Abstract
The global Coronavirus Disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has led to unprecedented social and economic consequences. The risk of morbidity and mortality due to COVID-19 increases dramatically in the presence of co-existing medical conditions while the underlying mechanisms remain unclear. Furthermore, there are no proven effective therapies for COVID-19. This study aims to identify SARS-CoV-2 pathogenesis, diseases manifestations, and COVID-19 therapies using network medicine methodologies along with clinical and multi-omics observations. We incorporate SARS-CoV-2 virus-host protein-protein interactions, transcriptomics, and proteomics into the human interactome. Network proximity measure revealed underlying pathogenesis for broad COVID-19-associated manifestations. Multi-modal analyses of single-cell RNA-sequencing data showed that co-expression of ACE2 and TMPRSS2 was elevated in absorptive enterocytes from the inflamed ileal tissues of Crohn's disease patients compared to uninflamed tissues, revealing shared pathobiology by COVID-19 and inflammatory bowel disease. Integrative analyses of metabolomics and transcriptomics (bulk and single-cell) data from asthma patients indicated that COVID-19 shared intermediate inflammatory endophenotypes with asthma (including IRAK3 and ADRB2). To prioritize potential treatment, we combined network-based prediction and propensity score (PS) matching observational study of 18,118 patients from a COVID-19 registry. We identified that melatonin (odds ratio (OR) = 0.36, 95% confidence interval (CI) 0.22-0.59) was associated with 64% reduced likelihood of a positive laboratory test result for SARS-CoV-2. Using PS-matching user active comparator design, melatonin was associated with 54% reduced likelihood of SARS-CoV-2 positive test result compared to angiotensin II receptor blockers or angiotensin-converting enzyme inhibitors (OR = 0.46, 95% CI 0.24-0.86).
Introduction
The ongoing global Coronavirus Disease 2019 (COVID-19) pandemic has led to 5 million confirmed cases and 327,000 deaths worldwide as of May 21, 2020. The United States alone has 1.58 million confirmed cases with a death toll of more than 93,000. Several retrospective studies have reported the clinical characteristics of individuals with symptomatic COVID-19 infection, and an emerging theme has been the significantly higher risk of morbidity and mortality among individuals with one or more comorbid health conditions, such as hypertension, asthma, diabetes mellitus, cardiovascular or cerebrovascular disease, chronic kidney disease and malignancy (1-6). However, these retrospective clinical studies are limited by small sample sizes and unmeasured confounders, leaving the underlying patho-mechanisms largely unknown. More specifically, it is unclear whether associations of disease manifestations and COVID-19 severity are merely a reflection of poorer health in general, or a clue to shared pathobiological mechanisms.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes COVID-19, is an enveloped virus that carries a single-stranded positive-sense RNA genome (6, 7). SARS-CoV-2 is a newly discovered member of the coronavirus (CoV) family (8). SARS-CoV-2 enters host cells via binding with its spike protein to the angiotensin converting enzyme 2 (ACE2) receptor on the surfaces of many cell types (9). This binding is primed by transmembrane protease serine 2 (TMPRSS2) (9) and the host cell protease furin (10) (Fig. 1A). Studies have shown that ACE2 and TMPRSS2 are highly co-expressed in alveolar type II (AT2) epithelial cells in the lung (11), nasal mucosa (12), bronchial secretory cells (13), and absorptive enterocytes in the ileum (14). Yet, much remains to be learned on how these critical human proteins involved in the infection and replication of SARS-CoV-2 are associated with various disease comorbidities and complications. Systematic identification of the host factors involved in the protein-protein interactions (PPIs) of SARS-CoV-2 and the human host will facilitate identification of therapeutic targets and advance understanding of the complications and comorbidities resulting from COVID-19 (15-17). Studies using transcriptomics (18), proteomics (19), and interactomics (20) (PPIs) methods have contributed to a better understanding of the SARS-CoV-2-host interactome, which enabled the investigation of the complications and comorbidities of SARS-CoV-2 and a facilitated search for effective treatment (Fig. 1B).
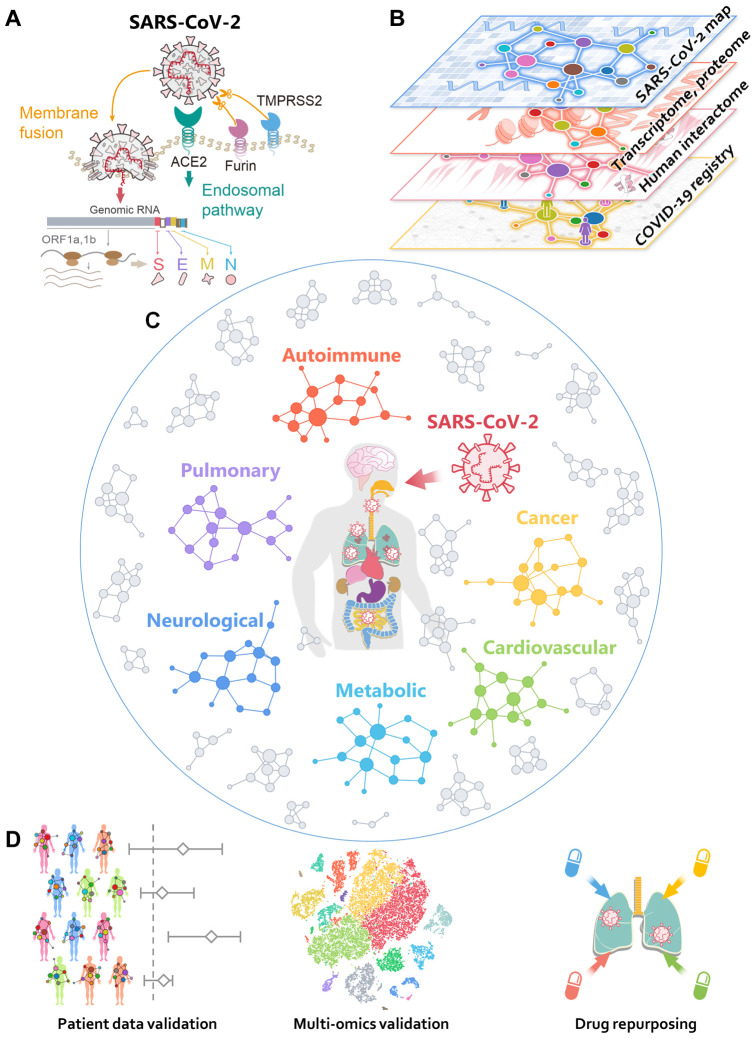
Fig. 1. Overall workflow of this study.
(A) A diagram illustrating the basic pathogenesis of SARS-CoV-2. (B) A diagram illustrating how to build a global interactome map for SARS-CoV-2. We compiled the SARS-CoV-2 human target gene/protein sets from multi-omics data from transcriptome, proteome, and human interactome, and validated network-based findings using patient data from the COVID-19 registry. (C) A diagram illustrating network-based measure of disease manifestations associated with COVID-19. We systematically evaluated the network proximities of the SARS-CoV-2 human target genes/proteins with 64 diseases across six main categories: autoimmune, cancer, cardiovascular, metabolic, neurological, and pulmonary. (D) A workflow illustrating validation of network-based findings. We performed single-cell analyses to further investigate the underlying mechanisms of COVID-19 with asthma and inflammatory bowel disease. We prioritized nearly 3,000 FDA approved/investigational drugs for their potential anti-SARS-CoV-2 effects from network-based findings and validated drug-COVID-19 outcomes using the institutional review board-approved COVID-19 patient registry.
Major efforts are underway to develop safe and effective drugs to treat COVID-19: preventive and therapeutic strategies currently being explored include vaccination, SARS-CoV-2-specific antibody, novel nucleoside analogues such as remdesivir, and repurposed drugs (21-25). The development of vaccination and antibody therapies takes significantly longer compared to drug repurposing. Many existing drugs are currently or have been tested in clinical trials, such as the antimalarial drug hydroxychloroquine and protease inhibitor combination lopinavir/ritonavir (26, 27). However, early results from these trials have not yet shown significant clinical benefits for COVID-19 patients (26, 27). We recently evaluated more than 2,000 FDA approved/investigational drugs using network-based method and prioritized 16 drug candidates and 3 drug combinations for COVID-19 (28). Yet, the answer to the key question why an approved drug originally documented for other diseases can be repurposed for COVID-19 remains unclear. One possible explanation is that COVID-19 shares common disease pathobiology or functional pathways elucidated by the human PPIs (24, 28, 29). Systematic identification of common disease pathobiological pathways shared by COVID-19 and other diseases would offer novel targets and therapies for COVID-19.
In this study, we present an integrative network medicine platform that quantifies the association of COVID-19 with other diseases across six categories, including autoimmune, malignant, cardiovascular, metabolic, neurological, and pulmonary (Fig. 1C). The rationale for these analyses rests on the notions that (i) the proteins that functionally associate with disease (such as COVID-19) are localized in the corresponding subnetwork within the comprehensive human PPI network (24, 29-31); and (ii) proteins that are associated with a specific disease may be directly targeted by the virus or are in the close vicinity of virus host proteins. We first performed network analysis followed by single-cell RNA-Seq data analysis to identify the underlying pathobiological relationships between COVID-19 and its associated comorbidities. Secondly, we used our network-medicine findings and patient data from a large COVID-19 clinical registry database to identify and prioritize existing FDA-approved drugs as potential COVID-19 drug candidates (Fig. 1D).
Results
A global map of the SARS-CoV-2 virus-host interactome
We assembled four host gene/protein sets for SARS-CoV-2 (see Methods) (Table S1): (1) SARS2-DEG, representing the differentially expressed genes (DEGs) from the transcriptomic data of SARS-CoV-2 infected primary human bronchial epithelial cells; (2) SARS2-DEP, representing the differentially expressed proteins (DEPs) from the proteomic data of SARS-CoV-2 infected human Caco-2 cells; (3) HCoV-PPI, representing the literature-based virus-host proteins across multiple human coronaviruses (HCoVs), including the SARS-CoV-1 (from the 2002-2003 pandemic) and MERS-CoV; and (4) SARS2-PPI (SARS-CoV-2 specific virus-host PPIs). Since HCoV-PPI and SARS2-PPI are both physical virus-host PPIs, we further combined them as the fifth data set, PanCoV-PPI.
We first performed functional enrichment analyses for the five different data sets. We found that while these data sets share several common pathways and ontology terms (Fig. 2A), such as phagosome, measles, apoptosis, NF-kappa B signaling pathway, neutrophil-related immunity, apoptotic processes, virus transport, viral genome replication, and response to interferon, they differ considerably in terms of their most significantly enriched pathways (Fig. S1-S5). This is especially noticeable for SARS2-DEP and SARS2-PPI. While SARS2-DEG (Fig. S1) and HCoV-PPI (Fig. S3) show more enrichment in immune responses and viral pathways; SARS2-DEP (Fig. S2) is more related to various cellular metabolic pathways; and SARS2-PPI (Fig. S4) is more enriched in DNA replication, RNA transcription, and protein translation. These observations suggest that these different SARS-CoV-2 data sets capture complementary aspects of the biological and cellular states of the viral life cycle and host immunity. Therefore, building a global virus-host map (including interactome, transcriptome, and proteome) that incorporates data from transcriptomics, proteomics, and physical virus-host PPIs are essential for a better understanding of the pathogenesis of COVID-19. This global virus-host map for SARS-CoV-2 can offer a more complete picture of the interconnected functional pathways involved in viral pathogenesis, thereby facilitating discovery of therapeutic targets.
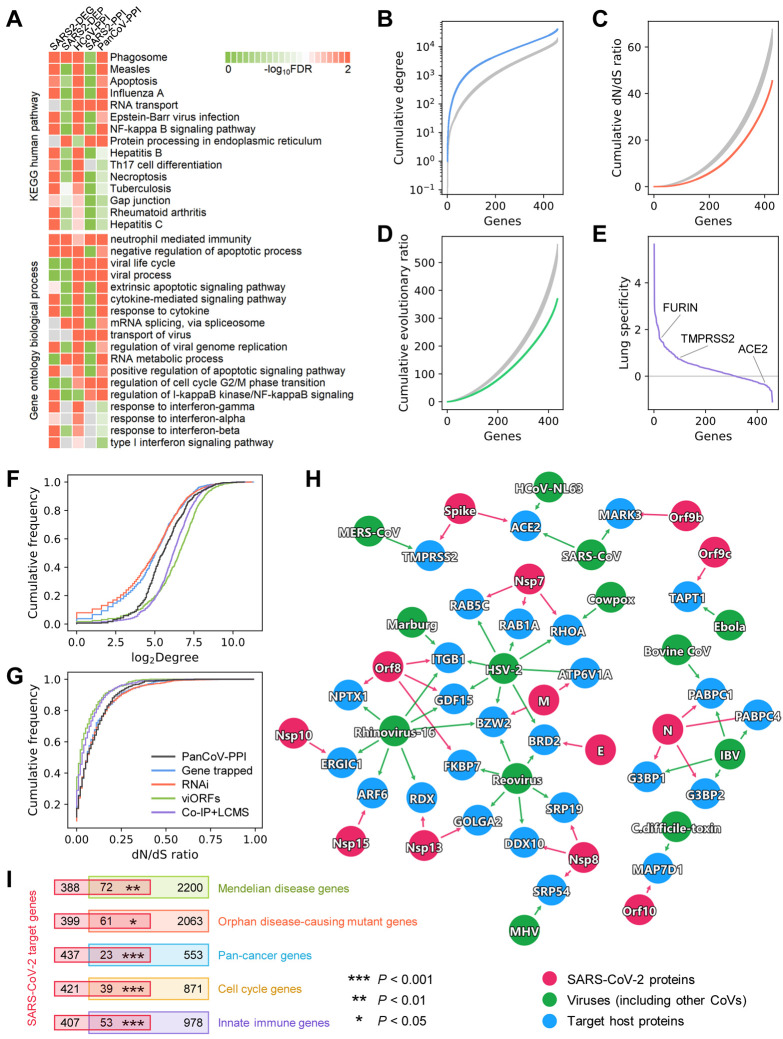
Fig. 2. Network and biological characteristics of the SARS-CoV-2 interactome map.
