Abstract
Messenger RNAs (mRNAs) are the templates for protein synthesis as the coding region is translated into the amino acid sequence. mRNAs also contain 3′ untranslated regions (3′UTRs) that harbor additional elements for the regulation of protein function. If the amino acid sequence of a protein is necessary and sufficient for its function, we call it 3′UTR-independent. In contrast, functions that are accomplished by protein complexes whose formation requires the presence of a specific 3′UTR are 3′UTR-dependent protein functions. We showed that 3′UTRs can regulate protein activity without affecting protein abundance, and alternative 3′UTRs can diversify protein functions. We currently think that the regulation of protein function by 3′UTRs is facilitated by the local environment at the site of protein synthesis, which we call the nurturing niche for nascent proteins. This niche is composed of the mRNA and the bound proteins that consist of RNA- binding proteins and recruited proteins. It enables the formation of specific protein complexes, as was shown for TIS granules, a recently discovered cytoplasmic membraneless organelle. This finding suggests that changing the niche for nascent proteins will alter protein activity and function, implying that cytoplasmic membraneless organelles can regulate protein function in a manner that is independent of protein abundance.
The influence of nurture on nature has classically been studied with respect to human behavior and is usually used to characterize the influence of external factors, such as parents, social relationships, and the surrounding culture on the development of a child (Collins et al. 2000). Nurture also affects nature during the development of organs, including the brain, where neuronal activity influences the construction of cortical networks (Ben-Ari 2002). Here, I propose that this principle is also true for protein functions regulated by 3′ untranslated regions (3′UTRs). We showed that proteins with the same amino acid sequence that are encoded from mRNA isoforms with alternative 3′UTRs have different functions (Berkovits and Mayr 2015; Ma and Mayr 2018; Lee and Mayr 2019). This suggests that protein functions are influenced by the local environment at the site of protein synthesis, which will be called the nurturing niche for nascent proteins. Proteins are born into a niche provided by the mRNA and its bound factors. In this analogy, 3′UTRs act as the ‘social relationships’ of nascent proteins as they influence certain protein functions. The influence of 3′UTRs will not entirely change the nature of a protein, as for example a kinase will remain a kinase, but 3′UTRs are able to recruit protein interactors that influence the function of the protein, thus changing its qualitative properties and fate. Furthermore, when a protein is translated within a larger RNA granule, the surrounding mRNAs can serve as the ‘surrounding culture’, again changing protein interactors and functions. In this review, I will summarize our current understanding of the regulation of protein functions by the nurturing niche provided by 3′UTRs (Fig. 1).
Figure 1. The 3′UTR with the bound RNA-binding proteins and recruited proteins creates a nurturing niche for nascent proteins.
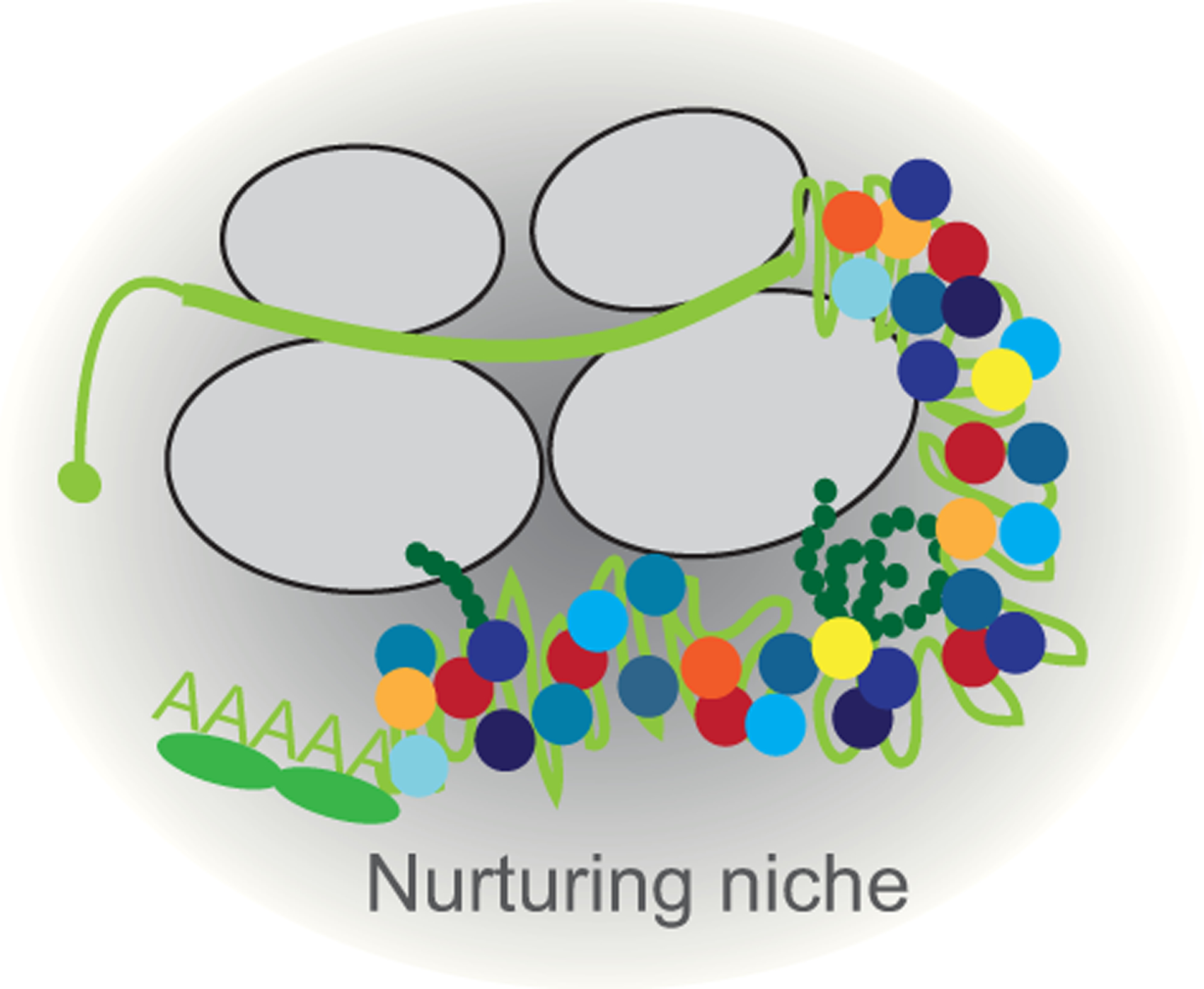
Shown are two ribosomes translating an mRNA (light green). The nascent peptide is depicted in dark green. The RNA-binding proteins bound to the 3′UTR are in yellow, orange, or red and the recruited proteins are in blue shaded colors.
3′UTR shortening does not correlate with increased mRNA levels in most transcriptome- wide studies
My lab studies the functions of 3′UTRs. When I started my lab the boundaries of human 3′UTRs were not well annotated. Therefore, we established a sequencing method called 3′-seq that allowed us to identify all 3′UTRs of the transcriptome (Lianoglou et al. 2013). The obtained reads overlap the junction between the 3′UTR and the poly(A) tail, thus providing singlenucleotide resolution of 3′UTR ends. Importantly, we validated the obtained data using an independent method and demonstrated that 3′-seq is quantitative. This means that in addition to identifying 3′UTR ends, we can quantify the reads and calculate the expression of individual 3′UTR isoforms.
We applied 3′-seq to a series of cell lines and normal human tissues and found that nearly half of human genes use alternative cleavage and polyadenylation to generate mRNA transcripts with alternative 3′UTRs (Lianoglou et al. 2013). Our validation experiments showed that in the vast majority of cases the coding region was identical and the only difference in the mRNA transcript was found in the 3′UTR. We were surprised that such a large number of mRNAs that generate short or long 3′UTRs encode proteins with identical amino acid sequence. This finding motivated us to study the functions of alternative 3′UTRs.
