Abstract
The bone marrow has been long known to host a unique environment amenable to colonization by metastasizing tumor cells. Yet, the underlying molecular interactions which give rise to the high incidence of bone metastasis (BM) in breast cancer patients have long remained uncharacterized. In our study, in vitro and in vivo assays demonstrated that Brachyury (Bry) could promote breast cancer BM. Bry drives epithelial–mesenchymal transition (EMT) and promotes breast cancer aggressiveness. As an EMT driver, SOX5 involves in breast cancer metastasis and the specific function in BM. Chromatin immunoprecipitation (ChIP) assays revealed SOX5 is a direct downstream target gene of Bry. ChIP analysis and reporter assays identified two Bry-binding motifs; one consistent with the classic conserved binding sequence and the other a new motif sequence. This study demonstrates for the first time that Bry promotes breast cancer cells BM through activating SOX5. In clinical practice, targeting the Bry-Sox5-EMT pathway is evolving into a promising avenue for the prevention of bone metastatic relapse, therapeutic resistance and other aspects of breast cancer progression.
Brachyury directly regulates the expression of SOX5 by binding to two motifs in its promoter region. The Bry-SOX5-EMT pathway may represent a potential target to develop treatments to prevent and treat bone metastasis from breast cancer.
Introduction
Breast cancer is the most prevalent form of cancer among women worldwide. Approximately 65–75% of patients with breast cancer develop bone metastases (BMs), leading to impaired quality of life and unfavorable outcomes (1,2). Various studies have investigated the mechanism of bone-specific metastasis, and certain signaling and transcription pathways have been identified as being involved in the process (3–6). Despite these advances, the specific molecular events that contribute to BM of breast cancer remain poorly defined.
Bry (short tail) gene, a gene mutation of the house mouse, known as Brachyury or T mutant, was first discovered in 1927 by Dobrovolskaia-Zavadskaia and encodes a protein of 435 amino acids, which functions as a transcription factor (TF) regulating posterior mesoderm formation and notochord differentiation, and is characterized by a highly conserved DNA-binding domain, designated as the T-domain (7,8). Bry (also known as T-box TF T) plays an important role in the regulation of epithelial–mesenchymal transition (EMT) in embryogenesis, during which epithelial cells change into mesenchymal cells (9–11). Bry can modulate cell migratory and adhesive behavior during the establishment of cell–matrix and cell–cell interactions and during the morphogenetic movements in a variety of multicellular organisms (12–14).
As an EMT driver, Bry has been reported to promote tumor cells of epithelial origin acquiring mesenchymal features (9,10) and to enhance tumor aggression (15). Survival analysis of 357 patients with breast cancer indicated that elevated levels of Bry were significantly associated with higher risk of recurrence and distant metastasis (11). Cell adhesion-mediated interactions between the cells and the extracellular matrix have important roles in the process of tumor metastasis and organ-specific colonization. Bry’s capacity to regulate cellular interactions with the extracellular bone matrix might promote and exacerbate BM of breast cancer cells. However, these speculations on Bry’s function remain to be experimentally confirmed.
Thus, in the present study, we aimed to explore the biological mechanism through which the TF Bry contributes to BM of breast cancer cells. The results showed that breast cancer samples with elevated Bry expression have a high risk of BM. In vitro and in vivo assays revealed that Bry promotes BM of breast cancer cells. Furthermore, SOX5, encoding SRY-Box 5, a Bry transcriptional target, mediates Bry-driven BM. The results of this study indicated that the Bry-SOX5 axis could be developed into a promising diagnostic and therapeutic target to manage and reduce BM in breast cancer.
Materials and Methods
Human samples and the ethics statement
Human breast cancer tissues were obtained from the Affiliated Suzhou Hospital of Nanjing Medical University (Suzhou, China). Tumor tissues were snap-frozen and embedded in paraffin for immunohistochemical (IHC) analyses. The study was approved by the Institutional Ethics Committee of the Nanjing Medical University and written consent was obtained from all participants.
Cell lines and cell culture
Human low-metastatic breast cancer cell line T47D, high-metastatic breast cancer cell line MDA-MB-231, osteoblast-like cell line MG-63 and mouse osteoblastic cell line MC3T3-E1 were purchased from the Shanghai Cell Bank (Shanghai, China). T47D cells were cultured in RPMI-1640 medium, MDA-MB-231 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM), MG-63 cells were cultured in minimal essential medium (MEM) and MC3T3-E1 cells were cultured in MEM-α medium. All cell lines were authenticated using PCR fingerprinting by the provider. All media (Hyclone, Logan, UT) were supplemented with 10% fetal bovine serum (FBS, Clark Bioscience, Claymont, DE) and 1% antibiotics (Penicillin-Streptomycin). Cells were incubated in a humidified atmosphere at 37°C and 5% CO2.
IHC analyses
The expression pattern of estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), marker of proliferation Ki-67 (MKI67), Bry and SOX5 in human breast cancer tissues was determined using immunohistochemistry, which was performed as described previously (16). The expressions of ER and PR were considered positive if immunoperoxidase staining of tumor cell nuclei was more than 5%. The expression of HER2 was scored on a qualitative scale from 0 to 3+ based on an interpretation of staining intensity: 0 and 1+ were classified as negative and 2+ and 3+ as positive. The MKI67 score was evaluated as the percentage of positively stained malignant cells and categorized into high (≥19% immunoreactive cells) and low (<19% immunoreactive cells) groups as described previously (17). Bry and SOX5 expression levels were scored blindly by two pathologists. Staining areas of ≤25, 26–50, 51–75 and >75% were scored as 1, 2, 3 and 4, respectively. The staining intensity was scored as follows: no staining: 0; pale yellow staining: 1; buffy staining: 2; strong brown staining: 3. The Bry expression levels were defined using a final score (staining area × staining intensity): low expression level (score ≤6) and high expression level (>6). SOX5 expression levels were defined using a final score (staining area × staining intensity): low expression level (score <6) and high expression level (≥6). The following antibodies were used: anti-ER, anti-PR, anti-HER2 and anti-MKI67 (ZSGB-BIO, Beijing, China), anti-Bry (Abcam, Cambridge, MA) and anti-SOX5 (Abcam).