(A) Pathway and gene ontology (biological process) enrichment analysis results of the SARS-CoV-2 host genes/proteins across five different data sets. We assembled five gene/protein data sets from SARS-CoV-2 host protein-protein interactions, transcriptomics, and proteomics (Table S1). (B, C, D) Network and biological characteristics of the SARS-CoV-2 host genes/proteins. The proteins in PanCoV-PPI have higher node degrees (B), lower non-synonymous to synonymous substitutions (dN/dS) ratios (C), and lower evolutionary ratios (D) compared to randomly selected proteins (grey, mean ± standard deviation of 100 repeats). (E) Among the 460 proteins in PanCoV-PPI, 450 (98%) are expressed in lungs, and 317 (69%) are lung specific expression (Z > 0). (F, G) The distribution of the node degrees in the human interactome and dN/dS ratios of PanCoV-PPI and four published virus-related host protein sets. (H) The shared target human proteins (blue) of SARS-CoV-2 (red) and other viruses (green). The detailed data are provided in Table S2. (I) SARS-CoV-2 target proteins overlap significantly with disease-associated genes (Mendelian disease and Orphan disease), cancer genes, cell cycle genes, and innate immune genes.
Network and biological characteristics of virus-host interactome for SARS-CoV-2
In addition to identifying the functions that these data sets represent, we next characterized the network patterns (node degree in the human PPI network) and bioinformatics features of these SARS-CoV-2 data sets, including ratio of non-synonymous to synonymous substitutions (dN/dS), evolutionary rate ratio, and lung expression specificity (Fig. 2B-G and Fig. S6). To find common as well as unique network and bioinformatic characteristics of SARS-CoV-2, we further compiled four additional virus-host gene/protein networks identified by different methods for comparisons (Table S2): (1) 900 virus-host interactions connecting 10 other viruses and 712 host genes identified by gene-trap insertional mutagenesis, (2) 2,855 known virus-host interactions connecting 2,443 host genes and 55 pathogens identified from RNA interference (RNAi), (3) 579 host proteins mediating translation of 70 innate immune-modulating viral open reading frames (viORFs), and (4) 1,292 host genes mediating influenza-host interactions identified by co-immunoprecipitation and liquid chromatography-mass spectrometry (Co-IP+LC/MS). We found that host proteins in PanCoV-PPI (Fig. 2B) and four other data sets (SARS2-DEG, SARS2-DEP, HCoV-PPI, and SARS2-PPI) (Fig. S6) were more likely to be highly connected (high degree or connectivity) in the human PPI network, including several hubs, such as JUN, XPO1, MOV10, NPM1, VCP, and HNRNPA1. PanCoV-PPI has a comparable degree distribution with host proteins/genes identified by viORFs and Co-IP+LC/MS, although being marginally lower than that identified by RNAi and gene-trap insertional mutagenesis assay (Fig. 2F).
Expression patterns of genes in a specific disease-related tissue play crucial role for elucidation of disease pathogenesis and drug discovery (24, 29). Given the major impact of SARS-CoV-2 on pulmonary function and lung injury (1), we inspected the lung-specific expression of genes in PanCoV-PPI using a Z score measure compared to other 30 tissues from the GTEx database (see Methods). We found that most host genes for SARS-CoV-2 have high expression in lung (Fig. 2E, Table S1) compared to other tissues; yet, ACE2 has a low expression in lung compared to other tissues. A recent study showed that ACE2 was primarily expressed in the epithelial cells in lungs, and only 3.8% of alveolar type 2 pneumocytes expressed both ACE2 and TMPRSS2, but it is significantly upregulated in smokers and 24-48 hours following SARS-CoV-2 infection (11, 32). Another study also showed that despite relatively low expression of ACE2 in the lung, ACE2 was expressed in multiple epithelial cell types along the airway (12). Therefore, it is important to understand the cell type-specific SARS-CoV-2 pathogenesis at single cell levels.
To inspect the evolutionary factors underlying the SARS-CoV-2-human PPIs, we investigated the selective pressure and evolutionary rates quantified by the non-synonymous versus synonymous substitution rate ratios (dN/dS ratios, see Methods) using human-mouse orthologous gene pairs. We found that PanCoV-PPI has a stronger purifying selection (lower dN/dS ratios (Fig. 2C) and evolutionary rate ratio (Fig. 2D)) compared to the same number of random genes. PanCoV-PPI is also comparable to other four virus-host genes/proteins identified by different assays (Fig. 2F, G) in terms of node degrees and dN/dS ratios. Altogether, these observations suggest that the virus-host PPIs assembled in this study offer a high-quality interactome map for SARS-CoV-2 for identifying pathogenesis and potential treatments for COVID-19.
To inspect shared viral pathways across different viruses and SARS-CoV-2, we further performed network overlap analysis of PanCoV-PPI with 712 host genes across 10 types of other viruses identified by gene-trap insertional mutagenesis assays (25). We found a significant overlap of the SARS-CoV-2 host proteins with non-coronaviruses (P < 0.002, Fisher's exact test, Fig. 2H). For example, BRD2, a transcriptional regulator which belongs to Bromodomain and Extra-Terminal motif family, is connected to SARS-CoV-2 and two other viruses, herpes simplex virus 2 (HSV-2) and reovirus. RHOA, encoding a small GTPase protein in the Rho family of GTPases, is connected to SARS-CoV-2, cowpox, and HSV-2 as well. RHOA has been reported to be involved in multiple human diseases, including cardiovascular disease (33) and cancer (34). These observations indicate possible disease manifestations associated with SARS-CoV-2.
SARS-CoV-2 cellular network perturbations of disease manifestations
Investigation of the relationships between human host proteins targeted by SARS-CoV-2 and disease susceptibility genes may offer crucial information for identifying COVID-19-associated disease manifestations. We thus inspected the overlap between SARS-CoV-2 host genes/proteins and the susceptibility gene sets implicated in different diseases and biological events (Fig. 2I). We found that host genes/proteins targeted by SARS-CoV-2 are significantly enriched in Mendelian disease (P = 0.002, Fisher's exact test), orphan disease (P = 0.044), and cancer (P < 0.001). Mechanistically, SARS-CoV-2 target host genes are significantly enriched in cell cycle genes (P < 0.001) and innate immune genes (P < 0.001).
Using the five SARS-CoV-2 host gene/protein sets, we next tried to identify potential COVID-19 comorbidities. To achieve this, we assembled the disease-protein network of COVID-19 and six disease categories (Table S3). For cancer, the driver genes for pan-cancer and individual cancer types were retrieved from the Cancer Gene Census (35) and a previous study (36). For autoimmune, pulmonary, neurological, cardiovascular, and metabolic categories, we extracted their associated genes/proteins from the Human Gene Mutation Database (37). Using the disease-protein network together with the SARS2-PPI (SARS-CoV-2 virus-host interactome) and HCoV-PPI (HCoV-host interactome) sets, we examined their overall connectivity in the human PPI network (Fig. 3A, Data File S1). To build the global network for the disease comorbidities, we extracted the PPIs from the human interactome for the virus target proteins and disease-associated proteins. Each small node indicates a virus target host protein (blue) or a disease-associated protein (green). Some disease-associated proteins can be directly targeted by the viruses, as shown in orange. Edges among these protein nodes indicate PPIs. For SARS-CoV-2, the targets of its individual viral proteins are shown based on the SARS2-PPI data set. Due to the tendency of having common disease-associated proteins, some disease categories tend to cluster closely, e.g., cancer and neurological. Diseases from other categories, such as autoimmune and pulmonary, are scattered. Most of the virus target proteins are connected with the disease-associated proteins, which suggests shared pathobiological pathways of COVID-19 and these diseases. Various cancer types formed a relatively distant module from the virus targets, compared to other disease categories.
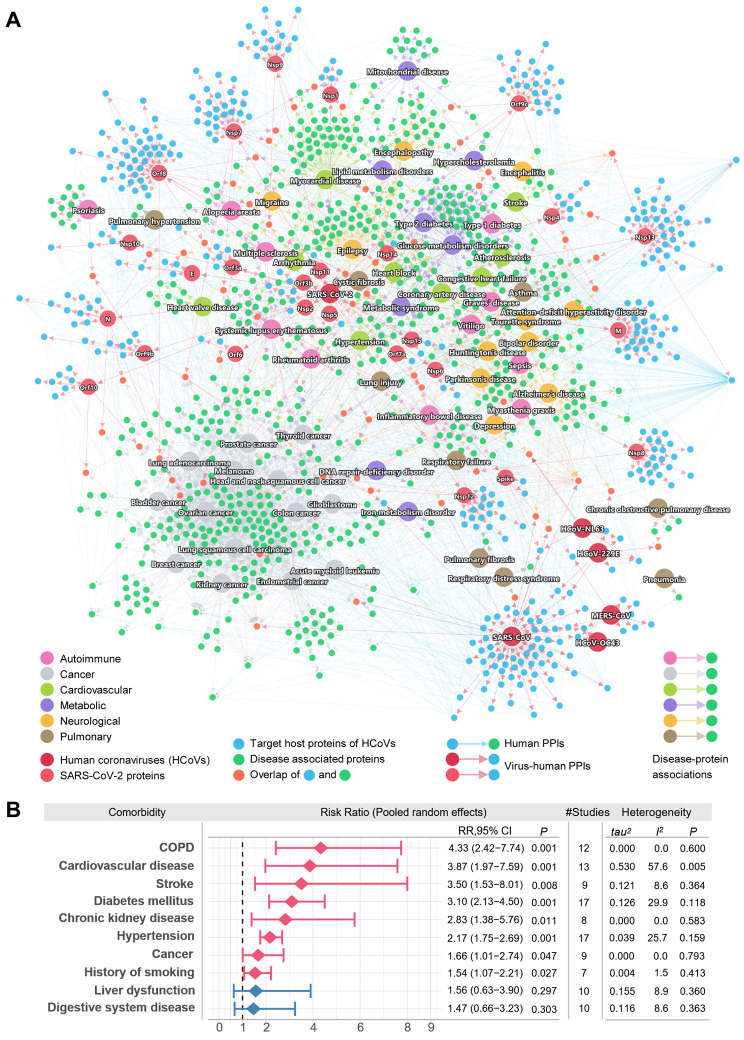
Fig. 3. A global network illustrating disease manifestations associated with human coronavirus.
(A) The target human proteins of SARS-CoV-2 are connected to the disease-associated proteins. Blue links (edges) indicate physical protein-protein interactions. For SARS-CoV-2, its viral proteins are shown by light red node. The target human proteins (blue) of the viruses are intricately connected to the disease-associated proteins (green). Human disease nodes are colored by different disease categories: autoimmune, cancer, cardiovascular, metabolic, neurological, and pulmonary. (B) Estimation of the pooled risk ratio using random effects meta-analysis for 10 comorbidities between severe versus non-severe COVID-19 patients. The tau2 and I2 statistics were calculated to quantify the heterogeneity among studies. I2 < 50% was considered as low heterogeneity among studies, 50% < I2 < 75% was considered as moderate heterogeneity, and I2 > 75% was considered as high heterogeneity.
Shown in Fig. 3A, these diseases can be targeted directly or interact with the targets of SARS-CoV-2 or other HCoVs. For example, among the four chronic obstructive pulmonary disease (COPD) associated proteins shown in the network, TGFB1 is the direct target of HCoV-229E, and all four proteins (TGFB1, DEFB1, SNAI1, and ADAM33) interact with at least one SARS-CoV-2 viral protein target. The risk of various cardiovascular diseases was found to be increased in COVID-19 patients, including heart block, coronary artery disease, and congestive heart failure, and arrhythmia, which is consistent with clinically reported myocardial injury (38) and cardiac arrest (39). These observations reveal common network relationship between COVID-19 and human diseases (Fig. 3A). We then performed meta-analysis of 34 COVID-19 clinical studies (Table S4) to evaluate the pooled risk ratios of 10 comorbidities among 4,973 COVID-19 positive patients (including 2,268 mild and 731 severe patients). The random effects model was used to estimate the pooled risk ratio of disease severity. The tau2 and I2 statistics were used to evaluate the heterogeneity among studies. We found that 8 comorbidities have significantly higher risks in severe COVID-19 patients (Fig. 3B). The overall pooled risk ratio of COPD was 4.33 (95% confidence interval (CI) 2.42-7.74, P = 0.001) in 12 low heterogeneous clinical studies (I2 = 0.0%, P = 0.6). The COVID-19 patients with cardiovascular diseases have a risk ratio of 3.87 (95% CI 1.97-7.59, P = 0.001), and there was a slightly higher heterogeneity across the 13 studies (I2 = 57.6%, P = 0.005). Patients with stroke, diabetes, chronic kidney disease, hypertension, cancer, and history of smoking were also found with higher risk ratios of severe COVID-19. We next turned to quantify the network-based relationships between COVID-19 and human diseases under the human interactome model using the network proximity measure.
