in the granular layer of the epidermis exhibits a skin barrier defect involving desmosome cleavage and cytokine production, including TSLP, through activation of PAR2 (13, 36). Consistent with its role in barrier function and type 2 immune responses, Klk5 deletion reverses the Netherthon’s syndrome phenotype in Spink5−/− mice, at least in part (20, 21). SPINK5 and SPINK7 are both highly expressed in the esophageal epithelium, and although SPINK5 expression is down-regulated in patients with EoE, SPINK7 expression is almost completely lost in patients with EoE compared to control individuals (6). Therefore, we suggest that both SPINK5 and SPINK7 countermeasure KLK5 activity in the human esophagus. We hypothesize that decreased expression of SPINKs in EoE unleashes uncontrolled KLK5 activity, which, in turn, disrupts epithelial junctional complexes, impairs the esophageal barrier function, and activates PAR2 (fig. S7).
Ex vivo stimulation of KLK5 on esophageal sections stimulated TSLP production. KLK5-induced TSLP production was inhibited by a PAR2 antagonist. PAR2 antagonism also inhibited cytokine release from SPINK7 gene–deleted epithelial cells in vitro and decreased allergen-induced esophageal eosinophilia in vivo. PAR2 is a G protein–coupled receptor expressed mainly by epithelial cells, endothelial cells, and sensory nerves. PAR2 activation leads to production of immunoregulatory cytokines, innate immune responses, and persistent inflammation, processes relevant to EoE (37), as well as hyperalgesia (38). We demonstrated that PAR2 expression is increased in biopsies from patients with EoE compared to control patients, and single-cell RNA sequencing analysis revealed a unique expression of F2RL1 (PAR2-encoding gene) in a subset of cells that express KLK5 in EoE biopsies, raising the possibility that a complex regulatory mechanism of PAR2 activation is controlled by extracellular proteases and protease inhibitors that remain in close proximity, probably by cross-linking of SPINKs to extracellular proteins (39). Single-cell RNA sequencing data revealed that KLK5 and TSLP are expressed by undifferentiated epithelial cells together with a set of genes that are specialized in posttranscriptional modification and intracellular transport of proteins. Chandramouleeswaran et al. (40) reported that TSLP protein is coexpressed with the differentiation marker KRT13 in the suprabasal and differentiated epithelium of patients with EoE. Because KLK5 and TSLP are secreted proteins, these proteins are not necessarily localized to the same cells that express these genes. Therefore, analysis of mRNA expression may not be comparable with protein expression.
Our data demonstrate that PAR2 antagonism inhibits polyI: C-induced Toll-like receptor (TLR) signaling. This finding is consistent with previous findings of cross-talk between these two receptors. These receptors are cellular sensors for detecting exogenous and endogenous ligands/agonists. Their expression is often similar; for example, both receptors are expressed by epithelial cells and sensory neurons which coexpress TPRV1 that is a signaling component of itch and pain (41). In addition, activation of both receptors initiates similar signaling pathways including activation of mitogen-activated protein kinase/activating protein 1 and nuclear factor κB (41). Rallabhandi et al. (42) revealed a direct association between TLR4 and PAR2 by coimmunoprecipitation. Other reports highlighted potential cross-talk between PAR2 and TLRs (43, 44).
Transmembrane mucins such as MUC4 have a critical role in transducing signaling pathways to the epithelium (45). We revealed that MUC4 was proteolytically overprocessed in patients with EoE compared to control patients, and we suggest that this is likely due to loss of SPINK inhibitors and increased KLK5 activity. The role of MUC4 and the consequences of its processing in EoE were not fully investigated in this study. In the esophagus, mucin proteins likely assemble a mucus layer that protects the epithelium by trapping or transporting bacteria and lubricates debris, similar to their role in the intestine (46, 47). Our results suggest that KLK5-mediated mucus remodeling may regulate the esophageal microbiome. Several studies have shown that the esophagus contains a diverse microbial population and that the esophageal microbiome is altered in EoE (48-50). In addition, microbial dysbiosis has been suggested to be one of the primary causes for food allergy (23, 51, 52). Klk5−/− mice exhibited microbial dysbiosis. Ruminococcaceae and Lachnospiraceae bacteria were less abundant in the Klk5−/− mice compared to Klk5+/+ mice; these bacteria are increased in the intestine of patients with milk allergy (23). Our results showed that human KLK5 regulates human MUC4 processing and MUC4 processing is possibly involved in the microbiome dysbiosis. In addition, in the skin, KLK5 is known to generate an arsenal of antimicrobial peptides derived from cathelicidin and defensin processing. Antimicrobial peptides regulate the killing of infecting microbes and simultaneously facilitate symbiotic interactions between the host and bacteria. In addition, antimicrobial peptides serve as alarmins of the immune system; therefore, dysregulation of these peptides or their processing can have a role in several disorders (10, 53-55). Our results suggest that in the absence of Klk5, mice may have alterations in the esophageal microbiome that may be protecting against food hypersensitivity.