At the time, we and others thought that one of the major functions of 3′UTRs is the regulation of protein abundance, mostly through regulation of mRNA stability (Barreau et al. 2005; Sandberg et al. 2008; Mayr and Bartel 2009; Bartel 2009). When we compared alternative 3′UTR isoform abundance and overall mRNA abundance of a gene from two different conditions or cell types, we expected to see a correlation between 3′UTR shortening and higher mRNA levels. However, in our dataset, we did not observe such a relationship (Fig. 2)(Lianoglou et al. 2013). Importantly, several other transcriptome-wide studies using different experimental systems agreed with our finding (Spies et al. 2013; Gruber et al. 2014; Zhang et al. 2016; Jia et al. 2017 Overall, from the mRNAs that significantly changed their 3′UTR isoform levels less than 20% also changed their mRNA abundance levels. Instead, across conditions and cell types the majority of mRNAs either changed their mRNA abundance levels or they changed their 3′UTR isoform expression (Fig. 2) (Lianoglou et al. 2013). This suggested that mRNA abundance and 3′UTR ratios are orthogonal measures of gene expression.
Figure 2. mRNAs either change their abundance or they change their 3′UTR ratio during B cell transformation.
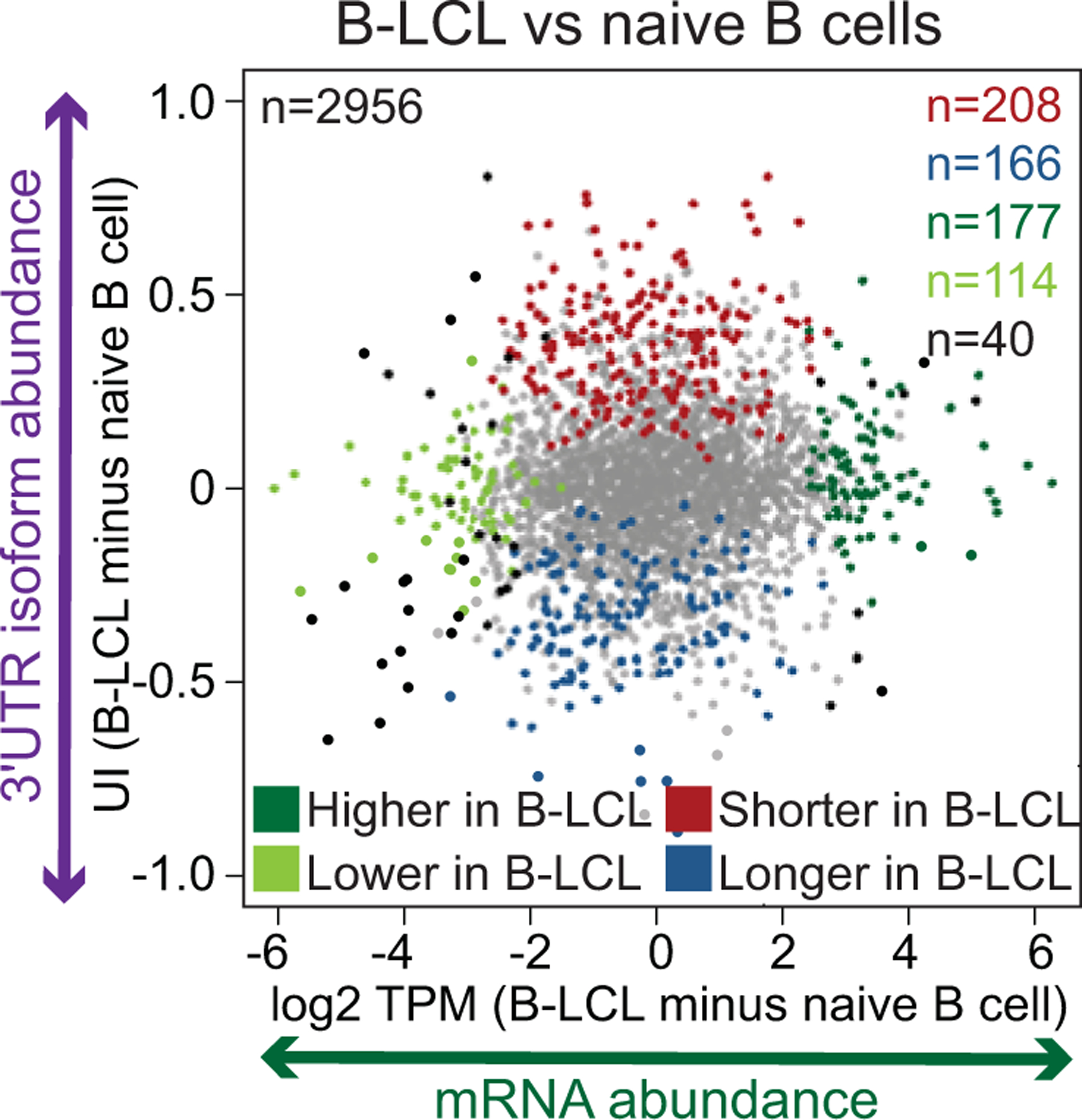
Shown are changes in mRNA levels (X-axis) versus changes in 3′UTR isoform abundance (Y-axis) in naive B cells before and after immortalization using Epstein-Barr virus transformation (called B lymphoblastic cells, B-LCL). Changes in 3′UTR isoform abundance are given as difference in UTR index (UI). The UI is the fraction of reads that map to short 3′UTR isoform out of all the reads mapping to the 3′UTR. Genes with significant mRNA or 3′UTR isoform changes are color-coded. Grey indicates no significant change and black indicates genes with a significant change in both mRNA levels and 3′UTR isoform abundance. Reprinted from Lianoglou et al., 2013.
In addition to the lack of correlation between 3′UTR shortening and mRNA abundance, also no association between 3′UTR isoform length and changes in protein levels was found during differentiation of embryonic stem cells. This study showed that 87% of genes with significant 3′UTR isoform changes did not show differences in protein abundance (Brumbaugh et al. 2018). However, it is worth pointing out that there are exceptions: several classes of genes with low mRNA transcript stability which include cell cycle regulators, cytokines, and oncogenes indeed use their 3′UTRs to change their protein levels substantially. This regulation is largely mediated by microRNAs and AU-rich elements (Kontoyiannis et al. 1999; Mayr and Bartel 2009; Herranz et al. 2015). Another exception happens in development during maternal to zygotic transition, where several groups showed that many mRNAs are cleared using elements found in 3′UTRs (Giraldez et al. 2006; Benoit et al. 2009).
The overall lack of a relationship between 3′UTR shortening and mRNA abundance then prompted some researchers to propose that alternative 3′UTR isoform expression may be random and may not matter (Spies et al. 2013; Neve and Furger 2014; Xu and Zhang 2018). This proposal, however, is inconsistent with the known roles of regulatory elements in 3′UTRs. First of all, 3′UTRs, including alternative 3′UTRs, are known to regulate mRNA localization (Lecuyer et al. 2007; An et al. 2008; Taliaferro et al. 2016; Mayr 2017; Tushev et al. 2018; Ciolli Mattioli et al. 2019). Second, early comparative genomic analyses found conserved 3′UTR sequences when comparing homologous genes across species, but noticed highly divergent 3′UTR sequences within an organism when comparing similar proteins. For example, they observed that actin family members that encode similar proteins have very different 3′UTRs. However, these 3′UTR sequences were conserved across organisms, suggesting that 3’UTRs contain additional genetic information to distinguish the functions of highly similar proteins (Yaffe et al. 1985). Although 3′UTRs are usually less conserved than coding regions, high sequence conservation of 3′UTR regulatory elements was confirmed in subsequent genome-wide studies (Siepel et al. 2005; Xie et al. 2005). Third, when comparing 3′UTR length of genes across species, we and others found that 3′UTR length has expanded substantially during evolution of more complex animals (Chen et al. 2012; Mayr 2016). And lastly, the use of replicates showed coordinated changes in 3′UTR isoform expression in different cell types and conditions which speaks against random 3′UTR isoform choice (Lianoglou et al. 2013; Singh et al. 2018; Lee et al. 2018).
Discovery of 3′UTR-dependent protein functions
To investigate functions of alternative 3′UTRs, we studied the CD47 gene which generates mRNA transcripts with short or long 3′UTRs and encodes a plasma membrane receptor that acts as a ‘don’t eat me signal’ (Jaiswal et al. 2009). We fused GFP to the coding region of CD47 and then added either its short or long 3′UTR. When we transfected these constructs into cells, we observed that CD47 protein encoded by the mRNA isoform with the long 3′UTR (CD47-LU) perfectly localized to the plasma membrane, whereas CD47 protein encoded by the mRNA isoform with the short 3′UTR (CD47-SU) mostly localized intracellularly (Berkovits and Mayr 2015). Interestingly, mRNA localization of the two constructs was similar and both of them localized to the endoplasmic reticulum (ER). This suggested that the alternative 3′UTRs of CD47 contain information for mRNA-independent protein localization.
