Abstract
Dietary phenolic compounds such as caffeic and chlorogenic acid exert an antiproliferative effect and modulate the gene-specific DNA methylation status in human breast tumor cells, but it remains unclear whether they interfere with global DNA methylation in human leukemia cells. We examined whether caffeic and chlorogenic acid (1-250 µM) exert antitumor action in human promyelocytic leukemia cells (HL-60) and human acute T-cell leukemia cells (Jurkat). Caffeic and chlorogenic acid did not reduce cell viability in the two cell lines, as assessed using the neutral red uptake and MTT assays. These phenolic acids (1-100 μM) neither induced DNA damage (comet assay) nor increased the micronuclei frequency (micronucleus assay) in HL-60 and Jurkat cells, indicating that they were not genotoxic or mutagenic. Analysis of global DNA methylation levels using a 5-mC DNA ELISA kit revealed that chlorogenic acid at a non-cytotoxic concentration (100 μM) induced global DNA hypomethylation in Jurkat cells, but not in HL-60 cells, suggesting that it exerts a cell-specific effect. Caffeic acid did not change global DNA methylation. As other phenolic compounds, chlorogenic acid probably modulates DNA methylation by targeting DNA methyltransferases. The hypomethylating action of chlorogenic acid can be beneficial against hematological malignances whose pathogenic processes involve impairment of DNA methylation.
Keywords: Polyphenols, comet assay, micronucleus, neutral red
Introduction
Coffee is one of the most popular beverages worldwide and has a great impact on the human body as a whole (Buldak et al., 2018). The phytochemical compounds present in coffee are able to inhibit oxidative stress and oxidative damage which is related to early process of the transformation of a normal cell into a malignant tumor (Grosso et al., 2017). Coffee has been demonstrated to play a role in cellular defense mechanisms, including regulating DNA repair and apoptosis, as well as having antiproliferative, antiangiogenic, and antimetastatic effects (Grosso et al., 2017). The coffee polyphenols caffeic acid and chlorogenic acid, exert antitumor effect through activation and inhibition of some important pathways of cancer metabolism (Lukitasari et al., 2018). Moreover, epidemiological studies revealed that high consumption of coffee was related to a lower risk of incidence of specific cancers and metabolic, digestive, hepatic, or disorders of neurological conditions (Buldak et al., 2018). The antitumor action of coffee polyphenols has been investigated in tumor cell lines, pre-clinical, and clinical studies (Rajagopal et al., 2018). Chlorogenic acid induces apoptosis and selectively inhibits proliferation of A498 human renal cancer cells but not of HEK293 human embryonic kidney cells (Wang et al., 2019). Caffeic acid inhibits proliferation of hepatocellular carcinoma cells and SK-Mel-28 human melanoma cancer cells by inducing apoptosis (Brautigan et al., 2018), alters gene expression in SK-Mel-28 cells (Pelinson et al., 2019), and attenuates the properties of cancer stem cell-like cells by increasing DNA methylating and inducing the expression of miR-148a (Li et al., 2015). Both caffeic acid and chlorogenic acid suppress DNA methylation in MCF-7 and MDA-MB-231 human breast cancer cells (Lee and Zhu 2006).
However, in vitro assays with polyphenols have controversial results. Chlorogenic acid inhibits the induction of oxidative damage in rat pheochromocytoma cells (Yao, et al., 2019), while caffeic acid protects human erythrocyte membranes from oxidizing species (Colina et al., 2019). Furthermore, caffeic acid at 100 μM induces DNA damage in human lymphocytes (assessed by the comet assay); at 500-1500 μM, it increases the micronucleus frequency in rat hepatoma tissue cells (Szeto et al., 2002; Maistro et al., 2011). The IARC Monographs classify caffeic acid in the group 2B as possibly carcinogenic to humans (IARC, 1993).
Epigenetic dysregulation, such as DNA hypo- or hypermethylation, is closely associated with the development and progression of hematological malignancies. Alterations in the cellular epigenome, such as impaired DNA methylation mediated by DNA methyltransferases, play important roles in tumorigenesis of acute myeloid leukemia and acute lymphoblastic leukemia (Dexheimer et al., 2017; Helmer et al., 2019).
A previous study demonstrated that habitual coffee consumption can alter DNA methylation sites in human genome. It revealed alterations in methylation status of CpG sites located near of 11 genes associated with habitual coffee consumption in peripheral blood mononuclear cells (Chuang et al., 2017).
To the best of our knowledge, there are no reports in the literature evaluating the effects of acid caffeic and acid chlorogenic effects on human leukemic cells. Therefore, in the present study, we examined the antitumor potential of caffeic acid and chlorogenic acid by assessing their cytotoxicity, genotoxicity and effects on global DNA methylation towards human promyelocytic leukemia cells (HL-60) and human acute T-cell leukemia cells (Jurkat).
Material and Methods
Chemicals
Acridine orange (CAS: 10127-02-3), caffeic acid (CAS: 331-39-5), chlorogenic acid (CAS: 327-97-9), cytochalasin B (CAS: 14930-96-2), dimethyl sulfoxide (DMSO, CAS: 67-68-5), 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT, CAS: 298-93-1), ethylenediamine tetraacetic acid (EDTA, CAS: 60-00-4), methyl methanesulfonate (MMS; CAS: 66-27-3), neutral red (CAS: 553-24-2), and trypan blue (CAS: 72-57-1) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Wizard® Genomic DNA Purification Kit was obtained from Promega (Madison, WI, USA). 5-mC DNA ELISA Kit (colorimetric) was obtained from Zymo Research (Irvine, CA, USA). Antibiotic mix (penicillin/streptomycin/neomycin), fetal bovine serum (FBS), RPMI (Roswell Park Memorial Institute) 1640 culture medium, and trypsin-EDTA were acquired from Gibco (Carlsbad, CA, USA). Low melting point (LMP) agarose (CAS: 39346-81-1) and normal melting point (NMP) agarose (CAS: 9012-36-6) were purchased from Invitrogen (Carlsbad, CA, USA). GelRed was acquired from Biotium (Hayward, CA, USA). The other reagents used were of analytical grade.
