Abstract
Bovine leptospirosis is a bacterial zoonotic disease caused by pathogenic Leptospira spp. The pathology and epidemiology of this infection are influenced by the numerous existing serovars and their adaptation to specific hosts. Infections by host-maintained serovars such as Hardjo are well documented, unlike those from the incidental ones. In July 2014, an emerging phenomenon of an increased incidence of icteric abortions associated with leptospiral infection occurred in southern Belgium. First-line serological analyses targeting cattle-adapted serovars failed at initial diagnosis. This study provides a comprehensive description of laboratory findings—at the level of necropsy, serology and molecular diagnosis—regarding icteric and non-icteric abortions (n = 116) recorded during this time (years 2014–2015) and associated with incidental infection by serovars such as Grippotyphosa, Australis and Icterohaemorrhagiae. Based on these tests, a diagnostic pathway is proposed for these types of infection in cattle to establish an affordable but accurate diagnosis in the future. These investigations add insights into the understanding of the pathogenesis of bovine leptospirosis associated with serovars classically described as non-maintenance.
Keywords: Leptospira spp., cattle, abortion, pathology, non-maintenance serovars, MAT, PCR, lfb1-phylogeny
1. Introduction
Bovine leptospirosis is a bacterial zoonotic disease caused by pathogenic Leptospira spp., bacteria classified in about 300 serovars and 64 genomospecies, of which 37 belong to the pathogenic clade [1,2,3]. The pathology and epidemiology of this bacterial disease are influenced by the existing serovars and their adaptation to specific hosts in wildlife or livestock [1]. Depending on the serovar–host relationship, incidental and host-maintained infections are described.
Leptospiral infections in cattle are known as incidental when involving non-maintenance pathogenic Leptospira spp. such as the serogroups Icterohaemorrhagiae, Hebdomadis, Grippotyphosa and Canicola [4,5,6]. These infections are transmitted through indirect or, more rarely, direct contact with an infected host, generally a wildlife reservoir (i.e., rodents for the serogroup Icterohaemorrhagiae [7,8]). On the other hand, cattle can act as the principal reservoir of the bacteria for the adapted L. borgepetersenii serovar Hardjobovis and L. interrogans serovar Hardjoprajitno. In this latter case, the transmission is more efficient and mainly occurs through direct contact with the contaminated urine or body fluids (milk or placental fluids) of infected animals residing in the same herd [9].
Infection by non-maintenance pathogenic Leptospira spp. is characterized by an acute form of the disease such as a high fever, hemolytic anemia, jaundice and multi-systemic illness [10]. Acute manifestation is rarely described for maintenance strains such as L. borgepetersenii serovar Hardjobovis and L. interrogans serovar Hardjoprajitno [11,12,13,14]. In this case, the host presents a sub-acute clinical phase of the disease that often goes unobserved. Bovine-adapted serovars follow a chronic course of infection that is associated with a prolonged renal carrier state of the adult animal associated with chronic renal diseases [15,16,17]. Chronically infected animals can excrete leptospires through urine, leading to increased environmental infection pressure. Economic issues can be important since these infections also cause reproductive failure, abortions, stillbirths, weak offspring and reduced milk production. Hence, it is a concern for public and animal health management. The management and diagnosis of infections due to L. borgepetersenii serovar Hardjobovis and L. interrogans serovar Hardjoprajitno in cattle are supported by numerous experimental and epidemiological studies [18,19,20,21,22,23,24]. L. borgepetersenii serovar Hardjobovis and L. interrogans serovar Hardjoprajitno have been shown to be causes of reproductive losses worldwide [25,26,27]. Instead, knowledge of the disease caused by non-maintenance serovars, other than from serological surveillance, is poor and linked to anecdotal reports [28,29,30]. The pathogenesis of reproductive disease due to Leptospira spp. remains poorly understood as well. Transplacental infection, occurring during the very limited time of maternal bacteremia, is supposed to be the sole cause of abortion [31].
In July 2014, an emerging phenomenon associated with an increased incidence of icteric abortions occurred in the southern region of Belgium [32]. Initial laboratory findings highlighted in dams serum high-level antibody titers against Leptospira serogroups Grippotyphosa and Australis, whilst a first-line ELISA-based serologic survey for L. borgepetersenii serovar Hardjobovis failed to identify the causative agent [32]. A definitive laboratory diagnosis was finally achieved at the animal national reference leptospirosis laboratory in the fall of 2014 by systematically associating positive serology in dams with the detection of pathogenic Leptospira spp. DNA in various organs of the abortuses. The conclusions were promptly reported to the national competent authorities (official national internal report). One epidemiological study thereafter conducted on prospective samples confirmed serological reactivity against Leptospira serogroups Grippotyphosa and Australis and provided genotypic indication in one sample of the same year, 2014 [33].
This study provides a comprehensive description of laboratory findings— at the level of necropsy, serology and molecular characterization—regarding the icteric and non-icteric abortions recorded during the years 2014–2015 and associated with leptospiral infection. Based on these tests and their relative intrinsic qualities, the goal of this work is to provide comprehensive tools for the clinical and laboratory diagnosis of bovine leptospirosis due to non-maintenance serovars. Besides, a diagnostic pathway is proposed to establish an affordable but accurate diagnosis of these cases in the future. Finally, these investigations add insights into the understanding of the pathogenesis and clinical manifestation of bovine leptospirosis associated with serovars classically described as non-maintenance.
2. Results
2.1. Characteristics at Necropsy of MAT Positive Abortions
A total of 116 fetuses were selected for the entire study. The gestational age varied from 3 to 9 months, with the majority of abortions occurring during the last trimester of gestation (Table 1). When considering the lesions assessed during necropsy, four entries were initially selected due to previously being observed in icteric abortions [32]: icterus (n = 52), splenomegaly (n = 59), coppery liver (n = 40) and peri-renal hemorrhage (n = 59). However, these entries accounted only partially for the microscopic agglutination test (MAT)-positive results observed (Table 2 and Table S1). Therefore, the presence of generalized hemorrhagic edema (n = 6) was additionally included to better understand the complete lesion panel associated with Leptospira infection. For each of these lesions, the association with a positive MAT result (regardless of the reactive serogroup) was evaluated. Four different seropositivity cut-offs for MAT were considered: 1/10, 1/100, 1/300 and 1/1000 (Table 2). A significant association was observed between seropositivity and the presence of icterus, splenomegaly or coppery liver for all of the considered cut-offs. A similar observation was made for the combination of these three lesions. By contrast, no link was established for the peri-renal hemorrhages, regardless of the MAT cut-off. An extended hemorrhagic pattern was associated with a positive MAT result for the cut-off of 1/1000 and, interestingly, observed only in the non-icteric abortion group (Table 2).
Table 1.
