Abstract
Both nitric oxide (NO) and strigolactone (SL) are growth regulating signal components in plants; however, regarding their possible interplay our knowledge is limited. Therefore, this study aims to provide new evidence for the signal interplay between NO and SL in the formation of root system architecture using complementary pharmacological and molecular biological approaches in the model Arabidopsis thaliana grown under stress-free conditions. Deficiency of SL synthesis or signaling (max1-1 and max2-1) resulted in elevated NO and S-nitrosothiol (SNO) levels due to decreased S-nitrosoglutathione (GSNO) reductase (GSNOR) protein abundance and activity indicating that there is a signal interaction between SLs and GSNOR-regulated levels of NO/SNO. This was further supported by the down-regulation of SL biosynthetic genes (CCD7, CCD8 and MAX1) in GSNOR-deficient gsnor1-3. Based on the more pronounced sensitivity of gsnor1-3 to exogenous SL (rac-GR24, 2 µM), we suspected that functional GSNOR is needed to control NO/SNO levels during SL-induced primary root (PR) elongation. Additionally, SLs may be involved in GSNO-regulated PR shortening as suggested by the relative insensitivity of max1-1 and max2-1 mutants to exogenous GSNO (250 µM). Collectively, our results indicate a connection between SL and GSNOR-regulated NO/SNO signals in roots of A. thaliana grown in stress-free environment. As this work used max2-1 mutant and rac-GR24 exerting unspecific effects to both SL and karrikin signaling, it cannot be ruled out that karrikins are partly responsible for the observed effects, and this issue needs further clarification in the future.
Keywords: Arabidopsis thaliana, nitric oxide, root, S-nitrosoglutathione reductase, strigolactone
Introduction
Strigolactones (SLs) have been first identified as germination inducers of parasite plants in the 1960s (Cook et al., 1966) and since then, they have been found to be phytohormones due to their multiple roles in regulating growth and developmental processes of higher plants (Gomez-Roldan et al., 2008; Umehara et al., 2008; Zwanenburg and Blanco-Ania, 2018; Bouwmeester et al., 2019).
SLs as terpenoid lactones can be categorized as canonical SLs containing ABC ring and noncanonical SLs lacking such a ring (Al-Babili and Bouwmeester, 2015; Waters et al., 2017). SLs are synthetized from carotenoids in the plastids with the involvement of enzymes such as beta-carotene-isomerase (D27), two carotenoid cleavage dioxygenases (CCD7/MAX3 and CCD8/MAX4), cytochrome P450 (MAX1), and LATERAL BRANCHING OXIDOREDUCTASE (Alder et al., 2012; Brewer et al., 2016). Following its transport into the cytoplasm, carlactone is converted into carlactonoic acid which is the common precursor of the naturally occurring SLs (Jia et al., 2019). Recently, the direct conversion of carlactonoic acid to orobanchol without passing through 4-deoxyorobanchol has been described (Wakabayashi et al., 2019). Moreover, a cytochrome P450 and a 2-oxoglutarate-dependent dioxygenase genes were identified being involved in SL synthesis in Lotus japonicus (Mori et al., 2020), and hydroxyl carlactone derivatives as relevant intermediaries in SL synthesis have been identified in Arabidopsis (Yoneyama et al., 2020). Despite the active research, our knowledge about the details of SL biosynthesis after carlactone is still limited (Bouwmeester et al., 2019). It has been shown that SLs are synthetized in both the root and the shoot and that the SL signal can spread from the root to the shoot system (Foo et al., 2001).
The perception of SLs involves the SL receptor DWARF14 (D14) protein having α/β fold hydrolase activity. The intact SL molecule promotes D14 activation which in turn deactivates bioactive SLs by the hydrolytic degradation following signal transmission (Seto et al., 2019). Consequently, the activated D14 can bind the MORE AXILLARY GROWTH2 (MAX2/D3) F-box type protein which assigns DWARF53 and SMXLs repressors for proteasomal degradation resulting in the induction of gene expression (Shabek et al., 2018; Bouwmeester et al., 2019). Recently, MAX2 was implicated as a regulator of karrikin (KAR) signaling (Nelson et al., 2011), and SMXL/D53, the downstream targets of MAX2 are responsible for the discrimination of SL and KAR signal pathways (Soundappan et al., 2015). The interference between SL and KAR signaling is further supported by the fact that rac-GR24 (racemic mixtures of GR24 stereoisomers) activates both signal pathways, thus exerts also non-SL-specific effects (Scaffidi et al., 2014; Li et al., 2016). The SL-induced gene expression manifests in physiological effects such as the inhibition of shoot branching, shaping of root system architecture, inducing leaf senescence (Pandey et al., 2016; Waters et al., 2017; Marzec and Melzer, 2018). Recently, Villaécija-Aguilar and co-workers (2019) added that root traits like root hair development, root skewing, straightness, and diameter are regulated by KAR signaling, while both KAR and SL pathways contribute to the regulation of lateral root density and epidermal cell length. Furthermore, SLs have been implicated in plant stress responses to diverse abiotic factors (reviewed by Mostofa et al., 2018) like nutrient deficiency (Kohlen et al., 2011), salinity and drought (Ha et al., 2014; Wang et al., 2019, reviewed by Mostofa et al., 2018) or chilling (Cooper et al., 2018).
Similar to SLs, research over the past 40 years has revealed that the gaseous signal molecule nitric oxide (NO) is a multifunctional growth regulator in plants (Kolbert et al., 2019a). While, the ability of SL synthesis is a unique feature of plants (Walker et al., 2019), any living organism is capable of the synthesis of NO. Algae utilize NO synthase (NOS)-like enzyme system for producing NO (Foresi et al., 2010; Foresi et al., 2015; Weisslocker-Schaetzel et al., 2017) while in higher land plants NOS gene homolog to animal gene has not been found (Jeandroz et al., 2016; Santolini et al., 2017; Hancock and Neill, 2019). The ability of NO liberation via NOS-system may be lost during the evolution of land plants (Fröhlich and Durner, 2011), which takes up high amounts of nitrate, and their physiological functions are greatly determined by nitrate acquisitions. A key process in nitrate-dependent NO synthesis of plants indirectly involves nitrate reductase (NR) activity which transfers electron from NAD(P)H to the NO-forming nitrite reductase (NOFNiR). This enzyme catalyzes the reduction of nitrite to NO (Chamizo-Ampudia et al., 2016; Chamizo-Ampudia et al., 2017). NO is synthetized endogenously within the plant body in a wide variety of tissues, and NO can also be taken up from the atmosphere or from the soil (Cohen et al., 2009). In biological systems, NO reacts with glutathione to form S-nitrosoglutathione (GSNO) being a less reactive and more stable molecule than NO. GSNO is able to release NO and can achieve long distance movement of NO signal via the xylem (Durner et al., 1999; Díaz et al., 2003; Barroso et al., 2006). Intracellular levels of GSNO are controlled by the activity of GSNO reductase (GSNOR) enzyme (Feechan et al., 2005; Lee et al., 2008; Chen et al., 2009) catalyzing the conversion of GSNO to GSSG and NH3 in the presence of NADH (Jahnová et al., 2019).
Unlike SLs, the signal of NO isn't perceived by specific receptor, but the transfer of NO bioactivity is achieved by direct modification of target proteins. Cysteine S-nitrosation, tyrosine nitration, and metal nitrosylation are three major NO-dependent posttranslational modifications being physiologically relevant (Astier and Lindermayr, 2012). Additionally, the link between NO-related signaling and Ca2+-, cGMP-, MAPK-, and PA-dependent signaling has also been revealed in diverse physiological processes (Pagnussat et al., 2004; Lanteri et al., 2008; Astier et al., 2011; Jiao et al., 2018). Like SLs, NO affects a range of physiological traits including seed development, vegetative and generative development like pollen tube growth, seed germination, root growth, gravitropism, flowering, fruit ripening (reviewed in Kolbert and Feigl, 2017). Additionally, NO also participates in responses of plants to abiotic stresses like salinity, drought, heavy metal, low oxygen availability, or temperature stresses (Fancy et al., 2017).
Based on the stimulating effect of NO on plant germination, vegetative growth or fruit ripening, NO-releasing substances such as nanoparticles could be effectively applied in agricultural practice (Rodríguez-Ruiz et al., 2019). Similarly, SLs and their agonists and antagonists may have a great potential for agricultural applications. Beyond plant protection, SLs may be used to improve the architecture of crops as well (Vurro et al., 2016; Takahashi and Asami, 2018).
