Abstract
Yeasts can adapt to a wide range of pH fluctuations (2 to 10), while Helicobacter pylori, a facultative intracellular bacterium, can adapt to a range from pH 6 to 8. This work analyzed if H. pylori J99 can protect itself from acidic pH by entering into Candida albicans ATCC 90028. Growth curves were determined for H. pylori and C. albicans at pH 3, 4, and 7. Both microorganisms were co-incubated at the same pH values, and the presence of intra-yeast bacteria was evaluated. Intra-yeast bacteria-like bodies were detected using wet mounting, and intra-yeast binding of anti-H. pylori antibodies was detected using immunofluorescence. The presence of the H. pylori rDNA 16S gene in total DNA from yeasts was demonstrated after PCR amplification. H. pylori showed larger death percentages at pH 3 and 4 than at pH 7. On the contrary, the viability of the yeast was not affected by any of the pHs evaluated. H. pylori entered into C. albicans at all the pH values assayed but to a greater extent at unfavorable pH values (pH 3 or 4, p = 0.014 and p = 0.001, respectively). In conclusion, it is possible to suggest that H. pylori can shelter itself within C. albicans under unfavorable pH conditions.
Keywords: Helicobacter pylori, Candida albicans, stress, pH, intracellular bacteria
1. Introduction
There is evidence showing that certain bacteria can become permanent or transitory endosymbionts of eukaryotic cells. This condition may have been acquired during evolution in order to serve as a specialized niche in which bacteria are protected from environmental stress and their transmission to a new host is facilitated [1,2,3]. Therefore, the endosymbiotic relationship between bacteria and hosts is a conserved phenomenon and has an important impact on the evolution of microorganisms [4]. For example, yeasts are highly sophisticated microorganisms with a notable capacity of change, which can adapt to environmental stress [5] and establish symbiotic relationships with certain microorganisms [6]. Furthermore, it has been found that Candida albicans may harbor Helicobacter pylori cells [3,6,7,8,9,10].
Candida is considered to be a genus of opportunistic pathogenic yeasts causing infection in immunocompromised individuals or in those with an altered health status, such as individuals with diabetes [11]. These yeasts can also cause infections in healthy individuals, such as women of fertile age [12,13]. Several Candida species share the ability to grow under diverse environmental conditions, including pH ranges from 2 to 10 in different anatomical and environmental niches, even when the availability of nutrients is restricted [14,15]. C. albicans is mainly associated with humans, growing on the surface of the mucosae of the gastrointestinal and genitourinary tracts and on the skin [16,17,18].
On the other hand, H. pylori is a facultative intracellular pathogenic microorganism. Some authors report that H. pylori may take advantage of this characteristic to protect itself from factors negatively affecting its viability [2,19]. The association between H. pylori and Candida was first proposed in 1998, when yeast colonies were found as contaminants of gastric biopsies in blood agar plates [20]. Optical microscopy showed the presence of bacteria-like bodies (BLBs) rapidly moving within the vacuoles of these gastric yeasts, which were purified and identified as Candida. Polymerase chain reaction (PCR) was used to reveal the bacterial nature of these BLBs [20]. Optical microscopy as well as the detection of H. pylori-specific genes and the immunodetection of proteins of this bacterium in yeasts of the genus Candida confirmed the intra-yeast presence of the pathogen H. pylori. These results supported the idea that C. albicans might act as a reservoir of H. pylori outside the human stomach [7,8,9,10,20,21,22]. Other authors reported the coexistence of C. albicans and H. pylori has a synergistic effect on the pathogenesis of giant gastric ulcers [23]. Further knowledge concerning the details of the interactions between these two microorganisms will eventually provide important information for the treatment of infectious diseases caused by H. pylori, particularly those showing resistance to antimicrobial agents [24].
H. pylori infection is directly related to the development of gastric pathologies, such as chronic gastritis, peptic ulcers, mucosa-associated lymphoid tissue lymphoma (MALT lymphoma), and gastric cancer [25], justifying the interest from many researchers to improve our knowledge regarding this pathogen. A further reason to study this microorganism is the fact that its transmission routes are not yet fully known. In general, it is accepted that the most probable H. pylori transmission routes are the fecal–oral, oral–oral, or gastro–oral ones [26]. Intrafamily dissemination has been proposed as the most common form to acquire the infection, the mother being the main factor participating in the dissemination of this pathogen. This asseveration is based on an investigation by Burucoa and Axon [27], in which, after studying children under 15 years of age whose parents were H. pylori positive, they found 10 children were infected with a strain whose genotype was the same as that of the mother, while only 2 children shared the genotype of the microorganism found in the father [28].
Moreover, it has been proposed that a concurrence of H. pylori and C. albicans indicates a more intimate relationship, which protects H. pylori from environmental stress [20]. Thus, it is necessary to evaluate the different factors that might favor the harboring of H. pylori within yeasts. One of these factors is the role of acidic pH on the relationship between both microorganisms. Therefore, the aim of this work was to evaluate, in vitro, the role of acidic pH as a trigger for the internalization of H. pylori into the vacuole of C. albicans yeast cells.