Construction of cell lines
To construct Bry overexpression/knockdown cell lines, viral particles containing a small interfering RNA (siRNA; GCGGTGACTGCTTATCAGA) targeting Bry or the human Bry coding region purchased from GenePharma (Suzhou, China) were utilized in MDA-MB-231 cells and T47D cells. The cell lines were constructed as described previously (16) and validated using western blotting. In addition, the knockdown of SOX5 in Bry-overexpression cell line (T47D) and the control cell line were established by transfection of siRNA (GCAAGAGACTTGTGGCCATAG) targeting SOX5 or the negative control (NC) siRNA (TTCTCCGAACGTGTCACGT), respectively.
Protein extraction and western blotting
Culture plates were washed with phosphate-buffered saline (PBS) solution and placed on ice. Total breast cancer cell proteins were extracted using mammalian protein extraction reagent (Thermo Fisher Scientific, Waltham, MA), containing a protease inhibitor cocktail (Thermo Fisher Scientific). Equal amounts of protein were electrophoresed using 10% sodium dodecyl sulfate-gel electrophoresis and transferred on to polyvinylidene fluoride membranes (Millipore, Billerica, MA). The membranes were blocked using Tris-buffered saline and 0.1% Tween 20, containing 5% non-fat dry milk for 1 h, and then incubated overnight with primary antibodies. The membranes were washed three times and incubated with the secondary antibodies (Multi-sciences Biotechnology, Zhejiang, China). The membranes were then visualized using a chemiluminescence (Multi-sciences Biotechnology) detection system. The primary antibodies were used as follows: anti-Bry (Abcam, ab20680, 1:2000), anti-SOX5 (Abcam, ab94396, 1:1000), anti-E-cadherin (Abcam, ab1416, 1:50), anti-N-cadherin (Abcam, ab18203, 1:1000), snail family transcriptional repressor 1 (anti-Snai1; Abcam, ab53519, 1:1000), anti-Vimentin (Abcam, ab8978, 1:500), epithelial cell adhesion molecule (anti-EpCAM; Abcam, ab213500, 1:1000), anti-Fibronectin (Abcam, ab32419, 1:1000) and anti-β-actin (Multi-sciences Biotechnology, 70-ab008-100, 1:2000).
Chemotaxis assays
The cellular chemotaxis capability was assessed using Boyden chamber inserts (Corning, Corning, NY). Breast cancer cells (3 × 104) were suspended in 200 µl of 2% FBS media and seeded in the upper chamber. The lower chamber was filled with 70–80% confluent MG-63 cells incubated in 600 µl of media containing 2% FBS. After 24 h (for MDA-MB-231 sh[short hairpin RNA]NC cells and shBry cells) or 48 h (for T47D NC cells and Bry cells, T47D Bry+shNC cells and Bry+shSOX5 cells), the cells that invaded into the lower surface of the filter were stained with Giemsa (NJJCTECH, China) and counted.
Adhesion assays
For the cell–osteoblast adhesion assays, 2 × 105 breast cancer cells containing the green fluorescent protein (GFP) were added to a 100% confluent MG-63 cell layer and then incubated for 15 min (for MDA-MB-231 shNC cells and shBry cells) or 20 min (for T47D NC cells and Bry cells, T47D Bry+shNC cells and Bry+shBry cells). To produce bone matrix, MC3T3-E1 cells were cultured in MEM-α media containing 10% FBS, 10 mM β-glycerophosphate (Sigma, St. Louis, MO) and 50 µg/ml ascorbic acid (Sigma) for 9 days. Then, the MC3T3-E1 cells were incubated in 20 mM NH4OH (Sigma) and 0.5% Triton X-100 (Solarbio, Beijing, China) for 5 min to remove the cells; the bone matrix remained in the wells. For cancer cell–bone matrix adhesion assays, 2 × 105 breast cancer cells containing GFP were added to the bone matrix layer and incubated for 15 min (for MDA-MB-231 shNC cells and shBry cells) or 20 min (for T47D NC cells and Bry cells, T47D Bry+shNC cells and Bry+shSOX5 cells). After aspirating off the floating cells, the remaining cells were washed with PBS and counted under a microscope.
Colony formation assays
Colony formation assays were used to observe and compare the breast cancer cell proliferation ability in the bone microenvironment. MG-63 cells were cultured in 24-well plates until they reached 80% confluence and the conditional media (CM) that mimicked the bone microenvironment was collected. Breast cancer cells (5 × 103) (MDA-MB-231 shNC cells and shBry cells, T47D NC cells and Bry cells, T47D Bry+shNC cells and Bry+shSOX5 cells) were suspended in 0.3% soft agar (Sigma,) with MG-63 CM, and then seeded on the surface of 0.6% soft agar with MG-63 CM in a six-well plate. After 10 days of culture, the colonies were counted under a microscope.
Xenograft mouse model
Four to six-week-old female nude mice were purchased from SLAC (Shanghai, China). Then, 2.5 × 105 MDA-MB-231 cells transfected with shBry or control cells suspended in 10 µl of PBS were injected into the anterior intercondylar area in the top of the right tibia. X-rays were taken weekly starting from day 14. After 4 weeks, lesions were visible and the mice were killed. The right legs were separated and the tumor lesions were exposed and measured (V = length × width2 × 0.52). Bone lesions were confirmed as tumors using hematoxylin and eosin (HE) staining. The expression of Bry, MKI67, E-cadherin and Vimentin were examined using IHC staining. The levels of Bry, E-cadherin and Vimentin were scored blindly by two pathologists. Staining areas of ≤25, 26–50, 51–75 and >75% were scored as 1, 2, 3 and 4, respectively. The staining intensity was scored as follows: no staining: 0; pale yellow staining: 1; buffy staining: 2; strong brown staining: 3. The expression levels were defined using a final score (staining area × staining intensity). MKI67 expression was evaluated as the percentage of positively stained malignant cells. The following antibodies were used: anti-E-cadherin (Abcam) and anti-Vimentin (ZSGB-BIO). The animal study was approved by the Ethics Committee of the Nanjing Medical University.