Cell culture conditions
All in vitro cell culture procedures were performed using a Class II and Type 1A laminar flow cabinet from Bioprotector (VecoFlow Ltda, Campinas, SP, Brazil). Human promyelocytic leukemia cells (HL-60) and human acute T-cell leukemia cells (Jurkat) were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured according to the procedures reported by Bal-Price and Coecke (2011). Cells were maintained in RPMI 1640 medium supplemented with 10% FBS, 1% antibiotic-antimycotic solution (5 mg/mL penicillin, 5 mg/mL streptomycin, and 10 mg/mL neomycin), and 0.024% (w/v) NaHCO3, in a CO2 incubator with 5% atmosphere, at 37 °C, and 95% relative humidity. The medium was changed every 2–3 days. All the experiments were conducted between the third and the eighth cell passage.
Determination of cell viability
Cell viability was assessed using the neutral red uptake assay described by Repetto et al. (2008) and the MTT assay reported by Mosmann (1983). First, HL-60 or Jurkat cells (5 104/mL) were seeded in 96-well plates, in complete culture medium, for 24 h. After stabilization, the cells were treated with caffeic acid or chlorogenic acid (1 – 250 μM), PBS pH 7.4 (negative control), DMSO (0.4% v/v; solvent control) or MMS (100 μM; positive control) for 24 h. Cell viability was determined using the neutral red uptake assay (Repetto et al., 2008). Absorbance of the samples was analyzed using an iMark™ Microplate Absorbance Reader (Agilent Technologies, Santa Clara, CA, USA), set at λ = 540 nm.
Second, cells (1 × 104/well) were seeded in 96-well culture plates and incubated for 24 h before treatment with three concentrations (1, 10, and 100 μM) of caffeic acid or chlorogenic acid, 100 μM MMS (positive control), DMSO (0.4% v/v; solvent control) or PBS pH 7.4 (negative control) for further 24 h. The reaction medium was later removed, a MTT solution (working solution: 10:100 μL v/v; 0.5 mg/mL) was added to the wells, and the plate was incubated for 4 h. Finally, the supernatant was replaced by DMSO, and absorbance was recorded in a spectrophotometer (Biotek Elx800; Winooski, VT, USA) set at λ = 570 nm. The percentage of viable cells was calculated after normalization with the negative control, which was considered as 100% cell viability.
Comet assay
HL-60 or Jurkat cells (1 × 105/mL) were seeded in 24-well plates, in complete culture medium, for 24 h. After stabilization, the cells were treated with caffeic acid or chlorogenic acid (1 – 100 μM), PBS pH 7.4 (negative control), DMSO (0.4% v/v; solvent control) or MMS (100 μM; positive control), for 4 h. Cell viability was determined by the trypan blue exclusion technique, using a Countess® Automated Cell Counter (Life Technologies, Carlsbad, USA). Cultures with cell viability > 80% were subjected to the comet assay, according to the protocol reported by Tice et al. (2000). Finally, the slides were stained with GelRed™ solution and analyzed in a fluorescence microscope (Carl Zeiss, Axiostar Plus, Jena, Germany) using an 515–560 nm excitation filter and a 590 nm filter barrier, with the aid of the Comet Assay IV™ software (Perceptive Instruments, Haverhil, UK).
Micronucleus assay
HL-60 or Jurkat cells (1 × 105/mL) were seeded in 6-well plates, in complete culture medium, and treated with caffeic acid or chlorogenic acid (1, 10 or 100 μM), PBS pH 7.4 (negative control), DMSO (0.4% v/v; solvent control) or 5-azacytidine (5 μM; positive control), for 24 h. Next, cytochalasin B (final concentration of 6.0 mg/mL) was added to the wells. Immediately before microscopy analysis (Carl Zeiss, Jena, Germany), cells were stained with acridine orange (0.1%; diluted 1:15 v/v in PBS). Binucleated cells with micronuclei (MNi) were scored at 1000 x magnification according to Fenech (2000). A total of 6,000 binucleated cells were analyzed for the presence of MNi. The fold-change in MNi frequency was expressed as the ratio between the mean value of each treatment and the mean value of the negative control (Fenech et al., 2016).
DNA isolation and global methylation assays
Genomic DNA was extracted from HL-60 or Jurkat cells (5 × 104/mL) treated with caffeic acid or chlorogenic acid (100 μM), PBS pH 7.4 (negative control), or DMSO (0.4% v/v; solvent control), for 72 h. DNA was extracted using the Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, USA) and quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Global DNA methylation was analyzed using the 5-mC DNA ELISA Kit (colorimetric) (Zymo Research, Irvine, CA, USA), according to the manufacturers' instructions, and absorbance was recorded at 405 nm (Biotek, Elx800 – Winooski, VT, USA). Global DNA methylation was expressed as % 5-mC.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5.0 software (La Jolla, CA, USA). All experiments were carried out in triplicate and the results were expressed as the mean ± standard deviation (SD). The positive and negative control groups were compared using the Student's t-test, while the test groups were compared with the negative control group using one-way analysis of variance (ANOVA) combined with the Dunnett's post-hoc test. p<0.05 was considered statistically significant.