2. Results
2.1. Acidic pH Stress Factor Assay
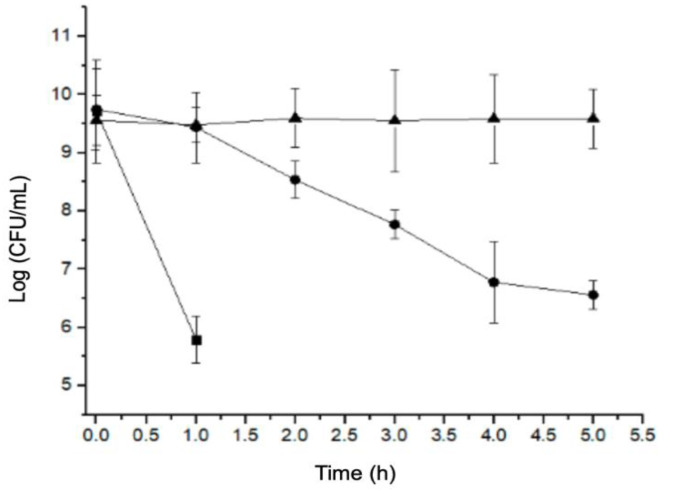
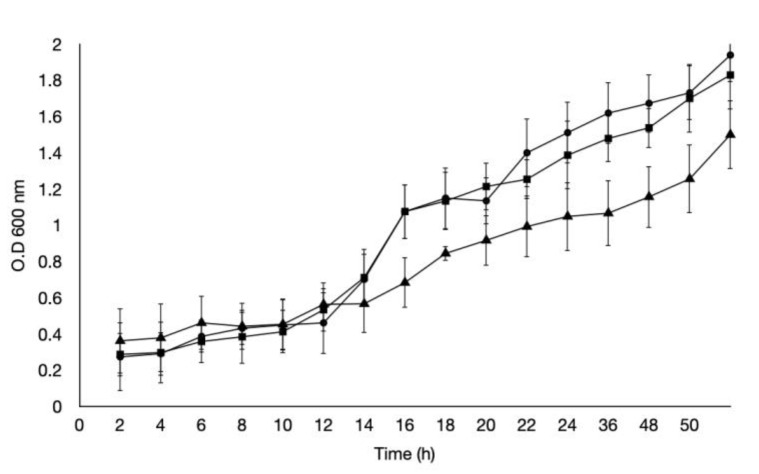
This assay allowed us to determine the viability of H. pylori and C. albicans exposed to different pH values. When exposed to pH 3, H. pylori showed an abrupt decrease in bacterial counts during the first hour of exposure. When exposed to pH 4, a gradual decrease in the bacterial counts was observed until 4 h; the counts then remained unchanged until 5 h, which was significantly different to the control (pH 7). Regarding H. pylori growth at pH 7, no changes in the bacterial cell counts were observed during the 5 h of the assay (Figure 1). Regarding the growth of C. albicans, under pH stress conditions, its growth at pH 3 was slower, not reaching the stationary phase even after 50 h of incubation (Figure 2).
Figure 1.
Growth curve of Helicobacter pylori J99 at pH 3 (■), pH 4 (●), and pH 7 (▲).
Figure 2.
Growth curve of Candida albicans at pH 3 (■), pH 4 (●), and pH 7 (▲). In vitro entry of H. pylori J99 into C. albicans ATCC 90028.
2.2. In Vitro Entry of H. pylori J99 into C. albicans ATCC 90028
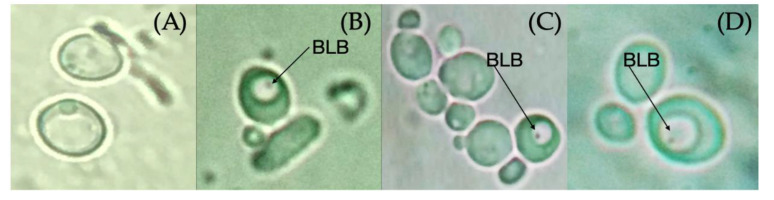
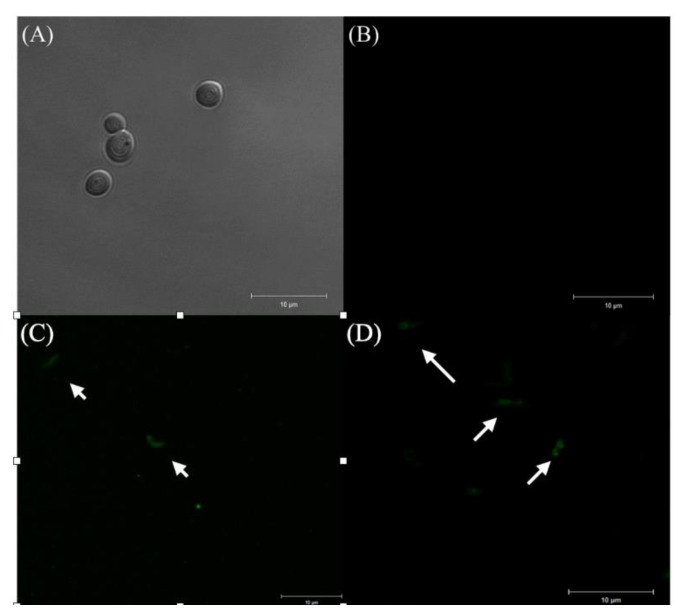
Regarding the entry of H. pylori into C. albicans, the first BLBs were observed as dots whose movement was restricted to the size of the vacuole during the first hour of co-culturing, as observed in (Figure 3). The immunofluorescent assay allowed us to identify intracellular H. pylori within yeasts (Figure 4). The percentage of C. albicans harboring bacteria observed by optical microscopy at different times and pH values are shown in Table 1. There was a significant difference in yeast harboring H. pylori at pH 3 or pH 4 when compared to pH 7, and at pH 4, the number of yeast harboring H. pylori was constant through time (Table 1). No significant difference regarding the entry of bacteria was observed when comparing pH 3 and pH 4, but the entry of bacteria into C. albicans at these two pH values showed a significant difference when compared to pH 7 at 24 and 48 h. The percentage of BLBs harboring yeasts showed variations according to the time of exposure to the stressing factor (pH); there were higher percentages of yeasts harboring BLBs during the first and last hour that the co-cultures were incubated at acidic pH values, showing a greater significance at pH 4 (p = 0.001) than at pH 3 (p = 0.014).
