Abstract
High-grade serous ovarian cancer (HGSOC) is thought to progress from a series of precursor lesions in the fallopian tube epithelium (FTE). One of the preneoplastic lesions found in the FTE is called a secretory cell outgrowth (SCOUT), which is partially defined by a loss of paired box 2 (PAX2). In the present study, we developed PAX2-deficient murine cell lines in order to model a SCOUT and to explore the role of PAX2 loss in the etiology of HGSOC. Loss of PAX2 alone in the murine oviductal epithelium (MOE) did not induce changes in proliferation, migration and survival in hypoxia or contribute to resistance to first line therapies, such as cisplatin or paclitaxel. RNA sequencing of MOE PAX2shRNA cells revealed significant alterations in the transcriptome. Silencing of PAX2 in MOE cells produced a messenger RNA expression pattern that recapitulated several aspects of the transcriptome of previously characterized human SCOUTs. RNA-seq analysis and subsequent qPCR validation of this SCOUT model revealed an enrichment of genes involved in estrogen signaling and an increase in expression of estrogen receptor α. MOE PAX2shRNA cells had higher estrogen signaling activity and higher expression of putative estrogen responsive genes both in the presence and absence of exogenous estrogen. In summary, loss of PAX2 in MOE cells is sufficient to transcriptionally recapitulate a human SCOUT, and this model revealed an enrichment of estrogen signaling as a possible route for tumor progression of precursor lesions in the fallopian tube.
We developed a murine oviductal model of secretory cell outgrowths and found an upregulation of estrogen signaling, a proposed pro-tumorigenic pathway in ovarian carcinogenesis. Additionally, this model provides a platform for further research into the critical steps for fallopian tube-derived HGSOC.
Introduction
Last year, an estimated 22 440 women were diagnosed with ovarian cancer, and 14 080 died of the disease (1), making ovarian cancer the most lethal gynecologic malignancy in the USA. High-grade serous ovarian cancer (HGSOC) is the most common and aggressive subtype of ovarian cancer. For many years, the ovarian surface epithelium was believed to be the only site of ovarian cancer progenitor cells, but recent evidence suggests that the fallopian tube epithelium (FTE) is also a common progenitor site (2,3). An FTE origin for HGSOC is supported by the discovery and characterization of precancerous lesions in the FTE, which are frequently described as ‘p53 signatures’ based on mutant p53 expression, which results in stabilization of the p53 protein (4,5). In fact, the prevailing paradigm in the origin of fallopian tube-derived HGSOC is that serous tubal intraepithelial carcinomas (STICs) in the tubal fimbriae are the direct precursors to HGSOC (4–7); however, a wide spectrum of premalignant lesions have been observed, each with its own hypothesized tumorigenic potential. Indeed, Soong et al. described the phenomena of ‘precursor escape’ linking benign tubal lesions to co-occurring HGSOC despite the absence of STICs, demonstrating the complexity of premalignant lesions in carcinogenesis (8).
Secretory cell outgrowths (SCOUTs) are thought to be the earliest FTE precursor lesion of HGSOC and have been reported to be in higher frequency in the fallopian tubes of HGSOC patients (9,10). A SCOUT constitutes an outgrowth of 30 or more secretory FTE cells, which have low or absent paired box 2 (PAX2) expression (9,11). Histological studies have further characterized SCOUTs by their lack of p76 and their positive expression for BCL2, STMN1 and EZH2 (12). Two distinct subtypes of SCOUTs have been described each with unique transcriptional profiles differentiating them from each other and from the FTE. Histologically, type 1 SCOUTs resemble normal fallopian epithelium, whereas type 2 SCOUTs appear less differentiated (11). PAX2-null SCOUTs are associated with age and are more prevalent in postmenopausal women and in women with HGSOC (10). Furthermore, SCOUTs can be found evenly distributed throughout the distal and proximal fallopian tube; however, their ratio to p53 signatures and STICs diminishes in the fimbriae (10,13). It is still unclear whether SCOUTs and HGSOC are directly connected via multistep tumorigenesis in a series of molecular insults or if SCOUTs develop independently and represent benign terminal lesions. Although it is not yet understood how SCOUTs may advance into these more aggressive precancerous lesions, it has been well documented that they overwhelmingly share a loss of PAX2 (3,9,10,14,15).
The paired box family are transcription factors that play an important role during embryogenesis. PAX2 is required for Müllerian duct formation (3,16). In adult tissues, PAX2 protein is present in normal FTE and oviductal epithelium (the oviduct is the murine equivalent of the human fallopian tube), but it is not expressed in the normal ovarian surface epithelium (17). Re-expressing PAX2 in serous ovarian cancer cell lines, a majority of which harbor a marked reduction of PAX2 expression, resulted in an attenuation of proliferation, highlighting PAX2’s tumor suppressive role (14,15,18). Loss of phosphatase and tensin homolog (PTEN) in murine oviductal epithelial (MOE) cells resulted in tumorigenesis along with a reduction of PAX2. Re-expression of PAX2 in PTEN-null MOE cell lines reduced tumor burden (19,20). Despite the mounting evidence for the role of PAX2 loss in the formation of early precancerous lesions in the FTE and in late-stage HGSOC tumor survival, little is known about the mechanisms by which loss of PAX2 facilitates these phenomena. Although PAX2 has been found to act as both a tumor suppressor and an oncogene in ovarian cancer as a whole, it has been reported that a majority of HGSOC cases harbor a marked reduction of PAX2. A collection of histological studies has found that PAX2 is lost in 89% of SCOUTs, 92% of p53 signatures, 67–75% of tubal intraepithelial carcinomas and 70% of high-grade fimbrial-ovarian carcinoma (3,10,11,14,15). Although loss of PAX2 is pervasive in serous subtypes of ovarian cancer, its role in malignancy remains elusive. PAX2 has been implicated in regulating PI3k signaling and transforming growth factor-β (TGF-β) signaling in models of HGSOC as a possible tumorigenic route (18,21). Further, recent publications have connected aberrations in hormonal signaling, a known risk factor for developing ovarian cancer, with changes in PAX2 expression (22,23).