Distribution of the autopsied fetuses according to their gestational age, presence of icterus (other lesions not included) and positivity rates of MAT (cut-off of 1/10).
| Month of Abortion | N Autopsied | Icteric (N Positive/ N Analyzed for Lesions) |
Positive MAT in Dam’s Sera (N Positive/N Tested) |
Positive MAT in Pleural Fluids (N Positive/N Tested) |
Positive PCR (N Positive/N Tested) |
|---|---|---|---|---|---|
| <5 | 8 | 0/8 | 5/7 | 0/1 | 0/0 |
| 5 | 2 | 0/2 | 1/2 | 0/0 | 0/1 |
| 6 | 5 | 0/5 | 4/5 | 0/0 | 0/2 |
| 7 | 7 | 2/6 | 4/7 | 0/0 | 0/5 |
| 8 | 43 | 18/40 | 30/42 | 1/9 | 13/25 |
| 9 | 51 | 32/50 | 31/45 | 0/16 | 19/37 |
| TOTAL | 116 | 52/111 | 75/108 | 1/26 | 32/70 |
Table 2.
Association between necropsy findings in bovine abortuses and results of MAT (positive reaction for any serogroup) performed on sera of the corresponding dams. P = presence; A = absence.
| Lesion | N Fetuses | Positivity Cut-Off of MAT | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1/10 | 1/100 | 1/300 | 1/1000 | |||||||
| % Positive MAT (n) | OR (95% IC) χ2 and p-Value |
% Positive MAT (n) | OR (95% IC) χ2 and p-Value |
% Positive MAT (n) | OR (95% IC) χ2 and p-Value |
% Positive MAT (n) | OR (95% IC) χ2 and p-Value |
|||
| Icterus | P | 44 | 90.91 (40) | 12.61 (3.94–40.40) χ2 = 20.99 p < 0.001 |
88.64 (39) | 13.55 (4.56–40.24) χ2 = 24,91 p < 0.001 |
77.27 (34) | 14.28 (5.33–38.29) χ2 = 30,04 p < 0.001 |
43.18 (19) | 7.14 (4.09-12.48) χ2 = 12.59 p < 0.001 |
| A | 52 | 44.23 (23) | 36.54 (19) | 19.23 (10) | 9.62 (5) | |||||
| Splenomegaly | P | 54 | 83.33 (45) | 6.67 (2.60–17.09) χ2 = 15.41 p < 0.001 |
77.78 (42) | 5.69 (2.33–13.91) χ2 = 13,94 p < 0.001 |
61.11 (33) | 4.43 (1.84–10.67) χ2 = 10.24 p = 0.001 |
33.33 (18) | 3.00 (1.12–8.04) χ2 = 3.61 p = 0.057 |
| A | 42 | 42.86 (18) | 38.10 (16) | 26.19 (11) | 14.29 (6) | |||||
| Coppery liver | P | 36 | 86.11 (31) | 5.43 (1.86–15.85) χ2 = 9.31 p = 0.002 |
80.56 (29) | 4.43 (1.68–11.66) χ2 = 8.47 p = 0.004 |
69.44 (25) | 5.99 (2.35–15.29) χ2 = 13.64 p < 0.001 |
44.44 (16) | 5.20 (2.83–9.56) χ2 = 10.01 p = 0.002 |
| A | 60 | 53.33 (32) | 48.33 (29) | 31.67 (19) | 13.33 (8) | |||||
| Icterus + splenomegaly + coppery liver | P | 29 | 89.66 (26) | 7.03 (1.94–25.49) χ2 = 9.16 p = 0.002 |
86.21 (25) | 6.44 (2.02–20.52) χ2 = 10.06 p = 0.002 |
75.86 (22) | 6.43 (2.38–17.33) χ2 = 13.41 p < 0.001 |
44.83 (13) | 4.14 (2.46–6.97) χ2 = 7.26 p = 0.007 |
| A | 67 | 55.22 (37) | 49.25 (33) | 32.84 (22) | 13.33 (11) | |||||
| Peri-renal hemorrhages | P | 54 | 64.81 (35) | 0.92 (0.39–2.16) χ2 = 0.0007 p = 0.978 |
57.41 (31) | 0.75 (0.33–1.72) χ2 = 0.22 p = 0.636 |
44.44 (24) | 0.88 (0.39–1.98) χ2 = 0.01 p = 0.918 |
25.93 (14) | 1.12 (0.44–2.85) χ2 = 0.007 p = 0.934 |
| A | 42 | 66.67 (28) | 64.29 (27) | 47.62 (20) | 23.81 (10) | |||||
| Extended hemorrhagic pattern | P | 6 | 83.33 (5) | 2.76 (0.31–24.65) p = 0.661 |
83.33 (5) | 3.49 (0.39–31.12) p = 0.397 |
83.33 (5) | 6.54 (0.73–58.26) p = 0.09 |
66.67 (4) | 7.00 (1.19–41.04) p = 0.033 |
| A | 90 | 64.44 (58) | 58.89 (53) | 43.33 (39) | 22.22 (20) | |||||
| TOTAL | 96 | 65.63 (63) | 60.42 (58) | 45.83 (44) | 25.00 (24) | |||||
2.2. Serological Pattern of Abortions
Serological analyses were performed on 88 individual dams’ sera, on 20 coupled dams’ sera and fetuses’ pleural fluids, and six individual fetuses’ pleural fluids. Pleural fluids were chosen as an alternative to missing post-mortem fetal sera because they reflect the serum antibody levels arising through passive diffusion [34]. These analyses allowed the obtaining of MAT profiles for all but two abortuses. Globally, a higher positivity rate was observed in abortuses with icteric (42/50, 80.7%) vs. non-icteric (33/64, 51.5%) patterns (Table S1). When both dam’s serum and fetus’ pleural fluid were tested and a positive serological MAT result was observed (18/20 abortions), positivity was observed only in the dam’s serum in 17 out of 18 cases (94.4%) (Table S1). For one single fetus, serologically positive reactions were observed in both the dam (high titer) and the pleural fluid of the fetus (low titer), with perfect agreement on the presumptive serogroup. The presumptive infecting Leptospira serogroup was defined in 65/75 (86.7%) of the MAT-positive abortuses, with the following frequencies: Grippotyphosa (n = 42), Australis (n = 13), Ballum (n = 4), Icterohaemorrhagiae (n = 2), Autumnalis (n = 1), Bataviae (n = 1), Pyrogenes (n = 1) and Tarassovi (n = 1). None of these serogroups belongs to the pathogenic Leptospira serovars classically described as maintenance in cattle. Cross-reactivities between the serogroups were observed, in order of decreasing frequency, between the serogroups Grippotyphosa and Ballum (18/75), Grippotyphosa and Autumnalis (14/75), Grippotyphosa and Australis (10/75), and Australis and Autumnalis (9/75) (Table S2).
2.3. Bacteriological Characteristics of Abortions
PCR was performed on 68 fetuses in available organs. A fetus was considered positive when a positive PCR result was reported in at least one organ. Again, clear high positivity rates were observed in abortuses with icteric (30/41, 73.17%) vs. non-icteric (1/27, 3.70%) patterns, as shown in Table 3. When associations were calculated between each lesion and a positive PCR result in abortuses, a significant link was only found for icterus, splenomegaly, coppery liver and the combination of these three lesions. No significant link was observed for peri-renal hemorrhages and general hemorrhagic pattern (Table 3). Based on the PCR results in the different organs, the placenta, spleen and liver appeared as the most appropriate of a fetus’ organs in which to look for non-maintenance Leptospira spp. colonization (Table 4). The positive rate was significantly higher in the placenta than in the kidney (χ2 = 6.15, p-value = 0.013). The median Ct (cycle threshold) values were averagely high in all the organs, and most of the Cts were above 30. For all but five of the tested fetuses (68), the MAT results for the dam’s serum were available to define the possible relations between PCR and MAT tests (Table S3). Cohen’s kappa coefficient, as well as the relative sensitivity and specificity towards PCR, were calculated for each cut-off of the MAT test. The cut-off of 1/300 showed the highest concordance value (kappa = 0.679, substantial agreement), with a relative sensitivity and a specificity approaching 85% (Table S3).