Figure 3.
Bacterium-like bodies (BLBs) within C. albicans ATCC 90028 vacuoles after 48 h of incubation at pH 3, pH 4, and pH 7, observed by optical microscopy using a 100× objective lens. (A) Negative control, C. albicans 90028 strain; (B) incubation at pH 3; (C) incubation at pH 4; and (D) incubation at pH 7.
Figure 4.
Immunofluorescent assay using Fluorescein Isothiocyanate (FITC)-labeled anti-H. pylori IgG polyclonal antibodies. (A) C. albicans ATCC 90028 not co-cultured with the bacterium (negative control) observed by differential interference contrast (DIC); (B) absence of fluorescence in the negative control; (C) presence of fluorescence emitted by H. pylori J99 (white arrows) (positive control); and (D) fluorescence emitted by the intracellular presence of H. pylori J99 (white arrows) in yeasts.
Table 1.
Percentage of yeasts harboring bacterium-like bodies (BLBs) within C. albicans ATCC 90028 at different times of incubation at pH 3, pH 4, and pH 7, observed by optical microscopy.
| Time (h) | pH 3 | pH 4 | pH 7 | p-Value |
|---|---|---|---|---|
| 0 | - | - | - | |
| 1 | 11.7% | 13.7% | 6.3% | 0.0559 |
| 3 | 3.0% | 3.3% | 2.7% | 0.9540 |
| 6 | 4.2% | 3.3% | 2.2% | 0.5773 |
| 12 | 7.1% | 6.4% | 2.2% | 0.0963 |
| 24 | 8.7% | 12.5% | 3.3% | 0.0031 |
| 48 | 11.1% | 18.7% | 4.1% | 0.0309 |
2.3. Amplification of The H. pylori J99 16S rDNA Gene from the Total DNA Extracted from C. albicans ATCC 90028
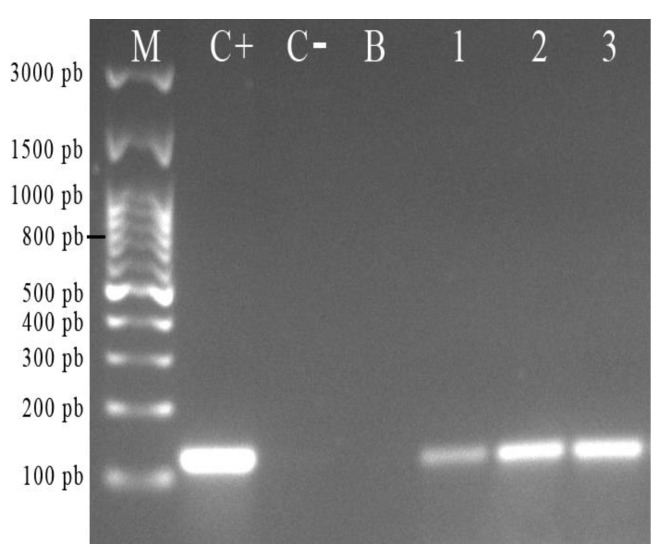
After agarose gel electrophoresis, it was possible to observe the presence of a band of 110 bp in the total DNA of bacterium yeast co-cultures incubated at pH 7, 4, and 3 (Figure 5, lanes 1–3) and also present in the positive H. pylori J99 control, but absent in the yeast not co-cultured with the bacterium. This band is compatible with the 16S rDNA gene of H. pylori in the total DNA extracted from C. albicans ATCC 90028 co-cultured with the bacteria at the three different pH values. This result confirmed the above-mentioned microscopic observations indicating the presence of H. pylori within C. albicans cells.
Figure 5.
Detection, in 2% agarose gel, of the 16S rDNA gene of H. pylori J99 in the total DNA extracted from C. albicans ATCC 90028 previously co-incubated with the bacterium for 48 h at pH 3, pH 4, and pH 7. (M) Molecular weight marker; (C+) positive control (H. pylori J99); (C−) negative control (C. albicans 90028 incubated in the absence of the bacterium); (B) blank (polymerase chain reaction (PCR) degree water); (1–3) amplicons from the total DNA of co-cultures incubated at pH 7, 4, and 3, respectively.