Results
Caffeic acid and chlorogenic acid are not cytotoxic towards HL-60 and Jurkat cells
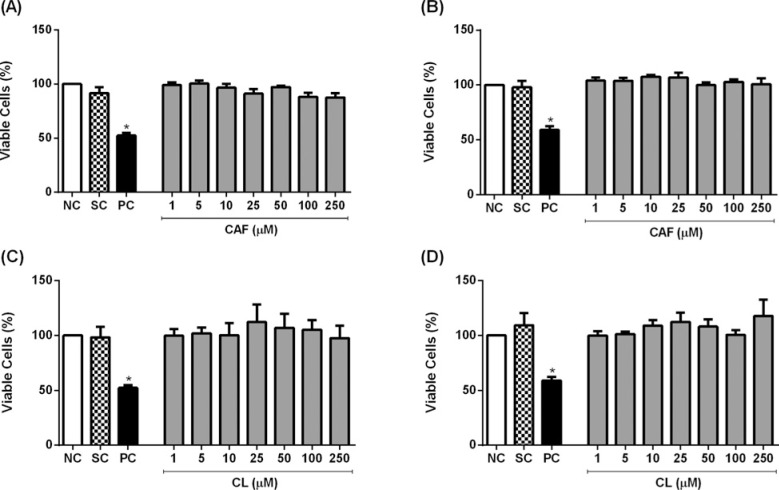
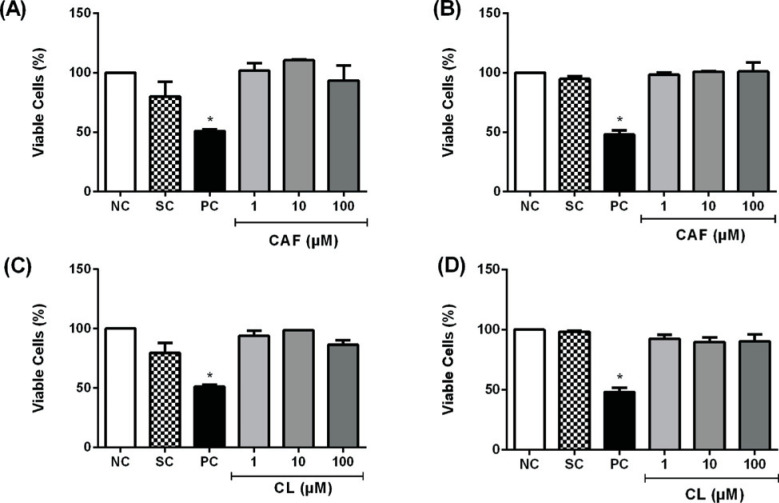
We examined whether caffeic acid and chlorogenic acid affected the cell viability of HL-60 and Jurkat cells, using the neutral red uptake and MTT assays. MMS, but not the phenolic acids at concentrations up to 250 μM, reduced cell viability assessed by the neutral red uptake assay (Figure 1). Similar results were obtained when the cells were treated with the phenolic acids at concentrations up to 100 μM, and cell viability was assessed using the MTT assay (Figure 2). Hence, caffeic acid and chlorogenic acid were not cytotoxic to HL-60 and Jurkat cells.
Figure 1. Cell viability of HL-60 (A, C) and Jurkat (B, D) cells treated with caffeic acid (CAF) and chlorogenic acid (CL) for 24 hours, as assessed using the neutral red uptake assay. Negative control (NC) = PBS; Solvent control (SC) = 0.4% DMSO; Positive control (PC) = 100 μM methyl methanesulfonate. Results are expressed as mean ± SD of three independent experiments.
*p<0.05 vs. negative control (Student's t test).
Figure 2. Cell viability of HL-60 (A, C) and Jurkat cells (B, D) treated with caffeic acid (CAF) and chlorogenic acid (CL) for 24 hours, as assessed using the MTT assay. Negative control (NC) = PBS; Solvent control (SC) = 0.4% DMSO; Positive control (PC) = 100 μM methyl methanesulfonate. Results are expressed as mean ± SD of three independent experiments.
*p <0.05 vs. negative control (Student's t test).
Caffeic acid and chlorogenic acid are not genotoxic to HL-60 and Jurkat cells
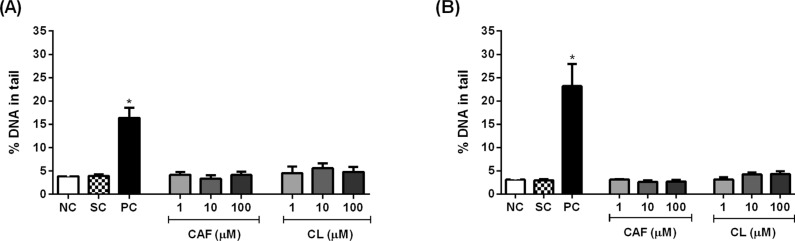
We used the comet assay to examine whether caffeic acid and chlorogenic acid induced DNA damage in HL-60 and Jurkat cells. MMS, but not the phenolic acids at concentrations up to 100 μM, increased the percentage of DNA in the tail in both cell lines (Figure 3). MMS induced a stronger DNA damage in Jurkat cells than in HL-60 cells, as demonstrated by the increased percentage of DNA in the tail.
Figure 3. Percentage of DNA in the tail in HL-60 (A) and Jurkat (B) cells treated with caffeic acid (CAF) and chlorogenic acid (CL) for 4 hours. Negative control (NC) = PBS; Solvent control (SC) = 0.4% DMSO; Positive control (PC) = 100 μM methyl methanesulfonate. Results are expressed as mean ± SD (n = 3).