We asked whether delivery of protease inhibitors would attenuate the pathogenesis of EoE. As a proof of concept, we systemically administered the broad-spectrum anti-serine protease A1AT, and this was sufficient to deliver functional A1AT to the esophagus. A1AT belongs to the serpin superfamily of serine protease inhibitors that cause irreversible conformational changes to disrupt the active site of target proteases (56). Consistent with previous reports, we demonstrate that A1AT has the capacity to inhibit KLK5 (57). We subsequently conjugated A1AT to a fluorescent probe and showed that A1AT can be delivered to the esophagus after intraperitoneal administration. We did not observe degradation products of A1AT in the esophagus, suggesting that A1AT mainly remains stable in the esophageal milieu. In addition, A1AT retained its bioactivity in the esophagus as assessed by ex vivo enzymatic activity after pharmacological delivery. Repeated injections of A1AT decreased OVA-induced esophageal eosinophilia without affecting IgE production or lung inflammation. These data highlight the importance of proteolytic regulation of the esophagus. In addition, A1AT decreased OVA-induced CCL24 production but not CCL11. These observations demonstrates a role for CCL24 in esophageal eosinophilia consistent with the role of CCL24 (but not CCL11) in regulating OVA-induced lung eosinophilia (58). It had been reported that A1AT has antiinflammatory properties in addition to its antiprotease activities (59). Therefore, the decreased eosinophilia that was observed after A1AT delivery may not solely stem from proteolytic inhibition. Regardless, A1AT delivery serves as a proof of concept that delivery of protease inhibitors may be potentially beneficial for the treatment of EoE.
Having established the capacity of proteolytic activity restriction to independently regulate esophageal eosinophilia, we asked whether inhibition of the downstream signaling of KLK5 would attenuate the pathogenesis of EoE. Murine esophageal eosinophilia was blocked after selective inhibition of PAR2, and IgE production was decreased, suggesting that PAR2 may be a pivotal mediator of EoE pathogenesis. Administration of the PAR2 antagonist did not affect the accumulation of inflammatory cells in the lungs. We suggest that PAR2 may have a different role in the lungs compared to the esophagus. It is notable that PAR2 signaling has been reported to attenuate a cockroach extract-induced airway hyperresponsiveness model (60), a house dust mite allergic model (61), and the OVA-induced allergic inflammation using PAR2-deficient mice (62). Unlike the OVA-induced allergic model that was used herein, both cockroach extract and house dust mite contain proteolytic enzymes that can directly activate PAR2. In contrast to the OVA-induced allergic model that was performed on PAR2-deficient mice (62), in our experiment, PAR2 was inhibited during the effector phase of allergic inflammation; whereas in the murine asthma studies, PAR2 was inhibited during the sensitization and effector phases together. Our data support a role of PAR2 in the esophagus, similar to what has been found in the skin, another squamous epithelium (63). In addition to KLK5, PAR2 activation can be induced by other proteases, including trypsin, KLK14, prostatin, matriptase, and mast cell tryptase (33). The nonredundant role of KLK5 in regulating esophageal eosinophilia is notable. Proteases are organized into activation cascades, and KLK5 is capable of amplifying the cascade by activating downstream protease targets (33). Loss of Klk5 may affect the activity of other proteases that might activate PAR2 and/or regulate the epithelial barrier function; therefore, Klk5 deficiency was sufficient to abrogate esophageal eosinophilia.
Limitations to this study include that the murine model used does not fully mimic the human disease and also that the murine esophagus is keratinized, whereas the human esophagus is not. In addition, the protein replacement studies were primarily limited to A1AT rather than SPINK7 which was not available in sufficient quantities for the in vivo studies. Last, although our study focused on KLK5, there are likely other serine proteases involved in EoE and also inhibited by SPINK7.
We suggest that protein replacement therapy with protease inhibitors is a particularly attractive strategy for treatment of EoE for three reasons: (i) proteolytic inhibition can restore the epithelial barrier, which, in turn, might restrain inflammation by preventing luminal stimuli from penetrating into the tissue; (ii) the potential for some protease inhibitors (such as A1AT) to act against a broad range of proteases, including PAR2-activating proteases; and (iii) inhibition of proteolytic activity can block several inflammatory processes, including processing and activation of cytokines (13, 64). In view of the reported collective observations, we suggest that disarming proteases by delivery of protease inhibitors that target KLK5 and PAR2 have the potential to be therapeutic for EoE.
