Abstract
Osteocytes, the most abundant cells in bone, were once thought to be inactive but are now known to have multifunctional roles in bone, including in mechanotransduction, regulation of osteoblast and osteoclast function and phosphate homeostasis. Because osteocytes are embedded in a mineralized matrix and are challenging to study, there is a need for new tools and cell models to understand their biology. We have generated two clonal osteogenic cell lines, OmGFP66 and OmGFP10, by immortalization of primary bone cells from mice expressing a membrane-targeted GFP driven by the Dmp1-promoter. One of these clones, OmGFP66, has unique properties compared to previous osteogenic and osteocyte cell models and forms 3-dimensional mineralized bone-like structures, containing highly dendritic GFP-positive osteocytes, embedded in clearly defined lacunae. Confocal and electron microscopy showed that structurally and morphologically, these bone-like structures resemble bone in vivo, even mimicking the lacunocanalicular ultrastructure and 3D spacing of in vivo osteocytes. In osteogenic conditions, OmGFP66 cells express alkaline phosphatase, produce a mineralized type-I-collagen matrix and constitutively express the early osteocyte marker, E11/gp38. With differentiation they express osteocyte markers, Dmp1, Phex, Mepe, Fgf23 and the mature osteocyte marker, Sost. They also express RankL, Opg and Hif1α, and show expected osteocyte responses to PTH, including downregulation of Sost, Dmp1 and Opg and upregulation of RankL and E11/gp38. Live-cell imaging revealed the dynamic process by which OmGFP66 bone-like structures form, the motile properties of embedding osteocytes and the integration of osteocyte differentiation with mineralization. The OmGFP10 clone showed an osteocyte gene expression profile similar to OmGFP66, but formed less organized bone nodule-like mineral, similar to other osteogenic cell models. Not only do these cell lines provide useful new tools for mechanistic and dynamic studies of osteocyte differentiation, function and biomineralization, but OmGFP66 cells have the unique property of modeling osteocytes in their natural bone microenvironment.
Keywords: Osteocyte, Osteoblast, Cell line, Bone, Mineralization, FGF23
INTRODUCTION
Osteocytes are terminally differentiated osteoblasts embedded within mineralized matrix. During the transition from osteoblast to osteocyte there is a striking change in morphology from a polygonal to a highly dendritic cell, accompanied by cytoskeletal rearrangements and reduced cell volume [reviewed in(1–3)]. Osteocytes are embedded in lacunae within the mineralized bone matrix and interconnect with neighboring osteocytes, surface osteoblasts, bone marrow, and blood vessels through their dendritic processes, which extend through canaliculi in the bone matrix [reviewed in(1–3)]. The osteocyte lacunocanalicular system is intimately connected to the circulation, allowing the osteocytes and their dendrites to be bathed in canalicular fluid that carries oxygen and nutrients to the osteocytes, exchanges minerals, and carries away waste(4).
Osteocytes have been challenging to study, in part because primary osteocytes are difficult to isolate in sufficient quantities for detailed biochemical studies and because they are terminally differentiated, post-mitotic cells. They also have a tendency to de-differentiate in culture when isolated from their natural mineralized environment. In the past two decades, following the identification of osteocyte-specific marker genes and the development of genetic tools to manipulate gene expression in osteocytes(5–7) as well as mice with osteocyte-targeted fluorescent reporters(8–11), major advances in our understanding of these unique cells has been possible. Osteocytes were previously viewed as passive cells, but it is now clear that they are active multifunctional cells that play a key role in regulating bone mass by controlling both osteoblast and osteoclast activity [reviewed in(1–3)]. They also serve as mechanosensors to regulate skeletal adaptation to mechanical loading(12–14) and function as endocrine cells in the regulation of phosphate homeostasis(15–17). Mature osteocytes are major producers of sclerostin, encoded by the SOST gene, a natural inhibitor of bone formation through inhibition of Wnt/β-catenin signaling(18,19). Inhibition of sclerostin has become a promising therapeutic target for development of bone anabolic agents for treatment of osteoporosis(20–23). To further understand the biology of osteocytes and to facilitate in vitro biochemical and mechanistic studies of their function, immortalized cell lines have been invaluable. However, to date only a handful of cell lines have been developed as models of osteocytes or osteocyte differentiation. These include the MLO-Y4 mouse osteocyte-like cell line (24), HOB-01-C1 human pre-osteocyte cell line(25), and the IDG-SW3(26) and Ocy454(27) cell lines.
The HOB-01-C1 human preosteocyte-like cell line was generated by immortalization of primary cells from human cancellous bone chips with a temperature sensitive SV40 T-antigen(25). They were designated as pre-osteocytes based on morphology, low expression of alkaline phosphatase, robust expression of type I collagen, osteocalcin and CD44, ability to form mineralized nodules and PTH-responsiveness. MLO-Y4 cells have been one of the most widely used osteocyte-like cell lines. This cell line was immortalized from the long bones of transgenic mice expressing SV40 T-antigen driven by the osteocalcin promoter(24). These clonal cells have a dendritic appearance and, compared to osteoblasts, express low levels of alkaline phosphatase and type I collagen. They express high levels of the early osteocyte marker, E11/gp38 as well as osteocalcin, connexin 43, CD44 and osteopontin(24,28). They also express high levels of RANKL and support osteoclast formation(29). An advantage of MLO-Y4 cells is that they express the early osteocyte-like phenotype constitutively and do not have to be induced to differentiate in vitro. However, a limitation is that they express low levels of some osteocyte markers, such as DMP1, PHEX, and SOST(28), and do not mineralize. Therefore, they model an early osteocyte rather than a mature osteocyte in its natural mineralized lacunar environment.
Two other widely used osteocyte-like cell lines are the IDG-SW3(26) and Ocy454 cell lines(27). These were generated by a similar approach of crossing Dmp1-GFP transgenic mice, which express green fluorescent protein (GFP) driven by the Dmp1 promoter(8) with the “immortomouse”, carrying a temperature-sensitive SV40-T-antigen(30). Both cell lines have the attractive feature that they carry the Dmp1-GFP reporter, which indicates the preosteocyte/early osteocyte phenotype and that the temperature-sensitive SV40 T-antigen is not expressed at 37°C, eliminating effects of transformation on differentiation. Primary cells were prepared from the long bones of these mice and immortalized single cell clones were isolated that showed osteocyte-like characteristics. IDG-SW3 cells provide an excellent model of osteoblast to osteocyte transition. When cultured in osteogenic medium with ascorbic acid and β-glycerophosphate, these cells progress through a late osteoblast to an early osteocyte phenotype over 7-14 days, expressing the Dmp1-GFP reporter and marker genes, such as Dmp1, E11/gp38, Phex and Mepe. They then mineralize and further differentiate over 14-28 days to express Fgf23 and the late osteocyte marker, Sost. An advantage over MLO-Y4 cells is that they can progress to the late osteocyte phenotype and are therefore useful for studying the transition from osteoblast to mature osteocyte. They may also better model the mineralized environment of osteocytes in vivo. Limitations are that they have to be differentiated in long term culture to acquire the osteocyte-like phenotype. Ocy454 cells have the advantage that they express the late osteocyte marker, Sost without the need for culturing in osteogenic media and they display a mature osteocytic phenotype in a shorter timeframe. These cells have proved very useful in studies examining regulation of Sost expression(31). They also express Dmp1 after 10-14 days in culture(27). Limitations are that they appear to express Sost before Dmp1 and that they do not model the mineralized microenvironment of the osteocyte. Furthermore, in both IDG-SW3 and Ocy454 cell lines, not all cells within the culture fully differentiate.
Due to their many strengths outlined above, the currently available osteocyte-like cell lines have proved to be valuable models and have been used in numerous studies that have provided key insights into osteocyte function. However, until now, the field has lacked a cell line that models osteocytes embedded within highly structured mineralized lacunae which mimics the 3D organization and spacing of osteocytes and their lacunocanalicular system in vivo. Here, we report the development of two novel osteogenic cell lines by immortalization from primary calvarial cells isolated from a transgenic mouse expressing a membrane-targeted green fluorescent protein (AcGFP1-mem) driven by the Dmp1 promoter. This reporter allows visualization of the osteocyte cell membrane by GFP fluorescence. One of these clonal cell lines, OmGFP66, has unique properties compared to previously published cell lines. In long term culture, OmGFP66 cells form highly organized 3 dimensional bone-like structures containing highly dendritic GFP-positive osteocytes, embedded in clearly defined mineralized lacunae. Structurally and morphologically, these bone-like structures resemble in vivo bone, even mimicking the 3D lacunocanalicular organization. We describe the development and characterization of these cell lines as novel tools for studying osteocyte differentiation and function.
MATERIALS AND METHODS
Antibodies and Reagents
Unless stated otherwise, reagents and chemicals were purchased from Sigma Aldrich (St. Louis, MO), or ThermoFisher Scientific (Waltham, MA). Antibodies included goat polyclonals against E11/gp38 (a.k.a. podoplanin) and sclerostin from R&D systems (Minneapolis, MN) [cat#s AF3244 and AF1589]. A hamster monoclonal (8.1.1) against E11/gp38(32) was a gift from Dr. Andrew Farr, (University of Washington, Seattle). Mouse monoclonal against SV40-T antigen was from Santa Cruz Biotechnology Inc. (Santa Cruz, CA) [cat# sc-58665]. Mouse monoclonal against GFP was from Clontech/Takara Bio USA (Mountain View, CA) [JL-8, cat# 632381]. The LF67 rabbit antiserum against type I collagen [α1(I) chain] was provided by Dr. Larry Fisher (NIDCR, Bethesda, MD)(33). Detection antibodies for Western blotting included peroxidase-conjugated anti-mouse and anti-goat antibodies from ThermoFisher [cat#s 31430 and 31402] and a peroxidase conjugated anti-hamster from Jackson Immunoresearch (West Grove, PA) [cat# 107-035-142]. For Western blot detection of β-actin a peroxidase conjugated anti-β-actin antibody was used [Sigma-Aldrich, cat# A3854]. Secondary antibodies for immunofluorescence included Cy3-conjugated anti-rabbit, anti-mouse and anti-hamster antibodies [Jackson Immunoresearch, cat#s 711-165-152, 715-165-150, and 107-005-142]. Rat tail collagen was from Becton-Dickinson (Franklin Lakes, NJ).
Cell culture
Unless stated otherwise, tissue culture reagents were obtained from Gibco (Life Technologies Inc., Grand Island, NY), GE Healthcare Life Sciences (Marlborough, MA), or Mediatech Inc. (Herndon, VA). Heat inactivated fetal bovine serum (FBS) was from Hyclone Laboratories (GE Healthcare Life Sciences) or Atlanta Biologicals (Flowery Branch, GA). OmGFP66 or OmGFP10 cells were routinely maintained in growth medium consisting of α-Minimum Essential Medium (α-MEM) containing 10% FBS, 2mM L-glutamine (LG) and 100U/ml penicillin/streptomycin (P/S) in a humidified 5% CO2 incubator at 37°C. The generation of these clonal cell lines is described below. For experiments, the cells were plated onto rat tail collagen-coated culture plates or lab-tek chamber slides (Nunc, Rosklilde, Denmark) at 2 × 104 cells/cm2 growth area in growth medium. At confluence, the medium was changed to osteogenic medium consisting of α-MEM supplemented with 5% or 3% FBS (optimized, depending on FBS lot), 100U/mL P/S, 2mM LG, 5mM β-Glycerophosphate (βGP) and 50μg/mL ascorbic acid, to promote osteogenic differentiation and matrix mineralization. The cells were cultured for up to 28 days, with media refreshed every 3 days and 24 hrs prior to harvesting at each time point. In some experiments, the OmGFP66 cells were cultured on 24-well plates coated with a collagen-hydroxyapatite (HA) gel. This was prepared by gelling a rat tail collagen solution at 2mg/ml according to manufacturer’s instructions (200μl/well), with the modification that the solution also contained a 1/20th volume of a 20% slurry of hydroxyapatite nanopowder in sterile water (final HA concentration = 1%). The plates were then dried and stored at −20°C, then rehydrated with PBS for 2h before use.