*p <0.05 vs. negative control (Student's t test).
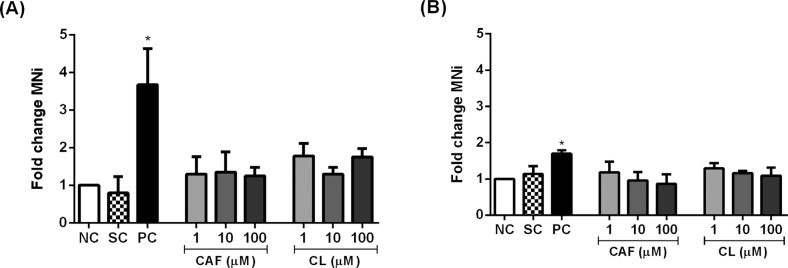
We expressed the MNi frequency as fold change relative to the negative control group because the MNi frequency in this group differed between HL-60 and Jurkat cells. The fold change in MNi frequency in both cell lines treated with caffeic acid and chlorogenic acid was similar to negative control (p > 0.05) (Figure 4).
Figure 4. Fold change in micronuclei (MNi) frequency per 1,000 binucleated HL-60 (A) and Jurkat (B) cells treated with caffeic acid (CAF) and chlorogenic acid (CL). Negative control (NC) = PBS; Solvent control (SC) = 0.4% DMSO; Positive control (PC) = 5 μM 5-azacytidine. Results are expressed as mean ± SD (n = 3).
*p <0.05 vs. negative control (Student's t test).
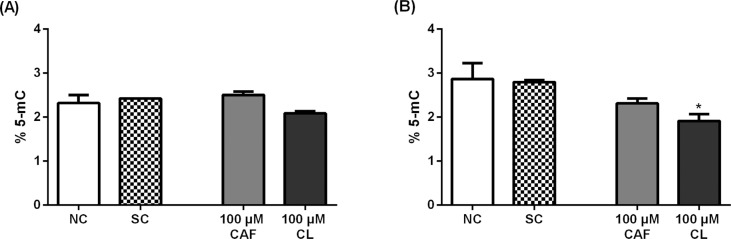
Chlorogenic acid decreases global DNA methylation in Jurkat cells
Compared with the negative control, the 72-h treatment with 100 μM chlorogenic acid, but not with caffeic acid, significantly decreased the levels of % 5-mC in Jurkat cells (p<0.05); in HL-60 cells, both phenolic acids did not alter the levels of % 5-mC (Figure 5).
Figure 5. Global DNA methylation (% 5-mc) in HL-60 (A) and Jurkat (B) cells treated with caffeic acid (CAF) and chlorogenic acid (CL) for 72 hours. Negative control (NC) = PBS; Solvent control (SC) = 0.4% DMSO. Results are expressed as mean ± SD (n = 3).
*p <0.05 vs. negative control (One-way ANOVA followed by the Dunnett's post-hoc test).
Discussion
No reports of toxicity were found in humans exposed to caffeic and chlorogenic acids. However, in a study performed with rats, the intraperitoneal dose of 11.29 mmol/kg of chlorogenic acid induced death in 4 of 6 treated animals, while doses less than 6.878 mmol/kg were nontoxic. The dose of 8.326 mmol/kg of caffeic acid induced death in 5 of 8 treated rats, but doses lower than 6.938 mmol/kg were nontoxic (Chaube and Swinyard, 1976).
In humans, the absorption was measured after the ingestion of 2.8 mmol of each acid, resulting in 33% of chlorogenic acid and 95% of caffeic acid. This study showed that about one third of chlorogenic acid and almost 100% of the caffeic acid were absorbed in the small bowel of humans (Olthof et al., 2001). Therefore, concentrations of 1 - 250 µM of caffeic and chlorogenic acids were chosen to be used in the present study.
In this study, we addressed whether caffeic acid and chlorogenic acid induced cytotoxicity, genotoxicity, and modify DNA global methylation in HL-60 and Jurkat cells. Both phenolic acids were not cytotoxic to the two cell lines, when tested at 1–250 μM and 1–100 μM using the neutral red uptake and MTT assays, respectively.
Caffeic acid suppresses proliferation of hepatocellular carcinoma cells by inducing apoptosis (Brautigan et al., 2018), inhibits migration of MCF-7 breast cancer cells (at 50-100 μM) (Kabala-Dzik et al., 2018), and suppresses proliferation and migration of A549 human lung cancer cells and HT29-D4 colon adenocarcinoma cells (Nasr Bouzaiene et al., 2015). Chlorogenic acid decreases viability of HCT116 and HT29 human colon cancer cells (Hou et al., 2017) and induces apoptosis in A2780 and A2780CP70 human ovarian cancer cells in vitro (Tai et al., 2010), but it has no significant effects on proliferation of HepG2 human hepatoma cells (Ramos et al., 2005).
Our findings corroborate a recent study that investigated the antiproliferative effect of six coffee compounds, including kahweol acetate, cafestol, caffeine, caffeic acid, chlorogenic acid, and trigonelline hydrochloride on LNCaP, LNCaP-SF, PC-3, and DU145 human prostate cancer cells (Iwamoto et al., 2019). Chlorogenic acid and caffeic acid at concentrations ranging from 5 to 50 µM do not inhibit proliferation and migration of human prostate cancer cells (Iwamoto et al., 2019). Caffeic acid is also not cytotoxic towards MDA-MB-231 cells, a triple-negative human caucasian breast adenocarcinoma cell line, affording IC50 values greater than 10,000 and 1,000 µM after 24 and 48 h of treatment, respectively (Kabala-Dzik et al., 2017).