MATERIALS AND METHODS
Study design
The aims of this study were to understand how the imbalance between proteases and inhibitors in the esophagus contributes to disease onset and specifically understand the mechanism by which the serine protease inhibitor SPINK7 mediates its function. We used a candidate approach to perform a functional screen to find direct targets of SPINK7 in vitro using recombinant proteases and their substrates. We elucidated KLK5 as one of the SPINK7 targets and predicted interaction between these proteins. We manipulated KLK5 expression by overexpressing KLK5 in esophageal epithelial cells and by CRISPR-Cas9 gene deletion of Klk5 in mice, thereby allowing us to examine a murine EoE model. Epithelial cells were differentiated and assessed by several experimental approaches, including FITC-dextran flux, epithelial resistance measurements, immunofluorescence analysis, proteolytic activity, and cytokine release. We induced an experimental EoE murine model and assessed the proteolytic activity, serum IgE, and MCPT1, CCL11, and CCL24 concentrations and the number of eosinophils in the esophagus, lungs, and blood in Klk5−/− mice compared to Klk5+/+ mice. To interfere with a KLK5 downstream pathway, we blocked PAR2 using a PAR2-selective antagonist ENMD-1068 in epithelial cells in vitro, ex vivo, and in vivo in an experimental EoE murine model. We also blocked serine protease activity in vivo by delivering A1AT to the esophagus. We conjugated A1AT to a fluorescent molecule and tracked its delivery, stability, and functionality in the esophagus. Last, we delivered A1AT in an experimental EoE murine model and assessed its effect on esophageal eosinophil accumulation. To avoid biased results, mice were assigned to treatment groups randomly, and analysis of samples was performed blindly. The number of mice per experiment was based on variability and magnitude of changes and is defined in figures. Primary data are reported in data file S1.
Human subjects
Control patients were defined as having no history of EoE diagnosis, having 0 eosinophils per high-power field (HPF) of esophageal biopsy samples, and having no evidence of esophagitis within esophageal biopsies, although they were presented with symptoms typical of EoE. Patients with EoE had ≥23 eosinophils per HPF in distal esophageal biopsies. Written informed consent was obtained for all human samples before a patient’s enrollment in the study, and all human studies were approved by the Cincinnati Children’s Hospital Medical Center (CCHMC) Institutional Review Board (protocol 2008-0090). Control and patients with EoE were age-matched.
Mice
Mice were obtained from the Jackson laboratory; generation of Klk5−/− mice is described in the Supplementary Materials. All the experiments were performed on age-matched (4- to 5-week-old mice) using males and females that were gender-matched, which were maintained in a pathogen-free barrier facility, and animals were handled according to institutional guidelines. A minimum of three mice per group was used in each experiment, and each experiment was conducted at least three times.
Statistical analyses
Statistical significance was determined using a t test (unpaired, two-tailed) or analysis of variance (ANOVA). Statistical analyses were performed using GraphPad Prism (GraphPad Software Incorporated). Statistical significance of gene ontology was calculated by extended versions of the PageRank and HITS algorithms and the K-Step Markov method using the ToppGene Suite. Variability analysis of the microbiome was performed by CLC Microbial Genomics Module (Qiagen) using PERMANOVA weighted test. Data from single-cell RNA sequencing was subjected to uniform manifold approximation and projection and shared nearest neighbor (SNN) modularity optimization–based clustering. Using principal components analysis and SNN modularity optimization–based clustering algorithm with a resolution of 0.5.
Supplementary Material
Fig. S1. SPINK7 inhibits KLK5 and KLK12.
Fig. S2. Generation of Klk5-deficient mice.
Fig. S3. A1AT inhibits KLK5 in vitro and trypsin-like proteolytic activity and CCL11 production in vivo.
Fig. S4. SPINK5 inhibits KLK5 in vitro.
Fig. S5. Esophageal KLK and F2RL1 expression profile.
Fig. S6. CCL11 production in the esophagus.
Fig. S7. A model of the SPINK7, KLK5, and PAR2 pathway in EoE.
Table S1. Prevalence rate of A1AD in patients with EoE compared to the general population.
Table S2. List of bacteria at the family level that are differentially abundant in the esophagus of Klk5-deficient mice.
Table S3. List of the most enriched genes in epithelial clusters 1, 2, and 3.
Data file S1. Primary data.
Acknowledgments:
We thank A. Rustgi (University of Pennsylvania) for the human telomerase reverse transcriptase–immortalized EPC2 cell line. We thank M. Brantly (Alpha-1 Foundation) for a gift of A1AT. We thank M.K. Mingler (CCHMC) for assistance with murine strains. We thank A. Munitz (Tel Aviv University) for reviewing of the manuscript and discussion. We also thank S. Hottinger (CCHMC) for editorial assistance, all of the participating families and the Cincinnati Center for Eosinophilic Disorders, and members of the Division of Allergy and Immunology.
Funding: This work was supported in part by NIH R37 AI045898, U19 AI070235, R01 AI057803, R01 DK107502, and P30 DK078392 (Gene and Protein Expression Core); the Campaign Urging Research for Eosinophilic Disease (CURED); the Sunshine Charitable Foundation and its supporters, Denise and David Bunning; and ADARE pharmaceuticals (to M.E.R.).