Furthermore, we noticed that the cells transfected with CD47-LU formed lamellipodia, whereas the cells that were transfected with CD47-SU did not. As lamellipodia formation is a sign of active RAC1, we examined the amount of RAC1-GTP (active RAC1) in the cells after transfection of either CD47-SU or CD47-LU. This revealed that transfection of CD47-LU increased RAC1 activation, whereas transfection of CD47-SU did not. This was a surprising observation as the protein that is generated from the constructs is identical. This finding indicated that the alternative 3′UTRs of CD47 can control CD47 protein function as only CD47-LU was able to activate a downstream signaling pathway. This was a remarkable discovery as it shows that 3′UTRs can change the qualitative property and the function of a protein (Berkovits and Mayr 2015).
We then investigated how the difference in protein function was regulated and found that the RNA-binding protein HuR only bound to the long, but not to the short 3′UTR of CD47. One of the most abundant interactors of HuR is the protein SET, a small acidic protein that is highly expressed in the nucleus and cytoplasm of nearly all cell types (Brennan and Steitz 2001). Despite the similarity in name, SET should not be confused with a SET domain, a domain found in methyltransferases. Instead, the protein SET has many different functions and was initially discovered as an inhibitor of the phosphatase PP2A (Li et al. 1996). It also acts as a histone chaperone that prevents histone acetylation and is regarded as a ‘reader’ of non-acetylated lysines (Seo et al. 2001; Wang et al. 2016).
We performed co-immunoprecipiation of GFP-tagged CD47-SU or CD47-LU and asked if endogenous SET binds. We found that SET only interacted with CD47-LU, but not with CD47-SU. Therefore, we regard the protein-protein interaction between SET and CD47-LU to be 3′UTR-dependent. Furthermore, the binding of SET to CD47 also depends on specific amino acids in the cytoplasmic domains of CD47. CD47 protein has five transmembrane domains and is translated on the surface of the ER. The cytoplasmic domains contain positively charged amino acids and mutation of 5/10 abrogated the binding of SET to CD47. As HuR binds to AU-rich and U-rich motifs in the 3′UTR of CD47 and SET binds to positively charged amino acids in CD47 protein, our data indicates that establishment of the 3′UTR-dependent interaction between SET and CD47 requires motifs in the mRNA as well as in the protein encoded by the mRNA (Berkovits and Mayr 2015).
Mass spectrometry identifies 3′UTR-independent and 3′UTR-dependent protein interactors of the E3 ubiquitin ligase BIRC3
In the case of CD47, we identified the 3′UTR-dependent protein interactor SET through literature search. However, this approach is not scalable. Therefore, we established an experimental strategy to identify 3′UTR-dependent protein interactors for basically any candidate. For our proof-of-principle experiments we chose BIRC3, a non-membrane bound E3 ligase that can be translated from an mRNA with a short or long 3′UTR. We generated constructs where we fused GFP to the coding region of BIRC3 and then added either the short or long BIRC3 3′UTR. Our goal was to identify 3′UTR-dependent BIRC3 protein interactors. We transfected the constructs into cells grown in SILAC media, followed by GFP coimmunoprecipitation, followed by quantitative mass spectrometry. This revealed that some protein interactors bound equally well to BIRC3 protein regardless if it was translated from the short or long 3′UTR isoform. We called these 3′UTR-independent interactors. However, we found many BIRC3 protein interactors that bound better to BIRC3 protein translated from the long 3′UTR isoform. We validated a number of interactors and were able to confirm 80% of the candidates that we set out to validate (Fig. 3A) (Lee and Mayr 2019). This experiment revealed that BIRC3 has 3′UTR-independent and 3′UTR-dependent interactors which could be separated into different functional categories. BIRC3, which is also called cIAP2, is known to regulate NFκB signaling and apoptosis (Srinivasula and Ashwell 2008; Beug et al. 2012). The regulation of cell death is consistent with the identification of DIABLO and XIAP, two 3′UTR-independent BIRC3 interactors and known regulators of cell death (Srinivasula and Ashwell 2008). In contrast, the 3′UTR-dependent interaction partners have known roles in the regulation of chromatin, mitochondria, and protein trafficking. These functions have previously not been associated with BIRC3 (Gyrd-Hansen and Meier 2010).
Figure 3. BIRC3 has 3′UTR-independent as well as 3′UTR-dependent interaction partners.
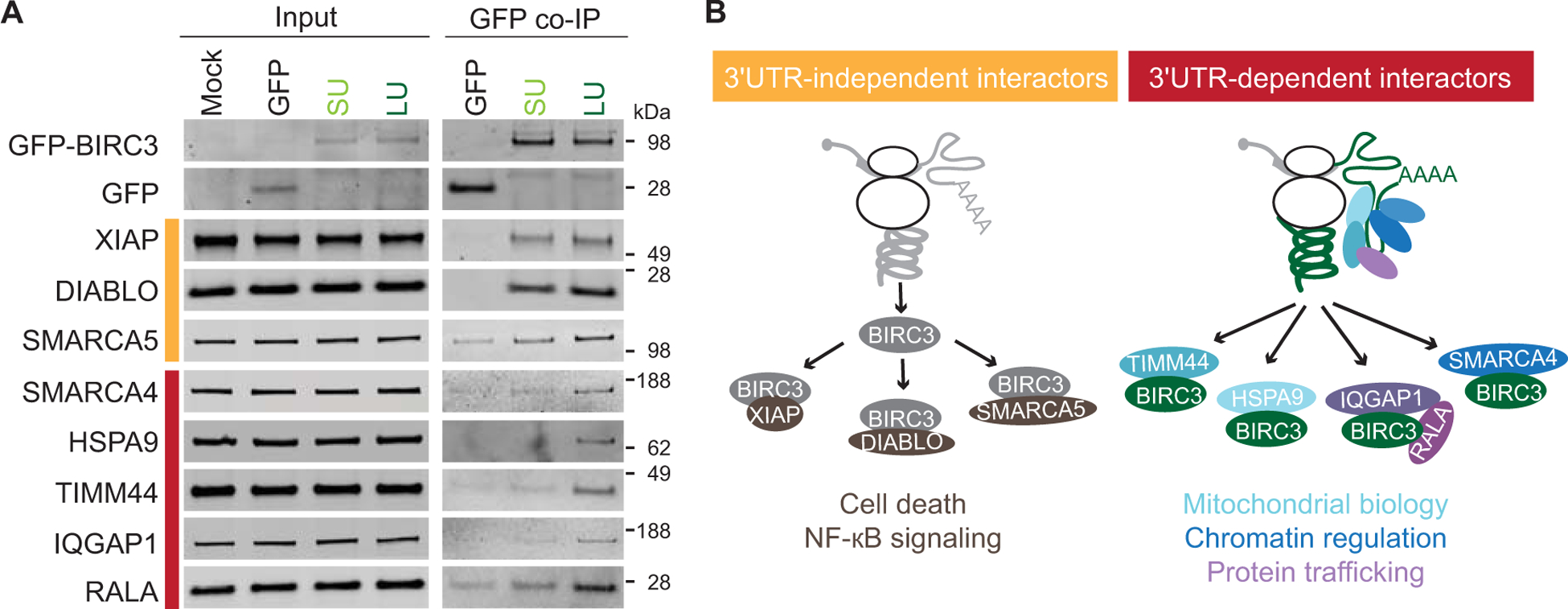
(A) Western blot validation of endogenous BIRC3 interactors. 3′UTR-independent (orange bar) and 3′UTR-dependent (red bar) BIRC3 protein interactors identified by GFP co-immunoprecipitation in HEK293T cells after transfection of constructs containing the coding region of BIRC3 fused to GFP and the short 3′UTR (SU) or the long 3′UTR (LU). 1% of input was loaded. (B) 3′UTR-independent BIRC3 functions are mediated by 3′UTR-independent protein interactors (brown), whereas 3′UTR-dependent BIRC3 functions are mediated by 3′UTR-dependent interactors (blue and purple colors). 3′UTR-dependent functions can only be accomplished by BIRC3-LU. Reprinted from Lee & Mayr, 2019.