Network-based measure of COVID-19-associated disease manifestations
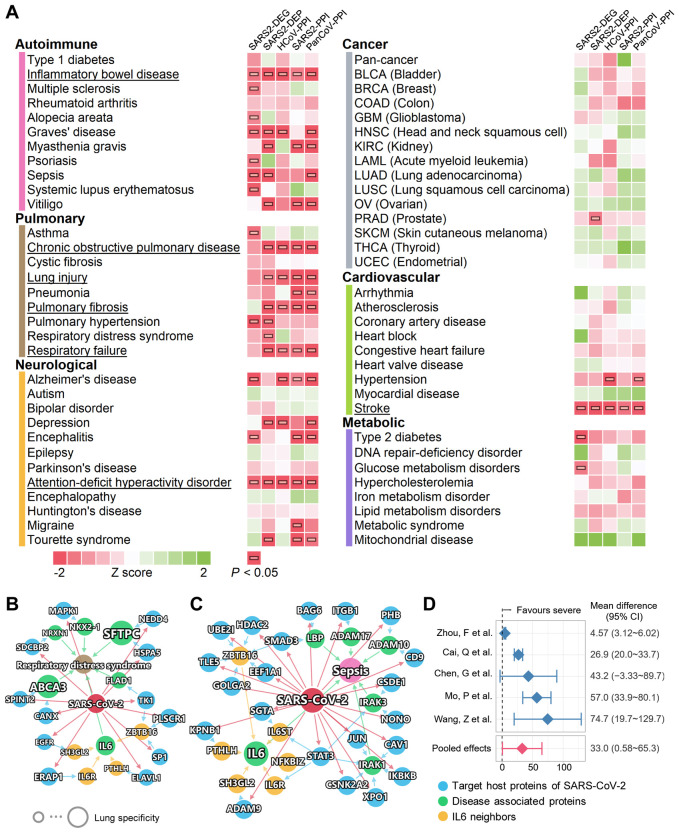
We systematically evaluated the network-based relationships of the 64 diseases across the 6 categories to COVID-19 (Fig. 4A). We used the state-of-the-art network proximity measure to evaluate the connectivity and the closeness of the disease proteins and SARS-CoV-2 host proteins, taking the topology of the human interactome network into consideration. To test the significance of the proximity, Z scores and P values were calculated based on the permutation tests and are shown in Fig. 4A. We found that each disease-disease pair has a well-defined network-based footprint. If the footprint between COVID-19 module and another disease module is significantly close (low Z score and P < 0.05), the magnitude of the proximity is indictive of their biological relationship: closer network proximity (Fig. 4A) of SARS-CoV-2 host genes/proteins with a disease module indicates higher potential of manifestation between COVID-19 and a specific disease. We first noticed that immunological, pulmonary, and neurological diseases show significant proximity to SARS-CoV-2 data sets more frequently than do cancer, cardiovascular, and metabolic diseases. Some diseases have significant proximities to more than one SARS-CoV-2 data sets, most notably inflammatory bowel disease (IBD), attention-deficit/hyperactivity disorder, and stroke, which achieved significant P value for all five SARS-CoV-2 protein sets. Pulmonary diseases, including COPD, lung injury, pulmonary fibrosis, and respiratory failure, achieved four significant proximities. Some diseases have significant proximities to certain SARS-CoV-2 data sets, indicating associations at certain levels, e.g., asthma (transcriptomic), respiratory distress syndrome (proteomic), and hypertension. Network visualization can further show the connections between SARS-CoV-2 and other diseases, for example, respiratory distress syndrome (Fig. 4B), sepsis (Fig. 4C), and COPD (Fig. S7). We found that multiple SARS-CoV-2 host proteins are directly connected with the disease-associated proteins (Fig. 4B). ABCA3 is a lipid transporter located in the outer membrane of lamellar bodies in AT2 cells; the mutations of the ABCA3 gene can disrupt pulmonary surfactant homeostasis and lead to inherited pulmonary diseases (40). Another membrane surface protein, the pulmonary-associated surfactant protein C encoded by SFTPC, can cause lung injury when misfolded (41). For sepsis, we noticed several inflammatory and immune-related proteins, such as IRAK1, IRAK3, IKBKB, and STAT3, suggesting overlap of the inflammatory response activated in COVID-19 and sepsis (Fig. 4C). It has been reported that an overzealous production of certain cytokines, such as IL-6, caused by dysregulation of innate immune responses to SARS-CoV-2 infection can result in a ‘cytokine storm’, better known as cytokine release syndrome (CRS) (42). The potential prognoses of acute respiratory distress syndrome and sepsis using IL-6 expression levels were also established (43-45). IL-6 also has a significantly increased expression level in the human bronchial epithelial cells infected with SARS-CoV-2 from the SARS2-DEG data set (18). In addition, it is also potentially affected by SARS-CoV-2 through multiple PPIs (Fig. 4B, 4C, IL-6 neighbors), such as IL-6R, endophilin A1 (encoded by SH3GL2), and parathyroid hormone like hormone (encoded by PTHLH). Our random effects meta-analysis of 5 clinical studies of COVID-19 revealed that there was an increase of IL-6 levels in severe COVID-19 patients compared to non-severe COVID-19 patients (Fig. 4D). The mean difference was 33.0 pg/ml (95% CI 0.58-65.3, Fig. 4D) with high literature heterogeneity (I2 = 94%, P < 0.001). These results indicate that IL-6 plays a critical role in COVID-19-associated respiratory distress syndrome and sepsis. Due to the importance of IL-6 in SARS-CoV-2 infection, IL-6 antagonists including tocilizumab (46) (NCT04315480) and sarilumab (NCT04327388) are being investigated in clinical trials for treatment of patients with severe COVID-19.
Fig. 4. A landscape of disease manifestations associated with COVID-19 quantified by network proximity measure.
(A) Heatmaps showing the network proximities of COVID-19 with 64 diseases across 6 categories. The network proximities of the disease modules and the five SARS-CoV-2 data sets were evaluated using the “closest” network proximity measure (see Methods). The magnitude of the proximity is indictive of their biological relationship: closer network proximity of SARS-CoV-2 host genes/proteins of a disease indicates higher potential of manifestation between COVID-19 and a specific disease. P < 0.05 computed by permutation test was considered significant (indicated by horizontal bars). Three categories, autoimmune, pulmonary, and neurological frequently show significant proximities to COVID-19. Inflammatory bowel disease, attention-deficit/hyperactivity disorder, and stroke achieved significance with all five SARS-CoV-2 data sets. (B, C) Highlighted subnetworks between SARS-CoV-2 host genes/proteins with the disease-associated proteins of respiratory distress syndrome (B) and sepsis (C). (D) Clinical data analyses showed an association of COVID-19 severity with IL-6 expression levels in patients. Meta-analysis of random effects model was performed using the mean difference in IL-6 (pg/ml). There was a high heterogeneity among these studies (I2 = 94%, P < 0.001).
As an illustration of the shared pathobiology and inflammatory pathways of COVID-19 with various disease manifestations (Fig. 4), we next turned to focus on two inflammation-driven diseases, asthma and IBD.
Inflammatory endophenotypes shared by COVID-19 and asthma
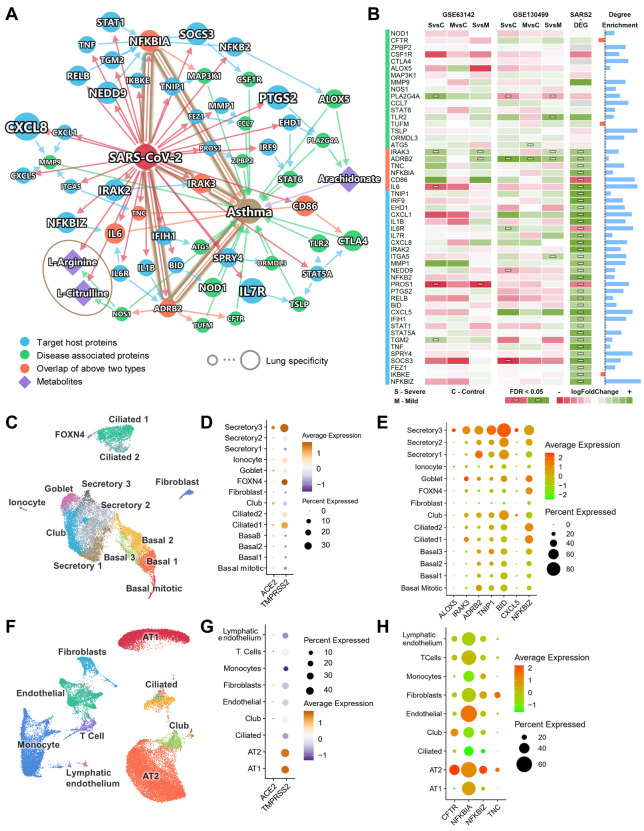
Patients with severe COVID-19 symptoms showed a higher prevalence of dyspnea (Fig. S8A, P < 0.001). To understand associations between COVID-19 and respiratory disease (including asthma), we adopted a multi-modal analysis utilizing metabolomics and transcriptomics data from two previous asthma cohorts (see Methods) under the human interactome network model. Fig. 5A shows the subnetwork of the connections among SARS-CoV-2 target host proteins and asthma-associated proteins. Most of these proteins have enriched connections within the subnetwork (Fig. 5B, blue bars) (more connections in the subnetwork than in a random network of the same size in the human interactome, see Methods). Six overlapped proteins (orange) from both groups were identified: NFKBIA, IRAK3, TNC, IL6, ADRB2, and CD86. A recent study showed that glucose metabolism plays a key role in influenza A-regulated cytokine storm (47). The unique plasma metabolome of asthmatics versus healthy controls also suggests activated inflammatory and immune pathways (48). Therefore, in addition to PPIs, we also integrated metabolomics data generated in a previously assembled asthma cohort (49). By matching the enzymes of the differential metabolites and the proteins in the PPI network, we found three key metabolites, including arachidonate, L-arginine, and L-citrulline. L-arginine and L-citrulline were decreased in the sera of COVID-19 patients (50). A previous study showed that these metabolites were also decreased in asthma patients (49). Arachidonate, the precursor of a variety of products that regulate inflammatory pathways (51), was found to have an increased level in the inflamed airways of asthmatics (52). Arachidonate can be converted by 5-lipoxygenase encoded by ALOX5 to leukotriene, which is release during an asthma attack and is responsible for the bronchoconstriction (53).
Fig. 5. Endophenotypes between asthma and COVID-19.
(A) A highlighted subnetwork between the asthma-associated genes, differential metabolites in asthma, and SARS-CoV-2 host genes, under the human interactome network model. (B) A heatmap highlighting differential gene expression analyses for the genes identified in asthma and COVID-19 subnetwork analysis (A). We performed differential gene expression analysis using two existing asthma cohorts (GSE63142 and GSE130499). SvsC, severe versus control; MvsC mild versus control; SvsM, severe versus mild. Blue bars show the node degree enrichment in the subnetwork (A) compared to a random network of the same size (see Methods). Black dotted line indicates enrichment = 1. (C) UMAP visualization for human bronchial epithelial cells. (D) Cell type-specific expression levels of ACE2 and TMPRSS2 across 14 cell types in human bronchial epithelial cells. (E) Cell type-specific expression levels of seven highlighted inflammatory genes (A and B) show elevated expression levels in secretory 3 cells compared to other cell types. (F) UMAP visualization for lung cells. (G) Cell type-specific expression levels of ACE2 and TMPRSS2 across 9 cell types in lung cells. (H) Cell type-specific expression levels of four highlighted inflammatory genes (A and B) show elevated expression levels in alveolar type II cells compared to other cell types.
We further examined the DEGs from two asthma cohorts from the Severe Asthma Research Program (54, 55). Utilizing two bulk RNA-Seq data sets from asthma patients and healthy controls, we identified elevated expression of IRAK3 and ADRB2 in SARS-CoV-2 infected human bronchial epithelial cells (Fig. 5B). IRAK-M, encoded by IRAK3, regulates the toll-like receptor/interleukin (IL)-1 receptor pathway and NF-κB pathway, and IRAK3 was identified as an asthma susceptibility gene (56). ADRB2 encodes the beta 2-adrenergic receptor. The polymorphisms of ADRB2 (p.Arg16Gly and p.Gln27Glu) increased the risk of asthma occurrence, and p.Gln27Glu was associated with asthma severity (57). Altogether, altered IRAK3 and ADRB2 may explain relationships between COVID-19 and asthma, though these findings require experimental and clinically validation in patients with these disorders.
To understand expressions of the proteins in the asthma-COVID-19 network across different cell types, especially those cells which express ACE2, we analyzed the single-cell RNA-Seq data from bronchial epithelium (Fig. 5C) and lung (Fig. 5F) (13). Consistent with previous studies, ACE2 and TMPRSS2 have a relatively higher expression in a subtype of the secretory cells (secretory 3 cells), compared to other bronchial epithelial cell types (Fig. 5D, Fig. S8C-F). In the lung, ACE2 and TMPRSS2 have a relatively higher expression in AT2 cells (Fig. 5G, Fig. S8H-K). We further examined the expression of the genes in the asthma-COVID-19 network (Fig. 5A) in these cell types (Fig. S9). Several genes were also more highly expressed in secretory 3 (Fig. 5E, ALOX5, IRAK3, ADRB2, TNIP1, BID, CXCL5, and NFKBIZ) and in AT2 (Fig. 5H, CFTR, NFKBIA, NFKBIZ, and TNC), than in other cell types. IRAK3 and ADRB2 are among the six overlapped genes, potentially implicating IRAK3 and ADRB2 in COVID-19-associated asthma at the single cell level as well.
Immune pathobiology shared by COVID-19 and inflammatory bowel disease
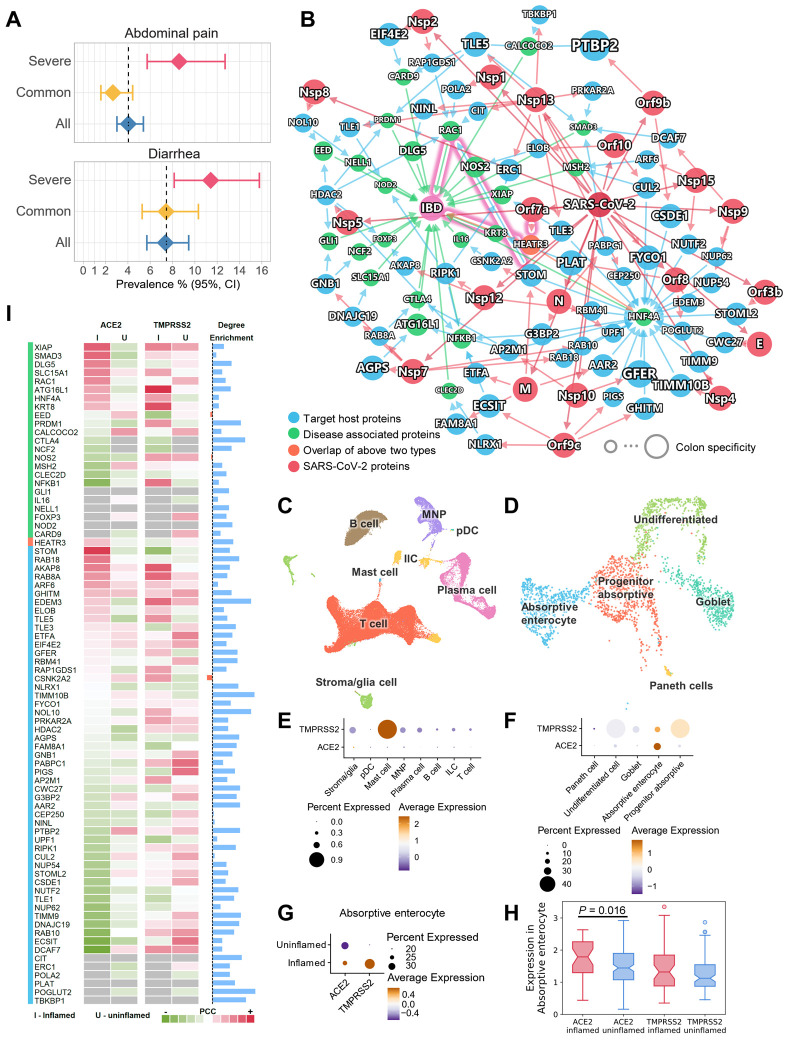
It has been shown that human small intestine is an additional SARS-CoV-2 target organ using confocal- and electron-microscopy (58). Diarrhea is now well-described as an occasional presenting symptom of COVID-19 (59). Our network proximity analysis showed a significant association of COVID-19 and IBD across all five SARS-CoV-2 data sets (Fig. 4A). In addition, severe COVID-19 patients had higher risks of abdominal pain and diarrhea (Fig. 6A, Fig. S10). To understand these associations at the cellular level, we integrated network analysis and single-cell RNA-Seq analysis using publicly available data (60). As shown in Fig. 6B, although only one IBD-associated protein, HEATR3, was found to be the target of the SARS-CoV-2 protein Orf7a, other IBD-associated proteins showed enriched number of connections to the SARS-CoV-2 target proteins (Fig. 6I, blue bars).