Footnotes
SUPPLEMENTARY MATERIALS
Competing interests: T.W.’s institution has received an Aimmune research grant for other works, and they have an Eosinophilic Esophagitis Diagnostic Panel patent owned by CCHMC. J.M.C. and M.E.R. are inventors of patents owned by CCHMC. M.E.R. and N.P.A. are inventors on a pending U.S. provisional patent application (62/126,814) submitted by CCHMC that covers serine protease inhibitors for the treatment of EoE. M.E.R. and T.W. are inventors on granted U.S. patent 9,928,344, owned by CCHMC that covers diagnostic methods of EoE based on esophageal transcriptome analysis. M.E.R.’s institution has received a grant from the NIH for this work. They have grants for other works from a U.S.-Israel Binational Grant and Patient-Centered Outcomes Research Institute. They have patent ownership by CCHMC. He has received consultancy fees from Pulm One, Spoon Guru, ClostraBio, Celgene, and Astra Zeneca and has an equity interest in the first three listed and royalties from reslizumab (Teva Pharmaceuticals), PEESSv2 (Mapi Research Trust), and UpToDate. The rest of the authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. 16S sequencing data are available at NCBI (accession no. PRJNA623362). Klk5−/− mice can be obtained with a material transfer agreement with CCHMC.
REFERENCES AND NOTES
- 1.Davis BP, Rothenberg ME, Mechanisms of disease of eosinophilic esophagitis. Annu. Rev. Pathol 11, 365–393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Shea KM, Aceves SS, Dellon ES, Gupta SK, Spergel JM, Furuta GT, Rothenberg ME, Pathophysiology of eosinophilic esophagitis. Gastroenterology 154, 333–345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dellon ES, Liacouras CA, Molina-Infante J, Furuta GT, Spergel JM, Zevit N, Spechler SJ, Attwood SE, Straumann A, Aceves SS, Alexander JA, Atkins D, Arva NC, Blanchard C, Bonis PA, Book WM, Capocelli KE, Chehade M, Cheng E, Collins MH, Davis CM, Dias JA, Di Lorenzo C, Dohil R, Dupont C, Falk GW, Ferreira CT, Fox A, Gonsalves NP, Gupta SK, Katzka DA, Kinoshita Y, Menard-Katcher C, Kodroff E, Metz DC, Miehlke S, Muir AB, Mukkada VA, Murch S, Nurko S, Ohtsuka Y, Orel R, Papadopoulou A, Peterson KA, Philpott H, Putnam PE, Richter JE, Rosen R, Rothenberg ME, Schoepfer A, Scott MM, Shah N, Sheikh J, Souza RF, Strobel MJ, Talley NJ, Vaezi MF, Vandenplas Y, Vieira MC, Walker MM, Wechsler JB, Wershil BK, Wen T, Yang GY, Hirano I, Bredenoord AJ, Updated international consensus diagnostic criteria for eosinophilic esophagitis: Proceedings of the AGREE conference. Gastroenterology 155, 1022–1033.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molina-Infante J, Gonzalez-Cordero PL, Arias A, Lucendo AJ, Update on dietary therapy for eosinophilic esophagitis in children and adults. Expert Rev. Gastroenterol. Hepatol 11, 115–123 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Rochman M, Travers J, Miracle CE, Bedard MC, Wen T, Azouz NP, Caldwell JM, Kc K, Sherrill JD, Davis BP, Rymer JK, Kaufman KM, Aronow BJ, Rothenberg ME, Profound loss of esophageal tissue differentiation in patients with eosinophilic esophagitis. J. Allergy Clin. Immunol 140, 738–749.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azouz NP, Ynga-Durand MA, Caldwell JM, Jain A, Rochman M, Fischesser DM, Ray LM, Bedard MC, Mingler MK, Forney C, Eilerman M, Kuhl JT, He H, Biagini Myers JM, Mukkada VA, Putnam PE, Khurana Hershey GK, Kottyan LC, Wen T, Martin LJ, Rothenberg ME, The antiprotease SPINK7 serves as an inhibitory checkpoint for esophageal epithelial inflammatory responses. Sci. Transl. Med 10, eaap9736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rochman M, Azouz NP, Rothenberg ME, Epithelial origin of eosinophilic esophagitis. J. Allergy Clin. Immunol 142, 10–23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furio L, Hovnanian A, Netherton syndrome: Defective kallikrein inhibition in the skin leads to skin inflammation and allergy. Biol. Chem 395, 945–958 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Jiang R, Shi Z, Johnson JJ, Liu Y, Stack MS, Kallikrein-5 promotes cleavage of desmoglein-1 and loss of cell-cell cohesion in oral squamous cell carcinoma. J. Biol. Chem 286, 9127–9135 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw JL, Petraki C, Watson C, Bocking A, Diamandis EP, Role of tissue kallikrein-related peptidases in cervical mucus remodeling and host defense. Biol. Chem 389, 1513–1522 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Sherrill JD, Kc K, Wu D, Djukic Z, Caldwell JM, Stucke EM, Kemme KA, Costello MS, Mingler MK, Blanchard C, Collins MH, Abonia JP, Putnam PE, Dellon ES, Orlando RC, Hogan SP, Rothenberg ME, Desmoglein-1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol. 7, 718–729 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samuelov L, Sarig