Estradiol is a steroid hormone that plays a critical role in the proliferation and differentiation of the reproductive organs, such as the uterus and mammary gland. Furthermore, estrogens exposure is one of the risk factors for developing ovarian cancer, and it is predicted to act primarily through its actions via estrogen receptors (ERs) (24–28). Increased estrogen exposure is associated with an increased incidence of neoplastic transformation through induction of proliferative signaling pathways, IL-6/ STAT3 mediated mitogenesis, antiapoptotic signaling and increased DNA damage (29–31). Histological studies have found that ER is expressed in 86% of ovarian tumor samples and up to 93% in serous subtype tumor samples (31). Experimental and epidemiologic studies have provided support for a connection between estrogen signaling and ovarian tumorigenesis; however, little has been studied connecting hormonal signaling with known premalignant lesions such as SCOUTs.
This study sought to transcriptionally recapitulate human SCOUTs in MOE cells using a PAX2shRNA and CRISPR-mediated PAX2 knockout. We compared these models to published data of the messenger RNA (mRNA) landscape of human SCOUTs. Further, the present study sought to leverage the newly created SCOUT models to explore the functional signaling mechanisms induced by loss of PAX2 in fallopian tube epithelial cell lines and identified ER signaling as one potentially dysregulated pathway.
Methods
Cell culture
MOE cells were donated by Barbara Vanderhyden, PhD, University of Ottawa in 2013 and validated as mycoplasma-free as described previously (32,33). MOE cells were maintained in complete medium containing α-modified Eagle’s medium (α-MEM; Corning #10-022-CV) supplemented with 10% v/v fetal bovine serum (FBS; Gemini #100–106), 2 mM L-glutamine (Gibco #25030081), 2 μg/ml epithelial growth factor (Peprotech #AF-100–15), 1 mg/ml gentamycin (Corning #30-005-CR), 50 U penicillin, 50 μg/ml streptomycin and 18.2 ng/ml β-estradiol. For experiments investigating estrogen, cells were cultured at least 48 h in ‘stripped media’ consisting of phenol red-free α-MEM (Life Technologies, Carlsbad, CA) supplemented with everything listed above without the β-estradiol and with charcoal-stripped FBS. Stable cell lines were generated by transfection with Lipofectamine 2000 followed by selection with 0.14 µg/ml puromycin [for PAX2 small hairpin RNA (shRNA) or scrambled shRNA]. For experiments investigating drug response, varying concentrations of cis-Diammineplatinum (II) dichloride (Cayman #13119) and Paclitaxel (Sigma #T7402) were used.
Guide RNA (gRNA) for CRISPR/Cas9 were designed using CRISPOR (http://crispor.tefor.net/; Supplementary Table S1, available at Carcinogenesis Online). The gRNAs (Integrated DNA Technologies, Coralville, IA) were cloned into pX330-U6-Chimeric_BB-CBh-hSpCas9 plasmid (gift from Dr Feng Zhang, Massachusetts Institute of Technology, Cambridge, MA; Addgene plasmid #42230) (34).The pX330 plasmid with gRNA was co-transfected with pPGKpuro plasmid (gift from Dr Rudolf Jaenisch, Massachusetts Institute of Technology, Cambridge, MA; Addgene plasmid # 11349) (35,36). Cells were treated with puromycin and single-cell clones were isolated. Genomic DNA was extracted from cells using genomic DNA extraction kit (#G170, Sigma–Aldrich, St Louis, MO) as per the manufacturer’s instructions, and the targeted exon was amplified and sequenced.
Cell viability
For sulforhodamine B (SRB) assays, MOE cells were plated at 5 × 103 cells/ml in a 96-well plate, then treated and incubated for 3–5 days followed by colorimetric assay as described previously (37). Absorbance at 505 nm was measured on a BioTek Synergy 2 microplate reader (BioTek, Winooski, VT).
Western Blot
Protein lysate (35 μg) was run on sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane. Blots were blocked with 5% milk or bovine serum albumin in tris-buffered saline-T and probed at 4°C overnight with primary antibodies (Supplementary Table S2, available at Carcinogenesis Online). Antirabbit horseradish peroxidase-linked secondary antibody (Cell Signaling #7074S) was used for 30 min in blocking buffer. Membranes were incubated and developed as described previously (20).
RNA isolation, complementary DNA synthesis and qantitative PCR (qPCR)
RNA extraction was performed using Trizol (Life Technologies, Grand Island, NY) and chloroform with isopropanol precipitation followed by ethanol washes and DNAse step. iScript™ complementary DNA synthesis kit (Bio-Rad #170–8841) and SYBR green (Thermo Fisher #AB1323A) were used according to manufacturer’s instructions. All qPCR measurements were performed using the CFX connect Real-Time PCR Detection System (Bio-Rad, Hercules, CA). Samples were normalized to the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Primers designed using NCBI Primer-Blast tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) (Supplementary Table S3, available at Carcinogenesis Online).
Migration assay
Cells were plated to confluence (1.5 × 105 cells/well) in a 24-well plate. A uniform wound was created through the cell monolayer and pictures were taken at 0 and 8 h after scratching using an AmScope MU900 with Toupview software (AmScope, Irvine, CA). The area of the scratch was analyzed with ImageJ NIH software. Percentage of closure was determined by measuring the final area of the wound relative to the initial area of the scratch.
RNA sequencing
RNA was isolated from cells using Qiagen RNAeasy Mini kit (#74104) as per manufacturer’s protocol. RNA libraries (three technical replicates) were created. RNA quality determination, mRNA enrichment, library construction, sequencing and transcriptome statistical analysis were performed at the Genomics Core Facility at Northwestern University as previously described (33).
Luciferase assay
Cells were trypsinized and plated in 24-well plate (3.5 × 104 cells/well) in phenol red-free and stripped FBS containing media. Cells were transfected with pERE-luciferase plasmid (100 ng/well) and RSV-β-galactosidase (100 ng/well using TransIT LT1 transfection reagent (0.9 μL per well, Mirus Bio, Madison, WI). Cells were treated with transfection mixture overnight in fresh media and then, if necessary, treated for 24 h with E2 or dimethyl sulfoxide (DMSO). Luciferase production and β-galactosidase activity (for transfection normalization) were measured as described previously (38).
Statistical analysis
Data are presented as mean ± standard error, with significance determined utilizing GraphPad Prism software (GraphPad, La Jolla, CA) for P ≤ 0.05. All data sets were analyzed for significant outliers by Grubbs’ test of deviation. All conditions were tested in three replicates in at least triplicate experiments. Statistical significance was determined by Student’s t-test, one-way analysis of variance (ANOVA), or two-way ANOVA with Tukey’s post hoc test. Specific statistical method used for each figure is specified in figure legend. P < 0.05 was considered statistically significant.