3. Discussion
It is known that the ecological niche of H. pylori is the human stomach, an acidic environment (pH 2.0 to 3.0) to which this bacterium has adapted. However, some H. pylori genes have also been reported in water (pH 6.5–7.4), milk (pH 6.6–6.8), and in the human oral cavity (pH 5.5–7.4), but viable bacteria have so far not been cultured from these samples. It is possible that the H. pylori strains present in these environments are dormant (i.e., viable but non-culturable). This assumption is based on the fact that H. pylori in drinking water or in the presence of stressing factors may acquire a coccoid morphology, associated with a state of dormancy [29,30].
Our study showed that H. pylori reduced its viability over time in an acidic pH, particularly pH 3, the lowest pH value analyzed (Figure 1). This is due to the absence of urea in the medium because, in the gastric environment, this bacterium neutralizes the pH through secretion of the urease enzyme that hydrolyzes urea to produce ammonium and CO2 [31]. In the absence of urea, this bacterium cannot raise the environmental pH and must seek other strategies to protect itself from stressors, such as its ability to invade and remain viable within eukaryotic cells. H. pylori has been reported not only to invade and survive within human cells, but also within free-living amoebas and yeasts isolated from the mouth of adults and newborns, as well as the vagina, yogurt, honey, and other environmental sources [2,19,32,33,34] The results of these studies suggest that eukaryotic organisms might protect H. pylori and be a transmission vehicle for this bacterium. In addition, C. albicans has a wide range of tolerance to pH, which was confirmed in this study: the C. albicans ATCC 90028 strain was not affected by the acidic pH values assayed (Figure 2), not reaching the stationary phase even after 50 h of incubation. Fungi have the capacity to adapt to or modify the environmental pH, secreting acids or alkalis, with some pathogenic fungi even secreting acids as a strategy to damage the tissues of the host [35]. Therefore, the wide range of pH tolerance showed by the C. albicans ATCC 90028 strain was expected.
When H. pylori and C. albicans were co-cultured at an acidic pH, the percentage of yeasts harboring bacteria was nearly twice that observed in the control (pH 7), demonstrating that although H. pylori enters the yeast without the presence of an apparent stressor, the pH is a factor capable of influencing the entry of H. pylori into yeasts. Regarding the difference between the yeasts harboring H. pylori at pH 4 and pH 3, a larger percentage of C. albicans harboring bacteria would be expected at lower pHs; nevertheless, our results showed the opposite. The cell wall of yeasts is normally composed of two layers, an external one composed of mannoproteins, called the fibrillar layer, and an inner layer composed of chitin and glucans [15]. Sherrington et al. [15] demonstrated that C. albicans exposed to acidic pHs (pH 2, 4, and 6) modified the structure of their cell wall, causing a significant loss of the fibrillar layer, and increasing the exposure of chitin and β-glucans at pH values below 4, as compared to those cultured at pH 8 or in yeast extract peptone dextrose (YPD) medium at pH 7 [36]. Therefore, it is possible to postulate that this change in the cell wall of the yeast might impede the entry of H. pylori at pH values below 4. It has also been reported that changes in pH also induce the remodeling of the plasmatic membrane of fungi. These modifications include changes in the concentration of fatty acids and ergosterol, turning the membrane into a more rigid structure [37,38]. Thus, this is another factor which may affect the entry of H. pylori into yeasts.
In addition, regarding the issue of H. pylori localization within vacuoles of eukaryotic cells, Siavoshi et al. were able to identify the presence of this bacterium within vacuoles of yeasts—the organelle where other viable bacterial cells were also observed [2,39]. It is known that the vacuole of yeasts plays important roles, including pathogenicity. It has been described that mutant C. albicans with severe defects for the biogenesis of vacuoles are avirulent, i.e., not able to form hyphae, which, for these fungi, are essential structures to invade and for its pathogenicity [40]. Other functions attributed to the vacuole are the classification of signals, osmoregulation, and storage of metabolites, such as Ca2+, phosphate, and amino acids [41]. Phosphate and amino acids may offer a sustaining environment for H. pylori within this yeast.
Although other authors have reported the presence of H. pylori within vacuoles of yeasts obtained from different sources, there is one study that proposes a stressing factor promotes the exit of intracellular bacteria from yeasts [6]. The present work is, to the best of our knowledge, the first report on the factors involved in the entry of H. pylori into yeasts, demonstrating that acidic pH values affect the entry of H. pylori into C. albicans. Since scientific reports with this focus are scarce, the results here described are of great importance for future research.
4. Materials and Methods
4.1. Strains and Culture Conditions
This work was done using the reference strain H. pylori J99 (ATCC 700824, urease+cagA+vacA+, isolated from a duodenal ulcer) and the C. albicans ATCC 90028 strain. H. pylori J99, used as a representative of the species H. pylori, was cultured in plates containing Columbia agar (OXOID, United Kingdom) supplemented with 5% horse blood and DENT (OXOID, Basingstoke, United Kingdom) at 37 °C for 72 h in an incubator (Thermo Scientific, Waltham, MA, USA) under microaerobic conditions (10% CO2). C. albicans ATCC 90028 was used as a representative of the genus Candida. It was cultured on Sabouraud agar (Merck, Darmstadt, Germany) supplemented with chloramphenicol (OXOID, Basingstoke, United Kingdom) and incubated at 37 °C for 24 h in an incubator (ZHICHENG, Shanghai, China) under aerobic conditions. H. pylori J99 strain is part of the culture collection of the Laboratory of Bacterial Pathogenicity of the University of Concepcion, Chile, the premises where this work was done. C. albicans ATCC 90028 was donated by Dr. Patricio Godoy, Institute of Clinical Microbiology, Universidad Austral de Chile, Valdivia, Chile.