Caffeic acid and chlorogenic acid (1 – 100 μM) were not genotoxic or mutagenic to HL-60 or Jurkat cells, since these phenolic acids did not induce DNA damage (assessed by the comet assay) or increased MNi frequencies (assessed by the micronucleus test). Caffeic acid does not cause DNA damage in human peripheral blood mononuclear cells and HL-60 cells (Fabiani et al., 2008), but induces DNA damage in human lymphocytes at concentrations as high as 1500 μM (Maistro et al., 2011), as assessed using the comet assay. This phenolic acid also reduces DNA damage in UVBirradiated lymphocytes, increases MNi frequency in Hep3B cells, but it does not alter MNi frequency in HepG2 cells, at concentrations ranging from 10 to 1000 μM (Majer et al., 2004; Prasad et al., 2009). Chlorogenic acid (20 µM) protects HaCaT human keratinocytes from UVB-induced oxidative damage (Cha et al., 2014).
The controversial cytotoxic and genotoxic effects of these phenolic acids may be related to the presence of transition metal ions and their pro-oxidant action (Bhat et al., 2007). In the present study, HL-60 and Jurkat cells were cultured in RPMI 1640 medium, which contains the reducing agent glutathione and high concentrations of vitamins. Recently, Grzesik et al. (2019) reported H2O2 generation associated with oxidation of phenolic compounds, including caffeic acid and chlorogenic acid, in Dulbecco's modified Eagle's medium (DMEM). DMEM contains iron while RPMI 1640 contains only trace concentrations of iron, suggesting that this transition metal promotes auto-oxidation of these compounds.
Downregulation of DNA methylation is an important molecular event that modulates the pathogenesis of hematological malignancies. Hypomethylating agents have been used as therapeutic alternatives to treat myelodysplastic syndromes and acute myeloid leukemia in elderly patients (Santini et al., 2013; Yun et al., 2016). Analysis of global DNA methylation revealed that non-treated HL-60 and Jurkat cells exhibited 2.3% and 2.9% of 5-mC DNA, respectively. The 5-mC levels in primary human fibroblasts ranged from 1.0% to 1.7% (Maierhofer et al., 2017), while the mean 5-mC level was 2.8% in peripheral blood of 100 male workers employed in automotive battery factories (Devoz et al., 2017), 3.82% in leukocytes from 390 volunteers with no known cancer (Shen et al., 2017), and 2.28% in non-treated LNCaP human prostate cancer cells in vitro (Wu et al., 2017).
A three-day treatment with caffeic acid (1 – 50 mM) and chlorogenic acid (1 – 20 mM) decreases the methylation-specific band for the retinoic acid receptor beta (RARB) but does not significantly change global DNA methylation in MCF-7 and MDA-MB-231 human breast cancer cells (Lee and Zhu, 2006). Tumor cells with elevated levels of DNA methyltransferases can be sensitive to treatment with hypomethylating agents, while cells deficient in these enzymes can be resistant to these agents (Juttermann et al., 1994).
In the present study, chlorogenic acid at a non-cytotoxic concentration (100 μM) induced global DNA hypomethylation in Jurkat cells but not in HL-60 cells, suggesting that this phenolic acid exerts a cell-specific action. The possible mechanism by which chlorogenic acid modulates DNA methylation is by targeting DNA methyltransferases (DNMTs), in a manner similar to that described for polyphenols (Li et al., 2016; Nasir et al., 2020).
Polyphenols compounds have a catechol group in their constitution, a key substrate for O-methylation of catecholamines mediated by the enzyme catechol-O-methyltransferase (COMT) (Lee et al., 2005; Kadayifci, et al., 2018). COMT increases the formation of S-adenosylhomocysteine (SAH), a well-known non-competitive inhibitor of DNMTs (Oliva et al., 1980; Lee et al., 2005). Global DNA methylation is decreased after DNMT1 inhibition (Christman, 2002). In fact, Lee et al. (2005) showed that caffeic and chlorogenic acids decreased the DNA methylation catalyzed by human DNMT1. Our results showed that chlorogenic acid decreased DNA methylation in Jurkat cells but not in HeLa cells. This suggests that the mechanisms by which chlorogenic acid acts could be cell-type specific and highlights the importance of using more than one cell line when studying the effects of compounds on DNA methylation.
The nucleoside analogues 5-azacytidine and 5-aza-2-deoxycytidine have been used as hypomethylating agents for over a decade in the chemotherapeutic treatment of acute myeloid leukemia (Leung et al., 2019). Considering the strong cytotoxicity of nucleoside analogues, additional studies are needed to develop innovative therapeutic strategies based on the use of non-nucleoside analogues. In this sense, plant-derived compounds are promising inhibitors of DNA methyltransferases (Schneider-Stock et al., 2012).
In conclusion, caffeic acid and chlorogenic acid are not cytotoxic or genotoxic towards HL-60 and Jurkat cells. Chlorogenic acid induces cell-specific hypomethylation in Jurkat cells, which can be beneficial against hematological malignances where impaired DNA methylation plays an important role in the pathogenic process.