Generation of Clonal Immortalized Cell Lines Expressing a Membrane Targeted GFP Variant in Osteocytes
The Dmp1-mGFP transgenic mouse has been described previously(10) and expresses a membrane-targeted GFP variant (AcGFP-mem, Clontech) in osteocytes driven by the Dmp1 promoter, hereafter referred to as Dmp1-mGFP. These mice were used in this study only as a tissue source for cell isolation. Euthanasia was performed under an approved IACUC protocol at the University of Missouri Kansas City (UMKC), and conformed to relevant federal guidelines. The UMKC animal facility is AAALAC approved and animal care and husbandry meets requirements in the Guide for the Care and use of Laboratory Animals (8th Ed.), National Research Council. Animals were group housed on a 12hr light/dark cycle with ad libitum food and water at 22°C constant temperature and 45-55% humidity.
Primary mouse calvarial cells were isolated from 7 day old Dmp1-mGFP transgenic mice by sequential trypsin/collagenase digestions using a modification of methods described previously for fetal rats(34,35). Briefly, the parietal and frontal bones were aseptically dissected and subjected to four 20 min digests in 0.2% collagenase/0.05% trypsin in α-MEM (no additives) on a shaker plate at 37°C. Digests 2-4 were kept as the osteoblast-enriched cell population and plated at 4x104 cells/cm2 in growth medium consisting of α-MEM supplemented with 10% FBS, 2mM LG, 100U/ml P/S and 30μg/ml gentamycin. The calvarial cells were immortalized by transfection with pSV3neo plasmid (American Type Culture Collection, Manassas, VA), which contains the SV40 T-antigen. G418 (neomycin)-resistant cells were cloned by limiting dilution and more than 115 single cell clones were initially screened for high expression of Dmp1-mGFP and ability to mineralize within a 2-3 wk period when cultured in osteogenic media. After two further rounds of screening, clones OmGFP10 and OmGFP66 were selected for further characterization based on high Dmp1-mGFP expression and ability to mineralize. Characterization of clones OmGFP10 and OmGFP66 is described below and followed similar approaches as described by Woo et al(26) for IDG-SW3 cells.
Fluorescence Imaging of mGFP Expression and Mineral Deposition
Cells were grown to confluence in collagen-coated 12 well plates and the media was changed to osteogenic media (day 0). To simultaneously monitor mineralization and mGFP expression, alizarin red (0.5μg/ml) was added to the medium 24h prior to harvest as a vital stain for calcium deposition. Mineralization and mGFP expression were evaluated at time points between day 3 and day 28 by fluorescence microscopy. The cultures were viewed and photographed on a Nikon TE2000E microscope under epifluorescence illumination with filter sets optimized for GFP or alizarin red and using a 4x 0.2 NA, a 10x 0.3 NA or 20x 0.45 NA objective. TIFF Images were acquired on a Photometrics Coolsnap HQ cooled CCD camera with 12-bit grey scale resolution interfaced with the “Metamorph” software (Molecular Devices, LLC, Sunnyvale, CA). Alternatively, the cultures were fixed in PBS + 4% paraformaldehyde and imaged on a Leica TCS Sp5 II scanning confocal microscope with a 20x 0.7 NA or 100x oil 1.44NA objective interfaced with LAS-AF software (version 3.2.0.9652, Leica Microsystems, Wetzlar, Germany). Image processing, such as contrast adjustments and red-green merging was done using Image J software (Rasband, W.S., ImageJ, NIH, Bethesda, Maryland, USA, http://imagej.nih.gov/ij/). For quantitation, fluorescence images were background subtracted and thresholded in Image J and the mineralized area or area of Dmp1-mGFP-positive cells was measured using the particle counting plugin. 3D volumetric rendering with shadowing was done using the 3D visualization module in the Leica LAS-AF software.
Alkaline phosphatase (ALP) staining and activity assay
For histochemical staining of alkaline phosphatase, cell layers were fixed for 10 min in neutral buffered formalin and washed twice in PBS. Alkaline phosphatase staining was performed using 5-bromo-4-chloro-3-indolyl phosphate nitro blue tetrazolium (BCIP-NBT) as a substrate (see supplementary methods for further details). For quantitation of alkaline phosphatase activity by colorimetric substrate assay, the cells were lysed in 0.05% Triton X-100. Alkaline phosphatase activity in cell lysates was determined by measuring conversion of ρ-nitrophenol phosphate to free ρ-nitrophenol as described previously(36,37). Protein content of the lysates was determined using the Bio-Rad Protein Assay kit according to manufacturer’s instructions (Bio Rad Laboratories, Hercules, CA). From linear regression analysis of protein and ρ-nitrophenol standards, the alkaline phosphatase specific activity was calculated as nM ρ-nitrophenol converted/μg protein/min.
Taqman real time qPCR
Total RNA was extracted using TRIzol reagent according to manufacturer’s instructions (Invitrogen/ThermoFisher, Waltham, MA). cDNA was synthesized from 2 μg of total RNA using a High Capacity cDNA Reverse Transcription Kit (#4368814, Applied Biosystems, Carlsbad, CA). Taqman primers/probe sets were designed to span exon-exon junctions (Themofisher/ Lifetechnologies) (see supplementary table 1 for details of the qPCR primers used). qPCR amplification was performed using a StepOnePlus real-time PCR system (Thermofisher/Applied Biosystems) and using TaqMan Gene Expression Master Mix kits according to manufacturers’ instructions (#4369016, Thermofisher/ Applied Biosystems). Each qPCR reaction contained cDNA transcribed from 50ng of total RNA as a template. Relative quantitation of target gene expression was calculated using the 2^-ΔΔCt method(38) with normalization to β-actin as a housekeeping gene. Using genomic DNA, a PCR-based method was also used to determine the gender of the cells(39) (see supplementary methods for details).
Western Blotting
Western blotting was performed as described previously(40,41) with modifications. Briefly, cell layers were rinsed twice in PBS and lysed in ice-cold RIPA buffer containing protease inhibitor cocktail (Sigma cat# P8340). 10 or 20μg total protein per sample was separated on 10% SDS-PAGE gels under reducing conditions and electroblotted onto PVDF membrane. Membranes were blocked in TBS + 5% BSA/1% milk at room temperature for 1 hr and incubated overnight at 4°C with primary antibodies followed by washing and incubation with peroxidase conjugated secondary antibodies. Immunoreactive bands were visualized using a SuperSignal West Dura or Femto Chemiluminescence kit (ThermoFisher) and imaged on a Fujifilm LAS 4000 gel documentation system with Multi-gauge software (Fujifilm, Tokyo, Japan). Blots were stripped in Restore stripping buffer (ThermoFisher) and re-probed with other antibodies of interest then lastly probed with HRP-anti-β-actin to confirm equal protein loading. Densitometry was performed using the Multi-gauge software.
Immunofluorescent Staining
For immunofluorescent staining, cells were grown in collagen-coated plastic 8-chamber lab-tek slides (Nunc/Thermofisher), and differentiated as described above. Immunostaining was performed as described previously(40,41). Briefly, the cells were fixed in PBS + 4% paraformaldehyde for 10 min at room temperature. Immunofluorescent staining was performed following decalcification in 10% EDTA solution, pH 7.4 for 48hr with or without permeabilization in 0.2% Triton X-100 in PBS for 10 min at room temperature, depending on the primary antibody. Background blocking was performed for 3 hr at room temperature in PBS + 1% donkey serum + 0.05% NaN3. Cells were incubated overnight at 4°C in primary antibodies diluted in blocking buffer. Non-immune serum or normal IgG of the same species and concentration as the primary antibody was used as a negative control. Cells were incubated with Cy3-conjugated secondary antibodies and stained slides were photographed on a Nikon TE2000E microscope with a 10x 0.45 NA or 20x 0.75 NA objective.
Histological Staining
For histology of mineralized OmGFP66 cells, the cells were washed twice in PBS, then individual bone-like structures were lifted from the culture well with fine forceps and transferred to a 1.5ml tube, then pelleted using a microfuge for 5 min at 3000 xg. The pelleted OmGFP66 samples or neonatal mouse calvaria for comparison were fixed in PBS/4% paraformaldehyde, dehydrated, embedded in methyl methacrylate and the undecalcified samples were sectioned using a standard protocol as previously described for mouse femur(42). The sections were deplasticized and rehydrated then stained using von Kossa tetrachrome staining using standard histological procedures.
Scanning and Transmission Electron Microscopy
Cells were fixed in 2.5% glutaraldehyde in 0.1M sodium cacodylate buffer for 1.5 hr. For scanning electron microscopy (SEM), the specimens were dehydrated in graded ethanol solutions with final treatment with HMDS (hexamethyldisilazane). Specimens were coated with gold-palladium and observed on a field-emission SEM XL30 (FEI, Hillsboro, OR). Secondary and backscattered electron images were obtained at accelerating voltages of 5kV and 30kV, respectively. For transmission electron microscopy (TEM) the fixed specimens were washed in sodium cacodylate buffer, post fixed with 1% osmium tetroxide for 1.5 hr and dehydrated in graded ethanol solutions. The specimens were infiltrated with epoxy resin (Embed-812, Electron Microscopy Sciences, Hatfield, PA) and incubated in an oven at 60°C for 48 hrs. Ultrathin sections were cut on an EM UC7 ultramicrotome (Leica Microsystems), stained with uranyl acetate and lead citrate and observed on a CM12 electron microscope (FEI, Hillsboro, OR) at 80 kV accelerating voltage.
Live Cell Imaging
For live cell imaging, cells were plated onto collagen-coated ibi-Treat chamber slides (Ibidi USA, Inc., Fitchburg, WI) and differentiated as described above. On the day of imaging, the media was refreshed. Live cell imaging was performed on an automated Nikon TE 2000E widefield epifluorescence microscope with precision motorized x, y, and z stage using a 20x 0.75 NA objective with filter sets optimized for each fluorophore. The “Metamorph” software (Molecular Devices, LLC, Sunnyvale, CA) controlled the microscope and multidimensional imaging parameters. Temperature was held constant at 37°C using a microscope incubator cabinet (“The Box” with “The Cube”, Life Imaging Systems, Reinach, Switzerland) and a humidified 5% CO2 atmosphere was maintained using a gas mixer (“The Brick”, Life Imaging Systems) in conjunction with a stage top chamber. Images were acquired for each time point under epifluorescent illumination using a Photometrics Coolsnap HQ cooled CCD camera with 12-bit grey scale resolution. Fields of 450 x 335μm were imaged at a spatial resolution of 696 x 520 pixels (with 2x2 binning) every 20 min for up to 5 days from 5 optical planes, with media refreshed every 48hr. Image stacks were processed in Metamorph using the “best focus” algorithm or by manually selecting the best focus plane and then exported as image stack (.stk) files. Alternatively, live imaging was performed on the Leica TCS Sp5 II confocal microscope with a 20x 0.7 NA objective interfaced with LAS-AF software (Leica Microsystems). Temperature was held constant at 37°C using a microscope incubator cabinet (“The Box” with “The Cube”, Life Imaging Systems) and a humidified 5% CO2 atmosphere was maintained using a Red-y-gas flow controller (Vogtlin, Aesch, Switzerland) in conjunction with a stage top chamber (PeCon GmbH, Erpach, Germany). Fields were imaged at a spatial resolution of 1024 x 1024 pixels every 30 min for up to 48 hr from 5 optical planes.