O, Harmon RM, Rapaport D, Ishida-Yamamoto A, Isakov O, Koetsier JL, Gat A, Goldberg I, Bergman R, Spiegel R, Eytan O, Geller S, Peleg S, Shomron N, Goh CSM, Wilson NJ, Smith FJD, Pohler E, Simpson MA, McLean WHI, Irvine AD, Horowitz M, McGrath JA, Green KJ, Sprecher E, Desmoglein 1 deficiency results in severe dermatitis, multiple allergies and metabolic wasting. Nat. Genet 45, 1244–1248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briot A, Deraison C, Lacroix M, Bonnart C, Robin A, Besson C, Dubus P, Hovnanian A, Kallikrein 5 induces atopic dermatitis-like lesions through PAR2-mediated thymic stromal lymphopoietin expression in Netherton syndrome. J. Exp. Med 206, 1135–1147 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jang H, Matsuda A, Jung K, Karasawa K, Matsuda K, Oida K, Ishizaka S, Ahn G, Amagai Y, Moon C, Kim SH, Arkwright PD, Takamori K, Matsuda H, Tanaka A, Skin pH Is the master switch of kallikrein 5-mediated skin barrier destruction in a murine atopic dermatitis model. J. Invest. Dermatol 136, 127–135 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Gouin O, L'Herondelle K, Buscaglia P, Le Gall-Ianotto C, Philippe R, Legoux N, Mignen O, Buhe V, Leschiera R, Sakka M, Kerfant N, Carre JL, Le Garrec R, Lefeuvre L, Lebonvallet N, Misery L, Major role for TRPV1 and InsP3R in PAR2-elicited inflammatory mediator production in differentiated human keratinocytes. J. Invest. Dermatol 138, 1564–1572 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Wen T, Aronow BJ, Rochman Y, Rochman M, Kc K, Dexheimer PJ, Putnam P, Mukkada V, Foote H, Rehn K, Darko S, Douek D, Rothenberg ME, Single-cell RNA sequencing identifies inflammatory tissue T cells in eosinophilic esophagitis. J. Clin. Invest 129, 2014–2028 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, Gober L, Kim C, Glessner J, Frackelton E, Thomas K, Blanchard C, Liacouras C, Verma R, Aceves S, Collins MH, Brown-Whitehorn T, Putnam PE, Franciosi JP, Chiavacci RM, Grant SF, Abonia JP, Sleiman PM, Hakonarson H, Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat. Genet 42, 289–291 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sleiman PM, Wang ML, Cianferoni A, Aceves S, Gonsalves N, Nadeau K, Bredenoord AJ, Furuta GT, Spergel JM, Hakonarson H, GWAS identifies four novel eosinophilic esophagitis loci. Nat. Commun 5, 5593 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kottyan LC, Davis BP, Sherrill JD, Liu K, Rochman M, Kaufman K, Weirauch MT, Vaughn S, Lazaro S, Rupert AM, Kohram M, Stucke EM, Kemme KA, Magnusen A, He H, Dexheimer P, Chehade M, Wood RA, Pesek RD, Vickery BP, Fleischer DM, Lindbad R, Sampson HA, Mukkada VA, Putnam PE, Abonia JP, Martin LJ, Harley JB, Rothenberg ME, Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat. Genet 46, 895–900 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furio L, Pampalakis G, Michael IP, Nagy A, Sotiropoulou G, Hovnanian A, KLK5 inactivation reverses cutaneous hallmarks of netherton syndrome. PLOS Genet. 11, e1005389 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasparek P, Ileninova Z, Zbodakova O, Kanchev I, Benada O, Chalupsky K, Brattsand M, Beck IM, Sedlacek R, KLK5 and KLK7 ablation fully rescues lethality of netherton syndrome-like phenotype. PLOS Genet. 13, e1006566 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deraison C, Bonnart C, Lopez F, Besson C, Robinson R, Jayakumar A, Wagberg F, Brattsand M, Hachem JP, Leonardsson G, Hovnanian A, LEKTI fragments specifically inhibit KLK5, KLK7, and KLK14 and control desquamation through a pH-dependent interaction. Mol. Biol. Cell 18, 3607–3619 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berni Canani R, Sangwan N, Stefka AT, Nocerino R, Paparo L, Aitoro R, Calignano A, Khan AA, Gilbert JA, Nagler CR, Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 10, 742–750 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sànchez-Solana B, Nueda ML, Ruvira MD, Ruiz-Hidalgo MJ, Monsalve EM, Rivero S, Garcia-Ramirez JJ, Diaz-Guerra MJ, Baladron V, Laborda J, The EGF-like proteins DLK1 and DLK2 function as inhibitory non-canonical ligands of NOTCH1 receptor that modulate each other's activities. Biochim. Biophys. Acta 1813, 1153–1164 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Vega ME, Giroux V, Natsuizaka M, Liu M, Klein-Szanto AJ, Stairs DB, Nakagawa H, Wang KK, Wang TC, Lynch JP, Rustgi AK, Inhibition of Notch signaling enhances transdifferentiation of the esophageal squamous epithelium towards a Barrett's-like metaplasia via KLF4. Cell Cycle 13, 3857–3866 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen G, Xu R, Yue B, Mei X, Li P, Zhou X, Huang S, Gong L, Zhang S, The expression of podoplanin protein is a diagnostic marker to distinguish the early infiltration of esophageal squamous cell carcinoma. Oncotarget 8, 19013–19020 