Chromatin immunoprecipitation and sequencing
Chromatin immunoprecipitation and sequencing (ChIP-seq) using wild-type MDA-MB-231 cells was performed to explore the underlying mechanisms of cancer migration, adhesion and colonization to bone tissue. The ChIP assay was performed using a ChIP assay kit (Millipore). The anti-Bry antibodies used in this assay were purchased from R&D Systems (Bio-Techne, Minneapolis, MN). The purity and concentration of DNA samples were determined using a Qubit® Fluorometer. DNA samples were end-repaired, tailed and adaptor ligated using TruSeq Nano DNA Sample Prep Kit (#FC-121–4002, Illumina, San Diego, CA). Fragments of ~200–1500 bp were selected using AMPure XP beads. The samples were then diluted to a final concentration of 8 pM and cluster generation was performed on the Illumina cBot using a HiSeq 3000/4000 PE Cluster Kit (#PE-410–1001, Illumina). Next, sequencing was performed on an Illumina HiSeq 4000 using HiSeq 3000/4000 SBS Kit (300 cycles; #FC-410–1003, Illumina). The data were then collected and analyzed.
Chromatin immunoprecipitation-PCR assay
After the ChIP assay was performed as above described, the putative Bry-binding site on SOX5 was amplified using the following primers:
F: 5′-GCTTCTGCTCAGGGCATCAC-3′; R: 5′-GCTGGGCTCCGACTCTTTCT-3′;
F: 5′-GTGTTGTTGCACTCTGGGTAATG-3′; R: 5′-TTTCCTCGCTGTAGCCTTCTC-3′;
F: 5′-TCCTTCCCTTCGGATGGATA-3′; R: 5′-CACCCTGACCTCCTTCTTGC-3′.
The PCR products were resolved electrophoretically on a 2% agarose gel.
Dual-luciferase reporter assay
MDA-MB-231 cells cultured in 96-well plates were transiently co-transfected with 50 ng of pGL6-SOX5 promoter or control vector and 30 ng of pcDNA3.0-Bry vector (GenePharma, Shanghai, China) for 24 h. The cells were then washed with PBS, subjected to lysis and the luciferase activities were measured using a dual-luciferase assay kit (Promega, Madison, WI). The results were normalized based on the Renilla luciferase luminescence intensity (Promega-GloMax).
Statistical analysis
Values were expressed as mean ± standard deviation. Statistical analyses were performed using SPSS17.0 software (IBM Corp., Armonk, NY). Spearman’s correlation test was used to determine the correlations between groups and Student’s t-test was used to compare the differences between two groups. A P value <0.05 was considered statistically significant.
Results
Bry expression correlates with BM in patients with breast cancer
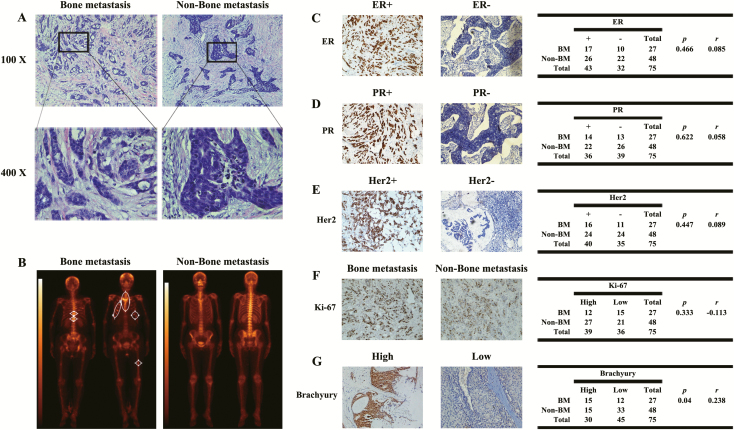
Seventy-five patients with operated breast cancer with at least 5 years of follow-up were retrieved from the archives of the Affiliated Suzhou Hospital of Nanjing Medical University (Suzhou, China). All the breast tumor tissue were removed thoroughly in the 75 patients. All the original diagnoses were established by two pathologists (Figure 1A). The clinical follow-up medical records were reviewed and the diagnoses of BM were based on the combination of a physical examination: computed tomography or magnetic resonance imaging or both and bone scintigraphy. Twenty-seven patients with breast cancer were diagnosed with BM (Figure 1B). Next, the paraffin-embedded samples of all 75 patients with and without BM were further analyzed. To evaluate the correlation of BM with ER, PR, HER2, MKI67 and Bry in breast cancer, we examined the expression of these proteins using immunohistochemistry. The results showed no correlation between ER, PR, HER2 or MKI67 and BM (Figure 1C–F). However, combined analysis of Bry expression and clinical data revealed that patients with high Bry expression had a tendency to suffer from BM (P = 0.04, r = 0.238; Figure 1G). Also, survival analysis of patients with breast cancer in The Cancer Genome Atlas (TCGA) database indicates that Bry expression level is prognostic. Patients with high Bry expression tend to have a low survival rate (P = 0.0293; Supplementary Figure S1, available at Carcinogenesis Online).
Figure 1.
Bry expression is associated with BM of breast cancer. (A) HE staining of breast cancer tissues. (B) BM of breast cancer was diagnosed using bone scintigraphy. (C–G) IHC analysis of ER, PR, HER2, MKI67 and Bry in breast cancer tissue samples; the expression of Bry was positively correlated with BM in patients with breast cancer (BM: n = 27; non-BM: n = 48) (P= 0.04, r= 0.238).
Bry promotes breast cancer cell invasion, migration, adhesion and colonization in the bone microenvironment in vitro
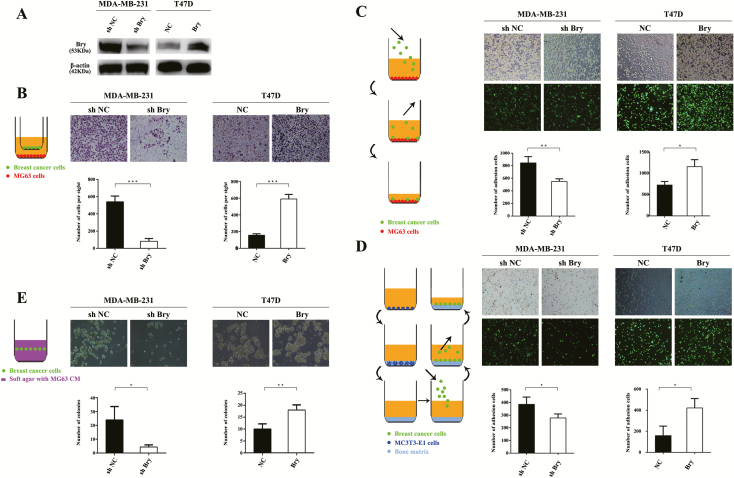
To identify the role of Bry in BM of breast cancer cells in vitro, chemotaxis, adhesion and colonization assays were performed. We chose the high-metastatic breast cancer cell line MDA-MB-231 to knock down Bry expression (MDA-MB-231 shBry cells) and the low-metastatic breast cancer cell line T47D to upregulate Bry expression (T47D Bry cells) (Figure 2A). Next, we investigated whether Bry could promote the above tumor cells to migrate and adhere to osteoblasts and colonize the bone microenvironment. The results of chemotaxis assays showed that the chemotactic migration of MDA-MB-231 shBry cells toward osteoblast-like MG-63 cells was significantly reduced compared with that of the shNC cells. Consistently, the chemotactic migration of T47D Bry cells toward MG-63 cells was significantly increased compared with that of the control cells (Figure 2B). Similarly, we evaluated the adhesive ability of breast cancer cells to osteoblasts and bone matrix, and found that the loss of Bry in MDA-MB-231 cells decreased their adhesion capability. In contrast, Bry overexpression in T47D cells enhanced their adhesion ability (Figure 2C and 2D). Next, we assessed the colony formation ability of breast cancer cells in MG-63 CM, which mimics the bone microenvironment in vitro. Similarly, the results showed that Bry promoted the ability of breast cancer cells to colonize the bone environment (Figure 2E).