Results
Silencing PAX2 does not alter proliferation, migration or drug response of MOE cells
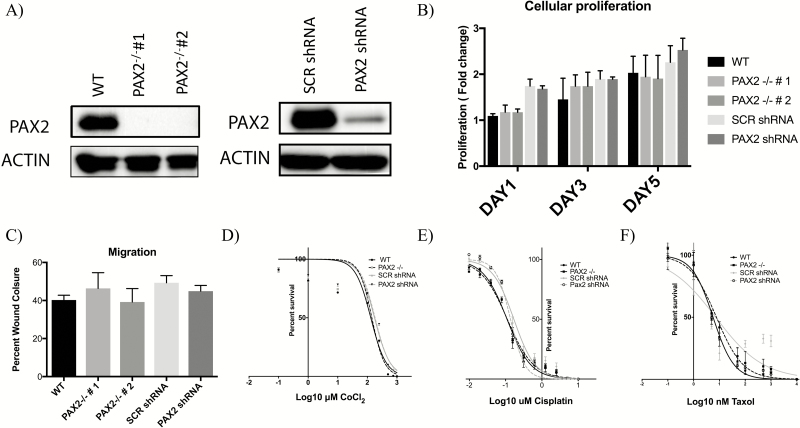
We have previously demonstrated that partial loss of PAX2 via stable transfection of PAX2shRNA (MOE:PAX2shRNA) does not significantly impact proliferation or migration despite evidence that re-expressing PAX2 in malignant cell lines is antiproliferative and reduces tumor burden in vivo (20). To expand our understanding of the role of PAX2 in altering preneoplastic lesion characteristics, a CRISPR knock out of PAX2 in MOE cells was developed (MOE:PAX2−/−). Two clones of this model were validated using western blot compared with wild-type control (Figure 1A). Proliferation of MOE:PAX2−/− clones after 1, 3 and 5 days was not significantly different than control (Figure 1B). Additionally, there was no significant change in migration of these cells as analyzed by scratch assay (Figure 1C). Hypoxic tumor microenvironments, through stabilization of hypoxia inducible factor 1 subunit alpha (HIF1⍺), have been shown to increase the aggressiveness of ovarian cancer, and survival in hypoxic conditions is a key adaptive trait for tumor progression (39). Therefore, we used CoCl2 as a hypoxia mimetic, which blocks the degradation of HIF1⍺. MOE:PAX2shRNA and MOE:PAX2−/− cell lines were exposed to a CoCl2 concentration gradient and their survival compared with control was assessed via SRB analysis. Neither PAX2 silencing in MOE:PAX2shRNA nor PAX2 knock out in MOE:PAX2−/− impacted the cells ability to survive in hypoxic-mimetic conditions (Figure 1D and Supplementary Table S4A, available at Carcinogenesis Online). Ovarian cancers are usually treated with a platinum compound or a taxane (3). MOE:PAX2shRNA and MOE:PAX2−/− cells were exposed to Cis-Diammineplatinum (II) dichloride (Cisplatin) and Paclitaxel and showed no change in IC50 survival compared with control (Figure 1E and F) (Supplementary Table S4B and S4C, available at Carcinogenesis Online).
Figure 1.
Silencing PAX2 does not alter proliferation, migration or drug response of murine fallopian tube epithelial cells. (A) Western blot analysis validating reduction of PAX2 expression in MOE:PAX2−/− clones 1 and 2 and in MOE:PAX2shRNA. (B) Relative growth of MOE cells after 1, 3 and 5 days as measured by SRB assay (N = 5, not significant, two-way ANOVA followed by Tukey’s post hoc). (C) Migration of MOE cells after 24 h as measured by scratch assay (N = 3, not significant, two-way ANOVA followed by Tukey’s post hoc). Cells were treated with varying concentrations of (D) CoCl2, (E) Cisplatin and (F) Taxol; survival after 3 days was measured by SRB assay. Data presented as ±standard error of the mean.
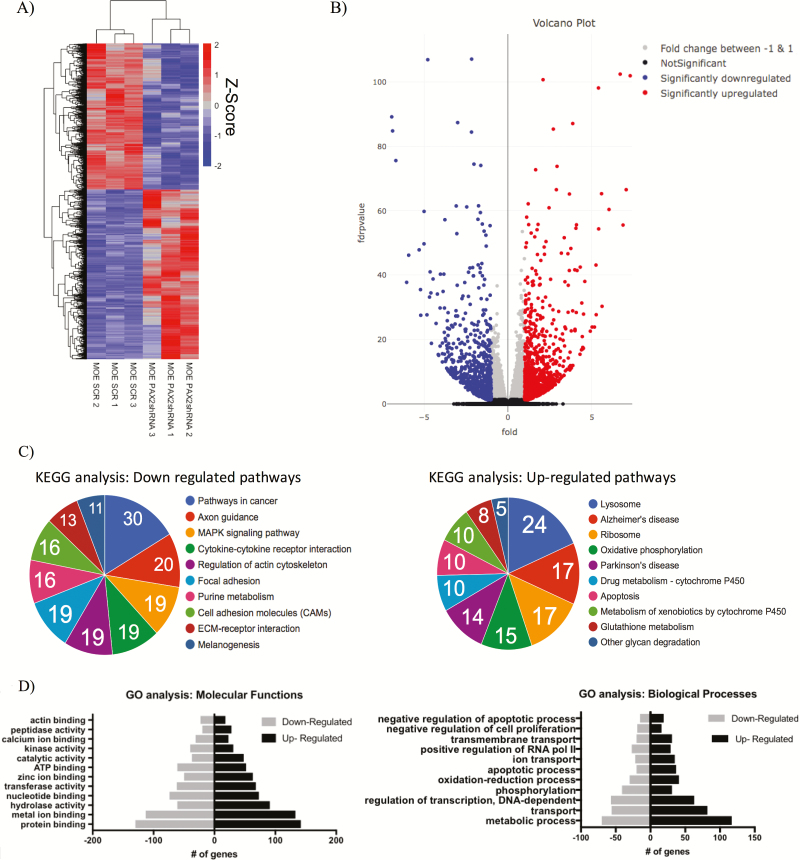
Genome-wide RNA sequencing analysis of PAX2-deficient murine oviductal cells revealed a significantly altered transcriptome
Given the consistent PAX2 suppression in epithelial ovarian cancer and its precursor lesions, we sought to define the impact of PAX2 deficiency on FTE gene expression. In order to get a global view of PAX2 loss, differentially expressed gene profiles were obtained by comparing RNA-seq data generated from three MOE:PAX2shRNA replicates and three MOE:SCRshRNA control replicates. Of the 16 527 transcripts identified by RNA-seq, 5567 genes were significantly changed (FDR-adjusted P-value of <0.05). Unsupervised hierarchal clustering of these 5567 genes across the six samples is illustrated in a representative heat map (Figure 2A). Nine hundred forty-four genes were significantly increased with a log2 fold change greater than 1 and 781 genes were significantly decreased with a log2 fold change less than −1 (Figure 2B). The top 10 genes, most significantly, upregulated and downregulated are listed in Tables 1 and 2, respectively. KEGG and gene ontology enrichment analyses identified significant expressional alterations of genes involved in a variety of cellular functions and metabolic processes (Figure 2C and D). Identical RNA-seq analysis was performed on MOE:PAX2−/− CRISPR clones in triplicate with MOE:WT cells as a control. Of the 16 414 transcripts detected, 5208 genes were significantly changed (FDR-adjusted P-value of <0.05). Unsupervised hierarchal clustering of the 5208 genes across the six samples is illustrated in a representative heat map (Supplementary Figure S1A, available at Carcinogenesis Online). As expected, many pathways found to be impacted by silencing of PAX2 were also found to be changed after CRISPR knock out. Cross comparison of gene set enrichment analyses (GSEA) of both RNA-seq data sets showed that complete loss of PAX2 in MOE cells changed the expression of genes that are involved in cell adhesion, p53 signaling and estrogen signaling compared with partial loss of PAX2 as measured by increased normalized enrichment score (Supplementary Figure S1B, available at Carcinogenesis Online).