It is sure that both NO and SL are important growth regulating signals of practical significance in plants. However, their interplay has been poorly examined. The majority of the few articles dealing with SL–NO interplay focus on the root system of crops like sunflower (Barthi and Bhatla, 2015), maize (Manoli et al., 2016), and rice (Sun et al., 2014) grown in the presence of different nutrient supplies. Collectively, these studies revealed that NO is an upstream regulator of SL signaling; however, the nature of the NO–SL relationship depends on the nutrient availability. During nitrate‐induced root elongation, NO reduces SL biosynthesis thus resulting in alterations of PIN‐mediated auxin transport leading to cell elongation. Exogenous SL induces NO production suggesting negative feedback regulation of SL levels (Manoli et al., 2016). Low N and P availability triggers NO formation which in turn induces the proteasomal‐degradation of D53 repressor protein and consequently intensifies SL signaling leading to root elongation (Sun et al., 2016). To clarify the role of SLs in root development, Marzec and Melzer (2018) recommended to perform experiments with plants grown during stress-free conditions. Because of the above reasons, this study aims to provide new evidence for the signal interplay between NO and SL in the formation of root system architecture using complementary pharmacological and molecular biological approaches in the model Arabidopsis thaliana grown under stress-free conditions.
Materials and Methods
Plant Material and Growth Conditions
Seeds of Arabidopsis thaliana wild-type (WT, Col-0), and their mutant lines gsnor1-3 (Chen et al., 2009), 35S:FLAG-GSNOR1 (Frungillo et al., 2014), max1-1, max2-1 (Stirnberg et al., 2002) were surfaced sterilized with 70% (v/v) ethanol for 1 min and with 30% sodium hypochlorite solution (1:3) for 15 min then washed five times with sterile distilled water. Seeds (approx. 30 seeds/Petri dish) were then transferred to half strength Murashige and Skoog medium (1% sucrose, 0.8% agar). Petri dishes were kept in a greenhouse under controlled conditions (photon flux density of 150 µmol m−2 s−1, 12/12 h light and dark cycle, relative humidity of 55–60%, temperature of 25 ± 2°C) for 7 days.
Treatments
Stock solution of rac-GR24 and TIS108 (both purchased from Chiralix B.V., Nijmegen, Netherlands) was prepared in acetone or in DMSO, respectively. Appropriate volumes of stock solutions were added to the medium following sterilization through sterile syringe yielding 2 µM GR24 or 5 µM TIS108 concentrations in the media. These concentrations were chosen in pilot experiments using several doses (1, 2, 5 µM for GR24 and 1, 5, 10 µM for TIS108). Stock solutions of GSNO and 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO) were prepared in DMSO and were diluted to the final concentrations (250 µM GSNO and 800 µM cPTIO) with distilled water. Four days after placing the seeds on the media, GSNO and cPTIO solutions were added to the surface of the agar containing the root system. One milliliter of GSNO or cPTIO was added per Petri dish using 2-ml syringe and sterile filter.
Morphological Measurements
Primary root (PR) lengths of Arabidopsis seedlings were measured and expressed in mm. Lateral roots within the primary root (smaller than stage VII) were considered as lateral root primordia (LRprim), whereas visible laterals which have already grown outside the PR were considered as emerged LRs (LRem, larger than stage VII, Malamy and Benfey, 1997; Feigl et al., 2019). The number of LRprim and LRem was determined by using Zeiss Axiovert 200 inverted microscope and 20× objective (Carl Zeiss, Jena, Germany). LR density (number mm−1) was calculated by dividing total number of LRs with PR length. The experiments were performed three times with 20 samples each (n = 60).
Detection of NO Levels
Levels of NO were detected with the fluorophore, 4-amino-5-methylamino-2′-7′-difluorofluorescein diacetate (DAF-FM DA). Arabidopsis seedlings were incubated in 10 µM dye solution for 30 min, in darkness, at room temperature and washed two times with TRIS-HCl buffer (10 mM, pH 7.4) according to Kolbert et al. (2012). Stained root samples were observed under Axiovert 200M (Carl Zeiss, Jena, Germany) fluorescent microscope equipped with digital camera (Axiocam HR) and filter set 10 (excitation 450–490 nm, emission 515–565 nm) Fluorescence intensities in the PRs were measured on digital images using Axiovision Rel. 4.8 software within circles of 38 µm radii. This analysis was carried out three times with 10 root tips examined (n = 10).
Determination of S-nitrosothiol Contents
The amount of SNO was quantified by Sievers 280i NO analyser (GE Analytical Instruments, Boulder, CO, USA) according to Kolbert et al. (2019b). Briefly, 250 mg of Arabidopsis seedlings was mixed with double volume of 1× PBS buffer (containing 10 mM N-ethylmaleimide and 2.5 mM EDTA, pH 7.4) and were grounded using Fast Prep ® Instrument (Savant Instruments Inc., Holbrook, NY). Samples were centrifuged twice for 15 min (20,000 g, 4°C). The supernatants were incubated with 20 mM sulphanilamide. 250 µl of the samples was injected into the reaction vessel filled with potassium iodide. SNO concentrations were quantified with the help of NO analysis software (v3.2). Measurement of SNO levels was performed on three separate plant generations with five technical replicates in each (n = 5).
Western Blot Analysis of GSNOR Protein Abundance
Whole Arabidopsis seedlings were grounded with extraction buffer (50 mM TRIS-HCl, pH 7.6–7.8) and centrifuged (4°C, 9300 g, 20 min). Protein extract was treated with 1% proteinase inhibitor and stored at −80°C. Protein concentrations were determined using the Bradford (1976) assay.
Fifteen microliters of denaturated protein extract was subjected to SDS-PAGE on 12% acrylamide gel. Proteins were transferred to PVDF membranes using the wet blotting procedure (25 mA, 16 h). After that, membranes were used for cross-activity assays with rabbit polyclonal antibody against GSNOR (1:2,000). Immunodetection was performed by using affinity, isolated goat anti-rabbit IgG-alkaline phosphatase secondary antibody at a dilution of 1:10,000, and bands were visualized by using the NBT/BCIP reaction. Protein bands were quantified by Gelquant software (provided by biochemlabsolutions.com). Western blot was carried out on three separate protein extracts from independent plant generations, at least two times per extract.
Spectrophotometric Measurement of GSNOR Activity
The specific activity of GSNOR was measured by monitoring the NADH oxidation in the presence of GSNO at 340 nm (Sakamoto et al., 2002). Plant homogenate was centrifuged (14,000 g, 20 min, 4°C), and 100 µg of protein extract was incubated in 1 ml reaction buffer (20 mM Tris-HCl pH 8.0, 0.5 mM EDTA, 0.2 mM NADH). Data are expressed as nmol NADH min−1 mg protein−1. This measurement was performed on three separate plant generations with five technical replicates in each (n = 5).
Quantitative Real Time PCR Analysis
The expression rates of Arabidopsis genes (NIA1, NIA2, GLB1, GLB2, GSNOR1, CCD7, CCD8, D14, MAX1, MAX2) were determined by quantitative real-time reverse transcription PCR (RT-qPCR). RNA was purified from 90 mg of 7-day-old seedlings by using a NucleoSpin RNA Plant mini spin kit (Macherey-Nagel) according to the manufacturer's instruction. Furthermore, an additional DNAase digestion and purifying step was applied (ZYMO Research), and cDNA was synthetized using RevertAid reverse transcriptase. Primer3 software was used for designing primers. The primers used for RT-qPCR analyses are listed in Table S1 . The expression rates of the NO- and SL associated genes were detected by quantitative real time PCR machine (qTOWER 2.0, Jena Instruments) using SYBR Green PCR Master Mix (Thermo Mix) (Gallé et al., 2009). Data were analyzed by using qPCRsoft3.2 software (Jena Instruments). Data were normalized to the transcript levels of the control samples; ACTIN2 (At3918780) and GAPDH2 (At1913440) were used as internal controls (Papdi et al., 2008). Each reaction was carried out in three replicates using cDNA synthesized from independently extracted RNAs. These analyses were performed on three separate plant generations with three technical replicates in each (n = 3).
Measurement of NO Liberation Capacity of GSNO
NO-sensitive electrode (ISO-NOP 2 mm, World Precision Instrument) was calibrated using a method of Zhang (2004). Donor solution (1 ml 250 µM GSNO in distilled water) was prepared and placed under illumination (150 µmol m−2 s−1) in the greenhouse in order to stimulate conditions similar to treatment conditions. To ensure constant mixing of the solution magnetic stirrer was applied during the measurement. NO concentration (nM) was calculated from a standard curve. The standard curve and the results are presented in Figure S2 . This measurement was carried out three times with three technical replicates in each (n = 3).