4.2. Exposure to Acidic pH Stress Assay
For this assay, H. pylori J99 and C. albicans ATCC 90028 were independently exposed to pH 3, 4, and 7 in order to choose a pH that did not affect C. albicans viability, but generated stress for H. pylori (pH 3 and 4). Cultures at pH 7 were used as controls. Firstly, both microorganisms were cultured until each culture reached its exponential phase. Then, each strain was independently cultured in 55 mL of Brain Heart Infusion (BHI) medium (Difco, Wokingham, United Kingdom) supplemented with 1% yeast extract, and the appropriate pH values were obtained using 10 M hydrochloric acid (HCl) or 10 M sodium hydroxide (NaOH) and a pH meter (Bante, Shanghai, China); the initial inoculum was adjusted to a turbidity equivalent to McFarland 7 (2 × 109 colony-forming units (CFU)/mL) and incubated for 72 h at 37 °C under microaerobic conditions. Every 4 h, a 200 µL aliquot was obtained from each culture and transferred to a well of 96-well plates, and the absorbance was measured spectrophotometrically (TECAN, Männedorf, Switzerland) at a wavelength of 600 nm, as reported by Huang et al. [30]. In parallel, microscopic observations (wet mount and Gram staining) were performed for each 4 h sampling time in order to observe the morphology of each microorganism (H. pylori J99 and C. albicans 90028). In addition, every 4 h, H. pylori bacterial counts were determined using the microdrop technique, wherein 100 µL of H. pylori culture was diluted with 900 µL saline solution, and five serial dilutions were prepared. Thereafter, 10 µL of the original dilution and of the five serial dilutions were seeded, by triplicate, in plates containing Columbia agar supplemented with DENT (OXOID, United Kingdom) and incubated at 37 °C for 36 h under microaerobic conditions. The number of CFUs was counted, and the results were expressed as CFU/mL using the formula shown below [30]. This assay was done in triplicate.
4.3. In Vitro Assessment of H. pylori J99 Entry into C. albicans ATCC 90028 Caused by Acidic pH
H. pylori J99 and C. albicans ATCC 90028 were co-cultured as described in the following. A 1 × 105 CFU/mL H. pylori suspension was prepared from a 36 h culture. In parallel, a 1 × 103 CFU/mL C. albicans ATCC 90028 suspension was prepared from a 20 h culture. Both suspensions were prepared in 15 mL Falcon tubes containing Brain Heart Infusion (BHI) broth supplemented with 1% yeast extract. Then, 100 µL of H. pylori suspension and 500 µL of C. albicans suspension were mixed in a Falcon tube containing 15 mL BHI broth supplemented with 1% yeast extract and incubated at 37 °C under microaerobic conditions. Aliquots of 20 µL of the co-culture were obtained at 0, 1, 3, 6, 12, 24, and 48 h and observed as wet mounts using immersion oil and the 100X objective lens of an optical microscope (Leica, Wetzlar, Germany) to search for the presence of yeasts harboring bacterium-like bodies (BLBs). The percentage of BLBs carrying yeasts was calculated after counting the number of yeasts harboring BLBs or not harboring them in 100 microscopic fields.
In order to have as many yeast cells harboring H. pylori as possible, the time with the higher percentage of intra-yeast BLBs for each pH assayed was selected for the following assays. Extracellular bacteria were eliminated as reported by Moreno-Mesonero et al. (2016) [19], with modifications. Each Falcon tube containing the co-culture was centrifuged at 5000× g for 3 min, the pellet was resuspended using 200 µL of 104 ppm sodium hypochlorite, and then it was incubated for 1 h to kill extracellular bacteria. Then, the suspension was centrifuged at 5000× g for 3 min, and the cells were washed thrice using 1.5 mL phosphate-buffered saline (PBS) and the same centrifugation protocol. Then, the collected yeasts were seeded on Sabouraud agar (Merck, Darmstadt, Germany) supplemented with chloramphenicol (OXOID, Basingstoke, United Kingdom) and incubated at 37 °C for 24 h in an incubator (ZHICHENG, Shanghai, China) under aerobic conditions. Next, three reseedings were done to eliminate possible persistent extracellular bacteria. At each reseeding, wet mounts were prepared and analyzed using an optical microscope to confirm the presence of intra-yeast BLBs. This assay was done in triplicate.
4.4. Detection of H. pylori J99 Entry into C. albicans ATCC 90028 by Immunofluorescence
This assay was done using Fluorescein Isothiocyanate (FITC) (Abcam, Cambrige, United Kingdom)—labeled rabbit polyclonal anti-H. pylori IgG antibodies at a concentration of 5 mg/mL (ab30954). C. albicans ATCC 90028 and H. pylori J99 strains were used as negative and positive controls, respectively. Samples and controls were diluted in Eppendorf tubes containing 1 mL PBS 1× and adjusted to a turbidity equivalent to McFarland 2 (6 × 108 UFC/mL). Using 96-well plates, 200 μL of each sample or control was placed in a well, and 1 μL of FITC-labeled anti-H. pylori IgG was added and incubated for 1 h at room temperature in darkness. After this time span, 10 μL was obtained from each well and placed on a slide observed at a wavelength of 528 nm using a confocal microscope (LSM780 NLO, ZEISS), with an Ar488 nm excitation and 490–560 nm emission. Transmitted light images, corresponding to fluorescence images with a 2–4 μm thickness, were obtained using the Zen 2012 software.