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Assis DM, Zalazar L, Juliano MA, De Castro R, Cesari A, Novel inhibitory activity for serine protease inhibitor Kazal type-3 (Spink3) on human recombinant kallikreins. Protein Pept. Lett 20, 1098–1107 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Nishimiya D, Kawaguchi Y, Kodama S, Nasu H, Yano H, Yamaguchi A, Tamura M, Hashimoto R, A protein scaffold, engineered SPINK2, for generation of inhibitors with high affinity and specificity against target proteases. Sci. Rep 9, 11436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brattsand M, Stefansson K, Hubiche T, Nilsson SK, Egelrud T, SPINK9: A selective, skin-specific Kazal-type serine protease inhibitor. J. Invest. Dermatol 129, 1656–1665 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Meyer-Hoffert U, Wu Z, Schroder JM, Identification of lympho-epithelial Kazal-type inhibitor 2 in human skin as a kallikrein-related peptidase 5-specific protease inhibitor. PLOS ONE 4, e4372 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kantyka T, Fischer J, Wu Z, Declercq W, Reiss K, Schroder JM, Meyer-Hoffert U, Inhibition of kallikrein-related peptidases by the serine protease inhibitor of Kazal-type 6. Peptides 32, 1187–1192 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Van Leeuw JB, Berra E, Gu Y, Petit T, Lacroix V, Lanthier N, Van Hoof M, Staessen JA, Persu A, Alpha-1 antitrypsin deficiency: A novel cause of isolated systolic resistant hypertension? J. Hypertens. 34, 1659–1661 (2016). [DOI] [PubMed] [Google Scholar]
- 33.de Veer SJ, Furio L, Harris JM, Hovnanian A, Proteases: Common culprits in human skin disorders. Trends Mol. Med 20, 166–178 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Paluel-Marmont C, Bellon N, Barbet P, Leclerc-Mercier S, Hadj-Rabia S, Dupont C, Bodemer C, Eosinophilic esophagitis and colonic mucosal eosinophilia in Netherton syndrome. J. Allergy Clin. Immunol 139, 2003–2005.e1 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Descargues P, Deraison C, Bonnart C, Kreft M, Kishibe M, Ishida-Yamamoto A, Elias P, Barrandon Y, Zambruno G, Sonnenberg A, Hovnanian A, Spink5-deficient mice mimic Netherton syndrome through degradation of desmoglein 1 by epidermal protease hyperactivity. Nat. Genet 37, 56–65 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Furio L, de Veer S, Jaillet M, Briot A, Robin A, Deraison C, Hovnanian A, Transgenic kallikrein 5 mice reproduce major cutaneous and systemic hallmarks of Netherton syndrome. J. Exp. Med 211, 499–513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramachandran R, Noorbakhsh F, Defea K, Hollenberg MD, Targeting proteinase-activated receptors: Therapeutic potential and challenges. Nat. Rev. Drug Discov 11, 69–86 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Vergnolle N, Bunnett NW, Sharkey KA, Brussee V, Compton SJ, Grady EF, Cirino G, Gerard N, Basbaum AI, Andrade-Gordon P, Hollenberg MD, Wallace JL, Proteinase-activated receptor-2 and hyperalgesia: A novel pain pathway. Nat. Med 7, 821–826 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Fischer J, Koblyakova Y, Latendorf T, Wu Z, Meyer-Hoffert U, Cross-linking of SPINK6 by transglutaminases protects from epidermal proteases. J. Invest. Dermatol 133, 1170–1177 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Chandramouleeswaran PM, Shen D, Lee AJ, Benitez A, Dods K, Gambanga F, Wilkins BJ, Merves J, Noah Y, Toltzis S, Yearley JH, Spergel JM, Nakagawa H, Malefyt R, Muir AB, Wang ML, Preferential secretion of thymic stromal lymphopoietin (TSLP) by terminally differentiated esophageal epithelial cells: Relevance to Eosinophilic Esophagitis (EoE). PLOS ONE 11, e0150968 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Widera D, Martinez Aguilar R, Cottrell GS, Toll-like receptor 4 and protease-activated receptor 2 in physiology and pathophysiology of the nervous system: More than just receptor cooperation? Neural Regen. Res 14, 1196–1201 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rallabhandi P, Nhu QM, Toshchakov VY, Piao W, Medvedev AE, Hollenberg MD, Fasano A, Vogel SN, Analysis of proteinase-activated receptor 2 and TLR4 signal transduction: A novel paradigm for receptor cooperativity. J. Biol. Chem 283, 24314–24325 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nhu QM, Shirey K, Teijaro JR, Farber DL, Netzel-Arnett S, Antalis TM, Fasano A, Vogel SN, Novel signaling interactions between proteinase-activated receptor 2 and Toll-like receptors in vitro and in vivo. Mucosal Immunol. 3, 29–39 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uehara A, Imamura T, Potempa J, Travis J, Takada H, Gingipains from Porphyromonas gingivalis synergistically induce the production of proinflammatory cytokines through protease-activated receptors with Toll-like receptor and NOD1/2 ligands in human monocytic cells. Cell. Microbiol 10, 1181–1189 (2008). [DOI] [PubMed] [Google Scholar]