Figure 2.
Genome-wide analysis of PAX2-deficient murine oviductal cells revealed a significantly altered transcriptome. (A) Representative heat map comparing RNA-seq data of MOE:PAX2shRNA compared with MOE:SCRshRNA. Plot shows 2469 of the 5567 significantly upregulated or downregulated genes shown (FDR-adjusted P-value <1E-4). (B) Volcano plot representing entire RNA-seq data set. (C) KEGG analysis performed on total RNA-seq highlighting pathways with the most upregulated or downregulated genes. (D) Gene ontology molecular function (left) and biological processes (right) analysis performed on total RNA-seq.
Table 1.
Top genes that were significantly upregulated in PAX2shRNA cells from RNA sequencing
| Gene | Description | Log2 fold change | FDR-adjusted P-value |
|---|---|---|---|
| Syt13 | Synaptotagmin XIII | 7.29 | 1.30E-102 |
| Efha1 | Mitochondrial calcium uptake 2 | 7.05 | 3.00E-67 |
| Itpkb | Inositol 1,4,5-trisphosphate 3-kinase B | 6.86 | 2.85E-56 |
| Rpl35 | Ribosomal protein L35A | 6.69 | 4.02E-103 |
| Mir682 | MicroRNA 682 | 6.03 | 4.19E-61 |
| Rnf130 | Ring finger protein 130 | 5.61 | 4.85E-31 |
| Sdc2 | Syndecan 2 | 5.58 | 5.31E-66 |
| Kdr | Kinase insert domain protein receptor | 5.41 | 4.50E-55 |
| Rpl19 | Ribosomal protein L19 | 5.39 | 7.89E-99 |
Table 2.
Top genes that were significantly downregulated in PAX2shRNA cells from RNA sequencing
| Gene | Description | Log2 fold change | FDR-adjusted P-value |
|---|---|---|---|
| Prom1 | Prominin 1 | −6.93 | 7.46E-90 |
| Serinc2 | Serine incorporator 2 | −6.88 | 1.70E-85 |
| Loxl2 | Lysyl oxidase-like 3 | −6.68 | 2.74E-76 |
| Lphn2 | Adhesion G protein-coupled receptor L2 | −6.04 | 1.74E-38 |
| H19 | H19, imprinted maternally expressed transcript | −5.92 | 6.49E-47 |
| Rai2 | Retinoic acid induced 2 | −5.29 | 1.43E-48 |
| Tbx15 | T-box 15 | −5.21 | 3.15E-36 |
| Tspan12 | Tetraspanin 12 | −5.20 | 2.93E-28 |
| Il22ra1 | Interleukin 22 receptor, alpha 1 | −5.00 | 1.93E-50 |
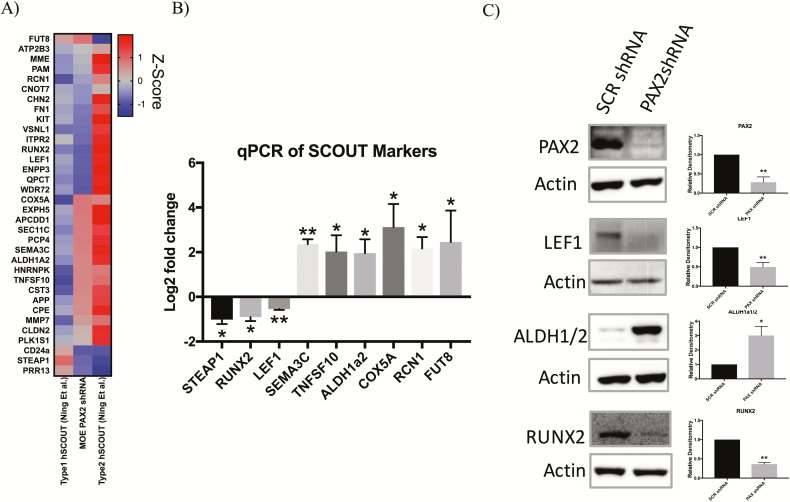
PAX2-deficient MOE cells model human SCOUTs
A genome-wide transcriptomic comparison was conducted in order to examine transcripts altered in PAX2-deficient MOE cells and microarray data from human laser-capture microdissected SCOUTs. Unsupervised hierarchical clustering analysis comparing RNA-seq data from the PAX2-deficient MOE cells to published microdissected human tissue of benign FTE and SCOUTs revealed similarities in transcriptional regulation between our MOE:PAX2shRNA and human SCOUTs (Supplementary Figure S2, available at Carcinogenesis Online). Ning et al. further published a subset of genes that uniquely characterize type 1 and type 2 SCOUTs (11). Side-by-side comparison of these transcriptional fingerprints to the mRNA profile of MOE:PAX2shRNA cells shows that this model has both type 1 and type 2 signatures (Figure 3A). A subset of these signature genes was validated with qPCR and western blotting (Figure 3B and C). qPCR validated RNA-seq findings with the exception of RCN1, which was downregulated in the RNA-seq (log2 fold change of −0.21; FDR-adjusted P-value = 0.469) but upregulated in the qPCR validation (log2 fold change of 2.165; P-value of 0.013). We performed RNA-seq on MOE:PAX2−/− cells and performed the same analysis described above. Although the MOE:PAX2−/− model similarly showed type 1 and type 2 SCOUT transcriptional characteristics, it did not show dysregulation of the same genes compared with the MOE:PAX2shRNA model (Supplementary Figure S3, available at Carcinogenesis Online).