Post acquisition image processing including contrast adjustments, color merging, conversion to 8-bit or RGB stacks, stack processing and compilation into movies was done using ImageJ software. Image stacks were registered using the “stackreg” plugin in ImageJ(43) to align the time series stacks and correct for rigid body motion. Aligned image stacks were assembled into movies using ImageJ.
Separation of Osteocyte-Enriched Bone-Like Structures from the Monolayer Culture
In long term culture, OmGFP66 cells form bone-like structures containing GFP-positive osteocytes (see results section). To determine if osteocyte markers were enriched in these bone-like structures compared to the surrounding monolayer cells, they were separated by a physical method that involved gently peeling the monolayer (which acted like a membrane) away, leaving the bone structures adhered to the culture plate. Alternatively, a digestion with 0.2% collagenase/0.05% trypsin in α-MEM was performed for 20 min, resulting in preferential removal of the monolayer cells, leaving the bone structures adhered to the plate. Western blotting and qPCR were performed on the separated cell populations.
Parathyroid hormone (PTH) treatment
At day 14, 21, and 28, OmGFP66 cells were treated with 50 or 100nM PTH, bovine fragment 1-34 (Sigma-Aldrich, cat# P3671) or vehicle for 24 hr. Total RNA was extracted and gene expression was quantified by Taqman qPCR as described above.
Data analysis
Data are expressed as the mean ± SEM. For comparisons between two experimental groups, Student’s t-test was used and for comparisons between multiple groups, one way analysis of variance (ANOVA) was used followed by Tukey’s post hoc test. Data were considered statistically significant with a p value ≤0.05.
RESULTS
OmGFP66 Cells Form in vitro Bone-like Structures That Resemble in vivo Bone
Screening of more than 115 G418-resistant immortalized calvarial single cell clones from Dmp1-mGFP mice resulted in selection of two clones, OmGFP66 and OmGFP10, for further study. The OmGFP66 (Fig. 1) and OmGFP10 clones (supplementary Fig. S1) strongly expressed the Dmp1-mGFP reporter, which reflects expression of Dmp1, a preosteocyte/early osteocyte marker. These clones produced a mineralized extracellular matrix when grown in osteogenic media. Timecourse studies in OmGFP66 cells showed that by day 7 in osteogenic media there were a few small foci of Dmp1-mGFP-positive cells starting to form, coinciding with areas of alizarin red staining, an indicator of mineralization (Fig 1A). Both Dmp1-mGFP expression and mineral deposition increased from day 7 through 28 (Fig. 1A). Merged images show that the Dmp1-mGFP-positive cells were exclusively located in the mineralized bone-like structures. These OmGFP66 mineralized structures had a trabecular-like appearance, as shown by the lower magnification view in Fig 1B, with about 25% of the culture area converting to bone-like structures. Quantitation showed that mineral deposition started between day 7 and 14 and increased up to day 28 (Fig 1 C). The area of Dmp1-mGFP-positive cells increased with a similar timecourse (Fig. 1D), suggesting increased numbers of osteocytes with time in culture. The OmGFP10 clone formed larger foci of Dmp1-mGFP-positive cells that coincided with alizarin red staining (supplementary Fig. S1A, B) and again Dmp1-mGFP expression and mineral deposition increased from day 7 through 28. Similar to OmGFP66, quantitation showed increased mineralization over 28 days in OmGFP10 cells (supplementary Fig S2B).
Figure 1: OmGFP66 cells form mineralized bone-like trabeculae over 7-28 days in culture.
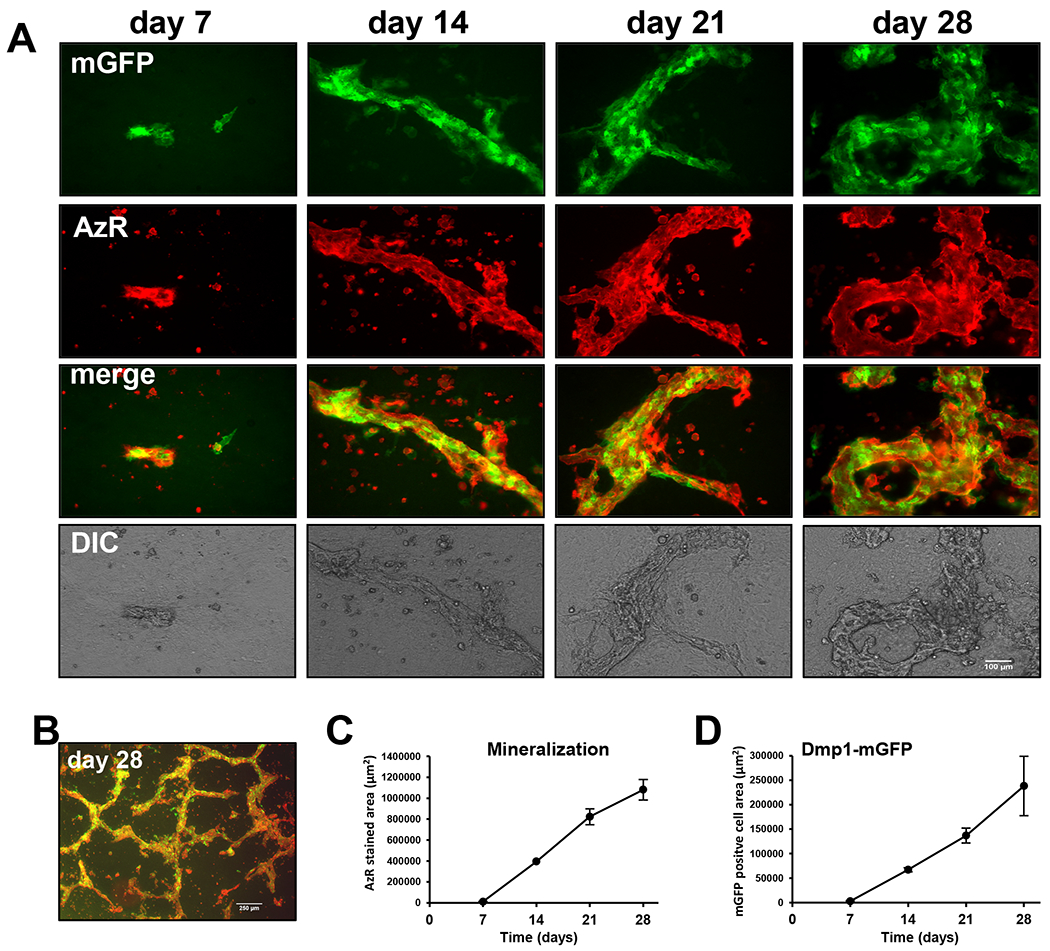
A) Time course in OmGFP66 cells showing expression of the Dmp1-mGFP reporter (green) and alizarin red fluorescence (red) to monitor calcium deposition. Note the initial formation of small bone-like structures containing mineral and Dmp1-mGFP-positive cells by day 7. These bone-like structures increase in number and size over 28 days, bar = 100μm. B) Lower power view showing the trabecular-like nature of the bone mineral, bar = 250μm. C, D) Quantitation of alizarin red stained area (C) and Dmp1-mGFP positive cell area (D) over 28 days (data are mean ± SEM, n= 4).
A striking and unique feature of the OmGFP66 clone was that it formed mineralized structures that closely mimicked in vivo bone, with a highly organized bone-like appearance and clearly defined lacunae (Fig. 2A). These bone-like structures are clearly distinct from the mineral deposited by many other osteogenic cell cultures, including the OmGFP10 clone, the previously published IDG-SW3 osteogenic cell line(26) and primary calvarial osteoblasts, all of which form more disorganized mineralized “nodules” that are much less structured and do not have clearly defined lacunae (see supplementary Fig S3 for comparison). Higher resolution confocal images of mGFP and alizarin red staining confirmed the organized structure of the OmGFP66 mineralized structures (Fig. 2B). The first sign that these bone-like structures were about to form was the appearance of cell condensations that formed 3-dimensional ridge-like structures, followed by induction of Dmp1-mGFP and mineral deposition, but at this stage the cells did not yet exhibit the highly dendritic morphology of osteocytes (not shown). By day 28, a high degree of organization was seen in which the bone-like structures contained distinct osteocyte lacunae occupied by mGFP-positive cells (Fig 2B and higher magnification images in Fig 2B1 and B2). The Dmp1-mGFP-positive cells were exclusively localized in the bone structures and their spacing, together with their highly dendritic morphology and interconnected dendrite networks resembled embedded osteocyte networks in calvarial bone in vivo (compare Fig 2B1 with Fig. 2C). The OmGFP10 clone differentiated to form Dmp1-mGFP-positive cells that had a stellate morphology but did not show the highly dendritic osteocyte-like morphology seen with OmGFP66 cells (supplementary Fig. S1B).
Figure 2: OmGFP66 cells form highly organized bone-like structures with a lacunar and osteocyte morphology resembling in vivo bone.
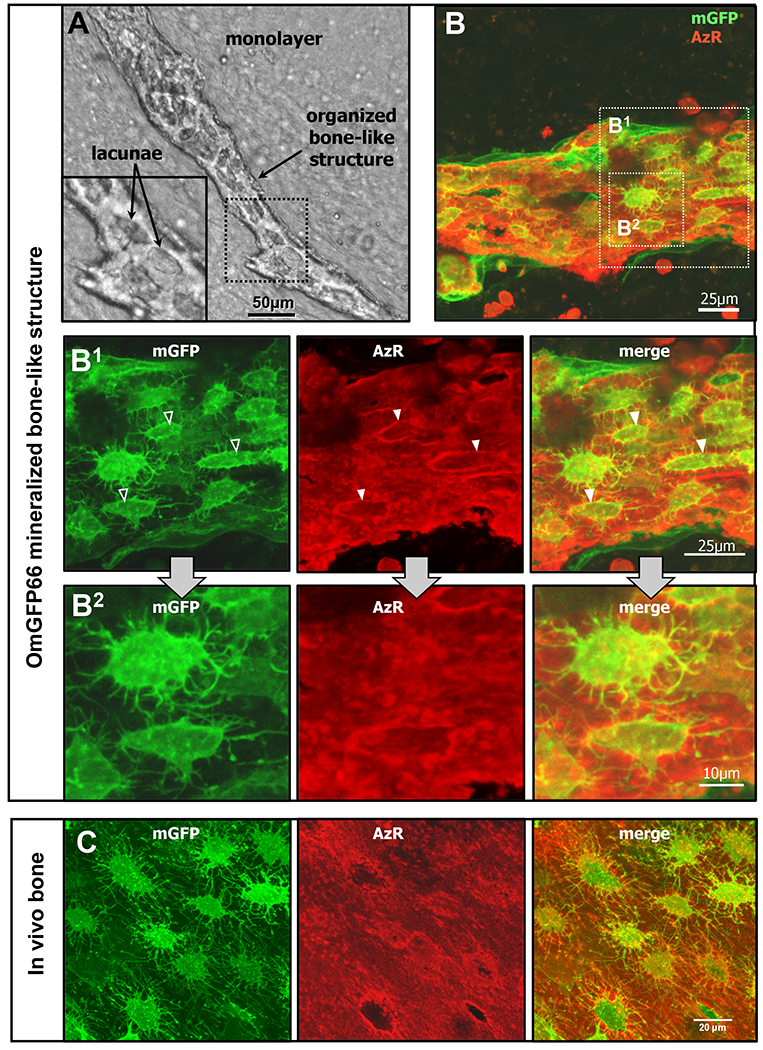
A) Brightfield image and enlarged inset showing an OmGFP66 bone-like structure. Note the clearly defined mineralized lacunae. B) Maximal Z-projected confocal image of a mineralized bone-like structure in a day 28 OmGFP66 culture. Dmp1-mGFP is shown in green and mineral is shown by alizarin red fluorescence (red). B1 and B2 show enlarged images from B depicting the highly dendritic osteocyte-like networks and show that the osteocytes are embedded within mineralized lacunae (arrowheads). C) Maximal Z-projected confocal images from a 7 day old Dmp1-mGFP transgenic mouse calvarium for comparison to the OmGFP66 mineralized structure in B1. Note the similarities in morphology and spacing of the Dmp1-mGFP-positive osteocytes and similar appearance of the mineralized lacunae by alizarin red fluorescence. (Bars: A = 50μm; B and B1 = 25μm; B2 = 10μm; C = 20μm).