Figure 2.
Bry promotes breast cancer cell invasion, migration, adhesion and colonization in the bone microenvironment. (A) The protein levels of Bry in Bry-knockdown cells or Bry-overexpression breast cancer cells compared with their respective control cells. (B) The chemotactic capacity of breast cancer cells toward osteoblast-like MG-63 cells was assessed using Transwell assays. (C) Assays to assess the adhesion capacity of breast cancer cells to osteoblast-like MG-63 cells. (D) Assays to assess the adhesion capacity of breast cancer cells to bone matrix. (E) The colonization ability of breast cancer cells in MG-63 CM was assessed using colony formation assays (*P < 0.05, **P < 0.01, ***P < 0.001).
Bry knockdown decreases the colonization and survival potential of breast cancer cells in bone tissue in vivo
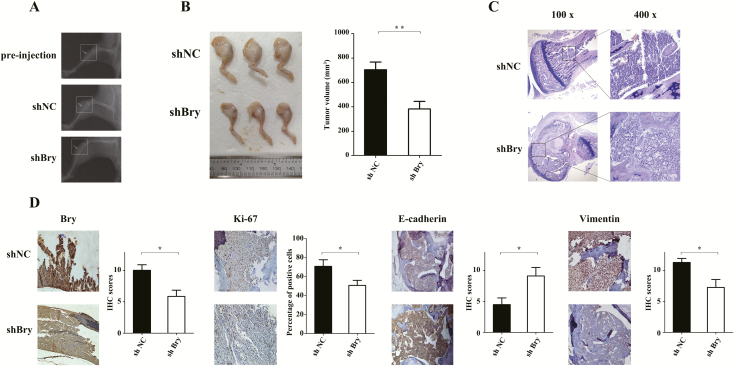
Although the intratibial injection mouse model we selected does not recapitulate the complete process of breast cancer BM, it can reflect several of critical steps in the end stage: the colonization of bone tissue and the induction tumor-related bone destruction (18,19). Four weeks after injecting Bry-downregulated and control MDA-MB-231 cells into tibias, the nude mice developed obvious swelling at the injection site. X-ray radiography examination revealed tumor-induced osteolytic lesions (Figure 3A). We observed significantly decreased tumor volumes in mice injected with Bry-knockdown MDA-MB-231 cells compared with that in mice injected with control cells (P < 0.01; Figure 3B). Furthermore, HE staining confirmed the bone lesions as tumors (Figure 3C) and IHC staining revealed that the expression of Bry and MKI67 in Bry-knockdown group was lower than that in the control group (Figure 3D). To verify the regulation by Bry of EMT in vivo, immunohistochemistry was also used to determine the activity of the EMT signal pathway (E-cadherin and Vimentin). As expected, the expression of E-cadherin was higher and the expression of Vimentin was decreased in the Bry-knockdown group compared with that in the control group (Figure 3D).
Figure 3.
Bry knockdown decreases colonization and survival potential in vivo. (A) The tumors that colonized bone tissues were examined by X-ray radiography. (B) After 4 weeks, the mice were killed, their right legs were separated and the tumor volumes were measured. (C) The tumors induced by injecting MDA-MB-231 cells into tibias were evaluated using HE staining. (D) IHC analysis of Bry, MKI67, E-cadherin and Vimentin in bone lesions (shNC: n = 6; shBry: n = 7; *P < 0.05, **P < 0.01, ***P < 0.001).
