Abstract
The clinical and epidemiologic management of the SARS-CoV-2 pandemic is critically dependent on molecular assays with short turn-around time. We validated the novel Xpert Xpress SARS-CoV-2 assay using a commercial nucleic acid testing (Roche Cobas 6800). We found an excellent concordance over a range of SARS-CoV-2 loads and across established human coronaviruses.
Keywords: SARS-CoV-2, COVID-19, GeneXpert, Respiratory infection, Viruses, Diagnostics, PCR, NAT, COBAS 6800
The current SARS-CoV-2/COVID-19 pandemic poses significant diagnostic challenges on each level of pre- to post-analytical steps (Tang et al., 2020). At the beginning of the pandemic, the experience with molecular diagnostic assays was limited, and more recently on overwhelming spectrum of new molecular assays being released on a weekly basis. Rapid and simple assays for SARS-CoV-2 detection with high sensitivity and specificity are very important for infection control counter measurements. Ideally, suspected cases should be assessed using nucleic acid testing (NAT) with high test performance and short turn-around times below a few hours. Currently, most testing strategies focus on high-throughput with capacities of 1000 or more samples per day using fully automated molecular assays. This strategy allows the management of diagnostic demands during the peak of a pandemic period.
In contrast, single cartridge-based assays are somewhat limited in this situation due to higher costs and a lower throughput capacity. Nevertheless, such diagnostic tests clearly have a place in a series of specific settings. For example, during the declining phase of the pandemic wave, once a more focused diagnostic approach is used and relevant. Cartridge-based diagnostic often allows the (i) diagnosis of critically ill cases in a short time period, (ii) assessment of suspected patients very rapidly, allowing for a specific epidemiological management, and (iii) transfer diagnostics to point-of-care scenarios including smaller laboratories. Rapid testing has been shown to provide important diagnostic information immediately improving patient management (O’Connell et al., 2020).
With this study, we aimed to evaluated the test performance of the new Xpert Xpress SARS-CoV-2 cartridge assay and compared the results to a commercial SARS-CoV-2 specific NAT.
We assessed the performance of the new available Xpert® Xpress SARS-CoV-2 (Cepheid) comparing the assay against the cobas® SARS-CoV-2 assay (Roche) on the COBAS 6800 system (Roche). The Xpert Xpress SARS-CoV-2 is a very recently released assay for use under the Emergency Use Authorization (EUA) only from U.S. Food & Drug Administration (FDA). The assay requires a sample load of 300μL, has a detection limit of 250 copies per mL, and a runtime of 45 min (Cepheid, 2020). This single-use disposable cartridge test is applied on the GeneXpert Dx instrument (Cepheid). The targets in the assay are the viral envelope E gene and the nucleocapsid N2 gene. The COBAS 6800 is a fully automated robotic system, which requires a sample load of 600μL. The viral targets in the COBAS 6800 assay are the E gene and the ORF1ab gene. Samples for this validation were aliquoted and frozen at −80 °C until batch-wise sample processing with the Xpert was performed.