OmGFP66 and OmGFP10 Cells Are Models of Osteocyte Differentiation
Further characterization of OmGFP66 and OmGFP10 revealed that both cell lines showed positive immunostaining for SV40 T-antigen, confirming their immortalized properties (Fig 3A and supplementary Fig. S2A). They also expressed alkaline phosphatase, consistent with their ability to mineralize. In OmGFP66 cells, alkaline phosphatase expression increased up to day 14-21 and then plateaued (Fig. 3B) and in OmGFP10 cells alkaline phosphatase was expressed constitutively over the 28 day timecourse (supplemental Fig. S2C).
Figure 3: OmGFP66 cells represent a model of osteocyte differentiation.
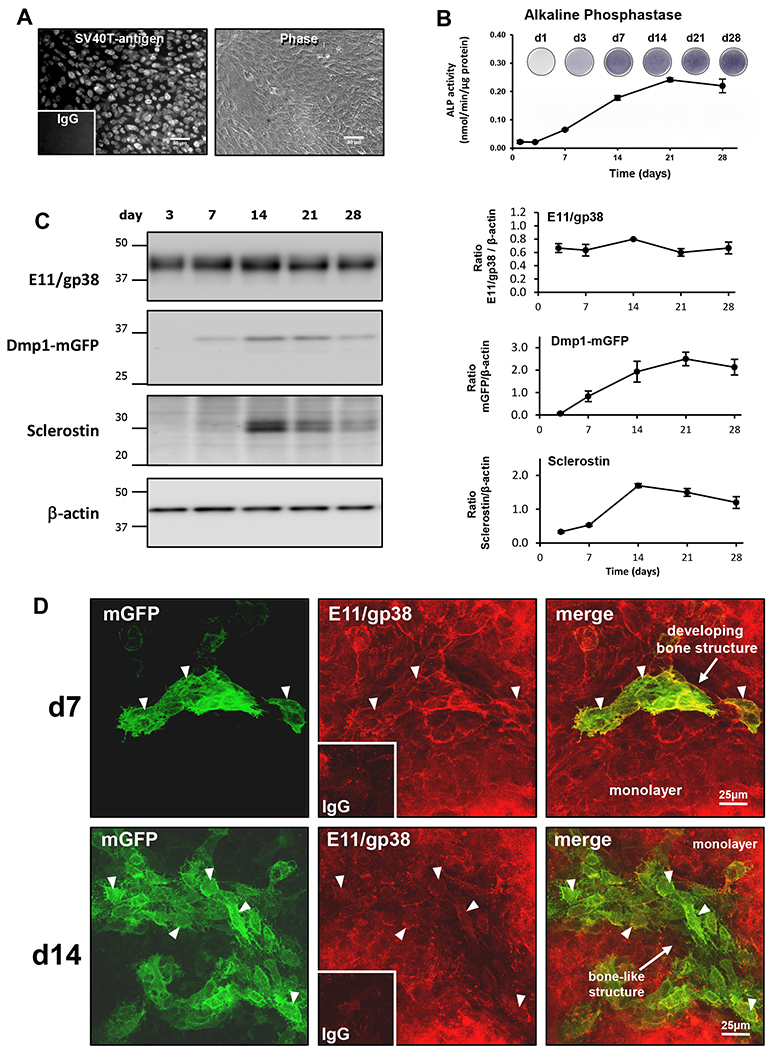
A) Immunostaining for SV40T-antigen in OmGFP66 cells alongside a phase contrast image of the cells. Inset shows the control stained with non-immune IgG, bar = 50μm. B) Timecourse of alkaline phosphatase staining and activity in OmGFP66 cells (data are mean ± SEM, n=3). C) Western blotting on OmGFP66 whole cell lysates showing the timecourse of expression of E11/gp38, Dmp1-mGFP and sclerostin, with β-actin as a control for equal loading of protein. Densitometric quantitation of the Western blots is shown at right (data are mean ± SEM, n=3). D) Maximal Z-projected confocal images of OmGFP66 cells at day 7 and 14 with immunostaining for E11/gp38. Insets show controls stained with non-immune IgG. Arrowheads indicate cells that are positive for both E11/gp38 (red) and mGFP (green). Bar = 25μm.
In order to use the OmGFP66 and OmGFP10 cell lines as new tools for studying osteocyte differentiation and function, it is important to confirm that they express established osteocyte markers. Western blotting on whole cell lysates revealed that OmGFP66 cells constitutively expressed the preosteocyte/osteocyte marker, E11/gp38, throughout the 28 day timecourse (Fig. 3C). Dmp1-mGFP, which reflects expression of the early osteocyte marker Dmp1, was first detectable at day 7 and increased up to day 21. Protein expression of the late osteocyte marker, sclerostin was first detected at day 14 and continued until day 28, with maximal expression between days 14-21. Similar to OmGFP66, the OmGFP10 clone showed constitutive E11/gp38 expression, with Dmp1-mGFP coming on at day 14 and sclerostin detected at day 21 with maximal expression at day 28 (supplemental Fig. S2D).
Immunostaining in OmGFP66 cells showed that at day 7, when the Dmp1-mGFP reporter was just starting to turn on in condensing ridges of cells prior to the formation of mineralized bone-like structures, these cells also stained positively for E11/gp38 (Fig. 3D, d7, arrowheads), but the E11/gp38 was also ubiquitously expressed in the surrounding monolayer cells. Note also that at this stage the mGFP-positive cells are not spaced apart and do not have an osteocytic morphology. By day 14 the Dmp1-mGFP-positive cells had become more highly organized, with a spacing similar to in vivo osteocytes and an extensive dendrite network connecting the cells. Many Dmp1-mGFP-positive osteocyte-like cells remained positive for E11/gp38 (Fig. 3D, d14, arrowheads), but the E11 expression in these osteocytes was declining, consistent with differentiation to a mature osteocyte phenotype. The monolayer cells continued to strongly express E11/gp38. By immunostaining, sclerostin-positive cells were detectable in OmGFP66 bone-like structures by day 14 (supplementary Fig. S4A), consistent with sclerostin detection by Western blotting by day 14 and also with differentiation to a mature osteocyte phenotype.
qPCR analysis in OmGFP66 cells confirmed expression of multiple osteocyte marker genes (Fig. 4A). E11/gp38, Dmp1, Phex and Sost were expressed robustly (Ct values for maximal expression <28), Fgf23 was expressed moderately (Ct value for maximal expression 28.9) and Mepe was weakly expressed (Ct value for maximal expression 30.7). Similar to results from Western blotting and immunostaining, E11/gp38 mRNA was constitutively expressed over time, with only a modest increase. The other osteocyte markers showed increased expression over time in culture, with Sost showing the highest fold induction (~5,000 fold) and Dmp1, Phex and Fgf23 showing fold increases of greater than 100 compared to day 1. OmGFP66 cells also expressed other genes known to be expressed by osteocytes, including RankL and Opg, whose expression increased with time in culture (Fig. 4B). They also expressed Cx43, Hif1α and Col1a1 constitutively throughout the timecourse at robust levels (Fig. 4B). Immunostaining for type I collagen confirmed an extensive network of collagen fibers throughout the extracellular matrix of both the OmGFP66 monolayer and bone-like structures (supplementary Fig. S4B). qPCR analysis of the OmGFP10 clone showed similar expression profiles for osteocyte marker genes (supplementary Fig. S5), again suggesting differentiation from an early to mature osteocyte phenotype.
Figure 4: Timecourse of expression of osteocyte markers by OmGFP66 Cells.
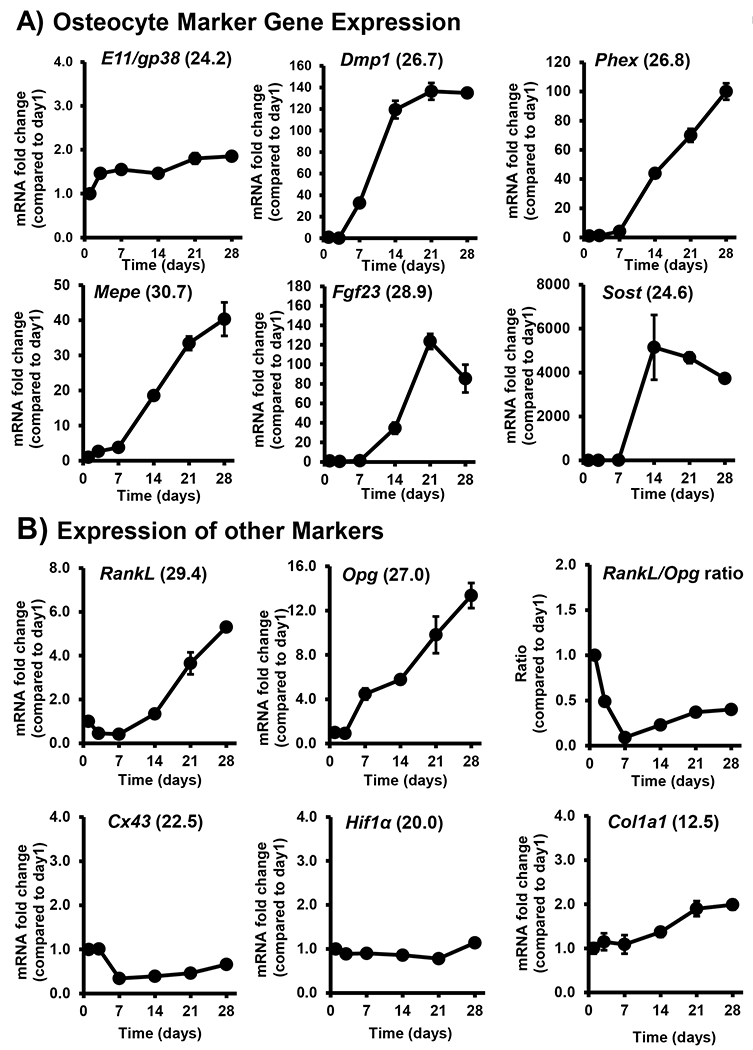
qPCR analysis of the timecourse of expression of A) osteocyte marker genes, E11/gp38, Dmp1, Phex, Mepe, Fgf23 and Sost by OmGFP66 cells and B) other markers, RankL, Opg, Cx43, Hif1α and Col1a1. Data were normalized to β-actin as a housekeeping control and are presented as the fold change compared to day 1 (mean ± SEM, n=3). Numbers in parentheses are the Ct values for the highest level of expression of each gene.