Figure 3.
PAX2-deficient MOE cells model human SCOUTs. (A) Heatmap comparing RNA-seq data of MOE:PAX2shRNA cells and mRNA expression of key genes deregulated in human SCOUTs (hSCOUT) as highlighted by Ning et al.(B) qPCR validation of subset of genes shown in heatmap. (C) Western blot validation of subset of genes shown in qPCR validation along with densitometry. Data are presented as mean ± standard error of the mean. Significant differences from control is represented by *P < 0.05 and **P < 0.01.
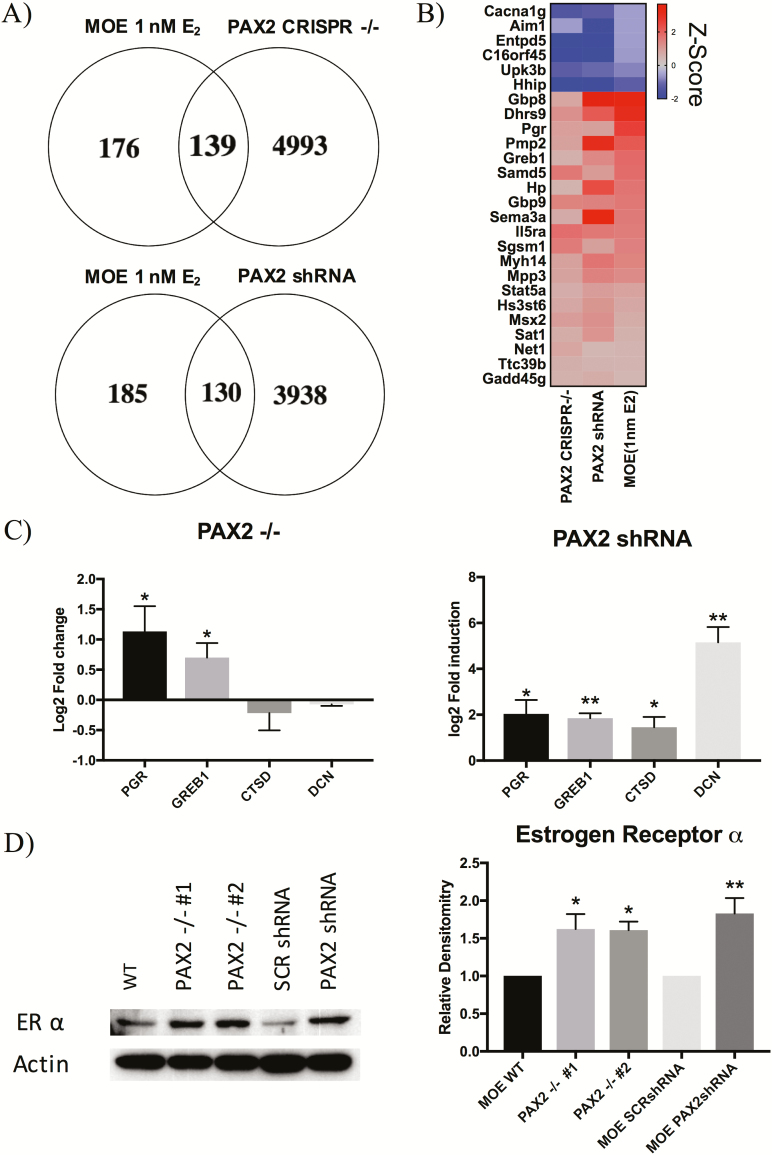
PAX2-deficient cells have increased expression of ER and genes involved in ER signaling
GSEA pathway analysis of both MOE:PAX2shRNA and MOE:PAX2−/− RNA-seq data sets revealed an enrichment of genes involved in early estrogen signaling (Supplementary Figure S1B, available at Carcinogenesis Online). To further investigate this pathway enrichment, we compared RNA-seq data of our PAX2shRNA and MOE:PAX2−/− clones to published RNA-seq data from MOE cells treated with 1 nM 17β-estradiol (E2) (40). MOE:PAX2shRNA cells had differential expression of 139 of the same genes altered in MOE cells treated with 1 nM E2. Similarly, 130 genes regulated by E2 exposure were found to be significantly altered in the MOE:PAX2−/− model (Figure 4A). The top 25 genes that were most significantly upregulated and downregulated in all three treatment groups (MOE:PAX2−/−, MOE:PAX2shRNA and 1 nM E2 treated MOE) are shown in the representative heatmap (Figure 4B). Among these genes are putative estrogen responsive genes, GREB1 and PGR (40,41). These genes, along with other known estrogen responsive genes, CTSD and DCN, were chosen for qPCR validation (40,41). MOE:PAX2shRNA cells showed an upregulation of all four selected genes, while MOE:PAX2−/− cells only showed an increase of PGR and GREB1 (Figure 4C). RNA-seq revealed an increase in expression of ESR1, the gene that encodes for estrogen receptor ⍺ (ER⍺) at a rate of 29.6% in MOE:PAX2shRNA (FDR-adjusted P-value = 0.0484) and 29.2% MOE:PAX2−/− (FDR-adjusted P-value = 3.34E-8). We confirmed an increase in ER⍺ protein expression in all models using western blot analysis (Figure 4D).
Figure 4.