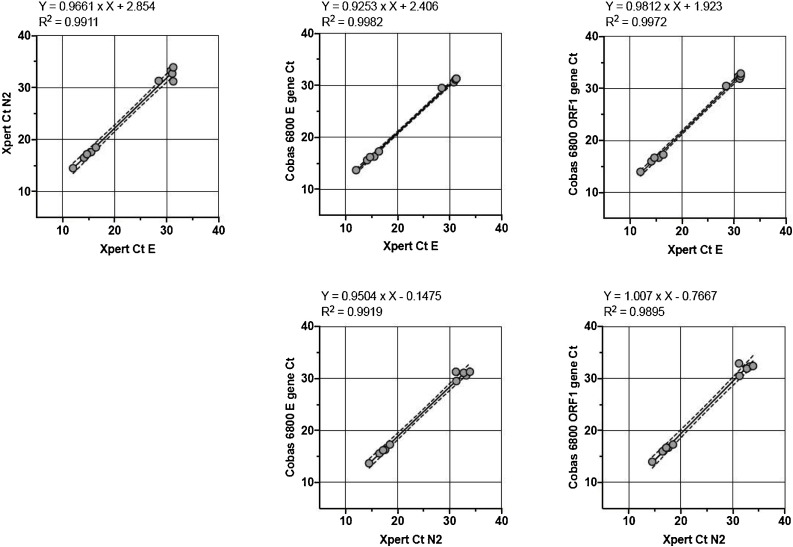
We used 19 nasopharyngeal swabs (Universal Transport Media and eSwab media, Copan) of patients with suspected COVID-19. The samples were from routine diagnostics and reflected 9 negative and 10 positive samples with a broad range of viral loads. These samples were collected within a week during the 2020 pandemic in Basel, Switzerland, and selected based on the COBAS 6800 results for validation of the Xpert system. We subsequently assessed the samples in parallel on both systems. A detailed comparison of the assay results can be seen in Table 1 . We observed an excellent concordance of the NAT results from both assays. The overall sample sensitivity and specificity were both 100 %. We observed very high coefficients of determination of the different viral gene targets (Fig. 1 ). Nine SARS-CoV-2 negative samples in the COBAS 6800 assay were also negative in the Xpert Xpress assay. We also included a total of eight samples positive for other coronaviruses (229E, OC43, NL63, and HUK1) in order to test assay specificity. These other coronaviruses positive samples were collected in 2019 and kept frozen at −80 °C until testing for SARS-CoV-2. We identified these samples using the Biofire® Filmarray® Respiratory Panel PCR (bioMérieux) covering 22 pathogens including coronaviruses (229E, OC43, NL63, and HUK1) and the Middle-East Respiratory Syndrome coronavirus (MERS-CoV) using the same nasopharyngeal swab material. The Biofire panel assay does not include a specific target for SARS-CoV-2. The Xpert Xpress SARS-CoV-2 assay showed no cross-reactivity with other already circulating coronaviruses (Table 1).
Table 1.
Comparison of different molecular assays for SARS-CoV-2 detection. Value with Ct-value showing positive result; “-“, PCR negative result.
| Sample ID | Virus | Xpert E Gene | Xpert N2 Gene | Cobas E Gene | Cobas ORF1 |
|---|---|---|---|---|---|
| 42218454 | SARS-CoV-2 | 28.5 | 31.3 | 29.5 | 30.5 |
| 42216999 | SARS-CoV-2 | 30.8 | 33.2 | 30.6 | 32.3 |
| 42217093 | SARS-CoV-2 | 30.5 | 33.7 | 31.1 | 31.9 |
| 42216810 | SARS-CoV-2 | 32.2 | 34.6 | 31.3 | 32.4 |
| 42212253 | SARS-CoV-2 | 30.1 | 32.1 | 31.3 | 32.9 |
| 42212277 | SARS-CoV-2 | 12 | 14.5 | 13.7 | 14.0 |
| 42212350 | SARS-CoV-2 | 14.1 | 16.5 | 15.6 | 16.0 |
| 42212053 | SARS-CoV-2 | 15.5 | 17.6 | 16.3 | 16.7 |
| 42217811 | SARS-CoV-2 | 14.7 | 17.2 | 16.2 | 16.7 |
| 42218290 | SARS-CoV-2 | 16.4 | 18.5 | 17.3 | 17.3 |
| 42212615 | None | – | – | – | – |
| 42217035 | None | – | – | – | – |
| 42217854 | None | – | – | – | – |
| 42218119 | None | – | – | – | – |
| 42212718 | None | – | – | – | – |
| 42211974 | None | – | – | – | – |
| 42212601 | None | – | – | – | – |
| 42212602 | None | – | – | – | – |
| 42212607 | None | – | – | – | – |
| 353599 | HKU1-CoV | – | – | – | – |
| 353396 | HKU1-CoV | – | – | – | – |
| 353165 | HKU1-CoV | – | – | – | – |
| 367715 | 229E-CoV | – | – | – | – |
| 353637 | NL63-CoV | – | – | – | – |
| 353333 | NL63-CoV | – | – | – | – |
| 353864 | OC43-CoV | – | – | – | – |
| 353355 | OC43-CoV | – | – | – | – |
Fig. 1.