Acknowledgments
This study was supported by the Brazilian funding agencies São Paulo Research Foundation (FAPESP; grant 2013/13733-6), National Council for Scientific and Technological Development (CNPq; research grant 302290/2015-0), and Coordination for the Improvement of Higher Education Personnel (CAPES; Finance Code 001). The authors are grateful to Dr Gustavo Rafael Mazzaron Barcelos from Federal University of São Paulo, for provided the 5-mC DNA Kit, and to Bs. Joana D'Arc Castania Darin and Dr. Regislaine Valéria Burim, both from Department of Clinical Analyses, Toxicology and Food Sciences at School of Pharmaceutical Sciences of Ribeirão Preto, University of São Paulo, Ribeirão Preto, SP, Brazil, for their helpful technical assistance.
Footnotes
Associate Editor: Carlos R. Machado
References
- Bal-Price A, Coecke S. Guidance on Good Cell Culture Practice (GCCP) In: Aschner M, Suñol C, Bal-Price A, editors. Cell Culture Techniques. Humana Press; New Jersey: 2011. pp. 1–25. [Google Scholar]
- Bhat SH, Azmi AS, Hadi SM. Prooxidant DNA breakage induced by caffeic acid in human peripheral lymphocytes: involvement of endogenous copper and a putative mechanism for anticancer properties. Toxicol Appl Pharmacol. 2007;218:249–255. doi: 10.1016/j.taap.2006.11.022. [DOI] [PubMed] [Google Scholar]
- Brautigan DL, Gielata M, Heo J, Kubicka E, Wilkins LR. Selective toxicity of caffeic acid in hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2018;505:612–617. doi: 10.1016/j.bbrc.2018.09.155. [DOI] [PubMed] [Google Scholar]
- Buldak RJ, Hejmo T, Osowski M, Buldak L, Kukla M, Polaniak R, Birkner E. The impact of coffee and its selected bioactive compounds on the development and progression of colorectal cancer in vivo and in vitro . Molecules. 2018;23:3309–3309. doi: 10.3390/molecules23123309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JW, Piao MJ, Kim KC, Yao CW, Zheng J, Kim SM, Hyun CL, Ahn YS, Hyun JW. The polyphenol chlorogenic acid attenuates UVB-mediated oxidative stress in human HaCaT keratinocytes. Biomol Ther (Seoul) 2014;22:136–142. doi: 10.4062/biomolther.2014.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaube S, Swinyard CA. Teratological and toxicological studies of alkaloidal and phenolic compounds from Solanum tuberosum L. Toxicol Appl Pharmacol. 1976;3:227–237. doi: 10.1016/0041-008x(76)90002-8. [DOI] [PubMed] [Google Scholar]
- Christman JK. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- Chuang YH, Quach A, Absher D, Assimes T, Horvath S, Ritz B. Coffee consumption is associated with DNA methylation levels of human blood. Eur J Hum Genet. 2017;25:608–616. doi: 10.1038/ejhg.2016.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colina JR, Suwalsky M, Manrique-Moreno M, Petit K, Aguilar LF, Jemiola-Rzeminska M, Strzalka K. An in vitro study of the protective effect of caffeic acid on human erythrocytes. Arch Biochem Biophys. 2019;662:75–82. doi: 10.1016/j.abb.2018.12.006. [DOI] [PubMed] [Google Scholar]
- Devoz PP, Gomes WR, Araujo ML, Ribeiro DL, Pedron T, Antunes LMG, Batista BL, Barbosa F, Barcelos GRM. Lead (Pb) exposure induces disturbances in epigenetic status in workers exposed to this metal. J Toxicol Environ Health A. 2017;80:1098–1105. doi: 10.1080/15287394.2017.1357364. [DOI] [PubMed] [Google Scholar]
- Dexheimer GM, Alves J, Reckziegel L, Lazzaretti G, Abujamra AL. DNA methylation events as markers for diagnosis and management of acute myeloid leukemia and myelodysplastic syndrome. Dis Markers. 2017:5472893–5472893. doi: 10.1155/2017/5472893. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabiani R, Rosignoli P, De Bartolomeo A, Fuccelli R, Servili M, Montedoro GF, Morozzi G. Oxidative DNA damage is prevented by extracts of olive oil, hydroxytyrosol, and other olive phenolic compounds in human blood mononuclear cells and HL60 cells. J Nutr. 2008;138:1411–1416. doi: 10.1093/jn/138.8.1411. [DOI] [PubMed] [Google Scholar]
- Fenech M. The in vitro micronucleus technique. Mutat Res. 2000;455:81–95. doi: 10.1016/s0027-5107(00)00065-8. [DOI] [PubMed] [Google Scholar]
- Fenech M, Knasmueller S, Bolognesi C, Bonassi S, Holland N, Migliore L, Palitti F, Natarajan AT, Kirsch-Volders M. Molecular mechanisms by which in vivo exposure to exogenous chemical genotoxic agents can lead to micronucleus formation in lymphocytes in vivo and ex vivo in humans. Mutat Res. 2016;770:12–25. doi: 10.1016/j.mrrev.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Grosso G, Godos J, Galvano F, Giovannucci EL. Coffee, caffeine, and health outcomes: An umbrella. Annu Rev Nutr. 2017;37:131–156. doi: 10.1146/annurev-nutr-071816-064941. [DOI] [PubMed] [Google Scholar]
- Grzesik M, Bartosz G, Stefaniuk I, Pichla M, Namiesnik J, Sadowska-Bartosz I. Dietary antioxidants as a source of hydrogen peroxide. Food Chem. 2019;278:692–699. doi: 10.1016/j.foodchem.2018.11.109. [DOI] [PubMed] [Google Scholar]