4.5. Amplification of The H. pylori 16S rDNA Gene from The Total DNA Extracted from C. albicans ATCC 90028
Extraction of the total DNA of C. albicans ATCC 90028 was done using the UltraClean Microbial DNA Isolation kit (M.O. BIO, Carlsbad, CA, USA), following the instructions of the manufacturer, and the extracted DNA was quantified by spectrophotometry (TECAN, Männedorf, Switzerland). The 16S rDNA gene of H. pylori was amplified using the SapphireAmptf Fast PCR Master Mix kit (TAKARA BIO INC, Japan). For each sample, 12.5 μL of Master Mix, 1 μL of the forward primer F- 5′CTC GAG AGA CTA AGC CCT CC 3′, 1 μL of the reverse primer R- 5′ATT ACT GAC GCT GAT GTG C 3′,5 μL of DNA of the sample, and 5.5 μL of PCR-grade water were added to obtain a final volume of 25 μL of PCR mixture. PCR conditions were as follows: initial denaturation 94 °C/1 min, denaturation temperature 98 °C/5 s, hybridization temperature 53 °C/5 s, and extension at 72 °C/40 s. Thirty cycles for each PCR reaction were done using a thermocycler (Eppendorf, Hauppauge, NY, USA). Amplification of the 16S rDNA gene was confirmed after 2% agarose gel electrophoresis (Lonza, Walkersville, MD, USA) run at 80 V for 90 min, and the gels were visualized under UV light using an Enduro model transilluminator (Labnet, Edison, NJ, USA) [22].
4.6. Statistical Analysis
To determine if there was a statistically significant difference in the percentage of yeasts harboring H. pylori at the three pH values assayed, the Chi-squared test was applied. p-values < 0.05 were considered as significant. Data were processed using the SPSS 24.0 software (IBM Company, Armonk, NY, USA).
5. Conclusions
The results suggest that H. pylori can shelter itself within C. albicans under unfavorable pH conditions.
Acknowledgments
We appreciate the important collaboration of the Advanced Microscopy Center, University of Concepción (Concepción, Chile), and of Patricio Godoy, Institute of Clinical Microbiology, Austral University of Chile (Valdivia, Chile), who provided us the C. albicans ATCC 90028 strain used to perform the assays.
Author Contributions
Conceptualization, K.S.-A. and A.G.-C.; methodology, K.S.-A., C.P.-S., S.V., A.G.-C.; validation, K.S.-A., C.P.-S., H.B., C.T.S., A.G.-C.; formal analysis, K.S.-A.; investigation, K.S.-A., C.P.-S., A.G.-C.; resources, H.B., A.G.-C.; data curation, K.S.-A., V.L.C.; writing—original draft preparation, K.S.-A., C.P.-S., H.B., A.G.-C.; writing—review and editing, K.S.-A.; C.P.-S., H.B., V.L.C., A.G.-C.; visualization, K.S.-A., C.P.-S.; supervision, A.G.-C.; project administration, A.G.-C.; funding acquisition, A.G.-C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Grant VRID-Enlace 218.036.047-1.0, University of Concepcion, Concepcion, Chile.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Berk S.G., Ting R.S., Turner G.W., Ashburn R.J. Production of respirable vesicles containing live Legionella pneumophila cells by two Acanthamoeba spp. Appl. Environ. Microbiol. 1998;64:279–286. doi: 10.1128/AEM.64.1.279-286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McFall-Ngai M., Hadfield M.G., Bosch T.C.G., Carey H.V., Domazet-Lošo T., Douglas A.E., Dubilier N., Eberl G., Fukami T., Gilbert S.F., et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Nat. Acad. Sci. USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Man S.M., Place D.E., Kuriakose T., Kanneganti T.D. Interferon-inducible guanylate-binding proteins at the interface of cell-autonomous immunity and inflammasome activation. J. Leukoc. Biol. 2017;101:143–150. doi: 10.1189/jlb.4MR0516-223R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siavoshi F., Heydari S., Shafiee M., Ahmadi S., Saniee P., Sarrafnejad A., Kolahdoozan S. Sequestration inside the yeast vacuole may enhance Helicobacter pylori survival against stressful condition. Infect. Genet. Evol. 2019;69:127–133. doi: 10.1016/j.meegid.2019.01.029. [DOI] [PubMed] [Google Scholar]
- 5.Gasch A.P., Werner-Washburne M. The genomics of yeast responses to environmental stress and starvation. Funct. Integr. Genomics. 2002;2:181–192. doi: 10.1007/s10142-002-0058-2. [DOI] [PubMed] [Google Scholar]
- 6.Heydari S., Siavoshi F., Ebrahimi H., Sarrafnejad A., Sharifi A.H. Excision of endosymbiotic bacteria from yeast under aging and starvation stresses. Infect. Genet. Evol. 2020;78:104141. doi: 10.1016/j.meegid.2019.104141. [DOI] [PubMed] [Google Scholar]