Fig. 6. Inflammatory endophenotype between inflammatory bowel disease (IBD) and COVID-19.
(A) Severe COVID-19 patients have higher risks of abdominal pain and diarrhea by meta-analysis. (B) A highlighted subnetwork between the IBD-associated genes and the SARS-CoV-2 virus proteins and virus-host (human) proteins under the human interactome network model. (C) UMAP visualization of non-epithelial cells from the ileal tissues of patients with Crohn’s disease. (D) UMAP visualization of epithelial cells from the ileal tissues of patients with Crohn’s disease. (E) Cell type-specific expression of ACE2 and TMPRSS2 in non-epithelial cells (C). (F) Cell type-specific expression of ACE2 and TMPRSS2 in epithelial cells (D). (G) The co-expression of ACE2 and TMPRSS2 are elevated in absorptive enterocytes of the inflamed ileal tissues compared to uninflamed tissues in patients with Crohn’s disease. (H) Box plot showing the expression of ACE2 and TMPRSS2 in absorptive enterocytes expressing ACE2 and TMPRSS2, respectively. (I) Co-expression analysis for the genes in the subnetwork with ACE2 and TMPRSS2. Heatmap shows the Pearson correlation coefficients (PCC) of ACE2 and TMPRSS2 with other genes (labeled in B) in the absorptive enterocytes. Blue bars show the degree enrichment of the genes in the subnetwork compared to a random network of the same size. Black dotted line indicates enrichment = 1.
Using single-cell data from the ileum (distal small bowel) in Crohn’s disease patients (60), we found that ACE2 and TMPRSS2 had low to undetectable expression in the non-epithelial cells (Fig. 6C, 6E, Fig. S11A-E). However, they showed higher expression levels in the epithelial cells, especially absorptive enterocytes (Fig. 6D, 6F, Fig. S11F-J). We further found that both ACE2 and TMPRSS2 had elevated expression levels in inflamed cells compared to uninflamed cells in the absorptive enterocytes (Fig. 6G, Fig. S11K, L). In absorptive enterocytes expressing ACE2, the expression of ACE2 was significantly increased (Fig. 6H, P = 0.016) in the inflamed ileal tissues of Crohn's disease patients compared to uninflamed tissues. These observations prompted us to investigate the co-expression of the network genes in the absorptive enterocytes (Fig. 6I). Several genes showed elevated co-expression with ACE2 in inflamed cells, including XIAP, SMAD3, DLG5, SLC15A1, RAC1, STOM, RAB18, and AKAP8.
We next turned to highlight two potential associations between COVID-19 and IBD. First, SARS-CoV-2 protein Orf7a can directly interact with HEATR3, whose variant was shown to be associated with increased risks of IBD by genome-wide association study (61). Second, SARS-CoV-2 infection may impact RAC1 signal transduction pathways. RAC proteins play important roles in many inflammatory pathways, and their dysregulation can be pathogenic. Increased RAC1 expression by single nucleotide polymorphisms promotes an inflammatory response in the colon (62). Mercaptopurine, an effective treatment for IBD, was found to lower RAC1 expression in IBD patients (63). Since our results show that RAC1 and ACE2 had higher co-expression in inflamed enterocytes (Fig. 6I), it is highly possible that these inflamed cells are more susceptible to SARS-CoV-2 infection, and that the infection could lead to an altered RAC1 expression level through PPIs with virus target proteins STOM, HDAC2, POLA2, CIT, and RAP1GDS1 (Fig. 6B). Notably STOM was also highly co-expressed with ACE2 in inflamed cells compared to uninflamed cells (Fig. 6I).
Network-based drug repurposing for COVID-19
Knowledge of the complex interplays between SARS-CoV-2 targets and human diseases indicate possibilities of drug repurposing, as the drugs that target other diseases could potentially target SARS-CoV-2 through the shared functional PPI networks. In addition, drug repurposing efforts may also reveal unrecognized biological connections between diseases that existing drugs treat and COVID-19. For example, the aforementioned anti-inflammatory drugs, tocilizumab and sarilumab that are now being tested for COVID-19 are originally used for rheumatoid arthritis. Although not significant, our network proximity results show that rheumatoid arthritis have small network proximities (negative Z scores) across all five SARS-CoV-2 data sets (Fig. 4A). Another drug, the thiopurine mercaptopurine which has been used to treat IBD (64), was one of the top repurposable drugs for COVID-19 proposed in our previous work (28).
Therefore, we next performed network-based drug repurposing using the existing knowledge of the drug-target network and the global map of the SARS-CoV-2 interactome built in this study. The basis for the network-based drug repurposing methodologies rests on the observation that for a drug with multiple targets to be effective against a disease, its target proteins should be within or in the immediate vicinity of the corresponding subnetwork of the disease in the human interactome, as we have demonstrated in multiple diseases previously (24, 29). Using our state-of-the-art network proximity framework, we measured the “closest” proximities of nearly 3,000 drugs and the four SARS-CoV-2 host gene/protein profiles (SARS2-DEG, SARS2-DEP, HCoV-PPI, and SARS2-PPI). Additionally, we performed gene set enrichment analysis (GSEA) using five gene/protein expression data sets, including one SARS-CoV-2 transcriptomics, one SARS-CoV-2 proteomics, one MERS-CoV and two SARS-CoV-1 transcriptomics data sets (see Methods). GSEA was used to evaluate the individual drugs for their potential to reverse the expression at the transcriptome or proteome level altered by the viruses.
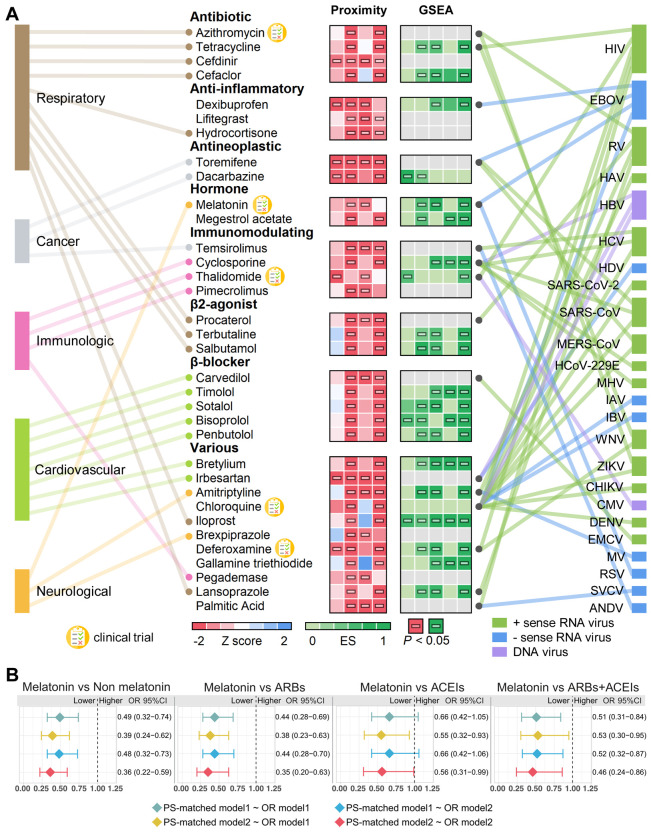
In total, we computationally identified 34 drugs that were associated (Z < −1.5 and P < 0.05, permutation test) with the SARS-CoV-2 data sets (SARS2-DEG, SARS2-DEP, HCoV-PPI, and SARS2-PPI). These drugs were significantly proximal to two or more SARS-CoV-2 host protein sets (Fig. 7A, Table S5). We manually curated their reported antiviral profiles. The disease categories that these drugs have been used to treat are also shown in the figure. Ten drugs have been used to treat respiratory-related diseases, and the most common categories for these drugs are antibiotic and β2 agonist. The next most popular disease category is cardiovascular diseases, for which seven drugs were predicted. Among the 34 drugs, three drugs achieved significant network proximity with all four SARS-CoV-2 data sets investigated here. These drugs are antibiotic drug cefdinir, which is a cephalosporin for the treatment of bacterial infections (65); antineoplastic drug toremifene, a selective estrogen receptor modulator shows striking activities in blocking various viral infections at low micromolar levels, including Ebola virus (66) (50% inhibitive concentration [IC50] = ~1 μM), MERS-CoV (67) (50% effective concentration [EC50] = 12.9 μM), SARS-CoV-1 (68) (EC50 =11.97 μM), and SARS-CoV-2 (69) (IC50 = 3.58 μM); and antihypertensive drug irbesartan, an angiotensin receptor block that can inhibit viral entry by inhibiting sodium/bile acid cotransporters (70).
Fig. 7. Network-based prediction and patient-based validation of drug repurposing for COVID-19.
(A) Thirty-four drugs from the top predicted list are highlighted with the disease category they are approved by U.S. FDA. We highlighted three types of evidences: (i) network proximities of drug’s targets across the four SARS-CoV-2 data sets (SARS2-DEG, SARS2-DEP, HCoV-PPI, and SARS2-PPI) in the human interactome; (ii) gene set enrichment analysis (GSEA) scores across five coronavirus transcriptomics and proteomics data sets (see Methods), and (iii) literature-reported antivirus profiles. GSEA scores shown in grey indicate that these drugs cannot be evaluated due to the lack of data. Five drugs that are currently being or have been tested in clinical trials are highlighted. Horizontal bars indicate P < 0.05. ES, enrichment score by GESA analysis (see Methods). HIV, human immunodeficiency virus. EBOV, Zaire ebolavirus. RV, rhinovirus. HAV, hepatitis A virus. HBV, hepatitis B virus. HCV, hepatitis C virus. HDV, hepatitis D virus. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2. SARS-CoV, severe acute respiratory syndrome coronavirus. MERS-CoV, Middle East respiratory syndrome coronavirus. HCoV-229E, human coronavirus 229E. MHV, mouse hepatitis virus. IAV, influenza A virus. IBV, influenza B virus. WNV, West Nile virus. ZIKV, Zika virus. ANDV, Andes virus. CHIKV, chikungunya virus. CMV, cytomegalovirus. DENV, dengue virus. EMCV, encephalomyocarditis virus. MV, measles virus. RSV, respiratory syncytial virus. SVCV, spring viremia of carp virus. (B) Observational studies highlighting that patients taking melatonin were associated with reduced risk of likelihood of a positive laboratory test result for SARS-CoV-2 from the COVID-19 registry. Melatonin usage also reduced the risk compared to angiotensin-converting enzyme inhibitors (ACEIs), angiotensin II receptor blockers (ARBs), and the pooled patient data of ARBs/ACEIs. PS, propensity score. OR, odds ratio. PS-matched model1 was matched using age, gender, race, and smoking; PS-matched model2 was matched using age, gender, race, smoking, coronary artery disease, diabetes, hypertension, and chronic obstructive pulmonary disease (COPD). OR model1, the odds ratios of COVID-19 in melatonin exposure; OR model2, the odds ratios of COVID-19 in melatonin exposure adjusted by age, gender, race, smoking, coronary artery disease, diabetes, hypertension, and COPD.
Validating drug-outcome relationships on COVID-19 using patient data
We next evaluated drug-outcome relationship using a large-scale patient data from the Cleveland Clinic COVID-19 patient registry (see Methods). We used subject matter expertise based on: (i) literature-reported antiviral effects for human coronaviruses or other types of human viruses; (ii) strength of network proximity prediction and GSEA analysis (Fig. 7A); and (iii) availability of sufficient patient data for meaningful evaluation (exclusion of infrequently used drugs). Applying these criteria resulted in identifying melatonin, a physiologic hormone common to many living organisms.
A retrospective COVID-19 cohort analysis was conducted to validate the potential prevention effect of melatonin (Fig. 7B). Among a total of 18,118 patients tested for COVID-19 in the Cleveland Clinic Health System in Ohio and Florida, 1,675 patients were diagnosed with COVID-19 between March 8 and April 16, 2020 (Table 1). We found that melatonin usage was associated with a 50-60% reduced likelihood of a positive laboratory test result for SARS-CoV-2 (OR = 0.36, 95% CI 0.22-0.59, Fig. 7B) after adjusting for age, sex, race, and various disease comorbidities (diabetes, hypertension, heart disease, and pulmonary conditions, see Methods) using a propensity score (PS) matching method. Angiotensin-converting enzyme inhibitors (ACEIs) and angiotensin II receptor blockers (ARBs) are two common types of drugs for treatment of hypertension. A recent study showed that inpatient use of ACEI/ARB was associated with lower risk of all-cause mortality compared with ACEI/ARB non-users hospitalized COVID-19 patients with hypertension (71). We further performed an observational study for three cohorts using user active comparator design and PS adjustment for confounding factors as described in our previous study (24). We found that melatonin usage was associated with reduced risk of likelihood of a positive laboratory test result for SARS-CoV-2 compared to ARBs (OR = 0.35, 95% CI 0.20-0.63) and ACEIs (OR = 0.56, 95% CI 0.31-0.99), and the pooled data of ARBs and ACEIs (OR = 0.46, 95% CI 0.24-0.86) (Fig. 7B). Altogether, network-based prediction (Fig. 7A) and multiple observational analyses (Fig. 7B) suggest potential of treatment for COVID-19 by melatonin; yet, randomized controlled clinically trials are urgently needed to test meaningfully the effect of melatonin for COVID-19 further.
Table 1.