Bry positively regulates the transcription of SOX5
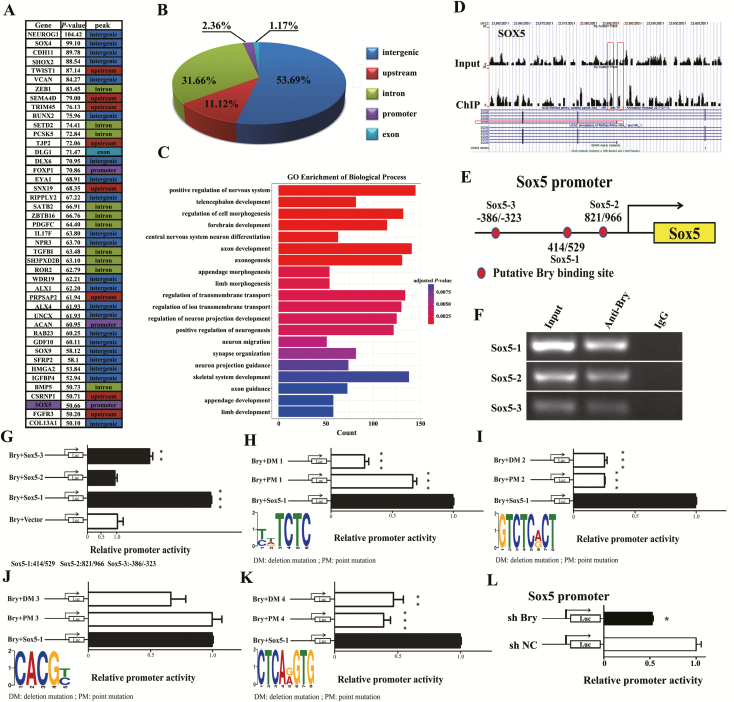
The above results showed that Bry could promote breast cancer BM. To explore the molecular mechanism, we performed a ChIP-seq assay. The results showed that the peaks were classified into five types that contained upstream, intergenic, intron, exon and promoter regions (Figure 4A and B). We further analyzed the gene ontology in the biological process category, including skeletal system development and regulation of cell morphogenesis (Figure 4C). We found that SOX5 may be the downstream target gene of Bry. SOX5 plays an important role in the regulation of EMT. A representative example of the ChIP-seq peaks for SOX5 is shown in Figure 4D. Our previous study showed that several downstream genes were significantly associated with Bry-binding events, including SOX5. Therefore, we investigated whether SOX5 was regulated by Bry. Indeed, through a sequence search of the SOX5 promoter, we found three potential Bry-binding fragments in the proximal promoter region of SOX5: 414 to 529 (SOX5-1), 801 to 966 (SOX5-2) and –386 to –323 (SOX5-3) (Figure 4E). A chromatin immunoprecipitation-PCR (ChIP-PCR) assay was then performed in MDA-MB-231 cells, and the SOX5 promoter (containing SOX5-1: 414 to 529, SOX5-2: 801 to 966 and SOX5-3: –386 to –323) was recruited by endogenous Bry (Figure 4F). We further detected the regulatory activity of Bry on the SOX5 promoter using dual-luciferase reporter assays and found that the SOX5 promoter activity was significantly increased in 293T cells co-transfected with the pcDNA3.0-Bry vector and pGL6-SOX5-1: 414/529 luciferase reporter vector but not in cells transfected with SOX5-2: 801/966 and SOX5-3: –386/–323 (Fig. 4G).
Figure 4.
Bry positively regulates the transcription of SOX5. (A) Forty-four genes containing upstream, intergenic, intron, exon and promoter regions are listed. (B) Pie charts indicating the simulated distribution of ChIP-seq peaks. (C) The gene ontology analysis, as listed by P value, and the lengths represented the counts of related genes (P < 0.05). (D) Representative ChIP-seq peaks surrounding Bry target genes (SOX5) (−10*log10 (P value) >50). (E) Schematic showing the potential Bry-binding sites in the SOX5 promoter. (F) The recruitment of endogenous Bry to the promoter of SOX5 was detected using a ChIP-PCR assay. (G) SOX5 promoter activity from three respective fragments was evaluated using dual-luciferase reporter assays. (H–K) SOX5 promoter activity with mutated (containing deletion and point mutations) Bry-binding sites was evaluated using dual-luciferase reporter assays. (L) SOX5 promoter activity in Bry-knockdown cells compared with that in control cells (*P < 0.05, **P < 0.01, ***P < 0.001).
Combining the analysis of ChIP-seq and the SOX5 promoter, we found four candidate Bry-binding sites in SOX5 (site 1: 472–479, TCTCTCTC; site 2: 471–478, GTCTCTCT; site 3: 485–489, CACGC and site 4: 495–502, CTCAAGTG). We further detected the activity of the SOX5 promoter-mutated Bry-binding sites (containing deletion and point mutations). The results study showed that the SOX5 promoter activity markedly decreased in cells transfected with pGL6-SOX5-1: 414/529 with a mutated Bry-binding site compared with the control cells transfected with pGL6-SOX5-1: 414/529 with the wild-type Bry-binding site (Figure 4H–K). Moreover, SOX5 promoter activity was decreased in MDA-MB-231 cells transfected with shBry (transfected with the total promoter) compared with that in the control cells (Figure 4L). Taken together, these results demonstrated that Bry regulates the expression of SOX5 by binding to a fragment at 414–529 (site 2 and site 4) of the promoter region.
Bry promotes breast cancer BM via the SOX5-EMT pathway
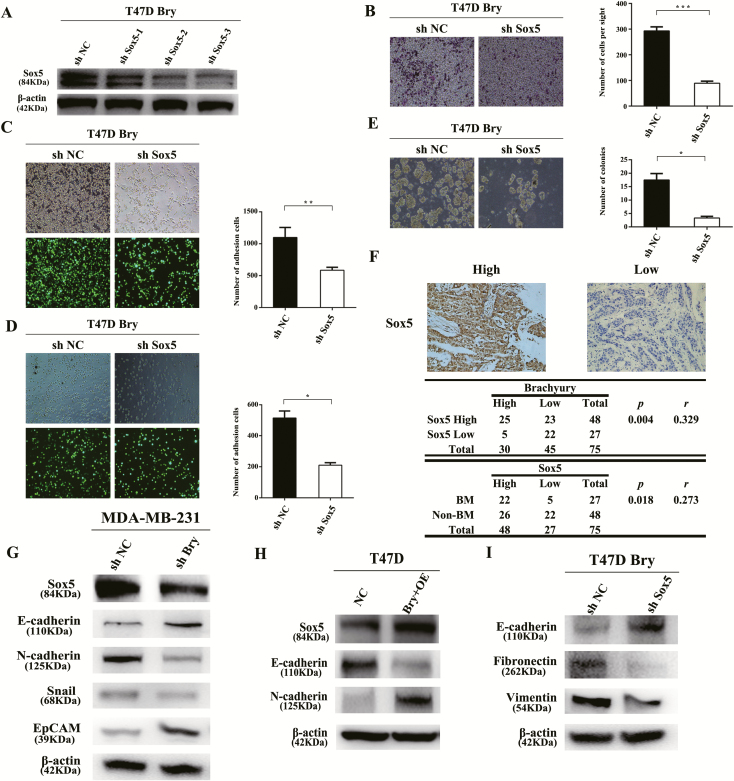
Considering the transcriptional regulation of SOX5 by Bry, we knocked down SOX5 (Bry+shSOX5) using three shRNAs (shSOX5-1, shSOX5-2 and shSOX5-3) in T47D Bry cells, and shSOX5-3 was selected for further experiments (Figure 5A). The results revealed that Bry+shSOX5 prevented the Bry-induced chemotactic migration phenotype toward MG-63 cells (Figure 5B).In addition, SOX5 knockdown in Bry-overexpressing cells reduced their adhesion ability to osteoblasts and bone matrix (Figure 5C and D). Furthermore, the colonization advantage of Bry-overexpressing cells in MG-63 CM was reversed by SOX5 knockdown (Figure 5E). Taken together, these results demonstrated that the transactivation of SOX5 by Bry increased the chemotaxis, adhesion and colonization abilities of cells in vitro. We also found that SOX5 expression correlated positively with Bry expression, as detected using IHC staining of the 75 cancer samples mentioned above (P = 0.004, r = 0.329; Figure 5F). Interestingly, the expression of SOX5 also correlated positively with BM in the 75 breast cancer samples. (P = 0.018, r = 0.273; Figure 5F). To investigate the potential functional mechanism underlying Bry-associated BM in breast cancer, the specific alterations along the SOX5-EMT signaling pathway under the impact of Bry were examined. Knockdown of Bry markedly decreased the expression levels of SOX5 and EMT-associated genes in MDA-MB-231 cells (Figure 5G). In contrast, overexpression of Bry increased the expression of SOX5 and EMT-associated genes in T47D cells (Figure 5H). Furthermore, knockdown of SOX5 in Bry-overexpressing cells prevented Bry-induced EMT (Figure 5I). These results demonstrated that Bry-induced BM from breast cancer is mediated by the SOX5-EMT pathway (Figure 6).