Statistical Analysis
All results are expressed as mean ± SE. Graphs were prepared in Microsoft Excel 2010 and in SigmaPlot 12. For statistical analysis, Duncan's multiple range test (one-way ANOVA, P ≤ 0.05) was used in SigmaPlot 12. For the assumptions of ANOVA, we used Hartley's Fmax test for homogeneity and the Shapiro–Wilk normality test.
Results and Discussion
Root System of GSNOR- and SL Mutant Arabidopsis Seedlings
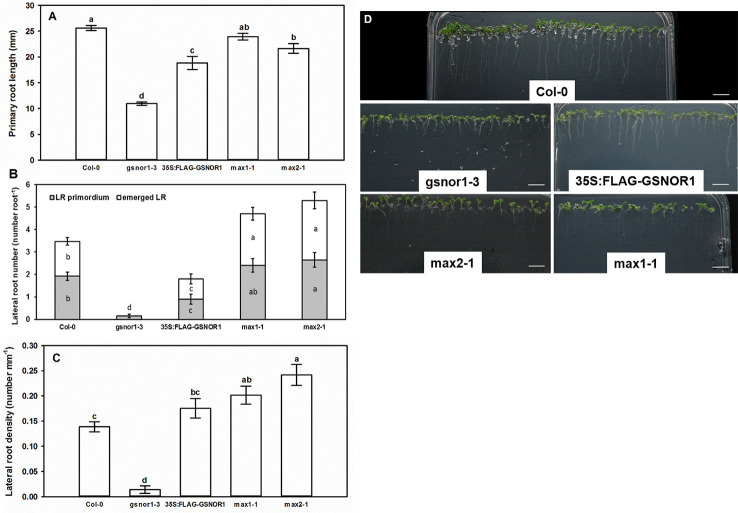
Compared to the wild-type (Col-0), the PR of gsnor1-3 mutant was by 57% shorter; its root system contained very few LRs, and consequently its LR density was low ( Figure 1 ) indicating that GSNOR activity is necessary for normal root development (Lee et al., 2008; Holzmeister et al., 2011; Kwon et al., 2012; Shi et al., 2015). Similarly, 35S:FLAG-GSNOR1 seedlings had shortened PRs and reduced numbers of laterals resulting in WT-like LR density, and the LR primordia to emerged LR ratio was similar to that of Col-0. As for the max1-1 mutant, WT-like PR length was accompanied by increased number of emerged LRs and by consequently enhanced LR density compared to Col-0. The PR of max2-1 mutant proved to be slightly (by 14%) shorter than in Col-0 and the LR number was significantly increased. The branched root systems of max1-1 and max2-1 suggest that MAX1-dependent SL biosynthesis and MAX2-associated SL-signaling inhibit LR development as was published previously by others (Kapulnik et al., 2011; Ruyter-Spira et al., 2011). The LRprim : LRem ratio was similar in Col-0 and the mutants suggesting that SLs similarly influence both the initiation and the emergence of LRs. However, max2-1 mutant has been proven to transmit both SL and KAR signals, thus the involvement of KAR in shaping root system architecture cannot be ruled out using this mutant (Villaécija-Aguilar et al., 2019).
Figure 1.
Primary root length (mm, A), lateral root number (number root−1, B) and lateral root density (number mm−1, C) in 7-day-old Col-0, GSNOR- and SL mutant Arabidopsis lines grown during stress-free conditions. Different letters indicate significant differences according to Duncan's test (n = 60, P ≤ 0.05). (D) Representative photographs taken from 7-day-old Arabidopsis seedlings of different mutant lines grown on ½ MS medium. Bars = 1 cm.
Levels of NO and SNO in GSNOR- and SL Mutant Arabidopsis Seedlings
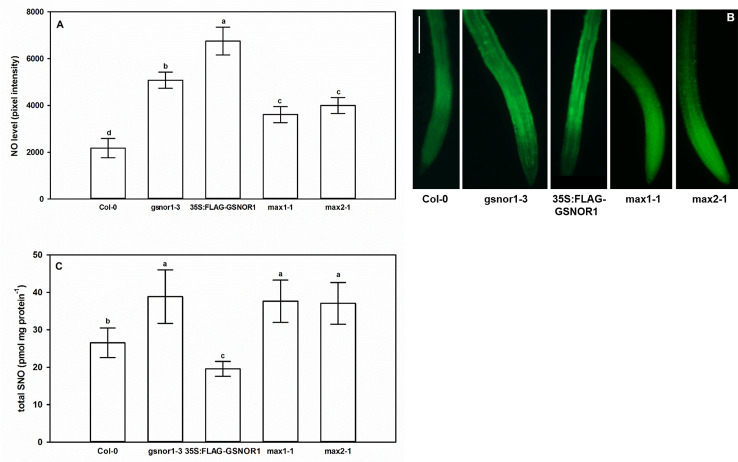
As shown in Figure 2 , the level of NO and SNO in gsnor1-3 was higher than in Col-0, while in 35S:FLAG-GSNOR1 plants, the increased endogenous NO level was accompanied by lower SNO levels than in the WT. The origin of the high NO level in the mutants is different. In 35S:FLAG-GSNOR1, elevated nitrate content and nitrate reductase activity were observed which may result in the enhanced NO level (Frungillo et al., 2014), while in gsnor1-3 the lack of GSNOR1 leads to enhanced SNO and consequently high NO contents. Based on these, applying 35S:FLAG-GSNOR1 mutant allows to draw conclusions about nitrate-derived NO while with the help of gsnor1-3 mutant we can get information about the role of GSNOR-dependent NO removal. Moreover, the similar root system of the GSNOR mutants ( Figure 1 ) can be explained by their high NO contents which are known to reduce auxin maximum and consequently cause PR shortening (Fernández-Marcos et al., 2011; Shi et al., 2015). In max1-1 and max2-1 significantly increased NO level and SNO content were detected compared to Col-0 ( Figure 2 ).
Figure 2.
Nitric oxide levels (pixel intensity, A) and SNO levels (pmol mg protein−1, C) in Col-0, GSNOR- and SL mutant Arabidopsis seedlings grown during stress-free conditions for 7 days. Different letters indicate significant differences according to Duncan's test (n = 10 or 5, P ≤ 0.05). (B) Representative microscopic images showing DAF-FM DA-stained root tips of examined Arabidopsis lines. Bar = 100 µm.
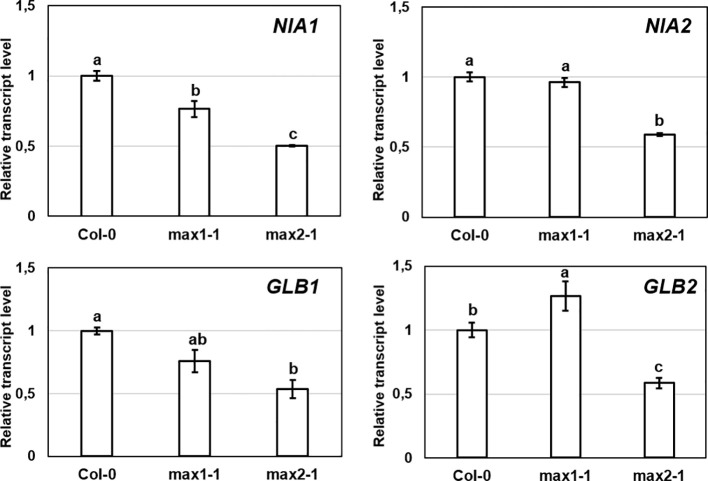
Expressions of genes involved in NO metabolism (NIA1, NIA2, GLB1, GLB2) in max1-1 mutants were similar to Col-0, but all examined genes were slightly down-regulated in max2-1 ( Figure 3 ). However, the changes were small and were not detectable in both max mutants, suggesting that these genes may not play a significant role in the regulation of NO in the absence of SLs.
Figure 3.
Relative transcript level of selected NO-associated genes (NIA1, NIA2, GLB1, GLB2) in control Col-0, max1-1 and max2-1 Arabidopsis seedlings. Different letters indicate significant differences according to Duncan's test (n = 3, P ≤ 0.05). Data were normalized using the A. thaliana ACTIN2 and GAPDH2 genes as internal controls. The relative transcript level in Col-0 control samples was arbitrarily considered to be 1 for each gene.