Baseline characteristics of the melatonin and carvedilol intaking patient groups from the COVID-19 registry.
| Melatonin | ||||||
|---|---|---|---|---|---|---|
| All patients | SARS-CoV-2 positive patients | |||||
| Melatonin − | Melatonin + | P | Melatonin − | Melatonin + | P | |
| Total patients | 17312 | 806 | 1635 | 40 | ||
| Age | 48.94 ± 20.49 | 62.84 ± 19.88 | < 0.001 | 53.63 ± 18.89 | 65.24 ± 18.44 | < 0.001 |
| Race other % | 1092 (6.3) | 43 (5.3) | 0.298 | 109 (6.7) | 1 (2.5) | 0.467 |
| Race black % | 3317 (19.2) | 202 (25.1) | < 0.001 | 380 (23.2) | 13 (32.5) | 0.239 |
| Race white % | 11922 (68.9) | 538 (66.7) | 0.219 | 1044 (63.9) | 23 (57.5) | 0.51 |
| Gender (male) % | 6833 (39.5) | 396 (49.1) | < 0.001 | 785 (48.0) | 21 (52.5) | 0.688 |
| Smoking % | 2222 (12.8) | 129 (16.0) | 0.258 | 91 (5.6) | 3 (7.5) | 0.743 |
| Diabetes % | 3222 (18.6) | 333 (41.3) | < 0.001 | 336 (20.6) | 13 (32.5) | 0.456 |
| Hypertension % | 6524 (37.7) | 594 (73.7) | < 0.001 | 662 (40.5) | 27 (67.5) | 0.029 |
| Coronary artery disease % | 1897 (11.0) | 268 (33.3) | < 0.001 | 152 (9.3) | 12 (30.0) | 0.001 |
| COPD & emphysema % | 740 (4.3) | 119 (14.8) | < 0.001 | 76 (4.6) | 5 (12.5) | 0.07 |
| ACEIs (%) | 1182 (6.8) | 156 (19.4) | < 0.001 | 93 (5.7) | 5 (12.5) | 0.141 |
| ARBs (%) | 900 (5.2) | 123 (15.3) | < 0.001 | 96 (5.9) | 8 (20.0) | 0.001 |
| Carvedilol | ||||||
| All patients | SARS-CoV-2 positive patients | |||||
| Carvedilol − | Carvedilol + | P | Carvedilol − | Carvedilol + | P | |
| Total patients | 17567 | 551 | 1647 | 28 | ||
| Age | 49.06 ± 20.59 | 65.46 ± 16.22 | < 0.001 | 53.67 ± 18.94 | 68.16 ± 13.52 | < 0.001 |
| Race other % | 1102 (6.3) | 33 (6.0) | 0.856 | 109 (6.6) | 1 (3.6) | 0.794 |
| Race black % | 3333 (19.0) | 186 (33.8) | < 0.001 | 379 (23.0) | 14 (50.0) | 0.002 |
| Race white % | 12134 (69.1) | 326 (59.2) | < 0.001 | 1054 (64.0) | 13 (46.4) | 0.086 |
| Gender (male) % | 6916 (39.4) | 313 (56.8) | < 0.001 | 789 (47.9) | 17 (60.7) | 0.248 |
| Smoking % | 2243 (12.8) | 108 (19.6) | 0.001 | 90 (5.5) | 4 (14.3) | 0.108 |
| Diabetes % | 3235 (18.4) | 320 (58.1) | < 0.001 | 333 (20.2) | 16 (57.1) | < 0.001 |
| Hypertension % | 6601 (37.6) | 517 (93.8) | < 0.001 | 662 (40.2) | 27 (96.4) | < 0.001 |
| Coronary artery disease % | 1871 (10.7) | 294 (53.4) | < 0.001 | 148 (9.0) | 16 (57.1) | < 0.001 |
| COPD & emphysema % | 791 (4.5) | 68 (12.3) | < 0.001 | 74 (4.5) | 7 (25.0) | 0.001 |
| ACEIs (%) | 1190 (6.8) | 148 (26.9) | < 0.001 | 93 (5.6) | 5 (17.9) | 0.02 |
| ARBs (%) | 884 (5.0) | 139 (25.2) | < 0.001 | 93 (5.6) | 11 (39.3) | < 0.001 |
COPD, chronic obstructive pulmonary disease. ACEIs, angiotensin-converting enzyme inhibitors. ARBs, angiotensin II receptor blockers. Ages are shown as mean ± standard deviation. All other characteristics are shown as number of cases (percentage). P values were calculated by Fisher's exact test. +: drug usage; −: non-drug usage.
Discussion
Recent studies indicated that SARS-CoV-2 infection was detected in multiple organs in addition to lungs, including heart, pharynx, liver, kidneys, brain, and intestine (58, 72). SARS-CoV-2 RNA was also found in patient stool (73). Therefore, investigation of how SARS-CoV-2 associates with other diseases could help reveal and understand its impact on systems and organs in addition to lungs. In this study, we systematically evaluated 64 diseases across 6 categories for their potential manifestations with COVID-19. We started with assembling and characterizing five SARS-CoV-2 data sets representing different cellular event levels including transcriptome, proteome, and interactome. Using state-of-the-art network proximity measure, we identified broad manifestations (such as autoimmune, neurological, and pulmonary, Fig. 4A) associated with COVID-19. Retrospective meta-analyses using the clinical data of 4,973 patients across 34 studies confirmed our network-based findings. We further investigated the molecular mechanisms of the association of COVID-19 and the comorbidities by multi-modal analyses of large-scale single cell data. We identified the cell types that have the highest expression levels of ACE2 and TMPRSS2: the lung AT2 cells, secretory bronchial epithelial cells, and absorptive enterocytes in ileum. We examined the expression of asthma- and IBD-associated genes in their relevant cell types. Combining these results with the results of differential expression analysis, network connectivity, and differential metabolites, we identified several key pathogenic pathways (including IRAK3 and ADRB2) that can be altered by the viral infection for asthma.
For asthma, our network-based findings suggest several possible shared pathobiological pathways associated with COVID-19. First, SARS-CoV-2 infection might alter the expression of several key inflammatory genes: IRAK3, which is associated with asthma (56, 74, 75); ADRB2, which is an essential genetic factor for asthma (57, 76); and NFKBIA, which showed critical transcriptional responses in childhood asthma (77). These genes show high expression in secretory 3 and AT2 cell types, suggesting a higher susceptibility to be impacted by SARS-CoV-2 infection through ACE2. SARS-CoV-2 increased the expression of IRAK3 and ADRB2, which lead to a higher risk of asthma (Fig. 5B). Second, decreased levels of L-arginine and L-citrulline were found in SARS-CoV-2 infected patients (50), while it has been shown that higher levels of these metabolites are protective against asthma (49). L-arginine can be converted to nitric oxide by the nitric oxide synthases, and it was shown that nitric oxide protects against viral infection through multiple potential mechanisms (78).
Our network proximity results show a strong connection between IBD and COVID-19 (Fig. 4A). We have also showed IBD-related pathways that can potentially be affected by SARS-CoV-2 infection (Fig. 6B). However, we should also note that the meta-analysis does not show a significant risk ratio of digestive system disease in COVID-19 patients (Fig. 3B), while two specific symptoms, abdominal pain and diarrhea, showed increased risks in severe COVID-19 patients (Fig. 6A). Digestive system disease covers a broad range of gastrointestinal diseases. Future studies are needed to reveal and validate the associations of COVID-19 and individual gastrointestinal diseases.
Finally, we computationally prioritized nearly 3,000 FDA approved/investigational drugs for their potential anti-SARS-CoV-2 effects using network proximity measure and GSEA. A list of 34 repurposable drugs with their reported antiviral profiles are highlighted, among which five drugs are currently in clinical trials (Fig. 7B). We further explored drug-disease outcome for melatonin using a large-scale COVID-19 registry. We found that among individuals who received testing for SARS-CoV-2, melatonin usage was associated with a 50-60% reduced likelihood of a positive laboratory test result for SARS-CoV-2 (Fig. 7B). Using user active comparator design, we further discovered that melatonin usage was associated with reduced risk of likelihood of a positive laboratory test result for SARS-CoV-2 compared to ARBs and ACEIs as well. Exogenous melatonin may be of benefit in older patients with COVID-19 given aging-related reduction of endogenous melatonin and vulnerability of the elderly to lethality of SARS-CoV-2 (79), the latter potentially due to declining immunity, i.e. immunosenescence (80). Moreover, melatonin suppresses NLRP3 inflammasome activation induced by cigarette smoking and attenuates pulmonary inflammation (81), not only via reduction of nuclear factor kappa-β p65 (NF-κB p65) and tumor necrosis factor-α (TNF-α) expression, but also via increase in anti-inflammatory cytokines such as IL-10 or IL-6 which can also have anti-inflammatory effects (82, 83). Thus, large-scale observational studies and randomized controlled trials are needed to validate the clinical benefit of melatonin for patients with COVID-19. It would be important, however, that the trials are designed with the understanding of the mechanism of the drug to be repurposed. For example, it would be obvious that drugs that decrease viral entry, e.g., part of melatonin’s action, would be beneficial in preventing infection or very early in the COVID-19 course, but may be inconsequential when utilized in severe or end-stage infection.
We acknowledge several potential limitations. First, although we integrated data from multiple sources to build the human interactome and the drug target network, they are still incomplete. Second, this study relied on the SARS-CoV-2 target host genes/proteins data sets, and their quality may influence the performance of our network analysis. Third, the patient-data analysis is retrospective and may have selection bias. Although we performed multiple types of PS-matching observations, residual confounding is possible despite high-dimensional covariate adjustment. For example, carvedilol, approved for both hypertension and heart failure, was identified for potential treatment of COVID-19 by network proximity measure (Fig. 7A). We found that carvedilol usage reduces the likelihood of a positive laboratory test result for SARS-CoV-2 (OR = 0.43, 95% CI 0.27-0.70, Fig. S12) after adjusting for age, gender, race, smoking history using PS method. Yet, this association does not meet statistical significance after further adjusting for multiple disease comorbidities (including coronary artery disease, diabetes, hypertension, and COPD) (OR = 0.58, 95% CI 0.32-1.05, Fig. S12). There are two possible explanations: (1) the small number of carvedilol usage may lack the statistical power for current observational study; (2) carvedilol may only reduce the likelihood of a positive laboratory test result for SARS-CoV-2 for patients having existing health conditions, such as hypertension. Replication of the associations and causal inference using large databases may rule out treatment effect heterogeneity and possible confounding further. Finally, although we made the intriguing observation that the use of melatonin was much less prevalent in individuals testing positive for SARS-CoV-2, we recognize that many asymptomatic or minimally symptomatic persons with the virus were not tested and therefore, their use of melatonin was not evaluable; melatonin use in this latter group might also have been high. It should therefore be noted that all drugs we identified as therapeutic candidates in this study must be validated using experimental assays and randomized clinical trials before they can be recommended for use in patients with COVID-19.
In conclusion, our study provides a powerful, integrative, network medicine strategy for advancing understanding of COVID-19-associated comorbidities and facilitating the identification of drug candidates for COVID-19. This approach also promises to address the translational gap between genomic studies and clinical outcomes, which poses a significant problem when rapid development of effective therapeutic interventions is critical during a pandemic. From a translational perspective, if broadly applied, the network-medicine tools applied here could prove helpful in developing effective treatment strategies for other complex human diseases as well, including other emerging infectious diseases.
Methods and Materials
Building the data sets of SARS-CoV-2 target host genes/proteins
We assembled four SARS-CoV-2 data sets of target host genes/proteins: (1) 246 differentially expressed genes in human bronchial epithelial cells infected with SARS-CoV-2 (18) (GSE147507), denoted as SARS2-DEG; (2) 293 differentially expressed proteins in human Caco-2 cells infected with SARS-CoV-2 (19), denoted as SARS2-DEP; (3) 134 strong literature evidence-based pan-human coronavirus target host proteins from our recent study (28) with 15 newly curated proteins, denoted as HCoV-PPI; (4) 332 proteins involved in the protein-protein interactions with 26 SARS-CoV-2 viral proteins identified by affinity purification-mass spectrometry (20), denoted as SARS2-PPI. Finally, due to the interactome nature of HCoV-PPI and SARS2-PPI, we combined these data sets as the fifth SARS-CoV-2 data set, which has 460 proteins and is denoted as PanCoV-PPI.
Building the disease gene profiles
We compiled the disease-associated gene sets from various sources. All databases were accessed on March 26, 2020.
Cancer.
The pan-cancer driver genes were retrieved from the Cancer Gene Census (35). Driver genes for individual cancer types were from a previous study (36).
Mendelian disease genes (MDGs).
A set of 2,272 MDGs were retrieved from the Online Mendelian Inheritance in Man (OMIM) database (84).
Orphan disease-causing mutant genes (ODMGs).
A set of 2,124 ODMGs were retrieved from a previous study (85).
Cell cycle genes.
910 human cell cycle genes were downloaded from a previous study identified by a genome-wide RNAi screening (86).
Innate immune genes.
1,031 human innate immunity genes were collected form InnateDB (87).
Genes associated with autoimmune, pulmonary, neurological, cardiovascular, and metabolic diseases.
The disease-associated genes/proteins were extracted from the Human Gene Mutation Database (HGMD) (37). A systematic search was manually performed for each disease using keywords such as disease name, alias, and symptoms to identify the disease terms in the HGMD. All disease terms were then verified for their relevance.
Functional enrichment analysis
We performed Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) biological process enrichment analyses to reveal the biological relevance and functional pathways of the five SARS-CoV-2 data sets. All functional enrichment analyses were performed using Enrichr (88).
Selective pressure and evolutionary rates characterization
We calculated the dN/dS ratio (89) and the evolutionary rate ratio (90) as described in our previous study (91).
Tissue specificity analysis
The RNA-Seq data (transcripts per million, TPM) of 31 tissues from the GTEx V8 release (accessed on March 31, 2020, https://www.gtexportal.org/home/) were downloaded. Genes with count per million (CPM) ≥ 0.5 in over 90% samples in a tissue were considered tissue-expressed genes and otherwise as tissue-unexpressed. To quantify the expression specificity of gene i in tissue t, we calculated the mean expression Ei and the standard deviation σi of a gene’s expression across all considered tissues. The significance of gene expression specificity in a tissue is defined as:
| (1) |
Risk ratio analysis for COVID-19 patients
PubMed, Embase and Medrxiv databases were searched for publications as of April 25th, 2020 (Fig. S13). The search was limited to English articles describing the demographic and clinical features of SARS-CoV-2 cases. We used the search term (“SARS-COV-2” OR “COVID-19” OR “nCoV 19” OR “2019 novel coronavirus” OR “coronavirus disease 2019”) AND (“clinical characteristics” OR “clinical outcome” OR “comorbidities”). Only research articles were included; reviews, case reports, comments, editorials, and expert opinions were excluded. Three criteria were used to select studies from a total of 1,054 initial hits: (1) studies that have ≥ 20 COVID-19 patients; (2) studies that grouped the outcomes with varying degree of severity of COVID-19 (e.g., severe vs. non-severe) according to the American Thoracic Society guidelines for community-acquired pneumonia; and (3) studies that were from different institutions. Two criteria were used for exclusion: (1) studies that focus on specific populations (e.g., only death cases, pregnant women, children or from family clusters); and (2) basic molecular biology research. Finally, 34 studies meeting these criteria were used for further analyses.