We then set out to find a biologically relevant 3′UTR-dependent function of BIRC3. This is a function of BIRC3 that can only be accomplished by BIRC3 protein translated from the long 3′UTR isoform (BIRC3-LU). We identified CXCR4-dependent B cell migration as such a function because we observed impaired B cell migration in BIRC3 knock-out cells and in cells with knock-down of the long 3′UTR isoform. As these cells are unable to mediate migration, despite the expression of BIRC3 protein translated from the short 3′UTR isoform (BIRC3-SU), our data indicates that migration is a 3′UTR-dependent function of BIRC3. We further showed that two of the validated 3′UTR-dependent BIRC3 interactors, IQGAP1 and RALA, were both required for migration and were mediators for one of the 3′UTR-dependent functions of BIRC3 (Lee and Mayr 2019).
In summary, we found that the E3 ligase BIRC3 has 3′UTR-independent and 3′UTR-dependent interactors (Fig. 3B). It seems that the 3′UTR-independent interactors form complexes with BIRC3 mostly in a post-translational manner as protein abundance of BIRC3 or the interactors was a crucial determinant for interaction. In contrast, protein abundance does not seem to be the most important factor to establish 3′UTR-dependent interactions. This interpretation is based on the observation that knock-down of the long BIRC3 3′UTR isoform did not change overall BIRC3 protein levels, suggesting that only a minor fraction of total BIRC3 protein consists of BIRC3-LU which was sufficient to mediate the 3′UTR-dependent functions. Importantly, BIRC3 uses the 3′UTR-dependent protein interactors to diversify its functions in a way that is independent of protein localization (Lee and Mayr 2019).
3′UTRs control protein activity without affecting protein abundance
So far, we had only studied the functions of alternative 3′UTRs. However, we reasoned that 3′UTR-mediated protein complex formation is likely to happen at the majority of 3′UTRs and not only at alternative 3′UTRs. Therefore, as next candidate, we chose the 3′UTR of the TP53 gene which generates a single 3′UTR isoform (Lianoglou et al. 2013). Moreover, we previously had only used GFP-tagged expression constructs and decided to use CRISPR to delete the 3′UTR at the endogenous locus (Mayr 2019). Our goal was to delete the majority of 3′UTR regulatory elements while keeping mRNA processing intact in order to not disturb protein production. To do so, we used two guide RNAs and deleted the DNA sequence encoding the 3′UTR between the stop codon and approximately 150 base pairs upstream of the polyadenylation signal. We generated clones with a homozygous deletion of the TP53 3′UTR in a human cell line and found that the 3′UTR deletion did not influence overall mRNA or protein levels of p53 (S. Mitschka, C. Mayr, unpubl.). This is in contrast to the widely accepted opinion that one of the major functions of 3′UTRs is the regulation of mRNA or protein abundance (Barreau et al. 2005; Bartel 2009). Despite no difference in p53 protein levels, in the cells with 3′UTR deletion, we observed a phenotypic difference that is consistent with premature activation of p53 (S. Mitschka, C. Mayr, unpubl.). This indicates that the TP53 3′UTR regulates the activity of p53 protein.
We used CRISPR-mediated manipulation to investigate the 3′UTR-dependent functions of a second candidate. Also in the case of PTEN, the 3′UTR did not affect overall mRNA or protein levels, but instead seemed to alter the enzymatic activity of the PTEN phosphatase (B. Kwon, S.H. Lee, C. Mayr, unpubl.). 3′UTRs were also manipulated using CRISPR by others. Again, deletion of the long 3′UTR of Dscam1 did not change Dscam1 protein levels, but impaired axon outgrowth in flies (Zhang et al. 2019). In our hands, all so far tested 3′UTRs controlled protein activity without affecting protein abundance, indicating that this function of 3′UTRs is widespread. This has also been suggested by others (Fernandes 2019; Ribero 2019).
How is information transferred from 3′UTRs to proteins?
We showed that 3′UTRs can control protein functions (Berkovits and Mayr 2015; Lee and Mayr 2019), suggesting that 3′UTRs contain genetic information for the regulation of protein function. We set out to investigate how the information is transferred from 3′UTRs to proteins. We showed previously that CD47 function is regulated by the 3′UTR-dependent binding of SET to CD47 protein. SET binds to both the long CD47 3′UTR (as it binds HuR and HuR binds to the long 3′UTR) as well as to CD47 protein. After the establishment of the SET-CD47 interaction, the CD47 3′UTR is no longer necessary, suggesting that SET is transferred from the 3′UTR to the protein. As the information contained in the long CD47 3′UTR is transferred to the protein through SET binding, we set out to study how SET transfer is regulated.
The RNA-binding protein HuR is important in this process as knock-down of HuR abrogated binding of SET to CD47 protein. However, HuR was not sufficient as SET interacted better with CD47 when it was translated from an mRNA with the entire long CD47 3′UTR compared to a 3′UTR that only consisted of HuR binding sites (Berkovits and Mayr 2015). Therefore, we hypothesized that a second RNA-binding protein cooperates with HuR to accomplish SET transfer and we set out to identify this protein.
As the 3′UTR of CD47 is 4,194 nucleotides long, we started with the identification of a region that contains the majority of information for SET transfer. We generated three 3′UTR pieces and tested their capacity to mediate CD47 cell surface localization, one of the read-outs for SET binding (Berkovits and Mayr 2015). We found that all three pieces contained some, but none of them contained the majority of information (Fig. 4) (W. Ma, C. Mayr, unpubl.). This suggested the presence of a repeated motif that is required for SET transfer. To identify it, we generated several short artificial 3′UTRs that contained different repeated motifs and tested their capacity for CD47 cell surface localization. This led to the discovery of ‘artificial UTR 5’ (aUTR5), derived from the TNFα AU-rich element which contains six AU-rich elements (Kontoyiannis et al. 1999). This short 3′UTR fragment was able to fully recapitulate the function of the long CD47 3′UTR with respect to SET binding and CD47 cell surface localization (Ma and Mayr 2018).
Figure 4. All fragments of the CD47 3′UTR contain information for SET transfer.
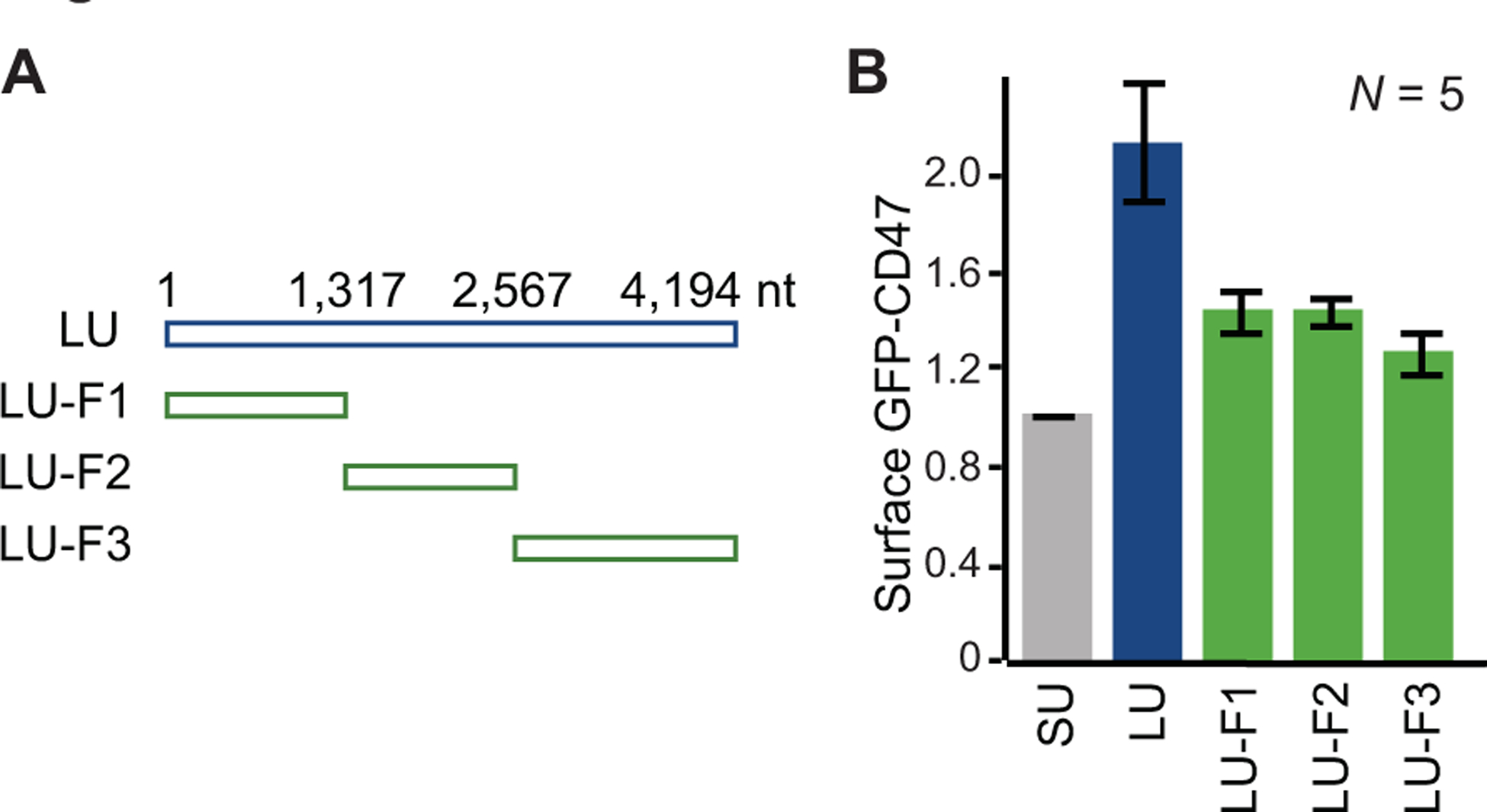
(A) Schematic showing the full-length CD47 3′UTR (LU) together with the three fragments (LU-F1, LU-F2, LU-F3) tested. The nucleotide positions for the boundaries of the LU fragments are shown. (B) A read-out for SET binding is surface expression of GFP-CD47. Shown are flow cytometry results of the indicated samples. SU is the short CD47 3′UTR and is used as negative control, whereas LU is the positive control.