PAX2-deficient cells have increased expression of ER and genes involved in ER signaling. (A) Venn diagram comparing genes that are upregulated or downregulated in the RNA-seq of MOE cells treated with 1 nM E2 compared with RNA-seq of MOE:PAX2-/- cells (top) and MOE:PAX2shRNA cells (bottom). (B) Representative heat map of genes most deregulated in all three RNA-seq data sets (MOE:PAX2−/−, MOE:PAX2shRNA and MOE:1nM E2). (C) qPCR analysis of putative estrogen responsive genes in MOE:PAX2−/− (left) and MOE:PAX2shRNA (right). (D) Representative western blot of ER expression in SCOUT model cells along with relative densitometry.
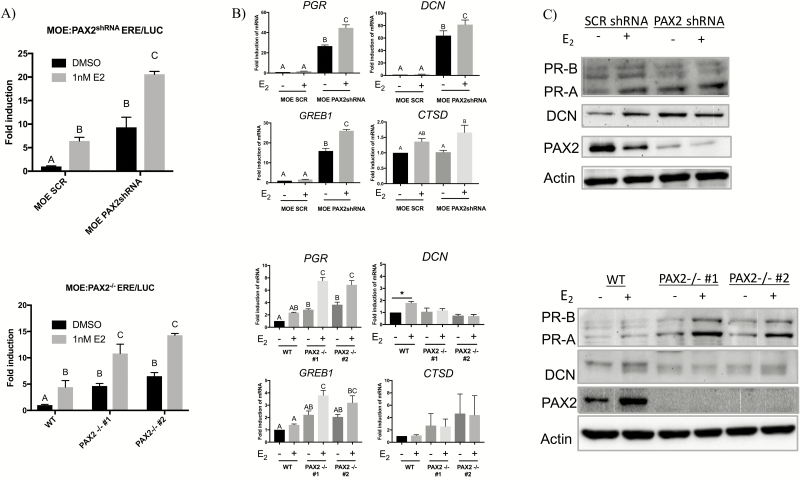
In order to link the increase in ER⍺ expression to the observed increase in expression of estrogen responsive genes, an ER responsive firefly luciferase assay (ERE/LUC) was performed in steroid-free media after which either DMSO or 1 nM E2 was added (Figure 5A). As expected, both MOE:PAX2shRNA and MOE:PAX2−/− clones showed an increase in luciferase activity after stimulation with 1 nM E2; however, both cell lines also showed significant luciferase induction in the absence of exogenous E2. Confirmatory qPCR and western blot analysis was also conducted targeting estrogen responsive genes in similar conditions (Figure 5B and C). MOE:PAX2shRNA cells had higher expression of PGR, GREB1 and DCN in both steroid-free and E2-supplemented conditions. PGR and GREB1 expression increased significantly in MOE:PAX2shRNA cells after treating with 1nM E2. Similarly, MOE:PAX2−/− cells showed an increase in PGR and GREB1 expression but not DCN or CTSD expression in steroid-free conditions or E2-supplemented conditions.
Figure 5.
Loss of PAX2 increases estrogen signaling in the fallopian tube independent of exogenous ligand. (A) estrogen signaling activity after treatment with 1 nM E2 or DMSO control in MOE:PAX2shRNA cells (top) and MOE:PAX2−/− cells (bottom) as measured by firefly luciferase assay in cells transfected with ERE/LUC. (B) qPCR analysis of putative estrogen responsive genes in after treatment with 1 nM E2 or DMSO control in MOE:PAX2shRNA cells (top) and MOE:PAX2−/− cells (bottom). (C) Western blot validation of subset of estrogen responsive genes shown in (B).
Discussion
PAX2-null SCOUTs represent one of the earliest proposed lesions in the fallopian tube and are thought to precede p53 signatures and STICS, although the exact role for PAX2 loss in the continuum of HGSOC development is still unclear (9,11,12). Ning et al. were the first to establish a link between SCOUTs, STINs and HGSOC in their expression profiling of key genes where they were able to establish a transcriptional differentiation between type 1 and type 2 SCOUTs (11). The aim of the current study was to develop a cellular model of a SCOUT, derived in this case from a MOE cell line, and use it to elucidate the possible functional changes and signaling alterations from PAX2 loss. We previously reported that re-expression of PAX2 in tumorigenic MOE:PTENshRNA cells reduced proliferation in vitro and reduced tumor formation in vivo (20). Here, we expanded on the role of PAX2 in FTE-derived HGSOC by exploring the effect of PAX2 loss in the context of otherwise normal oviductal cells.
Despite significant transcriptional changes in both our MOE:PAX2shRNA cells and MOE:PAX2−/− cells, our models remained functionally benign. We have previously shown that MOE:PAX2shRNA cells have no change in proliferation or migration; the present study confirms these data and expands on this observation by showing that complete loss of PAX2 in MOE:PAX2−/− cells similarly does not change proliferation or migration. Furthermore, we demonstrate that loss of PAX2 does not change the cells ability to survive in hypoxic conditions or resist first line therapies, cisplatin or paclitaxel. These results suggest that loss of PAX2 alone is not sufficient to induce tumorigenesis and is consistent with the benign classification of SCOUTs.
Comparison of RNA-seq data between partially PAX2-deficient MOE:PAX2shRNA cells and published microarray data from human SCOUTs allowed for an unbiased, genome-wide approach of confirming previous targets in fallopian tube cells regulated by PAX2 (Supplementary Figure S2, available at Carcinogenesis Online). The genes highlighted by Ning et al. that differentiated between the two types of SCOUTs, however, were mixed in our model. Human type 1 SCOUTS have a downregulation for LEF1, RCN1, RUNX2 and EZH2 compared with type 2. Type 2 also have an upregulation of ALDH1a2. MOE:PAX2shRNA cells had a downregulation of RUNX2 and LEF1 accompanied by an upregulation of ALDH1a2. These data show that loss of PAX2 alone may afford oviductal secretory cells the potential to become either type 1 or type 2 SCOUTs; however, further molecular insults or changes in the microenvironment may be needed to force definitive lineage differentiation. In addition to the MOE:PAX2shRNA SCOUT model, the current study also introduced a complementary MOE:PAX2−/− CRISPR model to better define how complete loss of PAX2 may impact the secretory oviductal epithelium. RNA-seq and subsequent qPCR validation revealed that, while MOE:PAX2−/− cells were still transcriptionally reminiscent of human SCOUTs, they diverged significantly from the MOE:PAX2shRNA model. Unexpectedly, MOE:PAX2−/− cells showed more type 1 character with downregulation of LEF1, RCN1, RUNX2 and ALDH1a2. This suggests that there is concentration-dependent regulation by PAX2 on these gene’s expression. This is important considering it is not understood how, or the extent to which, PAX2 is initially lost in the fallopian tube; but it is known that hallmarks of oviductal neoplastic lesions such as mutation of p53 and loss of PTEN reduce PAX2 expression and may do so in a compounding fashion (20). Transcription factors have varying affinities for the cis elements in promoters of genes under their control (42,43). Indirectly, partial or complete loss of PAX2 may have differing transcriptional outcomes because of activation of compensatory pathways or feedback loops triggered by complete loss but not partial loss. Direct chromatin binding studies followed by determining oligonucleotide affinity of PAX2 to specific promoters would be necessary to ascertain PAX2’s direct regulation of putative SCOUT markers.