Since expression of the osteocyte phenotype in OmGFP66 cells was closely integrated with mineralization, we also examined if culturing the cells on a collagen/mineral substrate promoted osteocyte differentiation. OmGFP66 cells were cultured in osteogenic media on a collagen-HA gel. By 3 days of culture, Dmp1-mGFP expression was induced (not shown), with an even stronger induction by 5 days (supplementary Fig. S6). In contrast, control cultures did not express Dmp1-mGFP at these early timepoints, suggesting that collagen-HA can induce differentiation towards an osteocyte-like phenotype. Using genomic DNA, a PCR-based method(39) was used to determine the gender of the OmGFP66 cell line, which indicated that OmGFP66 cells are female (data not shown).
The OmGFP66 cell line has a stable and reproducible phenotype and has been successfully differentiated by six different individuals in our laboratory. We have found it to be stable up to at least passage 20. The cell line was also sent to an independent laboratory at Indiana University, which reproduced the differentiation characteristics, cellular morphology and in vitro bone formation results (see supplementary figure S7). The OmGFP66 cells have also been subcloned. As is typical for subcloning, there was variation among single cell subclones, but many subclones retained the properties of the parental clone (data not shown). The OmGFP10 clone was less extensively characterized than OmGFP66 but has been successfully differentiated by three different individuals in our laboratory out to at least passage 7.
Electron Microscopy and Confocal Microscopy Reveal the 3-Dimensional Nature of OmGFP66 Bone-like Structures and Show That They Mimic in vivo Bone Ultrastructure
Histological analysis of OmGFP66 bone-like structures by von Kossa tetrachrome staining revealed osteocyte-like cells embedded in a mineralized matrix (Fig. 5A). This histological appearance is very similar to bone in vivo (Fig. 5I), in which embedded osteocytes can be seen, surrounded by mineralized bone matrix in a 7 day old neonatal mouse calvarium. The thickness of the OmGFP66 bone structures was ~30μm, also similar to the thickness of a neonatal mouse calvarium. Examination of the OmGFP66 bone ultrastructure by transmission electron microscopy (TEM) revealed a highly organized mineralized bone matrix (Fig. 5B, D), with the mineral clearly deposited on collagen fibrils that had a characteristic cross-striated banding pattern (Fig. 5C, inset). Figure 5B shows an osteocyte that appears to be embedding into the mineralized matrix. Strikingly, within the matrix of the OmGFP66 bone-like structures, canaliculi could even be seen, with osteocyte dendritic processes located within them that appeared very similar to osteocyte canaliculi in bone in vivo (Fig. 5D, arrowheads). The TEM ultrastructural appearance of the OmGFP66 bone matrix closely resembled the ultrastructure of mineralized bone in vivo (compare Fig. 5B with 5J; compare Fig. 5C with 5K and compare Fig. 5D with 5L). Scanning electron microscopy (SEM) using secondary electrons revealed the 3-dimensionality of the ridge-like bone structures, which were clearly raised from the cell monolayer (Fig. 5E, F). SEM imaging using backscattered electrons confirmed the mineralized nature of the bone-like structures (Fig. 5G) and showed the presence of lacunae within the mineralized matrix (Fig. 5G inset, arrowheads). An extensive network of collagen fibrils was seen in areas between cells (Fig. 5H). The 3-dimensional nature of the OmGFP66 bone-like structures is further emphasized by 3D confocal imaging (Fig. 6). Fig 6A shows the front view of a 3D render of Dmp1-mGFP positive osteocytes within an OmGFP66 bone-like structure, together with a pseudocolored image indicating the depth profile of the different osteocytes and a side view showing multiple layers of osteocytes. Fig 6B shows front and back angled 3D rendered views of the same field showing Dmp1-mGFP positive cells embedded in mineralized matrix, as indicated by alizarin red staining, and showing several osteocytes at different levels within the mineralized matrix.
Figure 5: Three dimensional nature of OmGFP66 bone-like structures and ultrastructural similarity to bone in vivo.
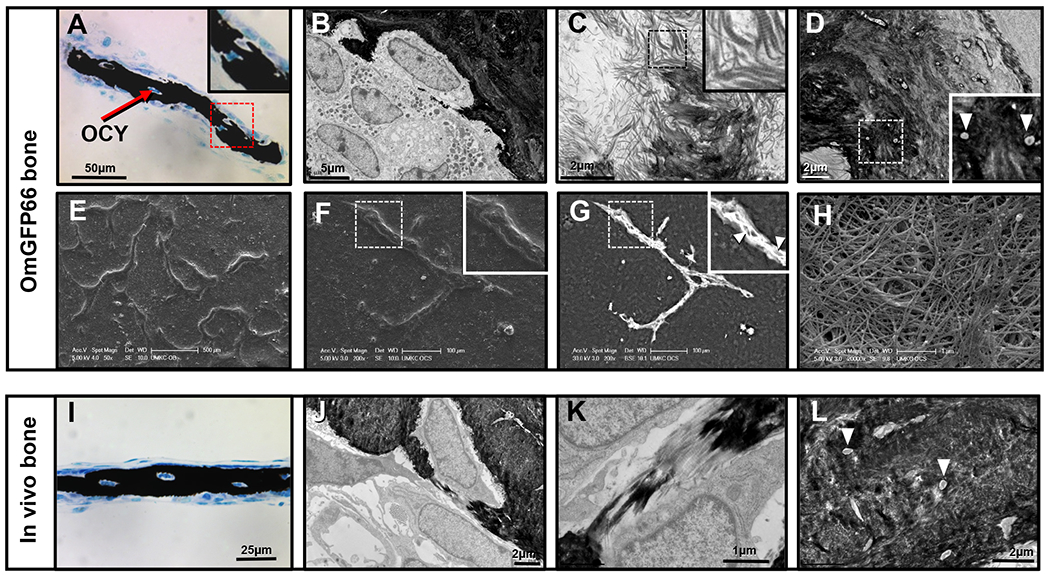
A) Von Kossa tetrachrome staining of an OmGFP66 mineralized bone structure in cross section. Note the mineralized bone (black) and embedded osteocytes. B-D) TEM micrographs of OmGFP66 cell cultures: B) shows an osteocyte embedding into the mineralized bone matrix; C) shows mineral deposition on collagen fibrils; D) shows canaliculi containing osteocyte dendritic processes within the mineralized bone matrix (inset, arrowheads). E–H) SEM micrographs of OmGFP66 cells: E) and F) are secondary electron images depicting the three-dimensional nature of the OmGFP66 bone-like structures; G) is a backscatter electron image showing the mineral within the bone matrix; H) shows abundant collagen fibrils present between cells. I-L) show images of bone in vivo to illustrate the many similarities with OmGFP66 cells. I) Von Kossa Tetrachrome staining in a 7 day mouse calvarium [compare with A]. J-L) are TEM micrographs of a 7 day old mouse calvarium: J) shows an osteocyte embedding [compare with B]; K) shows mineral deposition on collagen fibrils [compare with C]; L) shows osteocyte canaliculi containing dendritic processes (arrowheads) [compare with D]. Bars: A) = 50μm, B) = 5μm, C), D), J), L) = 2μm, E) = 500μm, F), G) = 100μm, H), K) = 1μm, I) = 25μm,
Figure 6: Confocal microscopy confirms the three dimensional nature of OmGFP66 bone-like structures and shows layers of interconnected osteocytes.
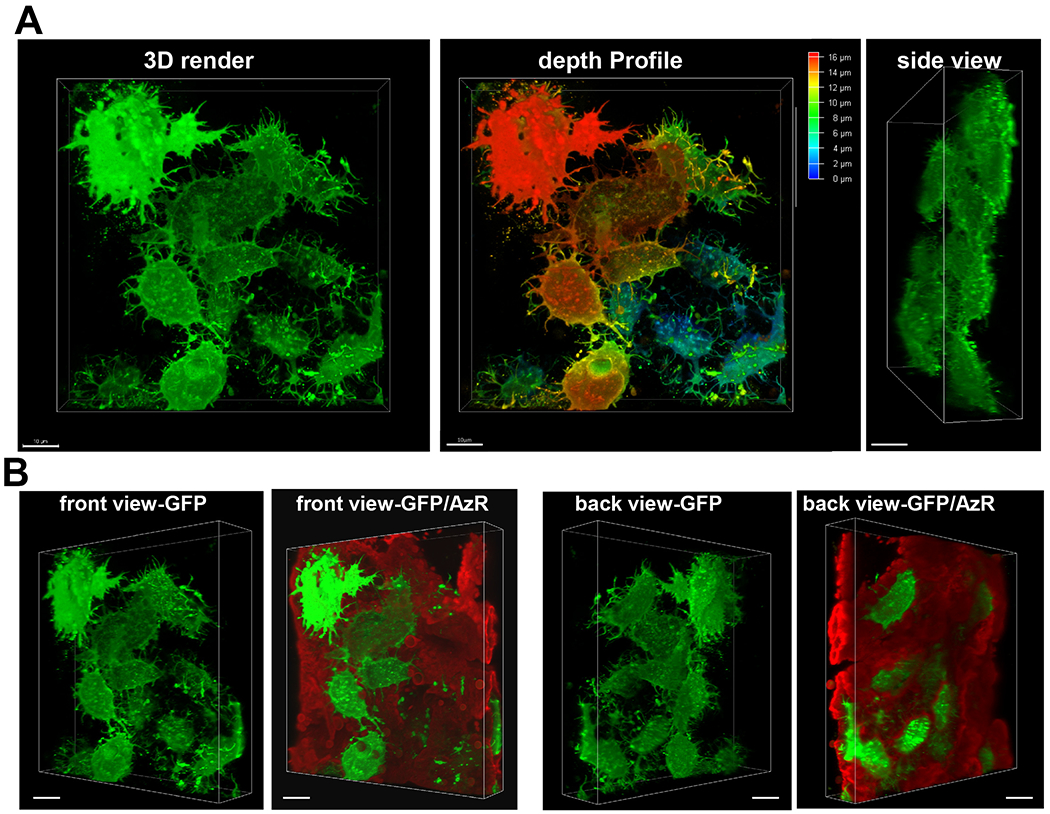
A) Front view of a 3D render of Dmp1-mGFP positive osteocytes within an OmGFP66 mineralized bone-like structure, together with a pseudocolored image indicating the depth profile of the different osteocytes and a side view showing multiple layers of osteocytes. B) Front and back angled 3D rendered views of the same field showing Dmp1-mGFP positive cells (green) embedded in mineralized matrix (red), as indicated by alizarin red staining, and showing several osteocytes at different levels within the mineralized matrix. (132 planes with 0.126 μm Z plane separation, 100x oil/1.44 NA objective, 1.7x digital zoom). Bar = 10μm.