With the aUTR5, we performed RNA affinity pull-down experiments and identified the RNA binding proteins that bind specifically to this 3′UTR compared with a size-matched control derived from the short 3′UTR of CD47. Using mass spectrometry analysis, we identified HuR and TIS11B as specific interactors of the aUTR5 (Ma and Mayr 2018). TIS11B is an RNA-binding protein also known as ZFP36L1 or BRF1 that binds to the canonical AU-rich element AUUUA (Stoecklin et al. 2002; Lai et al. 2002; Stumpo et al. 2004; Lykke-Andersen and Wagner 2005; Herranz et al. 2015). TIS11B is known to bind to the 3′UTRs of cytokines and destabilizes their mRNAs (Stoecklin et al. 2002; Lykke-Andersen and Wagner 2005; Herranz et al. 2015). Knock-down of TIS11B abrogated the 3′UTR-dependent binding of SET to CD47 and decreased CD47 cell surface localization. However, it did not change expression of the proteins involved in 3′UTR-mediated surface expression of CD47, including CD47, HuR, and SET, that all have several AU-rich elements in their 3′UTRs, suggesting that not all AU-rich elements have destabilizing capacity. This raised the question of how TIS11B would mediate SET transfer (Ma and Mayr 2018).
We hypothesized that TIS11B might generate a ‘special’ environment for SET transfer and performed imaging of TIS11B. We found that endogenous TIS11B formed granular assemblies in the vicinity of the ER. Live cell confocal imaging with Airyscan mode showed that TIS11B assembles into a tubule-like network that is intertwined with the ER and covers a large portion of the ER (Ma and Mayr 2018). TIS11B assemblies are RNA granules as they contain specific mRNAs and proteins. Therefore, we called them TIS granules. TIS granules were detected under steady-state cultivation conditions and in the absence of stress in all cell types investigated, including in primary cells directly isolated from human skin (M. Pan, W. Ma, C. Mayr, J. Young, unpubl.). Not all TIS11B molecules assemble into TIS granules, because we also detected soluble TIS11B protein in the cytoplasm.
To start to investigate the role of TIS11B or TIS granules in SET transfer, we visualized CD47 mRNAs. Using RNA-FISH, we observed that CD47-LU mRNA transcripts localized to TIS granules, whereas CD47-SU mRNA transcripts localized to the ER but mostly to regions not covered by TIS granules. As CD47 protein was detected at the site of mRNA localization, we concluded that CD47-LU is translated on the ER in the region of TIS granules whereas CD47-SU is translated on a different domain of the rough ER that is not associated with TIS granules (Ma and Mayr 2018). This indicates that one function of the alternative 3′UTRs of CD47 is the regulation of local translation on different subdomains of the ER.
Next, we investigated in more detail how the protein-protein interaction between SET and CD47 is formed. Usually, when two proteins are able to bind to each other, overexpression of the proteins results in increased interaction; a principle that is often used when proteins are tagged with FLAG and HA to test their interaction. However, overexpression of SET and CD47 did not result in increased interaction (Ma and Mayr 2018). Instead, we observed that overexpression of the RNA-binding protein TIS11B resulted in better interaction between SET and CD47-LU (Fig. 5). This raised the question if overexpression of TIS11B protein or the presence of TIS granules is crucial. To distinguish between the two scenarios we needed a mutant of TIS11B that behaves like the wild-type protein, but is unable to assemble into TIS granules. Through hypothesis-driven mutagenesis of TIS11B we obtained such a mutant. This mutant revealed that the presence of TIS granules is required for the binding of SET to CD47-LU (Fig. 5) (Ma and Mayr 2018). This observation allowed us to predict the following: CD47-SU is a protein that is translated on the ER outside of the TIS granule region and does not bind to SET. If translation in the TIS granule region is required for the interaction of SET and CD47, CD47-SU will bind to SET if CD47-SU is translated in the TIS granule region. We set up an experiment where we forced CD47-SU to be translated in the TIS granule region and were able to recapitulate the prediction. This finding indicates that certain protein-protein interactions can only be established
Figure 5. TIS granules are necessary for SET binding to CD47.
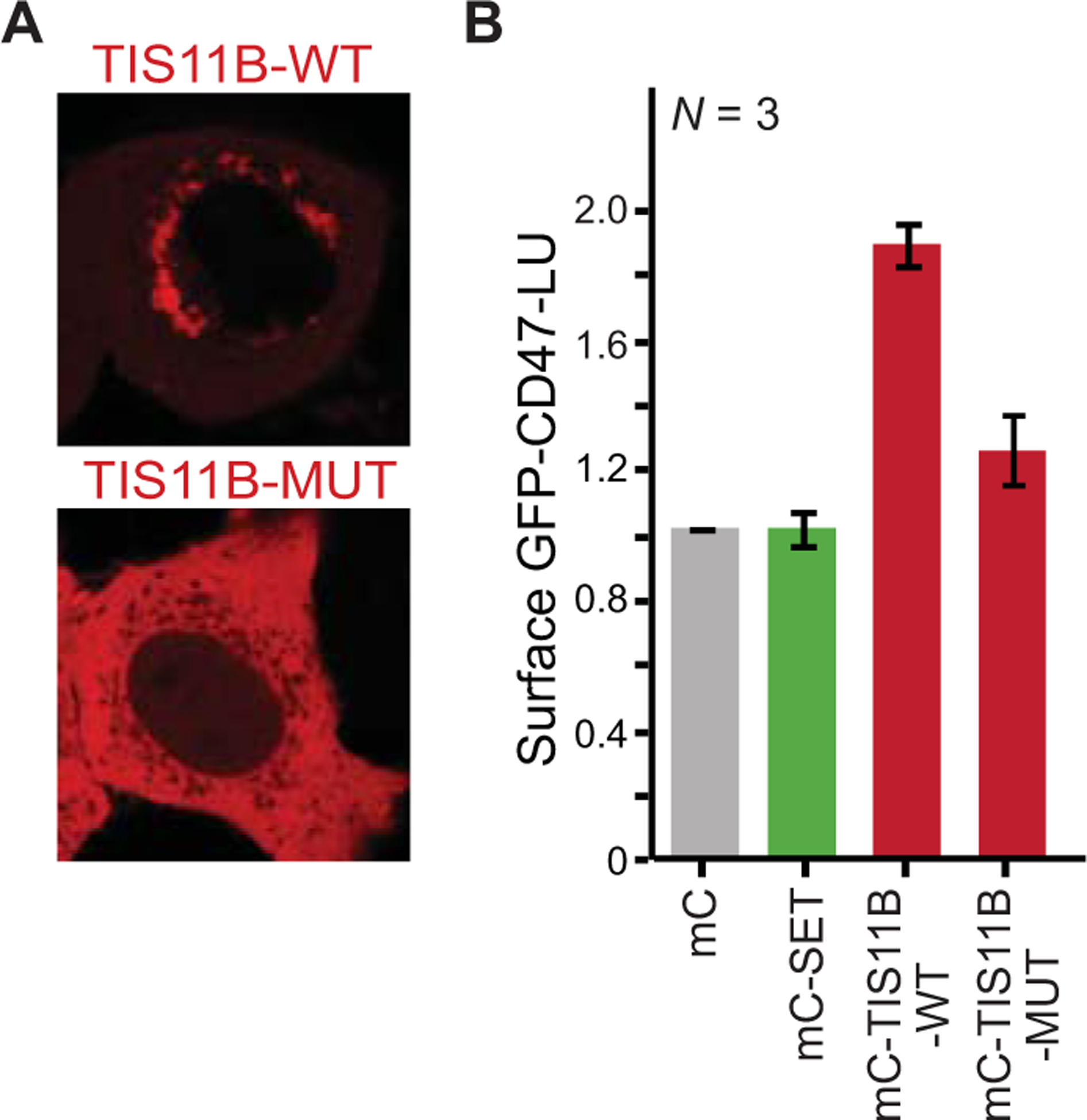
(A) Wild-type TIS11B (TIS11B-WT) forms TIS granules, whereas mutant TIS11B (TIS11B-MUT) does not. (B) Presence of TIS11B-WT substantially increases SET binding, but expression of TIS11B that is unable to form TIS granules (TIS11B-MUT) has a small effect. mC, mCherry. Shown as in Figure 4B. Adapted from Ma & Mayr, 2019.