- Helmer JA, Iraburu R, Tirado CA. Epigenetics of BALL. J Assoc Genet Technol. 2019;45:10–13. [PubMed] [Google Scholar]
- Hou N, Liu N, Han J, Yan Y, Li J. Chlorogenic acid induces reactive oxygen species generation and inhibits the viability of human colon cancer cells. Anticancer Drugs. 2017;28:59–65. doi: 10.1097/CAD.0000000000000430. [DOI] [PubMed] [Google Scholar]
- IARC . Some naturally occurring substances: Food items and constituents, heterocyclic aromatic amines and mycotoxins. International Agency for Research on Cancer; Lyon: 1993. 599 p [Google Scholar]
- Iwamoto H, Izumi K, Natsagdorj A, Naito R, Makino T, Kadomoto S, Hiratsuka K, Shigehara K, Kadono Y, Narimoto K, et al. Coffee diterpenes kahweol acetate and cafestol synergistically inhibit the proliferation and migration of prostate cancer cells. Prostate. 2019;79:468–479. doi: 10.1002/pros.23753. [DOI] [PubMed] [Google Scholar]
- Juttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2'-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci U S A. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabala-Dzik A, Rzepecka-Stojko A, Kubina R, Jastrzebska-Stojko Z, Stojko R, Wojtyczka RD, Stojko J. Comparison of two components of propolis: caffeic acid (CA) and caffeic acid phenethyl ester (CAPE) induce apoptosis and cell cycle arrest of breast cancer cells MDA-MB-231. Molecules. 2017;22:1554–1554. doi: 10.3390/molecules22091554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabala-Dzik A, Rzepecka-Stojko A, Kubina R, Wojtyczka RD, Buszman E, Stojko J. Caffeic acid versus caffeic acid phenethyl ester in the treatment of breast cancer MCF-7 cells: migration rate inhibition. Integr Cancer Ther. 2018;17:1247–1259. doi: 10.1177/1534735418801521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadayifci FZ, Zheng S, Pan YX. Molecular mechanisms underlying the link between diet and DNA methylation. Int J Mol Sci. 2018;19:4055–4055. doi: 10.3390/ijms19124055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WJ, Zhu BT. Inhibition of DNA methylation by caffeic acid and chlorogenic acid, two common catechol-containing coffee polyphenols. Carcinogenesis. 2006;27:269–277. doi: 10.1093/carcin/bgi206. [DOI] [PubMed] [Google Scholar]
- Lee WJ, Shim JY, Zhu BT. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol Pharmacol. 2005;68:1018–1030. doi: 10.1124/mol.104.008367. [DOI] [PubMed] [Google Scholar]
- Leung KK, Nguyen A, Shi T, Tang L, Ni X, Escoubet L, MacBeth KJ, DiMartino J, Wells JA. Multiomics of azacitidine-treated AML cells reveals variable and convergent targets that remodel the cell-surface proteome. Proc Natl Acad Sci U S A. 2019;116:695–700. doi: 10.1073/pnas.1813666116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Guo Y, Zhang C, Wu R, Yang AY, Gaspar J, Kong AN. Dietary phytochemicals and cancer chemoprevention: A perspective on oxidative stress, inflammation, and epigenetics. Chem Res Toxicol. 2016;29:2071–2095. doi: 10.1021/acs.chemrestox.6b00413. [DOI] [PubMed] [Google Scholar]
- Li Y, Jiang F, Chen L, Yang Y, Cao S, Ye Y, Wang X, Mu J, Li Z, Li L. Blockage of TGFbeta-SMAD2 by demethylation-activated miR-148a is involved in caffeic acid-induced inhibition of cancer stem cell-like properties in vitro and in vivo. FEBS Open Bio. 2015;5:466–475. doi: 10.1016/j.fob.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukitasari M, Nugroho DA, Widodo N. Chlorogenic acid: The conceivable chemosensitizer leading to cancer growth suppression. J Evid Based Integr Med. 2018;23 doi: 10.1177/2515690X18789628. 2515690X18789628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maierhofer A, Flunkert J, Dittrich M, Muller T, Schindler D, Nanda I, Haaf T. Analysis of global DNA methylation changes in primary human fibroblasts in the early phase following X-ray irradiation. PLoS One. 2017;12:e0177442. doi: 10.1371/journal.pone.0177442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maistro EL, Angeli JP, Andrade SF, Mantovani MS. In vitro genotoxicity assessment of caffeic, cinnamic and ferulic acids. Genet Mol Res. 2011;10:1130–1140. doi: 10.4238/vol10-2gmr1278. [DOI] [PubMed] [Google Scholar]
- Majer BJ, Mersch-Sundermann V, Darroudi F, Laky B, Wit K, Knasmuller S. Genotoxic effects of dietary and lifestyle related carcinogens in human derived hepatoma (HepG2, Hep3B) cells. Mutat Res. 2004;551:153–166. doi: 10.1016/j.mrfmmm.2004.02.022. [DOI] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity as-says. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Mukherjee N, Kumar AP, Ghosh R. DNA methylation and flavonoids in genitourinary cancers. Curr Pharmacol Rep. 2015;1:112–120. doi: 10.1007/s40495-014-0004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir A, Bullo MMH, Ahmed Z, Imtiaz A, Yaqoob E, Jadoon M, Ahmed H, Afreen A, Yaqoob S. Nutrigenomics: epigenetics and cancer prevention: a comprehensive review. Crit Rev Food Sci Nutr. 2020;60:1375–1387. doi: 10.1080/10408398.2019.1571480. [DOI] [PubMed] [Google Scholar]