Correlation of Ct-values between different assays and targeted viral genes for the n = 10 positive samples. Formula for best fit linear curve is shown (Graph Pad Prism version 8.4.0). Linear regression analysis was used to calculate R (O’Connell et al., 2020).
Overall, the handling of the Xpert Xpress SARS-CoV-2 cartridge was easy, requiring minimal hands-on-time. There was no handling problem reported by the laboratory technician. The required input sample volume of 300μL is reasonable as most nasopharyngeal swabs are provided in 1–2 mL of transport media. This would allow a repeated test in the case of a non-conclusive result, or subsequent quantification of the viral load with an alternative method.
The new Xpert Xpress SARS-CoV-2 assay provided very good results in comparison to our available molecular diagnostic assays and is in line with very recently published data (Moran et al., 2020). We noted an excellent specificity (100 %) and sensitivity (100 %). However, our validation has a few limitations. The number of samples included from routine diagnostics was small, but included samples from all important diagnostic scenarios. We validated samples from nasopharyngeal swabs only (nasal and pharyngeal combined) and no other respiratory material. Both, nasopharyngeal (NP) and oropharyngeal (OP) swabs are the recommended sample type for screening or diagnosis of early infection (Pan et al., 2020). Other sample types (e.g. bronchial lavage, tracheal secretion or sputum) should be validated separately as the higher viscosity may reduce the assay performance, and if pre-processing reagents such as sputasol were required, this may influence assay performance (Yu et al., 2018).
The usage of a cartridge-based system for detection of SARS-CoV-2 clearly needs a critical evaluation of the laboratory workflow. Although the runtime of the Xpert Xpress SARS-CoV-2 is only 45 min, a cartridge-based assay may not be the ideal choice during the peak of a pandemic – at least not for a core laboratory, when hundreds of samples need to be tested per day. First, the throughput of the Xpert system is lower in comparison to a high-throughput system. Second, the costs per sample of a single-use cartridge test is significantly higher. However, the cartridge-based diagnostics may be very helpful for rapid case-by-case evaluations e.g. for a surveillance setting with relative low sample numbers per day, medical emergency situations such as surgery to rapidly evaluate the patient, for smaller laboratories, and at the frontline rather as a point-of-care system. Smaller and less exposed laboratories should carefully validate each new molecular assay. The validation includes the whole process from sampling to result and allows critical review of the diagnostic process. In case of uncertainty, reference laboratories should be involved.
Declaration of Competing Interest
All authors report no conflicts of interest relevant to this article.
Acknowledgement
We thank Yoann Mahiddine (University Hospital Basel) for excellent technical assistance.
References
- Cepheid . U.S. Food and Drug Administration website; 2020. Xpert Xpress SARS-CoV-2 Test: package insert.https://www.fda.gov/media/136314/download [cited 2020 24.04.2020]. Available from: [Google Scholar]
- Moran A., Beavis K.G., Matushek S.M., Ciaglia C., Francois N., Tesic V. The detection of SARS-CoV-2 using the Cepheid Xpert Xpress SARS-CoV-2 and Roche cobas SARS-CoV-2 assays. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00772-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell S., Conlan C., Reidy M., Stack C., Mulgrew A., Baruah J. The impact of point-of-care testing for influenza A and B on patient flow and management in a medical assessment unit of a general hospital. BMC Res. Notes. 2020;13(1):143. doi: 10.1186/s13104-020-04986-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.W., Schmitz J.E., Persing D.H., Stratton C.W. The laboratory diagnosis of COVID-19 infection: current issues and challenges. J. Clin. Microbiol. 2020;58(6):1–9. doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Qiu T., Zeng Y., Wang Y., Zheng S., Chen X. Comparative evaluation of three preprocessing methods for extraction and detection of influenza a virus nucleic acids from sputum. Front. Med. (Lausanne) 2018;5:56. doi: 10.3389/fmed.2018.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]