Table 3.
Association between necropsy findings in bovine abortuses and PCR results for the diagnosis of leptospirosis. P = presence; A = absence.
| Lesions | N Fetuses | % Positive PCR (n) | OR (95% IC) χ2 and p-Value |
|
|---|---|---|---|---|
| Icterus | P | 41 | 73.17 (30) | 70.91 (8.57–586.89) χ2= 31.67 (p < 0.001) |
| A | 27 | 3.70 (1) | ||
| Splenomegaly | P | 45 | 64.44 (29) | 19.03 (3.95–91.81) χ2= 16.89 (p < 0.001) |
| A | 23 | 8.70 (2) | ||
| Coppery liver | P | 31 | 70.97 (22) | 7.60 (2.58–22.38) χ2= 12.97 (p < 0.001) |
| A | 37 | 24.32 (9) | ||
| Icterus + splenomegaly + coppery liver | P | 28 | 75.00 (21) | 9.00 (2.95–27.45) χ2= 14.65 (p < 0.001) |
| A | 40 | 25.00 (10) | ||
| Peri-renal hemorrhages | P | 49 | 46.94 (23) | 1.22 (0.42–3.54) χ2= 0.008 (p =0.930) |
| A | 19 | 42.11 (8) | ||
| Extended hemorrhagic pattern | P | 3 | 33.33 (1) | 0.58 (0.05-6.76) p = 1 |
| A | 65 | 46.15 (30) | ||
| TOTAL | 68 | 45.59 (31) | ||
Table 4.
PCR results obtained in various organs of the autopsied fetuses* (n = 68).
| Organ | N Positive/N Tested | Positivity (%) | Median Ct Value | Range Min–Max |
|
|---|---|---|---|---|---|
| Spleen | 18/45 | 40.00 | 36.67 | 31.06 | 41.45 |
| Placenta a | 17/32 | 53.13 | 34.14 | 27.52 | 41.41 |
| Kidney a | 4/21 | 19.05 | 33.92 | 28.10 | 38.64 |
| Adrenal glands | 1/15 | 6.67 | 36.60 | 36.60 | 36.60 |
| Liver | 5/14 | 35.71 | 34.58 | 31.90 | 37.60 |
| Lung | 2/10 | 20.00 | 34.61 | 32.30 | 36.92 |
| Brain | 0/5 | 0.00 | / | / | / |
| Hepatic lymph nodes | 2/3 | 66.67 | 37.45 | 36.70 | 38.20 |
* One abortus might be positive for several organs. a significantly different (p = 0.013).
2.4. Molecular Typing of Leptospira spp.
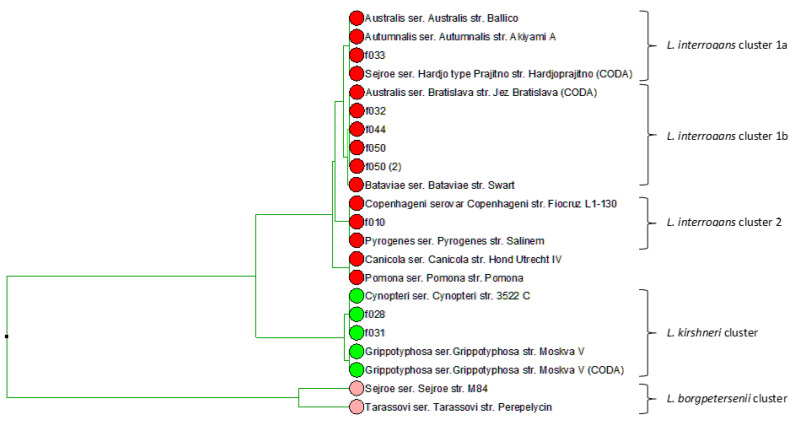
To better understand the epidemiology of Leptospira-induced abortions in this study and to identify, at the species level, the Leptospira infecting strain, the analysis of the sequence polymorphism in the fibronectin-binding protein gene (lfb1) gene was performed by using both high-resolution melting analysis (HRMA) and amplicon sequencing. The melting curve analysis of eight fetuses indicated the presence of two main types of PCR amplicon, with different melting temperatures (Tm), highlighting the presence of at least two Leptospira species causing abortions. The first cluster, with an average 81.8 °C Tm, was assigned, by comparison with the Tm of reference strains, to the Leptospira interrogans group; the second cluster, with a one-degree difference in Tm (average of 82.8 °C) was assigned to the Leptospira kirschneri group (Figure S1). Phylogenetic analyses of the lfb1 sequence in seven fetuses and reference strains (ours or those available in databases) corroborated observations by HRMA. The results, supported by the amplification of a minimum of 238 bp of sequence (accession numbers KY373222 to KY373229), evidenced the heterogeneity of the species involved in abortions: three Leptospira interrogans clusters (1a, 1b and 2) including the serovars Australis Ballico, Autumnalis and Hardjoprajitno (cluster 1a); the serovars Australis Bratislava and Bataviae (cluster 1b); the serovars Pyrogenes and Copenhageni (cluster 2); one Leptospira kirschneri cluster including the reference sequences of the serovars Cynopteri and Grippotyphosa (Figure 1). Except for the fetus f044, where the presumptive serogroup could not be identified by MAT, the Leptospira genospecies based on molecular typing is compatible with the presumptive serogroups determined by MAT in dams (Table 5).
Figure 1.
lbf1-derived phylogeny of Leptospira-induced icteric abortions and reference strains. Red, green and rose circles indicate strains of L. interrogans, L.kirschneri and L. borgepetersenii species, respectively. Reference strains are listed with their serogroup, serovar and strain name. Abortuses are indicated with their fetus ID as in Table S1.
Table 5.
Comparison of results in autopsied fetuses according to high-resolution melting analysis (HRMA), lfb1 phylogeny and serology in dams.