Figure 5.
Bry promotes breast cancer BM via the SOX5-EMT pathway. (A) Western blotting was used to detect the expression level of SOX5. (B) SOX5-knockdown in T47D Bry cells subjected to chemotaxis assays. (C) The results of cell–osteoblast adhesion assays. (D) The results of cell–bone matrix adhesion assays. (E) The results of colony formation assays. (F) IHC analysis of SOX5 in 75 breast cancer samples. The expression of Bry correlated with that of SOX5 in breast cancer samples and SOX5 expression correlated positively with BM (BM: n = 27; non-BM: n = 48). (G) The expression of SOX5 and EMT genes (E-cadherin, N-cadherin, Snail and EpCAM) was detected in Bry-knockdown and control cells. (H) The expression of SOX5 and EMT genes (E-cadherin and N-cadherin) was detected in Bry-overexpression (Bry+OE) and control cells. (I) The expression of EMT genes (E-cadherin, Fibronectin and Vimentin) was detected in Bry+shSOX5 and Bry+shNC cells (*P < 0.05, **P < 0.01, ***P < 0.001).
Figure 6.
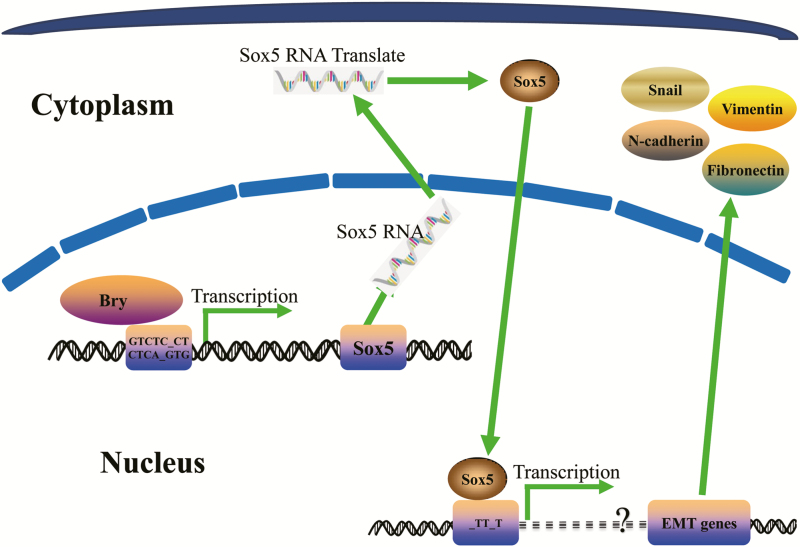
Schematic representation of the role of Bry in breast cancer cells. Bry transactivates SOX5 and further initiates the EMT process, which is involved in BM of breast cancer.
Discussion
BM of breast cancer always results in skeletal-related events such as hypercalcemia, pathological fractures, bone pain, spinal compression and reduced survival (20), which is one of the main causes of elevated morbidity and mortality in patients with breast cancer patients. The present study provides direct clinical and experimental evidence supporting the role of Bry in promoting breast cancer BM, in which Bry directly activates its downstream target, the SOX5 gene and initiates EMT.
It has long been recognized that different types of cancer metastasize to distant sites with a preference for a certain organ (21). Breast cancers show a strong predilection for BM (22). The classic ‘seed-and-soil’ hypothesis has been developed by many studies focusing on the interaction of disseminated tumor cells with host cells in the bone microenvironment (23). Breast cancer cells express a number of genes whose encoded proteins may act as homing determinants to facilitate their migration to bone, allowing tumor cells to extravasate from capillaries within bone marrow and adhere to the bone matrix and promoting communication with the host bone environment to establish BM. Different factors secreted by tumor cells, osteoclasts, osteoblasts and stromal cells regulate the activity and differentiation of colonized tumors cells and host cells (23,24), thereby escalating osteolysis.
Different gene expression signatures have been identified to mediate organ-specific metastasis of breast cancer (e.g. to the brain, lungs and bone) (25–27). Certain gene expression profiles that regulate bone-specific metastasis were identified using breast cancer cell lines or tissue samples (5,28–30) and were observed to involve adhesion, signal transduction, transcription and translation to create a specific microenvironment that favored BM. However, these metastasis signature genes identified by differential expression analysis were largely inconsistent. TFs, as master regulators that activate the metastasis genes and control multiple signal pathways, make better biomarkers and therapeutic targets than the signature genes themselves (31). Several TFs have been identified to promote bone colonization and metastasis of breast cancer cells (31–33). In the present study, the expression of TF Bry, along with known markers associated with the prognosis of breast cancer (ER, PR, HER2 and MKI67) (34), were detected in 75 patients with primary breast cancer. Bry expression correlated positively with BM, whereas ER, PR, HER2 and MKI67 expression showed no apparent correlation. This result supported Bry as a new candidate biomarker to predict breast cancer BM.