Epidemiological data show that ovarian cancer may be related to lifetime exposure to estrogens. Women who receive estrogen as hormone replacement therapy have an increased risk for developing ovarian cancer (44). Estrogen has also been shown to promote ovarian tumor growth in mice and promote proliferation of human ovarian cancer and ovarian surface epithelial cell lines (25,26,28). One study of postmenopausal women concluded that exogenous estrogens were associated with an increased risk of HGSOC, but not associated with nonserous tumors (27). Although there is an abundance of experimental data showing the impact of estrogens on ovarian cancer, and mixed clinical observations, little is known about how estrogen signaling may be altered in putative preneoplastic lesions of the FTE. Our results suggest loss of PAX2 in the oviduct upregulates ER and, thus, the expression of downstream targets of the ER. Increased expression of ESR1 mRNA and higher observed ERα protein further supported a ligand-dependent regulation. Our initial RNA-seq screen and qPCR validation performed in complete media led us to believe loss of PAX2 was simply upregulating estrogen responsive genes by upregulating ERα and its co-activators, leaving PAX2-deficient cells more susceptible to stimulation by exogenous estrogens. Indeed, loss of PAX2 in MOE:PAX2shRNA cells caused an upregulation of known co-activators GREB1, SRC1, SRC2 and SRC3 (TRAM1) (45,46). Surprisingly, ERE/LUC analysis revealed that, in the absence of exogenous estrogens, PAX2-deficient cells maintained significantly higher estrogen signaling activity than control. Subsequent qPCR and western blot validation confirmed that MOE:PAX2shRNA and MOE:PAX2−/− cells continued to express putative estrogen responsive genes in steroid-free media (40,41). These data suggest a ligand-independent mechanism of estrogen signaling induction in PAX2-deficient cells. These results present the possibility of a known subset of HGSOC precursor lesions having a predisposition for heightened ER signaling in the estrogen-rich environment of the distal fallopian tube. In context, this may provide one route for tumor progression by ovulatory exposure to follicular fluid in premenopausal women and unopposed hormone replacement therapy in postmenopausal women.
In summary, we developed a murine oviductal model of secretory cell outgrowths and found an upregulation of estrogen signaling, a proposed pro-tumorigenic pathway in ovarian carcinogenesis. Our SCOUT model is transcriptionally reminiscent of published human SCOUTs and carries both type 1 and type 2 characteristics while maintaining benign phenotypes in vitro. Given the heterogeneity in ovarian cancer etiology, it is unlikely that increased estrogen signaling is the only pathway for SCOUT progression; however, it may provide one route for predisposition for the observed random somatic mutations that drive precursor lesion progression. This model will provide a platform for further research into the critical steps for fallopian tube-derived HGSOC from PAX2-deficient SCOUTS.
Funding
This work was supported by National Institutes of Health R21 CA208610, UG3 ES029073 and CA240301 (to J.E.B.), and the University of Illinois at Chicago Provost’s graduate research award.
Conflict of interest: The authors have no conflict of interest to declare.
Supplementary Material
Acknowledgements
RNA-sequencing analysis was performed at the NUSeq Core Facility, which is supported by the Northwestern University Center for Genetic Medicine, Feinberg School of Medicine and Shared and Core Facilities of the University’s Office for Research.
Glossary
Abbreviations
- α-MEM
α-modified Eagle’s medium
- ANOVA
analysis of variance
- DMSO
dimethyl sulfoxide
- ER
estrogen receptor
- FBS
fetal bovine serum
- FDR
false discovery rate
- FTE
fallopian tube epithelium
- gRNA
guide RNA
- GSEA
gene set enrichment analyses
- HGSOC
high-grade serous ovarian cancer
- MOE
murine oviductal epithelium
- mRNA
messenger RNA
- KEGG
Kyoto encyclopedia of genes and genomes
- shRNA
small hairpin RNA
- PAX2
paired box 2
- SCOUT
secretory cell outgrowth
- SRB
sulforhodamine B
- STIC
serous tubal intraepithelial carcinoma
- TGF-β
transforming growth factor-β
References
- 1. Torre L.A. et al. (2018) Ovarian cancer statistics, 2018. CA Cancer J. Clin., 68, 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bell D.A. (2005) Origins and molecular pathology of ovarian cancer. Mod. Pathol., 18 (Suppl 2), S19–S32. [DOI] [PubMed] [Google Scholar]
- 3. Hardy L.R., et al. (2018) UnPAXing the divergent roles of PAX2 and PAX8 in high-grade serous ovarian cancer. Cancers (Basel), 10, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Singh N. et al. (2016) Primary site assignment in tubo-ovarian high-grade serous carcinoma: consensus statement on unifying practice worldwide. Gynecol. Oncol., 141, 195–198. [DOI] [PubMed] [Google Scholar]
- 5. Soong T.R. et al. (2018) Evidence for lineage continuity between early serous proliferations (ESPs) in the Fallopian tube and disseminated high-grade serous carcinomas. J. Pathol., 246, 344–351. [DOI] [PubMed] [Google Scholar]
- 6. Perets R. et al. (2013) Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca;Tp53;Pten models. Cancer Cell, 24, 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee Y. et al. (2007) A candidate precursor to serous carcinoma that originates in the distal fallopian tube. J. Pathol., 211, 26–35. [DOI] [PubMed] [Google Scholar]
- 8. Soong T.R. et al. (2019) The fallopian tube, “precursor escape” and narrowing the knowledge gap to the origins of high-grade serous carcinoma. Gynecol. Oncol., 152, 426–433. [DOI] [PubMed] [Google Scholar]
- 9. Chen E.Y. et al. (2010) Secretory cell outgrowth, PAX2 and serous carcinogenesis in the Fallopian tube. J. Pathol., 222, 110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Quick C.M. et al. (2012) PAX2-null secretory cell outgrowths in the oviduct and their relationship to pelvic serous cancer. Mod. Pathol., 25, 449–455. [DOI] [PubMed] [Google Scholar]