| HRMA rtPCR | lfb1 Sequencing | Serology of Dam | Necropsy | |||
|---|---|---|---|---|---|---|
| Fœtus ID | Sample ID | Tm | Species Group Attribution | Species Group Attribution | MAT | Group |
| f050 | 15095924-1509927-159915 | 81.92 | interrogans | interrogans cluster 1b | 1/500 Australis | Icteric |
| f033 | 14040522.1-1610225.2 | 81.93 | interrogans | interrogans cluster 1a | 1/1000 Icterohaemorrhagiae | Icteric |
| f010 | 14048419.1-1610225.3 | 81.69 | interrogans | interrogans cluster 2 | 1/500 Australis | Icteric |
| f032 | 14045546 | 81.71 | interrogans | interrogans cluster 1b | - | Icteric |
| f044 | 14057925-1417554.1-1417819 | 81.72 | interrogans | interrogans cluster 1b | ND (1/100 Ballum 1/100 Grippothyphosa) | Icteric |
| f028 | 15050216-1506066.2 | 82.96 | kirschneri | kirschneri cluster | 1/100 Grippotyphosa | Icteric |
| f007 | 14047394.1-1610225.1 | 82.82 | kirschneri | - | 1/500 Grippotyphosa | Icteric |
| f031 | 14044550 | 82.76 | kirschneri | kirschneri cluster | - | Icteric |
3. Discussion
Abortion is a well-known manifestation of leptospirosis in cattle. Nevertheless, extended anatomo-pathological descriptions of fetuses infected with leptospires are rarely available, especially for serovars other than L. borgepetersenii serovar Hardjobovis. The emergence of abortions due to non-maintenance leptospires observed in 2014 in southern Belgium showed that general icterus was the most striking lesion reported in the abortuses [32]. Jaundice is an acute manifestation commonly reported in newborn calves infected by Leptospira spp., due to hemolysis following hemolysin production. This lesion was also reported in fetuses in cases of experimental abortions with L. borgepetersenii serovar Hardjobovis [35]. On the basis of the abortuses included in this study, coppery liver and splenomegaly, besides icterus, were significantly associated with a positive result for Leptospira, for both serology and bacteriology. These lesions in abortuses are not pathognomonic, and their appreciation depends on the state of conservation of the cadaver. Less distinct lesions such light icterus could be missed by an unaware or untrained eye. Despite these minor drawbacks, necropsy of the fetuses remains, as strongly confirmed in this study, an efficient tool in the global context of abortion surveillance. As of the Belgian experience, such monitoring proved its efficacy during the re-emergence episodes of the Schmallenberg virus and, to a lesser extent, brucellosis [36,37]. In addition to the three mentioned lesions, generalized hemorrhagic edema has been observed in some abortuses and was shown to be associated with high antibody titers in dams. No specific serovar was associated with this lesion. Petechial and ecchymotic hemorrhages on internal organs completed the picture in most of these cases, whereas jaundice was absent in these fetuses except in one. The high Leptospira titers associated with generalized hemorrhagic edema suggested a per-acute phenomenon in this case. This feature might correspond to the first stage of the disease leading to an icteric pattern or even represent a completely different pathogenesis of bovine leptospirosis. Indeed, the outcome of Leptospira infection varies depending on the infecting serovars but also on host specificities or host–pathogen interaction [38].
Globally, serological analyses indicated that non-maintenance Leptospira spp. serogroups were, in different proportions, the causes of the icteric-hemorrhagic abortions observed in this study, namely serogroups Grippotyphosa, Australis, Ballum, Icterohaemorrhagiae, Autumnalis, Bataviae, Pyrogenes and Tarassovi. The seropositive results could not be attributed to vaccination, as a vaccine for cattle is not marketed in Belgium. The comparison of our results with European trends is difficult, although our data are in line with the marked increase in leptospirosis infection observed in the year 2014 [39,40]. However, a complete epidemiological picture of Leptospira spp. serogroups in cattle across Europe is unavailable. The reasons for this are various; seroprevalence studies are often restricted to a specific timeframe, data are not centralized at the European level for animals and the standardization of the MAT analyses is subject to constraints. The Grippotyphosa, Australis, Icterohaemorrhagiae and—to a lesser extent—Autumnalis and Bataviae serogroups have been observed in clinically relevant cattle herds in France [41]. Although this in line with our data, information is missing to ascertain whether the reproductive disorders described in France were similar to the ones observed in our study. Seroprevalence studies performed in cattle herds in Italy show that the non-maintenance serogroups Australis and Icterohaemorrhagiae are observed at high titer levels, together with the maintenance serovars Hardjo and Pomona, while Ballum, Canicola, Grippotyphosa and Tarassovi are rare and at minimal titers [42]. In Spain, the relevant cattle serovars are Bratislava, Hardjo and Pomona [43,44]. Limited seroprevalence data are present in Belgium to understand the relationship between the serogroups found in this study and their possible presence/spread in other (reservoir) animals; Australis and Grippotyphosa are the main serogroups found in dogs [45], and they were also detected in muskrats [34,46]. In humans, a marked increase in autochthonous cases was reported in 2014 [47], as observed in other parts of Europe [39], but a univocal association of the serovars involved in bovine icteric abortions in 2014 with human cases could not be established (National Reference Laboratory personal communication).
Molecular diagnosis, supported by serology, highlighted two mains genospecies responsible for abortions in this study: L. interrogans and L. kirschneri. Although this was relatively clear from our previous investigations [32], here, we demonstrated by lfb1 phylogeny that infections could be sourced back to genetically diverse strains (for instance, at least three clusters for the L. interrogans species) and not to a unique “outbreak” clone. This could probably explain the heterogenous serological response against Leptospira agglutinins and variation observed with the MAT analyses. Although the L. interrogans cluster included sequences that were very closely related to the L. interrogans serovar Hardjoprajitno, the involvement of this serovar was excluded in our conclusion as it was not supported by serological analyses operated both by ELISA [32] and MAT.
Together with the above observations allowing a better understanding of the Leptospira pathogenesis in cattle, this study provided additional insights for the laboratory diagnosis of bovine leptospirosis. First, antibodies against Leptospira spp. were detected by MAT in the dam’s serum and not in the pleural fluid of the fetuses, despite positive PCR results for fetus organs. This indicated that the dam’s serum was the most appropriate sample type for serodiagnosis in this study. Although we previously demonstrated that pleural fluids, in absence of post-mortem sera, are informative for leptospiral infective status [34], these samples taken from the bovine fetus are possibly not relevant in case of cattle infection with leptospires. It can also be speculated that we could not identify seropositive reactions because the fetuses were in bacteremia, which is associated in humans with an absent or very low serological antibody response [48]. Second, the cut-off for MAT was lowered to as low as 1/10 dilution. In classical serological diagnosis, the MAT cut-off is kept at 1/100 (or at 1/400 for higher specificity); the OIE manual suggests lowering this threshold in the case of serosurveillance studies [49]. We suggest lowering this threshold also in case of clustered cases recorded in a defined time-lapse and linked to unusual phenomena. Third, cross-reactivities between serovars were likely to occur, and, with the exception of few cases, they did not hamper the final main serogroup presumptive identification, which was in high agreement with the genospecies attribution. Fourth, leptospiral antigens in abortuses were found in additional organs other than the placenta, kidneys, liver and adrenal glands [50]. As suggested by our study, Leptospira spp. are likely to be also detected by PCR in the spleen. This is not surprising since this organ acts like a blood filter and thus is susceptible to sequestering bacteria. Last, this study strongly supports, in the case of poorly loaded DNA samples, the use of lfb1 phylogeny with the HRMA or sequencing methodology for outbreak-clustered case investigations to provide enough discriminatory genetic power for difficult samples [51,52].
Some limitations could be present in our study. A positive PCR result in the abortus indicates that the bacteria reached the gravid uterus, suggesting a strong link between the presence of Leptospira and abortion. However, most of the Ct results from PCR were high, suggesting a low quantity of bacteria in fetal tissues, and some cases could have be missed due to the limit of detection of the method. In addition, in our study, a negative PCR result could not allow the exclusion of Leptospira from the abortion causes because of the non-systematic testing of all the organs. For example, the placenta was not systematically transmitted with the fetus for analysis. Additionally, a defect in the collection of the lesional data during necropsy cannot be excluded, especially at the beginning of the emergence of leptospiral abortions (01/09/14) when the pathologists were less aware of the suspicious lesions. One last limitation is related to the number of cases upon which this study is based to determine the associations between lesions and MAT results. This was mainly due to the epidemic course, with a drop in the icteric cases in October 2014.