when a protein is translated within a particular subcellular compartment and not outside the compartment (Ma and Mayr 2018). As 3′UTRs contribute to the local environment within TIS granules (W. Ma, C. Mayr, unpubl.), our finding indicates that one function of mRNAs is to create a niche during protein synthesis.
Interestingly, in the TIS granule region we found a paradoxical relationship between SET binding and SET concentration: the interaction between SET and CD47 was increased despite lower abundance of SET in the TIS granule region compared to the cytoplasm (Fig. 6) (Ma and Mayr 2018). A similar observation was recently obtained with a different experimental system in vitro. Within a CAPRIN1 phase-separated environment the activity of the CNOT7 deadenylation enzyme was seven-times higher than in buffer, despite a lower concentration of CNOT7 in the condensate than outside (Kim et al. 2019). The activity increase did not happen in other phase-separated environments and was specific for CAPRIN1. The authors suggested that the protein solvent environment modulates the enzymatic activity of CNOT7 (Kim et al. 2019).
Figure 6. The environment in TIS granules is different from the cytoplasm.
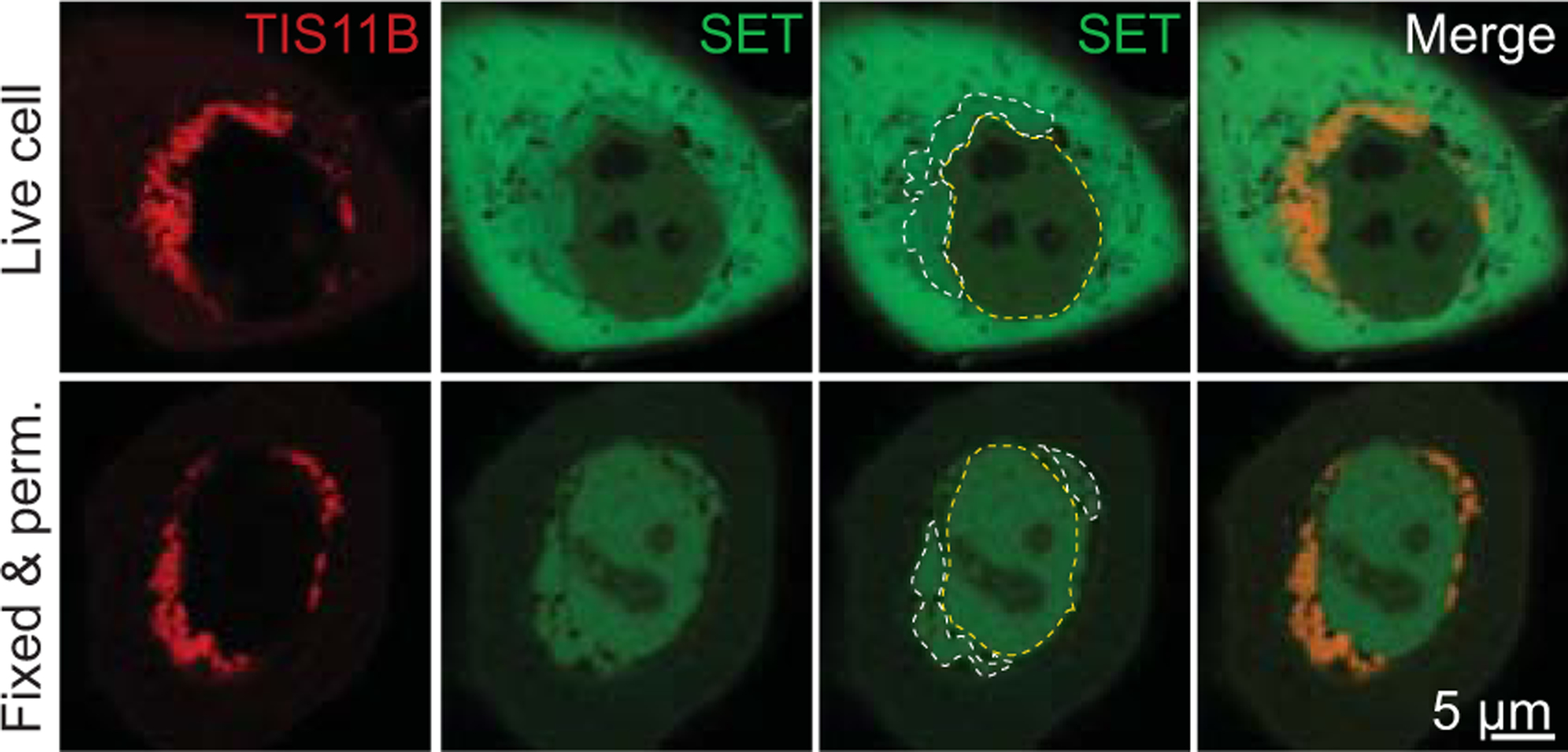
Confocal imaging of HeLa cells after transfection of GFP-TIS11B (red) and mCherry-SET (green). Top: SET is relatively depleted from the TIS granule region (demarcated by the white dotted line). The yellow dotted line demarcates the nucleus. Bottom: After fixing and permeabilization of cells, SET disappears from the cytoplasm but is retained in the TIS granule region suggesting that the biochemical environment in the TIS granule region is different from the cytoplasm. Adapted from Ma & Mayr, 2018.
Conclusions
These findings have several important implications. (1) The most straight-forward way how membraneless organelles and condensates may influence reactions is through partitioning and the concentration of molecules for reactions (Banani et al. 2017; Shin and Brangwynne 2017; Kato and McKnight 2018). This has been beautifully illustrated in signaling puncta formed by phosphorylated LAT. Within these LAT clusters, kinases are enriched and phosphatases are excluded, thus facilitating signaling reactions (Su et al. 2016).
(2) In addition to regulating partitioning, membraneless organelles were also shown to provide a different biochemical environment that can affect physical and structural properties of biomolecules: In buffer, a short piece of double-stranded DNA remained double-stranded,
whereas within a Ddx4 phase-separated environment it was destabilized, leading to sort of unwinding of the double-stranded DNA, possibly through cation-pi interactions acting on the backbone of the DNA (Nott et al. 2016).
(3) More recently, it was shown that the solvent environment provided by a CAPRIN1 phase-separated condensate increased the enzymatic activity of CNOT7 deadenylase. This was a remarkable result as the solvent was more important for the regulation of the enzymatic activity than the concentration of the enzyme (Kim et al. 2019). This finding is reminiscent of the observation that SET interacts with CD47 better in the TIS granule region than outside, despite a lower concentration of SET in the TIS granule region (Ma and Mayr 2018). These two examples provide a major conceptual advance in our understanding of how cellular reactions are regulated. So far, most of the time, it has been assumed that a high overall or local concentration is a key determinant of enzymatic activity, but the examples show that the solvent or the local environment created by the mRNA together with the bound proteins strongly influence reactions.