We performed random effects meta-analysis to estimate the pooled risk ratio with 95% CI of 10 comorbidities between severe versus non-severe COVID-19 patients. Mantel-Haenszel method was used to estimate the pooled effects of results (92). DerSimonian-Laird method was used to estimate the variance among studies (93). The continuous data such as IL-6 levels were transformed to mean and standard deviations first using Wan’s approach based on sample size, median and interquartile range (94). Next, we used inverse variance method to estimate the pooled mean difference and estimated the variance among studies by DerSimonian-Laird method. We estimated the pooled prevalence of 3 COVID-19 symptoms (abdominal pain, diarrhea, dyspnea) and 1 comorbidity (COPD) in three COVID-19 patient groups (severe, non-severe, and all) respectively. Random intercept logistic regression model was used to estimate pooled prevalence and maximum-likelihood estimator was used to quantify the heterogeneity of studies (95). The tau2 and I2 statistics were calculated for the heterogeneity among studies. We considered I2 < 50% as low heterogeneity among studies, 50%< I2 < 75% as moderate heterogeneity, and I2 > 75% as high heterogeneity. All meta-analyses were conducted by meta and dmetar packages in the R v3.6.3 platform.
Building the human protein-protein interactome
A total of 18 bioinformatics and systems biology databases were assembled to build a comprehensive list of human PPIs with five types of experimental evidences: (1) binary, physical PPIs from protein three-dimensional (3D) structures; (2) binary PPIs tested by high-throughput yeast-two-hybrid (Y2H) systems; (3) kinase-substrate interactions by literature-derived low-throughput or high-throughput experiments; (4) signaling network by literature-derived low-throughput experiments; and (5) literature-curated PPIs identified by affinity purification followed by mass spectrometry (AP-MS), Y2H, or by literature-derived low-throughput experiments. Inferred PPIs based on gene expression data, evolutionary analysis, and metabolic associations were excluded. Genes were mapped to their Entrez ID based on the NCBI database (96). The official gene symbols were based on GeneCards (https://www.genecards.org/). The final human protein-protein interactome used in this study included 351,444 unique PPIs (edges or links) connecting 17,706 proteins (nodes). Detailed descriptions for building the human protein-protein interactome are provided in our previous studies (24, 29, 30, 97).
Network proximity measure
We used the “closest” network proximity measure throughout this study. For two gene/protein sets A and B, their “closest” distance dAB was calculated as:
| (2) |
where d(a, b) is the shortest distance of a and b in the human interactome. To evaluate the significance, we performed a permutation test using randomly selected proteins from the whole interactome that were representative of the two protein sets being evaluated in terms of their degree distributions. We then calculated the Z score as:
| (3) |
where and σr were the mean and standard deviation of the permutation test. All network proximity permutation tests in this study were repeated 1,000 times.
Network-based comorbidity analysis
To reveal potential COVID-19 comorbidities, we computed the network proximity of the disease-associated proteins for each disease and the five SARS-CoV-2 data sets. SARS-CoV-2 target proteins with a non-negative tissue specificity in lung were used in the computation. The degree enrichment for protein i in a subnetwork was calculated as:
| (4) |
where di is the degree of i in the subnetwork, n is number of nodes in the subnetwork, Di is the degree in the complete human protein interactome, and N is the total number of nodes in the interactome.
Bulk and single-cell RNA-Seq data analysis
Bulk RNA-Seq data sets for asthma patients were retrieved from the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/) using the accession number GSE63142 (54) and GSE130499 (55). Differential expression of three comparisons, severe vs. control, mild vs. control, and severe vs. mild were performed using the GEO2R function. All single-cell data analyses and visualizations were performed with the R package Seurat v3.1.4 (98). GSE134809 (60) was downloaded from the NCBI GEO database. This data set contains 67,050 inflamed and uninflamed cells from the ileal samples of 8 patients with Crohn’s disease. Single-cell data of normal lung and primary human bronchial epithelial cells were downloaded from https://data.mendeley.com/datasets/7r2cwbw44m/1. These data sets contain 39,778 lung cells and 17,451 bronchial epithelial cells. Qualifying cells based on the criteria from the original paper were used for the analysis. “NormalizeData” was used to normalize the data. “FindIntegrationAnchors” and “IntegrateData” functions were used to integrate cells from different samples. UMAP was used as the dimension reduction method for visualization. Cell type markers from the original paper were used to label the cell types.
Building the drug-target network
To evaluate whether a drug is closely associated with SARS-CoV-2 target proteins in the human interactome, we gathered the drug-target interaction information from several databases: DrugBank database (v4.3) (99), Therapeutic Target Database (TTD) (100), PharmGKB database, ChEMBL (v20) (101), BindingDB (102), and IUPHAR/BPS Guide to PHARMACOLOGY (103). We included the interactions that have binding affinities Ki, Kd, IC50 or EC50 ≤ 10 μM and a unique UniProt accession number with “reviewed” status. The details for building the experimentally validated drug-target network can be found in our recent studies (24, 29, 30).
Network-based drug repurposing
We computed the “closest” network proximity as described before for 2,938 FDA approved or investigational drugs and the five SARS-CoV-2 data sets. For prioritization, we ranked the drugs by their distance to the data sets (D < 2) and Z score (Z < −1.5) from the network proximity analysis. The antiviral profiles of the highlighted drugs were manually curated. COVID-19 related clinical trials were retrieved on May 10, 2020.
Gene set enrichment analysis (GSEA)
The gene set enrichment analysis was conducted as described in our recent work (28) as an additional evidence for drug repurposing. Briefly, for each drug and coronavirus target gene set, we computed an enrichment score to indicate whether a drug can reverse the effect of SARS-CoV-2 at the transcriptome or proteome level. Gene expression profiles for the drugs were retrieved from the Connectivity Map (CMAP) database (104). Five gene sets were evaluated: (1) the differentially expressed genes in human bronchial epithelial cells infected with SARS-CoV-2 (18) (GSE147507); (2) the differentially expressed proteins in human Caco-2 cells infected with SARS-CoV-2 (19); (3, 4) two transcriptome data sets of SARS-CoV-1 infected samples from patient’s peripheral blood (105) (GSE1739) and Calu-3 cells (106) (GSE33267) respectively; (5) the differentially expressed genes in MERS-CoV infected Calu-3 cells (107) (GSE122876).
The enrichment score ES was calculated for up- and down- regulated genes separately first. The overall ES was calculated as:
| (5) |
where ESup and ESdown were calculated using aup/down and bup/down as:
| (6) |
| (7) |
j = 1,2, ⋯ , s were the genes in the gene sets sorted in ascending order by their rank in the drug profiles. V(j) indicates the rank of gene j, where 1 ≤ V(j) ≤ r, with r being the number of genes from the drug profile. Then, ESup/down was set to aup/down if aup/down > bup/down, and was set to −bup/down if bup/down > aup/down. Permutation test was performed to evaluate the significance. Drugs were considered to have potential treatment effect if ES > 0 and P < 0.05.
Patient data validation of the network-identified drugs using a COVID-19 registry
We used the institutional review board–approved registry COVID-19 registry data, including 18,118 individuals (1,675 positive) tested during March 8 to April 16, 2020 from the Cleveland Clinic Health System in Ohio and Florida. Data include COVID-19 test results, baseline demographic information, medications, and all recorded disease conditions and others. We conducted a series of retrospective case-control studies with an active comparator new user design to test the drug-outcome relationships for COVID-19. Data were extracted from electronic health records (EPIC Systems) and were manually checked by a study team trained on uniform sources for the study variables. We collected and managed all patient data using REDCap electronic data capture tools. The exposures of drugs (including carvedilol and melatonin) were used as recorded in the medication list in the electronic medical records at the time of testing for SARS-CoV-2. A positive laboratory test result for COVID-19 was used as the primary outcome. A propensity score (PS) was calculated for melatonin and carvedilol intake by multivariable logistic regression models with two sets of covariates: (1) PS-matched model1, age, gender, race and smoking; (2) PS-matched model2, age, gender, race, smoking, coronary artery disease, diabetes, hypertension and COPD. The nearest-neighbor algorithm was used to match the melatonin and carvedilol subjects and references subjects on the basis of PS ratio of 1:4. Two odds ratios (OR) models were used to estimate the COVID-19 positive risk between melatonin or carvedilol intaking groups and non-intaking groups: (1) OR model1, COVID-19 ~ melatonin/carvedilol; (2) OR model2 (adjusted OR model), COVID-19 ~ melatonin/carvedilol + age, gender, race, smoking, coronary artery disease, diabetes, hypertension and COPD. All analyses were conducted by matchit package in the R v3.6.3 platform.
Statistical analysis and network visualization
Statistical tests were performed with the Python package SciPy v1.3.0 (https://www.scipy.org/). P < 0.05 was considered statistically significant throughout this study. Networks were visualized using Gephi 0.9.2 (https://gephi.org/).
Acknowledgements
We thank all helpful discussions and critical comments regarding this manuscript from the COVID-19 Research Intervention Advisory Committee members at the Cleveland Clinic. SCE is the Alfred Lerner Memorial Chair of Innovative Research and CE is the Sondra J. and Stephen R. Hardis Endowed Chair of Cancer Genomic Medicine at the Cleveland Clinic.
Funding: This work was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health (NIH) under Award Number R00 HL138272 and the National Institute of Aging under Award Number R01AG066707 to F.C. This work was supported, in part, by the VeloSano Pilot Program (Cleveland Clinic Taussig Cancer Institute).
Footnotes
Competing interests: The authors declare that they have no conflict of interest. The content of this publication does not necessarily reflect the views of the Cleveland Clinic.
Data and code availability: The bulk and single-cell RNA-Seq data used in this study were downloaded from the NCBI GEO database with accession numbers GSE63142, GSE130499, and GSE134809. The lung and human bronchial epithelial single-cell data were downloaded from https://data.mendeley.com/datasets/7r2cwbw44m/1. Source code and other supporting data can be downloaded from https://github.com/ChengF-Lab/COVID-19_Map.