- Nasr Bouzaiene N, Kilani Jaziri S, Kovacic H, Chekir-Ghedira L, Ghedira K, Luis J. The effects of caffeic, coumaric and ferulic acids on proliferation, superoxide production, adhesion and migration of human tumor cells in vitro . Eur J Pharmacol. 2015;766:99–105. doi: 10.1016/j.ejphar.2015.09.044. [DOI] [PubMed] [Google Scholar]
- Oliva A, Galletti P, Zappia V, Paik WK, Kim S. Studies on substrate specificity of S-adenosylmethionine: protein-carboxyl methyltransferase from calf brain. Eur J Biochem. 1980;104:595–602. doi: 10.1111/j.1432-1033.1980.tb04463.x. [DOI] [PubMed] [Google Scholar]
- Olthof MR, Hollman PC, Katan MB. Chlorogenic acid and caffeic acid are absorbed in humans. J Nutr. 2001;131:66–71. doi: 10.1093/jn/131.1.66. [DOI] [PubMed] [Google Scholar]
- Pelinson LP, Assmann CE, Palma TV, Cruz IBM, Pillat MM, Manica A, Stefanello N, Weis GCC, Alves AO, Ulrich H, et al. Antiproliferative and apoptotic effects of caffeic acid on SK-Mel-28 human melanoma cancer cells. Mol Biol Rep. 2019;46:2085–2092. doi: 10.1007/s11033-019-04658-1. [DOI] [PubMed] [Google Scholar]
- Prasad NR, Jeyanthimala K, Ramachandran S. Caffeic acid modulates ultraviolet radiation-B induced oxidative damage in human blood lymphocytes. J Photochem Photobiol B. 2009;95:196–203. doi: 10.1016/j.jphotobiol.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Rajagopal C, Lankadasari MB, Aranjani JM, Harikumar KB. Targeting oncogenic transcription factors by polyphenols: A novel approach for cancer therapy. Pharmacol Res. 2018;130:273–291. doi: 10.1016/j.phrs.2017.12.034. [DOI] [PubMed] [Google Scholar]
- Ramos S, Alia M, Bravo L, Goya L. Comparative effects of food-derived polyphenols on the viability and apoptosis of a human hepatoma cell line (HepG2) J Agric Food Chem. 2005;53:1271–1280. doi: 10.1021/jf0490798. [DOI] [PubMed] [Google Scholar]
- Repetto G, Del Peso A, Zurita JL. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat Protoc. 2008;3:1125–1131. doi: 10.1038/nprot.2008.75. [DOI] [PubMed] [Google Scholar]
- Santini V, Melnick A, Maciejewski JP, Duprez E, Nervi C, Cocco L, Ford KG, Mufti G. Epigenetics in focus: pathogenesis of myelodysplastic syndromes and the role of hypomethylating agents. Crit Rev Oncol Hematol. 2013;88:231–245. doi: 10.1016/j.critrevonc.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Schneider-Stock R, Ghantous A, Bajbouj K, Saikali M, Darwiche N. Epigenetic mechanisms of plant-derived anticancer drugs. Front Biosci (Landmark Ed) 2012;17:129–173. doi: 10.2741/3919. [DOI] [PubMed] [Google Scholar]
- Shen J, Song R, Gong Y, Zhao H. Global DNA hypomethylation in leukocytes associated with glioma risk. Oncotarget. 2017;8:63223–63231. doi: 10.18632/oncotarget.18739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto YT, Collins AR, Benzie IFF. Effects of dietary antioxidants on DNA damage in lysed cells using a modified comet assay procedure. Mutat Res. 2002;500:31–38. doi: 10.1016/s0027-5107(01)00298-6. [DOI] [PubMed] [Google Scholar]
- Tai J, Cheung S, Chan E, Hasman D. Antiproliferation effect of commercially brewed coffees on human ovarian cancer cells in vitro. Nutr Cancer. 2010;62:1044–1057. doi: 10.1080/01635581.2010.492083. [DOI] [PubMed] [Google Scholar]
- Tice RR, Agurell E, Anderson D, Burlinson B, Hartmann A, Kobayashi H, Miyamae Y, Rojas E, Ryu JC, Sasaki YF. Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ Mol Mutagen. 2000;35:206–221. doi: 10.1002/(sici)1098-2280(2000)35:3<206::aid-em8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Wang X, Liu J, Xie Z, Rao J, Xu G, Huang K, Li W, Yin Z. Chlorogenic acid inhibits proliferation and induces apoptosis in A498 human kidney cancer cells via inactivating PI3K/Akt/mTOR signalling pathway. J Pharm Pharmacol. 2019;71:1100–1109. doi: 10.1111/jphp.13095. [DOI] [PubMed] [Google Scholar]
- Wu JH, Chen J, Wang Y, Xia B, Wang R, Zhao Y, Wang QX, Song Q, Yao SH, Zhang YH, et al. Effect of Mono-2-ethyhexyl phthalate on DNA methylation in human prostate cancer LNCaP cells. Biomed Environ Sci. 2017;30:641–648. doi: 10.3967/bes2017.085. [DOI] [PubMed] [Google Scholar]
- Yao J, Peng S, Xu J, Fang J. Reversing ROS-mediated neurotoxicity by chlorogenic acid involves its direct antioxidant activity and activation of Nrf2-ARE signaling pathway. Biofactors. 2019;45:616–626. doi: 10.1002/biof.1507. [DOI] [PubMed] [Google Scholar]
- Yun S, Vincelette ND, Abraham I, Robertson KD, Fernandez-Zapico ME, Patnaik MM. Targeting epigenetic pathways in acute myeloid leukemia and myelodysplastic syndrome: a systematic review of hypomethylating agents trials. Clin Epigenetics. 2016;8:68–68. doi: 10.1186/s13148-016-0233-2. [DOI] [PMC free article] [PubMed] [Google Scholar]