Enrichment for Expression of Osteocyte Markers in OmGFP66 Bone-Like Structures Compared to the Cell Monolayer
Since about 25% of the OmGFP66 cell layer appears to convert to mineralized bone-like structures (Fig 1B), we next set out to determine whether the increase in expression of osteocyte markers was localized within the osteocyte-enriched bone structures or whether expression was upregulated in the entire cell population. The OmGFP66 bone-like structures and cell monolayer were physically separated (see methods for details), leaving the bone adhered to the culture plate (Fig 7A). Western blotting showed strong enrichment for expression of Dmp1-mGFP and sclerostin in the bone derived fraction compared to the cell monolayer and also compared to the whole cell layer (including both bone-like structures and monolayer cells) (Fig. 7B). Consistent with immunostaining results from Fig. 3D, the E11/gp38 protein was expressed equally in all three cell populations (Fig. 7B). qPCR analysis showed a similar enrichment in mRNA expression for osteocyte markers Dmp1, Phex, Fgf23 and Sost in the bone-derived fraction compared to the monolayer and whole cell layer, with Fgf23 showing the highest (~20-fold) enrichment (Fig. 7C). Consistent with Western blot and immunostaining data, the early osteocyte marker, E11/gp38 was expressed equally in all three cell populations. RankL and Opg were also expressed at higher levels in the osteocyte enriched bone-like structures compared to the monolayer. Identical results were obtained using differential digestion with 0.2% collagenase/0.05% trypsin to separate the bone structures from the monolayer cells, with the exception that E11/gp38 protein was not detectable by Western blot after digestion, presumably because the extracellular domain recognized by the antibody is removed during digestion (data not shown).
Figure 7: Enrichment for osteocyte marker protein and gene expression in OmGFP66 bone-like structures.
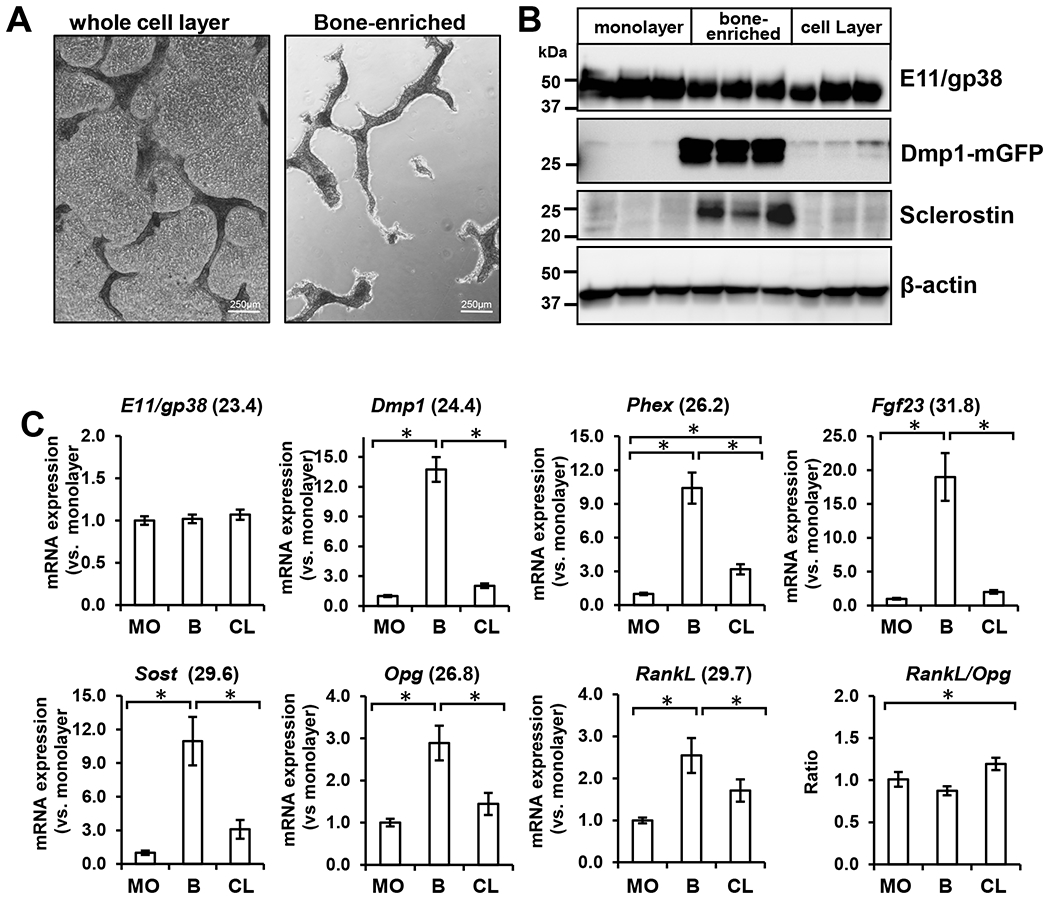
A) Phase contrast micrographs of OmGFP66 cell cultures before (whole cell layer) and after enrichment for bone-like structures (bone-enriched), bar = 250μm. B) Western blotting of lysates from the OmGFP66 monolayer and bone-enriched fractions compared to the whole cell layer. Note that E11/gp38 is equally expressed in all three fractions but that Dmp1-mGFP and sclerostin are highly enriched in the bone-enriched fraction compared to the monolayer and whole cell lysate. C) qPCR analysis showing expression of osteocyte marker genes, E11/gp38, Dmp1, Phex, Fgf23, Sost, Opg, and RankL by OmGFP66 cells in the monolayer (MO) and bone-enriched fraction (B) compared to the whole cell layer (CL). Data were normalized to β-actin as a housekeeping control and are presented as the fold change compared to the monolayer fraction. (mean ± SEM, n=3; * = p≤ 0.05, ANOVA/Tukey’s). Numbers in parentheses are the Ct values for the highest level of expression of each gene.
OmGFP66 Cells are PTH Responsive
To further establish OmGFP66 cells as a valid model of osteocyte differentiation, we next confirmed that they showed the expected changes in expression of osteocyte-related marker genes in response to stimulation with PTH. Treatment of OmGFP66 cells at day 14, 21 or 28 with 50 or 100nM PTH for 24h resulted in a dramatic downregulation of Dmp1 and Sost mRNA expression (Fig. 8), as expected, and the early osteocyte marker, E11/gp38 was slightly upregulated. PTH treatment also induced expression of RankL and downregulated Opg.
Figure 8: PTH regulation of osteocyte-expressed genes in OmGFP66 Cells.
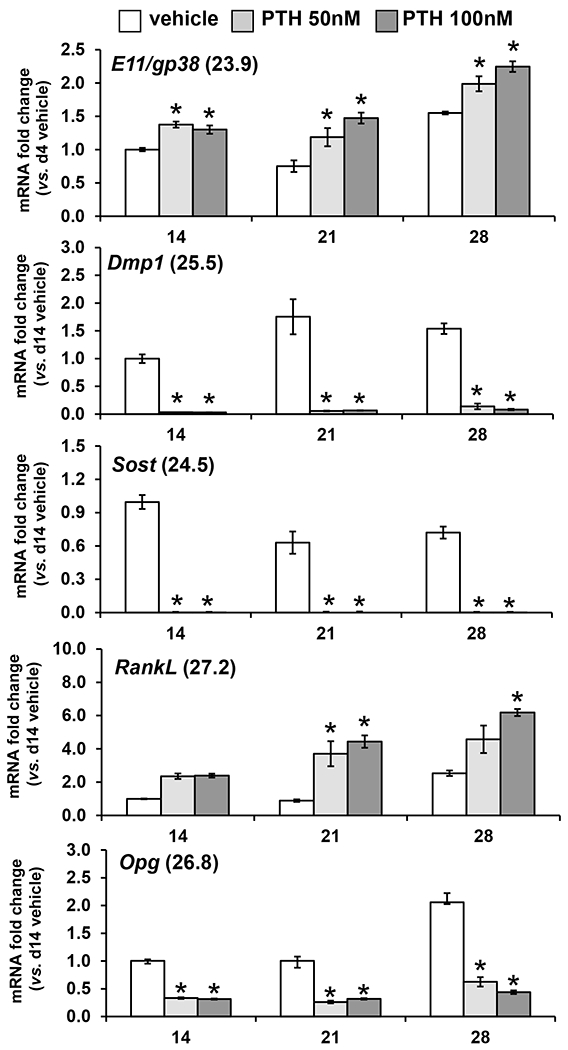
qPCR analysis showing expression of osteocyte marker genes, E11/gp38, Dmp1, Sost, RankL and Opg by OmGFP66 cells treated for 24h with vehicle or PTH (50nM and 100nM) on day 14, 21 or 28. Data were normalized to β-actin as a housekeeping control and are presented as the fold change compared to vehicle control (mean ± SEM, n=3; * = p≤ 0.05 vs. vehicle control ANOVA/Tukey’s). Numbers in parentheses are the Ct values for the highest level of expression of each gene.
Live Cell Imaging Shows that OmGFP66 Cells Provide a New Model for Investigating Osteocyte Differentiation, Embedding and Osteocyte Cell Dynamics
Timelapse imaging was performed on OmGFP66 cultures starting at day 8 when Dmp1-mGFP- positive bone-like structures started to form. Widefield epifluorescence imaging over an approximately 5 day duration, together with phase contrast imaging, showed that the mineral was deposited within ridges of Dmp1-mGFP positive cells that are closely associated with mineral deposition (see supplementary movie 1 and still images in supplementary Fig. S8). Growth of the bone-like structures appeared to start with cells switching on Dmp1-mGFP, followed by mineral deposition. Phase contrast imaging showed that the cells in the monolayer were constantly in motion (supplementary movie 1). Many of the Dmp1-mGFP-positive cells in the growing bone structure also initially showed motile properties, including moving along the surfaces of the forming bone and extending and retracting pseudopodia and dendritic processes. Embedding of Dmp1-mGFP positive cells and mineralization of the bone matrix was associated with a strong increase in Dmp1-mGFP expression and adoption of a dendritic morphology. By 11-12 days, in the oldest region of the bone, many of the cells switched off their expression of Dmp1-mGFP (bracketed area in supplementary Fig. S8), consistent with maturation to a late osteocyte phenotype.
Timelapse imaging was also performed on the confocal microscope over shorter periods of up to 2 days for clearer visualization of individual cells (see supplementary movie 2 and still images in supplementary Fig. S9). Note that the early bone forming at 8 days has condensations of Dmp1-mGFP-positive cells that are loosely packed together. Many of them are weakly GFP-positive, have a polygonal morphology and are motile. As the bone-like structure matures over 2 days of imaging, many of the cells become embedded, increase their expression of Dmp1-mGFP and adopt a dendritic osteocyte-like morphology with appropriate spacing within lacunae. Some cells appear to switch on Dmp1-mGFP expression in situ within their lacunae (arrows). Interestingly, we saw no examples of GFP-positive cells undergoing division, suggesting that they are post-mitotic cells.
DISCUSSION
Osteocytes have been historically viewed as quiescent, inactive cells with little function in bone other than as “placeholders”. This view of the osteocyte has changed dramatically in the last 15-20 years following the discoveries that osteocytes act as multifunctional cells that control osteoblast and osteoclast function, regulate phosphate homeostasis and play an important role in mechanotransduction in the skeleton [reviewed in(1–3)]. Interest in osteocyte biology and function has therefore surged in the past decade and there has been a need for new tools and cell models that can be used to further understand the function of these unique cells. Here we have presented two new osteogenic cell lines, OmGFP66 and OmGFP10, to expand and enhance the toolkit for investigations of osteocyte biology and function.