References
- 1.Bhatraju P. K., Ghassemieh B. J., Nichols M., Kim R., Jerome K. R., Nalla A. K., Greninger A. L., Pipavath S., Wurfel M. M., Evans L., Kritek P. A., West T. E., Luks A., Gerbino A., Dale C. R., Goldman J. D., O'Mahony S., Mikacenic C., Covid-19 in Critically Ill Patients in the Seattle Region - Case Series. The New England journal of medicine, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan W. J., Ni Z. Y., Hu Y., Liang W. H., Ou C. Q., He J. X., Liu L., Shan H., Lei C. L., Hui D. S. C., Du B., Li L. J., Zeng G., Yuen K. Y., Chen R. C., Tang C. L., Wang T., Chen P. Y., Xiang J., Li S. Y., Wang J. L., Liang Z. J., Peng Y. X., Wei L., Liu Y., Hu Y. H., Peng P., Wang J. M., Liu J. Y., Chen Z., Li G., Zheng Z. J., Qiu S. Q., Luo J., Ye C. J., Zhu S. Y., Zhong N. S., Clinical Characteristics of Coronavirus Disease 2019 in China. The New England journal of medicine, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z., Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. Jama, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y., Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. The Lancet. Respiratory medicine, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B., Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet (London, England) 395, 1054–1062 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L., Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet (London, England) 395, 507–513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou P., Yang X. L., Wang X. G., Hu B., Zhang L., Zhang W., Si H. R., Zhu Y., Li B., Huang C. L., Chen H. D., Chen J., Luo Y., Guo H., Jiang R. D., Liu M. Q., Chen Y., Shen X. R., Wang X., Zheng X. S., Zhao K., Chen Q. J., Deng F., Liu L. L., Yan B., Zhan F. X., Wang Y. Y., Xiao G. F., Shi Z. L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., Tan W., China Novel Coronavirus I., Research T., A Novel Coronavirus from Patients with Pneumonia in China, 2019. The New England journal of medicine 382, 727–733 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N.-H., Nitsche A., Müller M. A., Drosten C., Pöhlmann S., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271–280.e278 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann M., Kleine-Weber H., Pöhlmann S., A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Molecular Cell, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziegler C. G. K., Allon S. J., Nyquist S. K., Mbano I. M., Miao V. N., Tzouanas C. N., Cao Y., Yousif A. S., Bals J., Hauser B. M., Feldman J., Muus C., Wadsworth M. H., Kazer S. W., Hughes T. K., Doran B., Gatter G. J., Vukovic M., Taliaferro F., Mead B. E., Guo Z., Wang J. P., Gras D., Plaisant M., Ansari M., Angelidis I., Adler H., Sucre J. M. S., Taylor C. J., Lin B., Waghray A., Mitsialis V., Dwyer D. F., Buchheit K. M., Boyce J. A., Barrett N. A., Laidlaw T. M., Carroll S. L., Colonna L., Tkachev V., Peterson C. W., Yu A., Zheng H. B., Gideon H. P., Winchell C. G., Lin P. L., Bingle C. D., Snapper S. B., Kropski J. A., Theis F. J., Schiller H. B., Zaragosi L.-E., Barbry P., Leslie A., Kiem H.-P., Flynn J. L., Fortune S. M., Berger B., Finberg R. W., Kean L. S., Garber M., Schmidt A. G., Lingwood D., Shalek A. K., Ordovas-Montanes J., SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., Worlock K. B., Yoshida M., Barnes J. L., Banovich N. E., Barbry P., Brazma A., Collin J., Desai T. J., Duong T. E., Eickelberg O., Falk C., Farzan M., Glass I., Gupta R. K., Haniffa M., Horvath P., Hubner N., Hung D., Kaminski N., Krasnow M., Kropski J. A., Kuhnemund M., Lako M., Lee H., Leroy S., Linnarson S., Lundeberg J., Meyer K. B., Miao Z., Misharin A. V., Nawijn M. C., Nikolic M. Z., Noseda M., Ordovas-Montanes J., Oudit G. Y., Pe’er D., Powell J., Quake S., Rajagopal J., Tata P. R., Rawlins E. L., Regev A., Reyfman P. A., Rozenblatt-Rosen O., Saeb-Parsy K., Samakovlis C., Schiller H. B., Schultze J. L., Seibold M. A., Seidman C. E., Seidman J. G., Shalek A. K., Shepherd D., Spence J., Spira A., Sun X., Teichmann S. A., Theis F. J., Tsankov A. M., Vallier L., van den Berge M., Whitsett J., Xavier R., Xu Y., Zaragosi L.-E., Zerti D., Zhang H., Zhang K., Rojas M., Figueiredo F., Network H. C. A. L. B., SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nature Medicine, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukassen S., Chua R. L., Trefzer T., Kahn N. C., Schneider M. A., Muley T., Winter H., Meister M., Veith C., Boots A. W., Hennig B. P., Kreuter M., Conrad C., Eils R., SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J, e105114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H., Kang Z., Gong H., Xu D., Wang J., Li Z., Li Z., Cui X., Xiao J., Zhan J., Meng T., Zhou W., Liu J., Xu H., Digestive system is a potential route of COVID-19: an analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut, gutjnl-2020–320953 (2020). [Google Scholar]
- 15.Yang S., Fu C., Lian X., Dong X., Zhang Z., Understanding Human-Virus Protein-Protein Interactions Using a Human Protein Complex-Based Analysis Framework. mSystems 4, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu C., Ma Y., Zhao J., Nussinov R., Zhang Y., Cheng F., Zhang Z., Computational network biology: Data, models, and applications. DOI: 10.1016/j.physrep.2019.12.004 Physics Reports, (2020). [DOI] [Google Scholar]
- 17.Zumla A., Chan J. F., Azhar E. I., Hui D. S., Yuen K. Y., Coronaviruses - drug discovery and therapeutic options. Nat Rev Drug Discov 15, 327–347 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanco-Melo D., Nilsson-Payant B. E., Liu W.-C., Møller R., Panis M., Sachs D., Albrecht R. A., tenOever B. R., SARS-CoV-2 launches a unique transcriptional signature from in vitro, ex vivo, and in vivo systems. bioRxiv, 2020.2003.2024.004655 (2020). [Google Scholar]
- 19.Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., Cinatl J., Munch C., Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordon D. E., Jang G. M., Bouhaddou M., Xu J., Obernier K., White K. M., O'Meara M. J., Rezelj V. V., Guo J. Z., Swaney D. L., Tummino T. A., Huettenhain R., Kaake R. M., Richards A. L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B. J., Braberg H., Fabius J. M., Eckhardt M., Soucheray M., Bennett M. J., Cakir M., McGregor M. J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z. Z. C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I. T., Melnyk J. E., Chorba J. S., Lou K., Dai S. A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C. J. P., Perica T., Pilla K. B., Ganesan S. J., Saltzberg D. J., Rakesh R., Liu X., Rosenthal S. B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X. P., Liu Y., Wankowicz S. A., Bohn M., Safari M., Ugur F. S., Koh C., Savar N. S., Tran Q. D., Shengjuler D., Fletcher S. J., O'Neal M. C., Cai Y., Chang J. C. J., Broadhurst D. J., Klippsten S., Sharp P. P., Wenzell N. A., Kuzuoglu D., Wang H. Y., Trenker R., Young J. M., Cavero D. A., Hiatt J., Roth T. L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R. M., Frankel A. D., Rosenberg O. S., Verba K. A., Agard D. A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H. S., Fujimori D. G., Ideker T., Craik C. S., Floor S. N., Fraser J. S., Gross J. D., Sali A., Roth B. L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., Garcia-Sastre A., Shokat K. M., Shoichet B. K., Krogan N. J., A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature, doi: 10.1038/s41586-020-2286-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng F., In Silico Oncology Drug Repositioning and Polypharmacology. Methods Mol Biol 1878, 243–261 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Cheng F., Hong H., Yang S., Wei Y., Individualized network-based drug repositioning infrastructure for precision oncology in the panomics era. Brief Bioinform 18, 682–697 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Cheng F., Murray J. L., Rubin D. H., Drug Repurposing: New Treatments for Zika Virus Infection? Trends Mol Med 22, 919–921 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Cheng F., Desai R. J., Handy D. E., Wang R., Schneeweiss S., Barabasi A. L., Loscalzo J., Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat Commun 9, 2691 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng F., Murray J. L., Zhao J., Sheng J., Zhao Z., Rubin D. H., Systems Biology-Based Investigation of Cellular Antiviral Drug Targets Identified by Gene-Trap Insertional Mutagenesis. PLoS Comput Biol 12, e1005074 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F. G., Horby P. W., Zhang D., Wang C., A Trial of Lopinavir–Ritonavir in Adults Hospitalized with Severe Covid-19. New England Journal of Medicine, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jun L. D. CHEN, Li LIU,Ping LIU,Qingnian XU,Lu XIA,Yun LING,Dan HUANG,Shuli SONG,Dandan ZHANG,Zhiping QIAN,Tao LI,Yinzhong SHEN,Hongzhou LU, A pilot study of hydroxychloroquine in treatment of patients with common coronavirus disease-19 (COVID-19). J Zhejiang Univ (Med Sci) 49, 0–0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F., Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov 6, 14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng F., Lu W., Liu C., Fang J., Hou Y., Handy D. E., Wang R., Zhao Y., Yang Y., Huang J., Hill D. E., Vidal M., Eng C., Loscalzo J., A genome-wide positioning systems network algorithm for in silico drug repurposing. Nat Commun 10, 3476 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng F., Kovacs I. A., Barabasi A. L., Network-based prediction of drug combinations. Nat Commun 10, 1197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gysi D. M., Valle Í. D., Zitnik M., Ameli A., Gan X., Varol O., Sanchez H., Baron R. M., Ghiassian D., Loscalzo J., Barabási A.-L., Network Medicine Framework for Identifying Drug Repurposing Opportunities for COVID-19. arXiv, 2004.07229 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li G., He X., Zhang L., Ran Q., Wang J., Xiong A., Wu D., Chen F., Sun J., Chang C., Assessing ACE2 expression patterns in lung tissues in the pathogenesis of COVID-19. J Autoimmun, 102463 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimokawa H., Sunamura S., Satoh K., RhoA/Rho-Kinase in the Cardiovascular System. Circ Res 118, 352–366 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Zaoui K., Boudhraa Z., Khalife P., Carmona E., Provencher D., Mes-Masson A. M., Ran promotes membrane targeting and stabilization of RhoA to orchestrate ovarian cancer cell invasion. Nat Commun 10, 2666 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forbes S. A., Bindal N., Bamford S., Cole C., Kok C. Y., Beare D., Jia M., Shepherd R., Leung K., Menzies A., Teague J. W., Campbell P. J., Stratton M. R., Futreal P. A., COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 39, D945–950 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu C., Zhao J., Lu W., Dai Y., Hockings J., Zhou Y., Nussinov R., Eng C., Cheng F., Individualized genetic network analysis reveals new therapeutic vulnerabilities in 6,700 cancer genomes. PLoS Comput Biol 16, e1007701 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stenson P. D., Ball E. V., Mort M., Phillips A. D., Shiel J. A., Thomas N. S., Abeysinghe S., Krawczak M., Cooper D. N., Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat 21, 577–581 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Clerkin K. J., Fried J. A., Raikhelkar J., Sayer G., Griffin J. M., Masoumi A., Jain S. S., Burkhoff D., Kumaraiah D., Rabbani L., Schwartz A., Uriel N., Coronavirus Disease 2019 (COVID-19) and Cardiovascular Disease. Circulation, (2020). [DOI] [PubMed] [Google Scholar]
- 39.Baldi E., Sechi G. M., Mare C., Canevari F., Brancaglione A., Primi R., Klersy C., Palo A., Contri E., Ronchi V., Beretta G., Reali F., Parogni P., Facchin F., Bua D., Rizzi U., Bussi D., Ruggeri S., Oltrona Visconti L., Savastano S., Out-of-Hospital Cardiac Arrest during the Covid-19 Outbreak in Italy. New England Journal of Medicine, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beers M. F., Mulugeta S., The biology of the ABCA3 lipid transporter in lung health and disease. Cell Tissue Res 367, 481–493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nogee L. M., Abnormal expression of surfactant protein C and lung disease. Am J Respir Cell Mol Biol 26, 641–644 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Tay M. Z., Poh C. M., Rénia L., MacAry P. A., Ng L. F. P., The trinity of COVID-19: immunity, inflammation and intervention. Nature Reviews Immunology, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swaroopa D., Bhaskar K., Mahathi T., Katkam S., Raju Y. S., Chandra N., Kutala V. K., Association of serum interleukin-6, interleukin-8, and Acute Physiology and Chronic Health Evaluation II score with clinical outcome in patients with acute respiratory distress syndrome. Indian J Crit Care Med 20, 518–525 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hou T., Huang D., Zeng R., Ye Z., Zhang Y., Accuracy of serum interleukin (IL)-6 in sepsis diagnosis: a systematic review and meta-analysis. Int J Clin Exp Med 8, 15238–15245 (2015). [PMC free article] [PubMed] [Google Scholar]
- 45.Song J., Park D. W., Moon S., Cho H.-J., Park J. H., Seok H., Choi W. S., Diagnostic and prognostic value of interleukin-6, pentraxin 3, and procalcitonin levels among sepsis and septic shock patients: a prospective controlled study according to the Sepsis-3 definitions. BMC Infectious Diseases 19, 968 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu X., Han M., Li T., Sun W., Wang D., Fu B., Zhou Y., Zheng X., Yang Y., Li X., Zhang X., Pan A., Wei H., Effective treatment of severe COVID-19 patients with tocilizumab. Proceedings of the National Academy of Sciences, 202005615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Q., Fang P., He R., Li M., Yu H., Zhou L., Yi Y., Wang F., Rong Y., Zhang Y., Chen A., Peng N., Lin Y., Lu M., Zhu Y., Peng G., Rao L., Liu S., O-GlcNAc transferase promotes influenza A virus–induced cytokine storm by targeting interferon regulatory factor–5. Science Advances 6, eaaz7086 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Comhair S. A., McDunn J., Bennett C., Fettig J., Erzurum S. C., Kalhan S. C., Metabolomic Endotype of Asthma. J Immunol 195, 643–650 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holguin F., Grasemann H., Sharma S., Winnica D., Wasil K., Smith V., Cruse M. H., Perez N., Coleman E., Scialla T. J., Que L. G., L-Citrulline increases nitric oxide and improves control in obese asthmatics. JCI Insight 4, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., Quan S., Zhang F., Sun R., Qian L., Ge W., Liu W., Liang S., Chen H., Zhang Y., Li J., Xu J., He Z., Chen B., Wang J., Yan H., Zheng Y., Wang D., Zhu J., Kong Z., Kang Z., Liang X., Ding X., Ruan G., Xiang N., Cai X., Gao H., Li L., Li S., Xiao Q., Lu T., Zhu Y. J., Liu H., Chen H., Guo T., Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. medRxiv, 2020.2004.2007.20054585 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samuelsson B., Arachidonic acid metabolism: role in inflammation. Z Rheumatol 50 Suppl 1, 3–6 (1991). [PubMed] [Google Scholar]