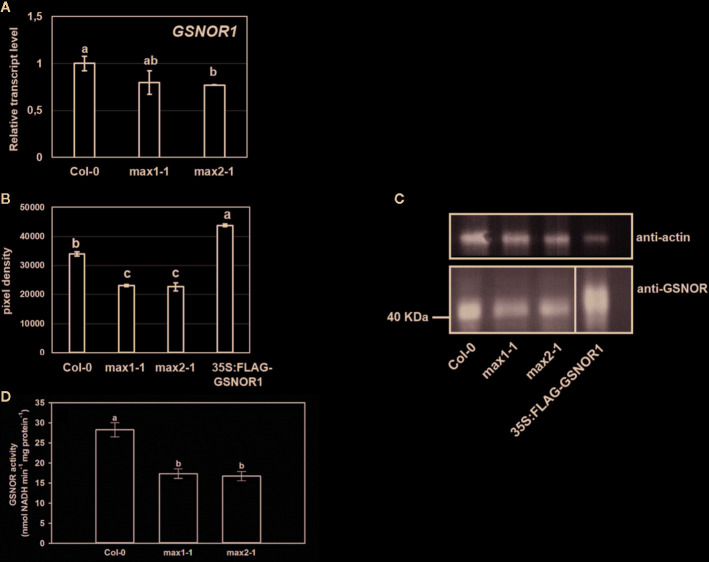
Higher NO levels of the max mutants may be associated with higher SNO levels. GSNOR is a key regulator of SNO metabolism (Lindermayr, 2018), thus we assumed that max mutants show differences in association with GSNOR enzyme. Although, there were no relevant differences in the rates of GSNOR1 expression in the plant lines ( Figure 4A ), the GSNOR protein abundance was significantly lower in max mutants compared to Col-0 ( Figures 4B, C ), and also the activity of the enzyme was decreased in max1-1 and max2-1 mutant seedlings ( Figure 4D ) which may provide the explanation for the elevated SNO and NO levels ( Figure 3 ). These results indicate that SL (and/or possibly KAR) deficiency posttranscriptionally influence GSNOR enzyme resulting in decreased SNO/NO levels. As NO acts through SLs (and/or possibly KAR) to regulate root development, the effect of SL on GSNOR-regulated NO levels may be considered as compensatory feedback mechanism. Next, we examined the responses of GSNOR deficient and -overexpressing Arabidopsis lines to exogenous application of SL analog GR24 and SL synthesis inhibitor TIS108.
Figure 4.
Relative transcript level (A) of GSNOR1 in Col-0, max1-1 and max2-1 seedlings. (B, C) Protein abundance of GSNOR in max mutants and 35S:FLAG-GSNOR1 (as a positive control). Anti-actin was used as a loading control. (D) GSNOR activity (nmol NADH min−1 mg protein−1) in Col-0, max1-1 and max2-1 seedlings. Different letters indicate significant differences according to Duncan's test (n = 3 or 5, P ≤ 0.05).
The Effect of SL Analog and Inhibitor on Root System and NO-Associated Genes in Arabidopsis
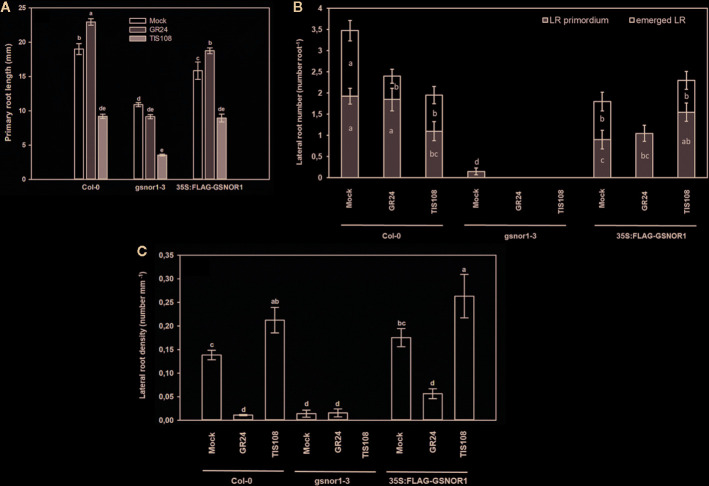
Similar to previously published results, GR24 treatment induced PR elongation in Col-0 Arabidopsis plants (Ruyter-Spira et al., 2011; Sun et al., 2014; Marzec, 2016), while TIS108 caused 50% inhibition of it ( Figure 5A ). To prove the SL-specific and non-toxic effect of TIS108 on Arabidopsis root, we applied GR24 together with TIS108 on Col-0 and we included max1-1 mutant as a TIS108-resistant line ( Figure S1 ). The max1-1 mutant proved to be less sensitive to the root growth inhibiting effect of TIS108 compared to the wild-type ( Figure S1A ), and GR24 partly reversed the root shortening effect of TIS108 in Col-0 ( Figure S1B ). These indicate that the applied concentration of TIS108 is not toxic and exerts its biological effect through SLs. In case of gsnor1-3, SL analog did not trigger PR elongation and TIS108 reduced PR length by 67% compared to the control. These suggest that the root system of gsnor1-3 is more sensitive to modifications of SL levels meaning that functional GSNOR enzyme is needed to control NO/SNO levels and to the positive effect of GR24 on PR elongation. Presumably, in case of GSNOR deficiency, NO/SNO levels are not properly regulated and high NO/SNO levels may cause PR shortening instead of elongation (Fernández-Marcos et al., 2011). The root elongation response of 35S:FLAG-GSNOR1 to SL analog or inhibitor did not differ from that of Col-0 indicating that overexpressing GSNOR enzyme or nitrate-derived NO has no effect on SL-induced elongation ( Figure 5A ). Treatment with GR24 resulted in reduced LRem number and unchanged LRprim number ( Figure 5B ) suggesting that SLs influence LR emergence but not LR initiation. In GSNOR overexpressing line, GR24-induced inhibition of LR emergence proved to be more pronounced than in Col-0. Additionally, in the stunted root system of gsnor1-3, the number of LR primordia was completely reduced by GR24. These results regarding the inhibitory effect of SL analog GR24 support previously published results (Kapulnik et al., 2011; Ruyter-Spira et al., 2011; Arite et al., 2012; De Cuyper et al., 2015; Marzec, 2016). However, without using different GR24 stereoisomers we cannot exclude the possibility that rac-GR24 may interact with KAI2 thus interfering KAR signal transduction (Scaffidi et al., 2014) and consequently influencing root development (Villaécija-Aguilar et al., 2019). In Col-0 roots, TIS108 decreased the number of both staged-LRs, but in 35S:FLAG-GSNOR1 it increased the number of LR primordia. Based on these we can assume that in case of normal GSNOR level reduced SL level inhibits LR initiation, while in the presence of increased GSNOR activity or nitrate-derived NO SL inhibition leads to the induction of LR initiation. These signal interactions may be complex and the knowledge of other contributing factors would be necessary to fully explain the observed effects. It can be a concern that the effect of the analog and the inhibitor is not always the opposite. At the same time, it is conceivable that an optimal SL level is needed for normal root growth. Increasing (by the addition of GR24) or lowering (by the addition of TIS108) the optimal SL level may result in similarly inhibited growth processes.
Figure 5.
Primary root length (mm, A), lateral root number (number root−1, B) and lateral root density (number mm−1, C) in Col-0, gsnor1-3 and 35S:FLAG-GSNOR1 Arabidopsis seedlings grown in the absence (Mock) or in the presence of GR24 (2 µM) or TIS108 (5 µM). Different letters indicate significant differences according to Duncan's test (n = 60, P ≤ 0.05).
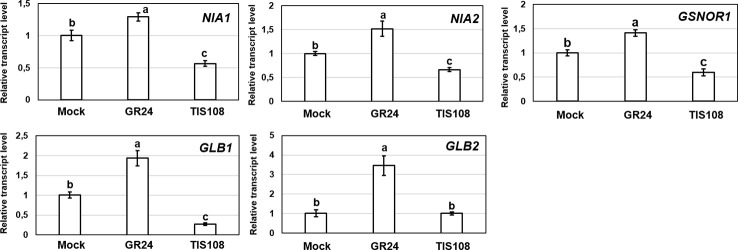
Treatment with GR24 resulted in significantly increased NO content in Arabidopsis roots (Kolbert, 2019). As for NO-associated genes, the expressions of NIA1 and NIA2 as well as GSNOR1 didn't show any relevant modification in the presence of GR24 ( Figure 6 ). In contrast, nitrogen regulatory protein P-II homolog (GLB1) and non-symbiotic hemoglobin 2 (GLB2) genes were upregulated by GR24. The GLB genes encode plant hemoglobins which may act as NO scavengers (Hebelstrup and Jensen, 2008; Hebelstrup et al., 2012; Mira et al., 2015). In this experimental system; however, GLB1 and GLB2 upregulation induced by GR24 did not lead to NO scavenging, but instead GR24 induced NO production (Kolbert, 2019). This seems to be an interesting contradiction that needs further research.