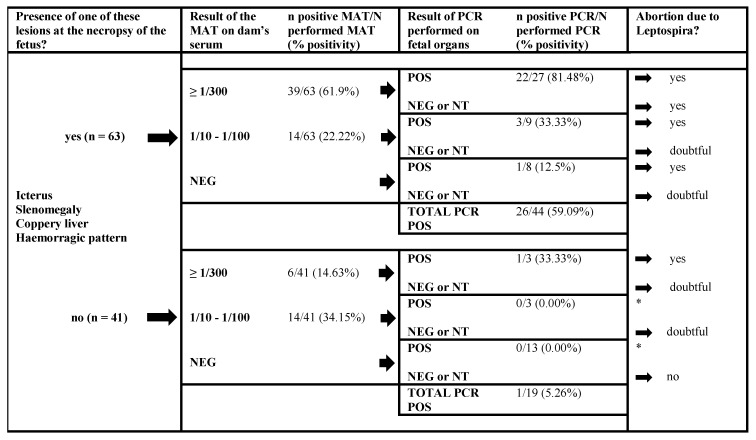
Based on overall our findings, an algorithm is proposed to diagnose, in first-line settings, leptospiral infections in cattle in an accurate and economically affordable manner (Figure 2). This approach is particularly reliable in contexts where no vaccination strategies in cattle are in place. In the case of a fetus showing suspicious lesions, MAT would be an initial preferred choice. In two thirds of the cases, it gave a positive result above 1/300 that can likely assign the abortion to an infection of the dam by Leptospira spp. PCR on fetal organs was not as efficient for attesting Leptospira as a causal agent; only about one quarter of the fetuses showed a positive result. However, using PCR in parallel to MAT allows the confirmation of more cases and the targeting of samples on which culture or molecular typing can be done. In the case of a doubtful diagnosis, complementary tests should be considered, i.e., a PCR on a dam’s vaginal discharge/urine [53,54] or a paired sampling of the dam’s serum to ascertain an increase in the serological titer [49]. This laboratory approach—knowing the history of vaccination, reproduction and clinical symptoms in the dams—enables the accurate etiological diagnosis of bovine leptospirosis caused by non-maintenance Leptospira spp.
Figure 2.
Recommended diagnostic approach for abortuses according to the results of necropsy, MAT on a dam’s serum and PCR on fetal organs performed in series, for a conclusion about Leptospira spp. as the cause of the abortion. NT: non tested, * did not occur in this study.
4. Materials and Methods
4.1. Study Protocol
The bovine abortuses selected for this study (116) were declared in southern Belgium (Walloon Region) between 27 August 2014 and 15 June 2015 and sampled within the framework of a national surveillance program of abortions in cattle. The study period relates to a time where unusual manifestations of congenital jaundice cases in bovine fetuses were firstly observed in Belgium [30]. In order to have equal representations of icteric and non-icteric abortuses, non-icteric cases were randomly selected among those declared in the same timeframe. The herds where these abortions occurred were not vaccinated as no Leptospira vaccine has marketing authorization in Belgium.
4.2. Necropsy and Sample Collection
All abortions were subjected to a standardized necropsy. The thoracic, abdominal and pelvic cavities were examined, as were the brain, the content of the abomasum and—if present—the placental cotyledons. Any lesion was systematically recorded, including (1) icterus, corresponding to a yellow coloring of the connective tissues, especially the visceral adipose ones; (2) splenomegaly, defined as an enlargement of the spleen with a muddy consistency of the organ; (3) coppery liver, the yellow coloration of the hepatic parenchyma with the absence of the modification of the liver volume or its consistency; (4) peri-renal hemorrhages, bleeding under the renal capsule; and (5) generalized hemorrhagic edema (subcutaneous and cavitary) associated, in some cases, with multifocal hemorrhages at the surfaces of organs. The estimation of the gestational ages of the abortuses was made based on the fetal length and external characteristics [55]. Pleural fluid from abortuses (n = 26) was sampled during necropsy, and serum from the dams (n = 108) was collected around the time of the abortion. Samples were stored at 4 °C or at −20 °C for subsequent processing if the tests could not be performed in the immediate days after necropsy.
4.3. Strain and Culture Conditions
Leptospira strains used for the MAT serological test were maintained in liquid Ellinghausen-McCullough-Johnson-Harris (EMJH) medium supplemented with 0.2% w/v yeast extract and 10% fetal calf serum (PAA Laboratories GmbH). The cultures were grown at 29 °C and inoculated weekly by 1:50 dilution. Strains were quality controlled twice-yearly against a panel of hyperimmune positive serovar-specific sera (AMC, Amsterdam).
4.4. MAT
MAT was performed on fetal pleural fluids and the dams’ sera as recommended by the OIE manual [49]. The Leptospira strains used in the MAT analysis of this study included pathogenic isolates belonging to twelve different serogroups as previously described [34]. Briefly, the sera were diluted in PBS and incubated with live strains at the desired threshold dilution in single well of a 96-well plate. The agglutination reaction was achieved following a 1 h incubation at 37 °C. A loopful of the reaction mix was placed on a slide, and a reading acquired under an upright microscope at 20x magnification (Olympus, Japan). Serum was considered positive if it presented agglutination in at least one of the tested serogroups. Endpoint titers were determined by starting from an initial dilution of 1:10 and using a three-fold dilution until the last condition showing 50% agglutination. The presumptive serogroup was identified by taking the highest agglutination titer of the serum with one particular pathogenic Leptospira strain.
4.5. DNA Extraction and Diagnostic Real Time PCR
For nucleic acid extraction, available tissue samples (spleen, placenta, kidneys, adrenal glands, liver, lung, brain or hepatic lymph nodes) were macerated in physiological water, and 200 µL of this homogenate was mixed with 235 µL of lysis-binding solution (MagMax, Applied Biosystem, Paisley, UK) and supplemented with lysozyme at a final concentration of 1 mg/mL. After incubation for one hour at 37 °C, samples were processed for extraction as defined by the manufacturer (MagMax nucleic acid extraction kit, Applied Biosystem). Alternatively, about 20 mg of the roughly chiseled organ was mixed with 180 μL of solution NM1 (LSI MagVet™ Universal Isolation Kit, Thermo Fisher Scientific, Paisley, UK) and, after 1 min of agitation, incubated for 30 min at 70 °C +/− 5 °C. The lysate was then agitated and centrifuged for 1 min at 6000 g. The DNA purification was performed with an automat KingFisher Flex 96TM according to manufacturer’s instructions. The diagnostic PCR, targeting the Lipl32gene, was performed with in house primers [56] or with the LSI TaqVetTM PathoLeptTM (Thermo Fisher Scientific, Merelbeke, BE) as defined by the manufacturer. For in house settings, the reaction mix consisted of 2x absolute mix (Applied Biosystem), 300 µM each of forward and reverse primers and 100 µM of the TaqMan probe. The thermal conditions were those defined elsewhere [56]. The PCR reactions were performed in triplicate on an ABI7500 thermocycler (Thermo Fisher Scientific). Samples showing amplification in at least one PCR reaction (Ct < 45) were considered as positive.