- 7.Siavoshi F., Salmanian A.H., Akbari F., Malekzadeh R., Massarrat S. Detection of Helicobacter pylori-specific genes in the oral yeast. Helicobacter. 2005;10:318–322. doi: 10.1111/j.1523-5378.2005.00319.x. [DOI] [PubMed] [Google Scholar]
- 8.Salmanian A.H., Siavoshi F., Akbari F., Afshari A., Malekzadeh R. Yeast of the oral cavity is the reservoir of Heliobacter pylori. J. Oral Pathol. Med. 2008;37:324–328. doi: 10.1111/j.1600-0714.2007.00632.x. [DOI] [PubMed] [Google Scholar]
- 9.Saniee P., Siavoshi F., Nikbakht Broujeni G., Khormali M., Sarrafnejad A., Malekzadeh R. Immunodetection of Helicobacter pylori-specific proteins in oral and gastric Candida yeasts. Arch. Iran. Med. 2013;16:624–630. [PubMed] [Google Scholar]
- 10.Saniee P., Siavoshi F., Nikbakht Broujeni G., Khormali M., Sarrafnejad A., Malekzadeh R. Localization of H.pylori within the vacuole of Candida yeast by direct immunofluorescence technique. Arch. Iran. Med. 2013;16:705–710. [PubMed] [Google Scholar]
- 11.Nett J.E. Special Issue: Candida and Candidiasis. J. Fungi (Basel) 2018;4:74. doi: 10.3390/jof4030074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zisova L.G., Chokoeva A.A., Amaliev G.I., Petleshkova P.V., Miteva-Katrandzhieva T., Krasteva M.B., Uchikova E.H., Kouzmanov A.H., Ivanova Z.V. Vulvovaginal Candidiasis in Pregnant Women and its Importance for Candida Colonization of Newborns. Folia. Med. (Plovdiv) 2016;58:108–114. doi: 10.1515/folmed-2016-0018. [DOI] [PubMed] [Google Scholar]
- 13.Yano J., Sobel J.D., Nyirjesy P., Sobel R., Williams V.L., Yu Q., Noverr M.C., Fidel P.L., Jr. Current patient perspectives of vulvovaginal candidiasis: Incidence, symptoms, management and post-treatment outcomes. BMC Womens Health. 2019;19:48. doi: 10.1186/s12905-019-0748-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miramón P., Lorenz M.C. A feast for Candida: Metabolic plasticity confers an edge for virulence. PLoS Pathog. 2017;13:e1006144. doi: 10.1371/journal.ppat.1006144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherrington S.L., Sorsby E., Mahtey N., Kumwenda P., Lenardon M.D., Brown I., Ballou E.R., MacCallum D.M., Hall R.A. Adaptation of Candida albicans to environmental pH induces cell wall remodelling and enhances innate immune recognition. PLoS Pathog. 2017;13:e1006403. doi: 10.1371/journal.ppat.1006403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odds F.C. Candida infections: An overview. Crit. Rev. Microbiol. 1987;15:1–5. doi: 10.3109/10408418709104444. [DOI] [PubMed] [Google Scholar]
- 17.Soll D.R., Galask R., Schmid J., Hanna C., Mac K., Morrow B. Genetic dissimilarity of commensal strains of Candida spp. carried in different anatomical locations of the same healthy women. J. Clin. Microbiol. 1991;29:1702–1710. doi: 10.1128/JCM.29.8.1702-1710.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soll D.R. Candida commensalism and virulence: The evolution of phenotypic plasticity. Acta. Trop. 2002;81:101–110. doi: 10.1016/S0001-706X(01)00200-5. [DOI] [PubMed] [Google Scholar]
- 19.Moreno-Mesonero L., Moreno Y., Alonso J.L., Ferrús M.A. DVC-FISH and PMA-qPCR techniques to assess the survival of Helicobacter pylori inside Acanthamoeba castellanii. Res. Microbiol. 2016;167:29–34. doi: 10.1016/j.resmic.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Siavoshi F., Nourali-Ahari F. Yeast protects Helicobacter pylori against the environmental stress. Arch. Iran. Med. 1998;1:2–821. [Google Scholar]
- 21.Salmanian A.-H., Siavoshi F., Beyrami Z., Latifi-Navid S., Tavakolian A., Sadjadi A. Foodborne yeasts serve as reservoirs of Helicobacter pylori. J. Food Safety. 2012;32:152–160. doi: 10.1111/j.1745-4565.2011.00362.x. [DOI] [Google Scholar]
- 22.Matamala-Valdés L., Sánchez-Alonzo K., Parra C., Sáez K., Aguayo-Reyes A., García A. Detection of intracellular Helicobacter pylori in Candida. SPP from neonate oral swabs. Rev. Assoc. Med. Bras. 2018;64:928–935. doi: 10.1590/1806-9282.64.10.928. [DOI] [PubMed] [Google Scholar]
- 23.Ince A.T., Kocaman O., Ismailova M., Tozlu M., Gücin Z., Iraz M. A rare co-existence of Helicobacter pylori, Candida albicans and Candida keyfir in a giant gastric ulcer. Turk. J. Gastroenterol. 2014;25:435–436. doi: 10.5152/tjg.2014.3401. [DOI] [PubMed] [Google Scholar]
- 24.Pittet D., Li N., Wenzel R.P. Association of secondary and polymicrobial nosocomial bloodstream infections with higher mortality. Eur. J. Clin. Microbiol. Infect. Dis. 1993;12:813–819. doi: 10.1007/BF02000400. [DOI] [PubMed] [Google Scholar]