Figure 6.
Relative transcript level of selected NO-associated genes (NIA1, NIA2, GSNOR1, GLB1, GLB2) in Col-0 Arabidopsis grown under without (Mock) or with GR24 (2 µM) or TIS108 (5 µM). Different letters indicate significant differences according to Duncan's test (n = 3, P ≤ 0.05). Data were normalized using the A. thaliana ACTIN2 and GAPDH2 genes as internal controls. The relative transcript level in Col-0 control samples was arbitrarily considered to be 1 for each gene.
The Effect of NO Donor and Scavenger on SL-Associated Genes and Root System of Arabidopsis
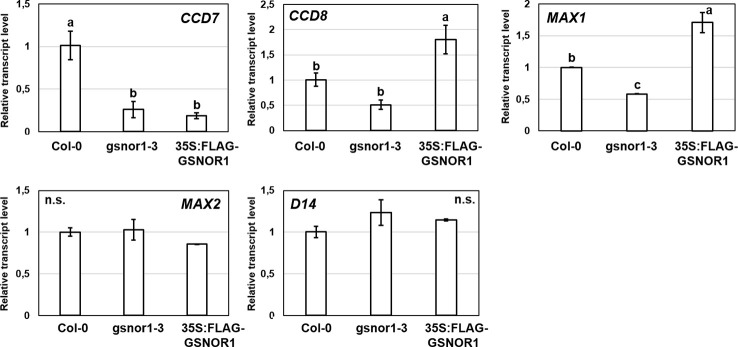
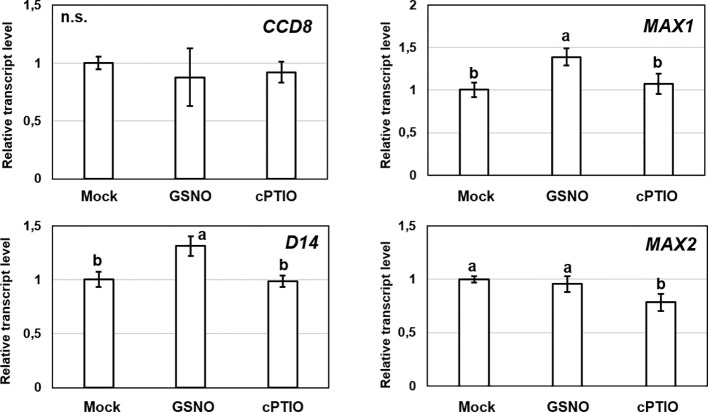
We were interested also in reverse interplay, i.e., whether under- or overproduction of GSNOR enzyme affects the expression of SL-associated genes ( Figure 7 ). The examined genes (CCD7, CCD8, MAX1) involved in the synthesis of SLs showed down-regulation in GSNOR-deficient Arabidopsis compared to Col-0. This indicates that in case of low GSNOR activity, SL biosynthesis is inhibited. This further supports the interaction between GSNO metabolism and SL production in Arabidopsis. In addition, CCD7 was down-regulated also in GSNOR overproducing 35S:FLAG-GSNOR1 seedlings. In contrast, the expressions of SL signaling genes (D14 and MAX2) were not altered by GSNOR deficiency or overproduction. However, this was not supported by pharmacological treatments (GSNO or cPTIO), because we didn't observe relevant up- or downregulation of SL-associated genes (CCD7, CCD8, MAX1, MAX2, D14) in the presence of NO donor (GSNO) or scavenger (cPTIO) treatments ( Figure 8 ). However, Castillo et al. (2018) observed larger induction in the expression of MAX1 and MAX2 in Arabidopsis seedlings due to NO treatment. From the applied 250 µM GSNO solution approx. 220 nM NO liberated over 15 min during the same circumstances as the plant treatments took place ( Figure S2 ).
Figure 7.
Relative transcript level of selected SL-associated genes in Col-0, gsnor1-3 and 35S:FLAG-GSNOR1 Arabidopsis seedlings grown during stress-free conditions. Different letters indicate significant differences according to Duncan's test (n = 3, P ≤ 0.05). Data were normalized using the A. thaliana ACTIN2 and GAPDH2 genes as internal controls. The relative transcript level in Col-0 control samples was arbitrarily considered to be 1 for each gene.
Figure 8.
Relative transcript level of selected SL-associated genes (CCD7, CCD8, MAX1, MAX2, D14) in Col-0 Arabidopsis grown in the absence (Mock) or in the presence of GSNO (250 µM) or cPTIO (800 µM). Different letters indicate significant differences according to Duncan's test (n = 3, P ≤ 0.05). Data were normalized using the A. thaliana ACTIN2 and GAPDH2 genes as internal controls. The relative transcript level in Col-0 control samples was arbitrarily considered to be 1 for each gene.
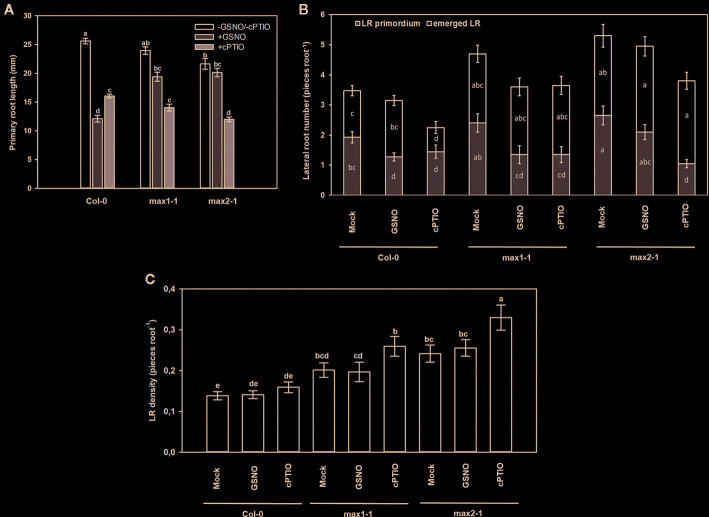
To further investigate this interaction, GSNO and cPTIO treatments were applied, and the responses of max mutants were examined ( Figure 9 ). Exogenous GSNO treatment resulted in 50% root shortening in Col-0, whereas this effect was absent in max mutants suggesting that the examined SL (and KAR) mutants are GSNO-insensitive and that SLs (and/or possibly KAR) are needed for GSNO-induced root shortening. Similar results were obtained in Arabidopsis hypocotyls, where NO-triggered shortening was not observed in max1, max2 and max4 mutants (Castillo et al., 2018). According to Fernández-Marcos et al. (2011) GSNO inhibits root meristem activity through the reduction of PIN1-dependent auxin transport. Since SLs were proved to negatively regulate PIN proteins in Arabidopsis roots (Ruyter-Spira et al., 2011), we can assume that GSNO may exert its effect on PINs via inducing SL (and/or possibly KAR) synthesis and/or signaling; although the link between NO, PINs and SL (and KAR) should be clarified by future research. The NO scavenger cPTIO shortened PRs to a similar extent in all three plant lines (Col-0, max1-1, max2-1). Moreover, GSNO inhibited LR initiation and slightly increased LR emergence of Col-0, while cPTIO supplementation decreased the number of both types of LR. In max1-1 and max2-1 seedlings, LR emergence seemed to be insensitive to NO donor or scavenger. However, GSNO treatment caused reduction in the number of LR primordia of the max1-1 mutant, and cPTIO treatment decreased LR initiation in both max mutants. Just like the matching effects of SL analog and inhibitor, the effects of NO donor and scavenger proved also to be often similar to each other, indicating the necessity of an optimal NO level for optimal root development.
Figure 9.
Primary root length (mm, A), lateral root number (number root−1, B) and lateral root density (number mm−1, C) in Col-0, max1-1, max2-1 Arabidopsis seedlings grown in the absence (Mock) or in the presence of GSNO (250 µM) or cPTIO (800 µM) for 3 days. Different letters indicate significant differences according to Duncan's test (n = 20, P ≤ 0.05).