4.6. High-Resolution Melting Analysis
The molecular typing of positive diagnostic PCR samples was based on the polymorphism of the fibronectin-binding protein gene (lfb1) [51,52]. This method has been proven as reliable to distinguish genomic Leptospira species and to investigate outbreaks, because it does not require samples to have as high bacterial loads as other methods. Polymorphism analysis was achieved with both high-resolution melting analysis (HRMA) and, if PCR amplicons were sufficiently loaded, by lfb1 gene fragment sequencing. PCR amplifications were achieved using primers previously described [51,52] on a Light Cycler 480 II (Roche) using the LightCycler FastStart DNA Master SYBR Green I (Roche Applied Science). The cycling conditions included a holding step at 95 °C for 10 min; a real-time run of 95 °C for 8 s, 60 °C for 5 s, and 72 °C for 12 s for 45 cycles; and a melting step of 30 °C (from 65 °C to 95 °C) with an acquisition rate of 5 °C. Melting curve analyses were performed with the Light Cycler 480 software release 1.5.1.62 by using default settings.
4.7. Sequencing
PCR amplicons were collected and sent to GENEWIZ (United Kingdom) for outsourcing the sequencing. The subcontracted service included PCR amplicon purification and sequencing by the dideoxy chain-termination procedure. The quality of the chromatograms was analyzed with the Bionumerics software package V 6.6 (Applied Maths, Belgium). An assembly between forward and reverse sequences was obtained with default settings. The sequences obtained in this study have been deposited in GenBank (accession numbers KY373222 to KY373229). Multiple alignments were done, setting the gap penalty at 0%, and cluster analysis was performed using the unweighted-pair group method with an arithmetic mean algorithm (UPGMA).
4.8. Statistical Analysis
Associations between necropsy findings and MAT/PCR results were evaluated with the Chi-square test, or Fisher’s exact test in cases of any expected frequency inferior to 5 (significance level of p < 0.05). The statistical tests were performed with SigmaPlot 11.0 (Systat Software, San Jose, CA). In addition, for the analysis of the lesional data, odds ratios were calculated with 95% confidence intervals (95% CI). For the agreement between the diagnostic tests (MAT and PCR), Cohen’s kappa statistic with 95% confidence intervals was calculated. The interpretation for the kappa statistic was as follows: <0.2, slight agreement; 0.2–0.4, fair agreement; 0.4–0.6, moderate agreement; 0.6–0.8, substantial agreement; and >0.8, almost perfect agreement [57]. The kappa coefficients were calculated using WIN EPISCOPE 2.0 [58]. In addition to the kappa coefficients, the relative sensitivities and specificities of the MAT towards PCR were calculated with 95% "exact" Clopper–Pearson confidence intervals.
Acknowledgments
The authors thank Behaeghel I., Forrez E., Charrot J., Evrard J. and Quinet C. for their collaborative support. Gratitude is also expressed to Marin M., Vannoorenberghe P. and the rest of the technical laboratory staff of the Veterinary Bacteriology and to Cuvelier P. from the laboratory staff of ARSIA for the technical help.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/9/6/413/s1, Table S1. Detailed results of necropsy, the MAT and the real-time PCR performed on 116 abortuses for the diagnosis of leptospirosis. SPL: splenomegaly; CL: coppery liver; PRH: peri-renal hemorrhages; SD: serum of the dam; PLF: pleural fluid. Dilution*: agglutination less than 50%. Table S2: Cross-reactions between serovars of Leptospira spp. observed on 75 dams’ sera submitted to MAT (cut-off of 1/10). Table S3: Comparison of results of PCR performed on fetuses and results of MAT performed on the serum of the corresponding dams. Figure S1: Melting curves derived from analysis of the lfb1 sequence polymorphisms in Leptospira spp. icteric abortions by HRMA.
Author Contributions
Conceptualization, F.G. and M.M.; Data curation, F.G., T.P., S.B., L.D., M.S. and M.M.; Formal analysis, F.G., R.B., S.B., L.D. and M.M.; Investigation, F.G., R.B., T.P., S.B., L.D., D.F., M.S. and M.M.; Resources, F.G., R.B., T.P., S.B., L.D., M.S. and M.M.; Supervision, F.G. and M.M.; Writing—original draft, F.G. and M.M.; Writing—review & editing, F.G., R.B., T.P., S.B., L.D., D.F., M.S. and M.M. All authors have read and agree to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Ethics statement
This study is a retrospective investigation of samples collected by the first line and the reference laboratory during the national surveillance program of abortions in cattle for brucellosis and now characterized for leptospiral infection.
References
- 1.Vincent A.T., Schiettekatte O., Goarant C., Neela V.K., Bernet E., Thibeaux R., Ismail N., Khalid M.K.N.M., Amran F., Masuzawa T., et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 2019;13:e0007270. doi: 10.1371/journal.pntd.0007270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Picardeau M. Diagnosis and epidemiology of leptospirosis. Med. Mal. Infect. 2013;43:1–9. doi: 10.1016/j.medmal.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Lehmann J.S., Matthias M.A., Vinetz J.M., Fouts D.E. Leptospiral pathogenomics. Pathogens. 2014;3:280–308. doi: 10.3390/pathogens3020280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellis W.A., Michna S.W. Bovine leptospirosis: A serological and clinical study. Vet. Rec. 1976;99:387–391. doi: 10.1136/vr.99.20.387. [DOI] [PubMed] [Google Scholar]
- 5.Hassanpour A., Mousavi G.H. A case report of leptospira grippotyphosa in the Azerbaijan buffalo in Iran. Ital. J. Anim. Sci. 2007;6:893–895. doi: 10.4081/ijas.2007.s2.893. [DOI] [Google Scholar]
- 6.Bahari A., Abdollahpour G., Sadeghi-Nasab A., Sattari Tabrizi S., Yavari M., Dadmehr B. A serological survey on leptospirosis in aborted dairy cattle in industrial farms of Hamedan suburb, Iran. Iran. J. Vet. Res. 2011;12:337–339. [Google Scholar]