Both the OmGFP66 and OmGFP10 cell lines express a membrane-targeted GFP driven by the Dmp1 promoter, which provides a convenient marker to monitor osteocyte differentiation and allows clear visualization of the cell membrane and dendrites. Of these two cell lines, OmGFP66 is particularly valuable as a new research tool for osteocyte biology, as it has the unique property of forming highly organized three dimensional bone-like structures in vitro with clearly defined lacunae in which the Dmp1-mGFP cells are embedded. Confocal and electron microscopy showed that the 3D structure of these bone-like trabeculae mimics in vivo bone, with 2-3 layers of interconnected osteocytes that have a spacing and three dimensional organization resembling osteocytes in their natural in vivo environment. In addition, the Dmp1-mGFP positive osteocytes formed by this cell line have a highly dendritic morphology, mimicking osteocytes in vivo. Not only do OmGFP66 osteocytes resemble real osteocytes morphologically, but ultrastructurally their bone matrix closely resembles the extracellular matrix of real bone tissue, even down to the presence of canaliculi running through the bone matrix containing osteocyte dendritic processes. To our knowledge, no previous osteocyte-like or other osteogenic cell line forms bone-like structures with such a high degree of structural organization reminiscent of bone in vivo.
In addition to the structural similarities of OmGFP66 cells compared to in vivo bone, both cell lines constitutively expressed the early osteocyte marker, E11/gp38 at the mRNA and protein level, and progressively mineralized, expressing osteocyte markers, Dmp1, Phex, Mepe, and Fgf23, followed by the late osteocyte marker, Sost. The expression of Sost/sclerostin, which is normally seen in mature, deeply embedded osteocytes, indicates that OmGFP66 and OmGFP10 osteogenic cells can progress all the way to the late stages of osteocyte differentiation. In OmGFP66 cells, all of these markers, except E11/gp38, were expressed at higher levels in the osteocyte-enriched bone-like structures compared to the cell monolayer. Similar to IDG-SW3 cells, OmGFP66 cells also expressed alkaline phosphatase and type I collagen and required culturing in osteogenic media to induce osteocyte differentiation. They also expressed other genes known to be expressed by osteocytes, including RankL, Opg and Hif1α. Based on these data, OmGFP66 and OmGFP10 should be added to the list of available cell models for studying osteocyte differentiation and function, including the HOB-01-C1 human pre-osteocyte cell line(25), the MLO-Y4 mouse early osteocyte(24), the IDG-SW3 mouse model of osteoblast to osteocyte transition(26) and the Ocy454 mature osteocyte-like cell line(27). The striking osteocyte-like morphology of Dmp1-mGFP-positive cells within OmGFP66 bone mineral, together with their expression of multiple osteocyte marker genes suggests that the OmGFP66 cell line in particular provides a new and unique cell model that will be very useful for determining the mechanisms regulating osteocyte gene expression and differentiation.
The expression of Fgf23 by OmGFP66 cells may be especially significant, as there are few available cell models that produce sufficiently high levels of FGF23 protein. IDG-SW3 cells have been used in studies showing FGF23 upregulation by pro-inflammatory cytokines(44) and showing post translational proteolytic processing of FGF23(45). We have measured FGF23 protein in the culture supernatant of OmGFP66 cells by ELISA at day 10 and detected levels of 75pg/ml, which increased as high as 1543pg/ml following treatment by 1,25 dihydroxyvitamin D3 (data not shown). This is consistent with in vivo observations showing FGF23 regulation by vitamin D(46). Therefore, OmGFP66 cells should prove to be a valuable new model for examining regulation of FGF23 expression and processing. Osteocytes are thought to be the major source of circulating FGF23 in vivo, which is released in response to elevated serum phosphate. FGF23 then acts on the kidney to decrease phosphate reabsorption and return serum phosphate to normal levels. Therefore, osteocytes in vivo play a critical endocrine role in regulating phosphate homeostasis. Consistent with this, we found that Fgf23 expression was much higher in the osteocyte enriched OmGFP66 bone-like structures compared to the cell monolayer, suggesting that osteocytes are the main source of FGF23 secreted into the culture medium. Patients with inherited hypophosphatemic rickets, vitamin D deficient rickets or chronic kidney disease have elevated serum FGF23 even though their serum phosphate is low, which leads to renal phosphate wasting and impaired mineralization(46–48). Therefore, a more complete understanding of the mechanisms regulating osteocyte FGF23 expression and processing is key to identifying potential therapeutic approaches for treatment of these patients.
OmGFP66 cells also expressed other genes known to be expressed by osteocytes, including Cx43, Hif1α, RankL and Opg. Expression of RankL was higher in the osteocyte enriched bone-like structures compared to the cell layer, and PTH treatment upregulated RankL, while downregulating Opg, consistent with in vivo studies suggesting that osteocytes provide a major source of RANKL for osteoclast formation in postnatal bone remodeling and in bone loss in secondary hyperparathyroidism induced by dietary calcium deficiency(49,50). The expression of high levels of Cx43 by OmGFP66 cells as well as the formation of an interconnected embedded osteocyte network with a canalicular structure, suggests that these cells could provide an advantageous in vitro model for studies of osteocyte gap junctional communication. Furthermore, the striking morphological similarity of OmGFP66 osteocyte networks to osteocytes in vivo, as well as their formation of a lacunocanalicular system suggests that they may provide a valuable new tool for mechanistically understanding osteocyte responses to mechanical loading and examining the fluid flow down the canaliculi.
Osteocytes are PTH-responsive and our data confirmed that many of the osteocyte-related genes in OmGFP66 cells were up or down-regulated in the expected manner by PTH, which is known to downregulate expression of Sost, Dmp1 and other osteocyte marker genes while upregulating expression of osteoblastic marker genes(26,51–53). This further validates OmGFP66 as a cell line that mimics osteocytes and suggests they can provide a valuable model for understanding PTH actions in bone cells and in osteocytes in particular. The downregulation of sclerostin by PTH is thought to play a key role in mediating the bone anabolic effect of intermittent PTH treatment(53). Since inhibition of sclerostin has become a major therapeutic target for development of bone anabolic treatments, understanding its regulation in osteocytes will be of major importance for identifying new therapeutic agents.
Although the OmGFP66 clone in particular has many attractive features, no cell line is perfect and this model also has limitations. Because only about 25% of the total cell area converts to bone-like structures, this cell model may be less useful for mRNA and protein expression studies on the whole cell population compared to IDG-SW3 and Ocy454 cells, where a larger percentage of the cell population appears to express the osteocyte phenotype. Our data suggests that this limitation can be overcome by using bone-enriched cultures in which the monolayer cells are digested away or physically separated from the mineralized bone structures. However, we believe the real advantage of the OmGFP66 cells will be their application for in situ imaging approaches, such as live cell imaging of osteocyte differentiation and embedding. Our own live cell imaging data from time-lapse movies of differentiating OmGFP66 cell cultures showed the close association of mineralization with expression of Dmp1-mGFP, suggesting that osteocyte differentiation and mineralization are dynamically integrated. Live imaging also showed the dynamic properties of the Dmp1-mGFP-positive cells before embedding and revealed a dramatic increase in Dmp1-mGFP expression associated with osteocyte embedding and adoption of a dendritic morphology. The OmGFP66 cell model should also prove useful for live in situ imaging of calcium signaling and gap junctional communication in the presence of stimuli such as mechanical loading, and for metabolic imaging of osteocyte mitochondria and mitochondrial dynamics.
An interesting observation from timelapse imaging was that the Dmp1-mGFP-positive cells appeared to arise exclusively from cells switching on the mGFP transgene rather than mGFP-positive cells undergoing cell division. This suggests that the mGFP-positive cells are post-mitotic, even in this immortalized cell line that expresses SV40 T-antigen. This is consistent with our observations from live cell imaging of primary calvarial osteoblasts from Dmp1-mGFP transgenic mice and live cell imaging of ex-vivo and in vivo (intravital) calvaria of Dmp1-mGFP transgenic mice, showing that Dmp1-mGFP positive cells are post-mitotic (data not shown). It also suggests that the SV40 T-antigen expression does not interfere with the normal osteocyte terminal differentiation process in this cell line.
In summary, we report on the establishment of two novel osteogenic cell lines that represent models of osteoblast to osteocyte differentiation. One of these has unique properties compared to existing osteogenic and osteocyte cell models. These cells appear to be capable of expressing a full differentiation profile from osteoblast to early osteocyte to mature osteocyte. During this process, they form three dimensional bone-like structures with an extensive interconnected dendritic network and a lacunocanalicular structure that mimics bone in vivo. These cells therefore provide a novel tool for investigations of osteocyte biology and because of the membrane targeted Dmp1-mGFP reporter, provide a novel tool for live imaging of the osteocyte differentiation process. The use of this cell line for studies of osteocytes should facilitate new discoveries on the molecular mechanisms regulating osteocyte differentiation and function.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grants P01-AG039355 (SLD and LFB), R21-AR054449 (SLD) and R21-AR071563 (SLD). We acknowledge use of the UMKC Confocal Microscopy Core, supported by NIH grants S10-RR027668 (SLD) and S10-OD021665 (SLD).