- 11. Ning G., et al. (2014) The PAX2-null immunophenotype defines multiple lineages with common expression signatures in benign and neoplastic oviductal epithelium. J. Pathol., 234, 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kobayashi H., et al. (2017) The conceptual advances of carcinogenic sequence model in high-grade serous ovarian cancer. Biomed. Rep., 7, 209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmoeckel E., et al. (2017) LEF1 is preferentially expressed in the tubal-peritoneal junctions and is a reliable marker of tubal intraepithelial lesions. Mod. Pathol., 30, 1241–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Song H. et al. (2013) PAX2 expression in ovarian cancer. Int. J. Mol. Sci., 14, 6090–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roh M.H., et al. (2010) High-grade fimbrial-ovarian carcinomas are unified by altered p53, PTEN and PAX2 expression. Mod. Pathol., 23, 1316–1324. [DOI] [PubMed] [Google Scholar]
- 16. Robson E.J.D., et al. (2006) A PANorama of PAX genes in cancer and development. Nat. Rev. Cancer, 6, 52–62. [DOI] [PubMed] [Google Scholar]
- 17. Dressler G.R. et al. (1990) Pax2, a new murine paired-box-containing gene and its expression in the developing excretory system. Development, 109, 787–795. [DOI] [PubMed] [Google Scholar]
- 18. Al-Hujaily E.M. et al. (2015) Divergent roles of PAX2 in the etiology and progression of ovarian cancer. Cancer Prev. Res. (Phila)., 8, 1163–1173. [DOI] [PubMed] [Google Scholar]
- 19. Russo A. et al. (2018) PTEN loss in the fallopian tube induces hyperplasia and ovarian tumor formation. Oncogene, 37, 1976–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Modi D.A. et al. (2017) PAX2 function, regulation and targeting in fallopian tube-derived high-grade serous ovarian cancer. Oncogene, 36, 3015–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alwosaibai K., et al. (2017) PAX2 maintains the differentiation of mouse oviductal epithelium and inhibits the transition to a stem cell-like state. Oncotarget, 8, 76881–76897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beauchemin D. et al. (2011) PAX2 is activated by estradiol in breast cancer cells of the luminal subgroup selectively, to confer a low invasive phenotype. Mol. Cancer, 10, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang M., et al. (2016) Paired box gene 2 is associated with estrogen receptor α in ovarian serous tumors: potential theory basis for targeted therapy. Mol. Clin. Oncol., 5, 323–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clinton G.M., et al. (1997) Estrogen action in human ovarian cancer. Crit. Rev. Oncol. Hematol., 25, 1–9. [DOI] [PubMed] [Google Scholar]
- 25. Bai W., et al. (2000) Estrogen stimulation of ovarian surface epithelial cell proliferation. In Vitro Cell. Dev. Biol. Anim., 36, 657–666. [DOI] [PubMed] [Google Scholar]
- 26. Nash J.D. et al. (1989) Estrogen and anti-estrogen effects on the growth of human epithelial ovarian cancer in vitro. Obstet. Gynecol., 73, 1009–1016. [DOI] [PubMed] [Google Scholar]
- 27. Shi L.F. et al. (2016) Hormone therapy and risk of ovarian cancer in postmenopausal women: a systematic review and meta-analysis. Menopause, 23, 417–424. [DOI] [PubMed] [Google Scholar]
- 28. Syed V. et al. (2001) Expression of gonadotropin receptor and growth responses to key reproductive hormones in normal and malignant human ovarian surface epithelial cells. Cancer Res., 61, 6768–6776. [PubMed] [Google Scholar]
- 29. Rodriguez C., et al. (2001) Estrogen replacement therapy and ovarian cancer mortality in a large prospective study of US women. JAMA, 285, 1460–1465. [DOI] [PubMed] [Google Scholar]
- 30. James V., et al. (2002) Menopausal hormone replacement therapy and risk of ovarian cancer. JAMA, 288, 334–341. [DOI] [PubMed] [Google Scholar]
- 31. Ho S.M. (2003) Estrogen, progesterone and epithelial ovarian cancer. Reprod. Biol. Endocrinol., 1, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Quartuccio S.M. et al. (2013) Conditional inactivation of p53 in mouse ovarian surface epithelium does not alter MIS driven Smad2-dominant negative epithelium-lined inclusion cysts or teratomas. PLoS One, 8, e65067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dean M. et al. (2019) Exposure of the extracellular matrix and colonization of the ovary in metastasis of fallopian-tube-derived cancer. Carcinogenesis, 40, 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haeussler M. et al. (2016) Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol., 17, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tucker K.L. et al. (1996) Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev., 10, 1008–1020. [DOI] [PubMed] [Google Scholar]
- 36. Cong L. et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Skehan P. et al. (1990) New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst., 82, 1107–1112. [DOI] [PubMed] [Google Scholar]
- 38. Dean M. et al. (2018) The flavonoid apigenin is a progesterone receptor modulator with in vivo activity in the uterus. Horm. Cancer, 9, 265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gomez-Roman N. et al. (2016) Hypoxia-inducible factor 1 alpha is required for the tumourigenic and aggressive phenotype associated with Rab25 expression in ovarian cancer. Oncotarget, 7, 22650–22664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moyle-Heyrman G. et al. (2016) Genome-wide transcriptional regulation of estrogen receptor targets in fallopian tube cells and the role of selective estrogen receptor modulators. J. Ovarian Res., 9, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lin C.Y. et al. (2004) Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells. Genome Biol., 5, R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Johnson K.D. et al. (2006) Differential sensitivities of transcription factor target genes underlie cell type-specific gene expression profiles. Proc. Natl. Acad. Sci. USA, 103, 15939–15944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Im H., et al. (2005) Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc. Natl. Acad. Sci. USA, 102, 17065–17070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Santen R.J., et al. ; Endocrine Society. (2010) Postmenopausal hormone therapy: an Endocrine Society scientific statement. J. Clin. Endocrinol. Metab., 95(7 Suppl 1), s1–s66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mohammed H., et al. (2013) Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep., 3, 342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Manavathi B. et al. (2014) Estrogen receptor coregulators and pioneer factors: the orchestrators of mammary gland cell fate and development. Front. Cell Dev. Biol., 2, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.