Conclusion
The majority of the articles dealing with SL–NO interplay uses pharmacological approach and focuses on the root system of crops grown with special nutrient supply (excess nitrate or nitrogen- or phosphor deficiency). This study combines molecular biological and pharmacological approaches in order to reveal interactions between NO and SLs as growth regulating signals in the model plant Arabidopsis thaliana grown in stress-free conditions. As this study used max2-1 mutant and rac-GR24, the observed effects might be non-specific to SL signaling, and the involvement of KAR signal pathway in this system cannot be ruled out. We observed for the first time that SL (and/or KAR)-deficiency resulted in elevated NO and SNO levels due to decreased GSNOR protein abundance and activity indicating that there is a signal interaction between SLs (and/or KAR) and GSNOR-regulated levels of NO/SNO. This was further supported by the down-regulation of SL biosynthetic genes (CCD7, CCD8 and MAX1) in gsnor1-3 containing elevated NO/SNO levels. Based on the more pronounced sensitivity of gsnor1-3 to GR24, we suspected that functional GSNOR is needed to control NO/SNO levels during SL (and/or KAR)-induced PR elongation. Furthermore, SLs (and/or KAR) may be involved in GSNO-regulated PR shortening as suggested by the relative insensitivity of max1-1 and max2-1 mutants to exogenous GSNO. Collectively, our results indicate for the first time a connection between SL (and/or KAR) and GSNOR-regulated NO/SNO signals in Arabidopsis thaliana roots. Future studies should reveal the SL- or KAR-specificity of interactions with NO using d14 and kai2 mutants and GR24 stereoisomers. In the future, the possible involvement of auxin signaling as a common interacting factor of NO and SL during root development should also be examined. Additional research efforts should focus on the possible role of NO-dependent posttranslational modifications (S-nitrosation, tyrosine nitration) in relation to SL-regulated plant development.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
DO performed the experiments and wrote the manuscript draft. GF performed the experiments and reviewed the manuscript. ÁM performed the experiments. AÖ performed experiments and reviewed the manuscript. ZK conceptualized the research, designed and directed the project, reviewed the manuscript draft, and wrote the final manuscript.
Funding
This work was financed by the National Research, Development and Innovation Fund [Grant no. NKFI-6, K120383]. ZK was supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences [Grant no. BO/00751/16/8]. DO was supported by UNKP-19-3-SZTE-201 New National Excellence Program of the Ministry for Innovation and Technology. Some of the experiments were carried out by ZK during a 3-month-long visit at the Institute of Biochemical Plant Pathology, Helmholtz Zentrum München supported by TEMPUS Foundation in the frame of the Hungarian Eötvös Scholarship (MAEÖ-1060-4/2017).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Éva Kapásné Török and Elke Mattes for her valuable assistance during the experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01019/full#supplementary-material
References
- Al-Babili S., Bouwmeester H. J. (2015). Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 66, 161–186. 10.1146/annurev-arplant-043014-114759 [DOI] [PubMed] [Google Scholar]
- Alder A., Jamil M., Marzorati M., Bruno M., Vermathen M., Bigler P., et al. (2012). The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science 335, 1348–1351. 10.1126/science.1218094 [DOI] [PubMed] [Google Scholar]
- Arite T., Kameoka H., Kyozuka J. (2012). Strigolactone Positively Controls Crown Root Elongation in Rice. J. Plant Growth. Regul. 31, 165–172. 10.1007/s00344-011-9228-6 [DOI] [Google Scholar]
- Astier J., Lindermayr C. (2012). Nitric oxide-dependent posttranslational modification in plants: An update. Int. J. Mol. Sci. 13, 15193–15208. 10.3390/ijms131115193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astier J., Besson-Bard A., Wawer I., Parent C., Rasul S., Jeandroz S., et al. (2011). “Nitric Oxide Signalling in Plants: Cross-Talk with Ca2+, Protein Kinases and Reactive Oxygen Species,” in Annual Plant Reviews book series, Volume 42: Nitrogen Metabolism in Plants in the Post-genomic Era (New York, USA: Wiley; ). [Google Scholar]
- Barroso J. B., Corpas F. J., Carreras A., Rodríguez-Serrano M., Esteban F. J., Fernández-Ocaña A., et al. (2006). Localization of S-nitrosoglutathione and expression of S-nitrosoglutathione reductase in pea plants under cadmium stress. J. Exp. Bot. 57, 1785–1793. 10.1093/jxb/erj175 [DOI] [PubMed] [Google Scholar]
- Bharti N., Bhatla S. C. (2015). Nitric oxide mediates strigolactone signaling in auxin and ethylene-sensitive lateral root formation in sunflower seedlings. Plant Signal. Behav. 10, e1054087. 10.1080/15592324.2015.1054087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester H. J., Fonne-Pfister R., Screpanti C., De Mesmaeke A. (2019). Strigolactones: Plant Hormones with Promising Features. Angew. Chem. Int. Ed. 58, 12778–12786. 10.1002/anie.201901626 [DOI] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Brewer P. B., Yoneyama K., Filardo F., Meyers E., Scaffidi A., Frickey T., et al. (2016). LATERAL BRANCHING OXIDOREDUCTASE acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. 113, 6301–6306. 10.1073/pnas.1601729113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo M.-C., Coego A., Costa-Broseta Á., León J. (2018). Nitric oxide responses in Arabidopsis hypocotyls are mediated by diverse phytohormone pathways. J. Exp. Bot. 69, 5265–5278. 10.1093/jxb/ery286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamizo-Ampudia A., Sanz-Luque E., Llamas Á., Ocaña-Calahorro F., Mariscal V., Carreras A., et al. (2016). A dual system formed by the ARC and NR molybdoenzymes mediates nitrite-dependent NO production in Chlamydomonas . Plant Cell Environ. 39, 2097–2107. 10.1111/pce.12739 [DOI] [PubMed] [Google Scholar]
- Chamizo-Ampudia A., Sanz-Luque E., Llamas A., Galvan A., Fernandez E. (2017). Nitrate reductase regulates plant nitric oxide homeostasis. Trend. Plant Sci. 22, 163–174. 10.1016/j.tplants.2016.12.001 [DOI] [PubMed] [Google Scholar]
- Chen R., Sun S., Wang C., Li Y., Liang Y., An F., et al. (2009). The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res. 19 (2009), 1377–1387. 10.1038/cr.2009.117 [DOI] [PubMed] [Google Scholar]
- Cohen M. F., Lamattina L., Yamasaki H. (2009). “Nitric Oxide Signaling by Plant-Associated Bacteria,” in Nitric Oxide in Plant Physiology. Eds. Hayat S., Mori M., Pichtel J., Ahmad A. (Weinheim, Germany: Wiley-VCH Verlag GmbH & Co.), 161–172. [Google Scholar]
- Cook C. E., Whichard L. P., Turner B., Wall M. E., Egley G. H. (1966). Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154, 1189–1190. 10.1126/science.154.3753.1189 [DOI] [PubMed] [Google Scholar]
- Cooper J. W., Hu Y., Beyyoudh L., Yildiz Dasgan H., Kunert K., Beveridge C. A., et al. (2018). Strigolactones positively regulate chilling tolerance in pea and in Arabidopsis. Plant Cell Environ. 41, 1298–1310. 10.1111/pce.13147 [DOI] [PubMed] [Google Scholar]
- Díaz M., Achkor H., Titarenko E., Martínez M. C. (2003). The gene encoding glutathione-dependent formaldehyde dehydrogenase/GSNO reductase is responsive to wounding, jasmonic acid and salicylic acid. FEBS Lett. 543, 136–139. 10.1016/S0014-5793(03)00426-5 [DOI] [PubMed] [Google Scholar]
- De Cuyper C., Fromentin J., Yocgo R. E., De Keyser A., Guillotin B., Kunert K., et al. (2015). From lateral root density to nodule number, the strigolactone analogue GR24 shapes the root architecture of Medicago truncatula . J. Exp. Bot. 66, 137–146. 10.1093/jxb/eru404 [DOI] [PubMed] [Google Scholar]
- Durner J., Gow A. J., Stamler J. S., Glazebrook J. (1999). Ancient origins of nitric oxide signaling in biological systems. Proc. Nat. Acad. Sci. U.S.A. 96, 14206–14207. 10.1073/pnas.96.25.14206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy N. N., Bahlmann A.-K., Loake G. J. (2017). Nitric oxide function in plant abiotic stress. Plant Cell Physol. 40, 462–472. 10.1111/pce.12707 [DOI] [PubMed] [Google Scholar]