- 45.van Putten JPM, Strijbis K, Transmembrane mucins: Signaling receptors at the intersection of inflammation and cancer. J. Innate Immun 9, 281–299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johansson ME, Hansson GC, Immunological aspects of intestinal mucus and mucins. Nat. Rev. Immunol 16, 639–649 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schroeder BO, Fight them or feed them: How the intestinal mucus layer manages the gut microbiota. Gastroenterol Rep 7, 3–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dellon ES, The esophageal microbiome in eosinophilic esophagitis. Gastroenterology 151, 364–365 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Harris JK, Fang R, Wagner BD, Choe HN, Kelly CJ, Schroeder S, Moore W, Stevens MJ, Yeckes A, Amsden K, Kagalwalla AF, Zalewski A, Hirano I, Gonsalves N, Henry LN, Masterson JC, Robertson CE, Leung DY, Pace NR, Ackerman SJ, Furuta GT, Fillon SA, Esophageal microbiome in eosinophilic esophagitis. PLOS ONE 10, e0128346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benitez AJ, Hoffmann C, Muir AB, Dods KK, Spergel JM, Bushman FD, Wang ML, Inflammation-associated microbiota in pediatric eosinophilic esophagitis. Microbiome 3, 23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feehley T, Plunkett CH, Bao R, Choi Hong SM, Culleen E, Belda-Ferre P, Campbell E, Aitoro R, Nocerino R, Paparo L, Andrade J, Antonopoulos DA, Berni Canani R, Nagler CR, Healthy infants harbor intestinal bacteria that protect against food allergy. Nat. Med 25, 448–453 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Azouz NP, Rothenberg ME, Mechanisms of gastrointestinal allergic disorders. J. Clin. Invest 130, 1419–1430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamasaki K, Schauber J, Coda A, Lin H, Dorschner RA, Schechter NM, Bonnart C, Descargues P, Hovnanian A, Gallo RL, Kallikrein-mediated proteolysis regulates the antimicrobial effects of cathelicidins in skin. FASEB J. 20, 2068–2080 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Two AM, Del Rosso JQ, Kallikrein 5-mediated inflammation in rosacea: Clinically relevant correlations with acute and chronic manifestations in rosacea and how individual treatments may provide therapeutic benefit. J. Clin. Aesthet. Dermatol 7, 20–25 (2014). [PMC free article] [PubMed] [Google Scholar]
- 55.Ahn CS, Huang WW, Rosacea Pathogenesis. Dermatol Clin 36, 81–86 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Song J, Matthews AY, Reboul CF, Kaiserman D, Pike RN, Bird PI, Whisstock JC, Predicting serpin/protease interactions. Methods Enzymol. 501, 237–273 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Yousef GM, Kapadia C, Polymeris ME, Borgono C, Hutchinson S, Wasney GA, Soosaipillai A, Diamandis EP, The human kallikrein protein 5 (hK5) is enzymatically active, glycosylated and forms complexes with two protease inhibitors in ovarian cancer fluids. Biochim. Biophys. Acta 1628, 88–96 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Pope SM, Fulkerson PC, Blanchard C, Akei HS, Nikolaidis NM, Zimmermann N, Molkentin JD, Rothenberg ME, Identification of a cooperative mechanism involving interleukin-13 and eotaxin-2 in experimental allergic lung inflammation. J. Biol. Chem 280, 13952–13961 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Jonigk D, Al-Omari M, Maegel L, Muller M, Izykowski N, Hong J, Hong K, Kim SH, Dorsch M, Mahadeva R, Laenger F, Kreipe H, Braun A, Shahaf G, Lewis EC, Welte T, Dinarello CA, Janciauskiene S, Anti-inflammatory and immunomodulatory properties of a1-antitrypsin without inhibition of elastase. Proc. Natl. Acad. Sci. U.S.A 110, 15007–15012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Asaduzzaman M, Davidson C, Nahirney D, Fiteih Y, Puttagunta L, Vliagoftis H, Proteinase-activated receptor-2 blockade inhibits changes seen in a chronic murine asthma model. Allergy 73, 416–420 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Davidson CE, Asaduzzaman M, Arizmendi NG, Polley D, Wu Y, Gordon JR, Hollenberg MD, Cameron L, Vliagoftis H, Proteinase-activated receptor-2 activation participates in allergic sensitization to house dust mite allergens in a murine model. Clin. Exp. Allergy 43, 1274–1285 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Schmidlin F, Amadesi S, Dabbagh K, Lewis DE, Knott P, Bunnett NW, Gater PR, Geppetti P, Bertrand C, Stevens ME, Protease-activated receptor 2 mediates eosinophil infiltration and hyperreactivity in allergic inflammation of the airway. J. Immunol 169, 5315–5321 (2002). [DOI] [PubMed] [Google Scholar]