- 7.Marquez A., Ulivieri T., Benoit E., Kodjo A., Lattard V. House mice as a real sanitary threat of human and animal leptospirosis: Proposal for integrated management. Biomed Res. Int. 2019;2019:3794876. doi: 10.1155/2019/3794876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayral F., Zilber A.L., Bicout D.J., Kodjo A., Artois M., Djelouadji Z. Distribution of leptospira interrogans by multispacer sequence typing in urban Norway rats (Rattus norvegicus): A survey in France in 2011–2013. PLoS ONE. 2015;10:e0139604. doi: 10.1371/journal.pone.0139604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanson L.E. Bovine leptospirosis. J. Dairy Sci. 1976;59:1166–1170. doi: 10.3168/jds.S0022-0302(76)84339-1. [DOI] [PubMed] [Google Scholar]
- 10.Adler B., de la Pena Moctezuma A. Leptospira and leptospirosis. Vet. Microbiol. 2010;140:287–296. doi: 10.1016/j.vetmic.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 11.Cordy D.R., Jasper D.E. The pathology of an acute hemolytic anemia of cattle in California associated with Leptospira. J. Am. Vet. Med. Assoc. 1952;120:175–178. [PubMed] [Google Scholar]
- 12.Hathaway S.C., Little T.W., Jones T.W., Stevens H., Butland R.W. Infection by leptospires of the Pomona serogroup in cattle and pigs in south west England. Vet. Rec. 1984;115:246–248. doi: 10.1136/vr.115.10.246. [DOI] [PubMed] [Google Scholar]
- 13.Pritchard G.C., Borland E.D., Wood L., Pritchard D.G. Severe disease in a dairy herd associated with acute infection with bovine virus diarrhoea virus, Leptospira harjo and Coxiella burnetii. Vet. Rec. 1989;124:625–629. doi: 10.1136/vr.124.24.625. [DOI] [PubMed] [Google Scholar]
- 14.Gummow B., Myburgh J.G., Thompson P.N., van der Lugt J.J., Spencer B.T. Three case studies involving Leptospira interrogans serovar pomona infection in mixed farming units. J. S. Afr. Vet. Assoc. 1999;70:29–34. doi: 10.4102/jsava.v70i1.747. [DOI] [PubMed] [Google Scholar]
- 15.Smith L.L. Chronic Leptospira hardjo and Leptospira hebdomadis infection in a fifty cow herd of dairy cattle, a case history. Proc. Annu. Meet. U. S. Anim. Heal. Assoc. 1969;73:181–183. [PubMed] [Google Scholar]
- 16.Ellis W.A., O’Brien J.J., Bryson D.G., Mackie D.P. Bovine leptospirosis: Some clinical features of serovar hardjo infection. Vet. Rec. 1985;117:101–104. doi: 10.1136/vr.117.5.101. [DOI] [PubMed] [Google Scholar]
- 17.Ellis W.A., O’Brien J.J., Cassells J.A., Neill S.D., Hanna J. Excretion of Leptospira interrogans serovar hardjo following calving or abortion. Res. Vet. Sci. 1985;39:296–298. doi: 10.1016/S0034-5288(18)31717-X. [DOI] [PubMed] [Google Scholar]
- 18.Ellis W.A., O’Brien J.J., Cassells J. Role of cattle in the maintenance of Leptospira interrogans serotype hardjo infection in Northern Ireland. Vet. Rec. 1981;108:555–557. doi: 10.1136/vr.108.26.555. [DOI] [PubMed] [Google Scholar]
- 19.Dhaliwal G.S., Murray R.D., Dobson H., Ellis W.A. Effect of Leptospira interrogans serovar hardjo infection on progesterone concentrations in heifers. Vet. Rec. 1997;140:19–20. doi: 10.1136/vr.140.1.19. [DOI] [PubMed] [Google Scholar]
- 20.Dhaliwal G.S., Murray R.D., Dobson H., Montgomery J., Ellis W.A. Effect of Leptospira interrogans serovar hardjo infection on milk yield in endemically infected dairy herds. Vet. Rec. 1996;139:319–320. doi: 10.1136/vr.139.13.319. [DOI] [PubMed] [Google Scholar]
- 21.Otaka D.Y., Martins G., Hamond C., Penna B., Medeiros M.A., Lilenbaum W. Serology and PCR for bovine leptospirosis: Herd and individual approaches. Vet. Rec. 2012;170:338. doi: 10.1136/vr.100490. [DOI] [PubMed] [Google Scholar]
- 22.Smyth J.A., Fitzpatrick D.A., Ellis W.A. Stillbirth/perinatal weak calf syndrome: A study of calves infected with Leptospira. Vet. Rec. 1999;145:539–542. doi: 10.1136/vr.145.19.539. [DOI] [PubMed] [Google Scholar]
- 23.Lilenbaum W., Souza G.N. Factors associated with bovine leptospirosis in Rio de Janeiro, Brazil. Res. Vet. Sci. 2003;75:249–251. doi: 10.1016/S0034-5288(03)00114-0. [DOI] [PubMed] [Google Scholar]
- 24.Dhaliwal G.S., Murray R.D., Ellis W.A. Reproductive performance of dairy herds infected with Leptospira interrogans serovar hardjo relative to the year of diagnosis. Vet. Rec. 1996;138:272–276. doi: 10.1136/vr.138.12.272. [DOI] [PubMed] [Google Scholar]
- 25.Langoni H., De Souza L.C., Da Silva A.V., Luvizotto M.C.R., Paes A.C., Lucheis S.B. Incidence of leptospiral abortion in Brazilian dairy cattle. Prev. Vet. Med. 1999;40:271–275. doi: 10.1016/S0167-5877(99)00020-3. [DOI] [PubMed] [Google Scholar]
- 26.Escamilla H.P., Martínez M.J.J., Medina C.M., Morales S.E. Frequency and causes of infectious abortion in a dairy herd in Queretaro, Mexico. Can. J. Vet. Res. 2007;71:314–317. [PMC free article] [PubMed] [Google Scholar]
- 27.Lucchese L., Benkirane A., Hakimi I., Idrissi A. El Seroprevalence study of the main causes of abortion in dairy cattle in Morocco. Vet. Ital. 2016;52:13–19. doi: 10.12834/VetIt.388.1813.1. [DOI] [PubMed] [Google Scholar]
- 28.Bahaman A.R., Ibrahim A.L., Stallman N.D., Tinniswood R.D. The bacteriological prevalence of leptospiral infection in cattle and buffaloes in West Malaysia. Epidemiol. Infect. 1988;100:239–246. doi: 10.1017/S0950268800067376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feresu S.B., Bolin C.A., Korver H., Van de Kemp H. Identification of leptospires of the Pomona and Grippotyphosa serogroups isolated from cattle in Zimbabwe. Res. Vet. Sci. 1995;59:92–94. doi: 10.1016/0034-5288(95)90038-1. [DOI] [PubMed] [Google Scholar]
- 30.Abdollahpour G., English A.W., Tasler J. Isolation of Leptospira interrogans serovar grippotyphosa from a heifer in New South Wales. Aust. Vet. J. 1996;73:109–110. doi: 10.1111/j.1751-0813.1996.tb09990.x. [DOI] [PubMed] [Google Scholar]
- 31.Ellis W.A. Animal leptospirosis. In: Adler B., editor. Leptospira and Leptospirosis. Springer; Berlin/Heidelberg, Germany: 2015. pp. 99–137. [Google Scholar]
- 32.Delooz L., Mori M., Petitjean T., Evrard J., Czaplicki G., Saegerman C. Congenital jaundice in bovine aborted foetuses: An emerging syndrome in Southern Belgium. Transbound. Emerg. Dis. 2015;62:124–126. doi: 10.1111/tbed.12326. [DOI] [PubMed] [Google Scholar]
- 33.Delooz L., Czaplicki G., Gregoire F., Dal Pozzo F., Pez F., Kodjo A., Saegerman C. Serogroups and genotypes of Leptospira spp. strains from bovine aborted foetuses. Transbound. Emerg. Dis. 2018;65:158–165. doi: 10.1111/tbed.12643. [DOI] [PubMed] [Google Scholar]