Furthermore, SOX5 was identified as the transcriptional target of Bry using ChIP assays. SOX5, a member of the sex-determining region Y (SRY) box (SOX) TF family, is required for embryonic notochord development and cell differentiation (35). Recent studies have shown that SOX5 is associated with tumor formation, progression and distant metastasis in various tumors via the EMT process (36–38). In the regulation of breast cancer progression, SOX5 induce EMT by transactivating TWIST1 via binding to a conserved SOX5-binding site (positions −133 to −138) in the TWIST1 promoter (39). Our study demonstrated that SOX5 is the downstream direct target gene of Bry in breast cancer cells. SOX5-knockdown in Bry-expressing breast cancer cells reversed the cells’ colonization and survival capabilities in bone matrix. These findings indicated that transactivation of SOX5 by Bry in breast cancer cells contributes to the development of BM, which is a preferential form of distant organ metastasis by human breast cancer cells.
In the embryonic development of vertebrates, Bry, a T-box TF gene, plays a vital role in the differentiation of axial midline mesoderm cells into notochord and gastrulation cells (40). A conserved Bry-binding motif, TNNCAC, was identified in mouse, Xenopus, zebrafish and human chordoma cell lines (41–45). This study discovered two Bry-binding motifs (Motif 2: GTCTCACT and Motif 4: CTCAGGTG) in the promoter of SOX5. Motif 2 is consistent with the abovementioned conserved binding sequence. Moreover, we identified another binding sequence CTCAGGTG (motif 4), which suggested that TF Bry has a more complex biological function.
Although some adjuvant bone-targeted treatments (bisphosphonates, zoledronate and denosumab) can inhibit the development of BM and decrease death from breast cancer (46), current therapeutic approaches mainly alleviate symptoms and slow disease progression. The identification of genes and molecules that drive breast cancer cells to colonize and grow in the bone microenvironment will provide novel therapeutic targets (47). The present study demonstrated that Bry could promote breast cancer metastasis to bone. SOX5 knockdown in Bry-expressing breast cancer cells resulted in a significant reduction in survival and tumorigenic capabilities in the bone microenvironment. The expression of Bry has a prognostic value in predicting the occurrence of BM. Furthermore, we found that Bry directly regulates the expression of SOX5 by binding to two motifs in its promoter region. The Bry-SOX5-EMT pathway may represent a potential target to develop treatments to prevent and treat BM from breast cancer.
Supplementary material
Supplementary data are available at Carcinogenesis online.
Figure S1. Brachyury expression level is prognostic. Survival rate of breast cancer patients with high and low Brachyury expression (P = 0.0293).
Funding: This work was supported by the Natural Science Foundation of Jiangsu Province of China (BK20151196), the Foundation of Social Development in Jiangsu—Clinical Frontier Technology (BE2017661).
Conflict of Interest Statement: The authors declare no potential conflicts of interest. No benefits in any form have been or will be received from a commercial party related directly or indirectly to the subject of this manuscript.
Glossary
Abbreviations
- BM
bone metastasis
- Bry
Brachyury
- ChIP-seq
Chromatin immunoprecipitation and sequencing
- CM
conditional media
- DMEM
Dulbecco’s modified Eagle’s medium
- EMT
epithelial–mesenchymal transition
- ER
estrogen receptor
- FBS
fetal bovine serum
- HE
hematoxylin and eosin
- HER2
human epidermal growth factor receptor 2
- IHC
immunohistochemical
- MEM
minimal essential medium
- MKI67
marker of proliferation Ki-67
- PBS
phosphate-buffered saline
- PR
progesterone receptor
- siRNA
small interfering RNA
- TF
transcription factor
References
- 1. Coleman R.E. (2001) Metastatic bone disease: clinical features, pathophysiology and treatment strategies. Cancer Treat. Rev., 27, 165–176. [DOI] [PubMed] [Google Scholar]
- 2. Wood S.L., et al. (2014) Omic-profiling in breast cancer metastasis to bone: implications for mechanisms, biomarkers and treatment. Cancer Treat. Rev., 40, 139–152. [DOI] [PubMed] [Google Scholar]
- 3. Deckers M., et al. (2006) The tumor suppressor Smad4 is required for transforming growth factor beta-induced epithelial to mesenchymal transition and bone metastasis of breast cancer cells. Cancer Res., 66, 2202–2209. [DOI] [PubMed] [Google Scholar]
- 4. Blanco M.A., et al. (2012) Global secretome analysis identifies novel mediators of bone metastasis. Cell Res., 22, 1339–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smid M., et al. (2006) Genes associated with breast cancer metastatic to bone. J. Clin. Oncol., 24, 2261–2267. [DOI] [PubMed] [Google Scholar]
- 6. Gilkes D.M. (2016) Implications of hypoxia in breast cancer metastasis to bone. Int. J. Mol. Sci., 17. doi:10.3390/ijms17101669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edwards Y.H., et al. (1996) The human homolog T of the mouse T(Brachyury) gene; gene structure, cDNA sequence, and assignment to chromosome 6q27. Genome Res., 6, 226–233. [DOI] [PubMed] [Google Scholar]
- 8. Palena C., et al. (2007) The human T-box mesodermal transcription factor Brachyury is a candidate target for T-cell-mediated cancer immunotherapy. Clin. Cancer Res., 13, 2471–2478. [DOI] [PubMed] [Google Scholar]
- 9. Shimoda M., et al. (2012) The T-box transcription factor Brachyury regulates epithelial-mesenchymal transition in association with cancer stem-like cells in adenoid cystic carcinoma cells. BMC Cancer, 12, 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fernando R.I., et al. (2010) The T-box transcription factor Brachyury promotes epithelial-mesenchymal transition in human tumor cells. J. Clin. Invest., 120, 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Palena C., et al. (2014) Overexpression of the EMT driver brachyury in breast carcinomas: association with poor prognosis. J. Natl. Cancer Inst., 106. doi:10.1093/jnci/dju054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Singer J.B., et al. (1996) Drosophila brachyenteron regulates gene activity and morphogenesis in the gut. Development, 122, 3707–3718. [DOI] [PubMed] [Google Scholar]
- 13. Scholz C.B., et al. (2003) The ancestral role of Brachyury: expression of NemBra1 in the basal cnidarian Nematostella vectensis (Anthozoa). Dev. Genes Evol., 212, 563–570. [DOI] [PubMed] [Google Scholar]