Footnotes
Disclosure- The authors of this manuscript have nothing to disclose
REFERENCES
- 1.Dallas SL, Prideaux M, Bonewald LF. The osteocyte: an endocrine cell … and more. Endocr Rev. October 2013;34(5):658–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prideaux M, Findlay DM, Atkins GJ. Osteocytes: The master cells in bone remodelling. Curr Opin Pharmacol. June 2016;28:24–30. [DOI] [PubMed] [Google Scholar]
- 3.Chen H, Senda T, Kubo KY. The osteocyte plays multiple roles in bone remodeling and mineral homeostasis. Med Mol Morphol. June 2015;48(2):61–8. [DOI] [PubMed] [Google Scholar]
- 4.Knothe Tate ML, Adamson JR, Tami AE, Bauer TW. The osteocyte. The international journal of biochemistry & cell biology. January 2004;36(1):1–8. [DOI] [PubMed] [Google Scholar]
- 5.Lu Y, Xie Y, Zhang S, Dusevich V, Bonewald LF, Feng JQ. DMP1-targeted Cre expression in odontoblasts and osteocytes. J Dent Res. April 2007;86(4):320–5. [DOI] [PubMed] [Google Scholar]
- 6.Bivi N, Condon KW, Allen MR, Farlow N, Passeri G, Brun LR, et al. Cell autonomous requirement of connexin 43 for osteocyte survival: consequences for endocortical resorption and periosteal bone formation. J Bone Miner Res. February 2012;27(2):374–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Powell WF Jr., Barry KJ, Tulum I, Kobayashi T, Harris SE, Bringhurst FR, et al. Targeted ablation of the PTH/PTHrP receptor in osteocytes impairs bone structure and homeostatic calcemic responses. J Endocrinol. April 2011;209(1):21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalajzic I, Braut A, Guo D, Jiang X, Kronenberg MS, Mina M, et al. Dentin matrix protein 1 expression during osteoblastic differentiation, generation of an osteocyte GFP-transgene. Bone. July 2004;35(1):74–82. [DOI] [PubMed] [Google Scholar]
- 9.Kalajzic I, Matthews BG, Torreggiani E, Harris MA, Divieti Pajevic P, Harris SE. In vitro and in vivo approaches to study osteocyte biology. Bone. June 2013;54(2):296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamel-ElSayed SA, Tiede-Lewis LM, Lu Y, Veno PA, Dallas SL. Novel approaches for two and three dimensional multiplexed imaging of osteocytes. Bone. July 2015;76:129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maye P, Stover ML, Liu Y, Rowe DW, Gong S, Lichtler AC. A BAC-bacterial recombination method to generate physically linked multiple gene reporter DNA constructs. BMC Biotechnol. March 13 2009;9:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein-Nulend J, van der Plas A, Semeins CM, Ajubi NE, Frangos JA, Nijweide PJ, et al. Sensitivity of osteocytes to biomechanical stress in vitro. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. March 1995;9(5):441–5. [DOI] [PubMed] [Google Scholar]
- 13.Thi MM, Suadicani SO, Schaffler MB, Weinbaum S, Spray DC. Mechanosensory responses of osteocytes to physiological forces occur along processes and not cell body and require alphaVbeta3 integrin. Proceedings of the National Academy of Sciences of the United States of America. December 24 2013;110(52):21012–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonewald LF, Johnson ML. Osteocytes, mechanosensing and Wnt signaling. Bone. April 2008;42(4):606–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng JQ, Ward LM, Liu S, Lu Y, Xie Y, Yuan B, et al. Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nature genetics. November 2006;38(11):1310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimada T, Kakitani M, Yamazaki Y, Hasegawa H, Takeuchi Y, Fujita T, et al. Targeted ablation of Fgf23 demonstrates an essential physiological role of FGF23 in phosphate and vitamin D metabolism. The Journal of clinical investigation. February 2004;113(4):561–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao Z, Huang J, Cao L, Liang Y, Han X, Quarles LD. Osteocyte-specific deletion of Fgfr1 suppresses FGF23. PLoS One. 2014;9(8):e104154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Zhang Y, Kang H, Liu W, Liu P, Zhang J, et al. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J Biol Chem. May 20 2005;280(20):19883–7. [DOI] [PubMed] [Google Scholar]
- 19.Ellies DL, Viviano B, McCarthy J, Rey JP, Itasaki N, Saunders S, et al. Bone density ligand, Sclerostin, directly interacts with LRP5 but not LRP5G171V to modulate Wnt activity. J Bone Miner Res. November 2006;21(11):1738–49. [DOI] [PubMed] [Google Scholar]
- 20.Padhi D, Allison M, Kivitz AJ, Gutierrez MJ, Stouch B, Wang C, et al. Multiple doses of sclerostin antibody romosozumab in healthy men and postmenopausal women with low bone mass: a randomized, double-blind, placebo-controlled study. Journal of clinical pharmacology. February 2014;54(2):168–78. [DOI] [PubMed] [Google Scholar]
- 21.Recker RR, Benson CT, Matsumoto T, Bolognese MA, Robins DA, Alam J, et al. A randomized, double-blind phase 2 clinical trial of blosozumab, a sclerostin antibody, in postmenopausal women with low bone mineral density. J Bone Miner Res. February 2015;30(2):216–24. [DOI] [PubMed] [Google Scholar]
- 22.McColm J, Hu L, Womack T, Tang CC, Chiang AY. Single- and multiple-dose randomized studies of blosozumab, a monoclonal antibody against sclerostin, in healthy postmenopausal women. J Bone Miner Res. April 2014;29(4):935–43. [DOI] [PubMed] [Google Scholar]
- 23.McClung MR, Grauer A, Boonen S, Bolognese MA, Brown JP, Diez-Perez A, et al. Romosozumab in postmenopausal women with low bone mineral density. The New England journal of medicine. January 30 2014;370(5):412–20. [DOI] [PubMed] [Google Scholar]
- 24.Kato Y, Windle JJ, Koop BA, Mundy GR, Bonewald LF. Establishment of an osteocyte-like cell line, MLO-Y4. J Bone Miner Res. 1997;12(12):2014–23. [DOI] [PubMed] [Google Scholar]
- 25.Bodine PV, Vernon SK, Komm BS. Establishment and hormonal regulation of a conditionally transformed preosteocytic cell line from adult human bone. Endocrinology. November 1996;137(11):4592–604. [DOI] [PubMed] [Google Scholar]
- 26.Woo SM, Rosser J, Dusevich V, Kalajzic I, Bonewald LF. Cell line IDG-SW3 replicates osteoblast-to-late-osteocyte differentiation in vitro and accelerates bone formation in vivo. J Bone Miner Res. November 2011;26(11):2634–46. Epub 2011/07/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spatz JM, Wein MN, Gooi JH, Qu Y, Garr JL, Liu S, et al. The Wnt Inhibitor Sclerostin Is Up-regulated by Mechanical Unloading in Osteocytes in Vitro. J Biol Chem. July 3 2015;290(27):16744–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang W, Harris MA, Heinrich JG, Guo D, Bonewald LF, Harris SE. Gene expression signatures of a fibroblastoid preosteoblast and cuboidal osteoblast cell model compared to the MLO-Y4 osteocyte cell model. Bone. January 2009;44(1):32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao S, Zhang YK, Harris S, Ahuja SS, Bonewald LF. MLO-Y4 osteocyte-like cells support osteoclast formation and activation. J Bone Miner Res. November 2002;17(11):2068–79. [DOI] [PubMed] [Google Scholar]
- 30.Jat PS, Noble MD, Ataliotis P, Tanaka Y, Yannoutsos N, Larsen L, et al. Direct derivation of conditionally immortal cell lines from an H-2Kb-tsA58 transgenic mouse. Proceedings of the National Academy of Sciences of the United States of America. June 15 1991;88(12):5096–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wein MN, Spatz J, Nishimori S, Doench J, Root D, Babij P, et al. HDAC5 controls MEF2C-driven sclerostin expression in osteocytes. J Bone Miner Res. March 2015;30(3):400–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wetterwald A, Hoffstetter W, Cecchini MG, Lanske B, Wagner C, Fleisch H, et al. Characterization and cloning of the E11 antigen, a marker expressed by rat osteoblasts and osteocytes. Bone. February 1996;18(2):125–32. Epub 1996/02/01. [DOI] [PubMed] [Google Scholar]
- 33.Bernstein EF, Fisher LW, Li K, LeBaron RG, Tan EM, Uitto J. Differential expression of the versican and decorin genes in photoaged and sun-protected skin. Comparison by immunohistochemical and northern analyses. Laboratory investigation; a journal of technical methods and pathology. June 1995;72(6):662–9. [PubMed] [Google Scholar]
- 34.Dallas SL, Miyazono K, Skerry TM, Mundy GR, Bonewald LF. Dual role for the latent transforming growth factor-beta binding protein in storage of latent TGF-beta in the extracellular matrix and as a structural matrix protein. J Cell Biol. October 1995;131(2):539–49. Epub 1995/10/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris SE, Bonewald LF, Harris MA, Sabatini M, Dallas S, Feng JQ, et al. Effects of transforming growth factor beta on bone nodule formation and expression of bone morphogenetic protein 2, osteocalcin, osteopontin, alkaline phosphatase, and type I collagen mRNA in long-term cultures of fetal rat calvarial osteoblasts. J Bone Miner Res. June 1994;9(6):855–63. [DOI] [PubMed] [Google Scholar]
- 36.Bonewald LF, Wakefield L, Oreffo RO, Escobedo A, Twardzik DR, Mundy GR. Latent forms of transforming growth factor-beta (TGF beta) derived from bone cultures: identification of a naturally occurring 100-kDa complex with similarity to recombinant latent TGF beta. Mol Endocrinol. June 1991;5(6):741–51. [DOI] [PubMed] [Google Scholar]
- 37.Dallas SL, Park-Snyder S, Miyazono K, Twardzik D, Mundy GR, Bonewald LF. Characterization and autoregulation of latent transforming growth factor beta (TGF beta) complexes in osteoblast-like cell lines. Production of a latent complex lacking the latent TGF beta-binding protein. J Biol Chem. March 04 1994;269(9):6815–21. [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. December 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
- 39.Clapcote SJ, Roder JC. Simplex PCR assay for sex determination in mice. BioTechniques. May 2005;38(5):702, 4,, 6 Epub 2005/06/11. [DOI] [PubMed] [Google Scholar]
- 40.Dallas SL, Keene DR, Bruder SP, Saharinen J, Sakai LY, Mundy GR, et al. Role of the latent transforming growth factor beta binding protein 1 in fibrillin-containing microfibrils in bone cells in vitro and in vivo. Journal of Bone & Mineral Research. 2000;15(1):68–81. [DOI] [PubMed] [Google Scholar]
- 41.Lu Y, Kamel-El Sayed SA, Wang K, Tiede-Lewis LM, Grillo MA, Veno PA, et al. Live Imaging of Type I Collagen Assembly Dynamics in Osteoblasts Stably Expressing GFP and mCherry-Tagged Collagen Constructs. J Bone Miner Res. February 20 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tiede-Lewis LM, Xie Y, Hulbert MA, Campos R, Dallas MR, Dusevich V, et al. Degeneration of the osteocyte network in the C57BL/6 mouse model of aging. Aging (Albany NY). October 26 2017;9(10):2190–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thevenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998;7(1):27–41. Epub 2008/02/13. [DOI] [PubMed] [Google Scholar]
- 44.Ito N, Wijenayaka AR, Prideaux M, Kogawa M, Ormsby RT, Evdokiou A, et al. Regulation of FGF23 expression in IDG-SW3 osteocytes and human bone by pro-inflammatory stimuli. Mol Cell Endocrinol. January 5 2015;399:208–18. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto H, Ramos-Molina B, Lick AN, Prideaux M, Albornoz V, Bonewald L, et al. Posttranslational processing of FGF23 in osteocytes during the osteoblast to osteocyte transition. Bone. March 2016;84:120–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Juppner H, Wolf M, Salusky IB. FGF-23: More than a regulator of renal phosphate handling? J Bone Miner Res. October 2010;25(10):2091–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marsell R, Jonsson KB. The phosphate regulating hormone fibroblast growth factor-23. Acta Physiol (Oxf). October 2010;200(2):97–106. [DOI] [PubMed] [Google Scholar]
- 48.Pereira RC, Juppner H, Azucena-Serrano CE, Yadin O, Salusky IB, Wesseling-Perry K. Patterns of FGF-23, DMP1, and MEPE expression in patients with chronic kidney disease. Bone. December 2009;45(6):1161–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiong J, Onal M, Jilka RL, Weinstein RS, Manolagas SC, O’Brien CA. Matrix-embedded cells control osteoclast formation. Nat Med. September 11 2011;17(10):1235–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiong J, Piemontese M, Thostenson JD, Weinstein RS, Manolagas SC, O’Brien CA. Osteocyte-derived RANKL is a critical mediator of the increased bone resorption caused by dietary calcium deficiency. Bone. September 2014;66:146–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prideaux M, Dallas SL, Zhao N, Johnsrud ED, Veno PA, Guo D, et al. Parathyroid Hormone Induces Bone Cell Motility and Loss of Mature Osteocyte Phenotype through L-Calcium Channel Dependent and Independent Mechanisms. PLoS One. 2015;10(5):e0125731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bellido T, Ali AA, Gubrij I, Plotkin LI, Fu Q, O’Brien CA, et al. Chronic elevation of parathyroid hormone in mice reduces expression of sclerostin by osteocytes: a novel mechanism for hormonal control of osteoblastogenesis. Endocrinology. November 2005;146(11):4577–83. [DOI] [PubMed] [Google Scholar]
- 53.Keller H, Kneissel M. SOST is a target gene for PTH in bone. Bone. August 2005;37(2):148–58. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