(4) Specific mRNAs are enriched in TIS granules and one of the determinants for their enrichment are the motifs located in their 3′UTRs (Ma and Mayr 2018). This implies that one of the functions of mRNAs is to compartmentalize the cytoplasm and to contribute to the generation of a local environment. The mRNA, its bound proteins, and the emergent properties acquired by their formation of higher-order assemblies create a local niche. In the future, it will be important to dissect the contribution of the enriched proteins and the enriched mRNAs to the material properties of TIS granules and other membraneless organelles.
(5) It is highly likely that in addition to TIS granules other membraneless organelles exist that allow translation and co-translational protein complex assembly. This would mean that the cytoplasm is much more compartmentalized than previously thought and it will be interesting to identify new cytoplasmic membraneless organelles and to determine the reactions that are promoted or inhibited. This may have important implications for synthetic and cell biology as we may be able to promote certain reactions through the generation of designer membraneless organelles (Reinkemeier et al. 2019).
(6) As shown for TIS granules, the local environment influences processes during protein translation and plays a role in peri-translational protein complex assembly (Ma and Mayr 2018; Natan et al. 2017). The niche formed at the site of protein synthesis may act as a filter to promote certain 3′UTR-dependent protein interactions over others (Fig. 7A–C). As most 3′UTRs contain thousands of nucleotides, many RNA-binding proteins are likely to bind to a specific 3′UTR. It is currently unknown how many RNA-binding proteins bind to a 3′UTR, but mass spectrometry analyses performed in C. elegans found approximately 10–30 different proteins bound per 1,000 nucleotides of mRNA (Theil et al. 2019). A complementary analysis in HeLa cells using high-resolution imaging estimated how often a single RNA-binding protein was bound to an mRNA and detected between 2–34 molecules with a median of 5–8 (Mateu-Regue et al. 2019). Each RNA-binding protein may recruit several effector proteins, thus generating a niche. It is conceivable that the composition of the niche may affect the binding kinetics of proteins with the nascent chain. This can be tested if a protein is forced to be translated in a different niche. One way to change the niche is to swap the 3′UTR or to recruit the mRNA into a different membraneless organelle (Fig. 7A–C).
Figure 7. The niche at the site of protein synthesis affects peri-translational protein complex formation.
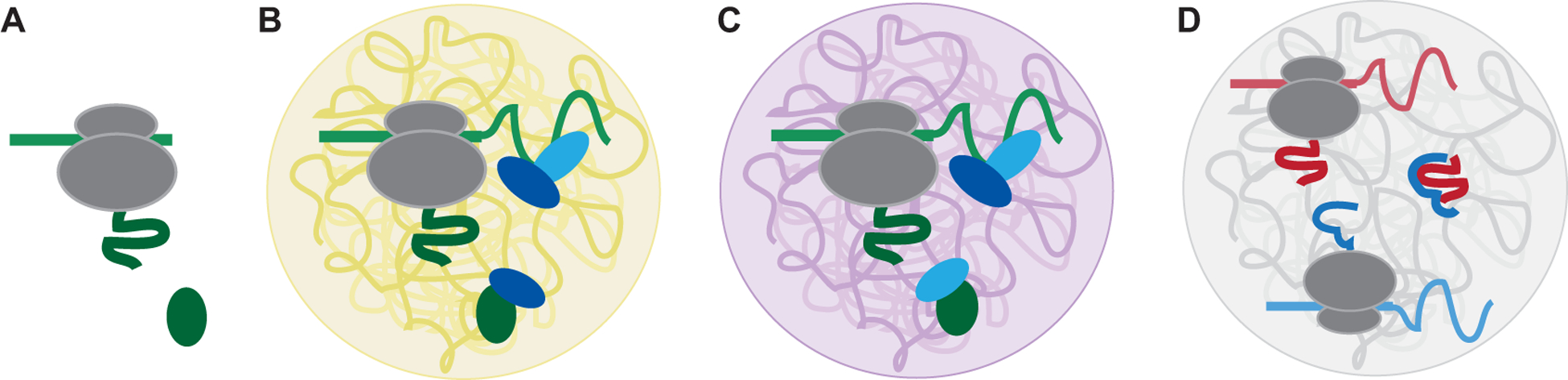
(A) Translation of an mRNA (light green) without 3′UTR may result in the lack of an RNA granule at the site of translation and may prevent co-translational protein complex assembly. The ribosome is shown in grey and the newly synthesized protein is shown in dark green. (B) As in (A), but the presence of a local RNA granule (yellow) allows a specific protein-protein interaction between a 3′UTR-recruited protein (dark blue) and the newly synthesized protein (dark green). (C) As in (B), but a different type of RNA granule (purple) allows the formation of a different 3′UTR-dependent protein complex. (D) Translation of two different mRNAs (light red, light blue) within a larger RNA granule (grey) containing a diversity of mRNAs may allow co-or peri-translational protein complex formation.
(7) It has long been thought that ‘mRNA regulons’ exist, where functionally related groups of mRNAs are co-regulated (Keene 2007). However, there is recent evidence that cytoplasmic mRNP (messenger RNA nucleoprotein complex) granules consist of single mRNAs which would preclude co-regulation of mRNAs (Mateu-Regue et al. 2019). However, formation of larger mRNA granules such as TIS granules that contain many mRNAs may enable the mingling of different mRNAs to allow co-regulation as well as co-or peri-translational protein complex assembly of transcripts that are translated in vicinity to each other (Fig. 7D) (Shiber et al. 2018; Mayr 2018; Schwarz and Beck 2019).
Acknowledgements
I thank W. Ma for coining the term ‘nurturing niche’. I thank several members of the Mayr lab, including W. Ma, E. Horste, X. Chen, and N. Robertson for critical comments on the manuscript and for helpful discussions. This work was funded by the NIH Director′s Pioneer Award (DP1-GM123454), the Pershing Square Sohn Cancer Research Alliance, and the NCI Cancer Center Support Grant (P30 CA008748).
Footnotes
The author declares no competing interests.