- 63.Frateschi S, Camerer E, Crisante G, Rieser S, Membrez M, Charles RP, Beermann F, Stehle JC, Breiden B, Sandhoff K, Rotman S, Haftek M, Wilson A, Ryser S, Steinhoff M, Coughlin SR, Hummler E, PAR2 absence completely rescues inflammation and ichthyosis caused by altered CAP1/Prss8 expression in mouse skin. Nat. Commun 2, 161 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Afonina IS, Müller C, Martin SJ, Beyaert R, Proteolytic processing of interleukin-1 family cytokines: Variations on a common theme. Immunity 42, 991–1004 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Morrison JF, Kinetics of the reversible inhibition of enzyme-catalysed reactions by tight-binding inhibitors. Biochim. Biophys. Acta 185, 269–286 (1969). [DOI] [PubMed] [Google Scholar]
- 66.Seeliger D, de Groot BL, Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des 24, 417–422 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mishra A, Schlotman J, Wang M, Rothenberg ME, Critical role for adaptive T cell immunity in experimental eosinophilic esophagitis in mice. J. Leukoc. Biol 81, 916–924 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Rubinstein E, Cho JY, Rosenthal P, Chao J, Miller M, Pham A, Aceves SS, Varki A, Broide DH, Siglec-F inhibition reduces esophageal eosinophilia and angiogenesis in a mouse model of eosinophilic esophagitis. J. Pediatr. Gastroenterol. Nutr 53, 409–416 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haeussler M, Schonig K, Eckert H, Eschstruth A, Mianne J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, Joly JS, Concordet JP, Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan CL, Hu YC, A transgenic core facility’s experience in genome editing revolution. Adv. Exp. Med. Biol 1016, 75–90 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Yang H, Wang H, Jaenisch R, Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat. Protoc 9, 1956–1968 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Mishra A, Hogan SP, Lee JJ, Foster PS, Rothenberg ME, Fundamental signals that regulate eosinophil homing to the gastrointestinal tract. J. Clin. Invest 103, 1719–1727 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jimenez-Garcia B, Pons C, Fernandez-Recio J, pyDockWEB: A web server for rigid-body protein-protein docking using electrostatics and desolvation scoring. Bioinformatics 29, 1698–1699 (2013). [DOI] [PubMed] [Google Scholar]
- 74.Tovchigrechko A, Vakser IA, GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 34, W310–W314 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tovchigrechko A, Vakser IA, Development and testing of an automated approach to protein docking. Proteins 60, 296–301 (2005). [DOI] [PubMed] [Google Scholar]
- 76.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen J, Bardes EE, Aronow BJ, Jegga AG, ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–W311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R, Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu D, Ahrens R, Osterfeld H, Noah TK, Groschwitz K, Foster PS, Steinbrecher KA, Rothenberg ME, Shroyer NF, Matthaei KI, Finkelman FD, Hogan SP, Interleukin-13 (IL-13)/IL-13 receptor α1 (IL-13Rα1) signaling regulates intestinal epithelial cystic fibrosis transmembrane conductance regulator channel-dependent Cl-secretion. J. Biol. Chem 286, 13357–13369 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Murphy SN, Mendis ME, Berkowitz DA, Kohane I, Chueh HC, Integration of clinical and genetic data in the i2b2 architecture. AMIA Annu. Symp. Proc, 1040 (2006). [PMC free article] [PubMed] [Google Scholar]
- 81.Mendis M, Phillips LC, Kuttan R, Pan W, Gainer V, Kohane I, Murphy SN, Integrating outside modules into the i2b2 architecture. AMIA Annu. Symp. Proc , 1054 (2008). [PubMed] [Google Scholar]
- 82.Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu TX, Collins MH, Putnam PE, Wells SI, Rothenberg ME, IL-13 involvement in eosinophilic esophagitis: Transcriptome analysis and reversibility with glucocorticoids. J. Allergy Clin. Immunol 120, 1292–1300 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Fulkerson PC, Rothenberg ME, Eosinophil development, disease involvement, and therapeutic suppression. Adv. Immunol 138, 1–34 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. SPINK7 inhibits KLK5 and KLK12.
Fig. S2. Generation of Klk5-deficient mice.
Fig. S3. A1AT inhibits KLK5 in vitro and trypsin-like proteolytic activity and CCL11 production in vivo.
Fig. S4. SPINK5 inhibits KLK5 in vitro.
Fig. S5. Esophageal KLK and F2RL1 expression profile.
Fig. S6. CCL11 production in the esophagus.
Fig. S7. A model of the SPINK7, KLK5, and PAR2 pathway in EoE.
Table S1. Prevalence rate of A1AD in patients with EoE compared to the general population.
Table S2. List of bacteria at the family level that are differentially abundant in the esophagus of Klk5-deficient mice.
Table S3. List of the most enriched genes in epithelial clusters 1, 2, and 3.
Data file S1. Primary data.