- Feechan A., Kwon E., Yun B. W., Wang Y., Pallas J. A., Loake G. J. (2005). A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. U. S. A. 102, 8054–8059. 10.1073/pnas.0501456102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigl G., Horváth E., Molnár Á., Oláh D., Poór P., Kolbert Zs. (2019). Ethylene-Nitric Oxide Interplay During Selenium-induced Lateral Root Emergence in Arabidopsis. J. Plant Growth. Regul. 38, 1481–1488. 10.1007/s00344-019-09950-9 [DOI] [Google Scholar]
- Fernández-Marcos M., Sanz L., Lewis D. R., Muday G. K., Lorenzo O. (2011). Nitric oxide causes root apical meristem defects and growth inhibition while reducing PIN-FORMED 1 (PIN1)-dependent acropetal auxin transport. Proc. Natl. Acad. Sci. U. S. A. 108, 18506–18511. 10.1073/pnas.1108644108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E., Turnbull C. G., Beveridge C. A. (2001). Long-distance signaling and the control of branching in the rms1 mutant of pea. Plant Physiol. 126, 203–209. 10.1104/pp.126.1.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresi N., Correa-Aragunde N., Parisi G., Caló G., Salerno G., Lamattina L. (2010). Characterization of a nitric oxide synthase from the plant kingdom: NO generation from the green alga Ostreococcus tauri is light irradiance and growth phase dependent. Plant Cell 22, 3816–3830. 10.1105/tpc.109.073510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresi N., Mayta M. L., Lodeyro A. F., Scuffi D., Correa-Aragunde N., García-Mata C., et al. (2015). Expression of the tetrahydrofolate-dependent nitric oxide synthase from the green alga Ostreococcus tauri increases tolerance to abiotic stresses and influences stomatal development in Arabidopsis . Plant J. 82, 806–821. 10.1111/tpj.12852 [DOI] [PubMed] [Google Scholar]
- Fröhlich A., Durner J. (2011). The hunt for plant nitric oxide synthase (NOS): is one really needed? Plant Sci. 181, 401–404. 10.1016/j.plantsci.2011.07.014 [DOI] [PubMed] [Google Scholar]
- Frungillo L., Skelly M. J., Loake G. J., Spoel S. H., Salgado I. (2014). S-nitrosothiols regulate nitric oxide production and storage in plants through the nitrogen assimilation pathway. Nat. Commun. 5, 5401. 10.1038/ncomms6401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallé Á., Csiszár J., Secenji M., Guóth A., Cseuz L., Tari I., et al. (2009). Glutathione transferase activity and expression patterns during grain filling in flag leaves of wheat genotypes differing in drought tolerance: response to water deficit. J. Plant Physiol. 166, 1878–1891. 10.1016/j.jplph.2009.05.016 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V., Fermas S., Brewer P. B., Puech-Pagès V., Dun E. A., Pillot J. P., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455 (7210), 189–194. 10.1038/nature07271 [DOI] [PubMed] [Google Scholar]
- Ha C. V., Leyva-González M. A., Osakabe Y., Tran U. T., Nishiyama R., Watanabe Y., et al. (2014). Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc. Natl. Acad. Sci. 111, 851–856. 10.1073/pnas.1322135111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock J. T., Neill S. (2019). Nitric Oxide: its generation and interactions with other reactive signaling compounds. Plants 8, E41. 10.3390/plants8020041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebelstrup K. H., Jensen E. O. (2008). Expression of NO scavenging hemoglobin is involved in the timing of bolting in Arabidopsis thaliana . Planta 227, 917–927. 10.1007/s00425-007-0667-z [DOI] [PubMed] [Google Scholar]
- Hebelstrup K. H., van Zanten M., Mandon J., Voesenek L. A. C. J., Harren F. J. M., Cristescu S. M., et al. (2012). Haemoglobin modulates NO emission and hyponasty under hypoxia-related stress in Arabidopsis thaliana. J. Ex. Bot. 63, 5581–5591. 10.1093/jxb/ers210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmeister C., Fröhlich A., Sarioglu H., Bauer N., Durner J., Lindermayr C. (2011). Proteomic analysis of defense response of wildtype Arabidopsis thaliana and plants with impaired NO- homeostasis. Proteom 11, 1664–1683. 10.1002/pmic.201000652 [DOI] [PubMed] [Google Scholar]
- Jahnová J., Luhová L., Petřivalský M. (2019). S-Nitrosoglutathione Reductase—The Master Regulator of Protein S-Nitrosation in Plant NO Signaling. Plants 8, 48. 10.3390/plants80200482019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeandroz S., Wipf D., Stuehr D. J., Lamattina L., Melkonian M., Tian Z., et al. (2016). Occurrence, structure, and evolution of nitric oxide synthase-like proteins in the plant kingdom. Sci. Signal. 9, re2. 10.1126/scisignal.aad4403 [DOI] [PubMed] [Google Scholar]
- Jia K.-P., Li C., Bouwmeester H. J., Al-Babili S. (2019). “Strigolactone Biosynthesis and Signal Transduction,” in Strigolactones - Biology and Applications. Eds. Koltai H., Prandi C. (Switzerland: Springer Nature; ), 1–45. [Google Scholar]
- Jiao C., Yang R., Wang P., Tian L., Gu Z. (2018). Mitogen-activated protein kinase mediates nitric oxide-induced isoflavone accumulation in soybean sprouts under UVB radiation. Can. J. Plant Sci. 98, 54–61. 10.1139/cjps-2016-0369 [DOI] [Google Scholar]
- Kapulnik Y., Delaux P. M., Resnick N., Mayzlish-Gati E., Wininger S., Bhattacharya C., et al. (2011). Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis . Planta 233, 209–216. 10.1007/s00425-010-1310-y [DOI] [PubMed] [Google Scholar]
- Kohlen W., Charnikhova T., Liu Q., Bours R., Domagalska M. A., Beguerie S., et al. (2011). Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis . Plant Physiol. 155, 974–987. 10.1104/pp.110.164640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbert Zs., Feigl G. (2017). “Cross-talk of Reactive Oxygen Species and Nitric Oxide in Various Processes of Plant Development,” in Reactive Oxygen Species in Plants: Boon Or Bane - Revisiting the Role of ROS. Eds. Singh V. P., Singh S., Tripathi D. K., Prasad S. M., Chauhan D. K. (Chichester, UK: John Wiley & Sons, Ltd; ), 261–289. [Google Scholar]
- Kolbert Zs., Pető A., Lehotai N., Feigl G., Ördög A., Erdei L. (2012). In vivo and in vitro studies on fluorophore-specificity. Acta Biol. Szeged. 56, 37–41. [Google Scholar]
- Kolbert Zs., Barroso J. B., Brouquisse R., Corpas F. J., Gupta K. J., Lindermayr C., et al. (2019. a). A forty-year journey: The generation and roles of NO in plants. Nitric. Oxide 93, 53–70. 10.1016/j.niox.2019.09.006 [DOI] [PubMed] [Google Scholar]
- Kolbert Zs., Molnár Á., Oláh D., Feigl G., Horváth E., Erdei L., et al. (2019. b). S-Nitrosothiol Signaling Is involved in Regulating Hydrogen Peroxide Metabolism of Zinc-Stressed Arabidopsis. Plant Cell Physiol. 60, 2449–2463. 10.1093/pcp/pcz138 [DOI] [PubMed] [Google Scholar]
- Kolbert Zs. (2019). Strigolactone-nitric oxide interplay in plants: The story has just begun. Physiol. Plant 165, 487–497. 10.1111/ppl.12712 [DOI] [PubMed] [Google Scholar]
- Kwon E., Feechan A., Yun B.-W., Hwang B.-H., Pallas J. A., Kang J.-G., et al. (2012). AtGSNOR1 function is required for multiple developmental programs in Arabidopsis . Planta 236, 887–900. 10.1007/s00425-012-1697-8 [DOI] [PubMed] [Google Scholar]
- Lanteri M. L., Laxalt A. M., Lamattina L. (2008). Nitric oxide triggers phosphatidic acid accumulation via phospholipase d during auxin-induced adventitious root formation in cucumber. Plant Physiol. 147, 188–198. 10.1104/pp.107.111815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee U., Wie C., Fernandez B. O., Feelisch M., Vierling E. (2008). Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis . Plant Cell 20, 786–802. 10.1105/tpc.107.052647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Chen L., Li Y., Yao R., Wang F., Yang M., et al. (2016). Effect of GR24 stereoisomers on plant development in Arabidopsis . Mol. Plant 9, 1432–1435. 10.1016/j.molp.2016.06.012 [DOI] [PubMed] [Google Scholar]
- Lindermayr C. (2018). Crosstalk between reactive oxygen species and nitric oxide in plants: Key role of S-nitrosoglutathione reductase. Free Rad. Biol. Med. 122, 110–115. 10.1016/j.freeradbiomed.2017.11.027 [DOI] [PubMed] [Google Scholar]
- Malamy J. E., Benfey P. N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana . Dev 124, 33–44. [DOI] [PubMed] [Google Scholar]