References
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B et al. 2008. Distinct role of long 3’ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell 134: 175–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK. 2017. Biomolecular condensates: organizers of cellular biochemistry. Nature reviews Molecular cell biology 18: 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB. 2005. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res 33: 7138–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Y 2002. Excitatory actions of gaba during development: the nature of the nurture. Nature reviews Neuroscience 3: 728–739. [DOI] [PubMed] [Google Scholar]
- Benoit B, He CH, Zhang F, Votruba SM, Tadros W, Westwood JT, Smibert CA, Lipshitz HD, Theurkauf WE. 2009. An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development 136: 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovits BD, Mayr C. 2015. Alternative 3’ UTRs act as scaffolds to regulate membrane proteinlocalization. Nature 522: 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beug ST, Cheung HH, LaCasse EC, Korneluk RG. 2012. Modulation of immune signalling by inhibitors of apoptosis. Trends in immunology 33: 535–545. [DOI] [PubMed] [Google Scholar]
- Brennan CM, Steitz JA. 2001. HuR and mRNA stability. Cellular and molecular life sciences : CMLS 58: 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbaugh J, Di Stefano B, Wang X, Borkent M, Forouzmand E, Clowers KJ, Ji F, Schwarz BA, Kalocsay M, Elledge SJ et al. 2018. Nudt21 Controls Cell Fate by Connecting Alternative Polyadenylation to Chromatin Signaling. Cell 172: 629–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Chen ST, Juan HF, Huang HC. 2012. Lengthening of 3’UTR increases with morphological complexity in animal evolution. Bioinformatics 28: 3178–3181. [DOI] [PubMed] [Google Scholar]
- Ciolli Mattioli C, Rom A, Franke V, Imami K, Arrey G, Terne M, Woehler A, Akalin A, Ulitsky I, Chekulaeva M. 2019. Alternative 3’ UTRs direct localization of functionally diverse protein isoforms in neuronal compartments. Nucleic Acids Res 47: 2560–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins WA, Maccoby EE, Steinberg L, Hetherington EM, Bornstein MH. 2000. Contemporary research on parenting. The case for nature and nurture. The American psychologist 55: 218–232. [PubMed] [Google Scholar]
- Fernandes N, Buchan JR 2019. RPS28B mRNA acts as a scaffold promoting cis-translational interaction of proteins driving P-body assembly. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. 2006. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312: 75–79. [DOI] [PubMed] [Google Scholar]
- Gruber AR, Martin G, Muller P, Schmidt A, Gruber AJ, Gumienny R, Mittal N, Jayachandran R, Pieters J, Keller W et al. 2014. Global 3’ UTR shortening has a limited effect on protein abundance in proliferating T cells. Nature communications 5: 5465. [DOI] [PubMed] [Google Scholar]
- Gyrd-Hansen M, Meier P. 2010. IAPs: from caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat Rev Cancer 10: 561–574. [DOI] [PubMed] [Google Scholar]
- Herranz N, Gallage S, Mellone M, Wuestefeld T, Klotz S, Hanley CJ, Raguz S, Acosta JC, Innes AJ, Banito A et al. 2015. mTOR regulates MAPKAPK2 translation to control the senescence-associated secretory phenotype. Nature cell biology 17: 1205–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Jamieson CH, Pang WW, Park CY, Chao MP, Majeti R, Traver D, van Rooijen N, Weissman IL. 2009. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell 138: 271–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Yuan S, Wang Y, Fu Y, Ge Y, Ge Y, Lan X, Feng Y, Qiu F, Li P et al. 2017. The role of alternative polyadenylation in the antiviral innate immune response. Nature communications 8: 14605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, McKnight SL. 2018. A Solid-State Conceptualization of Information Transfer from Gene to Message to Protein. Annu Rev Biochem 87: 351–390. [DOI] [PubMed] [Google Scholar]
- Keene JD. 2007. RNA regulons: coordination of post-transcriptional events. Nature reviews Genetics 8: 533–543. [DOI] [PubMed] [Google Scholar]
- Kim TH, Tsang B, Vernon RM, Sonenberg N, Kay LE, Forman-Kay JD. 2019. Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science 365: 825–829. [DOI] [PubMed] [Google Scholar]
- Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. 1999. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10: 387–398. [DOI] [PubMed] [Google Scholar]
- Lai WS, Kennington EA, Blackshear PJ. 2002. Interactions of CCCH zinc finger proteins with mRNA: non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J Biol Chem 277: 9606–9613. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131: 174–187. [DOI] [PubMed] [Google Scholar]
- Lee SH, Mayr C. 2019. Gain of Additional BIRC3 Protein Functions through 3’-UTR-Mediated Protein Complex Formation. Mol Cell 74: 701–712 e709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Singh I, Tisdale S, Abdel-Wahab O, Leslie CS, Mayr C. 2018. Widespread intronic polyadenylation inactivates tumour suppressor genes in leukaemia. Nature 561: 127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Makkinje A, Damuni Z. 1996. The myeloid leukemia-associated protein SET is a potent inhibitor of protein phosphatase 2A. J Biol Chem 271: 11059–11062. [DOI] [PubMed] [Google Scholar]
- Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. 2013. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev 27: 2380–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Wagner E. 2005. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev 19: 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Mayr C. 2018. A Membraneless Organelle Associated with the Endoplasmic Reticulum Enables 3’UTR-Mediated Protein-Protein Interactions. Cell 175: 1492–1506 e1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateu-Regue A, Christiansen J, Bagger FO, Winther O, Hellriegel C, Nielsen FC. 2019. Single mRNP Analysis Reveals that Small Cytoplasmic mRNP Granules Represent mRNA Singletons. Cell reports 29: 736–748 e734. [DOI] [PubMed] [Google Scholar]
- Mayr C 2016. Evolution and Biological Roles of Alternative 3’UTRs. Trends Cell Biol 26: 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C 2017. Regulation by 3’-Untranslated Regions. Annual review of genetics 51: 171–194. [DOI] [PubMed] [Google Scholar]
- Mayr C 2018. Protein complexes assemble as they are being made. Nature 561: 186–187. [DOI] [PubMed] [Google Scholar]
- Mayr C 2019. What Are 3’ UTRs Doing? Cold Spring Harb Perspect Biol 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. 2009. Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natan E, Wells JN, Teichmann SA, Marsh JA. 2017. Regulation, evolution and consequences of cotranslational protein complex assembly. Current opinion in structural biology 42: 90–97. [DOI] [PubMed] [Google Scholar]
- Neve J, Furger A. 2014. Alternative polyadenylation: less than meets the eye? Biochemical Society transactions 42: 1190–1195. [DOI] [PubMed] [Google Scholar]
- Nott TJ, Craggs TD, Baldwin AJ. 2016. Membraneless organelles can melt nucleic acid duplexes and act as biomolecular filters. Nature chemistry 8: 569–575. [DOI] [PubMed] [Google Scholar]
- Reinkemeier CD, Girona GE, Lemke EA. 2019. Designer membraneless organelles enable codon reassignment of selected mRNAs in eukaryotes. Science 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribero DM, Prod’homme A, Teixeira A, Zanzoni A, Brun C 2019. The role of 3’UTR-protein complexes in the regulation of protein multifunctionality and subcellular localization. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. 2008. Proliferating cells express mRNAs with shortened 3’ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz A, Beck M. 2019. The Benefits of Cotranslational Assembly: A Structural Perspective. Trends Cell Biol 29: 791–803. [DOI] [PubMed] [Google Scholar]
- Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. 2001. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104: 119–130. [DOI] [PubMed] [Google Scholar]
- Shiber A, Doring K, Friedrich U, Klann K, Merker D, Zedan M, Tippmann F, Kramer G, Bukau B. 2018. Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP. 2017. Liquid phase condensation in cell physiology and disease. Science 357. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh I, Lee SH, Sperling AS, Samur MK, Tai YT, Fulciniti M, Munshi NC, Mayr C, Leslie CS. 2018. Widespread intronic polyadenylation diversifies immune cell transcriptomes. Nature communications 9: 1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Burge CB, Bartel DP. 2013. 3’ UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res 23: 2078–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasula SM, Ashwell JD. 2008. IAPs: what’s in a name? Mol Cell 30: 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Colombi M, Raineri I, Leuenberger S, Mallaun M, Schmidlin M, Gross B, Lu M, Kitamura T, Moroni C. 2002. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J 21: 4709–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpo DJ, Byrd NA, Phillips RS, Ghosh S, Maronpot RR, Castranio T, Meyers EN, Mishina Y, Blackshear PJ. 2004. Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the Tristetraprolin family. Mol Cell Biol 24: 6445–6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, King DS, Taunton J, Rosen MK, Vale RD. 2016. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352: 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliaferro JM, Vidaki M, Oliveira R, Olson S, Zhan L, Saxena T, Wang ET, Graveley BR, Gertler FB, Swanson MS et al. 2016. Distal Alternative Last Exons Localize mRNAs to Neural Projections. Mol Cell 61: 821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil K, Imami K, Rajewsky N. 2019. Identification of proteins and miRNAs that specifically bind an mRNA in vivo. Nature communications 10: 4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tushev G, Glock C, Heumuller M, Biever A, Jovanovic M, Schuman EM. 2018. Alternative 3’ UTRs Modify the Localization, Regulatory Potential, Stability, and Plasticity of mRNAs in Neuronal Compartments. Neuron 98: 495–511 e496. [DOI] [PubMed] [Google Scholar]
- Wang D, Kon N, Lasso G, Jiang L, Leng W, Zhu WG, Qin J, Honig B, Gu W. 2016. Acetylation- regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature 538: 118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. 2005. Systematic discovery of regulatory motifs in human promoters and 3’ UTRs by comparison of several mammals. Nature 434: 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Zhang J. 2018. Alternative Polyadenylation of Mammalian Transcripts Is Generally Deleterious, Not Adaptive. Cell systems 6: 734–742 e734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe D, Nudel U, Mayer Y, Neuman S. 1985. Highly conserved sequences in the 3' untranslated region of mRNAs coding for homologous proteins in distantly related species. Nucleic Acids Res 13: 3723–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang KX, Tan L, Pellegrini M, Zipursky SL, McEwen JM. 2016. Rapid Changes in the Translatome during the Conversion of Growth Cones to Synaptic Terminals. Cell reports 14: 1258–1271. [DOI] [PubMed] [Google Scholar]
- Zhang Z, So K, Peterson R, Bauer M, Ng H, Zhang Y, Kim JH, Kidd T, Miura P. 2019. Elav-Mediated Exon Skipping and Alternative Polyadenylation of the Dscam1 Gene Are Required for Axon Outgrowth. Cell reports 27: 3808–3817 e3807. [DOI] [PMC free article] [PubMed] [Google Scholar]