- 25.Garcia A., Salas-Jara M.J., Herrera C., Gonzalez C. Biofilm and Helicobacter pylori: From environment to human host. World J. Gastroenterol. 2014;20:5632–5638. doi: 10.3748/wjg.v20.i19.5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mladenova I., Durazzo M. Transmission of Helicobacter pylori. Minerva. Gastroenterol. Dietol. 2018;64:251–254. doi: 10.23736/S1121-421X.18.02480-7. [DOI] [PubMed] [Google Scholar]
- 27.Burucoa C., Axon A. Epidemiology of Helicobacter pylori infection. Helicobacter. 2017;22(Suppl. 1) doi: 10.1111/hel.12403. [DOI] [PubMed] [Google Scholar]
- 28.Mamishi S., Eshaghi H., Mahmoudi S., Bahador A., Hosseinpour Sadeghi R., Najafi M., Farahmand F., Khodadad A., Pourakbari B. Intrafamilial transmission of Helicobacter pylori: Genotyping of faecal samples. Br. J. Biomed. Sci. 2016;73:38–43. doi: 10.1080/09674845.2016.1150666. [DOI] [PubMed] [Google Scholar]
- 29.She F.F., Lin J.Y., Liu J.Y., Huang C., Su D.H. Virulence of water-induced coccoid Helicobacter pylori and its experimental infection in mice. World J. Gastroenterol. 2003;9:516–520. doi: 10.3748/wjg.v9.i3.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azevedo N.F., Almeida C., Cerqueira L., Dias S., Keevil C.W., Vieira M.J. Coccoid form of Helicobacter pylori as a morphological manifestation of cell adaptation to the environment. Appl. Environ. Microbiol. 2007;73:3423–3427. doi: 10.1128/AEM.00047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ansari S., Yamaoka Y. Survival of Helicobacter pylori in gastric acidic territory. Helicobacter. 2017;22 doi: 10.1111/hel.12386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki N., Yoneda M., Naito T., Iwamoto T., Masuo Y., Yamada K., Hisama K., Okada I., Hirofuji T. Detection of Helicobacter pylori DNA in the saliva of patients complaining of halitosis. J. Med. Microbiol. 2008;57:1553–1559. doi: 10.1099/jmm.0.2008/003715-0. [DOI] [PubMed] [Google Scholar]
- 33.Agarwal S., Jithendra K.D. Presence of Helicobacter pylori in subgingival plaque of periodontitis patients with and without dyspepsia, detected by polymerase chain reaction and culture. J. Indian Soc. Periodontol. 2012;16:398–403. doi: 10.4103/0972-124X.100919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siavoshi F., Sahraee M., Ebrahimi H., Sarrafnejad A., Saniee P. Natural fruits, flowers, honey, and honeybees harbor Helicobacter pylori-positive yeasts. Helicobacter. 2018;23:e12471. doi: 10.1111/hel.12471. [DOI] [PubMed] [Google Scholar]
- 35.Vylkova S. Environmental pH modulation by pathogenic fungi as a strategy to conquer the host. PLoS Pathogens. 2017;13:e1006149. doi: 10.1371/journal.ppat.1006149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bahrami A.R., Rahimi E., Ghasemian Safaei H. Detection of Helicobacter pylori in city water, dental units’ water, and bottled mineral water in Isfahan, Iran. Sci. World J. 2013;2013:280510. doi: 10.1155/2013/280510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahumada-Rudolph R., Cajas-Madriaga D., Rudolph A., Reinoso R., Torres C., Silva M., Becerra J. Variation of sterols and fatty acids as an adaptive response to changes in temperature, salinity and pH of a marine fungus Epicoccum nigrum isolated from the Patagonian Fjords. Rev. Biol. Mar. Oceanogr. 2014;49:293–305. doi: 10.4067/S0718-19572014000200009. [DOI] [Google Scholar]
- 38.Valle-González E.R., Jackman J.A., Yoon B.K., Park S., Sut T.N., Cho N.J. Characterizing How Acidic pH Conditions Affect the Membrane-Disruptive Activities of Lauric Acid and Glycerol Monolaurate. Langmuir. 2018;34:13745–13753. doi: 10.1021/acs.langmuir.8b02536. [DOI] [PubMed] [Google Scholar]
- 39.Amieva M.R., Salama N.R., Tompkins L.S., Falkow S. Helicobacter pylori enter and survive within multivesicular vacuoles of epithelial cells. Cell. Microbiol. 2002;4:677–690. doi: 10.1046/j.1462-5822.2002.00222.x. [DOI] [PubMed] [Google Scholar]
- 40.Tournu H., Carroll J., Latimer B., Dragoi A.-M., Dykes S., Cardelli J., Peters T.L., Eberle K.E., Palmer G.E. Identification of small molecules that disrupt vacuolar function in the pathogen Candida albicans. PLoS ONE. 2017;12:e0171145. doi: 10.1371/journal.pone.0171145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klionsky D.J., Herman P.K., Emr S.D. The fungal vacuole: Composition, function, and biogenesis. Microbiol. Rev. 1990;54:266–292. doi: 10.1128/MMBR.54.3.266-292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]