- 34.Mori M., Van Esbroeck M., Depoorter S., Decaluwe W., Vandecasteele S.J., Fretin D., Reynders M. Outbreak of leptospirosis during a scout camp in the Luxembourg Belgian province, Belgium, summer 2012. Epidemiol. Infect. 2015;143:1761–1766. doi: 10.1017/S0950268814002763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellis W.A., Michna S.W. Experimental leptospiral abortion in cattle. Vet. Rec. 1974;94:255. doi: 10.1136/vr.94.12.255-a. [DOI] [PubMed] [Google Scholar]
- 36.Delooz L., Saegerman C., Quinet C., Petitjean T., De Regge N., Cay B. Resurgence of schmallenberg virus in belgium after 3 years of epidemiological silence. Transbound. Emerg. Dis. 2016;64:1641–1642. doi: 10.1111/tbed.12552. [DOI] [PubMed] [Google Scholar]
- 37.Comité Scientifique de l’Agence Fédérale pour la Sécurité de la Chaîne Alimentaire . AFSCA Réémergence de la brucellose bovine en Belgique entre 2010 et 2013 (SciCom N° 2011/10) AFSCA; Brussels, Belgium: 2016. [Google Scholar]
- 38.Picardeau M. Virulence of the zoonotic agent of leptospirosis: Still terra incognita? Nat. Rev. Microbiol. 2017;15:297. doi: 10.1038/nrmicro.2017.5. [DOI] [PubMed] [Google Scholar]
- 39.Pijnacker R., Goris M.G., te Wierik M.J., Broens E.M., van der Giessen J.W., de Rosa M., Wagenaar J.A., Hartskeerl R.A., Notermans D.W., Maassen K., et al. Marked increase in leptospirosis infections in humans and dogs in the Netherlands, 2014. Eurosurveillance. 2016;21 doi: 10.2807/1560-7917.ES.2016.21.17.30211. [DOI] [PubMed] [Google Scholar]
- 40.Institut de Santé Publique (WIV-ISP) Zoonoses et maladies à transmission vectorielles—Surveillance épidémiologique en Belgique, 2013 et 2014. Institut de Santé Publique; Ixelles, Belgium: 2015. [Google Scholar]
- 41.Ayral F.C., Bicout D.J., Pereira H., Artois M., Kodjo A. Distribution of Leptospira serogroups in cattle herds and dogs in France. Am. J. Trop. Med. Hyg. 2014;91:756–759. doi: 10.4269/ajtmh.13-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tagliabue S., Figarolli B.M., D’Incau M., Foschi G., Gennero M.S., Giordani R., Natale A., Papa P., Ponti N., Scaltrito D., et al. Serological surveillance of Leptospirosis in Italy: Two year national data (2010 2011) Vet. Ital. 2016;52:129–138. doi: 10.12834/VetIt.58.169.2. [DOI] [PubMed] [Google Scholar]
- 43.Alonso-Andicoberry C., García-Peña F.J., Pereira-Bueno J., Costas E., Ortega-Mora L.M. Herd-level risk factors associated with Leptospira spp. seroprevalence in dairy and beef cattle in Spain. Prev. Vet. Med. 2001;52:109–117. doi: 10.1016/S0167-5877(01)00249-5. [DOI] [PubMed] [Google Scholar]
- 44.Atxaerandio R., Aduriz G., Ziluaga I., Esteban J.I., Maranda L., Mainar-Jaime R.C. Serological evidence of Leptospira interrogans serovar Bratislava infection and its association with abortions in cattle in northern Spain. Vet. Rec. 2005;156:376. doi: 10.1136/vr.156.12.376. [DOI] [PubMed] [Google Scholar]
- 45.Behaeghel I., Butaye P., Goossens E. Evolution of leptospirosis in Belgian dogs from 2002 to 2009; Proceedings of the 54th BSAVA Annual Congress; Birmingham, UK. 31 March–3 April 2011. [Google Scholar]
- 46.Desmecht M., Colin G. Isolation in Belgium of Leptospira grippotyphosa from a muskrat. Ann. Med. Vet. 1988;132:693–696. [Google Scholar]
- 47.Lernout T., Van Esbroeck M. Surveillance Épidémiologique de la Leptospirose Leptospira spp.—2018. [(accessed on 9 April 2020)]; Available online: https://epidemio.wiv-isp.be/ID/diseases/Documents/Reports2018/Lepto_2018_fr.pdf.
- 48.Goris M.G.A., Leeflang M.M.G., Boer K.R., Goeijenbier M., van Gorp E.C.M., Wagenaar J.F.P., Hartskeerl R.A. Establishment of valid laboratory case definition for human leptospirosis. J. Bacteriol. Parasitol. 2011;3 doi: 10.4172/2155-9597.1000132. [DOI] [Google Scholar]
- 49.World Organisation for Animal Health (OIE) Manual for Diagnosis Tests and Vaccines for Terrestrial Animals. World Organisation for Animal Health; Paris, France: 2014. Chapter 2.1.9. Leptospirosis. [Google Scholar]
- 50.Dhaliwal G.S., Murray R.D., Dobson H., Montgomery J., Ellis W.A., Baker J.R. Presence of antigen and antibodies in serum and genital discharges of heifers after experimental intrauterine inoculation with Leptospira interrogans serovar hardjo. Res. Vet. Sci. 1996;60:157–162. doi: 10.1016/S0034-5288(96)90011-9. [DOI] [PubMed] [Google Scholar]
- 51.Perez J., Goarant C. Rapid Leptospira identification by direct sequencing of the diagnostic PCR products in New Caledonia. Bmc Microbiol. 2010;10:325. doi: 10.1186/1471-2180-10-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Naze F., Desvars A., Picardeau M., Bourhy P., Michault A. Use of a new high resolution melting method for genotyping pathogenic leptospira spp. PLoS ONE. 2015;10:e0127430. doi: 10.1371/journal.pone.0127430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loureiro A.P., Pestana C., Medeiros M.A., Lilenbaum W. High frequency of leptospiral vaginal carriers among slaughtered cows. Anim. Reprod. Sci. 2017;178:50e4. doi: 10.1016/j.anireprosci.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 54.Pinna A., Martins G., Loureiro A.P., Lilenbaum W. Detection of bovine carriers of leptospira by serological, bacteriological, and molecular tools. Trop. Anim. Health Prod. 2018;50:883–888. doi: 10.1007/s11250-018-1512-z. [DOI] [PubMed] [Google Scholar]
- 55.Njaa B.L. In: Kirkbride’s Diagnosis of Abortion and Neonatal Loss in Animals. 4th ed. Njaa B.L., editor. Wiley-Blackwell; West Sussex, UK: 2012. [Google Scholar]
- 56.Stoddard R.A., Gee J.E., Wilkins P.P., McCaustland K., Hoffmaster A.R. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn. Microbiol. Infect. Dis. 2009;64:247–255. doi: 10.1016/j.diagmicrobio.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 57.Petrie A., Watson P. Statistics for Veterinary and Animal Science. Blackwell Science; London, UK: 1999. [Google Scholar]
- 58.Thrusfield M., Ortega C., de Blas I., Noordhuizen J.P., Frankena K. WIN EPISCOPE 2.0: Improved epidemiological software for veterinary medicine. Vet. Rec. 2001;148:567–572. doi: 10.1136/vr.148.18.567. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.