- 14. Yamada A., et al. (2010) Highly conserved functions of the Brachyury gene on morphogenetic movements: insight from the early-diverging phylum Ctenophora. Dev. Biol., 339, 212–222. [DOI] [PubMed] [Google Scholar]
- 15. Pires M.M., et al. (2014) Brachyury: a new player in promoting breast cancer aggressiveness. J. Natl. Cancer Inst., 106. doi:10.1093/jnci/dju094 [DOI] [PubMed] [Google Scholar]
- 16. Zou S., et al. (2016) DNA polymerase iota (Pol ι) promotes invasion and metastasis of esophageal squamous cell carcinoma. Oncotarget, 7, 32274–32285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Viale G., et al. ; International Breast Cancer Study Group. (2008) Predictive value of tumor Ki-67 expression in two randomized trials of adjuvant chemoendocrine therapy for node-negative breast cancer. J. Natl. Cancer Inst., 100, 207–212. [DOI] [PubMed] [Google Scholar]
- 18. Campbell J.P., et al. (2012) Models of bone metastasis. J. Vis. Exp., (67), e4260. doi:10.3791/4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kretschmann K.L., et al. (2012) Mouse models of breast cancer metastasis to bone. Cancer Metastasis Rev., 31, 579–583. [DOI] [PubMed] [Google Scholar]
- 20. Weilbaecher K.N., et al. (2011) Cancer to bone: a fatal attraction. Nat. Rev. Cancer, 11, 411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rusciano D., et al. (1992) Why do cancer cells metastasize into particular organs? Bioessays, 14, 185–194. [DOI] [PubMed] [Google Scholar]
- 22.(1997) Skeletal complications of malignancy. Proceedings of a symposium. Bethesda, Maryland, April 19–20, 1997. Cancer, 80, 1527–1701. PMID: 9377495. [PubMed] [Google Scholar]
- 23. Ell B., et al. (2012) SnapShot: bone metastasis. Cell, 151, 690–690.e1. [DOI] [PubMed] [Google Scholar]
- 24. Coleman R.E. (2011) Bone cancer in 2011: prevention and treatment of bone metastases. Nat. Rev. Clin. Oncol., 9, 76–78. [DOI] [PubMed] [Google Scholar]
- 25. Bos P.D., et al. (2009) Genes that mediate breast cancer metastasis to the brain. Nature, 459, 1005–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Minn A.J., et al. (2005) Genes that mediate breast cancer metastasis to lung. Nature, 436, 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Valastyan S., et al. (2011) Tumor metastasis: molecular insights and evolving paradigms. Cell, 147, 275–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kang Y., et al. (2003) A multigenic program mediating breast cancer metastasis to bone. Cancer Cell, 3, 537–549. [DOI] [PubMed] [Google Scholar]
- 29. Klein A., et al. (2009) Identification of brain- and bone-specific breast cancer metastasis genes. Cancer Lett., 276, 212–220. [DOI] [PubMed] [Google Scholar]
- 30. Minn A.J., et al. (2005) Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J. Clin. Invest., 115, 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liang Y., et al. (2012) Transcriptional network analysis identifies BACH1 as a master regulator of breast cancer bone metastasis. J. Biol. Chem., 287, 33533–33544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mock K., et al. (2015) The EMT-activator ZEB1 induces bone metastasis associated genes including BMP-inhibitors. Oncotarget, 6, 14399–14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li X.Q., et al. (2016) RUNX2 promotes breast cancer bone metastasis by increasing integrin α5-mediated colonization. Cancer Lett., 380, 78–86. [DOI] [PubMed] [Google Scholar]
- 34. Sahin A.A., et al. (1991) Ki-67 immunostaining in node-negative stage I/II breast carcinoma. Significant correlation with prognosis. Cancer, 68, 549–557. [DOI] [PubMed] [Google Scholar]
- 35. Smits P., et al. (2003) Sox5 and Sox6 are required for notochord extracellular matrix sheath formation, notochord cell survival and development of the nucleus pulposus of intervertebral discs. Development, 130, 1135–1148. [DOI] [PubMed] [Google Scholar]
- 36. Ma S., et al. (2009) DNA fingerprinting tags novel altered chromosomal regions and identifies the involvement of SOX5 in the progression of prostate cancer. Int. J. Cancer, 124, 2323–2332. [DOI] [PubMed] [Google Scholar]
- 37. Chen X., et al. (2018) SOX5 predicts poor prognosis in lung adenocarcinoma and promotes tumor metastasis through epithelial-mesenchymal transition. Oncotarget, 9, 10891–10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang D.Y. et al. (2008) Transcription factor SOX-5 enhances nasopharyngeal carcinoma progression by down-regulating SPARC gene expression. J. Pathol., 214, 445–455. [DOI] [PubMed] [Google Scholar]
- 39. Pei X.H., et al. (2014) Sox5 induces epithelial to mesenchymal transition by transactivation of Twist1. Biochem. Biophys. Res. Commun., 446, 322–327. [DOI] [PubMed] [Google Scholar]
- 40. Hotta K., et al. (2008) Brachyury-downstream gene sets in a chordate, Ciona intestinalis: integrating notochord specification, morphogenesis and chordate evolution. Evol. Dev., 10, 37–51. [DOI] [PubMed] [Google Scholar]
- 41. Di Gregorio A., et al. (1999) Regulation of Ci-tropomyosin-like, a Brachyury target gene in the ascidian, Ciona intestinalis. Development, 126, 5599–5609. [DOI] [PubMed] [Google Scholar]
- 42. Tada M. et al. (2001) T-targets: clues to understanding the functions of T-box proteins. Dev. Growth Differ., 43, 1–11. [DOI] [PubMed] [Google Scholar]
- 43. Morley R.H. et al. (2009) A gene regulatory network directed by zebrafish No tail accounts for its roles in mesoderm formation. Proc. Natl. Acad. Sci. USA, 106, 3829–3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nelson A.C., et al. (2012) An integrated functional genomics approach identifies the regulatory network directed by brachyury (T) in chordoma. J. Pathol., 228, 274–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nibu Y., et al. (2013) From notochord formation to hereditary chordoma: the many roles of Brachyury. Biomed Res. Int., 2013, 826435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brown J., et al. (2018) Associations between serum bone biomarkers in early breast cancer and development of bone metastasis: results from the AZURE (BIG01/04) Trial. J. Natl. Cancer Inst., 110, 871–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tulotta C., et al. (2019) Endogenous production of IL1B by breast cancer cells drives metastasis and colonization of the bone microenvironment. Clin. Cancer Res., 25, 2769–2782. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.