- Manoli A., Trevisan S., Voigt B., Yokawa K., Baluška F., Quaggiotti S. (2016). Nitric oxide-mediated maize root apex responses to nitrate are regulated by auxin and strigolactones. Front. Plant Sci. 6, 1269. 10.3389/fpls.2015.01269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzec M., Melzer M. (2018). Regulation of root development and architecture by strigolactones under optimal and nutrient deficiency conditions. Int. J. Mol. Sci. 19, 1887. 10.3390/ijms19071887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzec M. (2016). Perception and signaling of strigolactones. Front. Plant Sci. 7, 1260. 10.3389/fpls.2016.01260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira M. M., Adel E.-S., Stasolla C. (2015). Ethylene is integrated into the nitric oxide regulation of Arabidopsis somatic embryogenesis. J. Genet. Eng. Biotechnol. 13, 7–17. 10.1016/j.jgeb.2015.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori N., Nomura T., Akiyama K. (2020). Identification of two oxygenase genes involved in the respective biosynthetic pathways of canonical and non-canonical strigolactones in Lotus japonicus . Planta 251, 40. 10.1007/s00425-019-03332-x [DOI] [PubMed] [Google Scholar]
- Mostofa M. G., Weiqiang Li W., Nguyen K. H., Fujita M., Phan Tran L.-S. (2018). Strigolactones in plant adaptation to abiotic stresses: An emerging avenue of plant research. Plant Cell Environ. 41, 2227–2243. 10.1111/pce.13364 [DOI] [PubMed] [Google Scholar]
- Nelson D. C., Scaffidi A., Dun E. A., Waters M. T., Flematti G. R., Dixon K. W., et al. (2011). F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana . Proc. Natl. Acad. Sci. 108, 8897–8902. 10.1073/pnas.1100987108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat G. C., Lanteri M. L., Lombardo M. C., Lamattina L. (2004). Nitric oxide mediates the indole acetic acid induction activation of a mitogen-activated protein kinase cascade involved in adventitious root development. Plant Physiol. 135, 279–286. 10.1104/pp.103.038554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A., Sharma M., Pandey G. K. (2016). Emerging Roles of Strigolactones in Plant Responses to Stress and Development. Front. Plant Sci. 7, 434. 10.3389/fpls.2016.00434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papdi C., Ábrahám E., Joseph M. P., Popescu C., Koncz C., Szabados L. (2008). Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol. 147, 528–542. 10.1104/pp.108.116897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Ruiz M., Zuccarelli R., Palma J. M., Corpas F. J., Freschi L. (2019). “Biotechnological Application of Nitric Oxide and Hydrogen Peroxide in Plants,” in Nitric Oxide and Hydrogen Peroxide Signaling in Higher Plants. Eds. Gupta D., Palma J., Corpas F. (Cham: Springer; ), 245–270. [Google Scholar]
- Ruyter-Spira C., Kohlen W., Charnikhova T., van Zeijl A., van Bezouwen L., de Ruijter N., et al. (2011). Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol. 155, 721–734. 10.1104/pp.110.166645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A., Ueda M., Morikawa H. (2002). Arabidopsis glutathione-dependent formaldehyde dehydrogenase is an S-nitrosoglutathione reductase. FEBS Lett. 515, 20–24. 10.1016/S0014-5793(02)02414-6 [DOI] [PubMed] [Google Scholar]
- Santolini J., Andre F., Jeandroz S., Wendehenne D. (2017). Nitric oxide synthase in plants: Where do we stand? Nitric. Oxide 63, 30–38. 10.1016/j.niox.2016.09.005 [DOI] [PubMed] [Google Scholar]
- Scaffidi A., Waters M. T., Sun Y. K., Skelton B. W., Dixon K. W., Ghisalberti E. L., et al. (2014). Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis . Plant Physiol. 165, 1221–1232. 10.1104/pp.114.240036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto Y., Yasui R., Kameoka H., Tamiru M., Cao M., Terauchi R., et al. (2019). Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat. Commun. 10, 191. 10.1038/s41467-018-08124-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabek N., Ticchiarelli F., Mao H., Hinds T. R., Leyser O., Zheng N. (2018). Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling. Nature 563 (7733), 652–656. 10.1038/s41586-018-0743-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y.-F., Wang D.-L., Wang C., Hendrickson Culler A., Kreiser M. A., Suresh J., et al. (2015). Loss of GSNOR1 function leads to compromised auxin signaling and polar auxin transport. Mol. Plant 8, 1350–1365. 10.1016/j.molp.2015.04.008 [DOI] [PubMed] [Google Scholar]
- Soundappan I., Bennett T., Morffy N., Liang Y., Stanga J. P., Abbas A., et al. (2015). SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis . Plant Cell 27, 3143–3159. 10.1105/tpc.15.00562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P., van De Sande K., Leyser H. M. O. (2002). MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Dev 129, 1131–1141. [DOI] [PubMed] [Google Scholar]
- Sun H., Tao J., Liu S., Huang S., Chen S., Xie X., et al. (2014). Strigolactones are involved in phosphate- and nitrate-deficiency-induced root development and auxin transport in rice. J. Exp. Bot. 65, 6735–6746. 10.1093/jxb/eru029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Bi Y., Tao J., Huang S., Hou M., Xue X., et al. (2016). Strigolactones are required for nitric oxide to induce root elongation in response to nitrogen and phosphate deficiencies in rice. Plant Cell Environ 39, 1473–1484. [DOI] [PubMed] [Google Scholar]
- Takahashi I., Asami T. (2018). Target-based selectivity of strigolactone agonists and antagonists in plants and their potential use in agriculture. J. Exp. Bot. 69, 2241–2254. 10.1093/jxb/ery126 [DOI] [PubMed] [Google Scholar]
- Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., et al. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200. 10.1038/nature07272 [DOI] [PubMed] [Google Scholar]
- Villaécija-Aguilar J. A., Hamon-Josse M., Carbonnel S., Kretschmar A., Schmidt C., Dawid C., et al. (2019). SMAX1/SMXL2 regulate root and root hair development downstream of KAI2-mediated signalling in Arabidopsis. PloS Genet. 15, e1008327. 10.1371/journal.pgen.1008327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vurro M., Prandi C., Baroccio F. (2016). Strigolactones: how far is their commercial use for agricultural purposes? Pest Manage Sci. 72, 2026–2034. 10.1002/ps.4254 [DOI] [PubMed] [Google Scholar]
- Wakabayashi T., Hamana M., Mori A., Akiyama R., Ueno K., Osakabe K., et al. (2019). Direct conversion of carlactonoic acid to orobanchol by cytochrome P450 CYP722C in strigolactone biosynthesis. Sci. Adv. 5, eaax9067. 10.1126/sciadv.aax9067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C. H., Siu-Ting K., Taylor, O'Connell A. M. J., Bennett T. (2019). Strigolactone synthesis is ancestral in land plants, but canonical strigolactone signalling is a flowering plant innovation. BMC Biol. 17, 70. 10.1186/s12915-019-0689-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Ni J., Shah F., Liu W., Wang D., Yao Y., et al. (2019). Overexpression of the Stress-Inducible SsMAX2 Promotes Drought and Salt Resistance via the Regulation of Redox Homeostasis in Arabidopsis. Int. J. Mol. Sci. 20, 837. 10.3390/ijms20040837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M. T., Gutjahr C., Bennett T., Nelson D. C. (2017). Strigolactone signaling and evolution. Annu. Rev. Plant Biol. 68, 291–322. 10.1146/annurev-arplant-042916-040925 [DOI] [PubMed] [Google Scholar]
- Weisslocker-Schaetzel M., Andre F., Touazi N., Foresi N., Lembrouk M., Dorlet P., et al. (2017). The NOS-like protein from the microalgae Ostreococcus tauri is a genuine and ultrafast NO-producing enzyme. Plant Sci. 265, 100–111. 10.1016/j.plantsci.2017.09.019 [DOI] [PubMed] [Google Scholar]
- Yoneyama K., Akiyama K., Brewer P. B., Mori N., Kawano-Kawada M., Haruta S., et al. (2020). Hydroxyl carlactone derivatives are predominant strigolactones in Arabidopsis . Plant Direct 4, e00219. 10.1002/pld3.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. (2004). Real time and in vivo monitoring of nitric oxide by electrochemical sensors from dream to reality. Front. Biosci. 9, 3434–3446. 10.2741/1492 [DOI] [PubMed] [Google Scholar]
- Zwanenburg B., Blanco-Ania D. (2018). Strigolactones: new plant hormones in the spotlight. J. Exp. Bot. 69, 2205–2218. 10.1093/jxb/erx487 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.