- 52.Calabrese C., Triggiani M., Marone G., Mazzarella G., Arachidonic acid metabolism in inflammatory cells of patients with bronchial asthma. Allergy 55 Suppl 61, 27–30 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Berger A., What are leukotrienes and how do they work in asthma? BMJ 319, 90 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Modena B. D., Tedrow J. R., Milosevic J., Bleecker E. R., Meyers D. A., Wu W., Bar-Joseph Z., Erzurum S. C., Gaston B. M., Busse W. W., Jarjour N. N., Kaminski N., Wenzel S. E., Gene expression in relation to exhaled nitric oxide identifies novel asthma phenotypes with unique biomolecular pathways. Am J Respir Crit Care Med 190, 1363–1372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weathington N., O'Brien M. E., Radder J., Whisenant T. C., Bleecker E. R., Busse W. W., Erzurum S. C., Gaston B., Hastie A. T., Jarjour N. N., Meyers D. A., Milosevic J., Moore W. C., Tedrow J. R., Trudeau J. B., Wong H. P., Wu W., Kaminski N., Wenzel S. E., Modena B. D., BAL Cell Gene Expression in Severe Asthma Reveals Mechanisms of Severe Disease and Influences of Medications. Am J Respir Crit Care Med 200, 837–856 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balaci L., Spada M. C., Olla N., Sole G., Loddo L., Anedda F., Naitza S., Zuncheddu M. A., Maschio A., Altea D., Uda M., Pilia S., Sanna S., Masala M., Crisponi L., Fattori M., Devoto M., Doratiotto S., Rassu S., Mereu S., Giua E., Cadeddu N. G., Atzeni R., Pelosi U., Corrias A., Perra R., Torrazza P. L., Pirina P., Ginesu F., Marcias S., Schintu M. G., Giacco G. S. D., Manconi P. E., Malerba G., Bisognin A., Trabetti E., Boner A., Pescollderungg L., Pignatti P. F., Schlessinger D., Cao A., Pilia G., IRAK-M Is Involved in the Pathogenesis of Early-Onset Persistent Asthma. The American Journal of Human Genetics 80, 1103–1114 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Paiva A. C. Z., Marson F. A. d. L., Ribeiro J. D., Bertuzzo C. S., Asthma: Gln27Glu and Arg16Gly polymorphisms of the beta2-adrenergic receptor gene as risk factors. Allergy, Asthma & Clinical Immunology 10, 8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lamers M. M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T. I., Ravelli R. B. G., Paul van Schayck J., Mykytyn A. Z., Duimel H. Q., van Donselaar E., Riesebosch S., Kuijpers H. J. H., Schippers D., van de Wetering W. J., de Graaf M., Koopmans M., Cuppen E., Peters P. J., Haagmans B. L., Clevers H., SARS-CoV-2 productively infects human gut enterocytes. Science, eabc1669 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D'Amico F., Baumgart D. C., Danese S., Peyrin-Biroulet L., Diarrhea During COVID-19 Infection: Pathogenesis, Epidemiology, Prevention, and Management. Clin Gastroenterol Hepatol, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin J. C., Chang C., Boschetti G., Ungaro R., Giri M., Grout J. A., Gettler K., Chuang L. S., Nayar S., Greenstein A. J., Dubinsky M., Walker L., Leader A., Fine J. S., Whitehurst C. E., Mbow M. L., Kugathasan S., Denson L. A., Hyams J. S., Friedman J. R., Desai P. T., Ko H. M., Laface I., Akturk G., Schadt E. E., Salmon H., Gnjatic S., Rahman A. H., Merad M., Cho J. H., Kenigsberg E., Single-Cell Analysis of Crohn's Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell 178, 1493–1508 e1420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang W., Hui K. Y., Gusev A., Warner N., Ng S. M., Ferguson J., Choi M., Burberry A., Abraham C., Mayer L., Desnick R. J., Cardinale C. J., Hakonarson H., Waterman M., Chowers Y., Karban A., Brant S. R., Silverberg M. S., Gregersen P. K., Katz S., Lifton R. P., Zhao H., Nunez G., Pe'er I., Peter I., Cho J. H., Extended haplotype association study in Crohn's disease identifies a novel, Ashkenazi Jewish-specific missense mutation in the NF-kappaB pathway gene, HEATR3. Genes Immun 14, 310–316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muise A. M., Walters T., Xu W., Shen–Tu G., Guo C. H., Fattouh R., Lam G. Y., Wolters V. M., Bennitz J., van Limbergen J., Renbaum P., Kasirer Y., Ngan B. Y., Turner D., Denson L. A., Sherman P. M., Duerr R. H., Cho J., Lees C. W., Satsangi J., Wilson D. C., Paterson A. D., Griffiths A. M., Glogauer M., Silverberg M. S., Brumell J. H., Single Nucleotide Polymorphisms That Increase Expression of the Guanosine Triphosphatase RAC1 Are Associated With Ulcerative Colitis. Gastroenterology 141, 633–641 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seinen M. L., van Nieuw Amerongen G. P., de Boer N. K. H., Mulder C. J. J., van Bezu J., van Bodegraven A. A., Rac1 as a Potential Pharmacodynamic Biomarker for Thiopurine Therapy in Inflammatory Bowel Disease. Therapeutic Drug Monitoring 38, 621–627 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Korelitz B. I., Expert opinion: experience with 6-mercaptopurine in the treatment of inflammatory bowel disease. World J Gastroenterol 19, 2979–2984 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perry C. M., Scott L. J., Cefdinir: A Review of its Use in the Management of Mild-to-Moderate Bacterial Infections. Drugs 64, 1433–1464 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Johansen L. M., Brannan J. M., Delos S. E., Shoemaker C. J., Stossel A., Lear C., Hoffstrom B. G., Dewald L. E., Schornberg K. L., Scully C., Lehar J., Hensley L. E., White J. M., Olinger G. G., FDA-approved selective estrogen receptor modulators inhibit Ebola virus infection. Sci Transl Med 5, 190ra179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cong Y., Hart B. J., Gross R., Zhou H., Frieman M., Bollinger L., Wada J., Hensley L. E., Jahrling P. B., Dyall J., Holbrook M. R., MERS-CoV pathogenesis and antiviral efficacy of licensed drugs in human monocyte-derived antigen-presenting cells. PLoS One 13, e0194868 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dyall J., Coleman C. M., Hart B. J., Venkataraman T., Holbrook M. R., Kindrachuk J., Johnson R. F., Olinger G. G. Jr., Jahrling P. B., Laidlaw M., Johansen L. M., Lear-Rooney C. M., Glass P. J., Hensley L. E., Frieman M. B., Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob Agents Chemother 58, 4885–4893 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jeon S., Ko M., Lee J., Choi I., Byun Y., Park S., Shum D., S. K., Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. bioRxiv, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang X. J., Hu W., Zhang T. Y., Mao Y. Y., Liu N. N., Wang S. Q., Irbesartan, an FDA approved drug for hypertension and diabetic nephropathy, is a potent inhibitor for hepatitis B virus entry by disturbing Na(+)-dependent taurocholate cotransporting polypeptide activity. Antiviral Res 120, 140–146 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Zhang P., Zhu L., Cai J., Lei F., Qin J.-J., Xie J., Liu Y.-M., Zhao Y.-C., Huang X., Lin L., Xia M., Chen M.-M., Cheng X., Zhang X., Guo D., Peng Y., Ji Y.-X., Chen J., She Z.-G., Wang Y., Xu Q., Tan R., Wang H., Lin J., Luo P., Fu S., Cai H., Ye P., Xiao B., Mao W., Liu L., Yan Y., Liu M., Chen M., Zhang X.-J., Wang X., Touyz R. M., Xia J., Zhang B.-H., Huang X., Yuan Y., Rohit L., Liu P. P., Li H., Association of Inpatient Use of Angiotensin Converting Enzyme Inhibitors and Angiotensin II Receptor Blockers with Mortality Among Patients With Hypertension Hospitalized With COVID-19. Circulation Research 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Puelles V. G., Lutgehetmann M., Lindenmeyer M. T., Sperhake J. P., Wong M. N., Allweiss L., Chilla S., Heinemann A., Wanner N., Liu S., Braun F., Lu S., Pfefferle S., Schroder A. S., Edler C., Gross O., Glatzel M., Wichmann D., Wiech T., Kluge S., Pueschel K., Aepfelbacher M., Huber T. B., Multiorgan and Renal Tropism of SARS-CoV-2. The New England journal of medicine, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H., Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology 158, 1831–1833 e1833 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakashima K., Hirota T., Obara K., Shimizu M., Jodo A., Kameda M., Doi S., Fujita K., Shirakawa T., Enomoto T., Kishi F., Yoshihara S., Matsumoto K., Saito H., Suzuki Y., Nakamura Y., Tamari M., An association study of asthma and related phenotypes with polymorphisms in negative regulator molecules of the TLR signaling pathway. Journal of Human Genetics 51, 284–291 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Pino-Yanes M., Sanchez-Machin I., Cumplido J., Figueroa J., Torres-Galvan M. J., Gonzalez R., Corrales A., Acosta-Fernandez O., Garcia-Robaina J. C., Carrillo T., Sanchez-Palacios A., Villar J., Hernandez M., Flores C., IL-1 receptor-associated kinase 3 gene (IRAK3) variants associate with asthma in a replication study in the Spanish population. J Allergy Clin Immunol 129, 573–575, 575 e571–510 (2012). [DOI] [PubMed] [Google Scholar]
- 76.Reihsaus E., Innis M., MacIntyre N., Liggett S. B., Mutations in the gene encoding for the beta 2-adrenergic receptor in normal and asthmatic subjects. Am J Respir Cell Mol Biol 8, 334–339 (1993). [DOI] [PubMed] [Google Scholar]
- 77.Ali S., Hirschfeld A. F., Mayer M. L., Fortuno E. S., Corbett N., Kaplan M., Wang S., Schneiderman J., Fjell C. D., Yan J., Akhabir L., Aminuddin F., Marr N., Lacaze-Masmonteil T., Hegele R. G., Becker A., Chan-Yeung M., Hancock R. E. W., Kollmann T. R., Daley D., Sandford A. J., Lavoie P. M., Turvey S. E., Functional Genetic Variation in NFKBIA and Susceptibility to Childhood Asthma, Bronchiolitis, and Bronchopulmonary Dysplasia. The Journal of Immunology 190, 3949–3958 (2013). [DOI] [PubMed] [Google Scholar]
- 78.Akaike T., Maeda H., Nitric oxide and virus infection. Immunology 101, 300–308 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Verity R., Okell L. C., Dorigatti I., Winskill P., Whittaker C., Imai N., Cuomo-Dannenburg G., Thompson H., Walker P. G. T., Fu H., Dighe A., Griffin J. T., Baguelin M., Bhatia S., Boonyasiri A., Cori A., Cucunuba Z., FitzJohn R., Gaythorpe K., Green W., Hamlet A., Hinsley W., Laydon D., Nedjati-Gilani G., Riley S., van Elsland S., Volz E., Wang H., Wang Y., Xi X., Donnelly C. A., Ghani A. C., Ferguson N. M., Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aw D., Silva A. B., Palmer D. B., Immunosenescence: emerging challenges for an ageing population. Immunology 120, 435–446 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang X., Bian Y., Zhang R., Liu X., Ni L., Ma B., Zeng R., Zhao Z., Song X., Liu C., Melatonin alleviates cigarette smoke-induced endothelial cell pyroptosis through inhibiting ROS/NLRP3 axis. Biochem Biophys Res Commun 519, 402–408 (2019). [DOI] [PubMed] [Google Scholar]
- 82.Shang Y., Xu S. P., Wu Y., Jiang Y. X., Wu Z. Y., Yuan S. Y., Yao S. L., Melatonin reduces acute lung injury in endotoxemic rats. Chin Med J (Engl) 122, 1388–1393 (2009). [PubMed] [Google Scholar]
- 83.Zhang J., Wang L., Xie W., Hu S., Zhou H., Zhu P., Zhu H., Melatonin attenuates ER stress and mitochondrial damage in septic cardiomyopathy: A new mechanism involving BAP31 upregulation and MAPK-ERK pathway. J Cell Physiol 235, 2847–2856 (2020). [DOI] [PubMed] [Google Scholar]
- 84.Hamosh A., Scott A. F., Amberger J. S., Bocchini C. A., McKusick V. A., Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 33, D514–517 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang M., Zhu C., Jacomy A., Lu L. J., Jegga A. G., The orphan disease networks. Am J Hum Genet 88, 755–766 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kittler R., Pelletier L., Heninger A.-K., Slabicki M., Theis M., Miroslaw L., Poser I., Lawo S., Grabner H., Kozak K., Wagner J., Surendranath V., Richter C., Bowen W., Jackson A. L., Habermann B., Hyman A. A., Buchholz F., Genome-scale RNAi profiling of cell division in human tissue culture cells. Nature Cell Biology 9, 1401–1412 (2007). [DOI] [PubMed] [Google Scholar]
- 87.Breuer K., Foroushani A. K., Laird M. R., Chen C., Sribnaia A., Lo R., Winsor G. L., Hancock R. E. W., Brinkman F. S. L., Lynn D. J., InnateDB: systems biology of innate immunity and beyond—recent updates and continuing curation. Nucleic Acids Res. 41, D1228–D1233 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kuleshov M. V., Jones M. R., Rouillard A. D., Fernandez N. F., Duan Q., Wang Z., Koplev S., Jenkins S. L., Jagodnik K. M., Lachmann A., McDermott M. G., Monteiro C. D., Gundersen G. W., Ma'ayan A., Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hirsh A. E., Fraser H. B., Wall D. P., Adjusting for selection on synonymous sites in estimates of evolutionary distance. Mol Biol Evol 22, 174–177 (2005). [DOI] [PubMed] [Google Scholar]
- 90.Bezginov A., Clark G. W., Charlebois R. L., Dar V. U., Tillier E. R., Coevolution reveals a network of human proteins originating with multicellularity. Mol Biol Evol 30, 332–346 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cheng F., Jia P., Wang Q., Lin C.-C., Li W.-H., Zhao Z., Studying Tumorigenesis through Network Evolution and Somatic Mutational Perturbations in the Cancer Interactome. Molecular Biology and Evolution 31, 2156–2169 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mantel N., Haenszel W., Statistical Aspects of the Analysis of Data From Retrospective Studies of Disease. JNCI: Journal of the National Cancer Institute 22, 719–748 (1959). [PubMed] [Google Scholar]
- 93.DerSimonian R., Laird N., Meta-analysis in clinical trials. Controlled Clinical Trials 7, 177–188 (1986). [DOI] [PubMed] [Google Scholar]
- 94.Wan X., Wang W., Liu J., Tong T., Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Medical Research Methodology 14, 135 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stijnen T., Hamza T. H., Özdemir P., Random effects meta-analysis of event outcome in the framework of the generalized linear mixed model with applications in sparse data. Statistics in Medicine 29, 3046–3067 (2010). [DOI] [PubMed] [Google Scholar]
- 96.N. R. Coordinators, Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 44, D7–19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smith I. N., Thacker S., Seyfi M., Cheng F., Eng C., Conformational Dynamics and Allosteric Regulation Landscapes of Germline PTEN Mutations Associated with Autism Compared to Those Associated with Cancer. Am J Hum Genet 104, 861–878 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Law V., Knox C., Djoumbou Y., Jewison T., Guo A. C., Liu Y., Maciejewski A., Arndt D., Wilson M., Neveu V., Tang A., Gabriel G., Ly C., Adamjee S., Dame Z. T., Han B., Zhou Y., Wishart D. S., DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res 42, D1091–1097 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang H., Qin C., Li Y. H., Tao L., Zhou J., Yu C. Y., Xu F., Chen Z., Zhu F., Chen Y. Z., Therapeutic target database update 2016: enriched resource for bench to clinical drug target and targeted pathway information. Nucleic Acids Res 44, D1069–1074 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gaulton A., Bellis L. J., Bento A. P., Chambers J., Davies M., Hersey A., Light Y., McGlinchey S., Michalovich D., Al-Lazikani B., Overington J. P., ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 40, D1100–D1107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu T. Q., Lin Y. M., Wen X., Jorissen R. N., Gilson M. K., BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 35, D198–D201 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pawson A. J., Sharman J. L., Benson H. E., Faccenda E., Alexander S. P. H., Buneman O. P., Davenport A. P., McGrath J. C., Peters J. A., Southan C., Spedding M., Yu W. Y., Harmar A. J., Nc I., The IUPHAR/BPS Guide to PHARMACOLOGY: an expert-driven knowledgebase of drug targets and their ligands. Nucleic Acids Res. 42, D1098–D1106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lamb J., Crawford E. D., Peck D., Modell J. W., Blat I. C., Wrobel M. J., Lerner J., Brunet J. P., Subramanian A., Ross K. N., Reich M., Hieronymus H., Wei G., Armstrong S. A., Haggarty S. J., Clemons P. A., Wei R., Carr S. A., Lander E. S., Golub T. R., The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006). [DOI] [PubMed] [Google Scholar]
- 105.Reghunathan R., Jayapal M., Hsu L.-Y., Chng H.-H., Tai D., Leung B. P., Melendez A. J., Expression profile of immune response genes in patients with Severe Acute Respiratory Syndrome. BMC Immunology 6, 2 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Josset L., Menachery V. D., Gralinski L. E., Agnihothram S., Sova P., Carter V. S., Yount B. L., Graham R. L., Baric R. S., Katze M. G., Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. mBio 4, e00165–00113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yuan S., Chu H., Chan J. F.-W., Ye Z.-W., Wen L., Yan B., Lai P.-M., Tee K.-M., Huang J., Chen D., Li C., Zhao X., Yang D., Chiu M. C., Yip C., Poon V. K.-M., Chan C. C.-S., Sze K.-H., Zhou J., Chan I. H.-Y., Kok K.-H., To K. K.-W., Kao R. Y.-T., Lau J. Y.-N., Jin D.-Y., Perlman S., Yuen K.-Y., SREBP-dependent lipidomic reprogramming as a broad-spectrum antiviral target. Nature Communications 10, 120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]